Abstract
Background
Circular RNAs (circRNAs) play crucial roles in the development and progression of various cancers, including cervical cancer. However, the role and regulatory mechanism of circ_0003221 in cervical cancer are still unclear.
Methods
The expression of circ_0003221, microRNA-758-3p (miR-758-3p), cytoplasmic polyadenylation element-binding protein 4 (CPEB4) was detected by quantitative real-time PCR (qRT-PCR). Cell Counting Kit-8 (CCK-8), colony formation, and 5-Ethynyl-2ʹ-deoxyuridine (Edu) assays were utilized to determine cell proliferation. Cell cycle distribution was analyzed by flow cytometry. Cell migration and invasion were detected by transwell assay. All protein levels were detected by Western blot assay. The interaction between miR-758-3p and circ_0003221 or CPEB4 was confirmed by dual-luciferase reporter, RNA immunoprecipitation (RIP) and RNA pull-down assays. Mice xenograft model of cervical cancer was established to verify the function of circ_0003221 in vivo.
Results
Circ_0003221 was upregulated in cervical cancer tissues and cells. Knockdown of circ_0003221 suppressed cell proliferation, migration, invasion, and EMT and induced cell cycle arrest in cervical cancer cells. MiR-758-3p was a direct target of circ_0003221, and miR-758-3p inhibition reversed the effects of circ_0003221 knockdown in cervical cancer cells. Moreover, CPEB4 was identified as a direct target of miR-758-3p, and miR-758-3p exerted its anti-cancer role by targeting CPEB4. Furthermore, circ_0003221 acted as a sponge of miR-758-3p to upregulate CPEB4 expression. In addition, circ_0003221 silence also suppressed tumor growth and EMT in vivo.
Conclusion
Circ_0003221 knockdown inhibited cervical cancer progression via modulating miR-758-3p/CPEB4 axis, which might suggest a new insight into the pathogenesis of cervical cancer.
Keywords: cervical cancer, circ_0003221, miR-758-3p, CPEB4
Introduction
Cervical cancer is one of the most commonly diagnosed cancers among females.1 Although outstanding advances have been made in therapeutic methods, the overall survival of cervical cancer patients remains unsatisfactory because of the recurrence and metastasis.2–4 Moreover, continuous infection of high-risk type human papillomavirus (HPV) has been confirmed as a vital pathogenic factor for the progression of cervical cancer, and environmental, immunological, genetic and epigenetic factors are also the causes of cervical cancer.5 High-risk HPV infection, such as HPV16/18, is easy to integrate HPV gene into the genome, interfering with the immune response of the body and the accumulation of microlesions, which eventually develop into cervical cancer. However, the detailed mechanisms of cervical cancer progression remain largely elusive. Therefore, it is urgent to better understand the underlying mechanisms of cervical cancer development and find a novel strategy for cervical cancer treatment.
In recent decades, non-coding RNAs (ncRNAs) have become a hotspot in the field of cancer.6 As a novel type of ncRNAs, circular RNAs (circRNAs) are generated from precursor mRNAs (pre-mRNAs) via back-splicing with neither 5ʹ-cap nor 3ʹ-end poly A tail.7,8 Currently, many reports have suggested that circRNAs can serve as oncogenes or tumor suppressors in multiple cancers, including cervical cancer.9,10 Recently, circRNAs are one of the new targets for cervical cancer, such as circUBAP2,11 hsa_circ_0000069,12 circ_0005576,13 circSLC26A414 have been identified to promote cervical cancer progression, suggesting the important role of circRNAs in cervical cancer tumorigenesis. As for circ_0003221 (also known as circPTK2, chr8:141856358–141900868), it has been demonstrated to be overexpressed in cervical cancer cells.15 Nevertheless, the role of circ_0003221 in cervical cancer remains largely unclear.
Large evidence has shown that circRNAs can serve as competing endogenous RNAs (ceRNAs), also known as microRNA (miRNA) sponges, via competitively binding to miRNA response elements, thereby regulating the expression of downstream target genes.16,17 Based on this regulation, circRNAs can function as miRNA sponges to regulate the expression of target genes, forming a regulatory circRNA-miRNA-mRNA network. MiR-758 has been identified to serve as a tumor-suppressing miRNA in cervical cancer.18 As a member of cytoplasmic polyadenylation element-binding protein (CPEB) family, CPEB4 has been revealed to be overexpressed in cervical cancer samples and involved in cervical cancer development.19 Based on the analysis of bioinformatics software, circ_0003221 and CPEB4 were found to have complementary binding sites with miR-758-3p, which stimulated us to assume the ceRNA regulatory network of circ_0003221/miR-758-3p/CPEB4 in cervical cancer.
In this research, the expression of circ_0003221 in cervical cancer tissues and cells was explored. Moreover, the function of circ_0003221 in vitro and in vivo as well as its underlying mechanisms were also investigated. We aimed to provide potential therapeutic targets for cervical cancer patients.
Materials and Methods
Specimen Collection
The clinical specimens including cervical cancer tissue samples (N=60) and adjacent normal tissue samples (N=60) were collected from the patients after surgical resection at The Second Affiliated Hospital University of South China. There are no patients received radiotherapy or chemotherapy before the operation. These samples were collected, promptly frozen in liquid nitrogen and then kept in a refrigerator at −80°C after surgical resection. Informed consent was signed by all subjects before enrollment in the research. This research was approved by the Research Ethics Committee of The Second Affiliated Hospital University of South China. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki, and that all subjects provided written informed consent.
Cell Culture and Transfection
Cervical cancer cells (SiHa and CaSki) and human cervical epithelial cells (ECT1/E6E7) were purchased from BeNa Culture Collection (Beijing, China) and cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen). All cells were maintained in a 5% CO2 incubator at 37°C.
The small interfering RNA (siRNA) against circ_0003221 (si-circ_0003221), miR-758-3p mimic (miR-758-3p), miR-758-3p inhibitor (anti-miR-758-3p), pcDNA-CPEB4 overexpression vector (pcDNA-CPEB4), and their matched controls (si-NC, miR-NC, anti-miR-NC mimic, and pcDNA) were all obtained from RiboBio (Guangzhou, China). Cell transfection was performed using Lipofectamine 3000 Reagent (Invitrogen).
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
After extracting total RNA using TRIzol reagent (Invitrogen), the cDNA was synthesized using the Primescript RT Reagent (TaKaRa, Kusatsu, Japan) for analysis of circRNA and mRNA, or miRNA reverse transcription PCR kit (RiboBio) for detection of miRNA. Then qRT-PCR was performed by using SYBR green PremixEx Taq II (Takara) on CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The primers for circRNA, miRNA, and mRNA were presented: circ_0003221 (forward 5ʹ-3ʹ: ATCATACTGGGAGATGCGGG; reverse 5ʹ-3ʹ: AGTTGGGGTCAAGGTAAGCA); miR-758-3p (forward 5ʹ-3ʹ: GCCGAGTTTGTGACCTGGTC; reverse 5ʹ-3ʹ: CAGTGCGTGTCGTGGAGT); CPEB4 (forward 5ʹ-3ʹ: GGATGGTTCACAGCCACTTGAC; reverse 5ʹ-3ʹ: CACACCTCCATATAGCCGATCC). β-actin (forward 5ʹ-3ʹ: GCCGGGACCTGACTGACTAC; reverse 5ʹ-3ʹ: TCTCCTTAATGTCACGCACGAT), U6 (forward 5ʹ-3ʹ: CTCGCTTCGGCAGCACATATACT; reverse 5ʹ-3ʹ: ACGCTTCACGAATTTGCGTGTC). The expression of genes was evaluated with 2-ΔΔCt method. β-actin was chosen as the reference gene for circ_0003221 and CPEB4, and U6 was served as an internal reference for miR-758-3p.
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 (Beyotime, Jiangsu, China) was utilized to detect cell viability. In short, 100 µL cell suspension (2×103 cells) was inoculated in 96-well plates At each indicated time point, CCK-8 solution (10 µL) was added into per well, followed by incubation for 2–3 h 37°C. Finally, 96-well plates were placed in a microplate reader (Bio-Rad) to determine the absorbance at 450 nm.
Colony Formation Assay
SiHa and CaSki cells were inoculated in a 6-well plate and then cultured for 14 days. After that, these colonies were fixed in paraformaldehyde (4%, Beyotime). Subsequently, these colonies were stained with crystal violet (0.1%, Beyotime) for 2 h. The number of the colonies was counted.
Flow Cytometry Analysis
SiHa and CaSki cells were inoculated in a 6-well plate. Following transfection, cells were collected and fixed with ice-cold 75% ethanol (Beyotime) for 12 h at −20°C. Next, fixed cells were collected and incubated with 25 μg/mL of propidium iodide (PI; Keygen Biotech, Nanjing, China) containing 50 µg/mL of RNase A (Keygen Biotech). After incubation for 20 min in the darkness, the cell cycle distribution was assessed with the flow cytometer (BD Biosciences, Franklin, NJ, USA).
5-Ethynyl-2ʹ-Deoxyuridine (Edu) Assay
Edu assay was performed using keyFluor488 Click-iT EdU detection kit (KeyGene, Nanjing, China) to test cell proliferation. In short, cells were was added into the 24-well plates. After transfection for 48 h, cells were indicated with 50 μM Edu for 2 h, and the cells were fixed in 4% paraformaldehyde and then incubated with Click-It reaction mixture. DAPI was used to stain the nucleic acids. Images were photographed with a fluorescence microscope (Olympus, Tokyo, Japan) at ×200 magnification.
Western Blot Assay
After isolation of total protein using RIPA lysis buffer (Beyotime), the protein samples were denatured by heating at 100°C for 3–5 min, and then quantified by a BCA protein assay kit (Tanon, Shanghai, China). After that, electrophoresis was performed with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; Beyotime) to separate protein, and protein was then blotted onto nitrocellulose membranes (Invitrogen). The membrane was blocked and then probed with primary antibody for 12 h at 4°C. The primary antibodies including proliferating cell nuclear antigen (PCNA) (ab18197, 1:1000), cyclinD1 (ab226977, 1:2000), E-cadherin (ab15148, 1:500), Snail (ab82846, 1:500), Vimentin (ab137321, 1:1000), and β-actin (ab227387, 1:5000) were purchased from Abcam (Cambridge, UK), and CPEB4 (PA5-25538, 1:1000) was purchased from Invitrogen. Subsequently, these membranes were continuously probed with secondary antibody (ab205718, 1:4000, Abcam). Lastly, blot signal was visualized by enhanced chemiluminescence reagent (Tanon).
Transwell Assay
SiHa and CaSki cells of each group suspended in serum-free medium (200 µL) was plated in the upper chamber (8μm pore size, Costar, Corning, NY, USA) with (invasion assay) or without (migration assay) Matrigel (BD Biosciences). Meanwhile, RPMI-1640 with 10% FBS was added into the bottom chamber. Twenty-four-hours later, the cells from the lower side of the membrane were fixed with paraformaldehyde (4%, Beyotime) for 0.5 h and stained by crystal violet solution (0.1%, Beyotime) for 1 h. At last, the stained cells were photographed under a microscope (Olympus) with a magnification of ×100.
Dual-Luciferase Reporter Assay
The candidate target miRNAs of circ_0003221 were predicted using Starbase 3.0 (http://starbase.sysu.edu.cn/), Circinteractome (https://circinteractome.nia.nih.gov/) and circBank (http://www.circbank.cn/). The candidate downstream targets of miR-758-3p were predicted by TargetScan (http://www.targetscan.org/), miRDB (http://mirdb.org/), Starbase 3.0, and OncomiR (http://www.oncomir.org). The wild-type (WT) luciferase reporter vector of circ_0003221 or CPEB4 (circ_0003221-WT- or CPEB4 WT-3ʹUTR) was synthesized via inserting the sequence of circFADS2 or CPEB4 containing the binding sites for miR-758-3p into pMIR-Report plasmid (Promega, Shanghai, China). Meanwhile, mutant (MUT) luciferase reporter vector of circ_0003221 or CPEB4 (circ_0003221-MUT or CPEB4 MUT-3ʹUTR) was generated by mutating the binding sites of miR-758-3p. Each of the above-mentioned plasmids and miR-758-3p/miR-NC were co-transfected into SiHa and CaSki cells for 48 h. Finally, dual-Luciferase Reporter Assay System (Promega) was employed to measure the firefly and Renilla luciferase activities.
RNA Immunoprecipitation (RIP) Assay
RIP was conducted with Magna RIP Kit (Millipore, Bedford, MA, USA). Briefly, SiHa and CaSki cells were lysed by RIP lysis buffer. After that, the lysate products were incubated with RIP buffer including magnetic beads conjugated with anti-immunoglobulin G (anti-IgG) or anti-Argonaute2 (anti-Ago2), and then rotated overnight. Next, immunoprecipitated RNA was extracted after incubation with Proteinase K. At last, qRT-PCR was conducted to detect the expression of circ_0003221, miR-758-3p and CPEB4.
RNA Pull-Down Assay
Biotin labeled miR-NC (Bio-miR-NC) and miR-758-3p (Bio-miR-758-3p) were obtained from RiboBio. Next, Bio-miR-NC and Bio-miR-758-3p were individually transfected into SiHa and CaSki cells for 48 h. Then, cells were collected and lysed using lysis buffer, followed by incubation with Dynabeads M-280 Streptavidin (Invitrogen) at 4°C for 2 h. The RNA complexes combining on the beads were washed and used for detecting the enrichment of circ_0003221 by qRT-PCR.
Tumor Formation Assay in vivo
BALB/c nude mice (female, 6 weeks old, n=10, weighing 20–25 g, Vital River, Beijing, China) were used to establish xenograft model. These mice were divided into two groups (n=5/group). Lentivirus-mediated shRNA targeting circ_0003221 (sh-circ_0003221) and its negative control (sh-NC) were provided by RiboBio. SiHa cells (1 × 106 cells, circ_0003221 silencing or control) were injected into nude mice. Tumor volume was examined with an external caliper and calculated according to the equation: 0.5 × length × width2. After 35 days, the mice were sacrificed, tumors were weighed and harvested to detect the abundance of circ_0003221, miR-758-3p and CPEB4. The animal experiments obtained the approval from the Animal Care and Use Committee of The Second Affiliated Hospital University of South China. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
All data from at least three independent experiments were displayed as mean ± standard deviation (SD) and analyzed by GraphPad Prism (GraphPad Software, San Diego California, USA). The significance of difference was evaluated with Student’s t-test (two groups), and one-way analysis of variance (ANOVA) was used when more than two groups were compared. The correlations among circ_0003221, miR-758-3p and CPEB4 were analyzed with Pearson’s correlation coefficient. Statistical significance was considered when P<0.05.
Results
Circ_0003221 Was Upregulated in Cervical Cancer Tissues and Cells
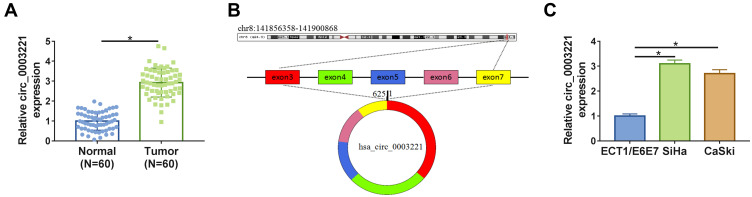
To explore whether the expression of circ_0003221 was dysregulated in cervical cancer, qRT-PCR was carried out. The results showed that the expression of circ_0003221 was increased in cervical cancer tissues compared to normal tissues (Figure 1A). Circ_0003221, with a length of 625 bp, is generated from exons 3–7 of PTK2 gene and is located on chromosome 8 (Figure 1B). Likewise, the expression of circ_0003221 was higher in cervical cancer cells (SiHa and CaSki) than that in ECT1/E6E7 cells (Figure 1C). These data indicated that circ_0003221 might play a critical role in cervical cancer.
Figure 1.
The expression of circ_0003221 was increased in cervical cancer tissues and cells. (A) The expression of circ_0003221 was detected by qRT-PCR in normal tissues and cervical cancer tissues. (B) The formation of circ_0003221 was presented. (C) The expression of circ_0003221 was measured by qRT-PCR in ECT1/E6E7, SiHa and CaSki cells. *P<0.05.
Knockdown of Circ_0003221 Suppressed Cell Proliferation and Metastasis and Induced Cell Cycle Arrest in Cervical Cancer Cells
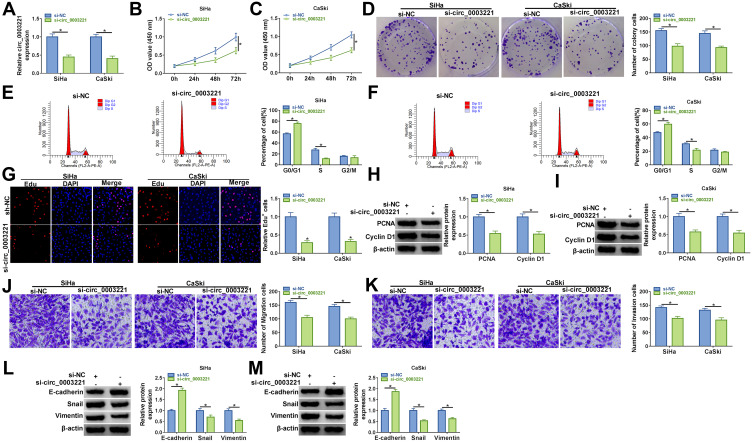
To investigate the potential biological functions of circ_0003221 in cervical cancer, we silenced circ_0003221 expression by transfection of si-circ_0003221. As presented in Figure 2A, transfection of si-circ_0003221 resulted in a significant decrease in the expression circ_0003221, suggesting that circ_0003221 was successfully knocked down. CCK-8 and colony formation assays showed that circ_0003221 downregulation suppressed SiHa and CaSki cell viability and colony formation ability (Figure 2B–D), indicating that circ_0003221 knockdown could inhibit cell proliferation. Cell cycle progression was analyzed using flow cytometry. Knockdown of circ_0003221 increased the percentage of SiHa and CaSki cells in G0/G1 phase, while decreased the percentage of SiHa and CaSki cells in S phase, indicating that the cell cycle was arrested at the G0/G1 phase (Figure 2E and F). Consistent with CCK-8 and colony formation results, Edu assay showed that circ_0003221 interference decreased the percentage of Edu positive (Edu+) cells (Figure 2G), indicating that circ_0003221 knockdown inhibited cell proliferation. PCNA is a cell cycle and growth regulated protein that required for DNA replication and damage repair.20 Cyclin D1 is required for cell cycle progression in G1 and is a growth-promoting protein.21 As illustrated in Figure 2H and I, the protein levels of PCNA and Cyclin D1 were reduced by knockdown of circ_0003221. Transwell assay indicated that silence of circ_0003221 inhibited SiHa and CaSki cell migration and invasion (Figure 2J and K). Epithelial-mesenchymal transition (EMT) is a key step toward cancer metastasis.22 Therefore, Western blot was conducted to measure the levels of EMT-related proteins. The results showed that downregulation of circ_0003221 increased the protein level of E-cadherin (an epithelial marker) and decreased the expression levels of Snail and Vimentin (mesenchymal markers) (Figure 2L and M), suggesting inhibition of the EMT process. Overall, circ_0003221 knockdown repressed the malignant behaviors of cervical cancer cells.
Figure 2.
Circ_0003221 silence inhibited cervical cancer cell progression in vitro. SiHa and CaSki cells were transfected with si-NC or si-circ_0003221. (A) The expression of circ_0003221 was examined by qRT-PCR. (B and C) CCK-8 assay was used for detecting cell viability. (D) Colony formation assay was performed to examine the number of colony cells. (E and F) Flow cytometry analysis was used to determine cell cycle distribution. (G) Cell proliferation ability was determined by Edu assay (×200). (H and I) Western blot assay was conducted to analyze the protein expression of PCNA and Cyclin D1. (J and K) Transwell assay was conducted to estimate cell migration and invasion (×100). (L and M) The protein levels of E-cadherin, Snail and Vimentin were determined by Western blot assay. *P<0.05.
Circ_0003221 Acted as a Sponge of miR-758-3p
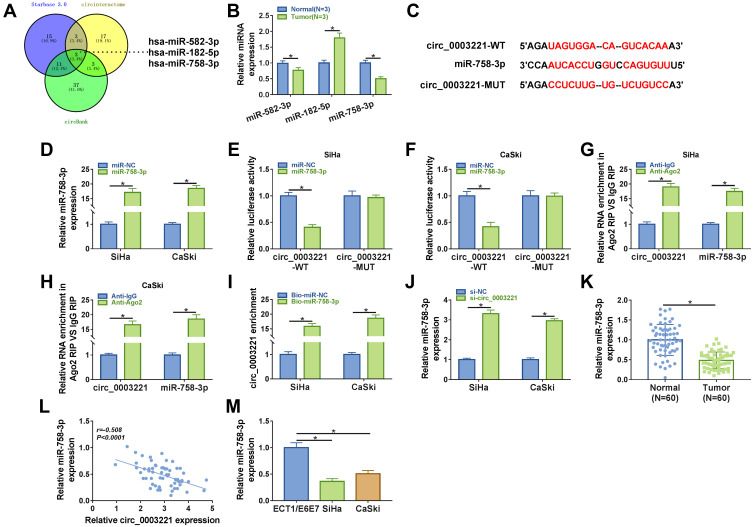
Increasing studies have verified that circRNAs can exert their functions by acting as ceRNAs to sponge miRNAs.23 Then, we further explored the molecular mechanism of circ_0003221 in cervical cancer. We identified three miRNAs (miR-582-3p, miR-182-5p, and miR-758-3p) from the overlap of three databases (Starbase 3.0, Circinteractome and circBank) as potential targets for circ_0003221 (Figure 3A). We found that the expression of miR-182-5p was upregulated in tumor tissues and the expression of miR-582-3p and miR-758-3p was downregulated in tumor tissues, and the expression of miR-758-3p decreased more (Figure 3B). Next, we choose miR-758-3p for further study. The complementary binding sites of miR-758-3p and circ_0003221 are shown in Figure 3C. Overexpression efficiency of miR-758-3p was confirmed by qRT-PCR in SiHa and CaSki cells (Figure 3D). To confirm the direct binding between miR-758-3p and circ_0003221, dual-luciferase reporter, RIP and RNA pull-down assays were performed. Dual-luciferase reporter assay manifested that miR-758-3p upregulation could reduce the luciferase activity of circ_0003221-WT, which could not be observed in circ_0003221-MUT group (Figure 3E and F). RIP assay showed that circ_0003221 and miR-758-3p were enriched by the Anti-Ago2 compared with that by Anti-IgG (Figure 3G and H). RNA pull-down experiments showed that transfection of Bio-miR-758-3p led to a significant increase in the enrichment of circ_0003221 in SiHa and CaSki cells (Figure 3I). Moreover, miR-758-3p expression in SiHa and CaSki cells transfected with si-circ_0003221 was significantly increased compared to si-NC group (Figure 3J). In cervical cancer tissues, miR-758-3p expression was decreased compared to normal tissues (Figure 3K). Furthermore, we found that miR-758-3p expression was negatively correlated with circ_0003221 expression in cervical cancer tissues (Figure 3L). In addition, the expression of miR-758-3p was also reduced in cervical cancer cells (SiHa and CaSki) compared to ECT1/E6E7 cells (Figure 3M). These results revealed that miR-758-3p was sponged by circ_0003221.
Figure 3.
MiR-758-3p was a direct target of circ_0003221. (A) 3 miRNAs (has-miR-582-3p, has-miR-182-5p, and has-miR-758-3p) that might be sponged by circ_0003221 were predicted by three databases (Starbase 3.0, Circinteractome and circBank). (B) The expression of miR-582-3p, miR-182-5p, and miR-758-3p was measured in normal tissues and cervical cancer tissues by qRT-PCR. (C) The binding sequences between miR-758-3p and circ_0003221 were presented. (D) The expression of miR-758-3p was detected by qRT-PCR in SiHa and CaSki cells transfected with miR-NC or miR-758-3p. (E–I) Dual-luciferase reporter assay, RIP assay and RNA pull-down assay were conducted to confirm the interaction between miR-758-3p and circ_0003221. (J) The level of miR-758-3p was detected by qRT-PCR in SiHa and CaSki cells transfected with si-NC or si-circ_0003221. (K) The expression of miR-758-3p was detected by qRT-PCR in normal tissues and cervical cancer tissues. (L) The correlation between miR-758-3p and circ_0003221 was analyzed in cervical cancer tissues. (M) QRT-PCR was used to examine the expression of miR-758-3p in ECT1/E6E7, SiHa and CaSki cells. *P<0.05.
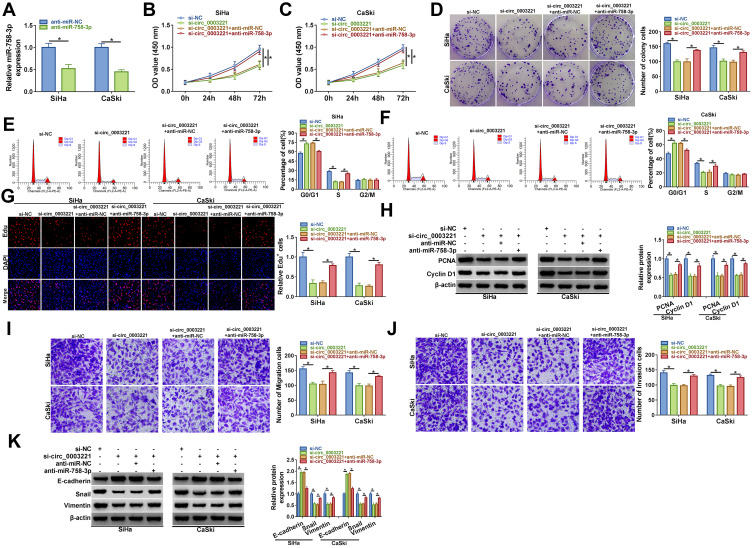
Inhibition of miR-758-3p Reversed the Effects of Circ_0003221 Knockdown on Proliferation, Cell Cycle and Metastasis in Cervical Cancer Cells
Inhibition efficiency of miR-758-3p was confirmed by transfection of miR-758-3p in SiHa and CaSki cells (Figure 4A). Next, our study explored whether circ_0003221 regulated the behavior of cervical cancer cells by sponging miR-758-3p. We found that miR-758-3p inhibition reversed the inhibitory effects of si-circ_0003221 on cell viability and colony-forming ability in SiHa and CaSki cells (Figure 4B–D). Meanwhile, downregulation of miR-758-3p abolished circ_0003221 knockdown-mediated impact on cell cycle progression (Figure 4E and F). Moreover, the effect of circ_0003221 silence on decrease of Edu+ cells was reversed by downregulating miR-758-3p (Figure 4G). Also, decreased miR-758-3p expression abated the suppressive influence of circ_0003221 silence on the levels of PCNA and Cyclin D1 (Figure 4H). In addition, miR-758-3p downregulation abated the anti-migration and anti-invasion effects caused by knockdown of circ_0003221 (Figure 4I and J). Besides, miR-758-3p could reverse the promotion of E-cadherin and reduction of Snail and Vimentin expression induced by si-circ_0003221 (Figure 4K). Taken together, these data suggested that circ_0003221 exerted its role by targeting miR-758-3p.
Figure 4.
Knockdown of circ_0003221 inhibited the progression of cervical cancer cells by upregulating miR-758-3p. (A) The expression of miR-758-3p was measured by qRT-PCR in SiHa and CaSki cells transfected with anti-miR-NC or anti-miR-758-3p. (B–K) SiHa and CaSki cells were transfected with si-NC, si-circ_0003221, si-circ_0003221 + anti-miR-NC, or si-circ_0003221 + anti-miR-758-3p. (B and C) CCK-8 assay was applied to detect cell viability. (D) Colony formation assay was used for measuring cell colony formation ability. (E and F) Cell cycle distribution was analyzed via flow cytometry analysis. (G) Cell proliferation ability was examined using Edu assay (×200). (H) Western blot assay was performed to detect the protein expression of PCNA and Cyclin D1. (I and J) Cell migration and invasion abilities were assessed by transwell assay (×100). (K) E-cadherin, Snail and Vimentin protein levels were determined by Western blot assay. *P<0.05.
CPEB4 Was a Direct Target of miR-758-3p
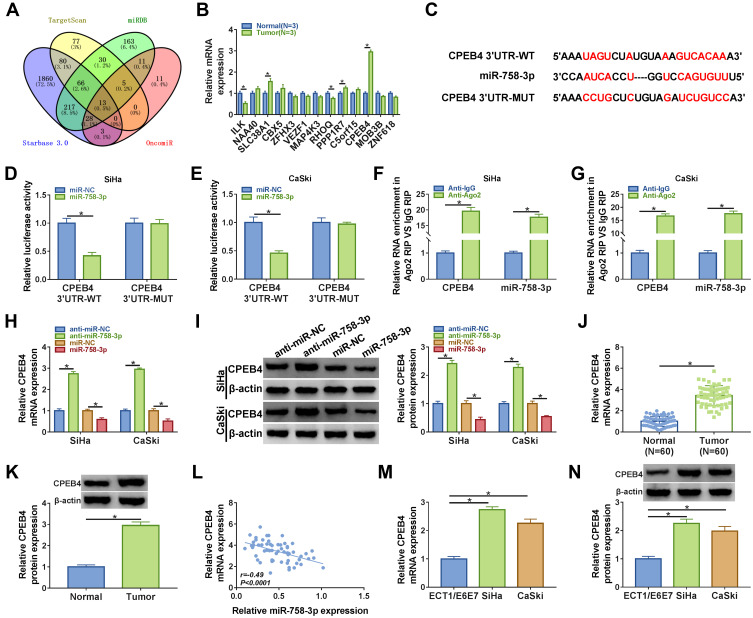
It is well accepted that miRNAs perform their functions mainly through the direct targeting of mRNAs, and we then aimed to identify the downstream targets of miR-758-3p. Four bioinformatics analysis tools, including TargetScan, miRDB, starbase 3.0, and OncomiR, showed that 13 predicted target mRNAs were found in the intersection part (Figure 5A). We found that ILK and RHOQ were downregulated, and SLC38A1, PPP1R7 and CPEB4 were upregulated in tumor tissues, and CPEB4 was upregulated the most (Figure 5B). Therefore, CPEB4 was selected for subsequent research. As presented in Figure 5C, there were several binding sites between miR-758-3p and CPEB4. Subsequently, dual-luciferase reporter assays confirmed that miR-758-3p overexpression markedly reduced the luciferase activity of CPEB4 3ʹUTR-WT but not the luciferase activity of CPEB4 3ʹUTR-MUT (Figure 5D and E). RIP assay indicated that the expression of CPEB4 and miR-758-3p was increased in Ago2-containing beads compared to IgG control group (Figure 5F and G). Moreover, we uncovered that the mRNA and protein expression of CPEB4 were increased by inhibiting miR-758-3p and reduced by overexpressing miR-758-3p in SiHa and CaSki cells (Figure 5H and I). In addition, CPEB4 mRNA and protein expression were upregulated in cervical cancer tissues in contrast to normal tissues (Figure 5J and K). Furthermore, the correlation between CPEB4 and miR-758-3p expression was analyzed in cervical cancer tissues. As shown in Figure 5L, a negative correlation between CPEB4 and miR-758-3p expression was observed. Furthermore, we found that CPEB4 mRNA and protein levels were upregulated in cervical cancer cells (SiHa and CaSki) compared to ECT1/E6E7 cells (Figure 5M and N). These data collectively indicated that miR-758-3p directly targeted CPEB4.
Figure 5.
CPEB4 was a downstream target of miR-758-3p. (A) Bioinformatics databases were used to predict the potential target genes of miR-758-3p. (B) The expression of potential target genes was detected by qRT-PCR in normal tissues and cervical cancer tissues. (C) The complementary binding sites of miR-758-3p and CPEB4 3ʹUTR were shown. (D–G) Dual-luciferase reporter assay and RIP assay were performed to verify the interaction between miR-758-3p and CPEB. (H and G) The mRNA and protein expression of CPEB4 were respectively determined by qRT-PCR and Western blot in in SiHa and CaSki cells transfected with anti-miR-NC, anti-miR-758-3p, miR-NC, or miR-758-3p. (J and K) CPEB4 mRNA and protein expression in normal tissues and cervical cancer tissues were detected by qRT-PCR and Western blot, respectively. (L) The correlation between CPEB4 mRNA expression and miR-758-3p expression was analyzed in cervical cancer tissues. (M and N) The mRNA and protein levels of CPEB4 in ECT1/E6E7, SiHa and CaSki cells were measured by qRT-PCR and Western blot, respectively. *P<0.05.
Overexpression of miR-758-3p Inhibited Cell Proliferation and Metastasis and Induced Cell Cycle Arrest in Cervical Cancer Cells by Targeting CPEB4
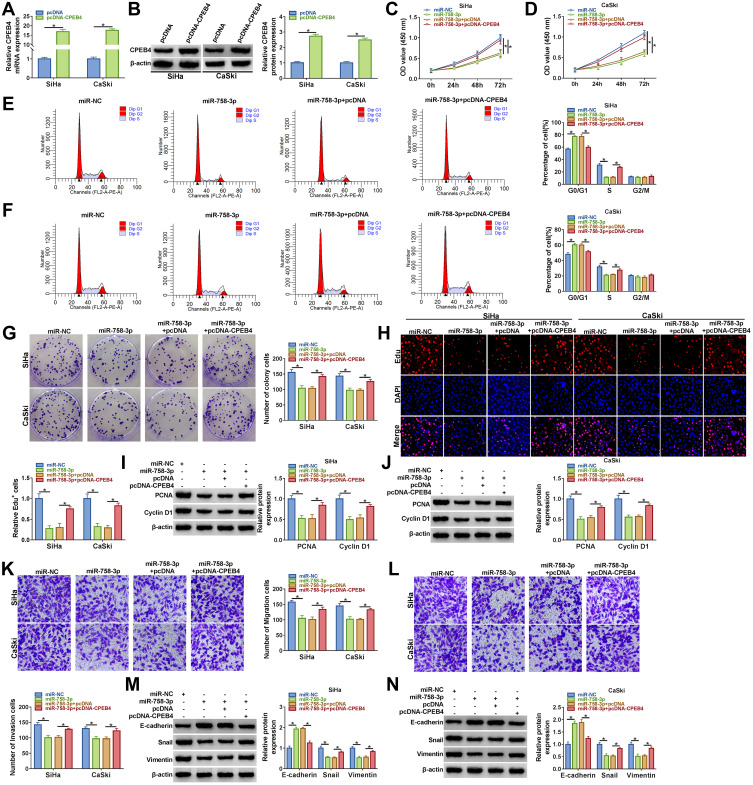
Overexpression efficiency of CPEB4 was determined by qRT-PCR and Western blot. The data indicated that transfection of CPEB4 increased the mRNA and protein expression of CPEB4 (Figure 6A and B), suggesting a high transection efficiency. To explore whether CPEB4 was required for miR-758-3p-mediated cervical cancer progression, cervical cancer cells (SiHa and CaSki) were transfected with miR-NC, miR-758-3p, miR-758-3p + pcDNA, miR-758-3p + pcDNA-CPEB4. We found that restoration of miR-758-3p reduced cell viability, colony-forming ability, and Edu+ cells and arrested cell cycle in the G0/G1 phase, while these effects were abated by upregulating CPEB4 (Figure 6C–H). Western blot showed that miR-758-3p overexpression reduced the protein expression of PCNA and Cyclin D1, which was reversed by elevating CPEB4 (Figure 6I and J). Moreover, enforced expression of miR-758-3p inhibited cell migration and invasion, while these inhibitory effects were reversed by addition of CPEB4 (Figure 6K and L). Meanwhile, the protein expression of E-cadherin was increased and the protein levels of Snail and Vimentin were reduced by upregulating miR-758-3p, which could be abolished by overexpression of CPEB4 (Figure 6M and N). These results indicated that miR-758-3p exerted anti-tumor role by targeting CPEB4 in cervical cancer cells.
Figure 6.
Overexpression of miR-758-3p repressed cervical cancer cell progression by downregulating CPEB4. (A and B) The mRNA and protein levels of CPEB4 in SiHa and CaSki cells transfected with pcDNA or pcDNA-CPEB4 were measured by qRT-PCR and Western blot, respectively. (C–N) SiHa and CaSki cells were transfected with miR-NC, miR-758-3p, miR-758-3p + pcDNA, miR-758-3p + pcDNA-CPEB4. (C and D) Cell viability was measured using CCK-8 assay. (E and F) Cell cycle distribution was measured using flow cytometry analysis. (G) Colony formation assay was performed to measure colony formation ability. (H) Edu assay was used to assess cell proliferation ability (×200). (I and J) Western blot assay was conducted to examine the protein levels of PCNA and Cyclin D1. (K and L) Cell migration and invasion were tested using transwell assay (×100). (M and N) The protein levels of E-cadherin, Snail and Vimentin were determined by Western blot assay. *P<0.05.
Circ_0003221 Sponged miR-758-3p to Regulate CPEB4 Expression in Cervical Cancer Cells
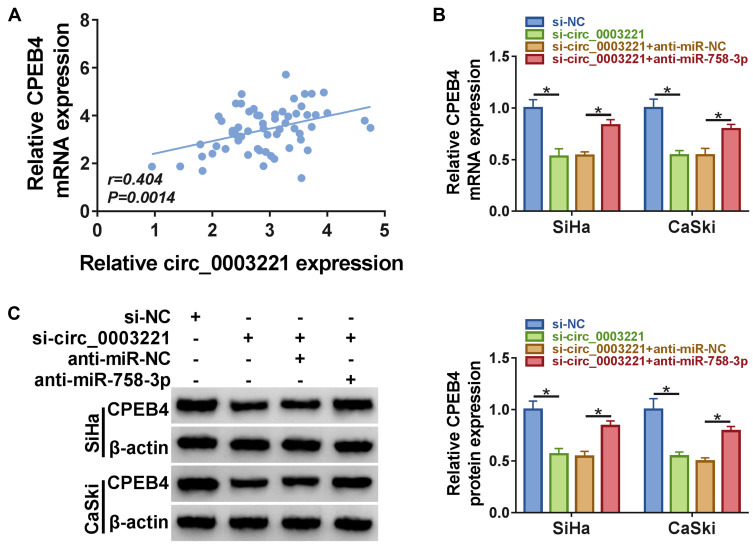
Next, the correlation between circ_0003221 and CPEB4 expression in cervical cancer tissues was analyzed. The results showed that CPEB4 mRNA level was positively correlated with circ_0003221 expression (Figure 7A). Subsequently, we explored whether circ_0003221 regulated CPEB4 expression by sponging miR-758-3p in cervical cancer cells. Results showed that CPEB4 mRNA and protein levels were decreased by silencing circ_0003221, which was restored by downregulating miR-758-3p (Figure 7B and C). These data indicated that circ_0003221 positively regulated CPEB4 expression by sponging miR-758-3p.
Figure 7.
Circ_0003221 regulated CPEB4 expression by sponging miR-758-3p. (A) The correlation between CPEB4 mRNA expression and circ_0003221 expression was analyzed in cervical cancer tissues. (B and C) SiHa and CaSki cells were transfected with si-NC, si-circ_0003221, si-circ_0003221 + anti-miR-NC, or si-circ_0003221 + anti-miR-758-3p. Then qRT-PCR and Western blot were used to measure the mRNA and protein expression of CPEB4. *P<0.05.
Interference of Circ_0003221 Inhibited Tumor Growth by Regulating miR-758-3p and CPEB4 Expression
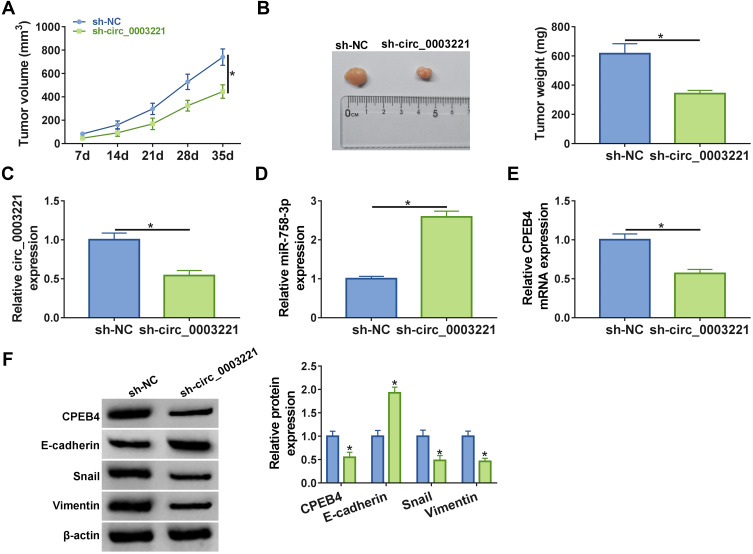
The mice xenograft model was established to assess whether circ_0003221 acted as a tumor promoter in vivo. Results showed that tumors volume and weight were markedly reduced in the sh-circ_0003221 group compared with sh-NC group (Figure 8A and B). Consistent with the in vitro results, knockdown of circ_0003221 reduced the expression of circ_0003221 and CPEB4 and enhanced miR-758-3p expression in tumor tissues (Figure 8C–E). Additionally, we found that downregulation of circ_0003221 reduced CPEB4, Snail and Vimentin protein expression and increased E-cadherin protein expression in excised tumor masses (Figure 8F). Altogether, these data proved that circ_0003221 knockdown repressed tumor growth in vivo by increasing miR-758-3p and decreasing CPEB4 expression.
Figure 8.
Depletion of circ_0003221 repressed tumor growth in vivo. SiHa cells transfected with sh-NC or sh-circ_0003221 were introduced into nude mice to establish mice xenograft model. (A) Tumor volume was assessed every 7 days after injection. (B) Representative photographs of dissected tumors were presented, and tumor weights were calculated after injection for 35 days. (C–E) The expression levels of circ_0003221, miR-758-3p and CPEB4 were determined by qRT-PCR in tumor tissues. (F) The protein expression of CPEB4, E-cadherin, Snail, and Vimentin in the collected tissues was detected by Western blot assay. *P<0.05.
Discussion
CircRNAs are highly stable and conserved among species due to their closed circular structures.24 Increasing evidence illustrates that circRNAs participate in the regulation of multiple physiological activities and diseases.25,26 The roles of circRNAs in tumorigenesis have recently attracted extensive attention. Herein, we found that circ_0003221 accelerated cell growth, migration, invasion, and EMT in cervical cancer. Mechanistically, circ_0003221 served as as a ceRNA to sponge miR-758-3p, thus upregulating CPEB4 expression.
Circ_0003221 has been suggested to function as a tumor-promoting circRNA in several cancers. For instance, Xu et al showed that circ_0003221 contributed to cell proliferation and migration of bladder cancer.27 Liu et al stated that circ_0003221 was overexpressed in gastric cancer and circ_0003221 promoted gastric cancer progression and radiosensitivity by regulating miR-369-3p/ZEB1 axis.28 Although the oncogenic role of circ_0003221 has been studied in some tumors, the role of circ_0003221 in cervical cancer has not been investigated. In our research, circ_0003221 was upregulated in cervical cancer tissues and cells, which was in line with previous finding.15 Further functional experiments revealed that silence of circ_0003221 could restrain cell proliferation and metastasis and could promote cell cycle arrest, implying that circ_0003221 might be a potential molecular target for cervical cancer treatment.
It is well known that circRNAs are involved in cancer progression via serving as ceRNAs for miRNAs.29 To investigate the molecular mechanism, bioinformatics tools were applied to predict the possible miRNA targets of circ_0003221. We demonstrated that circ_0003221 acted as a sponge for miR-758-3p. MiR-758-3p has been reported to function as a tumor suppressor via directly interacting with target genes in multiple cancers, such as hepatocellular carcinoma,30 bladder cancer,31 papillary thyroid cancer,32 clear cell renal cell carcinoma.33 Moreover, miR-758 was found to restrain cell proliferation and metastasis of cervical cancer via regulating HMGB3/WNT/β‑catenin signaling pathway.18 Furthermore, miR-758 was downregulated in cervical cancer, and miR-758 might regulate cervical cancer infiltration and invasion via targeting MEPE.34 In line with previous studies, we uncovered that miR-758-3p level was reduced in cervical cancer specimens and cell lines. Rescue assays indicated that miR-758-3p downregulation could abate the roles of circ_0003221 silence in cervical cancer cells. Therefore, we illustrated that circ_0003221 might promote cervical cancer progression via sponging miR-758-3p.
Next, we also researched the downstream targets of miR-758-3p. CPEB4 was demonstrated as a downstream target of miR-758-3p. CPEB4 has been demonstrated to be tightly related to tumor cell proliferation, migration, invasion, and interstitial transformation through various signaling pathways.35 Previous studies found that CPEB4 acted as an oncogene in breast cancer,36 gastric cancer,37 colorectal cancer,38 and lung cancer.39 In addition, CPEB4 has been reported to be upregulated in cervical cancer tissues, and EWSAT1 upregulated CPEB4 to promote cervical cancer development.19 Consistently, we also observed that CPEB4 was upregulated in cervical cancer tissues and cells. Moreover, we also identified that overexpression of CPEB4 could attenuate the anti-cancer role of miR-758-3p cervical cancer cells. In addition, we found that circ_0003221 acted as a ceRNA for miR-758-3p, thereby regulating the expression of downstream target CPEB4. Consistent with in vivo results, silencing circ_0003221 could suppress tumor growth via increasing miR-758-3p and decreasing CPEB4.
In summary, the data revealed that circ_0003221 was overexpressed in cervical cancer tissues and cells. Circ_0003221 knockdown inhibited cervical cancer cell proliferation and metastasis and induced cell cycle arrest via regulating miR-758-3p/CPEB4 axis. Our findings suggested that circ_0003221/miR-758-3p/CPEB4 axis might act as a new regulatory pathway and provide a promising target for cervical cancer intervention.
Funding Statement
This work was supported by the mechanism and clinical application of HPV mediating cervical cancer via CIP2A/autophagy pathway (No. 2018SK51505) and the study on the relationship between TMPRSS4 expression regulation and invasion and metastasis of cervical cancer (No. Hengkefa [2019] No. 47).
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Franco EL, Duarte-Franco E, Ferenczy A. Cervical cancer: epidemiology, prevention and the role of human papillomavirus infection. CMAJ. 2001;164(7):1017–1025. [PMC free article] [PubMed] [Google Scholar]
- 3.Small W, Bacon MA, Bajaj A, et al. Cervical cancer: a global health crisis. Cancer. 2017;123(13):2404–2412. doi: 10.1002/cncr.30667 [DOI] [PubMed] [Google Scholar]
- 4.Yee GP, de Souza P, Khachigian LM. Current and potential treatments for cervical cancer. Curr Cancer Drug Targets. 2013;13(2):205–220. doi: 10.2174/1568009611313020009 [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Batur P. Human papillomavirus in 2019: an update on cervical cancer prevention and screening guidelines. Cleve Clin J Med. 2019;86(3):173–178. doi: 10.3949/ccjm.86a.18018 [DOI] [PubMed] [Google Scholar]
- 6.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. doi: 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 9.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi: 10.1007/s12282-017-0793-9 [DOI] [PubMed] [Google Scholar]
- 10.Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng L, Jia X, Yu W, Wang C, Chen J, Liu F. Circular RNA UBAP2 contributes to tumor growth and metastasis of cervical cancer via modulating miR-361-3p/SOX4 axis. Cancer Cell Int. 2020;20:357. doi: 10.1186/s12935-020-01436-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Chen Z, Sun J, An N, Xi Q. CircRNA hsa_circRNA_0000069 promotes the proliferation, migration and invasion of cervical cancer through miR-873-5p/TUSC3 axis. Cancer Cell Int. 2020;20:287. doi: 10.1186/s12935-020-01387-5 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ma H, Tian T, Liu X, et al. Upregulated circ_0005576 facilitates cervical cancer progression via the miR-153/KIF20A axis. Biomed Pharmacother. 2019;118:109311. doi: 10.1016/j.biopha.2019.109311 [DOI] [PubMed] [Google Scholar]
- 14.Ji F, Du R, Chen T, et al. Circular RNA circSLC26A4 accelerates cervical cancer progression via miR-1287-5p/HOXA7 axis. Mol Ther Nucleic Acids. 2020;19:413–420. doi: 10.1016/j.omtn.2019.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Ma N, Zhang Y, et al. Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death Dis. 2020;11(5):399. doi: 10.1038/s41419-020-2607-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129. doi: 10.1016/j.omtn.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. doi: 10.1007/978-981-13-1426-1_6 [DOI] [PubMed] [Google Scholar]
- 18.Song T, Hou X, Lin B. MicroRNA-758 inhibits cervical cancer cell proliferation and metastasis by targeting HMGB3 through the WNT/β-catenin signaling pathway. Oncol Lett. 2019;18(2):1786–1792. doi: 10.3892/ol.2019.10470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Q, Xie Y, Wang L, Xu T, Gao Y. LncRNA EWSAT1 upregulates CPEB4 via miR-330-5p to promote cervical cancer development. Mol Cell Biochem. 2020;471(1–2):177–188. doi: 10.1007/s11010-020-03778-8 [DOI] [PubMed] [Google Scholar]
- 20.Prelich G, Stillman B. Coordinated leading and lagging strand synthesis during SV40 DNA replication in vitro requires PCNA. Cell. 1988;53(1):117–126. doi: 10.1016/0092-8674(88)90493-X [DOI] [PubMed] [Google Scholar]
- 21.Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993;7(5):812–821. doi: 10.1101/gad.7.5.812 [DOI] [PubMed] [Google Scholar]
- 22.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15(3):195–206. doi: 10.1016/j.ccr.2009.01.023 [DOI] [PubMed] [Google Scholar]
- 23.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011 [DOI] [PubMed] [Google Scholar]
- 24.Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Yang T, Xiao XJ. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi: 10.1186/s12943-018-0934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu ZQ, Yang MG, Liu HJ, Su CQ. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119(4):3317–3325. doi: 10.1002/jcb.26492 [DOI] [PubMed] [Google Scholar]
- 28.Liu Y, Yao K, Zhang K, Wang J, Dai Q, Wang R. Circular RNA PTK2 modifies the progression and radiosensitivity in gastric cancer via miR-369-3p/ZEB1 axis. RSC Adv. 2020;10(3):1711–1723. doi: 10.1039/C9RA08525D [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680. doi: 10.1002/1878-0261.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang D, Cho WC, Li Z, et al. MiR-758-3p suppresses proliferation, migration and invasion of hepatocellular carcinoma cells via targeting MDM2 and mTOR. Biomed Pharmacother. 2017;96:535–544. doi: 10.1016/j.biopha.2017.10.004 [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Chen B, Shi H, et al. miR-758-3p suppresses human bladder cancer cell proliferation, migration and invasion by targeting NOTCH2. Exp Ther Med. 2019;17(5):4273–4278. doi: 10.3892/etm.2019.7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Xu Z, Yu C, et al. MiR-758-3p regulates papillary thyroid cancer cell proliferation and migration by targeting TAB1. Pharmazie. 2019;74(4):235–238. doi: 10.1691/ph.2019.8933 [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, Liu Y. miR-758-3p inhibits proliferation, migration, and invasion of clear cell renal cell carcinoma and predicts favorable prognosis. Cancer Manag Res. 2020;12:9285–9295. doi: 10.2147/cmar.s261882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng X, Zhao Y, Wang J, Gao Z, Geng Q, Liu X. Regulatory roles of miRNA-758 and matrix extracellular phosphoglycoprotein in cervical cancer. Exp Ther Med. 2017;14(4):2789–2794. doi: 10.3892/etm.2017.4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu H, Liu B. CPEB4 is a candidate biomarker for defining metastatic cancers and directing personalized therapies. Med Hypotheses. 2013;81(5):875–877. doi: 10.1016/j.mehy.2013.08.030 [DOI] [PubMed] [Google Scholar]
- 36.Lu R, Zhou Z, Yu W, Xia Y, Zhi X. CPEB4 promotes cell migration and invasion via upregulating Vimentin expression in breast cancer. Biochem Biophys Res Commun. 2017;489(2):135–141. doi: 10.1016/j.bbrc.2017.05.112 [DOI] [PubMed] [Google Scholar]
- 37.Cao G, Chen D, Liu G, Pan Y, Liu Q. CPEB4 promotes growth and metastasis of gastric cancer cells via ZEB1-mediated epithelial- mesenchymal transition. Onco Targets Ther. 2018;11:6153–6165. doi: 10.2147/ott.s175428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong X, Xiao Y, Chen C, et al. MicroRNA-203-mediated posttranscriptional deregulation of CPEB4 contributes to colorectal cancer progression. Biochem Biophys Res Commun. 2015;466(2):206–213. doi: 10.1016/j.bbrc.2015.09.008 [DOI] [PubMed] [Google Scholar]
- 39.Hu J, Zhang L, Chen Q, et al. Knockdown of CPEB4 expression suppresses cell migration and invasion via Akt pathway in non-small cell lung cancer. Cell Biol Int. 2018;42(11):1484–1491. doi: 10.1002/cbin.10930 [DOI] [PubMed] [Google Scholar]