Abstract
Rice yellow mottle virus (RYMV), a mechanically transmitted virus that causes serious damage to cultivated rice plants, is endemic to Africa. Varietal selection for resistance is considered to be the most effective and sustainable management strategy. Standardized resistance evaluation procedures are required for the identification and characterization of resistance sources. This paper describes a protocol for mechanical inoculation of rice seedlings with RYMV and two methods of resistance evaluation – one based on a symptom severity index and the other on virus detection through double antibody sandwich-enzyme linked immunosorbent assay (DAS-ELISA).
Keywords: Rice, RYMV, Inoculation, Resistance, ELISA, Symptoms
Background
RYMV is a major biotic constraint for rice production in Africa ( Séré et al., 2013 ) and has been reported in most rice-growing countries in Africa and Madagascar. It is not transmissible through seed ( Konaté et al., 2001 ; Allarangaye et al., 2006 ) but by insect vectors (particularly beetles) and by contact during agricultural operations (Bakker, 1974; Traoré et al., 2006 ), especially while transplanting seedlings from seedbeds to the field. The virus is highly stable and capable of multiplying at high concentrations in its rice and wild Poaceae (Bakker, 1974) hosts.
Monitoring of RYMV incidence in seedbeds and varietal selection are the most efficient and sustainable ways of managing RYMV. There are two phenotypes of resistance – partial resistance, characterized by a delay in the appearance of symptoms ( Albar et al., 1998 ), and high resistance, characterized by the absence of virus detection using DAS-ELISA ( Ndjiondjop et al., 1999 ). Although partial resistance is widely distributed among Oryza sativa japonica varieties, only a few varieties from the cultivated rice species O. sativa and O. glaberrima express a high level of resistance to RYMV. Three major resistance genes – RYMV1, RYMV2 and RYMV3 – have been reported ( Ndjiondjop et al., 1999 ; Thiémélé et al., 2010 ; Pidon et al., 2017 ).
Evaluation of the level of resistance to RYMV in rice varieties or lines and the comparison of the outcomes of different experiments require the use of a standardized protocol. This paper describes such a protocol which is based on DAS-ELISA and symptom severity. Reference accessions (susceptible, partially and highly resistant) have been included in the protocol to enable the drawing of reliable conclusions by comparing the test entry with reference materials. The number of plants tested for each variety or line is, however, not specified as it depends on the genetic material being tested and the objective of the experiment.
Materials and Reagents
Fontainebleau sand (VWR, catalog number: VWRC27460.295)
Latex gloves
2 ml microcentrifuge tubes (Dominique DUTSCHER, catalog number: 033297)
5 mm-steel beads (Brammer)
500 ml-wash bottle
Plates 96 wells (VWR, catalog number: 735-0083)
10, 20, 200, and 1,000 μl tips
Paper towel
Rice seeds, including control accessions (susceptible, partially and highly resistant)
RYMV-infected rice leaves (fresh, dried or frozen at -20 °C)
Carborundum 0.037 mm used as an abrasive (VWR, catalog number: 22540.298)
Liquid nitrogen (if possible)
RYMV antibody (wide spectrum polyclonal antibody against purified RYMV virions – prepared as described in N’Guessan et al. (2000) and Afolabi et al. (2009) , or DSMZ, catalog number: AS-0732 or RT-0732)
Alkaline phosphatase-conjugated polyclonal antibody against RYMV (prepared as described in Clark and Adams (1977), or DSMZ, catalog number: RT-0732)
Skimmed milk powder
Substrate pNPP (para NitroPhenylPhosphate, Sigma-Aldrich, catalog number: N9389)
Polyvinylpyrrolidone (PVP-40), MW 40000 (VWR, catalog number: 26616.184)
Potassium phosphate (KH2PO4) (Sigma-Aldrich, catalog number: P0662)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S3264)
MilliQ water
Sodium carbonate (Na2CO3) (Sigma-Aldrich, catalog number: S7795)
Sodium hydrogen carbonate (NaHCO3) (Sigma-Aldrich, catalog number: S5761)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653)
Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333)
Tween 20 (Sigma-Aldrich, catalog number: P1379)
Diethanolamine (Fisher Scientific, catalog number: 10131470)
Hydrochloric acid (HCl) 10 N
Phosphate inoculation buffer (0.1 M pH 7.2) (see Recipes)
-
Buffers for ELISA test (see Recipes)
Buffer PBST 10x
Coating buffer pH 9.6
Substrate buffer pH 9.8 (see Recipes)
Equipment
Greenhouse or plant growth chamber
Mortars and pestles
Tissue lyser (QIAGEN, model: TissueLyser II)
10, 20, 200, and 1,000 μl volume pipettes
200 μl volume 8-channel pipette
Vortex
pH meter
Microcentrifuge
Microplate reader with 405 nm filter (TECAN, model: Infinite M200 Pro)
Procedure
-
Plant and virus preparation
Grow rice plants under the following greenhouse conditions during the whole experiment - approximately 12 h of light, 28 ± 1°C (day) and 25 ± 1°C (night), and 75% relative humidity. Perform inoculation at the 2 to 3-leaves development stage (Figure 1), roughly 2 weeks after inoculation (WAI). Apply a suitable fertilizer to the soil for optimal growth.
Use the following recommended rice varieties as checks: IR64, Bouake 189 or BG90-2 (susceptible O. sativa varieties); Azucena or Moroberekan (partially resistant O. sativa controls); accessions with any of the high resistance genes (RYMV1, RYMV2 or RYMV3; Thiémélé et al., 2010 ; Orjuela et al., 2013 ; Pidon et al., 2017 ) (resistant controls).
Propagate RYMV isolates on any of the susceptible control accessions – it is highly recommended to always use the same accession. Mechanically inoculate the plants (as described below) and harvest the leaves at 3 WAI when symptoms reach the 5 to 7 severity level (the symptoms severity scale is described below). Infected leaves may be kept at -20 °C for several months or years.
-
Inoculation
Cut the RYMV-infected leaves to be used as the source of inoculum into 0.5-1 cm pieces. Add Fontainebleau sand (about 200 mg to 1 g of leaves). Grind the mixture in a mortar with 0.1 M phosphate inoculation buffer, pH 7.2 (10 ml of buffer to 1 g of leaves) (Recipe 1; see Video 1). A minimum of 100 µl of inoculum per plant is required.
Add two pinches of carborundum and mix with a pestle.
Using gloves, rub leaves twice with ground inoculum as shown in Video 2. Keep at least 5 plants of the susceptible check variety as a non-inoculated control.
After 5 min, rinse the leaves with water to eliminate the excess carborundum (see Video 2).
-
Symptoms observation
Observation for symptoms can start between 10 and 15 days after inoculation (DAI), when symptoms first appear on susceptible control plants. The date of appearance of symptoms can be monitored on each plant on a daily basis. Symptoms are observed on newly emerged leaves.
-
The symptom severity scale on the leaves (Figure 2) is derived from the Standard Evaluation System for Rice (IRRI, 2002) but does not consider a reduction in plant height and delayed flowering:
1 = Leaves green (no apparent symptoms);
3 = Leaves green but with sparse dots or streaks;
5 = Leaves green or pale green with mottling;
7 = Leaves pale yellow or yellow;
9 = Leaves turn orange or show necrosis and sometimes die.
-
DAS-ELISA
Harvest samples of the last emerged leaves between 10 and 15 DAI. The samples can be used fresh, rapidly frozen at -20 °C, or dried in a herbarium.
For sample preparation with tissue lyser (QIAGEN), place 25 mg of fresh material (or 10 mg of dried material) in a 2 ml tube with a metal bead. Put the tubes in liquid nitrogen for 5 min. Grind in tissue lyser for 30 sec at an oscillation frequency of 30 Hz. Add 500 μl of 1x PBST. Vortex vigorously. Alternatively, grind the same quantity of samples with pestle and mortar with 500 µl of 1x PBST and then transfer to a 2 ml tube. Centrifuge for 5 min at 5,200 × g. Keep the samples for up to 24 h at 4 °C or frozen at -20 °C before using them for the ELISA test.
-
Coating
Dilute antibodies directed against purified RYMV virions in the coating buffer (Recipe 2) to a concentration of 1 µg/ml IgG or according to the dilution factor recommended by the provider. Add 100 μl of antibodies solution per well, except in the wells at the edges of the plate which should be filled to avoid edge effect. Cover the plate and incubate for 2 h at 37 °C in a moist environment (box with moist paper).
-
Washing
Discard the coating solution from the wells. Wash the plate by filling the wells with 1x PBST using a wash bottle and decant the washing buffer immediately (for a quick wash) or after 3 min. Proceed successively to a quick wash and three 3 min-washes. Remove residual buffer on a paper towel (Video 3).
-
Blocking
Add 200 μl of 3% (3 g by 100 ml) skimmed milk in 1x PBST per well. Cover the plate and incubate for 1 h at 37 °C.
-
Washing
Discard the solution and wash for 3 min with 1x PBST. Remove residual buffer on a paper towel.
-
Deposit of antigens
Add 100 µl of the sample supernatant to the wells. Cover the plate and incubate for 2 h at 37 °C. Always include positive controls (usually the RYMV inoculum) and negative controls (PBST buffer alone and non-inoculated plant) among samples. Two technical replicates of both controls and samples are recommended.
-
Washing
Discard the sample solution from the wells and remove residual sample solution on a paper towel to avoid inter-well contamination. Proceed to wash with 1x PBST as described in Step C2 above and remove residual buffer on a paper towel.
-
Addition of enzyme-conjugated antibodies
Dilute the antibody against RYMV conjugated with alkaline phosphatase in 1x PBST according to the recommendations of the provider or as determined experimentally according to Hill et al. (1981) . Add 100 μl of the conjugated antibodies solution per well. Cover the plates and incubate for 2 h at 37 °C.
-
Washing
Proceed to wash as described in Step C2 above.
-
Addition of substrate
Dissolve pNPP tablets in the substrate buffer (1 mg/ml; Recipe 3). As pNPP is light-sensitive, be careful to keep it in the dark as much as possible. Add 100 μl of substrate solution per well. Incubate in the dark and at room temperature for 1 h. Read the optical density (OD) at 405 nm on a microplate reader.
-
Figure 1. Two weeks old IR64 plants, ready for inoculation.

Video 1. Preparation of RYMV inoculum from infected rice leaves.
Video 2. Mechanical inoculation of RYMV on rice leaves.

Figure 2. Symptom severity scale on rice leaves.
Video 3. DAS-ELISA plate washing.

Data analysis
-
Symptom severity
The susceptible variety IR64 generally develops the first symptoms 10 to 15 DAI and the severity index rapidly reaches 7 or 9 on newly emerged leaves. The partially resistant variety Azucena develops symptoms about one week after IR64 does and often reaches a severity index of 5. These two varieties generally show clear mottling. On the contrary, other lines, in particular O. glaberrima accessions, can show less specific symptoms that may be difficult to distinguish from chlorosis caused by different abiotic stresses. Highly resistant varieties, such as Gigante, Tog5681, Tog7291 or Tog5307 ( Ndjiondjop et al., 1999 ; Thiémélé et al., 2010 ; Orjuela et al., 2013 ; Pidon et al., 2017 ), do not develop any symptoms except if a resistance-breaking isolate has been used as the source of inoculum or if resistance-breaking variants appear during the experiment. However, the appearance of resistance-breakdown is generally limited to a few plants in a short-term experiment. Therefore, it is advisable to stop the observation for symptoms 4 WAI to reduce the occurrence of resistance-breakdown cases. Figure 3 shows the symptoms of RYMV on the whole plant and its impact on plant development.
Contrary to what can be observed in other plant/virus pathosystems ( Ouibrahim et al., 2014 ; Poque et al., 2015 ), mechanical inoculation of RYMV is highly efficient – all susceptible or partially resistant plants are expected to be infected and symptom severity is expected to be homogenous among plants with the same genotype.
-
The symptoms severity indices corresponding to different observation dates on the same sample can be combined based on their arithmetic mean or on the Area Under Symptom Progression Curve (AUSPC) calculated as follows:
AUSPC = Σ [(Si +Si+1) x (ti+1 – ti)/2]
where, Si is the symptom severity index observed at date ti.
-
DAS-ELISA
-
After the OD reading, subtract the mean of negative controls (PBST) from all values. Calculate the means of technical replicates. Samples are considered positive when they are superior to 0.1 and twice the mean of non-inoculated controls. Table 1 provides an example of ELISA data analysis.
*ODcor corresponds to the OD405nm of the sample minus the mean of OD405nm of the PBST controls.
**Samples are considered positive if the mean (ODcor) is superior to 0.1 and twice the non-inoculated control (0.002 here).
Highly resistant controls are negative, except in the case of resistance-breakdown, whereas Azucena and IR64 are expected to be positive. Susceptible and partially resistant accessions can be differentiated using semi-quantitative analysis but preliminary tests are necessary to identify a dilution range that maximizes differences between samples.
-
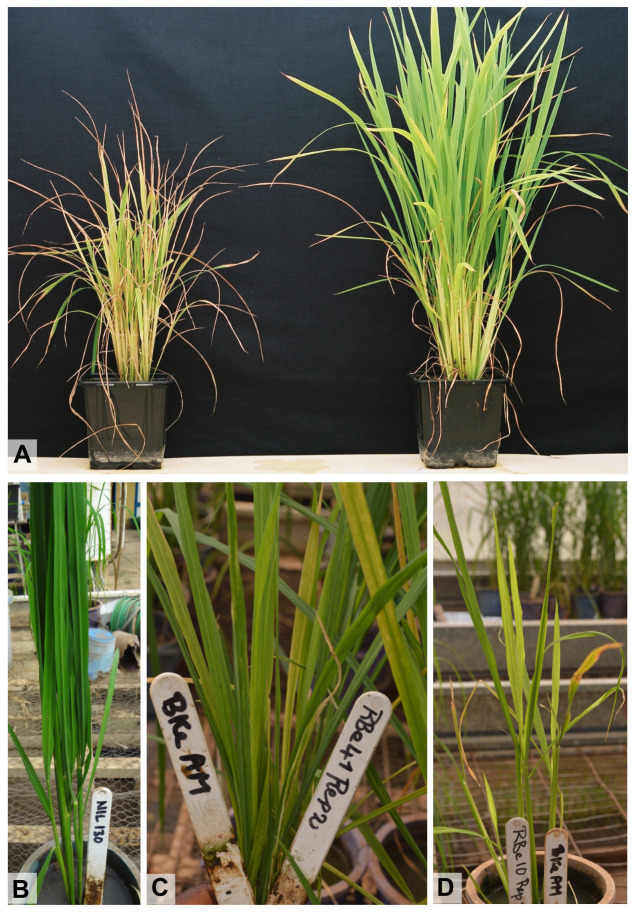
Figure 3. Symptoms of RYMV on the whole rice plant.
A. Inoculated IR64 plant (left) compared with a non-inoculated IR64 control (right) one month after inoculation. B-D. Plants with different resistance levels: highly resistant plant with a symptom severity index of 1 (B); susceptible plant with a symptom severity index of 5 (C); susceptible plant with a symptom severity index of 7 (D).
Table 1. Example of ELISA data analysis with two repeats per sample and four repeats for the PBST control .
| Sample | OD405nm | ODcor* | Mean (ODcor) | Conclusion** |
| PBS-T control | 0.079 | 0.00075 | 0 | |
| 0.077 | -0.00125 | |||
| 0.077 | -0.00125 | |||
| 0.08 | 0.00175 | |||
| Non inoculated control | 0.08 | 0.00175 | 0.001 | |
| 0.079 | 0.00075 | |||
| Sample A | 1.222 | 1.14375 | 1.194 | + |
| 1.323 | 1.24475 | |||
| Sample B | 0.09 | 0.01175 | 0.023 | - |
| 0.112 | 0.03375 | |||
| Sample C | 0.083 | 0.00475 | 0.003 | - |
| 0.081 | 0.00275 | |||
| Sample D | 1.321 | 1.24275 | 1.269 | + |
| 1.374 | 1.29575 | |||
| Sample E | 0.535 | 0.45675 | 0.518 | + |
| 0.658 | 0.57975 | |||
| Sample F | 0.985 | 0.90675 | 0.9992 | + |
| 1.156 | 1.07775 |
Notes
RYMV is a viral pathogen infecting rice and wild grasses and its manipulation may be restricted by biosafety concerns. Always manipulate the virus in accordance with your country’s legislation.
Resistance or susceptibility depends on specific interactions between plant and viral genotypes and the choice of resistant or susceptible controls may also depend on your RYMV isolates and the objectives of the experiment. If the highly resistant controls show RYMV symptoms before 15 DAI, your inoculum may contain a resistance-breaking genotype ( Hébrard et al., 2006 ; Traoré et al., 2010 ; Pinel-Galzi et al., 2017; Pinel- Galzi et al., 2016 ; Hébrard et al., 2018 ) and would be inappropriate for assessing the resistance phenotype of your material. In addition, to avoid false interpretations, it is highly recommend to check the presence of the resistance allele in each seed batch, using molecular markers linked to resistance alleles ( Thiémélé et al., 2010 ; Orjuela et al., 2013 ; Pidon et al., 2017 ).
Infected leaves to be used as a source of inoculum may be kept at -20 °C for several months or years. However multiplication of the virus on susceptible controls or a preliminary DAS-ELISA test is recommended when the source of inoculum is a leaf sample collected in the field or kept under drying conditions.
Incubation at 37 °C for 1 or 2 h during the DAS-ELISA test can be replaced by incubation overnight at 4 °C, provided the same procedure is used between experiments that have to be compared.
If required, background reactions in DAS-ELISA can be reduced by combining the following procedures: (i) add 2% Polyvinylpyrrolidone (PVP-40) to 1x PBST (sample preparation buffer); (ii) add 2% PVP-40 and 1% skimmed milk to the conjugate dilution solution (1x PBST).
Recipes
-
Phosphate inoculation buffer (0.1 M pH 7.2)
Prepare a solution of 0.1 M KH2PO4: 1.36 g in 100 ml of MilliQ water
Prepare a solution of 0.1 M Na2HPO4: 1.41 g in 100 ml of MilliQ water
Mix 32 ml of KH2PO4 0.1 M with 68 ml of Na2HPO4 0.1M
Control pH with a pH meter. It should be pH 7.2
Store at 4 °C
-
Buffers for ELISA test
-
Phosphate Buffer Saline Tween (PBST) concentrated 10x (1 L)
79.5 g NaCl
1.9 g KH2PO4
11.36 g Na2HPO4 anhydrous
1.93g KCl
Weigh and dissolve the above chemicals in 900 ml of MilliQ water, pH should be 6.8
Add 5 ml of Tween 20, Adjust volume to 1 L. Store at room temperature
For use, dilute 10 times in MilliQ water
-
Coating buffer pH 9.6 (500 ml)
0.79 g Na2CO3
1.425 g NaHCO3
Weigh and dissolve the above chemicals in 450 ml of MilliQ water, pH should be 9.6
Adjust the final volume to 500 ml with MilliQ water
Store at 4 °C
-
-
Buffer for substrate (diethanolamine)
Take 49 ml of diethanolamine, add 400 ml of MilliQ water, mix
Adjust pH to 9.8 with 10 N HCl
Complete up to 500 ml with MilliQ water
Store at 4 °C
Acknowledgments
This protocol was developed from procedures used previously in different laboratories (Fauquet and Thouvenel, 1977; Yassi et al., 1994 ; Konaté et al., 1997 ; N'Guessan et al., 2001; Albar et al., 1998 ). The work was funded by the Global Rice Science Partnership (GRiSP) as a New Frontiers Research program (project MENERGEP). The authors thank Dr. Ajayi for his help in manuscript correction. The authors declare no conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Afolabi S., Akator S. K., Abo E. M., Onasanya A. and Séré Y.(2009). Production of polyclonal antibodies to various strains of rice yellow mottle virus(RYMV) obtained across different agro-ecological zones in West Africa. Sci Res Essays 4: 306-309. [Google Scholar]
- 2. Albar L., Lorieux M., Ahmadi N., Rimbault I., Pinel A., Sy A. A., Fargette D. and Ghesquiere A.(1998). Genetic basis and mapping of the resistance to rice yellow mottle virus. I. QTLs identification and relationship between resistance and plant morphology. Theor Appl Genet 97: 1145-1154. [Google Scholar]
- 3. Allarangaye M. D., Traoré O., Traoré E. V. S., Millogo R. J. and Konaté G.(2006). Evidence of non-transmission of Rice yellow mottle virus through seeds of wild host species . J Plant Pathol 88: 307-313. [Google Scholar]
- 4. Bakker W.(1974). Characterisation and ecological aspects of rice yellow mottle virus in Kenya. Agricultural University, Wageningen, The Netherlands.
- 5. Clark M. F. and Adams A. N.(1977). Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol 34: 475-483. [DOI] [PubMed] [Google Scholar]
- 6. Fauquet C. and Thouvenel J.(1977). Isolation of the rice yellow mottle virus in Ivory Coast. Plant Disease Report 61: 443-446. [Google Scholar]
- 7. Hébrard E., Pinel-Galzi A., Bersoult A., Siré C. and Fargette D.(2006). Emergence of a resistance-breaking isolate of Rice yellow mottle virus during serial inoculations is due to a single substitution in the genome-linked viral protein VPg . J Gen Virol 87(5): 1369-1373. [DOI] [PubMed] [Google Scholar]
- 8. Hébrard E., Pinel-Galzi A., Oludare A., Poulicard N., Aribi J., Fabre S., Issaka S., Mariac C., Dereeper A., Albar L., Silue D. and Fargette D.(2018). Identification of a hypervirulent pathotype of Rice yellow mottle virus: A threat to genetic resistance deployment in West-Central Africa . Phytopathology 108(2): 299-307. [DOI] [PubMed] [Google Scholar]
- 9. Hill J. H., Bryant G. R. and Durand D. P.(1981). Detection of plant virus by using purified IgG in ELISA. J Virol Methods 3(1): 27-35. [DOI] [PubMed] [Google Scholar]
- 10. IRRI(International Rice Research Institute). (2002). Standard evaluation system for rice. Philippines: International Rice Research Institute, Manila, Philippines.
- 11. Konaté G., Sarra S. and Traoré O.(2001). Rice yellow mottle virus is seed-borne but not seed transmitted in rice seeds . Eur J Plant Pathol 107: 361-364. [Google Scholar]
- 12. Konaté G., Traoré O. and Coulibaly M. M.(1997). Characterization of rice yellow mottle virus isolates in Sudano-Sahelian areas. Arch Virol 142(6): 1117-1124. [DOI] [PubMed] [Google Scholar]
- 13. Ndjiondjop M. N., Albar L., Fargette D., Fauquet C. and Ghesquière A.(1999). The genetic basis of high resistance to rice yellow mottle virus(RYMV) in cultivars of two cultivated rice species. Plant Disease 83: 931-935. [DOI] [PubMed] [Google Scholar]
- 14. P. N’Guessan, Pinel A., Caruana M. L., Frutos R., Sy A., Ghesquière A. and Fargette D.(2000). Evidence of the presence of two serotypes of Rice yellow mottle sobemovirus in Côte d’Ivoire. Eur J Plant Pathol 106(2): 167-178. [Google Scholar]
- 15. P. N’Guessan, Pinel A., Sy A. A., Ghesquière A. and Fargette D.(2001). Distribution, pathogenicity, and interactions of two strains of Rice yellow mottle virus in forested and savanna zones of West Africa . Plant Dis 85: 59-64. [DOI] [PubMed] [Google Scholar]
- 16. Orjuela J., Deless E. F., Kolade O., Cheron S., Ghesquiere A. and Albar L.(2013). A recessive resistance to Rice yellow mottle virus is associated with a rice homolog of the CPR5 gene, a regulator of active defense mechanisms . Mol Plant Microbe Interact 26(12): 1455-1463. [DOI] [PubMed] [Google Scholar]
- 17. Ouibrahim L., Mazier M., Estevan J., Pagny G., Decroocq V., Desbiez C., Moretti A., Gallois J. L. and Caranta C.(2014). Cloning of the Arabidopsis rwm1 gene for resistance to Watermelon mosaic virus points to a new function for natural virus resistance genes . Plant J 79(5): 705-716. [DOI] [PubMed] [Google Scholar]
- 18. Pinel-Galzi A., Rakotomalala M., Sangu E., Sorho F., Kanyeka Z., Traoré O., Sérémé D., Poulicard N., Rabenantoandro Y., Séré Y., Konaté G., Ghesquière A., Hébrard E. and Fargette D.(2007). Theme and variations in the evolutionary pathways to virulence of an RNA plant virus species. PLoS Pathog 3(11): e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinel-Galzi A., Dubreuil-Tranchant C., Hebrard E., Mariac C., Ghesquiere A. and Albar L.(2016). Mutations in Rice yellow mottle virus polyprotein P2a involved in RYMV2 gene resistance breakdown . Front Plant Sci 7: 1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pidon H., Ghesquiere A., Cheron S., Issaka S., Hebrard E., Sabot F., Kolade O., Silue D. and Albar L.(2017). Fine mapping of RYMV3: a new resistance gene to Rice yellow mottle virus from Oryza glaberrima . Theor Appl Genet 130(4): 807-818. [DOI] [PubMed] [Google Scholar]
- 21. Poque S., Pagny G., Ouibrahim L., Chague A., Eyquard J. P., Caballero M., Candresse T., Caranta C., Mariette S. and Decroocq V.(2015). Allelic variation at the rpv1 locus controls partial resistance to Plum pox virus infection in Arabidopsis thaliana . BMC Plant Biol 15: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Séré Y., Fargette D., Abo M. E., Wydra K., Bimerew M., Onasanya A., Akator S. K.(2013). Managing the major diseases of rice in Africa. In: Wopereis, M. C. S., Johnson, D. E., Ahmadi, N., Tollens, E. and Jalloh, A.(Eds.). Realizing Africa's rice promise. CABI pp: 213-228. [Google Scholar]
- 23. Thiémélé D., Boisnard A., Ndjiondjop M. N., Chéron S., Séré Y., Aké S., Ghesquière A. and Albar L.(2010). Identification of a second major resistance gene to Rice yellow mottle virus, RYMV2, in the African cultivated rice species, O. glaberrima . Theor Appl Genet 121(1): 169-179. [DOI] [PubMed] [Google Scholar]
- 24. Traoré O., Pinel-Galzi A., Issaka S., Poulicard N., Aribi J., Aké S., Ghesquiere A., Séré Y., Konaté G., Hébrard E. and Fargette D.(2010). The adaptation of Rice yellow mottle virus to the eIF(iso)4G-mediated rice resistance . Virology 408(1): 103-108. [DOI] [PubMed] [Google Scholar]
- 25. Traoré O., Traoré M. D., Fargette D. and Konaté G.(2006). Rice seedbeds as a source of primary infection by Rice yellow mottle virus . Eur J Plant Pathol 115: 81-186. [Google Scholar]
- 26. Yassi M. N., Ritzenthaler C., Brugidou C., Fauquet C. and Beachy R. N.(1994). Nucleotide sequence and genome characterization of rice yellow mottle virus RNA. J Gen Virol 2): 249-257. [DOI] [PubMed] [Google Scholar]