Abstract
The human malaria parasite Plasmodium falciparum expresses variant PfEMP1 proteins on the infected erythrocyte, which function as ligands for endothelial receptors in capillary vessels, leading to erythrocyte sequestration and severe malaria. The factors that orchestrate the mono-allelic expression of the 45–90 PfEMP1-encoding var genes within each parasite genome are still not fully identified. Here, we show that the transcription factor PfAP2-O influences the transcription of var genes. The temporary knockdown of PfAP2-O leads to a complete loss of var transcriptional memory and a decrease in cytoadherence in CD36 adherent parasites. AP2-O-knocked-down parasites exhibited also significant reductions in transmission through Anopheles mosquitoes. We propose that PfAP2-O is, beside its role in transmission stages, also one of the virulence gene transcriptional regulators and may therefore be exploited as an important target to disrupt severe malaria and block parasite transmission.
Keywords: PfAP2-O, virulence factor, var, antigenic variation, epigenetic memory
Introduction
The human malaria parasite Plasmodium falciparum remains a risk to a significant portion of the world’s population. Despite the considerable success of control measures, P. falciparum still causes around 400,000, deaths per year, mainly in the sub-Saharan African countries, where most of the victims are either pregnant women or children under 5 years of age (World Health Organization, 2019). Common approaches for malaria control and elimination are artemisinin-based combination therapies (ACT) to treat blood stages in infected patients and insecticide-impregnated bed nets to prevent mosquito bites and parasite transmission. Although a significant reduction of malaria cases during the first decades of implementation of the aforementioned measures was achieved, recent studies have demonstrated the emergence of parasites resistant to almost all antimalarial treatments including artemisinin (Dondorp et al., 2009) and mosquitoes’ resistance to the most commonly-used insecticides—pyrethroids, organochlorines, carbamates, and organophosphates—across different endemic areas (WHO, 2020). Therefore, the study and characterization of new drug targets to interfere in parasite transmission are crucial to achieve long-term malaria eradication (Delves et al., 2018).
A major parasite virulence factor is PfEMP1 (Plasmodium falciparum erythrocyte membrane protein 1), which consists of proteins expressed on the infected red blood cell surface, encoded by the multicopy var gene family (Su et al., 1995). Based on features such as chromosomal location, protein domain structure, and sequence similarities, var genes have been classified into three major groups, termed (upsA, upsB, and upsC), two intermediate groups, (upsB/A and upsB/C), and two distantly related genes var1csa and var2csa (upsD and upsE respectively) (Lavstsen et al., 2003; Joergensen et al., 2010). The var genes are expressed in a timely, controlled manner so that only one or a few var genes are transcribed per parasite while all others are transcriptionally silenced, which means that only one PfEMP1 type is exported to the erythrocyte surface (Chen et al., 1998; Scherf et al., 1998). Eventually, by a mechanism known as switching, the active var gene is silenced and another one starts to be transcribed so that a different type of PfEMP1 is exported (Scherf et al., 1998). This phenomenon known as mutually allelic exclusion leads to the characterized PfEMP1 antigenic variation and is one of the most important strategies to avoid the host immune system (Chen et al., 1998).
The described mechanisms controlling changes in var transcription profiles, and how this switching is orchestrated, are mainly associated with chromatin modifications in var promoter regions. It has been shown that the active var gene is marked by the acetylation of lysine 9 (H3K9ac) and the presence of a special histone H2.AZ (Petter et al., 2011), while the repressed genes are marked by the trimethylation of the Histone 3 Lysine 9 (H3K9me3) (Lopez-Rubio et al., 2007). Additionally, the selected var allele that is transcribed in the next cycle (poised state) is epigenetically marked by demethylation of the Histone 3 lysine 4 (H3K4me3 to H3K4me2) (Lopez-Rubio et al., 2007). There are different proteins responsible for reading, marking, or removing these histone modifications. The histone deacetylases PfSIR2A and PfSIR2B were found to have a repressive role in upsA and upsB type genes, respectively (Duraisingh et al., 2005; Freitas et al., 2005; Tonkin et al., 2009). Also, the methyltransferases PfSET10 and PfSETvs are associated with var gene transcription. While PfSET10 seems to be responsible for the maintenance of the H3K4 methylation mark in the active var gene during the poised state (Volz et al., 2012), PfSETvs was found to be a repressor of var transcription when knockout parasites for this protein displayed virtually all var transcripts in the parasite nucleus (Jiang et al., 2013). The histone-mark-reading protein, Heterochromatin protein 1 (PfHP1) is associated with silenced chromatin around the promoter region of var, and also with genes involved in the production of sexual/transmission stages of the parasite, known as gametocytes (Pérez-Toledo et al., 2009; Brancucci et al., 2014). Accordingly, PfHP1 knockdown leads to transcriptional de-repression of var genes, disruption of allelic exclusion, and the induction of gametocytogenesis, indicating a link between var gene transcription and transmission (Brancucci et al., 2014). Regulatory elements in promoter regions of var genes are also involved in transcription regulation. It was shown that each var gene has two functional promoters, one in the exon I and the other within the intron, which seems to be necessary for repression of var transcription (Calderwood et al., 2003). Some long non-coding RNAs, (ncRNAs) that are transcribed from telomere repeats were proposed to influence var transcription, at least those in the subtelomeric region (Sierra-Miranda et al., 2012), and sense and antisense long ncRNAs transcribed from the var intron may have a regulatory effect (Epp et al., 2009). Furthermore, the exosome related RNAse PfRrP6 seems to fine-regulate the degradation of these RNAs (Fan et al., 2020), and the specific DNA helicase, RecQ1, was suggested to be involved in var transcription when its knockdown increased the var recombination levels and high rates of chimeric var genes were observed with actively transcribed var loci (Li et al., 2019). Finally, the chromatin remodeler, PfISWI, was suggested to play a key role in the activation of var genes (Bryant et al., 2020).
Members of the ApiAP2 family of plant-like transcription factor proteins have been associated with important processes throughout the entire life-cycle of Plasmodium. In the rodent malaria parasite P. berghei, parasites lacking the protein encoded by PbAP2-Sp (PF14_0633/PF3D7_1466400 P. falciparum ortholog) were unable to complete the sexual cycle inside the mosquito, as they could not form sporozoites (Yuda et al., 2010). Also in P. berghei, parasites lacking PbAP2-O (Pf11_0442/PF3D7_1143100 P. falciparum ortholog) developed as aberrant ookinetes that were unable to invade the Anopheles midgut and form oocysts (Yuda et al., 2009). In P. falciparum, the disruption of PfAP2-G (PF3D7_1222600) resulted in a complete loss of gametocyte formation, whereas increased expression led to the increased induction of gametocyte-commitment (Kafsack et al., 2014). Campbell and colleagues identified domains in three ApiAP2 members (Balaji et al., 2005) that showed affinity to specific domains in var 5′ upstream (5′ ups) regions (Campbell et al., 2010). One of them (PF3D7_0604100/PfSIP2) was shown to interact with upsB var sequences, and it was suggested that this protein has a role in tethering of chromatin (Flueck et al., 2010). For the second gene, Martins and colleagues demonstrated an upregulation of clonally-variant genes (stevor and rif, but not var) when the predicted protein, PfAP2-exp was truncated (Martins et al., 2017). The role of the third protein, PF3D7_1143100/Pf11_0442, (PbAP2-O ortholog) remains unknown in P. falciparum. Here, we addressed the function of this protein, named PfAP2-O, in the asexual and sexual cycles of P. falciparum and found that it influences transcription of var genes and is crucial for parasite transmission to Anopheles mosquitoes.
Material and Methods
P. falciparum Culture
P. falciparum NF54 parasites and transgenic strains were cultured in 4% hematocrit in RPMI supplemented with 0.25% Albumax 1 (Invitrogen)/5% human plasma under a 90% N2, 5% CO2, 5% O2 atmosphere. Culture medium was changed daily or every two days, and parasites were supplemented with fresh blood every 4 days or more often depending on the parasitemia in the culture. The strains containing a destabilizing domain were maintained in the presence of Shield-1 at 0.5 µM that stabilizes the target protein (de Azevedo et al., 2012).
In order to obtain synchronized cultures, mature stage parasites were frequently (every 3 weeks) purified using Voluven 6% (Fresenius Kabi) using the method published by Lelievre and colleagues (Lelièvre et al., 2005). When ring stage parasites were required, cultures with predominantly this form (up to 8 h after reinvasion) were treated for 10 min with 20 volumes (of the erythrocyte volume) 5% sorbitol solution at room temperature (Lambros and Vanderberg, 1979).
Molecular Cloning/Plasmid Construction
P. falciparum knock-in expression plasmids were constructed using the PfAP2-O 3′-end portion. This region was cloned into the transfection plasmid p_GFP_HA_DD24 ( Figure 1A ). First, genomic DNA (gDNA) was prepared from NF54 parasites using the Proteinase K/Phenol-Chloroform method described in Methods in Malaria Research (Ljungström et al., 2008). The final DNA pellet was resuspended in TE and used as a template for PCR using oligonucleotide primers PfAP2-O fw: 5′-AGATCTGAATTGTTCAGAACATTTAAATATAGTACC and PfAP2-O rev: 5′-CTGCAGTAAATTATTAAGGGGGATGTTATTATTAAC (restriction sites BglII and PstI underlined).
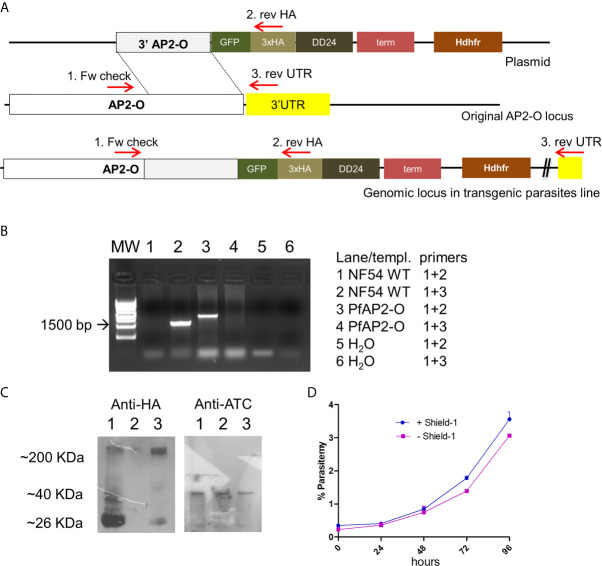
Figure 1.
PfAP2-O modification and subsequent partial knockdown leads to no discernible growth phenotype. (A) The plasmid structure and its integration of tags by homologous recombination after transfection and cloning are shown. (B) PCR analysis of genomic DNA from the recombinant parasite line confirms the integration of the plasmid and absence of wild type versions in the target gene. Combination of Oligonucleotides 3 + 1 results in the amplification of unmodified locus and the band corresponding to oligonucleotides 1 + 2 are the results of modified AP2-O gene. Note that there is no PCR fragment visible in lane 4 which would indicate residual non-modified AP2-O loci. (C) Western blot using an antiHA antibody to detect PfAP2-O-GFP expression in parasites submitted to temporary knockdown by the absence of Shield-1. Lane 1: Parasites grown in the presence of Shield-1. Lane 2: Parasites grown for two reinvasions without Shield-1. Lane 3: Parasites after re-establishment of PfAP2-O-GFP by re-addition of Shield-1 for two reinvasion cycles. As a loading control, a polyclonal antiATC (plasmodial aspartate transcarbamoylase) was used (see Supplementary Material for the full exposure). (D) Growth curves from NF54::PfAP2-O_GFP_HA_DD24 with and without 0.5 µM Shield-1 (data from triplicates).
The oligonucleotides were used in standard PCRs (30 cycles of 94°C, 40s, 50°C for 40s and 65°C for 90s, final polymerization of 10 min at 65°C). The PCR products were excised and purified via the glassmilk method (Boyle and Lew, 1995), ligated into pGEM T easy (Promega) and transformed in DH10B chemically competent E. coli cells. After plating and growth, colonies were grown in Terrific Broth—ampicillin (TB-amp) supplemented liquid medium and the plasmids extracted using a standard miniprep protocol (Green and Sambrook, 2012). Clones were sequenced by the dideoxy-dNTP method and checked for their integrity. The correct fragment was subcloned in the p_GFP_HA_DD24 vector via PstI/BglII. Recombinant plasmids used for transfection were retransformed in E. coli SURE cells and grown in 200 ml TB-amp cultures of which plasmids were recovered by the maxiprep protocol (Green and Sambrook, 2012). The knockout plasmid was constructed using pHHTK as a base vector (Duraisingh et al., 2002). As 5′ and 3′ homology regions flanking the hDHFR resistance cassette the above 3′ fragment was used. The 5′ fragment was created by PCR amplification of a 735 bp-fragment using the oligonucleotides forward 5′-ACTAGTGCCAAGATACTGTTATTGTTGATGT and reverse AGATCTCTTCCACCTTACCGCTATTCC. The amplified fragment was cloned in pGEM T easy, Sanger-sequenced and subcloned via SpeI and BglII in the corresponding sites in pHHTK, resulting in pHHTK-5′AP2-O. Then, the 3′ gene fragment from the pGEM clone described above was excised with EcoRI and subcloned in pHHTK-5′AP2-O. After checking for orientation of the insert, the resulting vector pHHTK-AP2-O-KO was grown out in higher quantities and transfected as described below (see also Supplementary Figure 1B ).
Transfection of Blood Stage Parasites
Empty erythrocytes were electroporated with 40 µg of maxiprep-purified plasmid in incomplete cytomix following the Hasenkamp protocol (Hasenkamp et al., 2012) with slight modifications. 2 * 107 mature parasites (schizonts) were concentrated up to 80% parasitemia by Voluven floating and mixed with the electroporated erythrocytes. After transfection, culture media were changed daily and parasites were submitted to drug pressure with 2.5 nM WR99210 (a gift from Jacobus Inc, USA) 48 h after transfection. Transfected parasites had their medium changed daily until day 6 when no more live parasites were visible. Once parasites reappeared (normally 16–25 days after transfection) and ring stage parasitemia reached 3–4%, several aliquots were frozen as backups. WR99210-resistant parasites were then cultivated for 20 days without WR99210 to promote the loss of episomal plasmids in parasites that did not recombine in the target locus. Then, WR99210 was re-added until parasitemia increased again and new aliquots were frozen. These drug on-off cycles were repeated three times to enrich recombinant parasites. Then, parasites were cloned by limiting dilution, and DNA was analyzed in order to identify clonal populations with the PfAP2-O locus modified using the oligonucleotides PfAP2-O “check” 5′-CATCAAAATGGATTTAATAATTGTTC and HA rev 5′-AGCGGCATAATCTGGAACATCGTAC.
RNA Purification and cDNA Synthesis
Total RNA was converted to cDNA following the protocol described in (Gölnitz et al., 2008). When parasitemias reached 5%, RNA from parasites with and without Shield-1 treatment were harvested using Trizol (Life Technologies) following the manufacturer’s instructions. The final RNA pellet was resuspended in nuclease free water. For reverse transcription, 10 μg of total RNA was digested three consecutive times by the enzyme DNAse I (Fermentas). The material obtained from this reaction was used for cDNA synthesis using RevertAid reverse transcriptase (Fermentas) using random hexamer oligonucleotides (N6). var genes’ transcript amounts were measured using the primers reported in Salanti et al. (2003). The relative transcript quantities of different genes (see a list of oligos used for RT-PCR in Supplementary Table 3 ) were then calculated by the 2−ΔCt method (Livak and Schmittgen, 2001) using the seryl tRNA ligase transcript as an endogenous control.
Pull-down of proteins associated with AP2-O_GFP_HA_DD24
A total protein extract from the NF54::AP2-O_GFP_HA_DD24 clone was prepared and the protein complex containing the HA tag was purified using the HA-Tag (C29F4) Rabbit mAb (Sepharose Beads conjugate, Cell Signaling Technology) kit. The obtained eluate was separated in a 12% SDS gel using immunoprecipitated NF54 wild-type parasites as a control. To detect proteins in the precipitated complex, the SDS gel was submitted to silver staining using the PlusOne Silver Staining Kit (GE Healthcare, Supplementary Figure 2 ). We observed three different protein bands in the extract from the modified parasites compared to the negative control. These bands and bands corresponding to the same size in the negative control were excised, trypsin-digested, and analyzed by mass spectrometry. Analyses were performed on LTQ-Orbitrap Velos ETD (Thermo) coupled with Easy nanoLC II (Thermo). The peptides were separated on a C18RP column on a 95 min gradient. The instrumental conditions were checked using 50 fmol of a tryptic digest of BSA as standard. The sample carryover was completely removed between runs. The peptide search was conducted against the protein database Plasmodium falciparum_reviewed Uniprot. The complete list of proteins detected is shown in Supplementary Table 2 and the analysis conditions are shown in Supplementary Data File 2 .
Adherent Phenotype Selection of Blood-Stage Parasites in Static Assays (Panning)
The selection of adhesive phenotypes of infected red blood cells (IRBCs) adherent to CHO-CD36 was done essentially as described in Gölnitz and colleagues (Gölnitz et al., 2008). Briefly, Voluven-floated trophozoite stage IRBCs (5 × 107–108, 50–80% parasitemia) were layered over confluent CHO-CD36 cells in RPMI/human plasma at pH6.8. Importantly, non-infected RBCs do not adhere to CHO-CD36 cells under these conditions [see also (Scherf et al., 1998; Gölnitz et al., 2008)]. The IRBCs were incubated over the cells for 1 h at 37°C and gently mixed every 15 min. After 1 h, the non-adherent IRBCs were aspired and the remaining adherent IRBCs were washed three times with RPMI at pH 6.8, and after the last washing step, the number of adherent IRBC was documented. The remaining IRBCs were then detached using RPMI/plasma at pH 7.2–7.4 and returned to the normal culture conditions. The process was repeated three times after which strongly adherent IRBCs predominantly expressing var gene PF3D7_0412400 (CHO-CD36 binding) were obtained. For the silencing assay, cells were grown in six-well plates, and central areas were marked in each well. For the cytoadherence readout, ten pictures for each condition were taken using the EVOS FL digital inverted microscope (AMG). The CHO cells and parasites were counted, and the results were plotted using GraphPad Prism 5 software.
Fluorescence Microscopy
For GFP expression analysis, parasites were fixed as described in Tonkin et al. (2004) followed by incubation with PBS/Saponin 0.01% and DAPI at a final concentration of 2 µg/ml at 37°C for 1 h. After that, the parasites were washed three times with PBS/Saponin 0.01% and incubated with the membrane marker Wheat Germ agglutinin (WGA) Texas Red™-X conjugate (Invitrogen) in PBS/Saponin 0.01% for 20 min at 37°C. Finally, parasites were washed three times with PBS/Saponin. Images were acquired in a fluorescence microscope Zeiss Axio Observer Z1 and processed using Photoshop version 5.
Nuclei Isolation and Fractionation
For the fractionation of nuclei, we followed the protocol described in Oehring et al. (2012). For this, parasites were first synchronized to detect PfAP2-O in each intraerythrocytic stage. Synchronized parasites were then lysed using saponin and washed three times with PBS. A second lysis was made using a hypotonic cytoplasmic lysis buffer CLB [20 mM HEPES (pH7.9), 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 0.65% NP-40, 1 mM DTT, Complete protease inhibitors (Roche)] followed by centrifugation for 5 min at 2,000 g; the supernatant was used as the cytoplasmic fraction. Seven washes with the CLB buffer were then made followed by the nuclei extraction with low salt buffer LSB [20 mM HEPES (pH7.9), 0.1 M KCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, protease inhibitors] during 20 min at 4°C. These extractions were used for western blot analysis.
Western Blot
Protein extracts were separated in 10% SDS polyacrylamide gels and transferred to a Hybond C nitrocellulose membrane (GE Healthcare). After blocking with 4% non-fat milk in 1× PBS/0.1% Tween20, membranes were incubated with the murine antiHA (for PfAP2-O detection), the Rabbit-Anti TPK (P. falciparum Thiamin pyrophosphokinase) or mouse antiATC (Aspartate trans-carbamoylase, both cytoplasmic markers), and Rabbit-AntiH3 (Nuclei marker, Sigma-Aldrich, 1:1,000) or mouse anti-Pfs16 followed by antiRabbit or antiMouse IgG-peroxydase antibodies (KPL, 1:5,000) incubation. Blots were washed with PBS/Tween between incubations and finally incubated with WesternPico Super signal substrate (Pierce/Thermo) for detection. As a loading control, a murine polyclonal antiATC antibody (1:1,000) was used. The signal intensities were measured using the ImageJ program (NIH).
P. falciparum Gametocyte Culture
P. falciparum wild type (NF54) and transgenic parasites PfAP2-O with and without Shield-1 were maintained under standard conditions to produce infectious gametocytes as described in Hyde et al. (2003). For this, two separate flasks of each strain were set up at 0.6% parasitemia and 6% hematocrit 14 and 17 days before the feeding using human blood obtained from the Scottish National Blood Transfusion Service. When parasitemia reached a high number and stressed parasites were observed in the blood smears the medium was increased by 50% and was changed every day until the day of mosquito feeding. Permission for the non-therapeutic use of human blood was obtained from the Scottish National Blood Transfusion Service Committee for the Governance of Blood and Tissue Samples for Non-Therapeutic Use, references 18–15.
Mosquito Infection With P. falciparum Gametocytes
For each parasite line, 100 mosquitoes of An. coluzzii Ngousso strain were infected using membrane feeding (Ranford-Cartwright et al., 2016), with a mixture of day 14 and day 17 gametocyte cultures of each parasite line. Of note, the NF54::AP2-O-GFP-DD24 line was cultivated in the absence of Shield-1 during gametocyte induction. Gametocytes were centrifuged and the pellet resuspended in the same volume of human serum. Then, they were mixed with 1 to 2 volumes of 40% hematocrit human blood prepared in human serum, to produce a final gametocytemia of 0.5–1%. The gametocyte blood meal was placed into individual membrane feeders heated to 37°C, and mosquitoes were allowed to feed for 20–30 min. Unfed mosquitoes were removed, and the blood-fed mosquitoes were maintained under standard insectary conditions (26 ± 1°C, 80% humidity, 12 h light:12 h dark cycle) and fed ad libitum on 5% glucose solution containing 0.05% (w/v) 4-aminobenzoic acid (PABA). Mosquitoes were dissected 10 days post-infection, and the midguts were examined microscopically (400× magnification) for the presence and number of oocysts. Permission for the non-therapeutic use of human blood was obtained from the Scottish National Blood Transfusion Service Committee for the Governance of Blood and Tissue Samples for Non-Therapeutic Use, references 18–15. This experiment was done three times.
Statistical Analysis of Mosquito Infection
To estimate the effect of the PfApiAP2-O knockout/knockdown in P. falciparum transmission, a generalized linear mixed-effect model GLMM was employed using the package lme4 (Bates et al., 2015) in R software version 3.5.3 (The R Foundation, 2013). To assess the effect on infection prevalence, a binomial GLMM with log link was used with fixed variables of parasite clone and gametocyte number present in the blood meal, with the replicate (infectious feed number) included as a random effect. A stepwise backward selection was used to identify the significant fixed effects (Burnham and Anderson, 2002), with optimal model selection performed using the Akaike Information Criterion (AIC) for which lower scores indicate better model fits, and likelihood ratio tests were used to compare the different models. Model diagnostics, including normality of residuals were done by visual inspection of residuals versus fitted values for each random effect group, and a Shapiro-Wilk test. Finally, to calculate how much of the variance was explained by the variables included in the final model, an R squared for the generalized linear mixed effect model r2glmm (Nakagawa et al., 2017) was applied. For this, the package MuMIn, version 1.41.0, was used, and the R2m indicates the variation in prevalence explained by the fixed effects, and the R2c included all the variables used in the model.
For intensity of infection (model 2), a zero-inflated negative binomial distribution was fitted to the data in a GLMM analysis using the package glmmTMB (Brooks et al., 2017) with the family nbinom 2 (log link). The final minimum model included fixed variables of clone and the number of gametocytes present in the blood meal, with feed number as a random effect: For model selection, diagnostics and to test overdispersion the same procedures from model 1 were used. As above, R software version 3.5.3 (R: The R Project for Statistical Computing) was used for data analysis and models fit, as additional packages were used lme4 (Bates et al., 2015), DHARMa 0.3.0 (Hartig, 2021) aods3 (Lesnoff et al., 2018) and MuMIn (v1.42.5) (Nakagawa et al., 2017).
Results
Decreased Quantities of PfAP2-O Do Not Interfere With P. falciparumBlood-Stage Growth
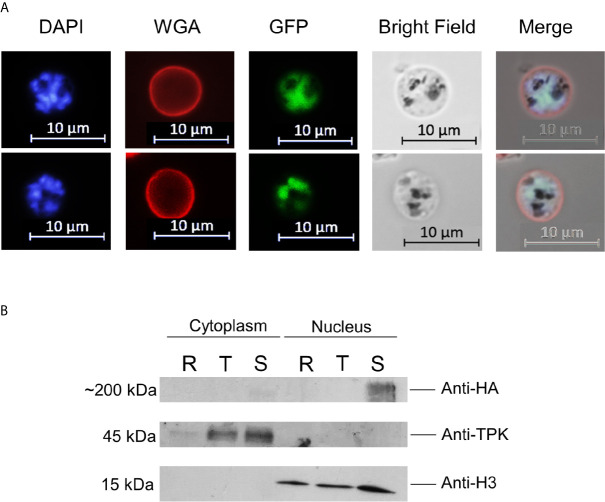
NF54 parasites were successfully modified by single crossover recombination to contain a destabilizing DD24 domain (de Azevedo et al., 2012) in the 3′-position of the PfAP2-O open reading frame ( Figures 1A, B and Supplementary Figure S1 ). Removal of Shield-1 from the cultures led to an ~85% depletion of the corresponding protein in mutant parasites ( Figure 1C and Supplementary Table 1 ). Similar to knockout experiments of the orthologous gene in P. berghei (Yuda et al., 2009), a significant decrease in the expression levels of this factor did not result in a lethal growth defect after 96 h in the absence of Shield-1 compared to control parasites ( Figure 1D ). Considering that a severe growth defect in piggy-bac insertion mutants of PfAP2-O (Zhang et al., 2018) was observed, we tried to delete the gene to confirm a deleterious phenotype. After several attempts, we were not able to obtain mutant parasites using the classic approach described by Duraisingh et al. [ Supplementary Figure S1 (Duraisingh et al., 2002)]. This indicates that PfAP2-O is essential for blood stages but that even small amounts PfAP2-O under DD24 mediated knockdown are sufficient to maintain asexual growth. Using the GFP and HA tags contained in the transgenic strain, we observed a stage-specific expression and exclusive nuclear localization of the protein by a combination of fluorescent microscopy and nuclei fractionation assays ( Figure 2 ). Of note, while PfAP2-O was only detected in the nuclear fraction of schizonts, Histone 3 was present in the ring, trophozoite, and schizont stage parasites. In contrast, Thiamin pyrophospho-kinase was visible exclusively in the cytosolic fraction of trophozoites and schizonts ( Figure 2 ).
Figure 2.
PfAP2-O-GFP-HA-DD24 is expressed in schizont stage parasites. (A) Fluorescence microscopy of late schizont parasites shows that GFP-tagged PfAP2-O colocalizes with the DAPI (nuclear stain) signal. WGA marks the surface of red blood cells. (B) Western blot of cytoplasmic and nuclear fractions obtained from PfAP2-O-GFP-HA-DD24 expressing parasites shows that AP2-O-GFP-HA-DD24 is found exclusively in the nucleus of schizonts. The membrane containing the transferred proteins was cut in three parts and was probed with monoclonal anti-HA (upper part), polyclonal anti-TPK (middle part), and monoclonal anti-Histone 3 antibodies (lower part). See Supplementary Figures for the full-size exposure.
PfAP2-O Knockdown Leads to Diversified Transcription Number of var Genes
In a previous study, the single AP2 domain of PfAP2-O was predicted to bind to elements found in the 5′ ups regions of var genes (Campbell et al., 2010). To elucidate any influence of PfAP2-O on var transcription, we selected clonal parasite populations to dominantly express a single var gene (PF3D7_0412400) by repeated panning of PfAP2-O-GFP-DD24 expressing trophozoite/schizont-infected RBCs over CHO-CD36 cells, as described previously (Gölnitz et al., 2008) ( Figure 3A ). After this, we performed the silencing of the PfAP2-O for 96 h, followed by re-establishment of the protein to observe var gene transcripts during knockdown and after the reestablishment of PfAP2-O. var genes are exclusively transcribed during the first 10–14 h after merozoite invasion. Therefore, RNA samples from synchronized ring stage-parasites were collected from three differently treated cultures: 1. RNA from panned parasites (initial sample), 2. RNA from parasites with PfAP2-O knockdown by the absence of Shield-1 for two reinvasions, and 3. RNA from parasites two reinvasions after the knockdown and reestablishment of PfAP2-O by addition of Shield-1 ( Figure 3 ).
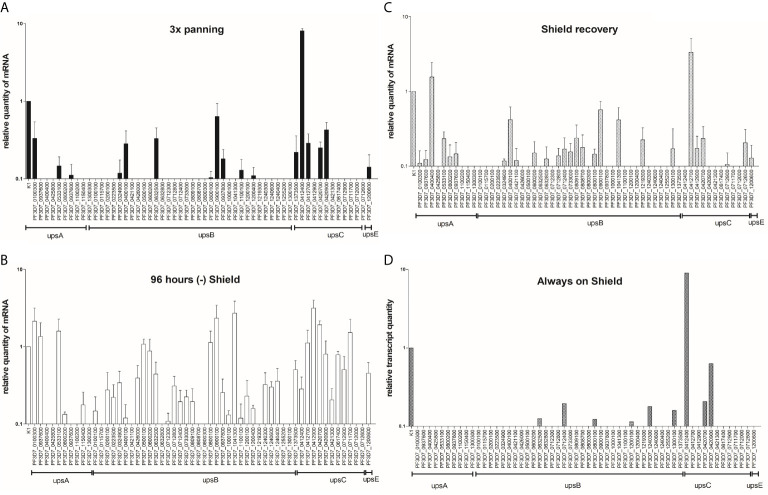
Figure 3.
var gene transcription analysis during a transient knockdown in PfAP2-O-GFP-HA-DD24 parasites. (A) RNAs extracted from NF54::PfAP2-O-GFP-DD24 parasites grown in the presence of Shield-1, and panned three times over CHO-CD36 cells were analyzed by RT-qPCR using t-seryl ligase (“K1”) as an internal control transcript, as described in Methods. (B) var transcript profile after 96 h incubation of parasites without Shield-1. (C) var transcript profile in parasites after re-establishment of PfAP2-O for two reinvasions (96 h). (D) var transcription profile in parasites never submitted to silencing of PfAP2-O (Always on Shield-1, at the end of the knockdown experiment). The data represent results from three independent experiments and error bars indicate the standard deviation between these.
In the absence of Shield-1, several var transcripts reproducibly showed increased relative amounts (PF3D7_0100300, PF3D7_0412900, PF3D7_0420700, PF3D7_0500100, PF3D7_0533100, PF3D7_0711700, PF3D7_0800100, PF3D7_0900100, PF3D7_0937600, PF3D7_1041300), while the initially dominant var gene transcript PF3D7_0412400 was less abundant ( Figure 3B ). When checking for the chromosomal location of the differently transcribed var genes, we observed transcripts from all var subgroups, upsA, upsB, and upsC. Furthermore, while most of the upregulated var genes showed the 5′ ups recognition motifs identified for subdomains of PfAP2-O (Campbell et al., 2010), others did not, such as PF3D7_0937600 and PF3D7_0800100. It appears that the depletion of PfAP2-O exerts mostly a de-repressing effect on var loci that were not exclusively related to any var gene subgroup or the predicted binding sites described by Campbell et al. (2010). Importantly, the continuous Shield-1 treatment itself does not alter var transcription or var transcription memory, since Shield-1-treated transfectant parasites used as control during these experiments showed similar var transcription patterns after selection on CHO-CD36 cells ( Figure 3D ).
Short-Term Silencing of PfAP2-O Leads to the Complete Deletion of var Transcription Memory
The var transcription memory over multiple reinvasions is believed to be maintained by factors that direct chromatin readers and writers to their respective sites of action. Transcriptional activity of genes is reversibly determined in the specific histone code of methylation and acetylation of histone 3 lysines and probably others (Lopez-Rubio et al., 2007). If PfAP2-O is acting hierarchically upstream in the events that result in the recruiting of silencing and activating factors, then its knockdown should erase the epigenetic memory of var gene transcription. To verify what influence a temporary knockdown of PfAP2-O had on var transcription, we observed the var transcript profile when the protein function was re-established by re-adding Shield-1. The pre-silencing most active var locus PF3D7_0412400 appeared to be silenced after the reestablishment of PfAP2-O. Instead, transcripts from its neighboring locus PF3D7_0412700 and upsA PF3D7_0400400 were dominantly detected, indicating that the transcriptional var memory had been erased. Again, cytoadherence-selected parasites that were cultivated on Shield-1 throughout the experiment were still dominantly expressing var gene PF3D7_0412400, confirming that no spurious global switching event occurred during the growth period ( Figures 3C, D ). This indicates that solely the retrieval and subsequent lower quantity of PfAP2-O accounted for the observed effect.
The Absence of PfAP2-O Modifies the Cytoadherence Phenotype
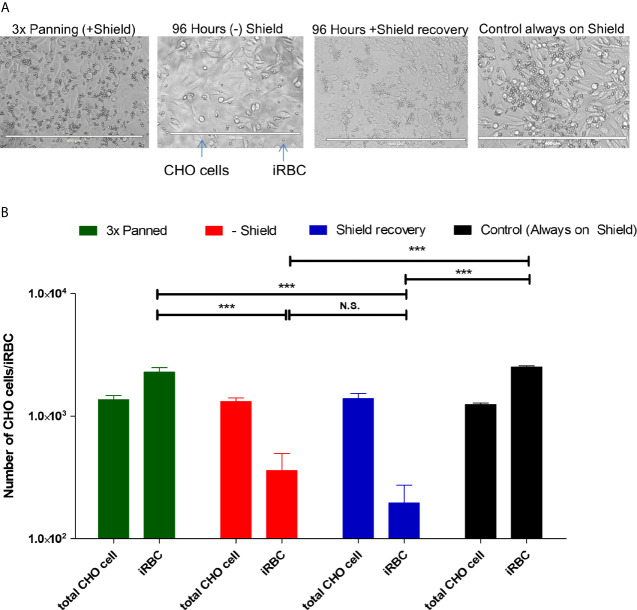
A landmark of different types of severe malaria is the adherence of infected red blood cells (IRBC) to determined receptors. Considering the observation that the PF3D7_0412400 var transcript was much less abundant in a transient knockdown of PfAP2-O, we tested if the parasites also lost the CHO-CD36 adherent phenotype. For this, a static cytoadherence procedure was conducted during and after the knockdown. Both CHO-CD36 cells and parasites were counted ( Figure 4 ). We observed that during and after silencing of PfAP2-O the number of parasites adhering to CHO-CD36 cells was significantly lower than the initially CHO-CD36 selected parasites and also the control parasite line, indicating that this factor may influence P. falciparum cytoadherence mediated by PfEMP1 proteins.
Figure 4.
Adherent phenotype analysis shows decreased cytoadherence of IRBC during and after knockdown. (A) Illustrative recording of each condition. From these pictures, cells and the infected red blood cells were counted. The white bar at the bottom of each picture indicates 400 µm. (B) Graphic representation of three independent experiments. The significance of difference in numbers of adhering iRBC was calculated using Student’s T-Test, and three asterisks indicate p values <0.005. N.S. is not significant. “Controls” are the 3× CHO-CD36 selected cultures that were cultivated with Shield-1 throughout the test and analyzed at the same timepoint as the knocked-down and Shield-1 re-added cultures.
PfAP2-O Seems to Interact With Histone Modifiers
PfAP2-O probably interacts with other factors to exert its biological role. To identify proteins that physically interact with PfAP2-O, a total protein extract from schizonts was prepared and PfAP2-O was immunoprecipitated via the HA tag and purified. Three protein bands at molecular weights of 70, 30, and 15 kDa (which did not appear in NF54 wildtype extracts immunoprecipitated in parallel) were detected and analyzed by mass spectrometry ( Supplementary Figure S2 ). In all of these extracts, fragments of a 167 kDa protein appeared, and this protein (PF3D7_0216700/PFB0765w) was predicted to be related to autophagy. Fragments of an essential 402 kDa protein with unknown function (PF3D7_0317300/PFC0650w) were detected in the 70 kDa and the 15 kDa fractions. We also observed that PfAP2-O associated with two histone modifiers namely histone-lysine N-methyltransferases SET1 (encoded by PF3D7_0629700) and SET2 (encoded by PF3D7_1322100, PfSET2/PfSET-VS) and another member of the ApiAP2 family (PF3D7_0420300, Supplementary Table 2 ). Intriguingly, no peptides from histones were identified in this assay, indicating that PfAP2-O may not directly interact with these.
The Role of PfAP2-O in P. falciparum Sexual Development
Several members of the ApiAP2 family have been associated with important processes in the Plasmodium life cycle, such as gametocyte commitment (PfAP2-G/PfAP2-G2) (Kafsack et al., 2014; Yuda et al., 2015), liver stage (Iwanaga et al., 2012), sporozoite (PbAP2-SP) (Yuda et al., 2010), and oocyst (PbAP2-O) (Yuda et al., 2009) development, and invasion ligand transcription (PfAP2-I) (Santos et al., 2017). Considering this, we tested if the knockdown of PfAP2-O regulated any gametocyte-related transcripts induced in the presence of PfAP2-G as well as the transcript of PfAP2-G itself.
The PfAP2-O silencing had a mixed effect on gametocyte-associated transcripts in trophozoite stage parasites. While no difference was discernible in the steady-state transcript quantities of PfAP2-G (PF3D7_1222600), several key gene transcripts previously associated with gametocyte development appeared in increased amounts ( Figure 5A ). The transcript of the early gametocyte-expressed gene Pfs16 (Pf3D7_0406200) appeared to increase up to four-fold when compared with the control. When the PfAP2-O protein was re-established, we observed that most of the transcripts returned to previous expression levels, except for Pfs16, which remained at high levels. To investigate if the Pfs16 transcript was indeed being translated, its corresponding protein was detected with a specific antibody. While during PfAP2-O-GFP knockdown Pfs16 was readily detected, re-establishment of PfAP2-O-GFP ( Figure 5B ) resulted in the disappearance of the protein.
Figure 5.
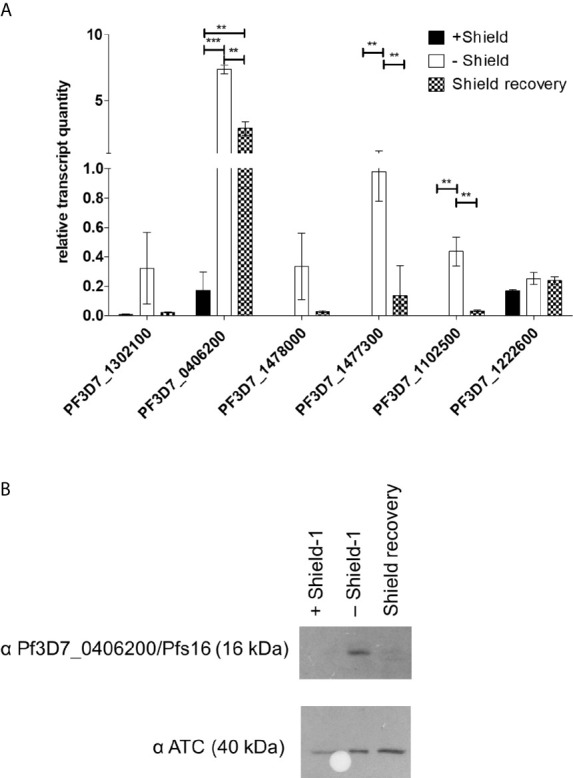
PfAP2-O-GFP-HA-DD24 is involved in the transcription/translation of gametocyte-related genes in P. falciparum. (A) Transcript abundance of gametocyte-stage specific genes in PfAP2-O-GFP-HA-DD24 expressing, knocked-down (two reinvasions without Shield-1), and recovered parasites (two reinvasion cycles without Shield-1, followed by two reinvasion cycles in the presence of Shield-1). The data were normalized to seryl-tRNA ligase (PF3D7_0717700), used as the internal control. For statistical evaluation, the ANOVA test with Bonferroni’s correction was used. ** is p < 0.01 and *** is p < 0.005. (B) Western blot detecting Pfs16/Pf3D7_0406200 in extracts of NF54::PfAP2-O_GFP_HA_DD24 parasites in the three conditions (on Shield-1/knocked-down/recovered parasites). As a control, an anti-ATC antibody was used as before. See Supplementary Figures for the full size exposure.
PfAP2-O Is Essential for P. falciparum Sexual Development
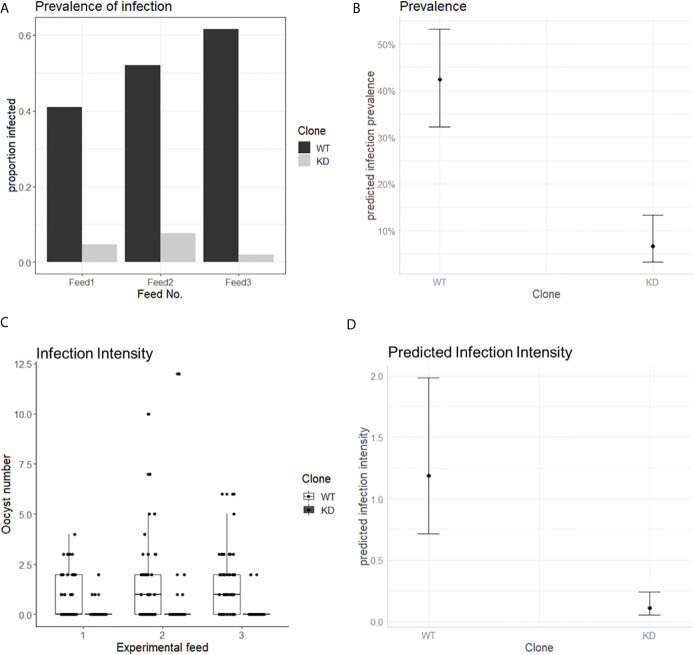
The murine P. berghei AP2-O protein (PbApiAP2-O) was associated with mosquito transmission since knockout parasites for this protein could not form functional ookinetes capable of invading the Anopheles midgut and thus completing the sporogonic cycle (Yuda et al., 2009). We then asked whether the PfAP2-O protein would have the same effect in P. falciparum. For this, Anopheles coluzzii mosquitoes were fed with the PfAP2-O-GFP-DD24 strain in the absence of Shield-1 (AP2-O knockdown, KD), and the prevalence of infected mosquitoes and oocyst number was measured and compared with the NF54 WT strain. In all experimental feedings, the infection prevalence was lower than 10% for the KD strain and higher than 40% for the WT ( Figure 6A ). Using a generalized linear mixed model (GLMM) ( Supplementary File 1 ) to predict the effects of PfAP2-O knockdown on parasite transmission, we observed a significantly lower oocyst prevalence in mosquitoes infected with the thePfAP2-O knockdown line compared to the NF54 wild type ( Figure 6B , P-value: 2.6 * 10−6), controlling for gametocyte numbers in the infectious blood meal which was positively correlated with infection (P = 0.0158). The odds ratio values indicated that mosquitoes fed blood meals containing knockdown parasites had a significantly lower probability of infection than those fed with the WT parasites (OR: 0.09, 95% CI). We also observed that a higher number of gametocytes in the infectious blood meal correlated with increased chances of infection (OR: 1.56, 95% CI).
Figure 6.
PfAP2-O is essential for P. falciparum transmission to Anopheles coluzzii. Mosquitoes were fed with gametocytes of NF54 and NF54::AP2-O-GFP-DD24, induced in the absence of Shield-1 (see Methods and Supplementary Figure 3 for details). (A) Prevalence of infected mosquitoes fed with ApiAP2-O knockdown and WT gametocytes in each experimental feeding. (B) Predicted effect of parasite clone on mosquito infection, bars indicate 95% confidence intervals. (C) number of oocysts observed in each experimental feed. (D) Predicted effect of each clone in oocyst number. Bars indicate 95% confidence intervals.
When the intensity of the infection (oocyst numbers) in mosquitoes was analyzed, we observed that in all experiments, the number of oocysts in mosquitoes fed with KD parasites was significantly lower compared to those fed with the NF54 WT parasites ( Figure 6C ). In a GLMM analysis, infection intensity ( Supplementary File 1 ) was significantly influenced by the parasite clone used (P = 5.61 * 10−14), but not by the gametocyte number in the blood meal (P: 0.0541) ( Figure 6D ).
These data show that the knockdown of PfApiAP2-O significantly decreased both the prevalence and the intensity of infection in mosquitoes and predict a positive relation between the expression of the protein and the transmission of P. falciparum from human to Anopheles mosquitoes.
Discussion
Transcriptional regulation in Plasmodium falciparum is typically related to chromatin modification and in this regard, histone methyltransferases, demethylases, a histone acetyltransferase, and deacetylases have a central role (Tonkin et al., 2009). These factors are believed to regulate the accessibility of transcription factors to promoter regions [revised in (Hollin et al., 2021)]. In the case of var genes, however, the hierarchy of events that finally leads to the selective activation of one promoter and the inactivation of the previously active one (switching) is unclear. We hypothesized that by using a temporary knockdown of factors putatively participating in these events, the hierarchy of elements playing a role in the maintenance or modification of epigenetic signatures of var transcription might be elucidated.
Members of the ApiAP2 TFs family seem to be key regulators in the Plasmodium life cycle since some of them have been associated with important processes such as transcription control of multigenic families, gametocyte commitment, and invasion of erythrocytes in P. falciparum (Kafsack et al., 2014; Martins et al., 2017; Santos et al., 2017), oocyst, and sporozoite formation in P. Berghei (Yuda et al., 2009; Yuda et al., 2010), and liver stage development also in P. Berghei (Iwanaga et al., 2012). Here we explored the function of PfApiAP2-O in P. falciparum asexual and sexual cycles with a particular focus on the control of var gene transcription. We successfully modified the target gene with various sequences that permitted to examine its function in different molecular and biological processes of the parasite life cycle. We observed a stage-specific expression and a nuclear localization through the GFP/HA tags, which is expected for DNA binding proteins. The knockdown assays showed that partial depletion of this factor has not a lethal effect on the asexual proliferation as we did not observe any defect or delay in parasite growth when the protein was absent.
In the murine malaria P. berghei, PbAP2-O has a crucial role in ookinete formation and invasion of mosquito midgut with no essential function during the intraerythrocytic cycle (Yuda et al., 2009). Similarly, normal levels of PfAP2-O seemed also not essential for asexual proliferation in P. falciparum, and long-term growth in the absence of Shield-1 revealed no change in parasite development or growth rate during the asexual intraerythrocytic cycle. However, several attempts to fully disrupt the PfAP2-O gene failed in our hands. Along with the growth defect shown in insertion-mutated versions of this protein showed by Zhang et al. (2018), these observations point to a lethal phenotype when this factor is completely absent during the intraerythrocytic cycle. As we observed that the knockdown was not complete, we hypothesize that the few functional PfAP2-O were still sufficient to maintain normal asexual growth. Moreover, we observed that PfAP2-O, in contrast to PbAP2-O (Yuda et al., 2009), is expressed in schizont stage parasites, already indicating additional roles besides the control of expression of female gametocyte- and ookinete-related genes.
The knockdown of PfAP2-O led to a swift and complete change of var transcription patterns, indicating that PfAP2-O is decisively involved in maintaining the transcriptional memory and acts as an important factor in var switching. Based on the results of previous works (Chookajorn et al., 2007; Lopez-Rubio et al., 2007; Salcedo-Amaya et al., 2009), we hypothesize that histone lysine 9 modifications (trimethylation or acetylation) are different at var loci before and after knockdown of PfAP2-O. In consequence, PfAP2-O seems to guide chromatin-modifying factors to silenced loci, since in the absence of PfAP2-O, several var loci—but not all—were actively transcribed. This is enforced by the fact that different histone modifiers were identified in the proteomic analysis. The transcript analysis also revealed that the same var loci were activated in three biological replicates. Thus, it appears that parasites with the activated var locus Pf3D7_0412400 preferentially switch to the same adjacent var (PF3D7_0412700 in this case) when PfAP2-O is temporarily unavailable. Perhaps small non-coding RNAs from the var locus-adjacent RUF6 loci are involved in this phenomenon (Wei et al., 2015; Fan et al., 2020).
The change of var transcription was also reflected in the change of cytoadherence patterns in parasites subjected to PfAP2-O protein depletion. Here, IRBC under PfAP2-O knockdown adhered significantly less to CHO-CD36 cells, probably due to the virtual disappearance of the PfEMP1 that most effectively binds CD36, encoded by Pf3D7_0412400. Taking into account that IRBC binding to CHO-CD36 occurred in fact via CD36 and not via other unrelated receptors found on CHO cells (Andrews et al., 2005), then interaction with the surface of CHO-CD36 cells with the PfEMP1 ectodomain present in Pf3D7_0412400 is stronger than that of the PF3D7_0412700. Both PfEMP1 ectodomains were predicted to be competent for CD36 binding, while the second most transcribed var PF3D7_0400400 encodes a CIDRα1.1 domain, which is not expected to interact with CD36 (Hsieh et al., 2016). The var PF3D7_0400400, upregulated after knockdown and recovery of PfAP2-O, is the so-called varsevere (Lavstsen et al., 2012) of the upsA var group of which expression is upregulated in severe malaria cases (Warimwe et al., 2009) and the onset of infection in non-immune individuals (Lavstsen et al., 2005). Another explanation of decreased cytoadherence under/after AP2-O knockdown may be that knob-associated proteins such as KAHRP or PfEMP3 were also influenced. In endemic areas, the disruption of cytoadherence remains a challenge in the treatment of patients with severe malaria (Mustaffa et al., 2017). The fact that the plant-like ApiAP2 proteins are not encoded in the human host may point PfAP2-O to a novel target of intervention to block the adhesion of infected red blood cells, perhaps alleviating life-threatening cytoadherence patterns.
Histone protein 1 (HP-1) has previously been shown to control transcription of gametocyte-associated genes by de-repression of PfAP2-G, but also that of variant genes such as var, rif, and Pfmc-2TM (Brancucci et al., 2014). Here, we demonstrated a repressive effect of the PfAP2-O factor, which may be independent of HP1, since no tight interaction between PfAP2-O and HP1 was observed by our proteomic analysis. Based on the upregulation of var and some gametocyte-associated genes when the PfAP2-O is depleted, we hypothesize that PfAP2-O indirectly triggers a repressive effect on its target genes. PfAP2-O may promote the recruitment of HP-1 to specific sites, which then leads to gene silencing; in the absence of PfAP2-O, the HP-1-guiding effect would then be impaired at least for some loci. The role of PfAP2-O on variant gene control is further supported by the observation that different histone modifiers such as PfSET1 and also PfSET2 co-precipitated with it and these have already been linked to var transcription (Jiang et al., 2013). Also, the factor identified as Pf08_0048, described as a putative Snf2-related CBP activator (chromatin remodeler) may be involved in var transcription, although the factor was not identified in the recent deadCas9-based approach to identify proteins associated to var loci (Bryant et al., 2020).
Regarding sexual commitment, we detected the increased presence of several early gametocyte-related transcripts in PfAP2-O-knocked-down parasites. Transcription of the early gametocyte gene Pfs16 (Pf3D7_0406200) was strongly increased upon PfAP2-O knockdown but did not return to pre-knockdown levels after re-exposure of the parasites to Shield-1. This may indicate the passing of a checkpoint (still undescribed) that leads to the persisting of Pfs16 transcription. However, despite the presence of Pfs16 transcript, Pfs16 protein was no longer detected after re-establishment of PfAP2-O, suggesting additional post-transcriptional control of this protein. Commitment to gametocyte development is known to be controlled by another member of the ApiAP2 family, PfAP2-G/PF3D7_1222600: Increases in PfAP2-G leads to increased transcription of gametocyte-related genes and conversion to gametocytes (Kafsack et al., 2014). The increased expression of PfAP2-G had no significant influence on the transcription of PfAP2-O (Kafsack et al., 2014), conversely, we observed that knockdown of PfAP2-O had no enhancing effect on the presence of the PfAP2-G transcript ( Figure 5A ).
We also investigated the role of PfAP2-O in parasite transmission through mosquito infection. There was no difference in the number of mature gametocytes obtained under standard culturing conditions for the WT and KD clones ( Supplementary Figure S3 ). Furthermore, the gametocytes produced by the PfAP2-O-knocked-down line did not present any visible difference in shape or size compared to the NF54 ( Supplementary Figure 3 ). Gametocytes produced under PfAP2-O knockdown were able to exflagellate, which indicates that this protein does not play an important role in gametocyte commitment and exflagellation. However, it is still uncertain whether these gametocytes have a biological defect that blocks the fertilization and mosquito midgut invasion, given that a number of gametocyte-related genes were deregulated (see Figure 5 ). When analyzing genomic features of the differentially regulated gametocyte-related genes, we found that all of them were centromeric, and only one gene (PF3D7_1477300, a PHIST gene) can be occupied by HP1, while the others (Pf3D7_1302100, Pf3D7_0406200, Pf3D7_ 1478000, Pf3D7_1102500) are not occupied by HP1 (Fraschka et al., 2018). This points to an additional layer of transcriptional control of these genes exerted directly or indirectly by AP2-O. In our experiments, we did not study fertilization and ookinete formation to assess this question due to the difficulties to induce them in vitro in P. falciparum. Our results showed that mosquitoes fed with the PfApiAP2-O strain under knockdown had a lower level of infection than those fed with the NF54 WT control, similar to the results shown for the PfAP2-O ortholog in P. berghei (Yuda et al., 2009). We showed that protein depletion of PfApiAP2-O led to a significant decrease in the prevalence of infected mosquitoes as well as a reduction of oocyst numbers. Thus, PfAP2-O appears to be essential for mosquito invasive stages, as was shown for its ortholog in P. berghei. These observations suggest a conservative role of this protein in human-mosquito transmission among Plasmodium species. Finally, the ApiAP2-O knockdown did not result in a complete blockade of mosquito infection: a minority of mosquitoes still was infected, with very low oocyst numbers. This could be explained by the fact that the knockdown was partial and small amounts of protein are still detected during Shield withdrawal.
Interactions between ApiAP2 proteins in P. berghei are involved in crucial processes during the parasite life cycle, including gametocyte development, and oocyst and sporozoite formation (Modrzynska et al., 2017). In our proteomic co-immunoprecipitation analysis another ApiAP2 member, PF3D7_0420300/PFD0985W, with no defined function, was also detected. Campbell and colleagues identified a significant number of target genes predicted for this protein, mainly associated with DNA replication and gene expression (Campbell et al., 2010). These observation suggests that these members may be part of a protein interaction network responsible for important steps in the P. falciparum life cycle, similar to that described in the murine parasite (Modrzynska et al., 2017).
In conclusion, we showed that PfAP2-O controls transcript levels of the var family, antigenic variation, and transcriptional memory in the P. falciparum asexual cycle. We also demonstrated a significant decrease in P. falciparum transmission to Anopheles mosquitos when this protein is absent. PfAP2-O apparently interacts with other transcription factors and histone modifiers in a dynamic protein complex involved in several key processes in P. falciparum biology. A further ChIPseq and RNAseq analyzing ring, trophozoite, and schizont stages may reveal further details about the interactors of AP2-O. Due to its influence on variant gene transcription and transmission, PfAP2-O may be an attractive drug target to alleviate virulence factor-induced pathogenesis, with additional impacts through blocking the passage of the parasite from an infected human to the invertebrate host.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: Wunderlich, Gerhard (2021), “DatasetsAP2-Oknockdown”, Mendeley Data, V1, doi: 10.17632/rrx9bsmyrv.1. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD026237.
Author Contributions
EC, LR-C, and GW conceived the study. EC, IO, WF, LR-C, and GW conducted experiments. EC, LR-C, and GW analyzed the data and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Research in GW’s lab is funded by the Fundação de Apoio à Pesquisa do Estado de São Paulo (FAPESP, grants 2015/17174-7 and 2017/24267-7). EC and WF were supported by FAPESP fellowships (2016/12659-5 and 2016/19145-7, respectively), and IO was supported by a Conselho Nacional de Pesquisa (CNPq) fellowship. GW is a CNPq Research fellow.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Professor Giuseppe Palmisano from the University of São Paulo for assistance with the mass spectrometry analysis, Wolfgang Fischer for technical support in University of São Paulo laboratories, and Dorothy Armstrong and Elizabeth Peat for the maintenance of the IBAHCM/Glasgow University mosquito insectaries.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.669088/full#supplementary-material
References
- Andrews K. T., Adams Y., Viebig N. K., Lanzer M., Schwartz-Albiez R. (2005). Adherence of Plasmodium Falciparum Infected Erythrocytes to CHO-745 Cells and Inhibition of Binding by Protein A in the Presence of Human Serum. Int. J. Parasitol. 35, 1127–1134. 10.1016/j.ijpara.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Balaji S., Babu M. M., Iyer L. M., Aravind L. (2005). Discovery of the Principal Specific Transcription Factors of Apicomplexa and Their Implication for the Evolution of the AP2-integrase DNA Binding Domains. Nucleic Acids Res. 33, 3994–4006. 10.1093/nar/gki709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Mächler M., Bolker B. M., Walker S. C. (2015). Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Software 67, 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Boyle J. S., Lew A. M. (1995). An Inexpensive Alternative to Glassmilk for DNA Purification. Trends Genet. 11, 8. 10.1016/S0168-9525(00)88977-5 [DOI] [PubMed] [Google Scholar]
- Brancucci N. M. B., Bertschi N. L., Zhu L., Niederwieser I., Chin W. H., Wampfler R., et al. (2014). Heterochromatin Protein 1 Secures Survival and Transmission of Malaria Parasites. Cell Host Microbe 16, 165–176. 10.1016/j.chom.2014.07.004 [DOI] [PubMed] [Google Scholar]
- Brooks M. E., Kristensen K., van Benthem K. J., Magnusson A., Berg C. W., Nielsen A., et al. (2017). glmmTMB Balances Speed and Flexibility Among Packages for Zero-Inflated Generalized Linear Mixed Modeling. R. J. 9, 378–400. 10.32614/rj-2017-066 [DOI] [Google Scholar]
- Bryant J. M., Baumgarten S., Dingli F., Loew D., Sinha A., Claës A., et al. (2020). Exploring the Virulence Gene Interactome With CRISPR/dCas9 in the Human Malaria Parasite. Mol. Syst. Biol. 16, e9569. 10.15252/msb.20209569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham K., Anderson D. (2002). Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. 2. edition (Berlin: Springer Verlag; ). [Google Scholar]
- Calderwood M. S., Gannoun-Zaki L., Wellems T. E., Deitsch K. W. (2003). Plasmodium Falciparum Var Genes are Regulated by Two Regions With Separate Promoters, One Upstream of the Coding Region and a Second Within the Intron. J. Biol. Chem. 278, 34125–34132. 10.1074/jbc.M213065200 [DOI] [PubMed] [Google Scholar]
- Campbell T. L., De Silva E. K., Olszewski K. L., Elemento O., Llinás M. (2010). Identification and Genome-Wide Prediction of DNA Binding Specificities for the ApiAP2 Family of Regulators From the Malaria Parasite. PloS Pathog. 6, e1001165. 10.1371/journal.ppat.1001165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Fernandez V., Sundstrom A., Schlichtherle M., Datta S., Hagblom P., et al. (1998). Developmental Selection of Var Gene Expression in Plasmodium Falciparum. Nature 394, 392–395. 10.1038/28660 [DOI] [PubMed] [Google Scholar]
- Chookajorn T., Dzikowski R., Frank M., Li F., Jiwani A. Z., Hartl D. L., et al. (2007). Epigenetic Memory At Malaria Virulence Genes. Proc. Natl. Acad. Sci. U. S. A. (United States) 104 (3), 899–902. 10.1073/pnas.0609084103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo M. F. M. F. M. F., Gilson P. R. P. R., Gabriel H. B. H. B., Simões R. F. R. F., Angrisano F., Baum J., et al. (2012). Systematic Analysis of FKBP Inducible Degradation Domain Tagging Strategies for the Human Malaria Parasite Plasmodium Falciparum. PloS One 7, e40981. 10.1371/journal.pone.0040981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delves M. J., Angrisano F., Blagborough A. M. (2018). Antimalarial Transmission-Blocking Interventions: Past, Present, and Future. Trends Parasitol. 34, 735–746. 10.1016/j.pt.2018.07.001 [DOI] [PubMed] [Google Scholar]
- Dondorp A. M., Nosten F., Yi P., Das D., Phyo P., Tarning J., et al. (2009). Artemisinin Resistance in Plasmodium Falciparum Malaria. N. Engl. J. Med. Mae Sot. Fam. Heal. Int. Churchill Hosp. N. J. W. N. Engl. J. Med. 3615361, 455–467. 10.1056/NEJMoa0808859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh M. T., Triglia T., Cowman A. F. (2002). Negative Selection of Plasmodium Falciparum Reveals Targeted Gene Deletion by Double Crossover Recombination. Int. J. Parasitol. 32, 81–89. 10.1016/S0020-7519(01)00345-9 [DOI] [PubMed] [Google Scholar]
- Duraisingh M. T., Voss T. S., Marty A. J., Duffy M. F., Good R. T., Thompson J. K., et al. (2005). Heterochromatin Silencing and Locus Repositioning Linked to Regulation of Virulence Genes in Plasmodium Falciparum. Cell 121, 13–24. 10.1016/j.cell.2005.01.036 [DOI] [PubMed] [Google Scholar]
- Epp C., Li F., Howitt C. A., Chookajorn T., Deitsch K. W. (2009). Chromatin Associated Sense and Antisense Noncoding RNAs are Transcribed From the Var Gene Family of Virulence Genes of the Malaria Parasite Plasmodium Falciparum. RNA 15, 116–127. 10.1261/rna.1080109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Shen S., Wei G., Tang J., Zhao Y., Wang F., et al. (2020). Rrp6 Regulates Heterochromatic Gene Silencing Via Ncrna RUF6 Decay in Malaria Parasites. MBio 11, e01110–e01120. 10.1128/mBio.01110-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck C., Bartfai R., Niederwieser I., Witmer K., Alako B. T. F., Moes S., et al. (2010). A Major Role for the Plasmodium Falciparum ApiAP2 Protein PfSIP2 in Chromosome End Biology. PloS Pathog. 6, e1000784. 10.1371/journal.ppat.1000784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschka S. A., Filarsky M., Hoo R., Niederwieser I., Yam X. Y., Brancucci N. M. B., et al. (2018). Comparative Heterochromatin Profiling Reveals Conserved and Unique Epigenome Signatures Linked to Adaptation and Development of Malaria Parasites. Cell Host Microbe 23, 407–420.e8. 10.1016/j.chom.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas L. H., Hernandez-Rivas R., Ralph S. A., Montiel-Condado D., Ruvalcaba-Salazar O. K., Rojas-Meza A. P., et al. (2005). Telomeric Heterochromatin Propagation and Histone Acetylation Control Mutually Exclusive Expression of Antigenic Variation Genes in Malaria Parasites. Cell 121 (1), 25–36. 10.1016/j.cell.2005.01.037 [DOI] [PubMed] [Google Scholar]
- Gölnitz U., Albrecht L., Wunderlich G. (2008). Var Transcription Profiling of Plasmodium Falciparum 3D7: Assignment of Cytoadherent Phenotypes to Dominant Transcripts. Malar. J. 7, 14. 10.1186/1475-2875-7-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M., Sambrook J. (2012). Molecular Cloning: A Laboratory Manual. 4 ed. (Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press; ). 10.3724/SP.J.1141.2012.01075 [DOI] [Google Scholar]
- Hartig F. (2021). DHARMa: residual diagnostics for hierarchical (multi-level/mixed) regression models. Available at: https://cran.r-project.org/web/packages/DHARMa/vignettes/DHARMa.html. [Google Scholar]
- Hasenkamp S., Russell K. T., Horrocks P. (2012). Comparison of the Absolute and Relative Efficiencies of Electroporation-Based Transfection Protocols for Plasmodium Falciparum. Malar. J. 11, 210. 10.1186/1475-2875-11-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollin T., Gupta M., Lenz T., Le Roch K. G. (2021). Dynamic Chromatin Structure and Epigenetics Control the Fate of Malaria Parasites. Trends Genet. 37, 73–85. 10.1016/j.tig.2020.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh F.-L., Turner L., Bolla J. R., Robinson C. V., Lavstsen T., Higgins M. K. (2016). The Structural Basis for CD36 Binding by the Malaria Parasite. Nat. Commun. 7, 12837. 10.1038/ncomms12837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J. E., Carter R., Ranford-Cartwright L., Alano P. (2003). The Culture and Preparation of Gametocytes of Plasmodium Falciparum for Immunochemical, Molecular, and Mosquito Infectivity Studies. Protoc. Mol. Parasitol. 27, 67–88. 10.1385/0-89603-239-6:67 [DOI] [PubMed] [Google Scholar]
- Iwanaga S., Kaneko I., Kato T., Yuda M. (2012). Identification of an AP2-family Protein That is Critical for Malaria Liver Stage Development. PloS One 7, e47557. 10.1371/journal.pone.0047557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Mu J., Zhang Q., Ni T., Srinivasan P., Rayavara K., et al. (2013). PfSETvs Methylation of Histone H3K36 Represses Virulence Genes in Plasmodium Falciparum. Nature 499, 223–227. 10.1038/nature12361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joergensen L., Bengtsson D. C., Bengtsson A., Ronander E., Berger S. S., Turner L., et al. (2010). Surface Co-Expression of Two Different PfEMP1 Antigens on Single Plasmodium Falciparum-Infected Erythrocytes Facilitates Binding to ICAM1 and PECAM1. PloS Pathog. 6, e1001083. 10.1371/journal.ppat.1001083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafsack B. F. C., Rovira-Graells N., Clark T. G., Bancells C., Crowley V. M., Campino S. G., et al. (2014). A Transcriptional Switch Underlies Commitment to Sexual Development in Malaria Parasites. Nature 507, 248–252. 10.1038/nature12920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambros C., Vanderberg J. P. (1979). Synchronization of Plasmodium Falciparum Erythrocytic Stages in Culture. J. Parasitol. 65, 418–420. 10.2307/3280287 [DOI] [PubMed] [Google Scholar]
- Lavstsen T., Magistrado P., Hermsen C. C., Salanti A., Jensen A. T. R., Sauerwein R., et al. (2005). Expression of Plasmodium Falciparum Erythrocyte Membrane Protein 1 in Experimentally Infected Humans. Malar. J. 4, 21. 10.1186/1475-2875-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavstsen T., Salanti A., Jensen A. T. R., Arnot D. E., Theander T. G. (2003). Sub-Grouping of Plasmodium Falciparum 3D7 Var Genes Based on Sequence Analysis of Coding and non-Coding Regions. Malar. J. 2, 27. 10.1186/1475-2875-2-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavstsen T., Turner L., Saguti F., Magistrado P., Rask T. S., Jespersen J. S., et al. (2012). Pnas Plus: Plasmodium Falciparum Erythrocyte Membrane Protein 1 Domain Cassettes 8 and 13 are Associated With Severe Malaria in Children. Proc. Natl. Acad. Sci. 109, 1791–1800. 10.1073/pnas.1120455109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelièvre J., Berry A., Benoit-Vical F., Lelievre J., Berry A., Benoit-Vical F. (2005). An Alternative Method for Plasmodium Culture Synchronization. Exp. Parasitol. 109, 195–197. 10.1016/j.exppara.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Lesnoff M., Lancelot R., Lancelot M. R. (2018). Package “Aods3” Title Analysis of Overdispersed Data Using S3 Methods. Available at: https://cran.r-project.org/web/packages/aods3/aods3.pdf. [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Li Z., Yin S., Sun M., Cheng X., Wei J., Gilbert N., et al. (2019). DNA Helicase RecQ1 Regulates Mutually Exclusive Expression of Virulence Genes in Plasmodium Falciparum Via Heterochromatin Alteration. Proc. Natl. Acad. Sci. U. S. A. 116, 3177–3182. 10.1073/pnas.1811766116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungström I., Perlmann H., Schlichtherle M., Scherf A., Wahlgren M. (2008). Methods in Malaria Research. 5th ed. Eds. Moll K., Ljungström I., Perlmann H., Scherf A., Wahlgren M. (Manassas, VA: MR4/ATCC, BioMalPar; ). [Google Scholar]
- Lopez-Rubio J. J., Gontijo A. M., Nunes M. C., Issar N., Hernandez Rivas R., Scherf A. (2007). 5’ Flanking Region of Var Genes Nucleate Histone Modification Patterns Linked to Phenotypic Inheritance of Virulence Traits in Malaria Parasites. Mol. Microbiol. 66, 1296–1305. 10.1111/j.1365-2958.2007.06009.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins R. M., Macpherson C. R., Claes A., Scheidig-Benatar C., Sakamoto H., Yam X. Y., et al. (2017). An ApiAP2 Member Regulates Expression of Clonally Variant Genes of the Human Malaria Parasite Plasmodium Falciparum. Sci. Rep. 7, 14042. 10.1038/s41598-017-12578-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrzynska K., Pfander C., Chappell L., Yu L., Suarez C., Dundas K., et al. (2017). A Knockout Screen of ApiAP2 Genes Reveals Networks of Interacting Transcriptional Regulators Controlling the Plasmodium Life Cycle. Cell Host Microbe 21, 11–22. 10.1016/j.chom.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustaffa K. M. F., Storm J., Whittaker M., Szestak T., Craig A. G. (2017). In Vitro Inhibition and Reversal of Plasmodium Falciparum Cytoadherence to Endothelium by Monoclonal Antibodies to ICAM-1 and CD36. Malar. J. 16, 279. 10.1186/s12936-017-1930-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Johnson P. C. D., Schielzeth H. (2017). The Coefficient of Determination R 2 and Intra-Class Correlation Coefficient From Generalized Linear Mixed-Effects Models Revisited and Expanded. J. R. Soc Interface 14, 20170213. 10.1098/rsif.2017.0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehring S. C., Woodcroft B. J., Moes S., Wetzel J., Dietz O., Pulfer A., et al. (2012). Organellar Proteomics Reveals Hundreds of Novel Nuclear Proteins in the Malaria Parasite Plasmodium Falciparum. Genome Biol. 13, R108. 10.1186/gb-2012-13-11-r108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Toledo K., Rojas-Meza A. P., Mancio-Silva L., Hernández-Cuevas N. A., Delgadillo D. M., Vargas M., et al. (2009). Plasmodium Falciparum Heterochromatin Protein 1 Binds to Tri-Methylated Histone 3 Lysine 9 and is Linked to Mutually Exclusive Expression of Var Genes. Nucleic Acids Res. 37, 2596–2606. 10.1093/nar/gkp115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter M., Lee C. C., Byrne T. J., Boysen K. E., Volz J., Ralph S. A., et al. (2011). Expression of P. Falciparum Var Genes Involves Exchange of the Histone Variant H2A.Z At the Promoter. PloS Pathog. 7, e1001292. 10.1371/journal.ppat.1001292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R: The R Project for Statistical Computing . [Google Scholar]
- Ranford-Cartwright L. C., McGeechan S., Inch D., Smart G., Richterová L., Mwangi J. M. (2016). Characterisation of Species and Diversity of Anopheles Gambiae Keele Colony. PloS One 11, e0168999. 10.1371/journal.pone.0168999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanti A., Staalsoe T., Lavstsen T., Jensen A. T. R., Sowa M. P. K., Arnot D. E., et al. (2003). Selective Upregulation of a Single Distinctly Structured Var Gene in Chondroitin Sulphate A-adhering Plasmodium Falciparum Involved in Pregnancy-Associated Malaria. Mol. Microbiol. 49, 179–191. 10.1046/j.1365-2958.2003.03570.x [DOI] [PubMed] [Google Scholar]
- Salcedo-Amaya A. M., van Driel M. A., Alako B. T., Trelle M. B., van den Elzen A. M., Cohen A. M., et al. (2009). Dynamic Histone H3 Epigenome Marking During the Intraerythrocytic Cycle of Plasmodium Falciparum. Proc. Natl. Acad. Sci. U. S. A. 106, 9655–9660. 10.1073/pnas.0902515106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J. M., Josling G., Ross P., Joshi P., Orchard L., Campbell T., et al. (2017). Red Blood Cell Invasion by the Malaria Parasite Is Coordinated by the PfAP2-I Transcription Factor. Cell Host Microbe 21, 731–741.e10. 10.1016/j.chom.2017.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf A., Hernandez-Rivas R., Buffet P., Bottius E., Benatar C., Pouvelle B., et al. (1998). Antigenic Variation in Malaria: in Situ Switching, Relaxed and Mutually Exclusive Transcription of Var Genes During Intra-Erythrocytic Development in Plasmodium Falciparum. EMBO J. 17, 5418–5426. 10.1093/emboj/17.18.5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra-Miranda M., Delgadillo D. M., Mancio-Silva L., Vargas M., Villegas-Sepulveda N., Martínez-Calvillo S., et al. (2012). Two Long non-Coding RNAs Generated From Subtelomeric Regions Accumulate in a Novel Perinuclear Compartment in Plasmodium Falciparum. Mol. Biochem. Parasitol. 185, 36–47. 10.1016/j.molbiopara.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. Z., Heatwole V. M., Wertheimer S. P., Guinet F., Herrfeldt J. A., Peterson D. S., et al. (1995). The Large Diverse Gene Family Var Encodes Proteins Involved in Cytoadherence and Antigenic Variation of Plasmodium Falciparum-Infected Erythrocytes. Cell 82, 89–100. 10.1016/0092-8674(95)90055-1 [DOI] [PubMed] [Google Scholar]
- The R Foundation (2013). R: A Language and Environment for Statistical Computing. Available at: https://www.r-project.org. [Google Scholar]
- Tonkin C. J., Carret C. K., Duraisingh M. T., Voss T. S., Ralph S. A., Hommel M., et al. (2009). Sir2 Paralogues Cooperate to Regulate Virulence Genes and Antigenic Variation in Plasmodium Falciparum. PloS Biol. 7, e84. 10.1371/journal.pbio.1000084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin C. J., van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., et al. (2004). Localization of Organellar Proteins in Plasmodium Falciparum Using a Novel Set of Transfection Vectors and a New Immunofluorescence Fixation Method. Mol. Biochem. Parasitol. 137, 13–21. 10.1016/j.molbiopara.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Volz J. C., Bártfai R., Petter M., Langer C., Josling G. A., Tsuboi T., et al. (2012). PfSET10, a Plasmodium Falciparum Methyltransferase, Maintains the Active Var Gene in a Poised State During Parasite Division. Cell Host Microbe 11, 7–18. 10.1016/j.chom.2011.11.011 [DOI] [PubMed] [Google Scholar]
- Warimwe G. M., Keane T. M., Fegan G., Musyoki J. N., Newton C. R. J. C., Pain A., et al. (2009). Plasmodium Falciparum Var Gene Expression is Modified by Host Immunity. Proc. Natl. Acad. Sci. U. S. A. 106, 21801–21806. 10.1073/pnas.0907590106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Zhao Y., Zhang Q., Pan W. (2015). Dual Regulatory Effects of non-Coding GC-Rich Elements on the Expression of Virulence Genes in Malaria Parasites. Infect. Genet. Evol. 36, 490–499. 10.1016/j.meegid.2015.08.023 [DOI] [PubMed] [Google Scholar]
- WHO (2020). Insecticide Resistance (Geneva: WHO; ). [Google Scholar]
- World Health Organization (2019). World Malaria Report 2019 (Geneva: WHO; ). [Google Scholar]
- Yuda M., Iwanaga S., Kaneko I., Kato T. (2015). Global Transcriptional Repression: An Initial and Essential Step for Plasmodium Sexual Development. Proc. Natl. Acad. Sci. U. S. A. 112, 12824–12829. 10.1073/pnas.1504389112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuda M., Iwanaga S., Shigenobu S., Kato T., Kaneko I. (2010). Transcription Factor AP2-Sp and its Target Genes in Malarial Sporozoites. Mol. Microbiol. 75, 854–863. 10.1111/j.1365-2958.2009.07005.x [DOI] [PubMed] [Google Scholar]
- Yuda M., Iwanaga S., Shigenobu S., Mair G. R., Janse C. J., Waters A. P., et al. (2009). Identification of a Transcription Factor in the Mosquito-Invasive Stage of Malaria Parasites. Mol. Microbiol. 71, 1402–1414. 10.1111/j.1365-2958.2009.06609.x [DOI] [PubMed] [Google Scholar]
- Zhang M., Wang C., Otto T. D., Oberstaller J., Liao X., Adapa S. R., et al. (2018). Uncovering the Essential Genes of the Human Malaria Parasite Plasmodium Falciparum by Saturation Mutagenesis. Science 360, eaap7847. 10.1126/science.aap7847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: Wunderlich, Gerhard (2021), “DatasetsAP2-Oknockdown”, Mendeley Data, V1, doi: 10.17632/rrx9bsmyrv.1. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository with the dataset identifier PXD026237.