Abstract
Fragile X syndrome (FXS) is the leading inherited cause of intellectual disability, resulting from the lack of functional fragile X mental retardation protein (FMRP), an mRNA binding protein mainly serving as a translational regulator. Loss of FMRP leads to dysregulation of target mRNAs. The Drosophila model of FXS show an abnormal circadian rhythm with disruption of the output pathway downstream of the clock network. Yet the FMRP targets involved in circadian regulation have not been identified. Here, we identified collapsing response mediator protein (CRMP) mRNA as a target of FMRP. Knockdown of pan-neuronal CRMP expression ameliorated the circadian defects and abnormal axonal structures of clock neurons (ventral lateral neurons) in dfmr1 mutant flies. Furthermore, specific reduction of CRMP in the downstream output insulin-producing cells attenuated the aberrant circadian behaviors. Molecular analyses revealed that FMRP binds with CRMP mRNA and negatively regulates its translation. Our results indicate that CRMP is an FMRP target and establish an essential role for CRMP in the circadian output in FXS Drosophila.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12264-021-00682-z.
Keywords: Fragile X syndrome, FMRP, CRMP, Circadian rhythm
Introduction
Circadian rhythms are present in diverse organisms, characterized by daily rhythmic oscillations in metabolism, physiological processes, cognition, and behaviors [1, 2]. The circadian clock system consists of three fundamental components: a central oscillator that drives endogenous rhythms, an input pathway that conveys environmental cues and entrains the central oscillator, and an output pathway that is governed by the central clock to generate overt rhythms. The circadian clock is driven by the interlocked transcription-translation feedback loops of clock genes, which regulate the expression of clock-controlled genes involved in various biological processes such as metabolism, redox homeostasis, inflammation, hormone secretion, and sleep [1, 3–6]. Evidence from both humans and animal models link circadian rhythms to sleep and normal physiology and behaviors. Circadian rhythms and sleep are tightly linked, and their causal interaction is difficult to delineate. The circadian clock regulates the timing and homeostat of sleep, and alterations in sleep also feed back on the circadian rhythm [7]. Sleep disorders and circadian disruption are commonly reported in neurodevelopmental diseases, including fragile X syndrome (FXS), Prader-Willi syndrome, and Angelman syndrome [8, 9]. Patients have perturbed sleep patterns, usually displaying altered sleep duration and abnormal awakenings at night [10, 11]. Sleep disorders are associated with impaired circadian clock function, and disrupted expression of circadian genes and deficient rhythms have been documented in patients and animal models of neuropsychiatric diseases [12–17].
FXS is the most common form of inherited autism spectrum disorders and intellectual disability, caused by loss of the fragile X mental retardation protein (FMRP) [18, 19]. FXS patients present cognitive disorders along with symptoms of autism and epilepsy. FXS animal models including mice and Drosophila exhibit typical phenotypes mimicking those in FXS patients, such as impaired cognition, social behavioral issues, and neuronal structural deficits as well as excessive protein synthesis and other abnormalities at the molecular level [20, 21]. Sleep disorders in FXS patients can also be well explored in animal models, as impaired sleep patterns and circadian rhythm defects have been reported in FXS mice and Drosophila, providing ideal approaches to further explore the underlying mechanisms. Both FXS mice and flies show significantly abnormal sleep profiles and impaired circadian rhythm [22–24]. As early as 2002, a defective locomotor activity rhythm and an aberrant eclosion rhythm have been reported in dfmr1 mutant flies. FXS flies fail to maintain a circadian locomotor activity rhythm in constant darkness and display an abnormal eclosion pattern compared with wild-type flies [14]. Although the circadian ventral lateral neurons (LNvs) of dfmr1 mutant flies display axonal structural defects, the core clock pacemaker neurons are normal [15, 25]. Elevated mGluR signaling has been thought to be the leading pathogenic mechanism of FXS, but genetic reduction of mGluR signaling or administration of its antagonist does not ameliorate the circadian activity defects in FXS Drosophila [26], excluding the possibility that the mGluR signaling pathway underlies regulation of the circadian rhythm in FXS. Recently, a study demonstrated that FMRP functions in the insulin-producing cells (IPCs), confirming a role of FMRP in the circadian output pathway [27]. Yet how FMRP regulates circadian behaviors and circadian output in Drosophila is still not fully understood.
FMRP is encoded by FMR1, the pathogenic gene of FXS. FMRP, mainly enriched in the nervous system, is an RNA binding protein that regulates its target genes and predominantly functions as a translation inhibitor. Besides this function, FMRP also plays an essential role in regulating dendritic mRNA transport and the stability of its target mRNAs [18]. To better understand the molecular mechanisms of FXS, researchers employed many methods to identify FMRP target mRNAs, such as high-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation (HITS-CLIP) and phospho-activatable ribonucleoside (PAR-CLIP) [28, 29]. In the mouse brain, 842 mRNAs associated with polyribosomes have been identified as potential target genes of FMRP [28]. Recently, 4,174 mRNAs were predicted to be FMRP targets at an early developmental stage in the mouse, including 1,610 new targets [30]. Among the hundreds of proposed FMRP targets, only a few have been validated by direct biochemical interaction and genetic manipulation in vivo in FXS animal models to illustrate their roles in its pathogenesis. The deregulated proteins encoded by mRNA targets identified in the last twenty years such as ADCY1, DGKκ, APP, and MAP1B have been shown to participate in the synaptic plasticity and cognitive defects in FXS [31–33]. So far, none of the known targets clarified have been reported to regulate the circadian rhythm in FXS.
Among the genes identified as potential targets of FMRP, collapsing response mediator protein 2 (CRMP2) is of particular interest due to its involvement in the regulation of circadian rhythm and neuronal development [34–36]. As an axonal guidance molecule, vertebrate CRMP2 mediates a variety of neuronal growth cone dynamics through signal transduction pathways [37]. CRMP2 promotes microtubule assembly and also participates in Numb-mediated endocytosis, facilitating the rate of axonal growth [38–40]. CRMP2-KO mice show developmental defects of hippocampal neurons in addition to abnormal sociability and impaired long-term potentiation [36]. Drosophila melanogaster has a single conserved CRMP gene, encoding a protein termed CRMP, which shares 46% amino-acid identity with human CRMP2. Loss of Drosophila CRMP results in defective olfactory memory and circadian rhythm deficits [34]. Considering the important role of CRMP in neuronal morphology and its potential function in Drosophila rhythm, we investigated whether CRMP is an FMRP target mRNA that regulates the circadian activity rhythm in FXS flies.
Here we found that reduction of CRMP repressed the circadian arrhythmicity and neural structural defects in FXS Drosophila and identified CRMP2 mRNA as a target of FMRP in cells. Our findings in Drosophila showed that genetic reduction of CRMP rescues the aberrant locomotor activity rhythm and LNv structural defects in the FXS Drosophila model. And specific knockdown of CRMP expression in IPCs but not in clock neurons ameliorates circadian behaviors. Further, results with mammalian and Drosophila cells showed that FMRP selectively binds with CRMP mRNA, negatively regulating its translation. Our findings suggest that FMRP regulates circadian output by suppressing CRMP translation in IPCs of the Drosophila brain.
Materials and Methods
Cell Culture
Human embryonic kidney (HEK) 293 cells obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich, St. Louis, USA, #D5546) containing 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Massachusetts, USA, #10099). To maintain N2a cells obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), DMEM and Opti-MEM medium (Thermo Fisher Scientific, Massachusetts, USA, #A4124802) were mixed at 1:1 and then 10% FBS was added. And Epstein-Barr virus-transformed lymphoblastoid cell lines were derived from a normal male and an FXS male patient with deletion of the FMR1 gene and cultured in RPMI-1640 medium (Thermo Fisher Scientific, Massachusetts, USA, #A4192301) supplemented with 20% FBS.
Ethics Approval and Consent to Participate
The experiments were undertaken with the understanding and written consent of each subject, and the study conformed with The Code of Ethics of the World Medical Association (Declaration of Helsinki), printed in the British Medical Journal (18 July 1964). This study was approved by the Ethics Committee of the Center for Medical Genetics, Central South University (approval number: 2013051201).
Drosophila Stocks and Maintenance
Fly strains containing dfmr13 and dfmr150M alleles were kind gifts from Dr. Peng Jin, (Emory University, School of Medicine, USA) and the Dilp2-Gal4 stock was from Dr. Luoying Zhang (Huanzhong University of Science and Technology, College of Life Science and Technology, China). The dfmr1 mutant flies used in this study were dfmr13/dfmr1Δ50M trans-heterozygotes. The elav-Gal4, and CRMPsupK1 stocks were from Bloomington Drosophila Stock Center (Indiana University Bloomington, IN, USA, stock numbers 458 and 40954). Fly strains carrying CRMP-RNAi-1 and CRMP-RNAi-2 RNA interference transgenes were from the Tsinghua Fly Center (Tsinghua University, Beijing, China, stock number TH02545.N) and the Vienna Drosophila Resource Center (Vienna, Austria, stock number v101510), respectively. Fly stocks were reared on standard cornmeal-molasses medium at 25°C. Flies were anaesthetized with CO2 to minimize suffering in the experiments.
RNA Co-immunoprecipitation
RNA co-immunoprecipitation was performed as described previously [41] with some modifications. Cells and Drosophila heads were lysed with lysis buffer (20 mmol/L Tris-HCl pH 7.4, 150 mmol/L NaCl, 5 mmol/L MgCl2, 1 mmol/L DTT, 1% Triton X-100) supplemented with RNase inhibitor (Takara, Kusatsu, Japan, #2313A) and proteinase inhibitor cocktail (Sigma-Aldrich, St. Louis, USA, #P8340). Cleared lysates with 1 mg total protein were incubated with Dynabeads® Protein G (Invitrogen, Paisley, UK, #10003D) coated by either anti-FMRP antibody [Millipore, Darmstadt, Germany, #MAB2160) for HEK293 and N2a cells; anti-dFMRP (Abcam, Cambridge, UK, #ab10299) for Drosophila S2 cells and brains] or normal mouse IgGs ( Sigma-Aldrich, Saint Louis, MO, USA, #I5381) overnight at 4°C, and 5% of the lysates were saved as input. About 30% of the beads were used for Western blot analysis and the rest for mRNA enrichment analysis. After addition of 10 pg of external control RNA from Caenorhabditis elegans into the input and RNA immunoprecipitation, RNA was extracted by TRIzol (Invitrogen, CA, USA, #15596-026) and reverse-transcribed using the RevertAid First Strand cDNA Synthesis Kit and random primers (Thermo Fisher Scientific, Waltham, USA, #K1622). Quantitative real-time PCR (qRT-PCR) was performed and the mRNA enrichment was calculated with C. elegans 18S rRNA as an external control and input for normalization. The primers used were as follows:
human-CRMP2-F: 5′-CTCGTTTCCAGATGCCTGAT-3′,
human-CRMP2-R: 5′-CTCAGGAACAACGTGGTCAA-3′,
mouse-CRMP2-F: 5′-ATTCCACGCATCACGAGCGA-3′,
mouse-CRMP2-R: 5′-GGTCTTCACCCCTCCTGGTA-3′,
fly-CRMP-F: 5′- CCAGAATCGCGTCTACATCAA-3′
fly-CRMP-R: 5′- TCCGCCAGGTATCGTTATTTC-3′
C. elegans 18S rRNA-F: 5′-TAGTGAGACGCCCAACAGC-3′,
C. elegans 18S rRNA-R: 5′-TGGCATCGTTTACGGTCAG-3′.
Polyribosome Profile Analysis
Polyribosome profiling was carried out according to the modified method described previously [41, 42]. FXS and normal lymphoblastoid cells were treated with 100 μg/mL cycloheximide ( Sigma-Aldrich, St. Louis, USA, #D5546) and incubated for 15 min at 37°C. Then cell lysates were prepared on ice in lysis buffer (10 mmol/L HEPES-KOH pH 7.4, 150 mmol/L KCl, 10 mmol/L MgCl2, 1 mmol/L DTT, 100 μg/mL cycloheximide, 1% Triton X-100, and RNase and proteinase inhibitors). The resulting supernatant of cytoplasmic lysate was loaded on a 15%–60% sucrose gradient prepared in the lysis buffer and spun at 45,000 rpm for 1 h at 4°C in an SW-55 rotor (Beckman Coulter,Inc., CA, USA). The sucrose gradients were separated into 11 fractions. C. elegans RNA (10 pg) was added into each fraction and total RNA was isolated, and the mRNA levels were quantified and analyzed by qRT-PCR as described above.
Immunohistochemistry and Sholl Analysis
Immunohistochemistry of adult brains and the analyses of LNv neurons were performed as previously described [25, 43]. The brains from adult flies (0–3 days old) were dissected in phosphate-buffered saline (PBS, pH 7.4) and then fixed with 4% paraformaldehyde in PBS for 60 min at 4°C. The brains were rinsed with PBS and then permeabilized with 0.1% triton X-100 in PBS (PBST) for 40 min. After blocking for 60 min with 5% goat serum (Thermo Fisher Scientific, Massachusetts, USA, #16210072) in 0.1% PBST at room temperature, the brains were incubated with anti-PDF (pigment-dispersing factor) antibody (C7 monoclonal, Developmental Studies Hybridoma Bank, University of Iowa, USA) at 1:100 dilution in 0.1% PBST overnight at 4°C. The brains were incubated with the secondary antibody mCy3 (Abcam, Cambridge, UK, #ab97035) at 1:200 for 2 h at room temperature in darkness. All images were captured on a Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems, Wetzlar, Germany). The posterior optic tract (POT) from the circadian large ventral lateral neurons (lLNvs) neurons was measured using the previous method [25], and POT splitting was defined as defasiculation of the POT for >25% of its length. The synaptic architecture of the small Lateral Neurons ventral (sLNvs) were analyzed using modified Sholl analysis to assess the arborization of their arbor terminals [43, 44]. Concentric rings in 10-μm steps were centered on the sLNv dorsal arbor bifurcation. PDF-positive puncta (diameter ≥1 μm) were counted in each ring to reflect the complexity of synaptic arborization. The terminal arborization of sLNv arbors were evaluated in both hemispheres and each hemisphere was represented as n = 1.
Circadian Behavior Analysis
Adult male flies were loaded into tubes for locomotor activity recording using the Drosophila Activity Monitoring system from Trikinetics (Los Angeles, CA, USA) as described previously [27, 45]. The experiment was conducted during the relative light time. Flies were entrained to a 12:12 h light/dark (LD) cycle for 4 days and then transferred to constant darkness for >7 days. More than 3 independent experiments were performed, and activity data from >20 flies per genotype were pooled and analyzed with Clocklab software (Actimetrics, Wilmette, USA). Circadian rhythm power is a measure of rhythmicity strength and flies with a power value ≥10 are defined as rhythmic. Relative rhythmicity power represents the average rhythmicity of each genotype compared to the average rhythmicity of the wild-type controls used in each experiment. Relative rhythmic power = powerexperimental/powerwild-type × 100.
Statistical Analysis
Statistical tests were performed and diagrams were constructed using GraphPad Prism 7 (RRID: RDG_1346427 GraphPad Software, lnc., San Diego, CA, USA). Unpaired two-tailed Student’s t-tests were performed for two conditions or samples. One-way ANOVA with two-tailed Tukey’s multiple comparison tests were performed for 3 or more conditions or samples. Two-way ANOVA was performed for Sholl analysis of PDF puncta distribution. All data obeyed normal distribution characteristics. All data are presented as the mean ± SEM; *P <0.05, **P <0.01, ***P <0.001.
Results
Knockdown of CRMP Restores the Circadian Activity Rhythm in FXS Drosophila
In previous studies, dfmr1 mutants displayed arrhythmic locomotor activity in constant darkness, revealing an essential role of FMRP in the regulation of the Drosophila circadian rhythm. To find potential FMRP target genes in the circadian regulation and to dissect the explicit underlying molecular pathway in Drosophila, we analyzed genes from the large-scale identification of mRNAs interacting with FMRP in FXS mouse brains that appeared in two independent studies using HITS-CLIP. We focused on those mediating functions in neural circuits and synaptic plasticity, and especially playing roles in the regulation of circadian rhythm. The microtubule-associated protein CRMP2 was noted due to its function in neuronal development and circadian regulation in Drosophila.
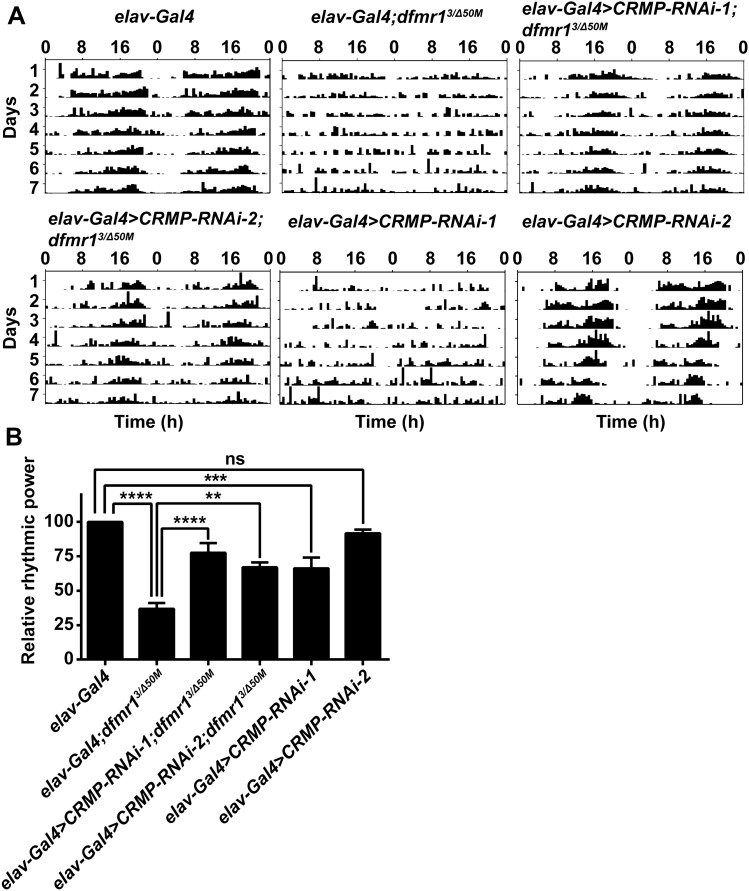
As CRMP-deficient flies also exhibited circadian rhythm defects, we speculated that CRMP may have a functional interaction with FMRP in regulating circadian behaviors, so we investigated the role of CRMP in the circadian activity rhythm of FXS Drosophila. Flies of all genotypes tested were entrained to a 12:12 LD cycle for 4 days prior to transfer to constant darkness for 7 days of activity recording and they had a normal circadian cycle of 23h–24 h (Table 1). The rhythm of free-running rest/activity were impaired in dfmr1 mutant flies with 34% of flies being arrhythmic, and the relative rhythmic power of dfmr1 mutants was significantly lower than in control flies (Fig. 1 and Table 1). Knockdown of CRMP expression with two CRMP RNAi transgene lines driven by the pan-neuronal elav-Gal4 showed decreases of approximately 70% and 45% in CRMP mRNA levels in the brain (Fig. S1A). The two CRMP RNAi lines both partially rescued the activity rhythm of dfmr1 mutants, with the percentages of arrhythmic flies reduced to 14% and 18%, respectively (Fig. 1 and Table 1). Actograms of representative flies showed that the dfmr1 mutants with CRMP knockdown had a normal circadian activity pattern (Fig. 1A). We also examined the rhythms in CRMP-knockdown lines and CRMPsupK1 mutants. The latter showed significant arrhythmic behaviors as reported previously [34] (Fig. S1 and Table S1). Flies with CRMP-RNAi-1 knockdown exhibited arrhythmic activity, while no significant change occurred in flies with another CRMP-knockdown line CRMP-RNAi-2 (Fig. 1 and Table 1), indicating that the knockdown efficiency of CRMP-RNAi-2 driven by elav-Gal4 was not sufficient to cause the arrhythmic phenotype. These results indicate that dysregulated CRMP expression due to the absence of FMRP in the central nervous system contributes to the abnormal locomotor activity rhythm in dfmr1 mutant Drosophila.
Table 1.
Circadian phenotypes in constant darkness
| Genotype | Number | Average period (h) | % Arrhythmic flies |
|---|---|---|---|
| elav-Gla4 | 107 | 23.70 ± 0.07 | 0.93 |
| elav-Gal4;dfmr13/Δ50M | 131 | 23.59 ± 0.09 | 34.35 |
| elav>CRMP-RNAi-1;dfmr13/Δ50M | 63 | 23.64 ± 0.06 | 14.29 |
| elav>CRMP-RNAi-2;dfmr13/Δ50M | 61 | 23.70 ± 0.14 | 18.03 |
| elav>CRMP-RNAi-1 | 93 | 23.40 ± 0.05 | 16.13 |
| elav>CRMP-RNAi-2 | 83 | 23.34 ± 0.05 | 1.20 |
Fig. 1.
Knockdown of CRMP in the central nervous system rescues circadian locomotor activity defects in dfmr1 mutants. A Representative actograms from flies of the indicated genotypes. dfmr1 mutants show significant defects of circadian locomotor activity compared to wild-type controls, while knockdown of CRMP partially ameliorates the arrhythmic patterns. Flies loaded in the monitors were entrained to an LD cycle for 4 days and their locomotor activity in constant darkness was recorded for 7 days and analyzed for the rhythmicity of individual flies. B Relative rhythmic power of the genotypes displayed in A. The dfmr1 mutant flies dfmr13/dfmr1Δ50M was shorted to dfmr13/Δ50M. dfmr1 mutants with both CRMP RNAi transgenes driven by elav-Gal4 show significant improvement in circadian locomotor activity relative to dfmr1 mutants. Some of CRMP-RNAi-1 transgene flies display arrhythmic activity the others show no circadian defects. **P <0.01, ***P <0.001, ****P <0.0001, one-way ANOVA. n >50 flies per genotype. Data are shown as the mean ± SEM. ns, not significant.
Knockdown of CRMP Ameliorates Structural Defects of LNv Neurons in FXS Drosophila
The circadian clock circuit in the Drosophila brain consists of 150 pacemaker neurons that are divided into 5 categories: dorsal lateral neurons, dorsal neurons (DN1, DN2 and DN3), lateral posterior neurons, and small and large ventral lateral neuron (sLNvs and lLNvs) that express the neuropeptide pigment-dispersing factor (PDF) [4]. LNvs have been shown to play an indispensable role in the regulation of circadian activity, and are reported to have structural deficits in both the axonal termini of sLNvs and the POT in dfmr1 mutant flies [15, 25].
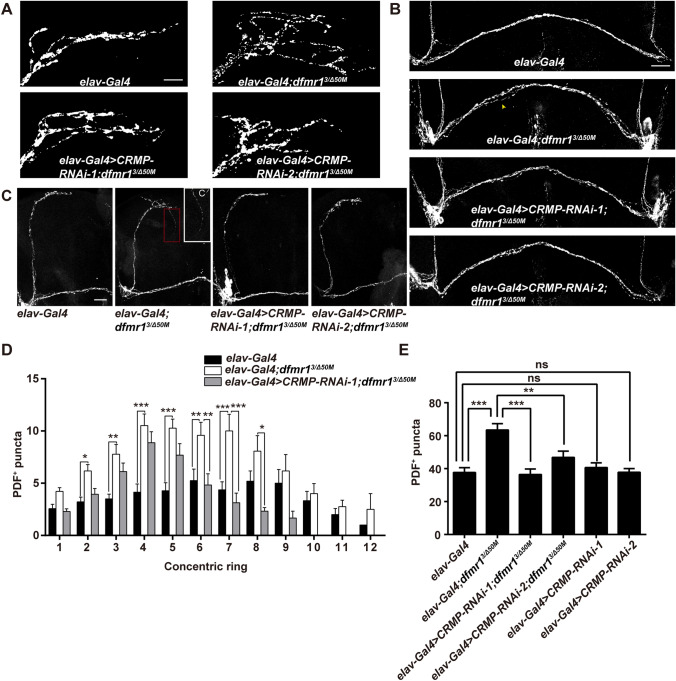
As CRMP2 is essential in neuronal development and axonal growth, we examined whether Drosophila CRMP plays a role in the development of LNv axons in FXS Drosophila. Axon terminals of sLNvs in the dorsal protocerebrum were over-elaborated and a split POT from the lLNvs was also observed in the dfmr1 mutants (Fig. 2A, B). Structural abnormalities in both lLNvs and sLNvs were ameliorated by genetic reduction of CRMP with two CRMP RNAi transgenes in the central nervous system. Over-elaborated terminals of sLNv axons indicate the elevated complexity of the synaptic architecture, which were assessed by PDF-positive puncta distributed at the terminals of sLNv arbors. Sholl analysis of PDF-positive boutons within each 10-μm ring showed that the numbers of PDF-positive puncta were increased in each ring of dfmr1 mutants compared to wild-type flies, and knockdown of CRMP significantly decreased the numbers of puncta in dfmr1 mutants (Fig. 2D and Fig. S2). The total number of PDF-positive puncta throughout the axonal arbor was also measured (Fig. 2). The total number was increased in dfmr1 mutant flies, and CRMP knock-down significantly rescued this phenotype (Fig. 2E; total puncta, 37.6 ± 2.94 control, 63.4 ± 3.88 dfmr13/dfmr1Δ50M 36.4 ± 3.31 elav-Gal4>CRMP-RNAi-1; dfmr13/dfmr1Δ50M, 46.9 ± 3.81 elav-Gal4>CRMP-RNAi-2; dfmr13/dfmr1Δ50M 40.6 ± 2.88 elav-Gal4>CRMP-RNAi-1, 37.8 ± 2.23 elav-Gal4>CRMP-RNAi-2; n >20 for all genotypes). Flies with a split POT from lLNvs were reduced to 27% and 29% in dfmr1 mutants with CRMP-RNAi-1 and CRMP-RNAi-2, respectively, compared to 69% in dfmr1 mutant flies (Fig. 2B and Table S2). As ectopic collateral branches arising from the sLNvs have been reported in dfmr1 mutant flies, we quantified the collateral branches in flies of all genotypes. Collateral branching was observed in 12% of dfmr1 mutants, while dfmr1 mutants with reduced CRMP expression displayed no such ectopic branching (Fig. 2C and Table S2). The structural morphology of sLNvs and lLNvs in CRMP-deficient flies was also documented. POT-splitting and ectopic collateral branches were observed in CRMPsupK1 mutants at higher percentages than in wild-type control flies, although the structure of LNv terminals was not altered (Fig. S3 and Table S2). These results show that CRMP plays a role in the maintenance of the normal morphology of both sLNvs and lLNvs and that excessive CRMP contributes to the defective axonal structures of LNvs in FXS Drosophila.
Fig. 2.
Genetic reduction of CRMP rescues structural deficits of LNv neurons in dfmr1 mutants. A Terminals of sLNvs at the dorsal protocerebrum of the depicted genotypes. dfmr1 mutants show increased branching complexity, while knockdown of CRMP with both RNAi transgenes in the nervous system reduces the over-elaborated axonal branches (scale bar, 10 μm). B Representative split POT phenotype (indicated by arrowhead) in the indicated flies. dfmr1 mutants show a POT defasiculation phenotype compared with the wild-type, and reduced CRMP expression rescues the POT splitting (scale bar, 75 μm). C Half brains of adult flies showing collateral branches. C’ Magnification of the collateral branch in a dfmr1 mutant Drosophila. Some of dfmr1 mutants have ectopic collateral branches (indicated by the red dash box), while mutant flies with two CRMP RNAi transgenes display no collateral branching, like wild-type flies (scale bar, 75 μm). D Sholl analysis of the distribution of PDF-reactive puncta throughout the sLNv axonal arbors. dfmr1 mutants harbor increased PDF puncta distributed in each ring, which is rescued by knockdown of CRMP. E Total numbers of PDF-positive (PDF+) puncta throughout the sLNv axonal arbors. Knockdown of CRMP with both RNAi transgenes rescues the significantly elevated total number of PDF puncta in dfmr1 mutants. *P <0.05, **P <0.01, ***P <0.001, two-way ANOVA for C and one-way ANOVA for D. n >20 hemispheres per genotype. Data are represented as the mean ± SEM. ns, not significant.
CRMP Participates in the Circadian Output Pathway in FXS Flies
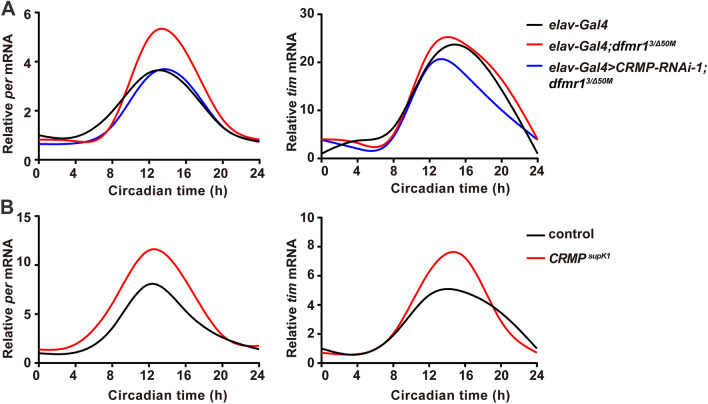
Despite severely impaired behavioral rhythmicity, the core molecular clock is not perturbed in the absence of FMRP. It has been shown that the circadian defects result from disrupted output pathways downstream of the core clock in dfmr1 mutants [15, 27]. We detected the circadian oscillations of the clock genes period (per) and timeless (tim) at the mRNA level in Drosophila brains. Similar to the previous findings, the circadian expression profiles of per and tim mRNA in dfmr1 mutant flies were indistinguishable from those of controls (Fig. 3A). And dfmr1 mutants with knockdown of CRMP exhibited the same mRNA profile (Fig. 3A). Next, we examined the molecular oscillations of per and tim mRNA in CRMP mutant flies to test whether CRMP influences the core clock, and found no irregularity in the mRNA cycling of either gene (Fig. 3B), indicating that CRMP does not affect the function of the core clock at the molecular level.
Fig. 3.
CRMP does not alter the circadian oscillations of per and tim mRNA expression in FXS flies. A Both dfmr1 mutant flies and elav-Gal4>CRMP-RNAi-1;dfmr13/Δ50M flies show normal circadian oscillations of per and tim mRNA like control flies. Circadian time (CT) starts at CT0. dActin was used as a reference gene. B mRNA expression of per and tim display normal circadian oscillations in CRMP mutant flies like control flies. Circadian time (CT) starts at CT0, dActin was used as a reference gene. n >10 heads per genotype at each time point.
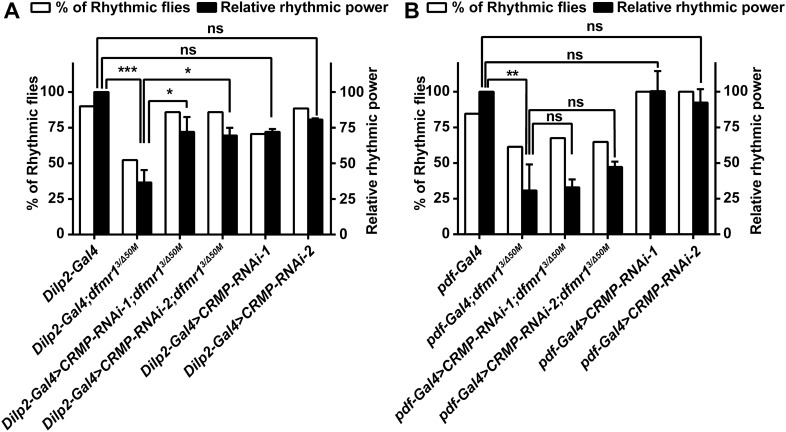
It has been reported that restoring dfmr1 expression in clock neurons does not improve circadian behavior, while selective expression of dfmr1 in the IPCs was able to significantly rescue the rhythmicity of FXS Drosophila. Combined with previous evidence, these findings demonstrate that FMRP functions in the IPCs to mediate the circadian locomotor activity rhythm in Drosophila [14, 15, 27]. We examined the interaction of FMRP and CRMP in the circadian output pathways, using Dilp2-Gal4 and pdf-Gal4 to direct expression in IPC cells and in the circadian pacemaker neurons, respectively. Most of the dfmr1 mutant flies with Dilp2-Gal4 were arrhythmic compared with the Dilp2-Gal4 controls. Knocking down CRMP in the circadian output IPC neurons with the specific Dilp2-Gal4, the free-running activity rhythm was significantly rescued in FXS flies, with elevated relative rhythmic power and an increased percentage of rhythmic flies (Fig. 4A). However, knockdown of CRMP by pdf-Gal4 in the circadian pacemaker LNv neurons, did not ameliorate the circadian defects (Fig. 4B). Taken together, our findings suggest that the translational regulation of CRMP by FMRP contributes to the circadian activity mediation in the output pathway in FXS Drosophila.
Fig. 4.
Knocking down CRMP in IPCs rescues locomotor rhythms in FXS flies. The panels show the percentage of rhythmic flies (white) and relative rhythmic power values (black) of the indicated genotypes. A dfmr1 mutants with both CRMP RNAi transgenes driven by Dilp2-Gal4 show significant improvement in circadian locomotor activity relative to dfmr1 mutants. B dfmr1 mutants with both CRMP RNAi transgenes driven by pdf-Gal4 show no alterations in circadian behaviors compared with dfmr1 mutants. *P <0.05, **P <0.01, ***P <0.001, one-way ANOVA. n >30 flies per genotype. Data are represented as the mean ± SEM. ns, not significant.
Interaction and Translational Regulation Between FMRP and CRMP2 mRNA
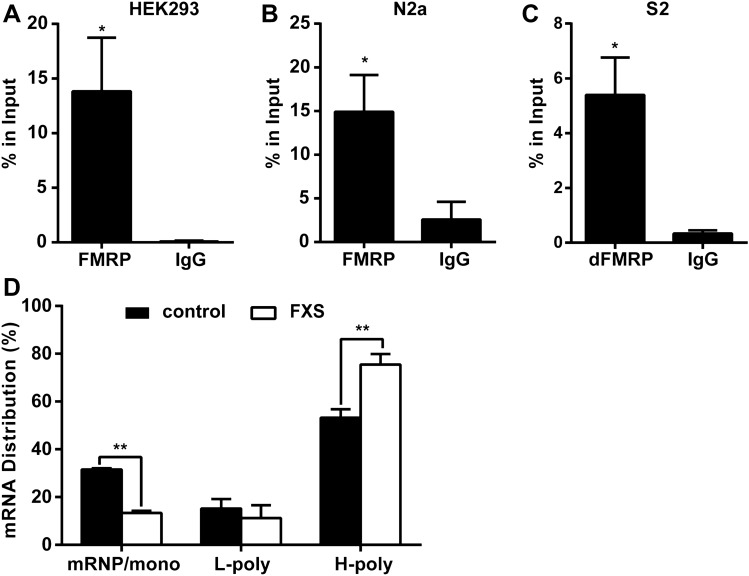
Results in Drosophila provided evidence for the in vivo interaction between FMRP and CRMP mRNA, and implied that CRMP mRNA may be a target of FMRP. To better illuminate how FMRP controls CRMP mRNA in the regulation of circadian behavior and neuronal structures in Drosophila, we further analyzed their interaction in human and mouse cells as well as Drosophila S2 cells. In three independent experiments performed in human HEK293 and mouse N2a cells, CRMP2 mRNA showed a strong association with FMRP. CRMP2 mRNAs were immunoprecipitated by FMRP but not the negative control IgG (Figs 5A, B and S4A). Experiments in Drosophila S2 cells showed similar results (Figs 5C and S4B), and the interaction of CRMP and FMRP was also verified in Drosophila brains (Fig. S4C).
Fig. 5.
FMRP interacts with CRMP2 mRNA and negatively regulates CRMP2 translation. A–C qPCR analysis of CRMP2 mRNA levels co-precipitated with FMRP in human HEK293 cells (A), mouse N2a cells (B), and Drosophila S2 cells (C). CRMP2 mRNA levels were normalized to the control C. elegans 18S rRNA and quantified by the relative value of percentage in input. CRMP2 mRNAs display an enrichment in FMRP antibody precipitates compared to the negative control IgG (*P <0.05, unpaired t-test; n = 3 independent experiments). Data are represented as the mean ± SEM. D qRT-PCR quantification analysis of CRMP2 mRNA levels in fractions of sucrose gradients from normal and FXS patient-derived lymphoblastoid cells. The sucrose gradient fractions are divided to three groups: fractions 1–5 mRNP/monosomes, fractions 6–7 light polysomes (L-poly), and fractions 8–11 heavy polysomes (H-poly). CRMP2 mRNAs are increased in the actively translating polyribosomes (H-poly) in FXS cells with a decreased distribution in the mRNP/mono fractions compared with the wild-type (*P <0.05, **P <0.001, unpaired t-test; n = 3 independent experiments). Data are represented as the mean ± SEM.
As FMRP is an mRNA-binding protein mainly regulating the translation of target mRNAs, we tested whether it controls the translation of CRMP2 mRNAs. We used polyribosome profile analysis to demonstrate the translation activity of CRMP2. Eleven fractions separated from the sucrose gradients were pooled into three groups: fractions 1–5 corresponding to messenger ribonucleoprotein (mRNP) and monosomes, fractions 6–7 corresponding to light polyribosomes, and fractions 8–11 corresponding to the most active translating pool of polysomes and associated mRNA (heavy polysomes). The polyribosome profile of CRMP2 revealed that CRMP2 mRNAs were associated with translationally-active polyribosomes in both normal and FXS patient-derived lymphoblastoid cells. And CRMP2 enrichment significantly increased in the polysomal fractions of FXS cells, indicating that the translation activity of CRMP2 mRNA was elevated in FXS due to the ablation of translational repression by FMRP (Figs 5D and S4D). CRMP2 protein levels and CRMP2 mRNA levels were assessed, and CRMP2 protein expression was elevated in FXS lymphoblastoid cells with no difference in mRNA expression between FXS and normal lymphoblastoid cells (Fig. S4E, F). Taken together, these results indicate that FMRP binds with its CRMP2 mRNA target and represses CRMP2 translational activity.
Discussion
FMRP binds with target mRNAs and participates in mRNA metabolism including translational regulation, mRNA stability, and dendritic transport. CRMP2 mRNA has been implicated as a candidate FMRP target in mouse brain and HEK293 cells [28–30], indicating a possible interaction between FMRP and CRMP2 mRNA. Our results first verified the genetic interaction between FMRP and CRMP in the Drosophila model of FXS, as demonstrated by the findings that knockdown of CRMP expression ameliorated the disrupted circadian activity rhythm and defective neuronal structures in FXS Drosophila. Furthermore, biochemical evidence from three different cell lines corroborated that CRMP2 mRNA was bound to FMRP, which repressed the translation of CRMP2 mRNA without altering its transcription. Our findings identified CRMP as a new target of FMRP, and showed that altered CRMP expression may contribute to the abnormal regulation of behavioral rhythms in FXS Drosophila.
FMRP functions in neuronal development and neuronal defects are found in human cortical tissue and FXS animal models. The density of dendritic spines in hippocampal neurons is significantly elevated in FXS mice, and axonal structures in pacemaker neurons are disrupted in FXS Drosophila [31, 36, 46–50]. CRMP2 also plays roles in the regulation of synaptic morphology and functions. Vertebrate CRMP2 is an important axonal guidance cue that participates in several signal transduction pathways regulating synaptic growth cone dynamics. CRMP2 promotes microtubule assembly by binding to tubulin heterodimers and promoting tubulin polymerization and facilitates the rate of axonal growth [38]. CRMP2 is also involved in Numb-mediated endocytosis of the neuronal cell adhesion molecule L1 at the growth cone, promoting the axon elongation [51]. CRMP2 has been shown to regulate neuronal development and neural function, in addition to its role in neuronal polarity and migration control. CRMP2-knockout mice have enlarged lateral ventricles, decreased dendritic spines, and defective synapse formation in the hippocampus [36]. In our study, CRMP-deficient flies exhibited axonal process growth defects in both pacemaker sLNvs and lLNvs. Consistent with this, axonal process defects in both sLNvs and lLNvs in FXS flies were rescued by reduction of CRMP, providing evidence that CRMP regulates synaptic morphology in FXS Drosophila.
Sleep problems have been reported in individuals with FXS. Patients mainly have difficulty falling asleep and experience frequent night awakenings, and some have snoring and obstructive sleep apnea [19, 52]. FXS mouse and Drosophila models also display sleep disorders, and besides significantly abnormal sleep profiles, impaired rhythmic locomotor activity has been reported in FXS mice and flies [22–24]. Several studies have corroborated the function of FMRP in the circadian output pathway to mediate rest/activity rhythms in Drosophila. Clock genes and proteins form the core transcription-translation feedback loop, regulating the output pathways to generate daily rhythms in organisms [1, 4]. The Drosophila clock genes per and tim display circadian oscillations at both the mRNA and protein levels. Loss of FMRP in Drosophila does not alter the circadian oscillations of both per and tim mRNA and proteins. While the core clock is intact and functions normally in FXS Drosophila, loss of FMRP disturbs circadian output, including the locomotor activity rhythm and the eclosion rhythm [15, 53, 54]. Furthermore, FMRP has been shown to function in the IPCs downstream of the clock neurons to regulate circadian behaviors in FXS flies [27]. This evidence, along with the current study, support the hypothesis that, instead of impacting the core circadian clock, FMRP mainly functions in the circadian output pathway to regulate the circadian activity rhythm in Drosophila. Previous studies have shown marked structural abnormalities in both lLNvs and sLNvs of FXS Drosophila. We found that FMRP regulates CRMP in the control of the synaptic morphology of LNvs, while reduction of CRMP in LNvs does not attenuate the rhythmic behavior deficits. Although FMRP controls the structural morphology of LNv axons, loss of FMRP has no evident effect on their rhythmic release of PDF neuropeptide. Moreover, Monyak et al. reported that FMRP functions mainly in the IPCs by restoring dfmr1 expression through Gal4 expressed in both clock neurons and downstream IPCs [27]. It will take further efforts to illustrate how the structural defects in LNvs contribute to the functional abnormality in downstream IPCs. While specifically knocking down CRMP expression in IPCs rescues the circadian activity rhythms, our findings show that dysregulated CRMP in IPCs contributes to the circadian output in FXS Drosophila.
Circadian rhythms are an emergent property of the intrinsic clock network. Interdependent clock neuron populations and output circuits are both required to maintain normal behavioral and physiological rhythms. Proper neuronal activity and synaptic connections are essential for the functional circadian neural circuit, which is fundamental to circadian rhythms in behavior [55–57]. The pars intercerebralis (PI) region of the Drosophila brain has been implicated in the circadian output regulation of locomotor behaviors. DH44+ SIFamide+ subsets of PI neurons comprise part of a circuit extending from sLNvs and passing through DN1 neurons, involved in the regulation of behavioral rhythms [55, 56, 58]. IPC cells are a cluster of insulin-producing neurons in the PI, and are also functionally connected to the clock circuit via synaptic contacts with DN1 [55, 59]. It has been reported that IPCs exhibit circadian patterns of neuronal activity, including firing frequency and proportion of bursting, and that the electrophysiological rhythm of IPCs is controlled by the circadian clock [59]. In line with previous evidence, our findings show that FMRP mediates behavioral rhythms through the translational regulation of CRMP mRNA in IPCs in FXS Drosophila.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Hunan Science and Technology major project of Birth Defect Cooperative Control (2019SK1010), the National Natural Science Foundation of China (81571253 and 81771385), the Hunan Provincial Natural Science Foundation (2016JJ3135), and the Changsha Municipal Natural Science Foundation (kq2007073).
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Wen Huang, Email: huangwen@sklmg.edu.cn.
Ranhui Duan, Email: duanranhui@sklmg.edu.cn.
References
- 1.Musiek ES, Holtzman DM. Mechanisms linking circadian clocks, sleep, and neurodegeneration. Science. 2016;354:1004–1008. doi: 10.1126/science.aah4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubowy C, Sehgal A. Circadian rhythms and sleep in drosophila melanogaster. Genetics. 2017;205:1373–1397. doi: 10.1534/genetics.115.185157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Top D, Young MW. Coordination between differentially regulated circadian clocks generates rhythmic behavior. Cold Spring Harb Perspect Biol 2018, 10. 10.1101/cshperspect.a033589. [DOI] [PMC free article] [PubMed]
- 4.Franco DL, Frenkel L, Ceriani MF. The underlying genetics of drosophila circadian behaviors. Physiology (Bethesda) 2018;33:50–62. doi: 10.1152/physiol.00020.2017. [DOI] [PubMed] [Google Scholar]
- 5.Bu B, He W, Song L, Zhang L. Nuclear envelope protein MAN1 regulates the drosophila circadian clock via period. Neurosci Bull. 2019;35:969–978. doi: 10.1007/s12264-019-00404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu Y, Zhu ZG, Ma QQ, Su YT, Han Y, Wang X, et al. A critical time-window for the selective induction of hippocampal memory consolidation by a brief episode of slow-wave sleep. Neurosci Bull. 2018;34:1091–1099. doi: 10.1007/s12264-018-0303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heyde I, Kiehn JT, Oster H. Mutual influence of sleep and circadian clocks on physiology and cognition. Free Radic Biol Med. 2018;119:8–16. doi: 10.1016/j.freeradbiomed.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Coulson RL, LaSalle JM. Epigenetics of circadian rhythms in imprinted neurodevelopmental disorders. Prog Mol Biol Transl Sci. 2018;157:67–92. doi: 10.1016/bs.pmbts.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 9.Allen CN, Nitabach MN, Colwell CS. Membrane currents, gene expression, and circadian clocks. Cold Spring Harb Perspect Biol 2017, 9. 10.1101/cshperspect.a027714. [DOI] [PMC free article] [PubMed]
- 10.Lassi G, Priano L, Maggi S, Garcia-Garcia C, Balzani E, El-Assawy N, et al. Deletion of the Snord116/SNORD116 alters sleep in mice and patients with Prader-Willi syndrome. Sleep. 2016;39:637–644. doi: 10.5665/sleep.5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buiting K, Williams C, Horsthemke B. Angelman syndrome - insights into a rare neurogenetic disorder. Nat Rev Neurol. 2016;12:584–593. doi: 10.1038/nrneurol.2016.133. [DOI] [PubMed] [Google Scholar]
- 12.Powell WT, Coulson RL, Crary FK, Wong SS, Ach RA, Tsang P, et al. A Prader-Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum Mol Genet. 2013;22:4318–4328. doi: 10.1093/hmg/ddt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi SQ, Bichell TJ, Ihrie RA, Johnson CH. Ube3a imprinting impairs circadian robustness in Angelman syndrome models. Curr Biol. 2015;25:537–545. doi: 10.1016/j.cub.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue S, Shimoda M, Nishinokubi I, Siomi MC, Okamura M, Nakamura A, et al. A role for the drosophila fragile X-related gene in circadian output. Curr Biol. 2002;12:1331–1335. doi: 10.1016/s0960-9822(02)01036-9. [DOI] [PubMed] [Google Scholar]
- 15.Dockendorff TC, Su HS, McBride SM, Yang Z, Choi CH, Siwicki KK, et al. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 16.Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A. 2013;110:9950–9955. doi: 10.1073/pnas.1305814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang XL, Yuan K, Zhang W, Li SX, Gao GF, Lu L. Regulation of circadian genes by the MAPK pathway: implications for rapid antidepressant action. Neurosci Bull. 2020;36:66–76. doi: 10.1007/s12264-019-00358-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis JK, Broadie K. Multifarious functions of the fragile X mental retardation protein. Trends Genet. 2017;33:703–714. doi: 10.1016/j.tig.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagerman RJ, Berry-Kravis E, Hazlett HC, Bailey DB, Jr, Moine H, Kooy RF, et al. Fragile X syndrome. Nat Rev Dis Primers. 2017;3:17065. doi: 10.1038/nrdp.2017.65. [DOI] [PubMed] [Google Scholar]
- 20.Berry-Kravis EM, Lindemann L, Jonch AE, Apostol G, Bear MF, Carpenter RL, et al. Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nat Rev Drug Discov. 2018;17:280–299. doi: 10.1038/nrd.2017.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahlhaus R. Of Men and Mice: Modeling the fragile X syndrome. Front Mol Neurosci. 2018;11:41. doi: 10.3389/fnmol.2018.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, Lee CC, et al. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am J Hum Genet. 2008;83:43–52. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sare RM, Harkless L, Levine M, Torossian A, Sheeler CA, Smith CB. Deficient sleep in mouse models of fragile X syndrome. Front Mol Neurosci. 2017;10:280. doi: 10.3389/fnmol.2017.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bushey D, Tononi G, Cirelli C. The drosophila fragile X mental retardation gene regulates sleep need. J Neurosci. 2009;29:1948–1961. doi: 10.1523/JNEUROSCI.4830-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeve SP, Bassetto L, Genova GK, Kleyner Y, Leyssen M, Jackson FR, et al. The drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15:1156–1163. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- 26.McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, Ferreiro D, et al. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a drosophila model of fragile X syndrome. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 27.Monyak RE, Emerson D, Schoenfeld BP, Zheng X, Chambers DB, Rosenfelt C, et al. Insulin signaling misregulation underlies circadian and cognitive deficits in a drosophila fragile X model. Mol Psychiatry. 2017;22:1140–1148. doi: 10.1038/mp.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ascano M, Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurin T, Lebrigand K, Castagnola S, Paquet A, Jarjat M, Popa A, et al. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 2018;46:6344–6355. doi: 10.1093/nar/gky267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sethna F, Feng W, Ding Q, Robison AJ, Feng Y, Wang H. Enhanced expression of ADCY1 underlies aberrant neuronal signalling and behaviour in a syndromic autism model. Nat Commun. 2017;8:14359. doi: 10.1038/ncomms14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007;5:e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, et al. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 34.Morris DH, Dubnau J, Park JH, Rawls JM., Jr Divergent functions through alternative splicing: the drosophila CRMP gene in pyrimidine metabolism, brain, and behavior. Genetics. 2012;191:1227–1238. doi: 10.1534/genetics.112.141101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ip JP, Fu AK, Ip NY. CRMP2: functional roles in neural development and therapeutic potential in neurological diseases. Neuroscientist. 2014;20:589–598. doi: 10.1177/1073858413514278. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, Kang E, Wang Y, Yang C, Yu H, Wang Q, et al. Brain-specific Crmp2 deletion leads to neuronal development deficits and behavioural impairments in mice. Nat Commun 2016, 7. 10.1038/ncomms11773. [DOI] [PMC free article] [PubMed]
- 37.Fang W, Gao G, Zhao H, Xia Y, Guo X, Li N, et al. Role of the Akt/GSK-3beta/CRMP-2 pathway in axon degeneration of dopaminergic neurons resulting from MPP+ toxicity. Brain Res. 2015;1602:9–19. doi: 10.1016/j.brainres.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 38.Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, et al. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura T, Fukata Y, Kato K, Yamaguchi T, Matsuura Y, Kamiguchi H, et al. CRMP-2 regulates polarized Numb-mediated endocytosis for axon growth. Nat Cell Biol. 2003;5:819–826. doi: 10.1038/ncb1039. [DOI] [PubMed] [Google Scholar]
- 40.Tan M, Cha C, Ye Y, Zhang J, Li S, Wu F, et al. CRMP4 and CRMP2 interact to coordinate cytoskeleton dynamics, regulating growth cone development and axon elongation. Neural Plast. 2015;2015:947423. doi: 10.1155/2015/947423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janusz A, Milek J, Perycz M, Pacini L, Bagni C, Kaczmarek L, et al. The Fragile X mental retardation protein regulates matrix metalloproteinase 9 mRNA at synapses. J Neurosci. 2013;33:18234–18241. doi: 10.1523/JNEUROSCI.2207-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gatto CL, Broadie K. Temporal requirements of the fragile X mental retardation protein in modulating circadian clock circuit synaptic architecture. Front Neural Circuits. 2009;3:8. doi: 10.3389/neuro.04.008.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernandez MP, Berni J, Ceriani MF. Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol. 2008;6:e69. doi: 10.1371/journal.pbio.0060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q, Li Y, Wang X, Qi J, Jin X, Tong H, et al. Fbxl4 Serves as a clock output molecule that regulates sleep through promotion of rhythmic degradation of the GABAA receptor. Curr Biol. 2017;27(3616–3625):e3615. doi: 10.1016/j.cub.2017.10.052. [DOI] [PubMed] [Google Scholar]
- 46.Hinton VJ, Brown WT, Wisniewski K, Rudelli RD. Analysis of neocortex in three males with the fragile X syndrome. Am J Med Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- 47.Galvez R, Greenough WT. Sequence of abnormal dendritic spine development in primary somatosensory cortex of a mouse model of the fragile X mental retardation syndrome. Am J Med Genet A. 2005;135:155–160. doi: 10.1002/ajmg.a.30709. [DOI] [PubMed] [Google Scholar]
- 48.Aloisi E, Le Corf K, Dupuis J, Zhang P, Ginger M, Labrousse V, et al. Altered surface mGluR5 dynamics provoke synaptic NMDAR dysfunction and cognitive defects in Fmr1 knockout mice. Nat Commun. 2017;8:1103. doi: 10.1038/s41467-017-01191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, et al. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang K, Li YJ, Guo Y, Zheng KY, Yang Q, Yang L, et al. Elevated progranulin contributes to synaptic and learning deficit due to loss of fragile X mental retardation protein. Brain. 2017;140:3215–3232. doi: 10.1093/brain/awx265. [DOI] [PubMed] [Google Scholar]
- 51.Arimura N, Menager C, Kawano Y, Yoshimura T, Kawabata S, Hattori A, et al. Phosphorylation by Rho kinase regulates CRMP-2 activity in growth cones. Mol Cell Biol. 2005;25:9973–9984. doi: 10.1128/MCB.25.22.9973-9984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kronk R, Bishop EE, Raspa M, Bickel JO, Mandel DA, Bailey DB., Jr Prevalence, nature, and correlates of sleep problems among children with fragile X syndrome based on a large scale parent survey. Sleep. 2010;33:679–687. doi: 10.1093/sleep/33.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, et al. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- 54.Xu S, Poidevin M, Han E, Bi J, Jin P. Circadian rhythm-dependent alterations of gene expression in drosophila brain lacking fragile X mental retardation protein. PLoS One. 2012;7:e37937. doi: 10.1371/journal.pone.0037937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavanaugh DJ, Geratowski JD, Wooltorton JR, Spaethling JM, Hector CE, Zheng X, et al. Identification of a circadian output circuit for rest: activity rhythms in drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cavey M, Collins B, Bertet C, Blau J. Circadian rhythms in neuronal activity propagate through output circuits. Nat Neurosci. 2016;19:587–595. doi: 10.1038/nn.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo C, Pan Y, Gong Z. Recent advances in the genetic dissection of neural circuits in drosophila. Neurosci Bull. 2019;35:1058–1072. doi: 10.1007/s12264-019-00390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dreyer AP, Martin MM, Fulgham CV, Jabr DA, Bai L, Beshel J, et al. A circadian output center controlling feeding: fasting rhythms in drosophila. PLoS Genet. 2019;15:e1008478. doi: 10.1371/journal.pgen.1008478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barber AF, Erion R, Holmes TC, Sehgal A. Circadian and feeding cues integrate to drive rhythms of physiology in drosophila insulin-producing cells. Genes Dev. 2016;30:2596–2606. doi: 10.1101/gad.288258.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.