Abstract
Background
Recently, differences in mortality rates of COVID-19 in different geographic areas have become an important subject of research because these different mortality rates appear to be associated with mutations that appeared in SARS-CoV-2. The part of the viral body called the spike protein plays a critical role in the viral attachment and entry of the virus into the host cell. Accordingly, we hypothesized that mutations in this area will affect viral infectivity.
Methods
A total of 193 sequences of spike SARS-CoV-2 were randomly retrieved from five different geographic areas and collection dates (from December 2019 until July 2020). Multiple sequence alignment for mutation and phylogenetic analyses was conducted using Bioedit, UniProt, and MEGA X.
Results
We found 169 total mutations with 37 different mutations across the included samples. The D614G is the first and most frequently established mutation in different regions including Europe, Asia, America, Africa and Australia with the number of mutations of 49, 33, 17, 16 and 4, respectively. Furthermore, we also found mutations in several important domains in this virus including NTD and CTR/RBD of S1 subunit and at S2 subunit area, namely the peptide fusion (FP), and both heptad repetition (HR1 and 2) domains that suggested this could influence virus binding and virus-host cell membrane fusion.
Conclusion
In summary, we concluded that mutation had generated diversity of spike SARS-CoV-2 sequences worldwide and is still growing. This analysis may provide important evidence that should be considered in vaccine development in different geographic areas.
Keywords: Spike protein, SARS-CoV-2, Mutation, Pandemic
At a glance of commentary
Scientific background on the subject
Regardless of the presence of comorbid conditions and provision of medical care, several recent reports suggest that to emerging mutations that have an effect on pathogenicity, and infectivity had lead increasing of mortality rate. The mutation of spike SARS-CoV-2 had reports in different region of the worldwide.
What this study adds to the field
The presents study found that the D614G mutation that located in SD2 is the first and most frequently established mutation in different regions worldwide. We also found mutations in several important domains of virus including NTD, CTR/RBD, FP, HR1 and HR2 domains that suggested affect virus binding and virus-membrane fusion.
In December 2019, the Chinese Health Authority announced a new pneumonia-related disease in Wuhan, Hubei Province, Central China, which was officially named as the Coronavirus Disease 2019 (COVID-19) [1,2]. The novel β-coronavirus, Severe Acute Respiratory Syndrome-Corona Virus 2 (SARS-CoV-2) was identified as the causative agent of the disease that has infected more than 24 million people and is responsible for as many as 827,730 deaths worldwide as of 29 August 2020 [[3], [4], [5]]. This virus is highly contagious with a baseline reproduction rate (R0) ranging between 2.2 and 3.9 with the estimated mortality varying from region to region, between 0.8% and 14.5% [6].
Regardless of the presence of comorbid conditions and provision of medical care, several recent reports suggest that the mortality rate due to this infection can be influenced by the region or geographic area. For example, that case fatality rate (CFR) of Italy is higher compared to other European countries or mortality rates differ between China and countries outside China [7,8]. However, there has been no concrete explanation for the causes of the large difference of mortality rates in different countries until recently [8,9]. Some researchers have attributed this CFR pattern to emerging mutations that have an effect on pathogenicity, and infectivity as occurred in several pre-SARS-CoV-2 outbreaks such as SARS-CoV-1 or Respiratory Syncytial Virus (RSV) [10].
Apart from affecting the infectivity and severity of the outbreak, the diversity of the coronavirus SARS-CoV-2 genomics can also affect the effectiveness of drugs and vaccines. We have learned from continuing antigenic drift in influenza viruses that this trend causes accumulation of mutations that lead to antibody resistance in infected populations. This causes the effectiveness of the vaccine given to decrease. Because this happens continuously from season to season, the influenza vaccine must also be renewed regularly in order to be effective in creating specific antibodies in patients who consume it [11,12].
Although there are reports stating that the diversity of the SARS-CoV-2 sequences is still very low [13], however, considering the persistence of outbreaks that have no visible end yet, the potential for the same trend as happened to the influenza virus over time is very likely to happen again with COVID-19. As an illustration, several previous studies demonstrated the presence of antigenic drift that occurred in the previously β-coronavirus outbreak, including SARS-CoV-1 and cold coronaviruses OC43 and 229E [6,14]. Some evidence of this event is the discovery of several mutations in SARS-CoV-1 both in the structural (spike (S), envelope (E), membrane (M) and nucleocapsid (N) protein) and non-structural proteins (such as Nsp1). Spike protein is an important structural part of the coronavirus and has been widely studied as a target in vaccine and drug development [14,15].
As the common β-coronavirus, SARS-CoV-2 uses its spike protein to bind to cellular receptors and mediates the membrane fusion and virus entry [16]. Structurally, the spike is a transmembrane protein that is processed into two subunits with complementary functions. The S1 subunit functions in receptor binding, while the S2 subunit is responsible for mediating virus-host cell membrane fusion [3,17]. Mutations in the SARS-CoV-1 spike protein have been reported to cause viral evasion of immune system recognition. For example, the P462L and D480 A/G mutations that occur in SARS-CoV induce viruses to escape to a monoclonal NAb [6,18].
In this study, we analyzed sequences of spike SARS-CoV-2 from patients/individuals in five geographic areas including Asia, Africa, America, Australia, and Europe with different collection dates to identify the mutation distribution in different areas over time and correlate their respective impacts.
Material and methods
Data spike SARS-CoV-2 sequence
Spike protein of SARS-CoV-2 reference sequence used for mutation analysis was the Wuhan-Hu-1 reference genome (NC_045512.2). Total of 193 nucleotides and amino acid sequences of about 3821 and 1273 Spike SARS-CoV-2 genomes were retrieved from the National Center for Biotechnology Information (NCBI) Virus Variation Resource repository database (https://www.ncbi.nlm.nih.gov/labs/virus/) between December to June 23rd, 2020 [19]. The sequences were collected from spike protein of SARS-CoV-2 genome which infected humans in various patients around the world, including China, Thailand, India, Bangladesh, Saudi Arabia, Bahrain, Japan, Taiwan, South Korea, Vietnam, Hongkong, Iran, Malaysia, Timor Leste, Kazakhstan, Egypt, Tunisia, Morocco, Nigeria, Australia, New Zealand, USA, Chile, Colombia, Jamaica, Puerto Rico, Brazil, Russia, Italy, Germany, Turkey, Spain, Netherlands, Poland, Serbia, Greece, France, Czech Republic, and Finland that are representative of different continental areas.
Alignment sequence
Multiple sequence alignments of all sequences were done by Bioedit, UniProt, and MEGA X [20] comparing reference sequences with isolated sequences from different samples to analyze mutations. Furthermore, the structures of all amino acid sequences (mutated and NA) were built by the Swiss Model and were subsequently visualized by USCF Chimera and Ez Mol.
Phylogenetic analysis
Multiple sequences of nucleic acid were aligned by MEGA X with MUSCLE method [21,22]. In setting of the cluster method, we used neighbor joining, and then with the default setting, phylogenetic analysis was conducted to identify evolutionary relationships across the spike of SARS-CoV2 genomes in different patients by using the MEGA X using neighbor joining for phylogenetic inference. We used 1000 bootstrap replicates for branch support and using the Kimura 2-parameter model for substitution model, the others were set to default [20,23,24].
Analysis data
All of the sequences were grouped into a specific region, including Asia, America, Europe, Africa and Australia. Furthermore, trend analyses were based on region and time collection date of samples to evaluate the rate of mutation.
Statistical analysis
Statistical analysis was performed using SPSS 22 (IBM Corp., Armonk, NY). Data are categorical variables expressed in terms of frequency and percentage. The non-parametric Kruskal Wallis test was used to compare the frequency of a single mutation, no mutation, and more than one mutation at the nucleotide and protein levels. The Dunn non-parametric test was used for post hoc analysis. The Mann Whitney non-parametric test was used for compared the frequency of mutations and non-mutations. All p-values were calculated using 0.05 as the level of significance.
Results
Phylogenetic analysis
The phylogenetic analysis was done to all spikes of SARS-CoV-2 including 193 samples to describe and comprehend show genomic diversity of the SARSCoV-2 spike in various geographic areas around the world which are presented in Fig. 1.
Fig. 1.
Phylogenetic tree of Spike SARS-CoV-2 sequence of the data included. (A) Bootstrap ConsensusTree and (B) is original tree. The tree analysis used the Neighbor-Joining (NJ) method, 1000 bootstrap replicates and Kimura 2-parameter model for the substitution model. It was built by MEGA X. Scale bar at bottom indicates 0.0005 nucleotide substitutions per site.
Identification of nucleotide and amino acids mutation based on geographic areas
Mutations of 193 complete genome sequences in the spike protein of SARS-SoV-2 were randomly collected in a database from December up to July 2020. We divided our database into five quadrants based on the following geographic areas: Asia, Europe, America, Australia and Africa. The Asia group consists of genome sequences from China, Thailand, Taiwan, India, Bangladesh, Saudi, Bahrain, Japan, Pakistan, Vietnam, Hongkong, Iran, South Korea, Timor Leste, South Korea, Malaysia, and Kazakhstan. The Europe group consists of genome sequences from Turkey, Spain, Netherlands, Russia, Italy, Poland, Serbia, Greece, France, Germany, Finland and Switzerland. The America group consists of genome sequences from USA, Chile, Colombia, Jamaica, and Puerto Rico. The Australia group includes genome sequences from Australian patients. The Africa group consists of genome sequences from Egypt, Morocco, Nigeria, Kenya and Tunisia. All genome sequences in each area were analyzed by comparing their alignment with the reference sequence. The reference sequences that we used as comparison were derived from the first reported sequence of China formerly called “Wuhan seafood market pneumonia virus” with ID number NC_045512.2 at https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.
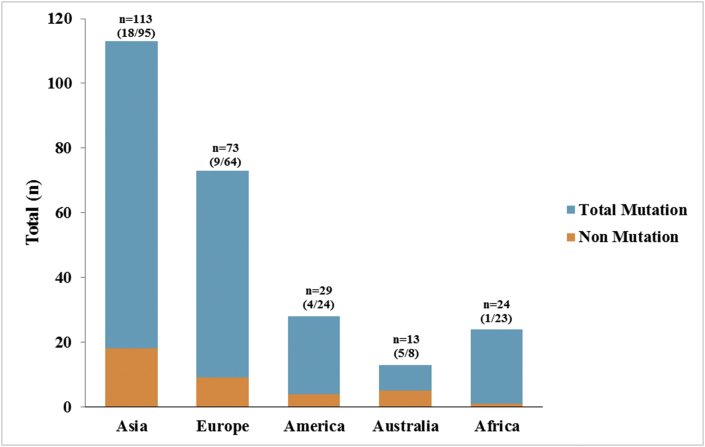
We evaluated the distribution of total mutation nucleotides in the protein spike of SARS-coV-2 [Fig. 2]. In this study, we found 214 total mutations with 61 types of mutations in the entire data. In Asia, there were 95 total mutations with 40 types of mutations which was the most mutations compared to other areas. Meanwhile, there were 15 types of mutations (n = 64) in Europe, followed by America that showed 6 types of mutations (n = 24), Australia had 4 types of mutations (n = 4) and there were 5 types of mutations in Africa (n = 23). The sequence location with the highest mutations is located at position A1841G. Total nucleotide mutations at A1841G were reported to occur the most in Europe, Asia, America, Africa and Australia with the number of mutations of 49, 37, 17, 16 and 4, respectively. In addition, in Asia there are several types of mutations with varying frequencies such as C882T (n = 6), C2367T (n = 5), G2485A (n = 3) and other types of mutations consisting of only one or two frequencies. In addition, the mutation types with the second most common frequencies in other areas were as follows: in Europe with C145T (n = 2), in America with C2472T (n = 3) and in Australia with C96T (n = 2) and the other mutation types had just one frequency in all areas.
Fig. 2.
Total nucleotides mutations and mutation type in spike protein of SARS-CoV-2. There were 214 total mutations divided into five geographic areas (Asia, Europe, America, Australia and Africa). The graphic shows that most of the mutations occurred in Asia (n = 95), Europe (n = 64), America (n = 24), Africa (n = 23) and Australia (n = 8). The type of mutation that had the highest number and occurred in all five areas was the A1841G mutation.
In order to compare total mutations and non-mutation nucleotides in the spike protein of SARS-CoV-2, we analyzed each genome from each geographic area based on whether or not the mutation occurred [Fig. 2]. We found a total of 37 spike genome samples without mutation out of the entire data included. The highest non-mutations occurred in Asia (n = 18) of 76 total samples, Europe (n = 9) of 64 total samples, America (n = 4) of 25 total samples, Australia (n = 5) of 12 total samples and Africa (n = 1) of 19 total samples [see Fig. 3].
Fig. 3.
Comparation of the total mutations and non-mutations nucleotides in the spike protein of SARS-CoV-2 based on geographic area.
Next, we analyzed amino acid mutations in the spike protein of SARS-Cov-2 [Fig. 4]. The total amino acids mutations were 169 with 37 different mutations and classified into 5 areas based on geographic location. We found that Asia has the highest mutations (n = 68) with 25 different mutations. Europe has number of the second total mutations (n = 59) with 10 different mutations. America has total mutations (n = 17) with 1 type mutation. Moreover, total mutations of Australia and Africa were 6 and 19, respectively, with 3 different mutations. In amino acid mutations, we found that the mutation located at position D614G has the highest number compared with other amino acid mutations. Total amino acids mutations at D614G reported occurred mostly in Europe, Asia, America, Africa and Australia with 49, 33, 17, 16 and 4, respectively. In addition, there were mutation types that have more than one case such as in Asia, the mutations A829T (n = 3), P25L (n = 2), V367F (n = 2), R682Q (n = 2), S13I (n = 2), and T22I (n = 2) while the other types have just one case. In Europe we found that most of the mutation types that have occurred in Europe have been just one type, namely H49Y (n = 2). In America, there is only one type of mutation that has occurred and it is located in D614G. The graph shows that Australia has three types of mutations and D614G mutation occurred more than other mutations. Additionally, in Africa there were three types of mutation, which are D614G (n = 16), T29I (n = 2) and Y279N (n = 1).
Fig. 4.
Total amino acids mutations and mutation type in the spike protein of SARS-CoV-2. There were 169 total mutations divided into five based on geographic areas (Asia, Europe, America, Australia and Africa). The graphic shows the most of total mutations is occurs in Asia (n = 68), Europe (n = 59), America (n = 17), Africa (n = 19) and Australia (n = 6). The type of mutation that had the highest number and occurred in all five areas was the D614G mutation.
According to the analysis, it is known that there is no difference in frequency between single mutated and non-mutated (p > 0.05 -Mann Whitney test). This study also analyzed the differences in amino acid mutations in one genome. The analysis showed that there was a significant difference between the group that had more than one mutation and a single mutation (p < 0.05 -Post hoc Dunn test), while the non-mutation group and the other groups were not statistically different (p > 0.05 -Post hoc Dunn test). The A1841G mutation which in this study is the dominant mutation and causes changes in the amino acid D614G.
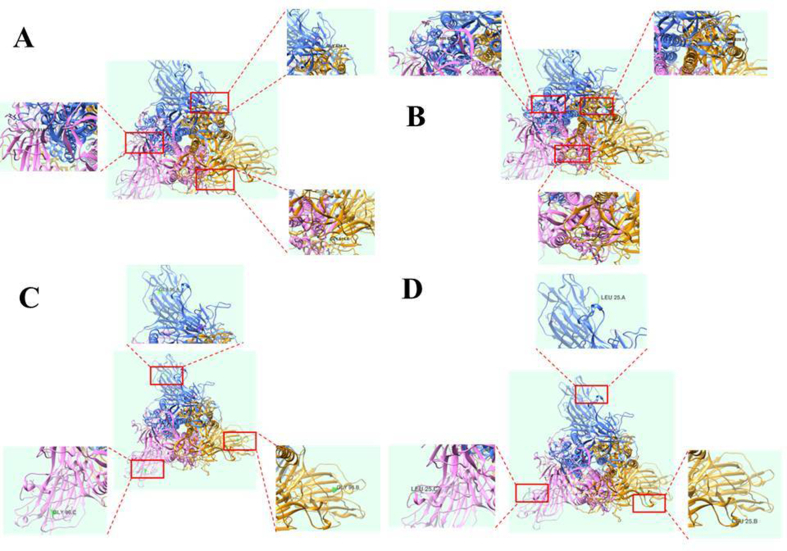
The mutations are distributed in different domains in the spike protein including N-terminal domain (NTD), (C-terminal domain/receptor binding domain (CTD/RBD), subdomains 1 and 2 (SD1 and SD2), as well as the Fusion protein (FP) heptad proteins (HR1 and HR2) which are presented in Fig. 5 and Table 1. This mutation also affects the structure of the spike residue as shown in Fig. 11, which presents four visualized representations of the structure of the mutated spike protein.
Fig. 5.
Schematic of Spike Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) genome.
Table 1.
Localization of different mutations in Spike SARS-CoV-2 genome.
| Type of Mutation | Domain |
|---|---|
| L5F and S13I | SP of S1 subunit |
| T22I, P25L, T29I, Y38C, H49Y, S50L, F86S, T95I, E96G, S151G, N185K, N188K, V213L, S221W, S247R, G261D, Y279N, | NTD of S1 subunit |
| Y453F and V367F | CTD/RBD of S1 subunit |
| T791I | FP of S2 subunit |
| A930V | HR1 of S2 subunit |
| N1187K | HR2 of S2 subunit |
| G1219V | TM of S2 subunit |
| D614G, E583D, T572I, R682Q, R682W, and T553N | S1 subunit |
| Q1002E, A829T and L1063F | S2 subunit |
Fig. 11.
Representative of visualized residue of Spike SARS-CoV-2 Mutated. (A) D614 Mutation (B) A829T (C) E96G and (D) P25L Mutation.
Furthermore, we also found 55 samples with nonmutation [Fig. 6]. From the whole 55 samples with non-mutation including Asia (n = 28) with 76 total samples, Europe (n = 10) with 64 total samples, America (n = 8) with 25 total samples, Australia (n = 7) with 12 total samples and Africa (n = 2) with 19 total samples.
Fig. 6.
Comparation of the total mutations and non-mutations amino acid in the spike protein of SARS-CoV-2 based on geographic area.
In this study, we also found association of Adenine 1841 Guanine (A1841G) mutation pattern with amino acid D614G mutation. We identified that A1841G is located in the coding region of the genome that encodes aspartate (D) (GAT). Substitution of Adenine to Guanine in nucleotide base 1841 leads to the change of base that encodes aspartate (GAT to GGT) which encodes glycine. This also is the same pattern in nucleotide mutation located at G2485A indicating an association with amino acid mutation located at A829T. It shows that G2485A (Guanine 2485 Adenine) that encodes Alanine (GCA) can have substitution with threonine (ACA). Interestingly, the additional mutation located at G2485A was only found in Asia (Thailand).
Identification of nucleotide and amino acids mutation over time
A total of 197 complete genome sequences of the spike protein of SARS-SoV-2 were randomly collected from NCBI database from December up to July 2020. Nucleotide mutations in the spike protein of SARS-CoV-2 were further categorized by collection date of sample to assess the rate of mutation periodically. In this research, the times of collection date were divided into every 8 months (December 2019, January, February, March, April, May, June and July 2020) in every continent, including Asia, Europe, America, Australia, and Africa. Based on the data in Fig. 7, it is known that the region with the most nucleotide mutations occurring during the first 8 months of the outbreak was Asia (n = 95), followed by Europe (n = 64), America (n = 24), Africa (n = 23), and Australia (n = 8).
Fig. 7.
SARS-CoV-2 Spike protein's nucleotide mutation overtime divided into five regions. There was no nucleotide mutations in December 2019. March 2020 is the month when most nucleotide mutations occurred with 85 numbers of mutations.
Unlike other regions, the occurrence of nucleotide mutations in Asia occurs every month, except in December [Fig. 7]. This pattern is presumably because this month is the first time the SARS-CoV-2 virus appeared, so the spike protein possessed by the virus is still the same as the initial template. This contrasts to Australia, which did not have any mutations (only in April, when the incidence of mutations reached six events).
In the graph below [Fig. 7], it is clear that on each continent there was an increase in the incidence of nucleotide mutations in March 2020 (yellow bar). This fact is very unique and also gives us a sign that in almost every region, the increase in COVID-19 cases and the virus’ ability to mutate occurred in the same month (March), even though the numbers were different. The increase of mutations can be seen particularly in America and Africa, followed by Europe. In the first three months (until February 2020), there were no reported incidents of nucleotide mutations in America and Africa. In the following months, there were mutations with a fairly high number. Slightly different from the occurrence in Europe, there were four incidents in January and February before finally there were high increments in nucleotide mutations up to 32 cases in March [Fig. 7]. Meanwhile in Australia, the new cases of nucleotide mutations spiked in April with the lower number (n = 6). This month also ranked second with the most nucleotide mutations among the five regions.
In this study, we also found that the A1841G mutation was identified in most cases in all areas (Asia, Europe, America, Australia and Africa). Here we describe how the distribution of the A1841G mutation in each area is presented in each month [Fig. 8]. Based on this graph, the most common occurrence of the A1841G mutation was in Europe (n = 49) from December 2019 to July 2020. In addition, based on our analysis, Europe was also the first region where the A1841G mutation occurred, precisely in January 2020 (n = 3). The cases of the A1841G mutation in Europe increased in March 2020 (n = 24) and April 2020 (n = 13) as seen in Fig. 8 (orange bar).
Fig. 8.
Monthly distribution of A1841G mutation in Spike protein of SARS-CoV-2 in different regions.
Since appearing in January 2020 in Europe, cases of the A181G mutation have begun to spread to various regions. A month after the first cases appeared in Europe, in February 2020, the A1841G mutation appeared in Asia (n = 3). Since its appearance, cases of the A1841G mutations in Asia (blue bar) have always appeared every month. From Asia, new cases emerged in March in America (gray bar), Australia (yellow bar), and Africa (dark blue bar). Until July 2020, cases of the A1841G mutation still appeared in Asia with four cases of mutations. Data in the graph [Fig. 8] show that in March 2020, all regions have had cases of the A1841G nucleotide mutation in the spike protein of the SARS-CoV-2 virus. This trend also supports the data in Fig. 4 which demonstrate that the highest mutation incidence occurred in March 2020 in all five regions (Asia, Europe, America, Australia, and Africa).
In accordance with the data presented in Fig. 9, it can be seen that the most cases of amino acids mutation from December 2019 to July 2020 was in Asia. In other regions, in certain months there were still some periods when there were no amino acid mutations. But in Asia, amino acid mutations cases were occurring in almost every month, except in the early part of the outbreak (December 2019) [see Fig. 10].
Fig. 9.
Amino acids mutation in Spike protein of SARS-CoV-2 over time. There was no mutation in December 2019 in the five areas (Asia, Europe, America, Australia, and Africa). First case appears in Asia, Europe, and Australia in January 2020 (orange bar). This was followed by America and Africa in March 2020 (yellow bar).
Fig. 10.
Monthly distribution of D614G mutation in Spike protein of SARS-CoV-2 in different region. There is no D614G amino acid mutation in December 2019. The first case appears on January 2020 with total 3 cases. Europe is the region that had the most D614G amino acid mutation cases over time (n = 49) and Australia is the region that had the least D614G amino acid mutation cases over time (n = 4). March 2020 had the most D614G amino acid mutations that occurred in all five regions.
As with nucleotide mutations, in this amino acid mutation, we can clearly know that the month with the highest number of amino acid mutations was March 2020. In this month, there were mutation increments in almost all areas, especially Europe (n = 29) and Asia (n = 20). Only in Australia data show the increasing in amino acid mutation cases soared in April (n = 1) [Fig. 9].
Fig. 4 describes the total mutations of amino acids that occurred in each region. Among the various forms of mutation, the most common occurrence of amino acid mutations was in D614G. From there, we regrouped the distribution of these mutation cases based on the order of the month of occurrence (December 2019–July 2020). Data are displayed in Fig. 9.
Based on the data in Fig. 9, it is known that there were no cases of mutation of the amino acid D614G in December 2019, at the beginning of the emergence of COVID-19. The first case of this mutation appeared in January 2020 only in Europe. After that, cases of the D614G mutation spread in February 2020 to Asia. As of March, all five regions reported cases of D614G amino acid mutation with the most cases in Europe (n = 24). In this month, the case of the D614G mutation jumped very rapidly (n = 49). Furthermore, there was a decrease in total mutations in the five regions over time (April 2020 (n = 32), May 2020 (n = 15), June 2020 (n = 14), and July 2020 (n = 4)). Even though it started in Europe, in June 2020, Europe no longer had cases of the D614G amino acid mutation. In the last 2 months (June–July 2020), the lone region that still had D614G mutation cases was Asia (light blue bar) [Fig. 9].
If we compare the incidence of nucleotide mutations and amino acid mutations in the spike protein of the SARS-CoV-2 virus that occurred in each region per month, even though they show the same graphical pattern, there are several mutations in nucleotides that are not in line with the number of mutations in amino acids.
Based on the comparison of Figs. 7 and 9, there are differences in numbers between the number of nucleotide mutations and the number of amino acid mutations in the SARS-CoV-2 virus in the five areas. The most noticeable difference from December 2019 to July 2020 is in Asia, with more total nucleotide mutations compared to the total mutations of amino acids. As an example of some of the prominent cases that can be described here are in March 2020, there were 30 nucleotide mutations [Fig. 7] while in the amino acid mutations there were only 20 types of mutations [Fig. 9]. In this month, there were mutations of nucleotides A2763G, C210T, G906T, and C2367T but there was no mutation (NA) in amino acids. In addition, there was also an occurrence of mutations of more than three types of nucleotides, but in the amino acids there was only one type of mutated amino acid. The contrasting difference was also seen in March 2020 in Europe with 32 types of nucleotide mutations, but only 29 types of mutations in amino acids.
In America there was a difference of four mutations between the types of mutations of nucleotides and amino acids in March 2020. This number is considered a large number compared to the following months (April, May 2020), where the difference was only 1–2 types of mutations. Based on existing data, this difference can occur because in one type of mutated amino acid there are two types of mutated nucleotide bases. For example, in April 2020 in America, the mutation of the amino acid D614G was always followed by a mutation of A1841G in the nucleotide base. However, there was one time when the mutation in D614G amino acid in spike protein of SARS-CoV-2 had two types of mutated nucleotide bases (A1841G and C1560T). In May 2020 data in America, it is known that mutations occurred in two types of nucleotides (T2115C and C2472T) but there were no mutations in their amino acids. This pattern shows that the occurrence of mutations in these two types of nucleotide bases does not affect the amino acid sequence.
In contrast to other regions, Africa shows a small difference in the number of mutations of nucleotides and amino acids (1–2 events). Only May 2020 had the same number of mutations (n = 2). This difference occurs because of the additional types of mutated nuclei bases, but the amino acids mutations are only in one type of amino acid.
However, not all graphs show differences in the number of mutations in nucleotide bases and amino acids. In certain months, there were cases where the number of mutations of nucleotides and amino acids were the same. This trend is shown by data from Europe in January and February 2020 in which both reported four types of mutations in nucleotides and amino acids. In addition, it can be seen in April 2020 in Europe (n = 15), and in January and March 2020 in Australia (n = 1).
The findings invite the presumption that mutations occur in the non-coding region in the spike protein of the SARS-CoV-2 virus. It could be that one of the mutated nucleotide bases detected is the nucleotide base from the non-coding region so that mutations are not found in the amino acid as well. However, this presumption needs further investigation.
Discussion
In this study, we analyzed mutations in the SARS-CoV-2 spike from different regions of the world over time to see the distribution of mutations across geographic areas. In total we retrieved 197 sequences of spike data from the NCBI database and compared them with their reference sequences in https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.
We noted that the nucleotide mutation A1841G is a mutation that was first introduced in January in the European region and subsequently spread so rapidly that it became the most common type of mutation in spike sequences worldwide over time. The mutation occurs in the coding sequence and is responsible for the change in D614 to G614. This D614G mutation does not change the antigenicity of SARS-CoV-2 because the position is not part of (the N-terminal domain [NTD] and the C-terminal domain [CTD]) which is the first priming of the SARS-CoV-2 antigen. The position of the mutation is in subdomain 2 (SD2), a subdomain that is conserved following NTD, CTD, and SD1 which can be done directly with the S2 subunit via disulfide linkage [25]. Although it does not have antigenicity, the D614G mutation is very ubiquitous with a higher mortality rate than from COVID-19 [26].
Based on the analysis of Toyoshima et al. (2020) the mutation is predicted to be in the HLA epitope area, namely S606-615, NQVAVLYQDV, and S612-620, YQDVNCTEV. They further found that this mutation was not associated with a high mortality rate in COVID-19 patients with the alleles HLA-A∗11: 01, HLA-A∗02: 06, and HLA-B∗54: 01.
Contrary to these findings, Becerra-Flores and Cardozo (2020) demonstrated that this mutation is implicated in more pathogenic SARS-CoV-2 strains leading to an increase in the CFR. One mechanism that was associated with the increasing of CFR is the presence of G614 variant which could destabilize the infectious form of the protein spike. This condition results in the closing of the receptor binding site of the host cell which is a mechanism expected to evade the immune system recognition. Additionally, another finding demonstrated that the increasing in CFR occurs as consequence of D614G mutation which results in an increase in the entry of virus cells that are harbored with this mutation leading to increased viral infectivity [27,28]. Based on statistical analysis, it is known that the genome group that has the A1841G mutation is more dominant than other mutations. This is also supported by the result that the frequency of one mutation is more dominant than more than one mutation. This study also shows that several genomes have a combination of mutations of A1841G and mutations of other types. The protein level also shows the same pattern, where the A1841G mutation causes a change in the aspartic acid at position 614 to Glycine. Also, the D614G mutation is the dominant mutation among other mutations. This pathogenicity occurs due to changes in the aspartic residues in position 614 which previously were able to bind firmly to the threonine and/or lysine residues by hydrogen bonds and salt bridges, respectively, becoming replaced with glycine residues which provide the increasing of flexibility space between adjacent protomers, making it easier for S1–S2 dissociation, and resulting in more ACE2 accessibility by RBD [27]. Apart from D614G, we also identified other mutations in the S1 subunit namely L5F and S13I which are in the signal peptide (SP) domain. However, it is still unknown how this domain affects the virus.
Furthermore, as previously reported by Lokman et al., in 2020 that found NTDs located in S1 subunit of SARS-CoV-2 are very prone to mutation, we found many types of mutations in the S1 N-terminal domain. We found 17 mutation types of total sequences located within the NTD including T22I, P25L, T29I, Y38C, H49Y, S50L, F86S, T95I, E96G, S151G, N185K, N188K, V213L, S221W, S247R, G261D and Y279N. S1 NTD is a unique domain in the SARS-CoV-2 spike protein sequence, which has a glycan binding site located at its primary apex and facing the target cell (host cell). This domain can recognize and bind to O-acetylated sialic acids (O-Ac-Sia) which are receptors on the surface of the host cell [17,29]. O-Ac-Sia is O-glycan required by the coronavirus for initial interaction with the ACE2 host cell receptor and resulting entry of SARS-CoV-2 into the epithelium of the main human airway [17].
Following the findings of mutations in NTD, mutations were also found in CTD/RBD of the SARS-CoV-2 spike sequence including Y453F and V367F. Together with NTD, CTD is responsible for the initial interaction of the virus with host cells so that this area has become the main target in drug and vaccine development at this time [25,30]. These mutations that occur in RBD may weaken the binding of antibodies that are generated at the time of initial interaction with SARS-CoV-2 in recovered or vaccinated patients because RBD contains an antigenic epitope which is important in viral antigen sensitization. For the vaccine response, the mutations will decrease the effectiveness of the vaccine [31]. However, the results of our study on the variation of amino acids in CTD and NTD can be considered for further validation of vaccines targeting this second domain to determine if one is more effective than the other target.
As known, the subunit S2 of the coronavirus contains four core domains including fusion peptide (FP), heptad repeat (HR1 and HR2) and the transmembrane domain at C-terminus [17,32,33]. These domains play an important role in virus-host cell membrane fusion. HR1 could refold and become a long α-helix which leads to shedding viral fusion peptides into the host membrane. HR1 will form a coiled-coil with HR2 and generate the fusion of the virus-host cell membrane [17,33]. Since we found several mutations that occur in these domains including T791I, A930V, N1187K and G1219V, the pattern suggests that they affect the membrane fusion process. Furthermore, besides playing a role in membrane fusion, previous findings demonstrated that mutations in heptad increased resistance to HR2 derived peptide entry inhibitors of hepatitis coronaviruses in mice models indicating that mutations occur in the HR area [34]. Accordingly, we are suggesting from this evidence that a mutation located in this repeated heptad would cause COVID-19 drug resistance. This analysis may provide important evidence that should be considered in vaccine development in different geographic areas.
Conclusions
In summary, we concluded that the mutation rate of the spike protein of SARS-CoV-2 that has occurred has resulted in an increasing diversity of this protein in various regions of the world overtime. D614G that is located in of the spike genome, has become the most common mutation worldwide and generates high infectivity of the virus and this finding suggested it is associated with CFR on a country by country basis. Furthermore, mutations were also found in several important domains in this virus including NTD and CTR/RBD which play a role in binding the virus to host cells. In addition, mutations were also found to occur in the S2 subunit area, namely the peptide fusion (FP), both heptad repetition (HR1 and 2) and transmembrane domains which suggest that they will affect the process of cell membrane-host cell membrane fusion. Although the impact of this mutation still lacks complete information and requires further investigations, however, these findings could be considered in vaccine development in different geographic areas.
Conflict of interest
There is no conflicts of interest.
Acknowledgements
The authors gratefully thank the staff of Klinik Bahasa Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada for grammatical and structural editing of this manuscript.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astuti I., Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diab Metabol Synd. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maitra A., Sarkar M.C., Raheja H., Biswas N.K., Chakraborti S., Singh A.K., et al. Mutations in SARS-CoV-2 viral RNA identified in Eastern India: possible implications for the ongoing outbreak in India and impact on viral structure and host susceptibility. J Biosci. 2020;45:76. doi: 10.1007/s12038-020-00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization [Internet]. Coronavirus (COVID-19). WHO; 2020 [cited August 29, 2020]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019?adgroupsurvey={adgroupsurvey}&gclid=Cj0KCQjwqKuKBhCxARIsACf4XuGYVBH2pYAS639DLUr_RQZz380uua0iViYoLAdD_cCCXuKvdRQxHjYaAtnlEALw_wcB/.
- 6.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khafaie M.A., Rahim F. Cross-country comparison of case fatality rates of COVID-19/SARS-COV-2. Osong Pub Health Res Perspect. 2020;11:74–80. doi: 10.24171/j.phrp.2020.11.2.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toyoshima Y., Nemoto K., Matsumoto S., Nakamura Y., Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J Hum Genet. 2020;65:1075–1082. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tal G., Mandelberg A., Dalal I., Cesar K., Somekh E., Tal A., et al. Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J Infect Dis. 2004;189:2057–2063. doi: 10.1086/420830. [DOI] [PubMed] [Google Scholar]
- 11.Boni M.F., Gog J.R., Andreasen V., Feldman M.W. Epidemic dynamics and antigenic evolution in a single season of influenza A. Proc Biol Sci. 2006;273:1307–1316. doi: 10.1098/rspb.2006.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., et al. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181:990–996. doi: 10.1016/j.cell.2020.04.021. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sui J., Aird D.R., Tamin A., Murakami A., Yan M., Yammanuru A., et al. Broadening of neutralization activity to directly block a dominant antibody-driven SARS-coronavirus evolution pathway. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Z., Li H., Wu X., Zhong Y., Zhang K., Zhang Y.P., et al. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol Biol. 2004;4:21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L., Fang Q., Deng F., Wang H., Yi C.E., Ba L., et al. Natural mutations in the receptor binding domain of spike glycoprotein determine the reactivity of cross-neutralization between Palm Civet Coronavirus and Severe Acute Respiratory Syndrome Coronavirus. J Virol. 2007;81:4694–4700. doi: 10.1128/JVI.02389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., et al. Virus variation resource: improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45:D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z.G., Xu S.P., Zhang Y.J., Bao Q.Y. Genetic distance of SARS coronavirus from the recent natural case. Vet Microbiol. 2007;120:167–172. doi: 10.1016/j.vetmic.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie Q., Li X., Chen W., Liu D., Chen Y., Li H., et al. Phylogenetic and phylodynamic analyses of SARS-CoV-2. Virus Res. 2020;287:198098. doi: 10.1016/j.virusres.2020.198098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Gobeil S., et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat Struct Mol Biol. 2020;27:925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int J Clin Pract. 2020;74 doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozono S., Zhang Y., Ode H., Seng T.T., Imai K., Miyoshi K., et al. Vol. 2020. bioRxiv; 2020. Naturally mutated spike proteins of SARS-CoV-2 variants show differential levels of cell entry. 06.15.151779. [Google Scholar]
- 28.Zhang L, Jackson CB, Mou H, Ojha A, Rangarajan ES, Izard T, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. BioRxiv 10.1038/s41467-020-19808-4 [Preprint]. 2020 [cited August 25, 2020]. Available from: https://www.biorxiv.org/content/10.1101/2020.06.12.148726v1.
- 29.Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q.K., et al. Human Coronavirus HKU1 spike protein sses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lokman S.M., Rasheduzzaman M., Salauddin A., Barua R., Tanzina A.Y., Rumi M.H., et al. Exploring the genomic and proteomic variations of SARS-CoV-2 spike glycoprotein: a computational biology approach. Infect Genet Evol: J Mol Epidemiol Evolution Genet Infect Dis. 2020;84:104389. doi: 10.1016/j.meegid.2020.104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ou J., Zhou Z., Dai R., Zhang J., Lan W., Zhao S., et al. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein. BioRxiv. 2020 https://www.biorxiv.org/content/10.1101/2020.03.15.991844v4 22020.03.15.991844 [Preprint]. [cited August 20, 2020]. Available from: [Google Scholar]
- 32.Wang D., Mai J., Zhou W., Yu W., Zhan Y., Wang N., et al. Immunoinformatic analysis of T- and B-cell epitopes for SARS-CoV-2 vaccine design. Vaccines. 2020;8:355. doi: 10.3390/vaccines8030355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahajan M., Chatterjee D., Bhuvaneswari K., Pillay S., Bhattacharjya S. NMR structure and localization of a large fragment of the SARS-CoV fusion protein: implications in viral cell fusion. Biochim Biophys Acta Biomembr. 2018;1860:407–415. doi: 10.1016/j.bbamem.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bosch B.J., Rossen J.W., Bartelink W., Zuurveen S.J., de Haan C.A., Duquerroy S., et al. Coronavirus escape from heptad repeat 2 (HR2)-derived peptide entry inhibition as a result of mutations in the HR1 domain of the spike fusion protein. J Virol. 2008;82:2580–2585. doi: 10.1128/JVI.02287-07. [DOI] [PMC free article] [PubMed] [Google Scholar]