Abstract
Background
While observational studies revealed an inverse association between serum 25(OH)vitamin D (25(OH)D) and the risk of attention deficit/hyperactivity disorder (ADHD), the causality of this relationship remains unclear.
Methods
We conducted a bidirectional two-sample Mendelian Randomization (MR) study to examine whether 25(OH)D has an effect on the risk to develop ADHD or vice versa. Information on single nucleotide polymorphisms (SNP) associated with serum 25(OH)D was obtained from a genome-wide association study (GWAS) considering phenotype data from 79,366 individuals of European ancestry. Data on risk for ADHD were derived from a GWAS analysis with 20,183 individuals diagnosed with ADHD and 35,191 controls. For our analysis, we considered effect sizes based on the European participants (19,099 cases and 34,194 controls).
Results
Single SNP analyses showed a causal effect of vitamin D on ADHD risk for only one SNP (rs12785878, p = 0.024). The overall MR estimates did not reveal a causal effect of 25(OH)D on risk for ADHD. In the reverse analysis, neither any single nor the multi-SNP MR analyses showed a causal effect of ADHD on 25(OH)D.
Conclusion
Results from this two-sample MR study did not confirm a causal effect of 25(OH)D on ADHD or vice versa. Accordingly, our study does not provide evidence that improving 25(OH)D via supplementation could reduce the risk of developing ADHD.
Electronic supplementary material
The online version of this article (10.1007/s00394-020-02439-2) contains supplementary material, which is available to authorized users.
Keywords: Vitamin D, 25(OH)vitamin D, ADHD, Attention, Mendelian randomization, Prevention
Introduction
Considering its impact on calcium and phosphate metabolism, vitamin D had initially been discussed in particular in the context of bone health [1]. While European scientific societies already recommend vitamin D supplementation during infancy [2], recent findings from observational studies on potential negative non-skeletal effects of vitamin D deficiency caused a debate on the meaningfulness of population-wide recommendations for vitamin D supplementation in childhood and adolescence [3]. Further, the role of vitamin D for mental health might be important since it is considered as one example of a nutrient supplement that could be beneficial in the management of mental disorders [4].
In a systematic literature search regarding childhood and adolescence, we recently identified a large number of (predominantly observational) studies dealing with the relationship between vitamin D and mental health [5]. Besides autism spectrum disorder, most of these studies focused on attention deficit/hyperactivity disorder (ADHD). Results from a recent genetic look-up analysis, based on data from large-scaled genome-wide associations studies (GWAS), revealed a genetic variant which is concomitantly associated with both vitamin D insufficiency and ADHD [6]. This genetic overlap might indicate a direct association.
In general, vitamin D effects on mental health seem plausible considering the fact that both vitamin D receptors and metabolizing enzymes are located in the brain [7]. With regard to ADHD, several pathways are currently discussed such as anti-inflammatory and anti-oxidative actions or an influence on neurotrophic factors and neurotransmitter metabolism [8]. A recent randomized controlled trial (RCT) in children with ADHD showed that supplementation of 2000 IU vitamin D per day for 12 weeks resulted in significantly increased serum dopamine levels in the intervention group compared to the placebo group while serum brain-derived neurotrophic factor and serotonin levels did not change [9].
A meta-analysis recently summarized results from four RCTs with vitamin D supplementation in ADHD patients and revealed a small, but significant improvement of ADHD symptoms [10]. However, all considered RCTs were conducted in children with an already manifest ADHD, enabling conclusions only on therapeutic, but not on preventive effects of vitamin D. However, RCTs to detect preventive effects would require very large sample sizes to provide sufficient statistical power as well as long-term follow-up periods [11] to cover a critical time frame of disorder pathogenesis.
In contrast, Mendelian randomization (MR) studies based on summary data from GWAS are a time-effective approach using genetic variants as instrumental variables (IVs) to examine the causal effect of blood concentrations of a specific nutrient (e.g. vitamin D) on a particular disorder (e.g. ADHD) [12]. The concept of MR studies implies a natural “quasi randomization”, since the individual composition of alleles and, thus, of IVs are determined randomly at conception, resulting in a reduced risk of confounding [13]. Bias from reverse causation, another limitation of observational studies, is also precluded in MR studies, as the individual genotype is determined at conception, and cannot be modified by the outcome of interest [13].
Genetic factors play an important role for levels of circulating vitamin D [14] with heritability estimates based on twin studies varying from 39 to 70% for Caucasians and 86% in a community-based study of adolescents (mean age 16 years) [14, 15]. Accordingly, MR studies seem to be particularly suitable to evaluate health effects of serum 25(OH) vitamin D levels (25(OH)D). Regarding the role of vitamin D for mental disorders, MR studies recently did not confirm the discussed effects on depression [16, 17]. More recently, a phenome-wide MR study using data from UK biobank revealed no evidence of causal effects of 25(OH)D on a large number of outcomes including depression, nonvertebral fracture, and all-cause mortality [18]. Since MR studies which focus on effects on ADHD have not been conducted, the aim of this two-sample MR study was to examine the effects of 25(OH)D on ADHD using summary-level data of recent large-scaled GWAS on 25(OH)D levels and ADHD. A reverse MR analysis on causal effects of ADHD on 25(OH)D was conducted to examine whether associations found in observational studies might represent reverse causation.
Methods
Data sources for MR analyses and selection of the genetic instruments
We conducted a two-sample MR analysis with 25(OH)D as exposure and ADHD as outcome variable and a reverse analysis with ADHD as exposure and level of 25(OH)D as outcome. Information on single nucleotide polymorphisms (SNPs) associated with serum 25(OH)D were obtained from the latest GWAS analysis at the time of our data analysis considering phenotype data from 79,366 individuals of European ancestry including 31 studies from epidemiological cohorts from Europe, Canada, and USA [19]. We used independent SNPs of all six loci with genome-wide significance for 25(OH)D as the genetic instrument. The effect estimates of these genetic variants on the exposure were derived from the publicly available summary statistics of the GWAS meta-analysis. The six 25(OH)D-associated SNPs explained 2.84% of the variance of serum 25(OH)D levels [19].
Data on risk for ADHD were derived from a GWAS analysis with 20,183 individuals diagnosed with ADHD and 35,191 controls from 12 cohorts [20]. Twelve loci with genome-wide significance were identified. The SNP-heritability was 21.6%. For our analysis, we considered effect estimates from European participants (19,099 cases and 34,194 controls). Two-sample MR assumes independent samples. The screening of the study groups in the GWAS for 25(OH)D and ADHD showed no sample overlap.
In case of unavailability of SNPs identified in the 25(OH)D GWAS in the ADHD GWAS or vice versa, we used proxy-SNPs as recommended [21]. For the search of proxies, SNPs with minimum linkage disequilibrium (LD) r2 ≥ 0.40, on the basis of GRCh37.p13, Ensembl version 87, 1000 genomes: phase 3 version 5 for European ancestry, were exported applying the in silico tool SNiPA [22] (http://www.snipa.org. Accessed on 19 August 2019). Post-hoc selection criteria for proxy-SNPs were defined: 1st highest r2, 2nd smallest distance to the lead SNP. In the sensitivity analysis for reverse MR, we selected the proxy SNPs with the following selection criteria: 1st highest r2, 2nd smallest distance to the lead SNP, 3rd no palindromic alleles. Because of the potential problems with the strand [21], we excluded SNPs, which were both palindromic and ambiguous.
During the review process of this paper, two additional GWAS on the impact of genetic variation on 25(OH)D concentrations revealed substantial larger numbers of genome-wide significant loci. Revez and colleagues used data from 417,580 European participants from UK Biobank and observed 143 independent loci [23]. Manousaki et al. combined data from 401,460 UK Biobank participants with data from 42,274 Europeans from a previous GWAS and observed 138 conditional independent SNPs in 69 independent loci [24]. These increased numbers of genome-wide significant SNPs could contribute to a stronger genetic instrument. However, Manousaki et al. concluded that several SNPs identified in their GWAS were mapped to genes not directly involved in 25(OH)D metabolism and could, thus, increase the risk of pleiotropic effects [24]. In contrast, four out of the six SNPs identified in the SUNLIGHT GWAS are related to genes involved in established pathways of vitamin D metabolism. Accordingly, we kept the results based on the SUNLIGHT GWAS as main analysis and provided findings based on genetic instruments derived from the two latter GWAS as additional sensitivity MR analyses.
For these two sensitivity analyses, we defined an alternative genetic instrument based on results from the respective GWAS on 25(OH)D and conducted separate MR analyses. Only those SNPs from the respective GWAS on 25(OH)D were considered which were both available in the ADHD GWAS and not ambiguous or palindromic. This requirement was met by 100 from 143 genome-wide significant SNPs identified by Revez et al. and 61 from 138 SNPs identified by Manousaki and colleagues.
Testing MR assumptions and statistical analysis
As recently suggested [25] we used several statistical methods for the calculation of an overall causal effect estimate of 25(OH)D on ADHD (and vice versa): Inverse Variance Weighted (IVW) and MR-Egger as main analyses [26] and Weighted Median [27] as well as mode-based estimators (MBE) [25] as sensitivity analyses. These methods make different assumptions and can, thus, be used to assess the robustness of MR results against violations of the instrumental variable assumptions. IVW assumes that all ratio estimates provide independent evidence on the causal effect and there is no pleiotropic effect. The IVW accordingly assumes that all genetic variants are valid instrumental variables. There is no intercept term in the regression model [26]. In MR-Egger the intercept term is estimated as part of the analyses and can be interpreted as the average pleiotropic effect of a genetic variant included in the analyses [26]. Weighted Median estimator is consistent even when up to 50% of the information comes from invalid instrumental variables [27]. The MBE is consistent even if the majority of instruments are invalid. Its power to detect a causal effect is smaller compared with the IVW and weighted median methods, but larger than that of MR-Egger regression [25].
To conduct a MR study, three core assumptions must be fulfilled [12]: (1) The genetic instrument must have a true association with the exposure. To fulfill this assumption we used independent genome-wide significantly associated SNPs for 25(OH)D (p < 5 × 10–8). For reverse analysis with ADHD as exposure, we used SNPs which were genome-wide significantly associated with ADHD (p < 5 × 10–8) with the risk of ADHD as the genetic instrument. (2) There is no association between the genetic variants and confounders of the relationship between risk factor and outcome. (3) Conditioning on the risk factor and possible confounders, there is no direct association between the genetic variants and outcome [12]. Thus, the effect of the genetic instrument on the outcome must be mediated exclusively by the exposure and there must be no direct effects [28].
From these assumptions, only the first assumption can be directly tested [28]. For this purpose, we conducted an F-test to test the weakness of the instrument. If F-statistic is less than ten, the instrument variable is considered weak [29]. For examination of assumptions 2 and 3, multiple approaches were applied. We examined horizontal pleiotropy by estimating the intercept of Egger's regression. If Eggers intercept is not significantly different from zero, it can be assumed that there is no horizontal pleiotropy [26]. Additionally, we used MR-PRESSO to identify horizontal pleiotropic outliers and to calculate an overall outlier-corrected causal estimate [30]. Genetic polymorphisms are sometimes associated with multiple aspects or dimensions of a single trait [28]. To test such heterogeneity of the instrument variable, we used Cochran’s Q-statistic with the null hypothesis is that the MR-Egger regression model describes the associations with the outcome with no excess heterogeneity [31]. This test examines whether causal estimates of genetic variants (SNPs) are comparative [32]. To investigate the relationship between study accuracy and effect size, we created a funnel plot [28]. Asymmetry in the funnel plot indicates that assumptions for MR are not met [33]. To examine whether an individual data point (SNP) has a large influence on the regression coefficients, we conducted a leave-one-out approach. For this analysis, we conducted the IVW regression by leaving each genetic variant out in turn [26].
Forest and scatter plots were used to visualize combined results of single and multi-SNP analyses. The scatter plots show the single SNP effects on the exposure against the single SNP effects on the outcome with corresponding standard deviations and estimated regression lines of the multi-SNP analyses. We performed a power analysis to estimate whether our analysis, given sample size, proportion of cases in the study, and the proportion of variance explained, provides sufficient statistical power to detect a true causal effect [34].
All tests were performed using the software “R “, version 3.5.2, and R-packages for performing 2-sample MR (https://github.com/MRCIEU/TwoSampleMR) and MR-PRESSO (https://github.com/rondolab/MR-PRESSO).
Results
MR assessing 25(OH)D effects on ADHD
We performed a MR analysis to investigate the causal effect of 25(OH)D levels on risk of ADHD. The data on the six SNPs associated with the genome-wide significance of p < 5 × 10–8 with 25(OH)D are presented in Table 1. For one of these SNPs, a proxy SNP (rs209955) was considered due to missing information in the ADHD GWAS (rs17216707). One ambiguous and palindromic SNP (rs8018720) was not included in the analysis resulting in five SNPs used for the definition of the IV (Table 1).
Table 1.
Genome-wide significant single nucleotide polymorphisms (SNPs) for natural log-transformed 25(OH)D levels and their association with ADHD
| SNP | 25(OH)D | ADHD | Palindromic + ambiguous | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | Eaf (A1) | Beta | SE | P value | A1 | A2 | OR | SE | P value | |||
| rs10741657 | CYP2R1 | A | G | 0.40 | 0.031 | 0.0022 | 2.05E–46 | A | G | 1.0048 | 0.0137 | 0.728 | False |
| rs10745742 | AMDHD1 | T | C | 0.40 | 0.017 | 0.0022 | 1.88E–14 | T | C | 1.0141 | 0.0138 | 0.311 | False |
| rs12785878 | NADSYN1-DHCR7 | T | G | 0.75 | 0.036 | 0.0022 | 3.80E–62 | T | G | 1.0313 | 0.0146 | 0.035 | False |
| rs17216707a | CYP24A1 | T | C | 0.77 | 0.026 | 0.0027 | 8.14E–23 | ||||||
| ars209955 | T | C | 0.31b | − 0.019 | 0.0024 | 2.49E−16 | T | C | 1.003 | 0.0156 | 0.848 | False | |
| rs3755967 | GC | T | C | 0.28 | − 0.089 | 0.0023 | 4.74E–343 | T | C | 1.0235 | 0.015 | 0.122 | False |
| rs8018720 | SEC23A | C | G | 0.82 | − 0.017 | 0.0029 | 4.72E–09 | C | G | 0.9958 | 0.0185 | 0.819 | True |
Eaf effect allele frequency, OR odds ratio, SE standard error, palindromic + ambiguous true this SNP is palindromic and ambiguous and will be excluded
ars209955 is the proxy for rs17216707: distance = − 9491, D′ = 0.7908, r2 = 0.4115; A1: Allele 1; A2: Allele 2
bEaf (European) from https://ldlink.nci.nih.gov/?tab=ldhap
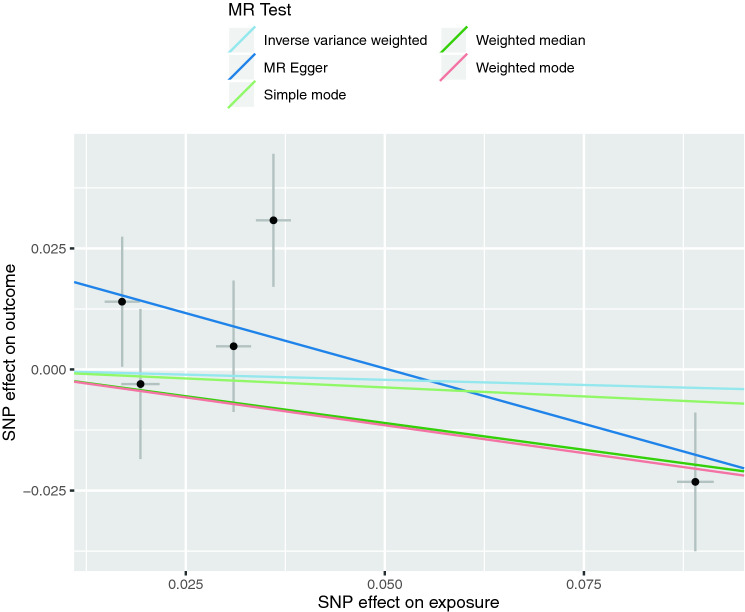
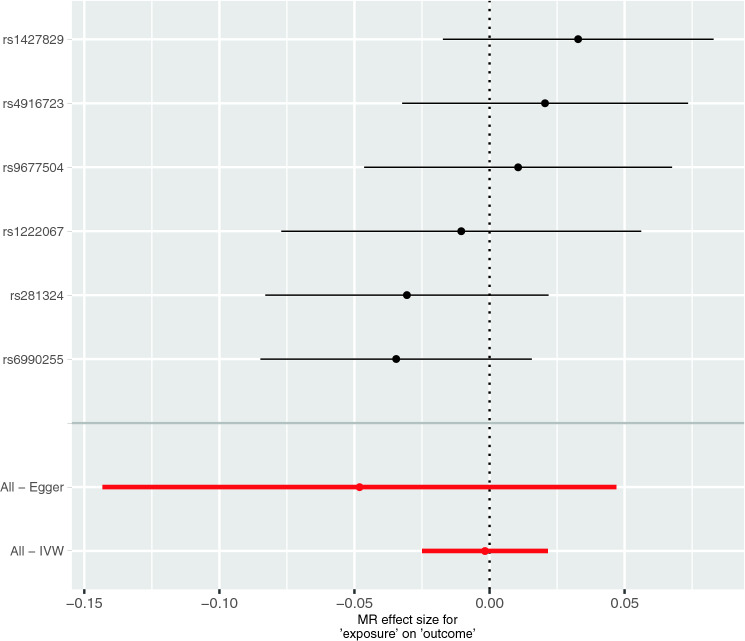
With F = 1558.762, the F-statistic indicates strong instrumental variables (variance explained by the six SNPs (2.84%) was used for the calculation). There was no evidence for pleiotropy (MR-Egger intercept: 0.0231, SE = 0.0139; p = 0.1955) or heterogeneity (Cochran's Q (df = 3) = 4.594, p = 0.204). MR-PRESSO also revealed no evidence for horizontal pleiotropy. Single SNP MR analyses showed that only SNP rs12785878 (NADSYN1-DHCR7) was associated with ADHD. The overall estimates calculated by IVW or MR-Egger, did not reveal an overall causal effect of 25(OH)D levels on the risk of ADHD (Table 2, Figs. 1, 2, and S1). Sensitivity analyses using Weighted Median, Simple Mode, and Weighted Mode confirmed the lack of associations (Table 2, Fig. 2). MR-PRESSO also did not show a significant association between 25(OH)D and ADHD (β = − 0.043, SD = 0.202, p = 0.843). However, the leave-one-out analysis showed that the exclusion of SNP rs3755967 (GC: group-specific component gene, GC protein is a major vitamin D-binding protein in plasma [35]) would lead to a significant overall positive effect of 25(OH)D on ADHD, i.e. higher 25(OH)D levels would cause higher ADHD risk (Figure S2). Power analyses revealed that our MR analyses had 80% power to detect an OR of 1.159 for ADHD per 1 standard deviation decrease in natural-log transformed 25(OH)D levels (Figure S3).
Table 2.
Results of single SNP MR analyses and the overall causal effect of natural-log transformed 25(OH)D levels on the risk of ADHD calculated using different statistical methods
| SNP | Beta | SE | P value |
|---|---|---|---|
| rs10741657 | 0.155 | 0.438 | 0.724 |
| rs10745742 | 0.824 | 0.789 | 0.297 |
| rs12785878 | 0.856 | 0.382 | 0.024 |
| rs209955 | − 0.155 | 0.803 | 0.847 |
| rs3755967 | − 0.261 | 0.161 | 0.105 |
| Inverse variance weighted | − 0.043 | 0.202 | 0.833 |
| MR Egger | − 0.457 | 0.301 | 0.227 |
| Weighted median | − 0.221 | 0.158 | 0.162 |
| Simple mode | − 0.074 | 0.466 | 0.881 |
| Weighted mode | − 0.230 | 0.161 | 0.226 |
Fig. 1.
Results of the single and multi-SNP analyses for the SNP effect of natural-log transformed 25(OH)D levels on ADHD. The black lines visualize the results of single SNP analyses; the red lines visualize the results of the multi-SNP analysis
Fig. 2.
Scatterplots of genetic associations with natural-log transformed 25(OH)D levels against risk for ADHD using different MR methods. The slopes of each line represent the causal association for each method
We conducted additional sensitivity MR analyses using alternative genetic instruments considering results from two recent GWAS on 25(OH)D [23, 24]. Both of these additional MR analyses confirmed the results from our main analysis since the overall estimates, calculated by IVW or MR-Egger, did not reveal an overall association of genetically predicted 25(OH)D levels on the risk of ADHD (Tables S1 and S2, Figures S4 and S5). Weighted Median, Simple Mode, and Weighted Mode confirmed the lack of associations (Table S1 and S2).
MR assessing ADHD effects on 25(OH)D
To test reverse causality, we performed a MR analysis where we considered the risk of ADHD as exposure and 25(OH)D as the outcome. Data on the association of the 12 selected SNPs with ADHD and with 25(OH)D is presented in Table 3. We used effect estimates and P-values that were calculated for Europeans. For seven SNPs, which were not included in GWAS for 25(OH)D we used proxy-SNPs. For SNPs rs11420276 and rs5886709 no proxies where available. Since we had to exclude four palindromic SNPs (Table 3: palindromic + ambiguous = true), we were able to include six SNPs in this MR analysis. With F = 21,867.14, the F-statistic indicates strong instrumental variables (for F-statistic calculation, the proportion of variance (R2 = 0.216) was considered as determined using those cohorts of European ancestry and all 12 genome-wide significant SNPs in the ADHD GWAS). There was no evidence for pleiotropy (MR-Egger intercept: 0.0043, SE = 0.0044; p = 0.379). Heterogeneity tests were not significant (Cochran's Q (df = 4) = 4.473, p = 0.346) and MR-PRESSO also revealed no significant outliers.
Table 3.
SNPs associated with ADHD (European ancestry) and natural-log transformed 25(OH)D levels
| SNP | Chr | Position | ADHD | 25(OH)D | Palindromic + ambigous | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A1 freq | OR | SE | P value | A1 | A2 | Beta | SE | P value | ||||
| rs11420276a | 1 | 44184192 | G | GT | 0.696 | 1.11305 | 0.0149 | 6.452e−13 | ||||||
| rs11591402 | 10 | 106747354 | A | T | 0.224 | 0.91174 | 0.0164 | 1.76e−08 | A | T | 0.0012 | 0.0025 | 0.6195 | True |
| rs1222063b | 1 | 96602440 | A | G | 0.328 | 1.10098 | 0.0174 | 3.068e−08 | ||||||
| brs1222067 | 1 | 96597502 | A | C | 0.688 | 0.92635 | 0.0155 | 7.908e−07 | A | C | 0.0008 | 0.0026 | 0.7464 | False |
| rs1427829 | 12 | 89760744 | A | G | 0.434 | 1.08567 | 0.0136 | 1.349e−09 | A | G | 0.0027 | 0.0021 | 0.1974 | False |
| rs212178c | 16 | 72578131 | A | G | 0.883 | 0.88950 | 0.0205 | 1.198e−08 | ||||||
| crs12596294 | 16 | 72587093 | A | T | 0.098 | 1.12008 | 0.0207 | 4.157e−08 | A | T | 0.0029 | 0.0034 | 0.3873 | True |
| rs281324 | 15 | 47754018 | T | C | 0.531 | 0.92450 | 0.0135 | 6.684e−09 | T | C | 0.0024 | 0.0021 | 0.2426 | False |
| rs28411770d | 4 | 31151456 | T | C | 0.651 | 1.08992 | 0.0151 | 1.152e−08 | ||||||
| drs7674790 | 4 | 31149277 | A | T | 0.610 | 1.07455 | 0.014 | 2.996e−07 | A | T | 0.0002 | 0.0022 | 0.9348 | True |
| rs4858241e | 3 | 20669071 | T | G | 0.622 | 1.08567 | 0.0143 | 8.172e−09 | ||||||
| ers17808771 | 3 | 20691823 | A | T | 0.758 | 1.07326 | 0.0153 | 3.773e−06 | A | T | 0.0008 | 0.0024 | 0.7341 | True |
| rs4916723 | 5 | 87854395 | A | C | 0.573 | 0.92515 | 0.0138 | 1.807e−08 | A | C | − 0.0016 | 0.0021 | 0.4515 | False |
| rs5886709f | 7 | 114086133 | G | GTC | 0.463 | 1.07993 | 0.0137 | 2.056e−08 | ||||||
| rs74760947g | 8 | 34352610 | A | G | 0.957 | 0.83560 | 0.0317 | 1.393e−08 | ||||||
| grs6990255 | 8 | 34126948 | T | C | 0.0586481 | 1.18270 | 0.0315 | 1.03e−07 | T | C | − 0.0058 | 0.0043 | 0.1779 | False |
| rs9677504 | 2 | 215181889 | A | G | 0.109 | 1.12019 | 0.0213 | 9.829e−08 | A | G | 0.0012 | 0.0033 | 0.7171 | False |
Chr chromosome, A1 allele 1, A2 allele 2, freq allele frequency, OR odds ratio, SE standard error; palindromic + ambiguous true this SNPs are palindromic and ambiguous (excluded after harmonization)
aNo Proxy-SNP available for rs11420276
bFor rs1222063: proxy rs1222067; distance: − 4938; R2:0.679350
cFor rs212178: proxy rs12596294; distance: 8962; R2: 0.924371
dFor rs28411770: proxy rs7674790; distance: − 2179; R2: 0.797643
ers4858241: proxy rs17808771; distance:22,752; R2: 0.646552
fNo proxy-SNP available for rs5886709
gFor rs74760947: proxy rs6990255; distance: − 225,662; R2: 0.947204
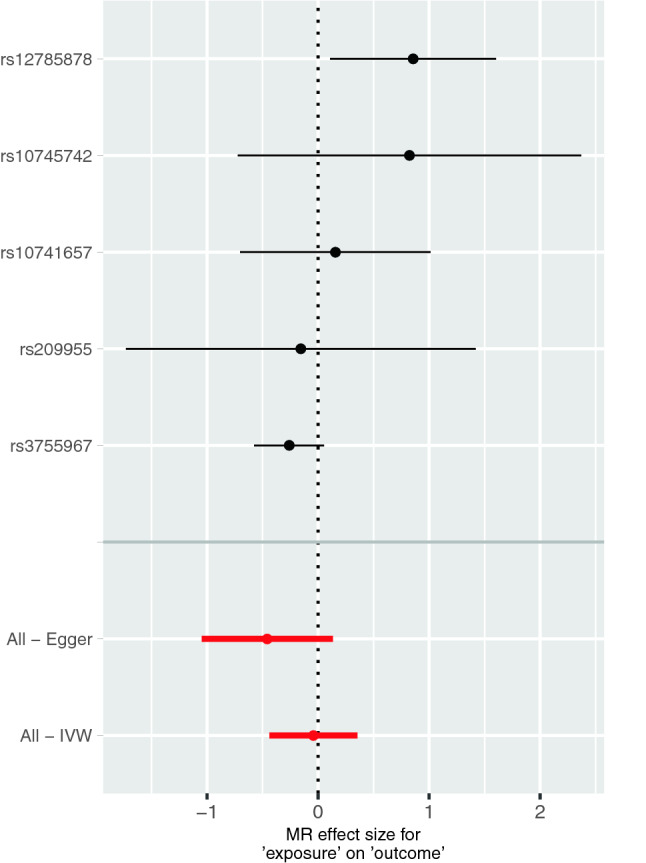
No single SNP showed a causal effect of ADHD on 25(OH)D. Further, the overall effect was not significant (IVW method: β = − 0.002, p = 0.887; MR Egger method: β = − 0.048, p = 0.377) (Table 4, Figs. 3, S6 and S7). Sensitivity analyses using Weighted Median, Simple Mode, and Weighted Mode (Table 4, Figures S6 and S7) and MR-PRESSO (β = − 0.001, SD = 0.019, p = 0.892) also revealed no significant overall effect. Leave-one-out analysis also showed no effect (Figure S8). Additionally, we performed a sensitivity analysis with proxy SNPs, which were neither palindromic nor ambiguous (Table S3). This analysis also showed no causal effect of ADHD on 25(OH)D (Table S4, Figure S9).
Table 4.
Results of single SNP MR analyses and the overall causal effect of ADHD on 25(OH)D levels using different statistical methods
| SNP | Beta | SE | P value |
|---|---|---|---|
| rs1222067 | − 0.010 | 0.034 | 0.758 |
| rs1427829 | 0.033 | 0.026 | 0.199 |
| rs281324 | − 0.031 | 0.027 | 0.253 |
| rs4916723 | 0.0206 | 0.027 | 0.446 |
| rs6990255 | − 0.035 | 0.026 | 0.177 |
| rs9677504 | 0.011 | 0.029 | 0.716 |
| Inverse variance weighted | − 0.002 | 0.012 | 0.887 |
| MR Egger | − 0.048 | 0.049 | 0.377 |
| Weighted median | 0.002 | 0.015 | 0.900 |
| Simple mode | 0.015 | 0.026 | 0.571 |
| Weighted mode | 0.019 | 0.027 | 0.514 |
Fig. 3.
Results of the single and multi-SNP analyses for the SNP effect of risk for ADHD on natural-log transformed 25(OH)D levels. The black lines visualize the results of single SNP analyses; the red lines visualize the results of the multi-SNP analysis
Discussion
The main finding of this MR study was that there was no genetic evidence for a causal effect of 25(OH)D levels on the risk of ADHD. Additionally, our analysis did also not indicate a causal relationship in the opposite direction, i.e. ADHD did not have an effect on 25(OH)D levels.
Interestingly, one of the five SNPs used for the definition of the IV (rs12785878) even indicated a significant positive effect of 25(OH)D on the risk of ADHD, i.e. higher risk for ADHD at higher 25(OH)D levels (p = 0.024). Also, one of the leave-one-out sensitivity analyses indicated a significant overall effect in the same direction, when rs3755967, indicating an inverse association, was excluded from the analysis. However, it has to be kept in mind that rs12785878 would not survive correction for multiple testing (five SNPs). Additionally, underlying pathophysiologic mechanisms of a harmful effect of 25(OH)D on ADHD are difficult to understand. Thus, this surprising finding underlines the fact that additional GWAS on 25(OH)D and ADHD are still urgently needed to increase the number of genome-wide significant SNPs and to further improve the genetic instrument for future MR studies.
During infancy, recommendations in Germany include routine vitamin D supplementation [36]. For children aged > 2 years, the German Society for Pediatrics and Adolescent Medicine recently concluded that such routine supplementation is not recommended for children who do not have risk factors and chronic diseases which are associated with calcium or vitamin D resorption disorders [3]. Findings from the meta-analyses of observational studies on ADHD might raise concern regarding this recommendation since 25(OH)D levels were found to be lower in ADHD patients compared to healthy controls [37, 38]. Additionally, low 25(OH)D cord blood levels were prospectively associated with higher ADHD scores in toddlers [39] indicating a potential role of vitamin D supply in the perinatal period and, thus, perhaps also for long-term prevention of this neuro-developmental disorder in later life. In general, RCTs are regarded as the gold standard to prove causality, but the few RCTs on vitamin D and ADHD were restricted to the therapeutic effects of an already manifest disorder. Considering results from four RCTs, a recent meta-analysis suggested that vitamin D supplementation could be one treatment option, but all included RCTs had some considerable limitations such as small sample size (35–96 subjects) and a low to very-low quality of evidence according to GRADE criteria [10]. Accordingly, the authors requested well-designed RCTs to confirm the efficacy of vitamin D supplementation for children and adults with ADHD [10].
Overall, current evidence seems to be in line with findings on 25(OH)D and depression: Here, observational studies also showed positive associations [40], while the potential effects of vitamin D supplementation might be restricted to subjects with clinically significant depressive symptoms [41]. While preventive effects on depression onset were only assessed in a few RCTs with follow-up periods of more than one year, MR studies on depression did not support the suggested preventive role of 25(OH)D [16, 17]. Now, results from our current MR study did also not confirm a causal effect of 25(OH)D on ADHD. In general, it has been argued that conflicting results between observational studies and MR studies might indicate bias from residual confounding in observational studies or reverse causation [12]. The latter would imply that suffering from the disease would have negative effects on 25(OH)D, e.g. due to social withdrawal and reduced sunlight exposure. While such reverse effects on 25(OH)D are plausible in internalizing disorders such as depression, it seems less obvious in the case of ADHD. Also, results from our reverse MR analysis did not reveal any evidence for reverse causality. Nevertheless, residual confounding in observational studies still seems reasonable since low 25(OH)D could be a marker of ill health in general [42].
To the best of our knowledge, our study is the first two-sample MR study on causal relationships between 25(OH)D levels and ADHD. A strength of our study is the consideration of recent, large-scaled GWAS on both vitamin D metabolism (n = 79,366) [19] and on ADHD (n = 20,183 cases) [20]. Accordingly, our study had sufficient statistical power which allowed the detection of OR > 1.27 per 1 standard deviation decrease in natural-log transformed 25(OH)D levels with a statistical power of 100% (OR > 1.159 with 80% power). However, we cannot rule out small preventive effects below this threshold. Despite using the latest available GWAS with the largest sample sizes and highest number of identified SNPs for our data analyses, it must be kept in mind that this field of genetic research has a substantial turnover and findings from our MR study should not be interpreted as final results. Actually, while this paper was under review, two GWAS on 25(OH)D concentrations, each with more than 400,000 European study participants, were published. These GWAS revealed 143 and 138 genome-wide significant SNPs, respectively [23, 24]. Sensitivity MR analyses considering these findings for the definition of the IV also showed no significant association between genetically predicted 25(OH)D concentrations and ADHD. Accordingly, our result that there is no evidence for a causal effect of 25(OH)D on ADHD appears robust.
A limitation of our MR study is that only six SNPs, which explain 2.8% of the variance of 25(OH)D levels, could be considered for the definition of the IV. One of these SNPs was ambiguous and palindromic and had thus had to be excluded. Thus, our instrument might be regarded as weak and bias the association with ADHD. However, the F-statistics indicated that our genetic instrument was sufficiently strong. Furthermore, our sensitivity analyses using a higher number of SNPs as genetic instrument confirmed results from our main analysis.
Another general issue of MR studies is horizontal pleiotropy, i.e. an association between the MR instrument and the outcome of interest via pathways other than the suggested exposure [12]. Even though this issue cannot be completely ruled out in MR studies, the MR Egger intercept and MR-PRESSO analyses revealed no indication of pleiotropy.
When interpreting the results, it is important to note that the vitamin D GWAS did not focus on individuals with low (or high) levels of 25(OH)D. Since our MR analysis assumed a linear relationship between exposure and outcome, the results might not apply to people with a severe 25(OH)D deficiency, but to the general population [11]. Additionally, it must be kept in mind that the instrumental variable was not derived from GWAS focussing on childhood 25(OH)D. In conclusion, our MR study using data from large-scaled genetic studies provides initial evidence that 25(OH)D might not have substantial preventive effects on ADHD. Well-designed, large-scaled RCTs would be important to definitely evaluate the preventive capacity of routine supplementation during infancy. Complex requirements regarding study design such as long intervention and follow-up periods, especially in diseases with low incidence [43], might be one explanation why such RCTs are still lacking.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Ethics approval
This MR study was conducted solely based on publicly available summary statistics of GWAS meta-analyses. URLs: https://ldlink.nci.nih.gov/?tab=ldhap [44]; http://www.snipa.org; https://www.med.unc.edu/pgc/; https://drive.google.com/drive/folders/0BzYDtCo_doHJRFRKR0ltZHZWZjQ; https://github.com/MRCIEU/TwoSampleMR; https://shiny.cnsgenomics.com/mRnd/; https://github.com/rondolab/MR-PRESSO.
References
- 1.Institute of Medicine (US), committee to review dietary reference intakes for vitamin D and calcium (2011) Dietary reference intakes for calcium and vitamin D. In. Washington, National Academies Press (US), [PubMed]
- 2.Braegger C, Campoy C, Colomb V, Decsi T, Domellof M, Fewtrell M, Hojsak I, Mihatsch W, Molgaard C, Shamir R, Turck D, van Goudoever J, Nutrition ECo, Vitamin D in the healthy European paediatric population. J Pediatr Gastroenterol Nutr. 2013;56(6):692–701. doi: 10.1097/MPG.0b013e31828f3c05. [DOI] [PubMed] [Google Scholar]
- 3.Reinehr T, Schnabel D, Wabitsch M, Bechtold-Dalla Pozza S, Buhrer C, Heidtmann B, Jochum F, Kauth T, Korner A, Mihatsch W, Prell C, Rudloff S, Tittel B, Woelfle J, Zimmer KP, Koletzko B (2019) Vitamin D supplementation after the second year of life: joint position of the Committee on Nutrition, German Society for Pediatric and Adolescent Medicine (DGKJ e.V.), and the German Society for Pediatric Endocrinology and Diabetology (DGKED e.V.). Mol Cell Pediatr 6(1):3. doi:10.1186/s40348-019-0090-0 [DOI] [PMC free article] [PubMed]
- 4.Sarris J, Logan AC, Akbaraly TN, Amminger GP, Balanza-Martinez V, Freeman MP, Hibbeln J, Matsuoka Y, Mischoulon D, Mizoue T, Nanri A, Nishi D, Ramsey D, Rucklidge JJ, Sanchez-Villegas A, Scholey A, Su KP, Jacka FN, International Society for Nutritional Psychiatry R Nutritional medicine as mainstream in psychiatry. Lancet Psychiatry. 2015;2(3):271–274. doi: 10.1016/S2215-0366(14)00051-0. [DOI] [PubMed] [Google Scholar]
- 5.Föcker M, Antel J, Ring S, Hahn D, Kanal O, Ozturk D, Hebebrand J, Libuda L. Vitamin D and mental health in children and adolescents. Eur Child Adolesc Psychiatry. 2017;26(9):1043–1066. doi: 10.1007/s00787-017-0949-3. [DOI] [PubMed] [Google Scholar]
- 6.Hebebrand J, Peters T, Schijven D, Hebebrand M, Grasemann C, Winkler TW, Heid IM, Antel J, Focker M, Tegeler L, Brauner L, Adan RAH, Luykx JJ, Correll CU, Konig IR, Hinney A, Libuda L. The role of genetic variation of human metabolism for BMI, mental traits and mental disorders. Mol Metab. 2018;12:1–11. doi: 10.1016/j.molmet.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eyles DW, Smith S, Kinobe R, Hewison M, McGrath JJ. Distribution of the vitamin D receptor and 1 alpha-hydroxylase in human brain. J Chem Neuroanat. 2005;29(1):21–30. doi: 10.1016/j.jchemneu.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Saedisomeolia A, Samadi M, Gholami F, Seyedi M, Effatpanah M, Hashemi R, Abdolahi M, Honarvar NM. Vitamin D's molecular action mechanism in attention-deficit/ hyperactivity disorder: a review of evidence. CNS Neurol Disord Drug Targets. 2018;17(4):280–290. doi: 10.2174/1871527317666180501111627. [DOI] [PubMed] [Google Scholar]
- 9.Seyedi M, Gholami F, Samadi M, Djalali M, Effatpanah M, Yekaninejad MS, Hashemi R, Abdolahi M, Chamari M, Honarvar NM. The effect of vitamin D3 supplementation on serum BDNF, dopamine, and serotonin in children with attention-deficit/hyperactivity disorder. CNS Neurol Disord Drug Targets. 2019;18(6):496–501. doi: 10.2174/1871527318666190703103709. [DOI] [PubMed] [Google Scholar]
- 10.Gan J, Galer P, Ma D, Chen C, Xiong T. The effect of vitamin D supplementation on attention-deficit/hyperactivity disorder: a systematic review and meta-analysis of randomized controlled trials. J Child Adolesc Psychopharmacol. 2019;29(9):670–687. doi: 10.1089/cap.2019.0059. [DOI] [PubMed] [Google Scholar]
- 11.Manousaki D, Richards JB. Commentary: Role of vitamin D in disease through the lens of Mendelian randomization-Evidence from Mendelian randomization challenges the benefits of vitamin D supplementation for disease prevention. Int J Epidemiol. 2019;48(5):1435–1437. doi: 10.1093/ije/dyz183. [DOI] [PubMed] [Google Scholar]
- 12.König IR, Greco FMD. Mendelian randomization: Progressing towards understanding causality. Ann Neurol. 2018;84(2):176–177. doi: 10.1002/ana.25293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng J, Baird D, Borges MC, Bowden J, Hemani G, Haycock P, Evans DM, Smith GD. Recent Developments in Mendelian Randomization Studies. Curr Epidemiol Rep. 2017;4(4):330–345. doi: 10.1007/s40471-017-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Kiel DP, Kraft P. The genetics of vitamin D. Bone. 2018 doi: 10.1016/j.bone.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Mills NT, Wright MJ, Henders AK, Eyles DW, Baune BT, McGrath JJ, Byrne EM, Hansell NK, Birosova E, Scott JG, Martin NG, Montgomery GW, Wray NR, Vinkhuyzen AA. Heritability of transforming growth factor-beta1 and tumor necrosis factor-receptor type 1 expression and vitamin D levels in healthy adolescent twins. Twin Res Hum Genet. 2015;18(1):28–35. doi: 10.1017/thg.2014.70. [DOI] [PubMed] [Google Scholar]
- 16.Libuda L, Laabs BH, Ludwig C, Buhlmeier J, Antel J, Hinney A, Naaresh R, Focker M, Hebebrand J, Konig IR, Peters T (2019) Vitamin D and the risk of depression: a causal relationship? Findings from a Mendelian Randomization Study. Nutrients 11(5). doi:10.3390/nu11051085 [DOI] [PMC free article] [PubMed]
- 17.Michaelsson K, Melhus H, Larsson SC (2018) Serum 25-hydroxyvitamin D concentrations and major depression: a mendelian randomization study. Nutrients 10(12). doi:10.3390/nu10121987 [DOI] [PMC free article] [PubMed]
- 18.Meng X, Li X, Timofeeva MN, He Y, Spiliopoulou A, Wei W, Gifford A, Wu H, Varley T, Joshi P, Denny JC, Farrington SM, Zgaga L, Dunlop MG, McKeigue P, Campbell H, Theodoratou E. Phenome-wide Mendelian-randomization study of genetically determined vitamin D on multiple health outcomes using the UK Biobank study. Int J Epidemiol. 2019;48(5):1425–1434. doi: 10.1093/ije/dyz182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, O'Reilly PF, Aschard H, Hsu YH, Richards JB, Dupuis J, Ingelsson E, Karasik D, Pilz S, Berry D, Kestenbaum B, Zheng J, Luan J, Sofianopoulou E, Streeten EA, Albanes D, Lutsey PL, Yao L, Tang W, Econs MJ, Wallaschofski H, Volzke H, Zhou A, Power C, McCarthy MI, Michos ED, Boerwinkle E, Weinstein SJ, Freedman ND, Huang WY, Van Schoor NM, van der Velde N, Groot L, Enneman A, Cupples LA, Booth SL, Vasan RS, Liu CT, Zhou Y, Ripatti S, Ohlsson C, Vandenput L, Lorentzon M, Eriksson JG, Shea MK, Houston DK, Kritchevsky SB, Liu Y, Lohman KK, Ferrucci L, Peacock M, Gieger C, Beekman M, Slagboom E, Deelen J, Heemst DV, Kleber ME, Marz W, de Boer IH, Wood AC, Rotter JI, Rich SS, Robinson-Cohen C, den Heijer M, Jarvelin MR, Cavadino A, Joshi PK, Wilson JF, Hayward C, Lind L, Michaelsson K, Trompet S, Zillikens MC, Uitterlinden AG, Rivadeneira F, Broer L, Zgaga L, Campbell H, Theodoratou E, Farrington SM, Timofeeva M, Dunlop MG, Valdes AM, Tikkanen E, Lehtimaki T, Lyytikainen LP, Kahonen M, Raitakari OT, Mikkila V, Ikram MA, Sattar N, Jukema JW, Wareham NJ, Langenberg C, Forouhi NG, Gundersen TE, Khaw KT, Butterworth AS, Danesh J, Spector T, Wang TJ, Hypponen E, Kraft P, Kiel DP. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat Commun. 2018;9(1):260. doi: 10.1038/s41467-017-02662-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demontis D, Walters RK, Martin J, Mattheisen M, Als TD, Agerbo E, Baldursson G, Belliveau R, Bybjerg-Grauholm J, Bækvad-Hansen M, Cerrato F, Chambert K, Churchhouse C, Dumont A, Eriksson N, Gandal M, Goldstein JI, Grasby KL, Grove J, Gudmundsson OO, Hansen CS, Hauberg ME, Hollegaard MV, Howrigan DP, Huang H, Maller JB, Martin AR, Martin NG, Moran J, Pallesen J, Palmer DS, Pedersen CB, Pedersen MG, Poterba T, Poulsen JB, Ripke S, Robinson EB, Satterstrom FK, Stefansson H, Stevens C, Turley P, Walters GB, Won H, Wright MJ, Albayrak Ö, Anney RJL, Arranz MJ, Banaschewski TJ, Bau C, Biederman J, Buitelaar JK, Casas M, Charach A, Crosbie J, Dempfle A, Doyle AE, Ebstein RP, Elia J, Freitag C, Föcker M, Gill M, Grevet E, Hawi Z, Hebebrand J, Herpertz-Dahlmann B, Hervas A, Hinney A, Hohmann S, Holmans P, Hutz M, Ickowitz A, Johansson S, Kent L, Kittel-Schneider S, Lambregts-Rommelse N, Lehmkuhl G, Loo SK, McGough JJ, Meyer J, Mick E, Middletion F, Miranda A, Mota NR, Mulas F, Mulligan A, Nelson F, Nguyen TT, Oades RD, O’Donovan MC, Owen MJ, Palmason H, Ramos-Quiroga JA, Renner TJ, Ribasés M, Rietschel M, Rivero O, Romanos J, Romanos M, Rothenberger A, Royers H, Sánchez-Mora C, Scherag A, Schimmelmann BG, Schäfer H, Sergeant J, Sinzig J, Smalley SL, Steinhausen H-C, Thompson M, Todorov A, Vasquez AA, Walitza S, Wang Y, Warnke A, Williams N, Witt SH, Yang L, Zayats T, Zhang-James Y, Smith GD, Davies GE, Ehli EA, Evans DM, Fedko IO, Greven CU, Groen-Blokhuis MM, Guxens M, Hammerschlag AR, Hartman CA, Heinrich J, Jan Hottenga J, Hudziak J, Jugessur A, Kemp JP, Krapohl E, Murcia M, Myhre R, Nolte IM, Nyholt DR, Ormel J, Ouwens KG, Pappa I, Pennell CE, Plomin R, Ring S, Standl M, Stergiakouli E, Pourcain BS, Stoltenberg C, Sunyer J, Thiering E, Tiemeier H, Tiesler CMT, Timpson NJ, Trzaskowski M, van der Most PJ, Vilor-Tejedor N, Wang CA, Whitehouse AJO, Zhao H, Agee M, Alipanahi B, Auton A, Bell RK, Bryc K, Elson SL, Fontanillas P, Furlotte NA, Hinds DA, Hromatka BS, Huber KE, Kleinman A, Litterman NK, McIntyre MH, Mountain JL, Northover CAM, Pitts SJ, Sathirapongsasuti JF, Sazonova OV, Shelton JF, Shringarpure S, Tian C, Vacic V, Wilson CH, Andreassen OA, Asherson P, Burton CL, Boomsma DI, Cormand B, Dalsgaard S, Franke B, Gelernter J, Geschwind D, Hakonarson H, Haavik J, Kranzler HR, Kuntsi J, Langley K, Lesch K-P, Middeldorp C, Reif A, Rohde LA, Roussos P, Schachar R, Sklar P, Sonuga-Barke EJS, Sullivan PF, Thapar A, Tung JY, Waldman ID, Medland SE, Stefansson K, Nordentoft M, Hougaard DM, Werge T, Mors O, Mortensen PB, Daly MJ, Faraone SV, Børglum AD, Neale BM, Consortium AWGotPG, Early L, Genetic Epidemiology C, andMe Research T Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2018 doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;45(6):1717–1726. doi: 10.1093/ije/dyx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmuller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics. 2015;31(8):1334–1336. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Revez JA, Lin T, Qiao Z, Xue A, Holtz Y, Zhu Z, Zeng J, Wang H, Sidorenko J, Kemper KE, Vinkhuyzen AAE, Frater J, Eyles D, Burne THJ, Mitchell B, Martin NG, Zhu G, Visscher PM, Yang J, Wray NR, McGrath JJ (2020) Genome-wide association study identifies 143 loci associated with 25 hydroxyvitamin D concentration. Nat Commun 11(1). doi:10.1038/s41467-020-15421-7 [DOI] [PMC free article] [PubMed]
- 24.Manousaki D, Mitchell R, Dudding T, Haworth S, Harroud A, Forgetta V, Shah RL, Luan J, Langenberg C, Timpson NJ, Richards JB. Genome-wide association study for vitamin D levels reveals 69 independent loci. Am J Hum Genet. 2020;106(3):327–337. doi: 10.1016/j.ajhg.2020.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46(6):1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377–389. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Davey Smith G. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr. 2016;103(4):965–978. doi: 10.3945/ajcn.115.118216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26(5):2333–2355. doi: 10.1177/0962280215597579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology (Cambridge, Mass) 2017;28(1):30–42. doi: 10.1097/EDE.0000000000000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katikireddi SV, Green MJ, Taylor AE, Davey Smith G, Munafo MR. Assessing causal relationships using genetic proxies for exposures: an introduction to Mendelian randomization. Addiction. 2018;113(4):764–774. doi: 10.1111/add.14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–1501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang F, Brune JL, Naylor SL, Cupples RL, Naberhaus KH, Bowman BH. Human group-specific component (Gc) is a member of the albumin familiy. Proc Natl Acad Sci. 1985;82:7994–7998. doi: 10.1073/pnas.82.23.7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koletzko B, Bauer CP, Cierpka M, Cremer M, Flothkötter M, Graf C. Ernährung und Bewegung von Säuglingen und stillenden Frauen—Aktualisierte Handlungsempfehlungen von "Gesund ins Leben—Netzwerk Junge Familie", eine Initiative von IN FORM. Monatsschr Kinderheilkd. 2016;164:S433–S457. doi: 10.1007/s00112-016-0173-0. [DOI] [Google Scholar]
- 37.Khoshbakht Y, Bidaki R, Salehi-Abargouei A. Vitamin D status and attention deficit hyperactivity disorder: a systematic review and meta-analysis of observational studies. Adv Nutr. 2018;9(1):9–20. doi: 10.1093/advances/nmx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotsi E, Kotsi E, Perrea DN. Vitamin D levels in children and adolescents with attention-deficit hyperactivity disorder (ADHD): a meta-analysis. Atten Defic Hyperact Disord. 2019;11(3):221–232. doi: 10.1007/s12402-018-0276-7. [DOI] [PubMed] [Google Scholar]
- 39.Mossin MH, Aaby JB, Dalgard C, Lykkedegn S, Christesen HT, Bilenberg N. Inverse associations between cord vitamin D and attention deficit hyperactivity disorder symptoms: a child cohort study. Aust N Z J Psychiatry. 2017;51(7):703–710. doi: 10.1177/0004867416670013. [DOI] [PubMed] [Google Scholar]
- 40.Anglin RE, Samaan Z, Walter SD, McDonald SD. Vitamin D deficiency and depression in adults: systematic review and meta-analysis. Br J Psychiatry J Mental Sci. 2013;202:100–107. doi: 10.1192/bjp.bp.111.106666. [DOI] [PubMed] [Google Scholar]
- 41.Shaffer JA, Edmondson D, Wasson LT, Falzon L, Homma K, Ezeokoli N, Li P, Davidson KW. Vitamin D supplementation for depressive symptoms: a systematic review and meta-analysis of randomized controlled trials. Psychosom Med. 2014;76(3):190–196. doi: 10.1097/PSY.0000000000000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Autier P, Boniol M, Pizot C, Mullie P. Vitamin D status and ill health: a systematic review. Lancet Diabetes Endocrinol. 2014;2(1):76–89. doi: 10.1016/S2213-8587(13)70165-7. [DOI] [PubMed] [Google Scholar]
- 43.Harroud A, Richards JB. Mendelian randomization in multiple sclerosis: a causal role for vitamin D and obesity? Mult Scler. 2018;24(1):80–85. doi: 10.1177/1352458517737373. [DOI] [PubMed] [Google Scholar]
- 44.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.