Abstract
Three-dimensional cell cultures (“organoids”) promise to better recapitulate native tissue physiology than traditional 2D cultures and are becoming increasingly interesting for disease modeling and compound screening efforts. While a number of protocols for the generation of neural organoids have been published, most protocols require extensive manual handling and result in heterogeneous aggregates with high sample-to-sample variation, which can hinder screening-based strategies. We have now developed a fast and efficient protocol for the generation and maintenance of highly homogeneous and reproducible midbrain organoids. The protocol is streamlined for use in fully automated workflows but can also be performed manually without the need for highly specialized equipment. It relies on the aggregation of small molecule neural precursor cells (smNPCs) in standard 96-well V-bottomed plates under static culture conditions without cumbersome matrix embedding. The result is ready-to-assay uniform 3D human midbrain organoids available in freely scalable quantities for downstream analyses in 3D cell culture.
Graphic abstract:
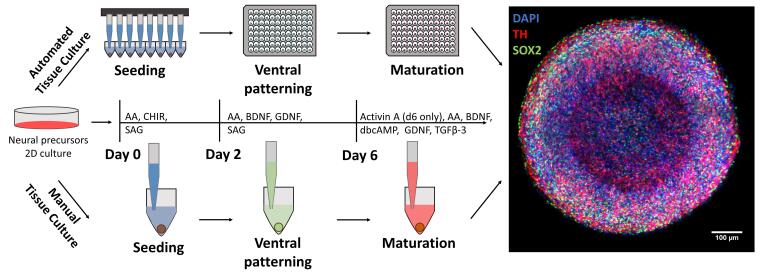
Automated midbrain organoid generation workflow and timeline
Keywords: Organoids, Midbrain, Stem cells, Neuroscience, Drug development, Automation, High Throughput Screening (HTS)
Background
While classical preclinical model systems including 2D cell culture and animal models have driven our understanding of human biology and disease, there is increasing evidence of human-specific processes that elude these standard model systems. Particularly, the human brain is unique in its structure and physiology and has proven difficult to approximate in research settings ( Kim et al., 2020 ). Organoids, i.e., stem cell-derived, self-organized, three-dimensional microtissues, may mimic aspects of human organ structure and function better than previous model systems ( Sato et al., 2009 ; Lancaster et al., 2013 ; Takasato et al., 2015 ; McCracken et al., 2014 ; Dye et al., 2015 ), thus promising to improve our understanding of uniquely human processes (Fatehullah and Barker 2016; Dutta et al., 2017 ).
Over recent years, different groups have focused on mimicking specific brain regions to better understand their tissue biology during development and disease. One region of specific interest is the midbrain because of its prominent early involvement in Parkinson’s disease, the second most common neurodegenerative disorder affecting 2-3% of individuals aged 65 years or older ( Poewe et al., 2017 ). While a number of different midbrain organoid systems have been developed ( Jo et al., 2016 ; Qian et al., 2016 ; Monzel et al., 2017 ), the high degree of homogeneity, reproducibility, and scale-up required for disease modeling and drug development have proven challenging.
We recently published the generation of automated midbrain organoids (AMOs), which were specifically developed to overcome these challenges ( Renner et al., 2020 ). AMOs are highly reproducible and homogeneous with regard to their size, morphology, gene and protein expression, functionality (i.e., neural activity), and response to compounds. They are generated from small molecule neural precursor cells (smNPCs), a cell type optimized for easy, small molecule-based handling as well as fast and robust neural differentiation ( Reinhardt et al., 2013 ). Combined with an organoid generation workflow that does not require any manual intervention (e.g., matrix embedding), this makes our protocol easily reproducible and results in highly homogeneous midbrain organoids. Thus, AMOs are ideally suited for all applications that require large numbers of comparable samples with clearly defined and predictable parameters, most notably compound screening. However, many other applications including disease modeling and developmental biology can also greatly benefit from homogeneous starting conditions without the need for industry-scale sample numbers. A predictable 3D model like AMOs facilitates the discrimination between normal biological variance and the influence of e.g., a single point mutation in a model for a hereditary disease. Thus, we developed the protocol to also be fully compatible with manual pipetting and standard lab equipment, making it available and useful to a large audience.
While the general workflow described herein can be applied to a number of different organoid protocols and starting cell types, we note that smNPCs are not capable of forming forebrain organoids. smNPCs are restricted to mid- and hindbrain fates, including the generation of motor neurons. For details, please see ( Reinhardt et al., 2013 ). Moreover, smNPCs do not form neural rosettes. Our midbrain organoids are optimized for homogeneity to facilitate screening strategies at the cost of complexity. If a strong internal tissue polarity is desired, we recommend starting the culture with other, less restricted cell types, including pluripotent stem cell (PSC)-derived NPCs of an earlier developmental stage than smNPCs or even PSCs at the risk of increasing resulting organoid heterogeneity.
Materials and Reagents
Screw cap tubes (Sarstedt, catalog numbers: 62.547.254 [50 ml] and 62.554.502 [15 ml])
Serological pipets (Falcon, catalog numbers: 356543 [5 ml], 356551 [10 ml], and 356525 [25 ml])
Pipet tips (StarLab, catalog numbers: S1120-1810 [20 μl], S1120-8810 [200 μl], and S1122-1830 [1,000 μl])
Standard tissue culture-treated 6-well plates (Sarstedt, catalog number: 83.3920)
Conical/V-bottomed 96-well plates (Thermo Fisher, catalog number: 277143)
Vacuum filtration system (Corning, catalog number: 431097)
Optional: Biomek liquid handler pipette tips AP96 P250 (Beckman Coulter, catalog number: 717252)
Small molecule neural precursor cells [smNPCs ( Reinhardt et al., 2013 )]
KnockOut (KO) DMEM/F-12 (Thermo Fisher, catalog number: 12660012)
Matrigel (BD Biosciences, catalog number: 354263)
DMEM/F-12 (Thermo Fisher, catalog number: 11320033)
Neurobasal medium (Thermo Fisher, catalog number: 21103-049)
B27 supplement without vitamin A (Thermo Fisher, catalog number: 12587010)
N2 supplement (Thermo Fisher, catalog number: 17502-048)
Penicillin-Streptomycin (Sigma-Aldrich, catalog number: P4333)
GlutaMAX (Thermo Fisher, catalog number: 35050061)
Bovine Serum Albumin (BSA, Thermo Fisher, catalog number: 15260037)
Human Serum Albumin (HSA, Biological Industries, catalog number: 05-720-1B)
Accutase (Sigma-Aldrich, catalog number: A6964)
Polyvinyl Alcohol (PVA; Sigma-Aldrich, catalog number: 363170)
Ascorbic acid (Sigma-Aldrich, catalog number: A4544)
CHIR-99021 (Biomol, catalog number: Cay13122)
Smoothened Agonist (SAG; Cayman Chemical, catalog number: Cay11914)
Brain Derived Neurotrophic Factor (BDNF; PeproTech, catalog number: 450-02)
Glial cell line-Derived Neurotrophic Factor (GDNF; PeproTech, catalog number: 450-10)
Activin A (PeproTech, catalog number: 120-14E)
Transforming Growth Factor Beta 3 (TGF-β3; PeproTech, catalog number: 100-36E)
N6,2’-O-Dibutyryladenosine 3’,5’-cyclic monophosphate (dbcAMP; Sigma-Aldrich, catalog number: D0627)
Basal medium (see Recipes)
smNPC medium (see Recipes)
Aggregation medium (see Recipes)
Patterning medium (see Recipes)
Maturation medium (days 6-X) (see Recipes)
Split medium (see Recipes)
Equipment
Water bath
Humidified CO2 incubator
Optional: CO2 incubator connected to an automated liquid handling system (ALHS)
Sterile tissue/cell culture hood
Fridge and freezers (4°C, -20°C, and -80°C)
Centrifuge (e.g., Eppendorf, model: Centrifuge 5702)
Stereomicroscope with camera (e.g., Leica MZ10 F [microscope] and Leica DFC425 C [camera], Leica Microsystems)
Mechanical pipets
Optional: Multichannel mechanical pipets
Optional: Automated liquid handling system (e.g., Biomek FXP Laboratory Automation Workstation, Beckman Coulter)
Software
Leica application suite v. 4.8 (LAS, Leica Microsystems)
Fiji/ImageJ ( Schindelin et al., 2012 )
Optional: Biomek Software v. 3.3 (controlling the automated liquid handler, Beckman Coulter)
Procedure
-
Preparation of matrigel-coated plates for 2D smNPC cell culture
Note: Before the final coating step, all plasticware and solutions containing matrigel must be kept on ice to prevent premature gelation during handling.
-
Dilute matrigel 1:100 in cold (4°C) KnockOut DMEM/F-12
Note: We recommend keeping a frozen stock of 1:4 diluted matrigel in KnockOut DMEM/F-12 and further diluting this 1:25 in KnockOut DMEM/F-12 immediately before use.
Quickly add 1 ml 1:100 matrigel solution per well of a 6-well plate to cover the entire bottom of the well. Distribute the solution evenly across the surface of the well by gently rocking the plate. If you imagine a coordinate system with the XY plane parallel to the ground, first tilt the plate along the Y-axis about ten degrees in either direction 5 times, then tilt the plate along the X-axis about ten degrees in either direction 5 times or until the entire plate bottom is covered by matrigel solution. Seal the plate with Parafilm to prevent evaporation.
-
Incubate at room temperature overnight. It is possible to store the plates at 4°C for approximately 2 weeks.
Note: If plates are needed on the same day, it is also possible to incubate them at 37°C for 3 h and use them immediately.
-
-
smNPC culture
Note: Generally, smNPCs are seeded and cultured in 2 ml medium per well in 6-well plates. As the cells become more confluent, carefully monitor the cell culture medium. If the pH indicator shows acidification (i.e., the commonly used phenol red will turn yellow), refresh medium every day and/or increase media volume by 50-100% up to 4 ml per well. Figure 1 includes examples of smNPC cultures at different stages as a reference.
Prepare smNPC medium (see Recipes, Table 2) from basal medium (see Recipes, Table 1) and prewarm it to 37°C in a water bath. Small molecules should be added to the medium immediately before use. Avoid heat-cycling the medium once reconstituted.
Aspirate old medium.
Add 1 ml accutase per well of a 6-well plate and incubate at 37°C for 10-15 min (check after 7-8 min whether cells are detached). Cells should detach without mechanical agitation.
Resuspend the cells 2-3 times as gently as possible using a 1,000 μl manual pipette to maximize cell harvest and to disperse small remaining aggregates. Transfer suspension into a tube with 6 ml split medium (see Recipes, Table 6) to dilute and inactivate accutase.
Centrifuge for 2 min at 220 × g.
Aspirate the supernatant. Take care not to aspirate the pellet.
Resuspend the pellet in smNPC medium and seed the cells at a ratio of approximately 1:10-1:20 (depending on cell line and experience).Note: Aspirate the supernatant from the matrigel-coated wells before adding the cell solution.
Distribute cells evenly across the surface of the well by gently rocking the plate in an XY-motion (see Step A2). Avoid swirling/circular motions as these tend to concentrate the cells in the center of the well. Place plate into a 37°C CO2 incubator.
Exchange medium every other day and maintain cells until they reach approximately 90-100% confluence before splitting them again/using them for organoid generation. See Figure 1d.
-
Organoid generation and maintenance
Note: See Figure 2 for representative brightfield images of organoids at different ages.
Day 0
Detach smNPCs from the matrigel-coated cell culture plate using accutase as described in Steps B3 and B4.
-
Collect cells in split medium (see Recipes, Table 6), count them, and transfer the appropriate number of cells (see note below) to a new Falcon tube. Then spin them down at 220 × g for 2 min.
Note on cell numbers and volumes: Seeding requires 9,000 cells in 150 μl medium per well of a 96-well plate. This is equivalent to: 9,000 cells/0.15 ml = 60,000 cells/ml. Thus, prepare the cell suspension at a concentration of 60,000 cells/ml. Make sure to prepare 10-20% surplus volume of the cell suspension to account for pipetting inaccuracies/dead volume.
-
Resuspend the pellet in the appropriate amount of aggregation medium (see Recipes, Table 3).
Note: Step 4 and all following feeding steps can be done either by hand or using the automated liquid handling system (ALHS)/pipetting robot.
-
Seed 9,000 cells in 150 μl aggregation medium per well of a 96-well V-bottomed plate and incubate at 37°C 5% CO2 for 48 h.
Day 2
Note: All media changes involving organoids should be performed carefully to avoid damaging the samples or accidentally removing them from the wells. To facilitate manual media aspiration, we recommend slightly tilting the plate and performing all liquid handling by pipetting (not by vacuum pumps or similar devices). Leave a small amount of old medium (≤ 10 μl) in the well so as to not damage the organoids by extensive direct contact with a pipet tip. A small number of organoids (< ca. 50) can be handled with standard single channel mechanical pipets. For larger numbers of samples (more than half a 96-well plate), we recommend using multichannel pipets (in our experience, a standard mechanical pipet with 200-μl capacity works well). Finally, further scale-up for high-throughput applications makes it feasible to use a pipetting robot / automated liquid handler. The same precautions as for manual handling apply – it is advisable to leave a small amount of old medium (≤ 10 μl) per well rather than damage organoids by more ambitious removal of liquids. While dependent on the exact setup, here, dead volumes are generally higher than for manual pipetting, and we advise preparing 5-10 ml surplus of all liquids.
-
1
Replace medium with 150 μl patterning medium per well (see Recipes, Table 4).
Day 4
-
2
Replace medium with 150 μl patterning medium per well.
Day 6
-
3
Replace medium with 150 μl maturation medium plus activin A per well (see Recipes, Table 5).
Day 8 + X
-
4
Replace medium with 150 μl maturation medium (same as day 6 without activin A) per well.
-
5
Feed aggregates every 2 days with the same medium until they reach the desired age for analysis/follow-up experiments.
Note: Maturation dynamics vary from cell line to cell line and need to be tested empirically by immunostaining or qPCR. Generally, organoids contain a large number of MAP2+ neurons by day 30. Glial cells including GFAP+ astrocytes arise later, around day 40 onward. For more details, please refer to the detailed analyses in our previous publication (Renner et al., 2020).
Note: Sometimes a few dead cells can be found at the bottom of the well (especially at earlier stages. Here, see day 15). As long as the aggregates themselves maintain their structure, this is not an issue. The dead cells will be washed away during the next feeding procedure.
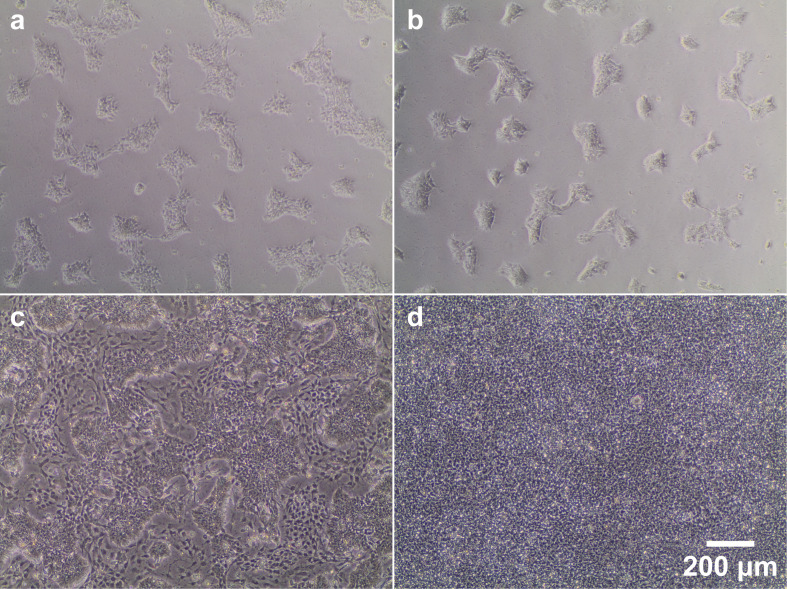
Figure 1. Phase contrast images of 2D smNPC culture at different stages of confluence and differentiation.
(a/b) Representative images illustrating the density of smNPCs 2 days after splitting at either 1:10 (a) or 1:20 (b). (c) Example image of a low-quality smNPC culture contaminated by large numbers of differentiated cells (very dark and large, located between colonies). Partially differentiated cultures may be rescued by maintaining high splitting ratios (at least 1:20) over several passages, as smNPCs usually proliferate faster than differentiated cells. These types of culture should not be used for generating organoids. (d) smNPCs at the optimal confluence for organoid generation or splitting.
Table 2. Composition of smNPC medium.
| Reagent | Supplier | Dilution factor | Product no. | Stock conc. | Final conc. |
|---|---|---|---|---|---|
| Basal medium | |||||
|
CHIR-99021 (in DMSO) |
Biomol | 2,000 | Cay13122 | 6 mM | 3 μM |
| SAG (in DMSO) | Cayman Chemical | 20,000 | Cay11914 | 10 mM | 0.5 μM |
Table 1. Composition of basal medium.
| Reagent | Amount for 100 ml | Supplier | Product no. | Dilution factor | Stock conc. |
|---|---|---|---|---|---|
| DMEM/F-12 | 48.6 ml | Thermo Fisher | 11320033 | 2 | |
| Neurobasal | 48.6 ml | Thermo Fisher | 21103049 | 2 | |
| N2 | 250 μl | Thermo Fisher | A1370701 | 400 | |
| B27 without Vit A | 500 μl | Thermo Fisher | 12587010 | 200 | |
|
Ascorbic Acid (in water) |
50 μl | Sigma-Aldrich | A4544 | 2,000 | 200 mM |
| PenStrep | 1,000 μl | Sigma-Aldrich | P4333 | 100 | |
| GlutaMax | 1,000 μl | Thermo Fisher | 35050061 | 100 |
Table 6. Composition of split medium.
| Reagent | Amount for 50 ml | Supplier | Product no. | Dilution factor | Stock conc. |
|---|---|---|---|---|---|
| DMEM/F-12 | 49.3 ml | Thermo Fisher | 11320033 | ||
| BSA | 667 μl | Thermo Fisher | 15260037 | 75 | 7.5% |
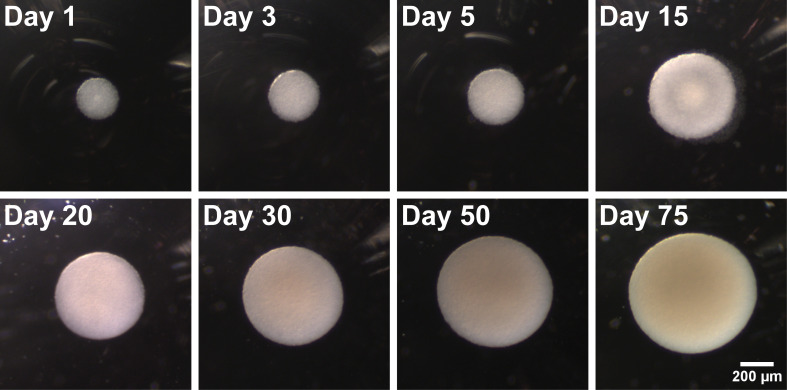
Figure 2. Representative brightfield images illustrating organoid growth and morphology over time.
Seeded on day 0 as a single cell smNPC suspension, cells aggregate to a spherical shape within one day. Over time, they maintain their homogeneous round shape and grow in size.
Table 3. Composition of aggregation medium.
| Reagent | Amount for 50 ml | Supplier | Dilution factor | Product no. | Stock conc. | Final conc. |
|---|---|---|---|---|---|---|
| smNPC medium | 45 ml | |||||
| Polyvinyl alcohol (PVA, in water)* | 5 ml | Sigma-Aldrich | 10 | 363170 | 4% (w/v) | 0.4% (w/v) |
Table 4. Composition of patterning medium.
| Reagent | Supplier | Dilution factor | Product no. | Stock conc. | Final conc. |
|---|---|---|---|---|---|
| Basal medium | |||||
|
BDNF (in PBS + 0.1% (v/v) HSA) |
PeproTech | 10,000 | 450-02 | 10 μg/ml | 1 ng/ml |
|
GDNF (in PBS + 0.1% (v/v) HSA) |
PeproTech | 10,000 | 450-10 | 10 μg/ml | 1 ng/ml |
| SAG (in DMSO) | Cayman Chemical | 10,000 | Cay11914 | 10 mM | 1 μm |
Table 5. Composition of maturation medium.
| Reagent | Supplier | Dilution factor | Product no. | Stock conc. | Final conc. |
|---|---|---|---|---|---|
| Basal medium | |||||
|
Activin A (day 6 only!) (in PBS + 0.1% (v/v) HSA) |
PeproTech | 2,000 | 120-14E | 10 μg/ml | 5 ng/ml |
|
BDNF (in PBS + 0.1% (v/v) HSA) |
PeproTech | 5,000 | 450-02 | 10 μg/ml | 2 ng/ml |
|
GDNF (in PBS + 0.1% (v/v) HSA) |
PeproTech | 5,000 | 450-10 | 10 μg/ml | 2 ng/ml |
|
TGFβ-3 (in PBS + 0.1% (v/v) HSA) |
PeproTech | 1,000 | 100-36E | 1 μg/ml | 1 ng/ml |
| dbcAMP (in water) | Sigma-Aldrich | 500 | D0627 | 50 mM | 100 μm |
Data analysis
During culture, monitor organoids regularly for normal growth and morphology using a stereomicroscope (see Figure 2 as a reference).
For a number of other possible downstream analyses and QC steps, including e.g., qPCR, whole mount staining and clearing, organoid viability, and functional analyses (calcium imaging and multi-electrode arrays), please refer to our previous publication ( Renner et al., 2020 ).
Notes
As outlined above, this protocol facilitates either manual or fully automated sample generation. Since each organoid is generated in a separate well and remains separate from all other organoids from seeding to maturation, they represent true biological replicates. Starting with a single cell suspension rather than partially dissociated colonies also maximizes homogeneity between different wells and plates of the same batch. Generating one organoid per well also greatly simplifies the downstream application of compounds and subsequent analysis. In our runs, we routinely generated organoid batches with a coefficient of variation (i.e., standard deviation divided by the mean) of less than 5% with regard to the area of their largest cross-section and around 5% for their viability. All these factors render AMOs ideally suitable for compound screening and toxicity testing since scale-up of fully automated workflows is comparably trivial. While many scientific lines of questioning (e.g., mechanistic analyses) often do not require sample numbers in the hundreds or thousands, the high homogeneity and reproducibility of AMOs still offer advantages, and we recommend testing our protocol with manual pipetting using a single or multichannel pipet.
Recipes
Note: All media without growth factors and small molecules can be stored for up to 2 weeks at 4°C in the dark. Growth factors and small molecules should always be added immediately before use.
Basal medium (Table 1)
smNPC medium (Table 2)
-
Aggregation medium (Table 3)
*PVA is purchased as a crystalline powder. We recommend preparing a 4% stock solution, which can be stored at 4°C for several months. To prepare the stock solution, dissolve PVA in water by heating to 60-70°C overnight with constant stirring. After the solution has cooled down, sterile filter it with e.g., a vacuum filtration system before use.
Patterning medium (days 2-4) (Table 4)
Maturation medium (days 6-X) (Table 5)
Split medium (Table 6)
Acknowledgments
This work was funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No [669168]). HR is supported by the International Max Planck Research School - Molecular Biomedicine, Münster, Germany. This protocol is based on our previous publication ( Renner et al., 2020 ).
Competing interests
The work presented here is the subject of the patent application EP 18 19 2698.0-1120 to the European Patent Office, where the authors are inventors.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Dutta D., Heo I. and Clevers H.(2017). Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol Med 23(5): 393-410. [DOI] [PubMed] [Google Scholar]
- 2. Dye B. R., Hill D. R., Ferguson M. A., Tsai Y. H., Nagy M. S., Dyal R., Wells J. M., Mayhew C. N., Nattiv R., Klein O. D., White E. S., Deutsch G. H. and Spence J. R.(2015). In vitro generation of human pluripotent stem cell derived lung organoids . Elife 4: e05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fatehullah A., Tan S. H. and Barker N.(2016). Organoids as an in vitro model of human development and disease. Nat Cell Biol 18(3): 246-254. [DOI] [PubMed] [Google Scholar]
- 4. Jo J., Xiao Y., Sun A. X., Cukuroglu E., Tran H. D., Goke J., Tan Z. Y., Saw T. Y., Tan C. P., Lokman H., Lee Y., Kim D., Ko H. S., Kim S. O., Park J. H., Cho N. J., Hyde T. M., Kleinman J. E., Shin J. H., Weinberger D. R., Tan E. K., Je H. S. and Ng H. H.(2016). Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 19(2): 248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim J., Koo B. K. and Knoblich J. A.(2020). Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol 21(10): 571-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lancaster M. A., Renner M., Martin C. A., Wenzel D., Bicknell L. S., Hurles M. E., Homfray T., Penninger J. M., Jackson A. P. and Knoblich J. A.(2013). Cerebral organoids model human brain development and microcephaly. Nature 501(7467): 373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McCracken K. W., Cata E. M., Crawford C. M., Sinagoga K. L., Schumacher M., Rockich B. E., Tsai Y. H., Mayhew C. N., Spence J. R., Zavros Y. and Wells J. M.(2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516(7531): 400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Monzel A. S., Smits L. M., Hemmer K., Hachi S., Moreno E. L., van Wuellen T., Jarazo J., Walter J., Bruggemann I., Boussaad I., Berger E., Fleming R. M. T., Bolognin S. and Schwamborn J. C.(2017). Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports 8(5): 1144-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poewe W., Seppi K., Tanner C. M., Halliday G. M., Brundin P., Volkmann J., Schrag A. E. and Lang A. E.(2017). Parkinson disease. Nat Rev Dis Primers 3: 17013. [DOI] [PubMed] [Google Scholar]
- 10. Qian X., Nguyen H. N., Song M. M., Hadiono C., Ogden S. C., Hammack C., Yao B., Hamersky G. R., Jacob F., Zhong C., Yoon K. J., Jeang W., Lin L., Li Y., Thakor J., Berg D. A., Zhang C., Kang E., Chickering M., Nauen D., Ho C. Y., Wen Z., Christian K. M., Shi P. Y., Maher B. J., Wu H., Jin P., Tang H., Song H. and Ming G. L.(2016). Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165(5): 1238-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reinhardt P., Glatza M., Hemmer K., Tsytsyura Y., Thiel C. S., Hoing S., Moritz S., Parga J. A., Wagner L., Bruder J. M., Wu G., Schmid B., Ropke A., Klingauf J., Schwamborn J. C., Gasser T., Scholer H. R. and Sterneckert J.(2013). Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One 8(3): e59252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Renner H., Grabos M., Becker K. J., Kagermeier T. E., Wu J., Otto M., Peischard S., Zeuschner D., TsyTsyura Y., Disse P., Klingauf J., Leidel S. A., Seebohm G., Scholer H. R. and Bruder J. M.(2020). A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. Elife 9: e52904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., van Es J. H., Abo A., Kujala P., Peters P. J. and Clevers H.(2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459(7244): 262-265. [DOI] [PubMed] [Google Scholar]
- 14. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P. and Cardona A.(2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takasato M., Er P. X., Chiu H. S., Maier B., Baillie G. J., Ferguson C., Parton R. G., Wolvetang E. J., Roost M. S., Chuva de Sousa Lopes S. M. and Little M. H.(2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526(7574): 564-568. [DOI] [PubMed] [Google Scholar]