Summary
Free-text clinical notes in electronic health records are more difficult for data mining while the structured diagnostic codes can be missing or erroneous. To improve the quality of diagnostic codes, this work extracts diagnostic codes from free-text notes: five old and new word vectorization methods were used to vectorize Stanford progress notes and predict eight ICD-10 codes of common cardiovascular diseases with logistic regression. The models showed good performance, with TF-IDF as the best vectorization model showing the highest AUROC (0.9499–0.9915) and AUPRC (0.2956–0.8072). The models also showed transferability when tested on MIMIC-III data with AUROC from 0.7952 to 0.9790 and AUPRC from 0.2353 to 0.8084. Model interpretability was shown by the important words with clinical meanings matching each disease. This study shows the feasibility of accurately extracting structured diagnostic codes, imputing missing codes, and correcting erroneous codes from free-text clinical notes for information retrieval and downstream machine-learning applications.
Keywords: clinical notes, cardiovascular disease, ICD-10 codes, natural language processing, interpretability
Highlights
-
•
Five NLP word vectorization models predict 8 ICD-10 codes with high AUROC and AUPRC
-
•
The best-performing TF-IDF models showed full interpretability with important words
-
•
The models showed high transferability when tested on the MIMIC-III ICU dataset
The bigger picture
The mining of the structured data in electronic health records such as diagnostic codes enables many clinical applications, but much clinical information is locked in the unstructured free-text clinical notes because they are more difficult to use in data mining. In addition, the structured diagnostic codes are often missing or even erroneous. To accurately structure the free-text notes in the form of diagnostic code for downstream usage, we used old and new natural language processing methods together with interpretable classification algorithms to extract eight diagnostic codes of common cardiovascular diseases. This work helps to structure free-text clinical notes, impute missing diagnostic codes, and correct erroneously diagnostic codes noted by clinicians to improve the data quality of diagnostic codes as the fundamental structured data for later information retrieval and downstream data-mining applications.
Unstructured data in electronic health records are difficult for data mining while structured diagnostic codes are often missing or even erroneous. This work used five old and new natural language processing techniques to extract eight cardiovascular diseases' diagnostic codes, which helps to structure free-text clinical notes, impute missing diagnostic codes, and correct erroneously diagnostic codes noted by clinicians to improve the data quality of diagnostic codes as the fundamental structured data for later information retrieval and downstream data-mining applications.
Introduction
The digitization of hospitals has enabled electronic health records (EHR) to become accessible to researchers for secondary usage that benefits healthcare research.1, 2, 3, 4 The analyses of EHR contribute to a better understanding of the clinical trajectories of patients,5 and improved patient stratification and risk evaluation.6,7 However, much of the information in the EHR is locked in free-text clinical notes.2,4 Typically, EHR include structured data such as age (e.g., 50), sex (e.g., M), and white blood cell count (e.g., 10.5 K/μL), and unstructured data such as free-text notes (“ … has the following active medical issues hx of afib …”). Analyzing these free-text clinical notes is challenging.1,2,8 Historically, the information in free-text clinical notes has been extracted mostly manually by clinical experts for archiving, retrieval, and analyses, and this has been particularly relevant to chronic disease because clinical notes dominate over structured data. More recently, natural language processing (NLP) and machine-learning methods have shown great promise to automatically analyze clinical notes.1,2,9,10
EHR data enable researchers and clinicians to perform information extraction and encode the information for later information retrieval and secondary usage.4 Based on these clinical notes, ICD-10 codes (i.e., the International Classification of Diseases, 10th Revision)11 are used by clinicians to encode diagnoses. Some typical applications of EHR have been using these diagnostic codes in downstream tasks, such as automatic information retrieval, risk prediction, and the prediction of disease subtypes.1,2,9,10 As the ICD-10 diagnostic codes form the basis, its quality determines the performance of downstream tasks. Furthermore, EHR data in structured format rather than in free-text format can be more easily used in machine-learning applications or combined with other data types.
Yet diagnostic codes are frequently missing in EHR or the recorded diagnostic codes may be inaccurate. Misclassification and inaccuracy in diagnostic codes have been reported in an increasing number of papers, for instance in cases related to myocardial infarction and stroke.12, 13, 14, 15 McCarthy et al.12 reported that a substantial percentage of patients who had myocardial injury were miscoded as having type 2 myocardial infarction, which may have serious consequences. Chang et al.13 found disagreement in stroke coding. The erroneous coding may negatively influence stroke case identification in epidemiological studies and hospital-level quality metrics. Goldstein14 found patients with indicated primary ICD-9-CM codes to have conditions other than acute ischemic stroke. Horsky et al.15 demonstrated that there were significant deficiencies in documented diagnostic codes with clinicians coding ICD-10 codes in simulated scenarios, which also suggested that additional training for clinicians was needed. Recent studies have focused on the problem of diagnostic code prediction.1,9 Although some good results have been produced, many of the previous diagnostic code prediction studies have applied deep-learning methods that make the models difficult to interpret.2,3,9 Because ICD-10 codes are usually the start for downstream tasks and clinicians attach significance to interpretable information extraction systems,4 interpretable models may have certain advantages over less-interpretable models in that they may not only enable accurate ICD-10 code imputation but also enable clinicians to readily understand the models and control the quality of the diagnostic codes with their expertise.
In this study, we propose the use of NLP word vectorization algorithms and logistic regression (LR) to predict eight ICD-10 codes related to common cardiovascular diseases from free-text outpatient progress notes (Figure 1). We compared both interpretable models and less-interpretable models with regard to their performances on the ICD-10 code prediction tasks. The proposed models show good classification performance on eight ICD-10 codes in two Stanford cohorts and the models generalized well on the MIMIC-III (Medical Information Mart for Intensive Care III) dataset. Additionally, the most interpretable models also showed the best performance on all datasets.16,17
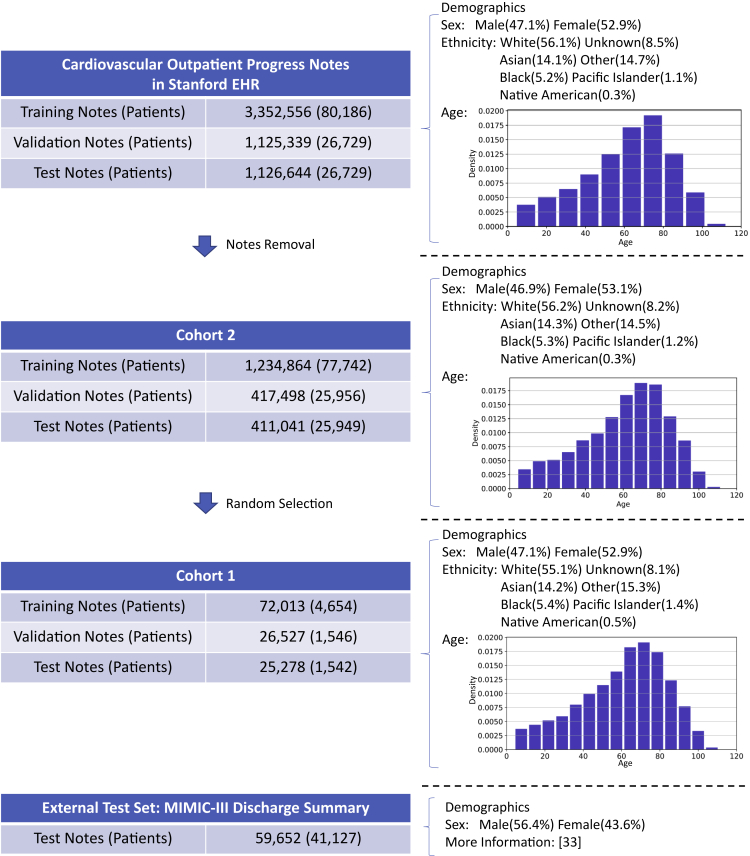
Figure 1.
Overview of the cohorts used in this study
Visualization of the cohorts used in this study with the number of notes and patients of the cardiovascular EHR at Stanford, two subsets (cohort 1 and cohort 2), and the MIMIC-III test set for model validation
In each cohort, the major demographic features including sex, ethnicity, and age are also shown.
Results
Data visualization
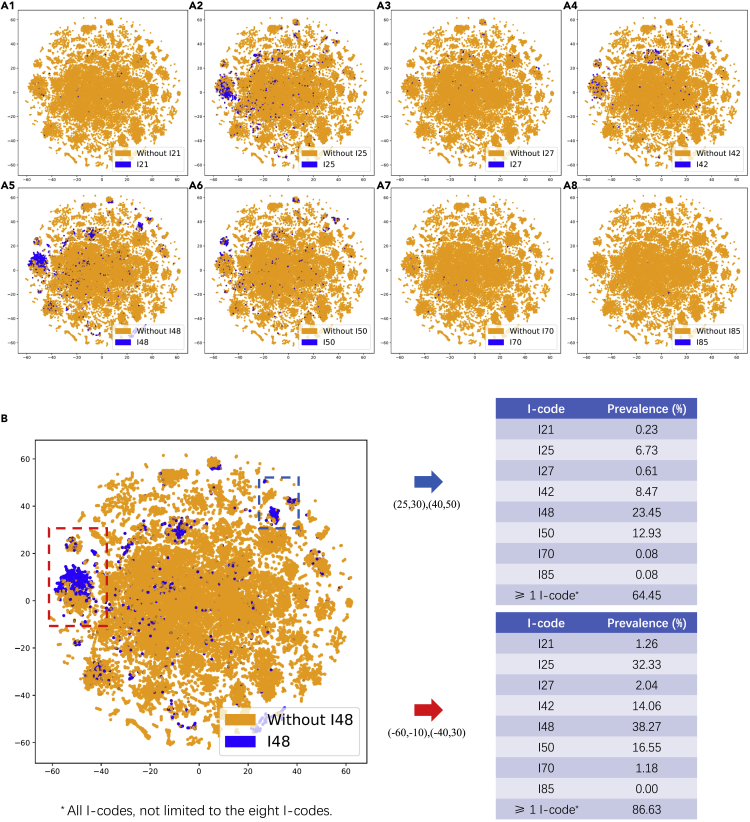
We first visualized the feature vectors with TF-IDF using t-distributed stochastic neighbor embedding (t-SNE) to explore the data in the clinical notes in the cohort 1 training set (Figure 2). Due to limited space, we presented the TF-IDF visualization as a demonstration because of its full interpretability. We found clusters related to several cardiovascular diseases. The selected clusters within the bounding boxes showed high prevalence in I-codes, suggesting that the feature vectors may be able to distinguish ICD-10 codes.
Figure 2.
t-SNE visualization of the training notes in the cohort 1 of eight I-codes based on TF-IDF
(A) t-SNE visualization of the TF-IDF word vectors of the cohort 1 training notes of eight I-code classification tasks.
(B) Prevalence of eight I-codes in the two selected regions with high prevalence of I-codes. Here, more than one I-code means all types of I-codes, not limited to the eight I-codes we investigated.
Prediction of ICD-10 codes
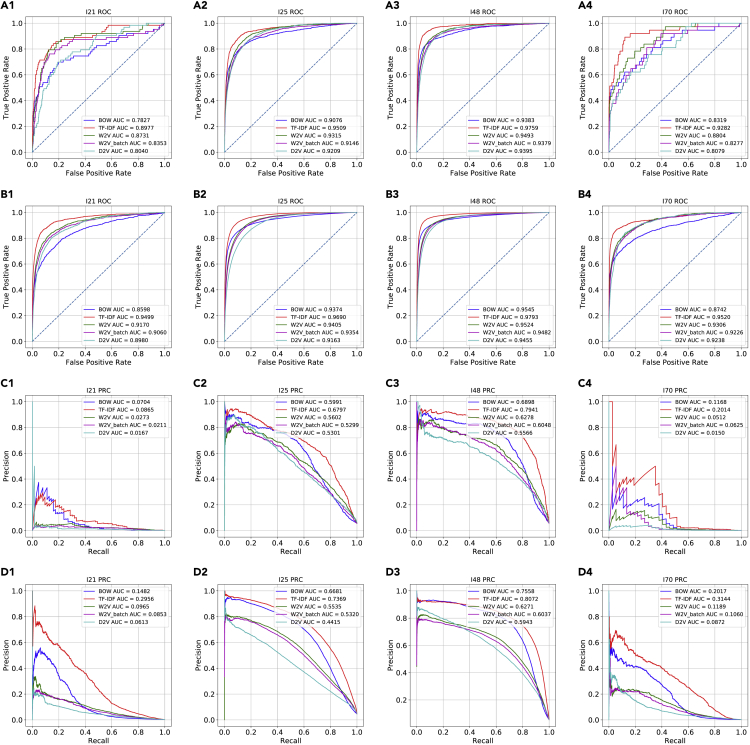
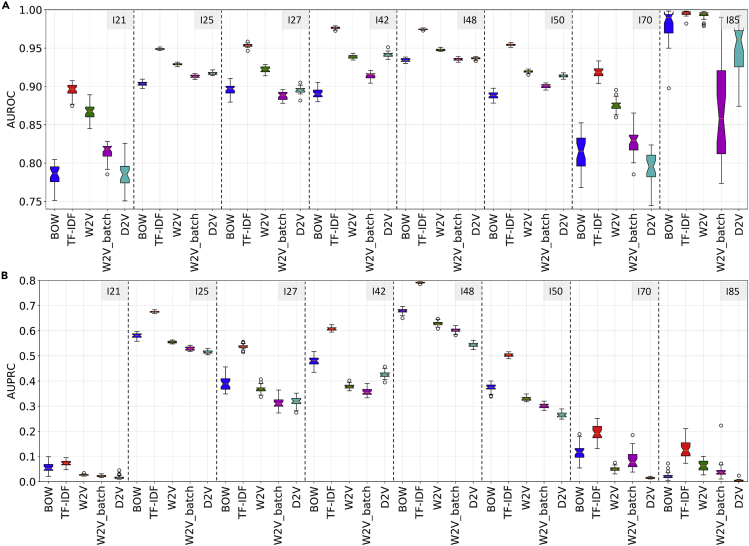
First, we tested our machine-learning workflow for predicting ICD-10 codes on cohort 1. These results showed that LR and the word vectors enabled the classification of the eight diagnostic codes related to cardiovascular diseases (I-code) with high prevalence on cohort 1 with both high area under the receiver-operating characteristic curve (AUROC) and high area under the precision recall curve (AUPRC) (Figures 3, S1, and S5). The AUROC values in all classification tasks were higher than 0.75, and term frequency-inverse document frequency (TF-IDF) outperformed the other four word vectorization methods, with AUROC values higher than 0.85 on four selected codes with different prevalence. There was a variance in the AUPRC among the codes with varying prevalence. For the codes with high prevalence such as I25 and I48, the AUPRC values were above 0.65 and 0.75 for TF-IDF. Additionally, the 30 bootstrapping experiments on cohort 1 showed the best performances given by TF-IDF on the majority of the codes (Figure 4).
Figure 3.
Predictive performance on two cohorts for four cardiovascular diseases
The Receiver-operating characteristic curves (ROC) and precision recall curves (PRC) of the logistic regression (LR) models trained on five different word vectorization methods and on four of the eight I-code classification tasks that represent different prevalence
Cohort 1, A1–A4 and C1–C4; cohort 2, B1–B4 and D1–D4.
Figure 4.
AUROC and AUPRC of classifiers based on different word vectorization methods in 30 bootstrapping experiments on cohort 1
(A) AUROC results. The best model in bootstrapping experiments based on AUROC was TF-IDF (mean AUROC (95% CI)): I21, 0.8952 (0.8768–0.9075); I25, 0.9487 (0.9470–0.9514); I27, 0.9537 (0.9505–0.9585); I42, 0.9763 (0.9735–0.9790); I48, 0.9745 (0.9731–0.9762); I50, 0.9543 (0.9522–0.9571); I70, 0.9185 (0.9046–0.9333); I85, 0.9951 (0.9918–0.9981).
(B) AUPRC results. The best model in bootstrapping experiments based on AUPRC was TF-IDF (mean AUPRC (95% CI)): I21, 0.0723 (0.0549–0.0951); I25, 0.6752 (0.6709–0.6830); I27, 0.5370 (0.5189–0.5557); I42, 0.6079 (0.5949–0.6240); I48, 0.7913 (0.7878–0.7948); I50, 0.5028 (0.4888–0.5161); I70, 0.1941 (0.1344–0.2514); I85, 0.1281 (0.0727–0.2108).
Second, on the larger cohort 2, with more data, the results showed that the LR models trained on the word vectorization methods classified the I-codes with an improvement in both AUROC and AUPRC, particularly on the codes with lower prevalence (Figures 3 and S2). TF-IDF outperformed the other word vectorization methods in terms of both AUROC and AUPRC. On the codes with lower prevalence (i.e., I21 and I70) the performances were significantly improved, with AUROC values around 0.95 and AUPRC values above 0.25 based on TF-IDF word vectors.
Interpretation of important words in classification
To interpret the models, we extracted the ten most important words in 30 bootstrapping experiments on cohort 1 (Table 1). The results showed that not only many important words that were found were overlapping in the bootstrapping experiments, but also that most words could be explained on the basis of the meanings related to the diagnostic codes. For example, for acute myocardial infarction, non-ST-elevation myocardial infarction, myocardial, myocardial infarction, thrombus, and infarction were found to be important; for chronic ischemic heart disease, coronary, coronary artery disease, artery/arterial, and angina were found to be important; for atrial fibrillation, flutter, fibrillation, atrial, fibrillation, atrial fibrillation, and paroxysm were found to be important. Meanwhile, the results based on the two metrics were similar, indicating that the importance of words was relatively stable over the 30 bootstrapping experiments. To conclude, the models based on TF-IDF and LR predicted I-codes not only showed high AUROC and AUPRC, but were also interpretable based on clinically meaningful terms determining the prediction.
Table 1.
The ten most important words found in 30 bootstrapping experiments based on ranking metric and coefficient metric with TF-IDF and LR
| Code | No. of words | Top ten words (ranking metric) | Top ten words (coefficient metric) |
|---|---|---|---|
| I21 | 90 | nstemi, myocardi, mi, thrombu, infarct | nstemi, myocardi, mi, infarct, thrombu |
| stemi, stent, plavix, jayden, bracken | stemi, stent, plavix, jayden, xarelto | ||
| I25 | 65 | coronari, cad, arteri, nativ, mi | coronari, cad, arteri, nativ, mi |
| angina, plavix, cabg, stent, lad | angina, plavix, cabg, stent, lad | ||
| I27 | 80 | pulmonari, hypertens, sildenafil, ph, revatio | pulmonary, hypertens, sildenafil, ph, revatio |
| echo, diastol, flolan, ex, shah | echo, diastol, ex, vinicio, fpah | ||
| I42 | 79 | cardiomyopathi, carvedilol, coreg, ef | cardiomyopathi, carvedilol, coreg, ef, lv |
| hypertroph, hcm, hocm, echo, icd | hypertroph, hcm, hocm, echo, icd | ||
| I48 | 71 | fibril, atrial, fib, afib, coumadin, af | fibril, atrial, fib, afib, coumadin, af |
| irregular, paroxysm, xarelto, digoxin | irregular, paroxysm, xarelto, digoxin | ||
| I50 | 91 | failur, chf, heart, lasix, congest | failur, chf, heart, lasix, congest |
| diastol, systol, bnp, coreg, spironolacton | diastol, systol, bnp, coreg, spironolacton | ||
| I70 | 94 | atherosclerosi, aorta, arteri, vascular, peripher | atherosclerosi, aorta, arteri, vascular, peripher |
| stenosi, dystroph, claudic, nail, renal | claudic, stenosi, dystroph, nail, renal | ||
| I85 | 63 | cirrhosi, varic, liver, transplant, ascit | cirrhosi, varic, liver, transplant, ascit |
| portal, esophag, lutchman, propranolol, hepat | portal, esophag, lutchman, propranolol, hepat |
The ranking metric ranks the important words by the sum of the rankings of the word importance in bootstrapping experiments, and the coefficient metric ranks the important words by the sum of LR coefficients in bootstrapping experiments. The words are shown after stemming.
False-positive analysis of the prediction
Next, to test whether there were missing diagnostic codes in the datasets that could be imputed by the I-code prediction models, we analyzed several randomly selected false-positive cases (Table 2). We found mentions of specific diagnostic terms in the notes of false-positive predictions, which indicated that some of the false-positive predictions were correct with the possible reason being that the ground-truth diagnostic codes might be missing. This analysis suggests that it is possible to impute missing I-codes based on the model predictions in a subset of cases. However, additional manual curation efforts are needed because the most accurate TF-IDF word vectorization was word based, which cannot handle negation, and personal or family history. For instance, an I-code might be predicted due to a patient's medical history but not necessarily noted down as the diagnostic code for that specific encounter.
Table 2.
Analysis of false-positive predictions based on TF-IDF
| Code | Evidence from note | T/F | Potential cause |
|---|---|---|---|
| I25 | “ … all negative for stress induced ischemia …” | F | negation |
| I48 | “ … has the following active medical issues hx of afib …” | T | |
| I48 | “ … paroxysmal atrial fibrillation is seen here …” | T | |
| I48 | “ … was found to be in atrial fibrillation …” | T | |
| I48 | “ … cardiac history of atrial flutter and atrial fibrillation …” | F | personal medical history |
| I48 | “ … for post hospital check after admission for atrial fibrillation …” | F | personal medical history |
The note predictions were manually extracted, analyzed, and labeled as true (T) or false (F) based on evidence in the notes, and the potential causes of erroneous predictions were analyzed.
Model transferability on the MIMIC-III dataset
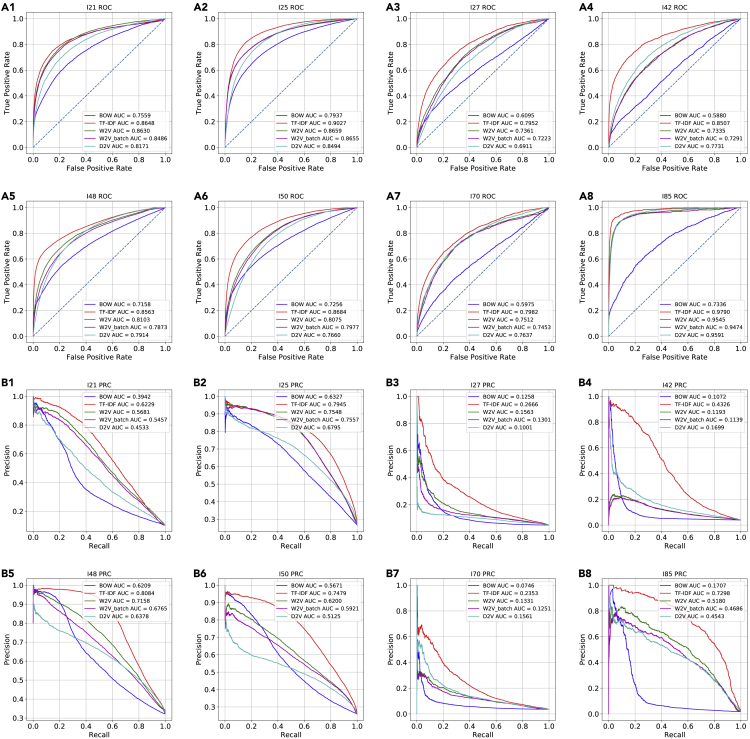
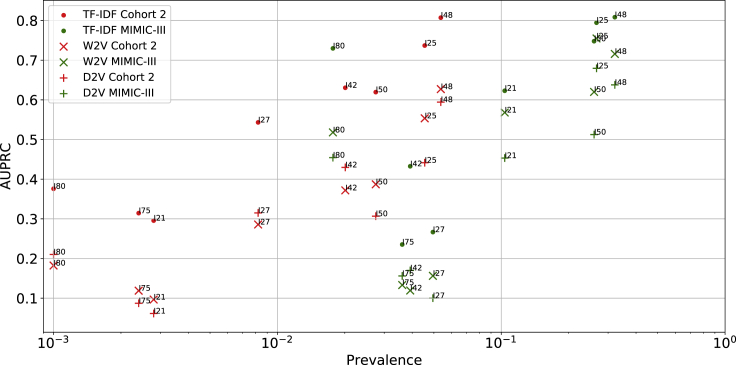
To test the model transferability, we extracted the discharge summaries in the MIMIC-III dataset and the corresponding ICD-9 diagnostic codes of each of the eight ICD-10 codes, and tested the pre-trained word vectorization models and classification models on the MIMIC-III dataset without any fine-tuning. The high AUROC and AUPRC values showed that all models (i.e., TF-IDF, word2vec [W2V], batch-word2vec [W2V_batch], and doc2vec [D2V]) models could be well transferred to the classification of the diagnostic codes in the MIMIC-III dataset (Figures 5 and 6; Table 3). On the same I-code prediction task, TF-IDF showed the best performances with the highest AUROC values and AUPRC values while Bag-of-words (BOW) performed the worst in terms of AUROC and AUPRC on the majority of the classification tasks. When compared with the test set of cohort 2, the TF-IDF models reached higher AUPRC values on I21, I25, I48, I50, and I85. Based on these results, we observed a positive relationship between AUPRC values and prevalence, where AUPRC is better for higher prevalence. In general, besides BOW models, all other models generalized well to the external cohort.
Figure 5.
Predictive performance for eight cardiovascular diseases on an external dataset
The Receiver-operating characteristic curves and precision recall curves in the classification of eight corresponding ICD-9 codes on the discharge summary in MIMIC-III dataset
A1–A8, the classification AUROC; B1–B8, the classification AUPRC.
Figure 6.
Area under the precision recall curves for TF-IDF, W2V, and D2V models on cohort 2 and the MIMIC-III dataset
Each marker shape denotes one vectorization method and each color denotes one dataset.
Table 3.
Prevalence and prediction performance (AUROC and AUPRC) on test sets of Stanford cohorts and MIMIC-III dataset based on TF-IDF, W2V, and D2V
| Method | Code | Cohort 1 |
Cohort 2 |
MIMIC-III |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Prevalence | AUROC | AUPRC | Prevalence | AUROC | AUPRC | Prevalence | AUROC | AUPRC | ||
| TF-IDF | I21 | 0.25% | 0.8977 | 0.0865 | 0.28% | 0.9499 | 0.2956 | 10.36% | 0.8648 | 0.6229 |
| I25 | 5.62% | 0.9509 | 0.6797 | 4.55% | 0.9690 | 0.7369 | 26.64% | 0.9027 | 0.7945 | |
| I27 | 1.26% | 0.9560 | 0.5391 | 0.82% | 0.9698 | 0.5432 | 4.95% | 0.7952 | 0.2666 | |
| I42 | 2.20% | 0.9790 | 0.6163 | 2.01% | 0.9810 | 0.6305 | 3.92% | 0.8507 | 0.4326 | |
| I48 | 5.79% | 0.9759 | 0.7941 | 5.37% | 0.9793 | 0.8072 | 32.16% | 0.8563 | 0.8084 | |
| I50 | 2.96% | 0.9567 | 0.5082 | 2.75% | 0.9732 | 0.6195 | 25.98% | 0.8684 | 0.7479 | |
| I70 | 0.15% | 0.9282 | 0.2014 | 0.24% | 0.9520 | 0.3144 | 3.61% | 0.7982 | 0.2353 | |
| I85 | 0.02% | 0.9975 | 0.1315 | 0.10% | 0.9915 | 0.3759 | 1.77% | 0.9790 | 0.7298 | |
| W2V | I21 | 0.25% | 0.8731 | 0.0273 | 0.28% | 0.9170 | 0.0965 | 10.36% | 0.8630 | 0.5681 |
| I25 | 5.62% | 0.9315 | 0.5602 | 4.55% | 0.9405 | 0.5535 | 26.64% | 0.8659 | 0.7548 | |
| I27 | 1.26% | 0.9268 | 0.3785 | 0.82% | 0.9407 | 0.2858 | 4.95% | 0.7361 | 0.1563 | |
| I42 | 2.20% | 0.9418 | 0.3836 | 2.01% | 0.9467 | 0.3718 | 3.92% | 0.7335 | 0.1193 | |
| I48 | 5.79% | 0.9493 | 0.6278 | 5.37% | 0.9524 | 0.6271 | 32.16% | 0.8103 | 0.7158 | |
| I50 | 2.96% | 0.9214 | 0.3355 | 2.75% | 0.9441 | 0.3871 | 25.98% | 0.8075 | 0.6200 | |
| I70 | 0.15% | 0.8804 | 0.0512 | 0.24% | 0.9306 | 0.1189 | 3.61% | 0.7512 | 0.1331 | |
| I85 | 0.02% | 0.9969 | 0.0628 | 0.10% | 0.9805 | 0.1823 | 1.77% | 0.9545 | 0.5180 | |
| D2V | I21 | 0.25% | 0.8040 | 0.0167 | 0.28% | 0.8980 | 0.0613 | 10.36% | 0.8171 | 0.4533 |
| I25 | 5.62% | 0.9209 | 0.5301 | 4.55% | 0.9163 | 0.4415 | 26.64% | 0.8494 | 0.6795 | |
| I27 | 1.26% | 0.9033 | 0.3423 | 0.82% | 0.9307 | 0.3149 | 4.95% | 0.6911 | 0.1001 | |
| I42 | 2.20% | 0.9468 | 0.4451 | 2.01% | 0.9530 | 0.4298 | 3.92% | 0.7731 | 0.1699 | |
| I48 | 5.79% | 0.9395 | 0.5566 | 5.37% | 0.9455 | 0.5943 | 32.16% | 0.7914 | 0.6378 | |
| I50 | 2.96% | 0.9185 | 0.2700 | 2.75% | 0.9271 | 0.3067 | 25.98% | 0.7660 | 0.5125 | |
| I70 | 0.15% | 0.8079 | 0.0150 | 0.24% | 0.9238 | 0.0872 | 3.61% | 0.7637 | 0.1561 | |
| I85 | 0.02% | 0.9673 | 0.0033 | 0.10% | 0.9822 | 0.2102 | 1.77% | 0.9591 | 0.4543 | |
Discussion
In this work, NLP methods were used to compare five different word vectorization methods from free-text outpatient clinical notes, whereby LR was shown to be effective in predicting the diagnostic codes of eight cardiovascular diseases. Among them, on both the smaller cohort 1 and the larger cohort 2 from the Stanford EHR dataset, the best word vectorization method according to AUROC and AUPRC was TF-IDF (Figures 3, 4, S1, and S2). From cohort 1 to cohort 2, the scalability of the models was shown that with more data, the classification performance could be improved (Figures 3 and S4). Additionally, the majority of the word vectorization models and classification models trained on the Stanford EHR dataset also showed transferability when applied to the MIMIC-III dataset (Table 3). The TF-IDF, W2V, W2V_batch, and D2V models performed well on the Stanford cohorts and generalized well on the different MIMIC-III dataset. In this study, we did not fine-tune the models on the MIMIC-III dataset and used it only as a test set, because the models already showed good performance when transferred to a different dataset. The simple BOW word vectors showed a sharp decrease in AUROC and AUPRC values on the different datasets, showing that the word vectorization models might have overfitted the Stanford EHR dataset because it directly used the word counts as features without any normalization, and the distribution of the word counts in different datasets is likely to be different. The word vectorization method with normalization on word counts (TF-IDF) and the word embeddings (W2V, D2V) that seeks a lower-dimension representation showed higher robustness in classification performance when transferred to a different dataset, potentially because the normalization and the reduced dimensionality may lead to smaller variance across datasets. In this study, we did not mean to hype old or new NLP methods but to test whether they work on the diagnostic code prediction task with good performance, robustness, and generalizability with a proper evaluation pipeline. We were surprised to manifest that the word-based vectorization method TF-IDF with simple LR was effective in accurately predicting diagnostic codes based on free-text clinical notes. The results imply that these models can be used in accurately predicting diagnostic codes and improving the quality of diagnostic codes at different clinical sites. Furthermore, although the new word embeddings (W2V, W2V_batch, and D2V) did not show higher AUROC and AUPRC when compared with TF-IDF, they were in lower dimensions (200/600) than TF-IDF and BOW (414,391), which could be helpful to significantly reduce computational costs with fair classification performance in AUROC and AUPRC.
Additionally, the interpretability of the models was shown in this work by important-word analysis and false-positive-case analysis. The important words found in each I-code prediction task were clinically meaningful (Table 1). The robustness of the important words was also shown by bootstrapping. In a previous study, Wei and Eickhoff1 applied a convolutional neural network (CNN) to predict diagnostic ICD-10 codes with good performance, but the deep-learning based models were hard to interpret. Sheikhalishahi et al.2 also mentioned in their review paper that the model interpretability was a significant issue for more complex methods. Wei and Eickhoff1 claimed that the simple word vectors do not give good results and showed that the CNN embedding with a support vector machine reached a precision value of 0.2162 and a recall value of 0.7732 in the prediction of diagnostic codes. Although direct benchmarking and comparison cannot be made due to differences in the prevalence of ICD-10 codes and datasets selected in our study, the simple word vectorization models and LR showed good predictive performances (Figure 3 and Table 3) while maintaining interpretability, and therefore could contribute to the diagnostic code prediction and quality control for clinicians.
False-positive case analysis showed that some of the false-positive predictions might be correct and could be applied to impute potential missing codes that do not have I-codes recorded by clinicians (Table 2). The false-positive predictions might not be wrong but are simply missing. However, among the false-positive cases, we also observed that certain mistakes were caused by negation, past medical history, and family history. Because the best model of TF-IDF is word based, it models the contents of the free text by each individual word, and these issues cannot be directly detected by the TF-IDF model. Therefore, to impute missing I-codes, the proposed classifiers here could be used to complete records, in combination with additional methods to assert negation, temporality, and identity of the experiencer.
More generally, an important use case of this work is to impute ICD-10 codes from free-text format. Diagnostic codes rich in clinical information can be missing and the noted diagnostic codes may also be inaccurate, which has been shown by recent studies for diagnostic codes related to myocardial infarction and stroke.12,13 This study proposes a method to impute missing diagnostic codes and potentially correct misclassified diagnostic codes based on model predictions. In addition, the model interpretability also enables clinicians to interpret the models and check whether particular imputation is correct. The improvement of the quality of diagnostic codes may help further machine-learning diagnosis because machine-learning algorithms typically require structured data. Many of the previous studies directly use the diagnostic codes for the subsequent downstream classification tasks.1,2,9,10 Improving the quality of diagnostic codes also could improve the data quality for further machine-learning processes.
Although this study has shown promising results for predicting diagnostic codes based on clinical notes, there are several points that warrant further study. First, our modification of segregating the texts into batches by windows did not improve the performances when compared with the conventional W2V model that took an average of all word vectors. The probable reason may be that the notes and sections are of different lengths. Roughly splitting the notes into fixed batches may not successfully partition the different sections. In the future, studies can be designed to automatically detect and partition sections to improve the classification performance.
Second, we have shown that our models developed on the basis of the Stanford EHR datasets worked well on both Stanford dataset and the MIMIC-III dataset, which has been regarded as the standard and publicly available dataset for clinical data-mining research. These results showed the potential of the models for cross-institute applications without extra fine-tuning. However, the models were built and evaluated on diagnostic codes without clinician curation, and there is a lack of highly curated datasets with cleaned correct code labels for diagnostic code prediction benchmarking. In the future, it would be worthwhile for researchers to collect and publish a better curated dataset that can serve as the standard baseline to assess any diagnostic predictors developed by researchers and thus benefit this field of research in the long term.
Third, we mainly applied the context-independent word vectorization methods (besides D2V) as features for notes in the diagnostic code prediction for better interpretability. In this study, we did not investigate the use of transformer-based models and recurrent-neural-network-based models, motivated for several reasons. First, recent reports have shown that transformer models did not show significant improvement over conventional models in recent studies.18, 19, 20 For example, Rishivardhan et al.21 applied four transformer models—BERT, RoBERTa, Electra, and XLNet—to predict ICD10-PCS codes and showed the highest precision of 0.025, the highest recall of 0.049, and the highest F1 score of 0.033. Second, language models are complex models that require a large corpus for training, which can be difficult in the clinical text-mining setting due to patient information protection protocols. Third, transformer models are not easily interpretable. In the future, more studies that use context-dependent sentence embeddings, including transformer-based models, to improve their performance in clinical applications are needed.
In this study, after the first step of data processing, 63.2% of notes were removed because they either did not have a diagnostic code or were shorter than 60 words. We used these dropped notes in training the D2V embedding models. Part of the unlabeled notes might still contain meaningful information related to classification. Methods such as semi-supervised learning22 and conformal predictions23,24 might hold potential to make use of these unlabeled data, which could potentially further improve the prediction performance. In addition, due to the class imbalance and low prevalence of certain diagnostic codes in the Stanford EHR dataset, the model performance could be further improved with more data collected across different institutes or via federated learning.
Furthermore, in this study we used the simple BOW word vectorization as the baseline model. Other baselines can be developed: for example, MedTagger25,26 can be used to tag medical concepts specific to each disease, such as indicators for negation, family medical history, and personal medical history, after which classifiers or rule-based diagnostic code-labeling models can be developed based on the tagging.
Finally, this work focused on the prediction of ICD-10 codes for structuring the free-text clinical notes, and the structured codes were not tested in downstream tasks such as phenotyping or outcome prediction with machine learning. This work might help subsequent prediction tasks. For example, the structured diagnostic codes based on the information from clinical notes can be combined with other data sources in data-fusion tasks including imaging data, genomics data, and laboratory test data to predict prognosis, patient outcome, and disease subtypes.27, 28, 29, 30
Experimental procedures
Resource availability
Lead contact
Further information and requests for data should be directed to the lead contact, Olivier Gevaert (ogevaert@stanford.edu).
Materials availability
This study did not use any reagents.
Data and code availability
The data and code used in this study are not shareable, as the data concern patient information. Please contact the corresponding author to discuss remote access to the data in a de-identified manner.
Data description
We used outpatient progress notes of 133,644 patients diagnosed with cardiovascular diseases at Stanford Health Care. The patients were partitioned into a training set (60%), a validation set (20%), and a test set (20%). All notes belonging to the same patient were partitioned into the same dataset to avoid information leakage across datasets. The dataset included 5,604,539 notes from 31,502 encounters dated from April 2000 to October 2016. The encounters link different structured data, such as age, demographics, and diagnostic codes, and unstructured data such as free-text clinical notes. Multiple different notes and diagnostic codes can be associated with a single encounter. The free-text clinical notes and diagnostic codes were both extracted from the encounters. The data were retrospectively collected and de-identified in accordance with approved IRB guidelines (Protocol: IRB-50033: Machine Learning of Electronic Medical Records for Precision Medicine) (Figures 1 and S6).
We focused on the following eight common cardiovascular diseases from clinical notes: acute myocardial infarction (I21), chronic ischemic heart disease (I25), pulmonary heart disease (I27), cardiomyopathy (I42), atrial fibrillation flutter (I48), heart failure (I50), atherosclerosis (I70), and esophageal varices (I85). As ICD-10 codes have a hierarchy to organize the more than 69,000 diagnostic codes, we aimed at predicting the three-letter prefixes of the ICD-10 diagnosis codes. The relationship between free-text clinical notes and ICD codes is a one-to-multiple relationship. Therefore, we designed eight binary classification tasks for each of the eight ICD codes independently. The positive/negative labeling was independently extracted and coded for each ICD code from the encounter. Furthermore, the eight selected diseases represent different levels of imbalance in the dataset with the largest prevalence (5.37%) being more than 50 times the lowest prevalence (0.10%), which could be used to evaluate the model performance over a high variance of prevalence (Table 3).
Data processing
Notes with fewer than 60 words (after checking the notes, we found that the descriptions of procedures which were irrelevant to diagnosis were generally shorter than 60 words, therefore we chose a threshold of 60 words to balance notes for modeling and removal of uninformative notes) and notes without any labeled ICD-10 code were excluded, resulting in the removal of 63.2% notes defining cohort 2. For prototyping and testing the scalability of the models, a smaller cohort, cohort 1, was built with randomly selected notes from cohort 2 (Figure 1 and Table S1).
Next, we processed the clinical notes by changing them to uncapitalized text and removing any special characters, punctuation, mathematical symbols, and URLs. Stop-words such as conjunctions were removed with Gensim,31 and the words were tokenized. Stemming was then used to reduce inflected words to word stems with the Porter stemming algorithm32 with the Natural Language Toolkit library.33
Word vectorization
We used four different vectorization algorithms to convert the free-text notes to numerical features (i.e., word vectors): BOW, TF-IDF, W2V, and D2V (supplemental experimental procedures). In addition, W2V_batch was introduced as a modified model based on W2V.
BOW34 and TF-IDF16 are word-count-based word vectorization algorithms. In this study, after applying BOW and TF-IDF to cohort 1 and cohort 2, the feature dimensions were 88,815 and 414,391, respectively.
W2V35 is another vectorization algorithm to obtain word embeddings based on shallow neural networks. In this work, a pre-trained W2V model was used: a biomedical W2V model trained on a corpus collected from PubMed and MIMIC-III36 with 16,545,452 terms and an embedding dimension of 200. After converting each term in a text into a 200-dimension embedding, an average of all the term embeddings was taken as the embedding for a note.
The progress notes we used can be divided into three general sections, describing patient history, description at presentation, and plan/billing. In addition to taking the average as a note embedding, a batched form of W2V was introduced in this study by splitting a note into several batches (n = 1, 3, 5) as an attempt to extract section-related contents. For instance, a note with a length of 1,000 words could be split into five batches and the first 200 word embeddings were averaged as the feature of the first batch. In this modified batch-word2vec model (W2V_batch), the embedding dimension was where n was the number of batches. The n was chosen to be 3 based on the average area under receiver-operating characteristic curve (AUROC) on the validation set in cohort 1.
D2V is based on W2V but further inputs the tagged document ID in the training of word vectors.37 In the training process, a word vector is trained for each term, and a document vector is generated for each document. In the inference process for prediction, all weights are fixed to calculate the document vector for a new document. In this study, to avoid overfitting we used the 63.2% dropped notes (neither in cohort 1 nor in cohort 2 because the notes were either shorter than 60 words or without any ICD-10 codes) to train our D2V model with 40 epochs and an embedding dimension of 200. The number of terms modeled was 327,113.
To visualize the data, we used the non-linear dimensionality reduction method, t-SNE.38 We used t-SNE to visualize the 400k feature vectors of BOW and TF-IDF motivated based on the successful use of t-SNE in a wide range of previous applications in capturing nonlinearities during dimensionality reduction analysis.39, 40, 41
Classification algorithm
Once we obtain the word vectors of the notes, the vectors become the input of a classification model to predict the diagnostic code. We used LR for ICD-10 code prediction considering model interpretability. LR42 applies the sigmoid function in combination with least squares regression for classification. In this study, we used a Python implementation of LR in the scikit-learn package.43 We used regularized LR based on model interpretability. LR is a linear classifier that usually does not suffer from overfitting to specific datasets and regularized LR involves fewer hyperparameters that require tuning. L2 regularization was used in this study and the penalty strength C was tuned based on the average AUROC on the validation set in cohort 1 (Figure S3). A 1:50 class weight was added to deal with the imbalanced cases, since the average prevalence of the eight I-codes was approximately 2%. In this study, eight LR classification models were built for the eight ICD codes independently.
Model assessment and interpretation
To assess the performances of different word vectorization methods, we used AUROC and AUPRC as the metrics to evaluate the word vectorization and the LR models in eight diagnostic code classification tasks. AUROC is the area under the curve with the x axis denoting the false-positive rate and y axis denoting the true-positive rate. AUPRC is the area under the curve with the x axis denoting the recall and y axis denoting the precision. AUROC has been a widely used metric in evaluating binary classifiers without dependence on the decision threshold on predicted class probability. The AUPRC was also used in this study because it is more sensitive to prevalence and can better reflect model performance in an imbalanced dataset.44 For cohort 1, bootstrapping45 was done on the training set 30 times to test the model’s robustness.
As BOW and TF-IDF are directly interpretable word-based vectorization algorithms, to interpret the models, we analyzed the LR coefficients to identify the important words in classification. The top ten most important words for decision were extracted after bootstrapping the training samples in 30 repeats. In each of the bootstrapping experiments, the 30 most important words were extracted as the candidates, and the final top ten most important words were selected based on two metrics: (1) the ranking metric: the sum of rankings of the important words over all bootstrapping results (smaller ranking sums mean higher importance); (2) the coefficient metric: the sum of LR coefficients of the important words over all bootstrapping results (larger coefficient sums mean higher importance).
Because the recorded diagnostic codes can be missing and inaccurate in clinical practice, to test whether it was possible to impute missing ICD-10 codes based on the model predictions, we randomly selected several false-positive cases and analyzed the corresponding notes.
External validation
Next, the model transferability was tested on the MIMIC-III dataset of de-identified health-related data of 40,000 intensive care unit stays at Beth Israel Deaconess Medical Center.46 The MIMIC-III dataset has been regarded as the benchmark dataset in many NLP tasks including time-of-stay prediction and diagnostic code prediction.9,47,48 We directly applied the word vectorization models (BOW, TF-IDF, W2V, W2V_batch, and D2V) and the corresponding LR classifiers trained on the training set of the larger cohort 2 of Stanford notes to predict the diagnostic codes of the discharge summary in MIMIC-III dataset (59,652 notes and 41,127 patients). The MIMIC-III dataset was only involved in model testing, and no model fine-tuning on the MIMIC-III dataset was done. As MIMIC-III uses the ICD-9 as diagnostic codes, the ground truth was set to the corresponding ICD-9 codes of the eight cardiovascular diseases. In this study, we matched the ICD-10 codes to the corresponding ICD-9 codes by matching the three-letter prefix and the highest hierarchy of the ICD-9 code that describes a specific disease. The matched ICD-9-ICD-10 codes49 of the diseases and the prevalence in the MIMIC-III discharge summary are: 410 (I21, acute myocardial infarction), 10.36%; 414 (I25, chronic ischemic heart disease), 26.64%; 416 (I27, pulmonary heart disease), 4.95%; 425 (I42, cardiomyopathy), 3.92%; 427 (I48, atrial fibrillation flutter), 32.16%; 428 (I50, heart failure), 25.98%; 440 (I70, atherosclerosis), 3.61%; 456 (I85, esophageal varices), 1.77%. Proportional Z tests showed statistically significant difference in the prevalence of the eight codes between the cohort 2 training set of Stanford data and the MIMIC-III data.
Acknowledgments
This research used data or services provided by STARR, the Stanford medicine research data repository, a clinical data warehouse containing live Epic data from Stanford Health Care, the University Healthcare Alliance, and Packard Children's Health Alliance clinics and other auxiliary data from hospital applications such as radiology PACS. The STARR platform is developed and operated by Stanford Medicine Research IT team and is made possible by Stanford School of Medicine Research Office. Research reported in this publication was partially funded by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health, R01 EB020527, and R56 EB020527. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work is also supported by the Stanford Department of Bioengineering.
Author contributions
X.Z., M.H.-D., P.M., and O.G. conceived the study. O.G. collected the data. X.Z. carried out the experiments, analyzed the data, and wrote the manuscript. M.H.-D., P.M., and O.G. supervised the work and revised the manuscript. O.G. provided the funding for this work.
Declaration of interests
X.Z., M.-H.-D., and P.M. declare no competing interests. O.G. reports grants from National Institutes of Health, grants from Onc. AI, grants from Lucence Health Inc., and grants from Nividien Inc., outside the submitted work; in addition, O.G. has a Provisional patent filed on related work pending to Stanford.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: June 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2021.100289.
Supplemental information
References
- 1.Wei X., Eickhoff C. Embedding electronic health records for clinical information retrieval. arXiv. 2018 1811.05402. [Google Scholar]
- 2.Sheikhalishahi S., Miotto R., Dudley J.T., Lavelli A., Rinaldi F., Osmani V. Natural language processing of clinical notes on chronic diseases: systematic review. JMIR Med. Inform. 2019;7:e12239. doi: 10.2196/12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi E., Bahadori M.T., Schuetz A., Stewart W.F., Sun J. Machine Learning for Healthcare Conference. PMLR; 2016. Doctor AI: predicting clinical events via recurrent neural networks; pp. 301–318. [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Wang L., Rastegar-Mojarad M., Moon S., Shen F., Afzal N., Liu S., Zeng Y., Mehrabi S., Sohn S., Liu H. Clinical information extraction applications: a literature review. J. Biomed. Inform. 2018;77:34–49. doi: 10.1016/j.jbi.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miotto R., Li L., Kidd B.A., Dudley J.T. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci. Rep. 2016;6:26094. doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen P.B., Jensen L.J., Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat. Rev. Genet. 2012;13:395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein B.A., Navar A.M., Pencina M.J., Ioannidis J. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J. Am. Med. Inform. Assoc. 2017;24:198–208. doi: 10.1093/jamia/ocw042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn L., Eickhoff C. Implicit negative feedback in clinical information retrieval. arXiv. 2016 1607.03296. [Google Scholar]
- 9.Mullenbach J., Wiegreffe S., Duke J., Sun J., Eisenstein J. Explainable prediction of medical codes from clinical text. arXiv. 2018 1802.05695. [Google Scholar]
- 10.Osler T.M., Glance L.G., Cook A., Buzas J.S., Hosmer D.W. A trauma mortality prediction model based on the ICD-10-CM lexicon: TMPM-ICD10. J. Trauma Acute Care Surg. 2019;86:891–895. doi: 10.1097/TA.0000000000002194. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization . World Health Organization; 2008. ICD-10 International Statistical Classification of Diseases and Related Health Problems. [Google Scholar]
- 12.McCarthy C., Murphy S., Cohen J.A., Rehman S., Jones-O’Connor M., Olshan D.S., Singh A., Vaduganathan M., Januzzi J.L., Wasfy J.H. Misclassification of myocardial injury as myocardial infarction: implications for assessing outcomes in value-based programs. JAMA Cardiol. 2019;4:460–464. doi: 10.1001/jamacardio.2019.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang T.E., Lichtman J.H., Goldstein L.B., George M.G. Accuracy of ICD-9-CM codes by hospital characteristics and stroke severity: Paul Coverdell national acute stroke program. J. Am. Heart Assoc. 2016;5:e003056. doi: 10.1161/JAHA.115.003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein L.B. Accuracy of ICD-9-CM coding for the identification of patients with acute ischemic stroke: effect of modifier codes. Stroke. 1998;29:1602–1604. doi: 10.1161/01.str.29.8.1602. [DOI] [PubMed] [Google Scholar]
- 15.Horsky J., Drucker E.A., Ramelson H.Z. American Medical Informatics Association; 2017. Accuracy and completeness of clinical coding using ICD-10 for ambulatory visits; pp. 912–920. (AMIA Annual Symposium Proceedings). [PMC free article] [PubMed] [Google Scholar]
- 16.Jones K.S. A statistical interpretation of term specificity and its application in retrieval. J. Doc. 1972;28:11–21. [Google Scholar]
- 17.Garcelon N., Neuraz A., Salomon R., Bahi-Buisson N., Amiel J., Picard C., Mahlaoui N., Benoit V., Burgun A., Rance B. Next generation phenotyping using narrative reports in a rare disease clinical data warehouse. Orphanet J. Rare Dis. 2018;13:85. doi: 10.1186/s13023-018-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagliardini M., Gupta P., Jaggi M. Unsupervised learning of sentence embeddings using compositional n-gram features. arXiv. 2017 1703.02507. [Google Scholar]
- 19.Wieting J., Bansal M., Gimpel K., Livescu K. Towards universal paraphrastic sentence embeddings. arXiv. 2015 1511.08198. [Google Scholar]
- 20.Arora S., Liang Y., Ma T. A simple but tough-to-beat baseline for sentence embeddings. In: Bengio Y., LeCun Y., editors. 5th International Conference on Learning Representations. ICLR; 2017. [Google Scholar]
- 21.Rishivardhan K., Kayalvizhi S., Thenmozhi D., Sachin Krishan T. Transformers in semantic indexing of clinical codes. In: Cappellato L., Eickhoff C., Ferro N., Névéol A., editors. Proceedings of the Conference and Labs of the Evaluation Forum CEUR Workshop. CLEF; 2020. pp. 1–6. [Google Scholar]
- 22.Lu Z., Leen T.K. Advances in Neural Information Processing Systems 17 (NIPS 2004) 2005. Semi-supervised learning with penalized probabilistic clustering; pp. 849–856. [Google Scholar]
- 23.Zhan X., Guan X., Wu R., Wang Z., Wang Y., Luo Z. Proceedings of the IET Doctoral Forum on Biomedical Engineering, Healthcare, Robotics and Artificial Intelligence 2018 (BRAIN 2018) IET; 2018. Online conformal prediction for classifying different types of herbal medicines with electronic noise. [DOI] [Google Scholar]
- 24.Zhan X., Wang Z., Yang M., Luo Z., Wang Y., Li G. An electronic nose-based assistive diagnostic prototype for lung cancer detection with conformal prediction. Measurement. 2020;158:107588. [Google Scholar]
- 25.Liu H., Bielinski S.J., Sohn S., Murphy S., Wagholikar K.B., Jonnalagadda S.R., Ravikumar K.E., Wu S.T., Kullo I.J., Chute C.G. An information extraction framework for cohort identification using electronic health records. AMIA Jt Summits Transl. Sci. Proc. 2013;2013:149. [PMC free article] [PubMed] [Google Scholar]
- 26.Wen A., Fu S., Moon S., El Wazir M., Rosenbaum A., Kaggal V.C., Liu S., Sohn S., Liu H., Fan J. Desiderata for delivering NLP to accelerate healthcare AI advancement and a Mayo Clinic NLP-as-a-service implementation. NPJ Digit. Med. 2019;2:130. doi: 10.1038/s41746-019-0208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheerla A., Gevaert O. Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics. 2019;35:i446–i454. doi: 10.1093/bioinformatics/btz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C., Cintra M., Brennan K., Zhou M., Colevas A.D., Fischbein N., Zhu S., Gevaert O. Development and validation of radiomic signatures of head and neck squamous cell carcinoma molecular features and subtypes. EBioMedicine. 2019;45:70–80. doi: 10.1016/j.ebiom.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee P., Zhou M., Lee E., Schicht A., Balagurunathan Y., Napel S., Gillies R., Wong S., Thieme A., Leung A., Gevaert O. A shallow convolutional neural network predicts prognosis of lung cancer patients in multi-institutional computed tomography image datasets. Nat. Mach. Intelligence. 2020;2:274–282. doi: 10.1038/s42256-020-0173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Q., Zhan X., Zhou Z., Li Y., Xie P., Zhang S., Li X., Yu Y., Zhou C., Zhang L., Gevaert O. AI-based analysis of CT images for rapid triage of COVID-19 patients. NPJ Digital Med. 2021;4:75. doi: 10.1038/s41746-021-00446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehurek R., Sojka P. Software framework for topic modelling with large corpora. In: Witte R., editor. Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks. NLP Frameworks; 2010. pp. 45–50. [Google Scholar]
- 32.Porter M.F. An algorithm for suffix stripping. Program. 1980;14:130–137. [Google Scholar]
- 33.Bird S., Klein E., Loper E. O’Reilly Media, Inc.; 2009. Natural Language Processing with Python: Analyzing Text with the Natural Language Toolkit. [Google Scholar]
- 34.Harris Z.S. Distributional structure. Word. 1954;10:146–162. [Google Scholar]
- 35.Mikolov T., Sutskever I., Chen K., Corrado G., Dean J. Distributed representations of words and phrases and their compositionality. arXiv. 2013 1310.4546. [Google Scholar]
- 36.Zhang Y., Chen Q., Yang Z., Lin H., Lu Z. BioWordVec, improving biomedical word embeddings with subword information and MeSH. Sci. Data. 2019;6:52. doi: 10.1038/s41597-019-0055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Q., Mikolov T. 31st International Conference on Machine Learning. Vol. 32. 2014. Distributed representations of sentences and documents; pp. 1188–1196. (PMLR). [Google Scholar]
- 38.Van der Maaten L., Hinton G. Visualizing data using t-SNE. J. Machine Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 39.Birjandtalab J., Pouyan M.B., Nourani M. 2016 IEEE-EMBS International Conference on Biomedical and Health Informatics (BHI) IEEE; 2016. Nonlinear dimension reduction for EEG-based epileptic seizure detection. pp. 595–598. [Google Scholar]
- 40.Liu L., Zhan X., Wu R., Guan X., Wang Z., Zhang W., Wang Y., Luo Z., Li G. Boost AI power: data augmentation strategies with unlabelled data and conformal prediction, a case in alternative herbal medicine discrimination with electronic nose. arXiv. 2021 2102.03088. [Google Scholar]
- 41.Li W., Cerise J.E., Yang Y., Han H. Application of t-SNE to human genetic data. J. Bioinform. Comput. Biol. 2017;15:1750017. doi: 10.1142/S0219720017500172. [DOI] [PubMed] [Google Scholar]
- 42.Cox D.R. The regression analysis of binary sequences. J. R. Stat. Soc. Ser. B (Methodol.) 1958;20:215–232. [Google Scholar]
- 43.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 44.Davis J., Goadrich M. The relationship between Precision-Recall and ROC curves. In: Cohen W., Moore A., editors. Proceedings of the 23rd International Conference on Machine Learning. Association for Computing Machinery; 2006. pp. 233–240. [Google Scholar]
- 45.Efron B. Bootstrap methods: another look at the jackknife. In: Kotz S., Johnson N.L., editors. Breakthroughs in Statistics. Springer; 1992. pp. 569–593. [Google Scholar]
- 46.Johnson A.E., Pollard T.J., Shen L., Li-Wei H.L., Feng M., Ghassemi M., Moody B., Szolovits P., Celi L.A., Mark R.G. MIMIC-III, a freely accessible critical care database. Sci. Data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Major V., Tanna M.S., Jones S., Aphinyanaphongs Y. American Medical Informatics Association; 2016. Reusable filtering functions for application in ICU data: a case study; pp. 844–853. (AMIA Annual Symposium Proceedings). [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J., Osorio C., Sy L.W. An empirical evaluation of deep learning for ICD-9 code assignment using MIMIC-III clinical notes. Comput. Methods Programs Biomed. 2019;177:141–153. doi: 10.1016/j.cmpb.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 49.Slee V.N. Centers for Disease Control and Prevention; 1978. The International Classification of Diseases: Ninth Revision (ICD-9) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and code used in this study are not shareable, as the data concern patient information. Please contact the corresponding author to discuss remote access to the data in a de-identified manner.