Abstract
Lipid transfer from host plants to arbuscular mycorrhiza fungi was hypothesized for several years because sequenced arbuscular mycorrhiza fungal genomes lack genes encoding cytosolic fatty acid synthase ( Wewer et al., 2014 ; Rich et al., 2017 ). It was finally shown by two independent experimental approaches ( Jiang et al., 2017 ; Keymer et al., 2017 ; Luginbuehl et al., 2017 ). One approach used a technique called isotopolog profiling ( Keymer et al., 2017 ). Isotopologs are molecules, which differ only in their isotopic composition. For isotopolog profiling an organism is fed with a heavy isotope labelled precursor metabolite. Subsequently, the labelled isotopolog composition of metabolic products is analysed via mass spectrometry. The detected isotopolog pattern of the metabolite(s) of interest yields information about metabolic pathways and fluxes ( Ahmed et al., 2014 ). The following protocol describes an experimental setup, which enables separate isotopolog profiling of fatty acids in plant roots colonized by arbuscular mycorrhiza fungi and their associated fungal extraradical mycelium, to elucidate fluxes between both symbiotic organisms. We predict that this strategy can also be used to study metabolite fluxes between other organisms if the two interacting organisms can be physically separated.
Keywords: Arbuscular mycorrhiza, Lotus japonicus, Isotopolog profiling, Stable isotope labelling, Rhizophagus irregularis, MSR medium, Inter-organismic lipid transfer, Root organ culture, Nurse plant system
Background
Arbuscular mycorrhiza fungi are biotrophic organisms. As such, they cannot be cultivated independently but rely on interaction with host plants to stay alive and complete their life cycle. This characteristic makes it challenging to study the two symbiotic organisms and especially the fungus separately.
To cultivate, treat and harvest the fungus separately from the host root, a 2-compartmented Petri dish system was developed and used for labelling studies in previous work (Bécard and Fortin, 1988; Pfeffer et al., 1999 ; Trépanier et al., 2005 ). This system is composed of two compartments; one containing a Ri (root-inducing) T-DNA transformed carrot root (Mosse and Hepper, 1975), which hosts the fungus (‘carrot compartment’) and another one, which contains only the fungus (fungal compartment), because the extraradical mycelium has grown across the border, which divides the two compartments. Kuhn et al. (2010) have advanced this setup to colonize Medicago truncatula tester plants in the fungal compartment.
In this protocol, we used this previous knowledge and further modified the system to our needs. Combining this growth system with stable isotopolog labelling and profiling ( Eisenreich et al., 2013 ) enables analysis of plant and fungal metabolites separately from each other to gain information about metabolite fluxes between the two symbionts.
Materials and Reagents
-
Growth system setup
Fine sandpaper (60 µm)
2-Compartmented Petri dish (diam. 9.4 cm) (Greiner Bio One International, catalog number: 635161)
Square Petri dishes (12 cm) (Greiner Bio One International, catalog number: 688161)
Pipette tips
Scalpel
Parafilm
2.0 ml Eppendorf tubes
1.5 ml Eppendorf tubes
Black card-sheet paper (50 x 70 cm; 150 g/m2)
Adhesive tape
Sterile Rhizophagus irregularis spores (Agronutrition, Carbonne, France; LOT: 000300195)
1% NaClO solution
Bacto agar (BD, BactoTM, catalog number: 214010)
EtOH
[U13C6]-Glucose (Sigma-Aldrich, catalog number: 389374)
10% KOH
Ink & vinegar staining solutions ( Vierheilig et al., 1998 )
Liquid nitrogen
MSR plate with carrot root organ culture (Bécard and Fortin, 1988)
MSR plate with colonized carrot root organ culture (Bécard and Fortin, 1988)
Sucrose
Gelrite (Duchefa Biochemie, catalog number: G1101)
Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: M2773)
Potassium nitrate (KNO3) (Sigma-Aldrich, catalog number: P8291)
Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541)
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655)
Calcium nitrate tetrahydrate (Ca(NO3)2·4H2O) (Sigma-Aldrich, catalog number: C2786)
Calcium pantothenate (B5) (Sigma-Aldrich, catalog number: 705837)
Biotin (B7) (Sigma-Aldrich, catalog number: B4639)
Nicotinic acid (B3) (Sigma Aldrich; catalog number: N0761)
Pyridoxine (B6) (Sigma-Aldrich, catalog number: P5669)
Thiamine (B1) (Sigma-Aldrich, catalog number: T1270)
Cyanocobalamine (B12) (Sigma-Aldrich, catalog number: V6629)
NaFeEDTA (Sigma-Aldrich, catalog number: E6760)
Manganese(II) sulfate tetrahydrate (MnSO4·4H2O) (Merck, catalog number: 102786)
Zinc sulfate heptahydrate (ZnSO4·7H2O) (Sigma-Aldrich, catalog number: Z1001)
Boric acid (H3BO3) (Sigma-Aldrich, catalog number: B6768)
Copper(II) sulfate pentahydrate (CuSO4·5H2O) (Sigma-Aldrich, catalog number: C8027)
Sodium molybdate dihydrate (Na2MoO4·2H2O) (Sigma-Aldrich, catalog number: M1651)
Ammonium molybdate tetrahydrate ((NH4)6Mo7O24·4H2O) (Sigma-Aldrich, catalog number: M1019)
Sodium citrate dihydrate (Sigma-Aldrich, catalog number: W302600)
Citric acid monohydrate (Sigma-Aldrich, catalog number: C1909)
-
MSR-medium (w/3% gelrite, w/10% sucrose) (see Recipes)
Solution 1: Macroelements
Solution 2: Calcium nitrate
Solution 3: Vitamins
Solution 4: NaFeEDTA
Solution 5: Microelements
-
MSR medium (w/ 3% gelrite, w/o 10% sucrose) (see Recipes and adjust accordingly)
Note: Refer to MSR-medium (w/ 3% gelrite, w/10% sucrose) in Recipe section for preparation of this medium.
-
MSR medium (w/o 3% gelrite, w/o sucrose) (see Recipes and adjust accordingly)
Note: Refer to MSR-medium (w/ 3% gelrite, w/ 10% sucrose) in Recipe section for preparation of this medium.
-
10 mM citrate buffer (see Recipes)
0.1 M sodium citrate
0.1 M citric acid
-
Isotopolog profiling
1.5 ml glass vials and screw caps for GC/MS autosampler (VWR, catalog numbers: 548-0018 and 548-0814)
MeOH containing 3 M HCl (Sigma-Aldrich, catalog number: 33050-U)
Hexane, anhydrous (Sigma-Aldrich, catalog number: 296090)
Equipment
-
Growth system setup
Mortar
Rotator
Incubator at 25 °C without lights
Plant growth chamber, set at 24 °C, 16/8 h day/night cycle
Clean bench
Bunsen burner
Microwave
25 ml flasks
Balance
Magnetic stirrer
Stir bars
Forceps
Pipettes
Microcentrifuge
Benchtop N2 container
-80 °C freezer
-
Isotopolog profiling
Freeze dryer (Christ, Osterode, Germany)
Drying oven
GCMS-QP 2010 plus with autosampler AOC20i and LabSolution software (Shimadzu, model: GCMS-QP 2010 Plus)
Silica capillary column (equity TM-5; 30 m by 0.25 mm, 0.25-µm film thickness; Sigma-Aldrich, Supelco, catalog number: 28089-U)
Software
LabSolution software: https://www.shimadzu.com/an/gc/advflowtech/sw-dl.html
Isotopo: http://www.tr34.uni-wuerzburg.de/software_developments/isotopo/
Procedure
-
Growth system setup
-
Cultivation of Lotus japonicus seedlings
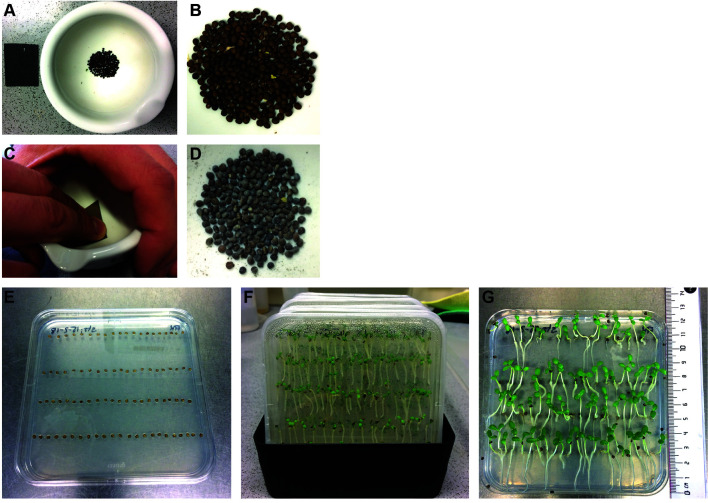
Lotus japonicus seeds are carefully scarified with sandpaper inside a mortar until the seeds turn slightly grey (Figures 1A-1D). They are then surface sterilized with 1% NaClO for 6 min, washed three times with sterile water and subsequently incubated in sterile water on a rotator for 180 min at room temperature. Imbibed seeds are placed in four rows, each row with 20 seeds, on square Petri dishes (Figure 1E) containing 0.8% Bacto agar and incubated in the dark at 24 °C for 3 days.
Subsequently, the germinated seedlings on square Petri dishes are cultivated for additional 10 days in the light (16 h/8 h day-night cycle) at 24 °C. Then they are used for the two-compartment setup (Figure 1G). The plates are placed into dark-colored flat boxes (Figure 1F) to provide a dark-stimulus to the roots from the bottom and encourage straight growth.
-
Cultivation of carrot root organ culture on 2-compartment test Petri dishes
All steps need to be performed in a clean bench to avoid contamination of root organ culture.
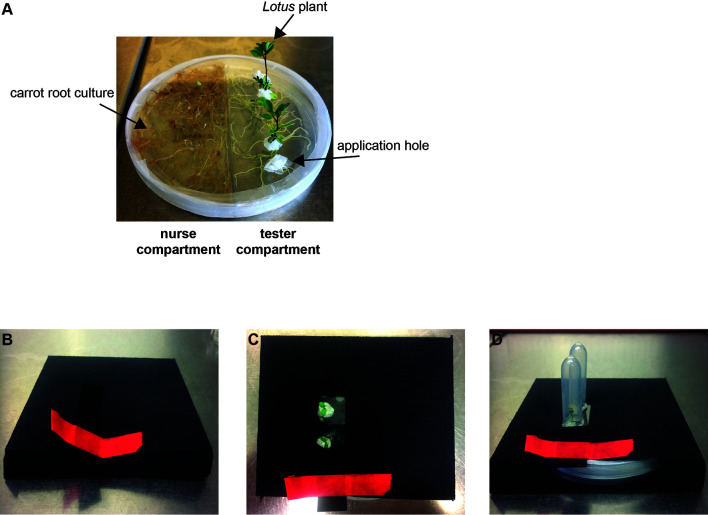
Both compartments (tester & nurse compartment, Figure 2A) of the spilt Petri dish are filled with MSR medium. However, only the MSR medium of the nurse compartment contains 10% sucrose, whereas the MSR medium of the tester compartment does not. Both compartments need to be filled with medium up to the top of the dish in order to assure the availability of sufficient substrate during the entire course of cultivation.
A fresh root piece (about 2.5 cm) of a carrot root organ culture is placed onto the medium in the nurse compartment. The Petri dish is closed, sealed with Parafilm and incubated for 1 week upside down in darkness at 25 °C.
7 days later, the carrot root culture in the nurse compartment is inoculated with R. irregularis. To this end, MSR-agar blocks (about 0.125 cm3) from another root organ culture containing fungal spores and mycelium are placed on two different spots on the growing carrot root culture in the nurse compartment. The plate is sealed and again incubated upside down in total darkness at 25 °C to allow the fungus to colonize the whole plate including the tester compartment.
Incubate for 4 to 6 weeks. The plates need to be checked regularly (about once per week) in order to assure that only the fungus crosses the compartment barrier but the carrot roots do not. If carrot roots grow across the barrier to the tester compartment they need to be pruned using sterile scissors.
When the fungal extraradical mycelium has spread over both compartments (both compartments are covered with fungal hyphae) you can proceed to the next stage.
-
Placement of two Lotus seedlings into the tester compartment
Clean the lid of the Petri dish with EtOH using a tissue paper to ensure maximum sterility.
Heat forceps in the flame of a Bunsen burner and use the hot forceps to melt three holes (approx. 0.5 cm diameter) into the lid of the Petri dish on the tester compartment side. Two of the holes are needed for Lotus growth and the third is later used for labelled substrate application.
13 days old Lotus seedlings are placed through the prepared lid holes in the tester compartment. Take care that the root is well-attached to the medium and is not hanging loose in the air.
Seal the Petri dish with Parafilm.
Carefully seal the two seedling holes as well as the application hole with Parafilm to avoid contamination.
Cover seedlings immediately with a 2 ml Eppendorf tube (like a mini-greenhouse) to prevent them from drying out. Keep seedlings covered for the first 5 days of incubation.
Build black envelopes yourself from commercially available cardboard sheets to cover the Petri dish such that the roots and fungus are kept in the dark and the Lotus shoots are in the light (Figures 2B-2D).
Incubate for 3 weeks at 24 °C, 16/8 h day/night cycle until the fungus has colonized the root.
-
Application of [U13C6]-glucose
Labelled glucose is applied 3 weeks after Lotus seedling placement.
100 mg of [U13C6]-glucose are dissolved in 2 ml MSR-medium (w/o sugar, w/o gelrite).
The 2 ml [U13C6]-glucose/MSR solution is carefully pipetted into the tester compartment via the application hole in the lid. Strictly avoid that the labelled glucose solution also drips in the nurse compartment and wait until the liquid has soaked into the agar before moving the plate.
-
After application of the labelled substrate, the application hole of the Petri dish is sealed again and the setup is incubated in the growth chamber for another week.
Note: One week after the stable isotope-labelled substrate has been added, the tester plants as well as the extraradical mycelium of the fungus are harvested.
-
Harvest of the tester plants
Both plants are carefully isolated from the plate. By slowly lifting the lid of the Petri dish the attached Lotus plants will be extricated from the medium. Make sure that no gelrite pieces remain on the roots.
Use one of the plants for quantification of root length colonization. Place the root of this plant in 10% KOH. Later stain the root with ink & vinegar ( Vierheilig et al., 1998 ) and quantify root length colonization using a modified gridline intersect method ( McGonigle et al., 1990 ).
The other root system is used for isotopolog profiling. Place it into a 2 ml Eppendorf tube and immediately shock freeze it in liquid nitrogen.
-
Extraction of the fungal extraradical mycelium
The MSR/gelrite medium of the tester compartment is cut into small pieces (about 0.125 cm3) and transferred into a 25 ml flask filled with citrate buffer.
The buffer containing gelrite pieces is stirred for 15 min using a magnetic stirrer (750 rpm). Within this time the gelrite is dissolved and the fungal mycelium knots together.
When knotted together, transfer the mycelium into a 1.5 ml Eppendorf tube by fishing it from the buffer with forceps.
Centrifuge the tube for 1 min at 9,391 × g and remove the residual liquid.
Subsequently shock freeze the sample in liquid nitrogen and keep it in a -80 °C freezer until mass spectrometry.
-
-
Isotopolog profiling
Freeze dry the samples.
Transfer samples to 1.5 ml glass vials.
Derivatize with 500 µl MeOH containing 3 M HCl (Sigma-Aldrich) at 80 °C for 20 h.
Dry the sample using a gentle stream of nitrogen gas.
Add 100 µl dry hexane to dissolve methyl esters of the fatty acids.
Analyze the samples by GC/MS: use a quadrupole GC/MS machine equipped with an autosampler and heated injection port (GCMS-QP 2010 plus; Shimadzu).
GC/MS setup.
Inject an aliquot of the solution in split mode (1:5) at an injector and interface temperature of 260 °C.
Hold the column at 170 °C for 3 min, then increase temperature by a gradient of 2 °C/min to a temperature of 192 °C, afterwards by a temperature gradient of 30 °C/min to a final temperature of 300 °C.
Perform one scan run for each sample, detecting a mass range from m/z 45-600, for unambiguous identification of the fatty acid and for confirmation of the retention times; this is especially important after a longer period, without this special analysis, and after a column change.
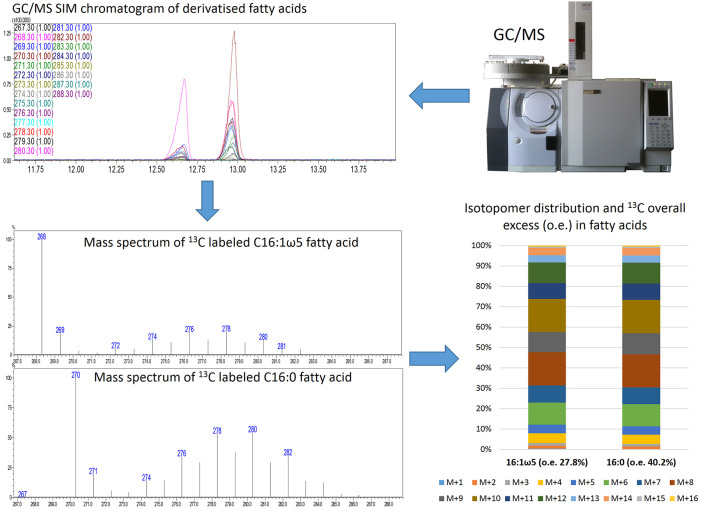
Analyse samples in SIM (single ion monitoring) mode (m/z values 267 to 288) at least three times. Retention times for fatty acids 16:1 ω5 (unlabeled m/z 268) and 16:0 (unlabeled m/z 270) are 12.87 min and 13.20 min, respectively. For the analysis of other fatty acids, determine retention times and m/z values in scan runs and implement the values into the SIM method.
Collect data with LabSolution software or the software connected to your GC/MS system.
Calculate overall 13C enrichment and isotopolog composition by comparison with an unlabeled sample according to Ahmed et al. (2014) . The software package is open source and can be downloaded by the following link: http://www.tr34.uni-wuerzburg.de/software_developments/isotopo/.
Compare overall 13C excess (average value of 13C atoms incorporated into 16:0/16:1 ω5 fatty acids) as well as isotopomer distribution (M + 1, M + 2, M + 3, … M + 16). For detailed explanation of nomenclature see Eisenreich et al. (2013) .
Perform at least three independent labeling experiments.
Data can be displayed as stacked columns as shown in Figure 3.
Figure 1. Cultivation of Lotus japonicus seedlings.
A and C. Lotus japonicus seeds are carefully scarified with sandpaper inside a mortar until the seeds turn slightly grey. Seeds before (B) and after (D) scarification. E. Imbibed seeds on 0.8% Bacto agar plates. F. Cultivation of seedlings in dark-coloured flat boxes (16 h light/8 h dark for 10 days). G. Seedlings ready for transfer to 2-compartement setups.
Figure 2. Growth system setup.
A. Cultivated Lotus japonicus plants in the split Petri dish growth system at 4 weeks post placement. B. Example of a black carton envelope to keep the plate with roots and fungus in the dark. The red adhesive tape serves to close the opening of the paper envelope. C-D. Completed setup ready for incubation without (C) or with (D) Eppendorf tubes to prevent the seedlings from drying out.
Figure 3. Schematic representation of the istotopolog profiling pipeline.
Analysis of samples via GC/MS SIM results in chromatograms of derivatized fatty acids. The individual mass spectra of 16:0 and 16:1 ω5 are extracted and the isotopomer distribution as well as the 13C overall excess (o.e) of these fatty acids is calculated. The isotopomer patterns can be displayed as stacked bars.
Data analysis
Statistical differences of overall 13C excess values for the different tested plant genotypes are analyzed via ANOVA followed by posthoc Tukey test in R, using at least 3 biological replicates per genotype.
Notes
Differences in root system development, distribution of labelled substrate on the Petri dish and the resulting differences in uptake of labelled substrate by the plant can lead to divergent isotopolog patterns among samples (see Keymer et al., 2017 ). However, when a metabolite (i.e., lipid) is transferred from the plant to the fungus, the isotopolog pattern among plant root and associated extraradical fungal mycelium is equivalent, notwithstanding the inter-sample variation.
Recipes
-
MSR-medium (w/ 3% gelrite, w/ 10% sucrose)
Medium preparation for 1 L:
Solution 1 10 ml
Solution 2 10 ml
Solution 4 5 ml
Solution 5 1 ml
Sucrose 10 g
Adjust to pH 5.5
Autoclave
Solution 3 5 ml
-
Solution 1: Macroelements
39.6 g MgSO4·7H2O
3.8 g KNO3
3.3 g KCl
0.21 g KH2PO4 in 500 ml H2O
-
Solution 2: Calcium nitrate
17.95 g Ca(NO3)2·4H2O in 500 ml H2O
-
Solution 3: Vitamins
90 mg calcium panthotenate (B5)
0.1 mg biotin (B7)
100 mg nicotinic acid (B3)
90 mg pyridoxine (B6)
100 mg thiamine (B1)
40 mg cyanocobalamine (B12) in 500 ml H2O
-
Solution 4: NaFeEDTA
0.4 g NaFeEDTA in 500 ml H2O
-
Solution 5: Microelements
(1) 1.225 g MnSO4·4H2O in 50 ml adjust to 100 ml
(2) 0.14 g ZnSO4·7H2O in 50 ml adjust to 100 ml
(3) 0.925 g H3BO3 in 50 ml adjust to 100 ml
(4) 1.1 g CuSO4·5H2O in 30 ml adjust to 50 ml
(5) 0.12 g Na2MoO4·2H2O in 50 ml adjust to 100 ml
(6) 1.7 g (NH4)6Mo7O24·4H2O in 50 ml adjust to 100 ml
Mix 100 ml (1) + 100 ml (2) + 100 ml (3) +5ml (4) + 1 ml (5) + 1 ml (6) and adjust to 500 ml to obtain Solution 5
-
-
10 mM citrate buffer
180 ml (0.1 M sodium citrate) + 820 ml (0.1 M citric acid) (you need 25 ml per compartment)
For 1.0 L stock solutions:
0.1 M sodium citrate: 29.41 g sodium citrate dehydrate (FW = 294.10 g/mol)
0.1 M citric acid: 21.01 g citric acid monohydrate (FW = 210.14 g/mol)
Acknowledgments
This protocol was developed for the work published in Keymer et al. (2017) financed by the Hans Fischer Gesellschaft e. V. to WE and by the Collaborative Research Center 924 (SFB924) of the Deutsche Forschungsgemeinschaft (DFG) ‘Molecular Mechanisms of Yield and Yield Stability in Plants’ (project B03) to CG. The authors have not conflicts of interest or competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Ahmed Z., Zeeshan S., Huber C., Hensel M., Schomburg D., Münch R., Eylert E., Eisenreich W., and Dandekar T.(2014).‘Isotopo’ a database application for facile analysis and management of mass isotopomer data. Database(Oxford): bau077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bécard G. and Fortin J. A.(1988). Early events of vesicular–arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytologist 108: 211-218. [DOI] [PubMed] [Google Scholar]
- 3. Eisenreich W., Huber C., Kutzner E., Knispel N. and Schramek N.(2013). Isotopologue profiling: towards a better understanding of metabolic pathways. In: Weckwerth, W. and Kahl, G.(Eds). The Handbook of Plant Metabolomics. Wiley-Blackwell 26-56. [Google Scholar]
- 4. Jiang Y., Wang W., Xie Q., Liu N., Liu L., Wang D., Zhang X., Yang C., Chen X., Tang D. and Wang E.(2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356: 1172-1175. [DOI] [PubMed] [Google Scholar]
- 5. Keymer A., Pimprikar P., Wewer V., Huber C., Brands M., Bucerius S. L., Delaux P. M., Klingl V., Ropenack-Lahaye E. V., Wang T. L., Eisenreich W., Dormann P., Parniske M. and Gutjahr C.(2017). Lipid transfer from plants to arbuscular mycorrhiza fungi. Elife 6: e29107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuhn H., Kuster H. and Requena N.(2010). Membrane steroid-binding protein 1 induced by a diffusible fungal signal is critical for mycorrhization in Medicago truncatula . New Phytol 185(3): 716-733. [DOI] [PubMed] [Google Scholar]
- 7. Luginbuehl L. H., Menard G. N., Kurup S., Van Erp H., Radhakrishnan G. V., Breakspear A., Oldroyd G. E. D. and Eastmond P. J.(2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356: 1175-1178. [DOI] [PubMed] [Google Scholar]
- 8. McGonigle T. P., Miller M. H., Evans D. G., Fairchild G. L. and Swan J. A.(1990). A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytol 115: 495-501. [DOI] [PubMed] [Google Scholar]
- 9. Mosse B. and Hepper C.(1975). Vesicular-arbuscule mycorrhizial infections in root organ cultures. Phys Plant Pathol 5: 215-223. [Google Scholar]
- 10. Pfeffer P. E., Douds Jr D. D., Becard G. and Shachar-Hill Y.(1999). Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol 120(2): 587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rich M. K., Nouri E., Courty P. E. and Reinhardt D.(2017). Diet of arbuscular mycorrhizal fungi: bread and butter? Trends Plant Sci 22(8): 652-660. [DOI] [PubMed] [Google Scholar]
- 12. Trépanier M., Becard G., Moutoglis P., Willemot C., Gagne S., Avis T. J. and Rioux J. A.(2005). Dependence of arbuscular-mycorrhizal fungi on their plant host for palmitic acid synthesis. Appl Environ Microbiol 71(9): 5341-5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vierheilig H., Coughlan A. P., Wyss U. and Piche Y.(1998). Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol 64: 5004-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wewer V., Brands M. and Dormann P.(2014). Fatty acid synthesis and lipid metabolism in the obligate biotrophic fungus Rhizophagus irregularis during mycorrhization of Lotus japonicus . Plant J 79: 398-412. [DOI] [PubMed] [Google Scholar]