Abstract
Cyclic GMP-AMP synthase (cGAS) is a key sensor responsible for cytosolic DNA detection. Here we report that GTPase-activating protein SH3 domain–binding protein 1 (G3BP1) is critical for DNA sensing and efficient activation of cGAS. G3BP1 enhanced DNA binding of cGAS by promoting the formation of large cGAS complexes. G3BP1 deficiency led to inefficient DNA binding by cGAS and inhibited cGAS-dependent interferon (IFN) production. The G3BP1 inhibitor epigallocatechin gal-late (EGCG) disrupted existing G3BP1–cGAS complexes and inhibited DNA-triggered cGAS activation, thereby blocking DNA-induced IFN production both in vivo and in vitro. EGCG administration blunted self DNA–induced autoinflammatory responses in an Aicardi–Goutières syndrome (AGS) mouse model and reduced IFN-stimulated gene expression in cells from a patient with AGS. Thus, our study reveals that G3BP1 physically interacts with and primes cGAS for efficient activation. Furthermore, EGCG-mediated inhibition of G3BP1 provides a potential treatment for cGAS-related autoimmune diseases.
The innate immune system senses danger signals, such as molecular patterns from pathogens or tissue damage, by a variety of germline-encoded pattern-recognition receptors (PRRs)1. The emergence of DNA in cytoplasm represents an important danger signal for pathogen infection and triggers robust immune responses1–3. cGAS is a key intracellular PRR that detects cytosolic microbial DNA or self DNA. The engagement of cGAS by DNA triggers cGAS activation and synthesis of the second messenger 2′ 3′-cyclic GMP-AMP (cGAMP)4. cGAMP binds to the endoplasmic reticulum protein STING (stimulator of interferon genes) and strongly activates the downstream pathway to produce type I IFN and other proinflammatory cytokines5,6.
Although sensing of foreign DNA is a fundamental mechanism for host defense, aberrant activation of cGAS by self DNA is a major cause of several severe autoimmune diseases. For example, the DNA 3′ repair exonuclease TREX1 is responsible for the degradation of cytosolic DNA, and deficiency in TREX1 in cells results in the accumulation of cytosolic DNA, which is believed to drive cGAS-mediated chronic inflammation7. It is noteworthy that loss-of-function mutations of TREX1 have been seen in patients suffering autoimmune diseases such as Aicardi–Goutières syndrome (AGS) and systemic lupus erythematosus8,9. The Trex1-null mice exhibit severe autoimmune responses similar to those observed in patients with AGS7,8. Previous publications showed that deletion of Cgas in Trex1−/− mice fully rescued the disease phenotype of Trex1−/− mice10,11. Thus, inhibition of cGAS provides a potential treatment for these autoimmune diseases.
To understand the regulation of cGAS activation, we identified the cGAS-interacting proteins in cells using liquid chromatography–mass spectrometry (LC–MS). G3BP1 (also known as stress granule assembly factor 1) was consistently identified as one of the coprecipitated proteins with high scores in cGAS pull-down experiments. It is known that G3BP1 is required for normal RNA stress granule (SG) assembly. SGs are cytoplasmic foci enriched with RNAs and proteins when the cell is under stress12. G3BP1 plays an important role in the intracellular RNA-sensing pathway, as RIG-I-like receptor–mediated signaling can be regulated by SGs13. Given the known function of G3BP1 in the RNA-sensing pathway, we suspected that it might also play a role in DNA-sensing pathways. Our data indicate that G3BP1 is critical for DNA sensing and for efficient activation of cGAS through promoting the formation of large cGAS complexes.
Results
G3BP1 is critical for cGAS-mediated IFN production.
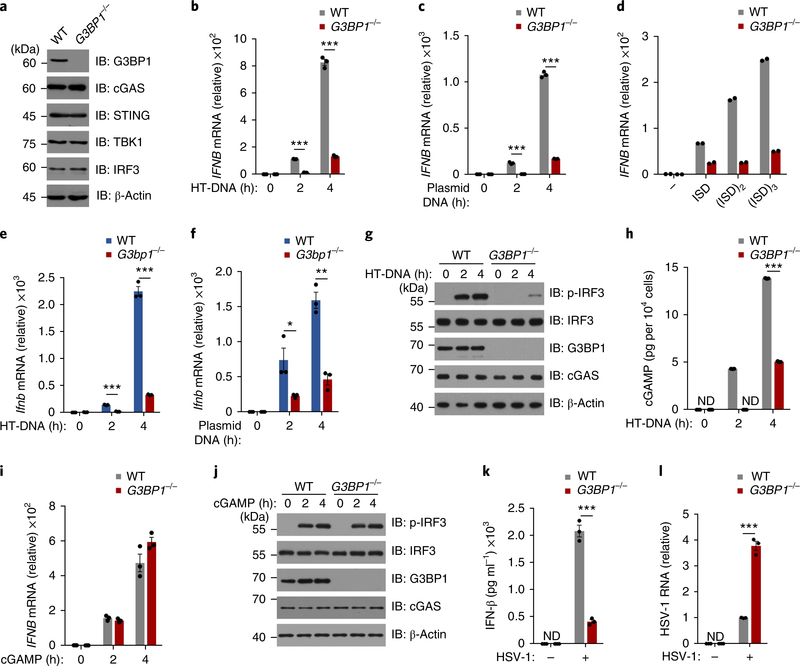
We used CRISPR/Cas9 to delete G3BP1 in U937 cells (human monocytic cell line) (Fig. 1a) and examined the role of G3BP1 in cGAS-mediated type I IFN production by measuring IFN-β expression. Compared with wild-type U937 cells, deficiency in G3BP1 resulted in a severe decrease in IFN-β production induced by different types of intracellular DNAs, including herring testis DNA (HT-DNA) and plasmid DNA (Fig. 1b,c and Supplementary Fig. 1a,b). In contrast, DNA-induced IFNB expression was not appreciably affected (Supplementary Fig. 1c,d) when we deleted the G3BP1 homolog G3BP2 in U937 cells (Supplementary Fig. 1e). We next tested the effect of DNA length using 1–3 concatenated interferon-stimulatory DNA (ISD, a 45-base-pair double-stranded DNA). Although the longer DNA activated cGAS more efficiently, as reported14, we found that G3BP1 was critical for cGAS-mediated DNA sensing regardless of DNA length (Fig. 1d). To further confirm the role of G3BP1 in DNA-induced interferon production, we used G3bp1−/− mice. As the deletion of G3bp1 is embryonically lethal15, we generated mouse embryo fibroblasts (MEFs) from both wild-type and G3bp1−/− embryos (Supplementary Fig. 1f). Consistent with our data in U937 cells, the DNA-induced expression of Ifnb was severely reduced in G3bp1−/− MEFs compared with wild-type controls (Fig. 1e,f and Supplementary Fig. 1g). In response to cytosolic DNA transfection, phosphorylation of interferon-regulatory factor 3 (IRF3) was greatly diminished in G3BP1-deficient cells, compared with that in wild-type cells, indicating that G3BP1 is required for the activation of cGAS/STING signaling (Fig. 1g and Supplementary Fig. 1h–j). Finally, RNAi-mediated G3BP1-knockdown experiments produced similar results (Supplementary Fig. 1k,l). Taken together, these data suggest that G3BP1 is critical for DNA-induced type I IFN production.
Fig. 1 |. G3BP1 is critical for cGAS-mediated type I interferon production.
a, Representative immunoblot (IB) of wild-type (WT) and G3BP1−/− U937 cells with indicated antibodies. b,c, qPCR analysis of IFNB mRNA expression in U937 cells transfected with HT-DNA (2 μg ml−1) (b) or plasmid DNA (2 μg ml−1) (c) for indicated time. d, qPCR analysis of IFNB mRNA expression in U937 cells transfected with DNA of different length (2 μg ml−1) (n = 2 independent experiments). e,f, qPCR analysis of Ifnb mRNA expression in WT and G3bp1−/− MEFs transfected with HT-DNA (0.5 μg ml−1) (e) or plasmid DNA (0.5 μg ml−1) (f) for indicated time. g,j, Immunoblot analysis of U937 cells treated with HT-DNA (g) or cGAMP (1 μg ml−1) (j). h, cGAMP production of HT-DNA-treated WT or G3BP1−/− U937 cells was analyzed by LC–MS/MRM. ND, not detected. i, qPCR analysis of IFNB mRNA expression in cGAMP-treated U937 cells. k,l, ELISA of secreted IFN-β (k) and qPCR analysis of HSV-1 RNA (l) in U937 cells that were untreated (−) or infected with HSV-1 (multiplicity of infection = 1) (+) for 24 h. β-Actin, loading control (a,g,j). *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed t-test (b,c,e,f,h,k,l). Data are representative of three experiments (a–c,e–l). Data are mean ± s.e.m. of triplicate samples in b,c,e,f,h,i,k,l. U937 cells were differentiated with PMA (phorbol 12-myristate 13-acetate) before any treatment, unless stated otherwise.
To further study the activity of cGAS in G3BP1-deficient cells, we established a liquid chromatography–mass spectrometry/multiple reaction monitoring (LC–MS/MRM)-based approach to determine the quantity of cGAMP produced in wild-type and G3BP1-deficient cells (Supplementary Fig. 2a,b). After cytosolic DNA challenge, cGAMP production was significantly reduced in G3BP1−/− U937 cells compared with wild-type cells (Fig. 1h). Similar results were obtained in G3bp1−/− MEFs (Supplementary Fig. 2c). G3BP1 seems to regulate the cGAS/STING pathway upstream of cGAMP production, since cGAMP induced similar expression of IFNB (Fig. 1i and Supplementary Fig. 2d) and IRF3 phosphorylation (Fig. 1j) in both wild-type and G3BP1-deficient cells. This conclusion was further confirmed by using c-di-GMP (Supplementary Fig. 2e), which is another STING activator downstream of cGAS16. Thus, G3BP1 regulates cGAS activation. cGAS is critical for immune defense against DNA viruses, such as herpes simplex virus-1 (HSV-1), or retroviruses, such as human immunodeficiency virus (HIV)17–19. We infected wild-type and G3BP1-deficient U937 cells with HSV-1 and found that the HSV-1-induced IFN-β production in G3BP1-deficient cells was greatly attenuated compared with that in wild-type U937 cells (Fig. 1k and Supplementary Fig. 3a). Consistent with this, we found that while entry of HSV-1 was not affected by G3BP1 deletion (Supplementary Fig. 3b), the abundance of viral RNA and titers in G3BP1-deficient U937 cells at 24 h after infection were much greater than those in wild-type cells (Fig. 1l and Supplementary Fig. 3c,d). We obtained similar results after infection of U937 cells with HIV-1 (Supplementary Fig. 3e). Furthermore, by detecting Toll-like receptor (TLR)-mediated Tnf expression, we showed that G3BP1 deficiency did not inhibit the TLR pathways (Supplementary Fig. 3f–h). Thus, G3BP1 contributes to cGAS activation and cGAS-mediated antiviral defense.
RNA stress granule does not regulate cGAS activation.
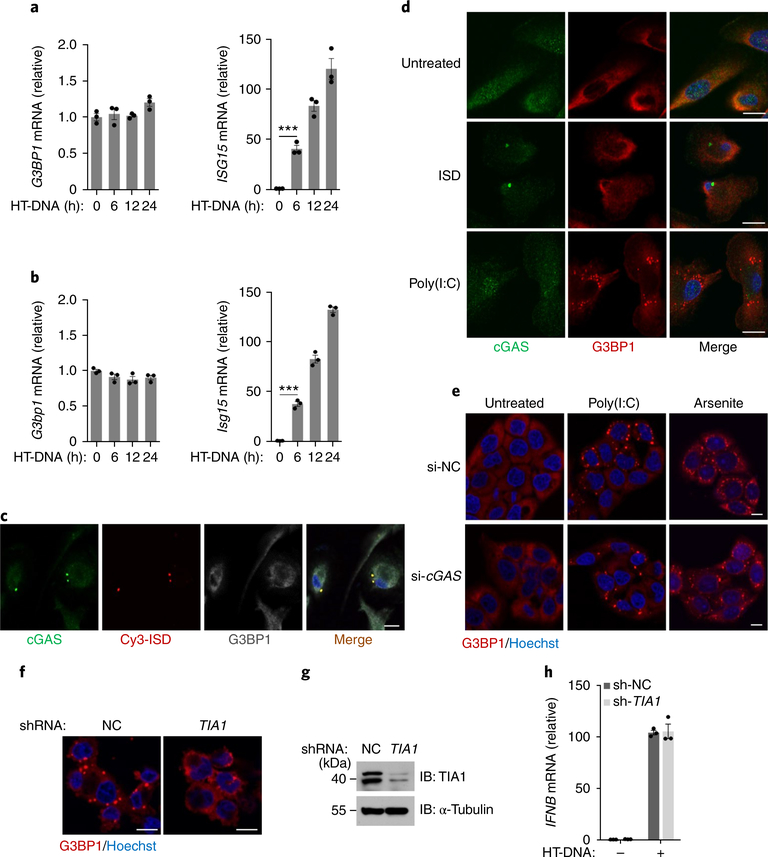
To understand how G3BP1 regulates cGAS activation, we first examined the expression pattern of G3BP1 and found that this protein was ubiquitously expressed in the tissues examined (Supplementary Fig. 4a). We also found that the mRNA expression of this gene was not inducible in response to DNA or interferon treatment (Fig. 2a,b and Supplementary Fig. 4b,c). G3BP1 is known to be required for normal RNA SG assembly13. We then investigated if SGs play a role in the cGAS-mediated DNA-sensing pathway. Using a fluorescence-labeled ISD to transfect cells, we found that DNA transfection did not induce the formation of RNA SGs but did trigger cGAS–DNA aggregation (Fig. 2c). Furthermore, cGAS was not localized on RNA SGs (Fig. 2d and Supplementary Fig. 4d,e), and cGAS deficiency did not affect the formation of SGs (Fig. 2e and Supplementary Fig. 4f,g). We also showed that disruption of the SG formation by depleting T cell–restricted intracellular antigen-1 (TIA1), another critical SG-associated protein20, did not affect DNA-induced cGAS-mediated IFNB expression (Fig. 2f–h). Therefore, the function of G3BP1 in cGAS regulation appears to be distinct from its well-established role in SG. It has also been reported that the autophagy protein Beclin-1 and initiation of autophagy negatively regulate cGAS activation21. However, G3BP1 deficiency did not enhance autophagy or cGAS–Beclin-1 interaction (Supplementary Fig. 4h,i). Thus, G3BP1 regulates cGAS independently of SGs or autophagy.
Fig. 2 |. SGs are not involved in G3BP1-mediated cGAS activation.
a,b, qPCR analysis of indicated mRNA expression in U937 cells (a) or primary MEF cells (b) transfected with HT-DNA for indicated time. ***P < 0.001, two-tailed t-test. c, Representative immunofluorescent staining of cGAS (green), Cy3-labeled ISD (red) and G3BP1 (gray) in human primary macrophages. d, Immunofluorescent staining of cGAS (green) and G3BP1 (red) in human primary macrophages left untreated or treated with ISD (1 μg ml−1) or poly(I:C) (1 μg ml−1) for 2 h. e, Immunofluorescent staining of G3BP1 (red) in HeLa cells transfected with non-targeting control small interfering RNA (si-NC) or CGAS-specific small interfering RNA (si-cGAS), followed by stimulation with poly(I:C) (2 μg ml−1) for 2 h or arsenite (500 μM) for 1 h. f, Representative images of SG formation, indicated by G3BP1 (red), in arsenite-stimulated U937 cells that stably expressed non-targeting control short hairpin RNA (sh-NC) or TIA1-specific short hairpin RNA (sh-TIA1). g, Knockdown of TIA1 in (f) was verified by immunoblotting. α-Tubulin, loading control. h, qPCR analysis of IFNB mRNA expression in U937 transfected with HT-DNA (2 μg ml−1) for 2 h. Hoechst (blue) (c–f), nuclear stain. Scale bars (c–f), 10 μm. Data are representative of three experiments (a–d,f–h) or two experiments (e). Data are mean ± s.e.m. of triplicate samples in a,b,h.
G3BP1 directly binds to cGAS.
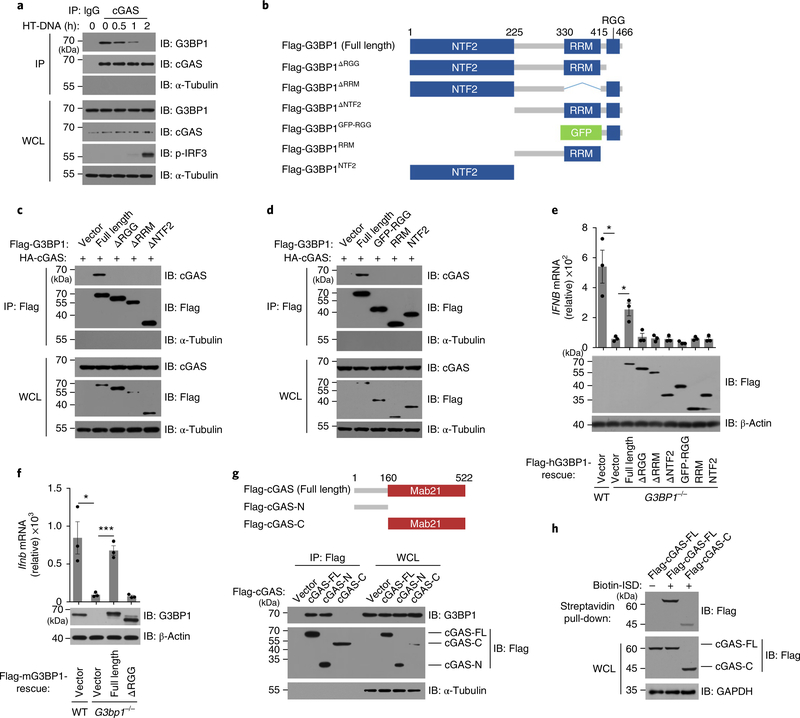
Since G3BP1 did not colocalize with cGAS after DNA transfection, we further investigated the interaction between endogenous cGAS and G3BP1 in response to DNA stimulation. We found that DNA transfection led to the gradual dissociation of cGAS and G3BP1 (Fig. 3a and Supplementary Fig. 5a,b). G3BP1 contains three conserved domains: nuclear transporter factor 2 (NTF2) domain, RNA-recognition module (RRM) and RGG (arginine–glycine–glycine) motif22. We generated several flag-tagged G3BP1 mutants, RGG-deleting (flag-G3BP1ΔRGG), RRM-deleting (flag-G3BP1ΔRRM), NTF2-deleting (flag-G3BP1ΔNTF2), RGG motif (flag-G3BP1RGG), RRM domain (flag-G3BP1RRM) and NTF2 domain (flag-G3BP1NTF2) (Fig. 3b), and found that only intact G3BP1 could bind to cGAS (Fig. 3c,d and Supplementary Fig. 5c,d). Since deletion of any domain of G3BP1 affected its binding to cGAS, we considered the possibility that the domain deletions led to a disordered mutant G3BP1 protein. Indeed, full-length G3BP1, but not any of the mutants, rescued DNA-induced IFNB expression in G3BP1-deficient cells (Fig. 3e,f). Thus, full-length G3BP1 protein is needed for its role in regulating cGAS activation.
Fig. 3 |. The interaction between G3BP1 and cGAS.
a, cGAS–G3BP1 interaction was analyzed by immunoprecipitation with IgG or cGAS antibody in human primary macrophages treated as indicated. b, Schematic drawing of the domains of G3BP1. c,d, The G3BP1–cGAS interaction was analyzed by immunoprecipitation with anti-flag M2 beads in HEK293T cells expressing HA-tagged cGAS and flag-tagged full-length and truncated G3BP1. e, qPCR analysis of IFNB mRNA expression in HT-DNA-treated WT and the indicated rescued cells (U937) (top). The expression of rescued proteins was analyzed by immunoblotting (bottom). f, qPCR analysis of Ifnb mRNA expression in HT-DNA-treated WT and the indicated rescued cells (MEF) (top). Immunoblot analysis of the rescued proteins are shown (bottom). g, Schematic drawing of the domains of cGAS (top). Plasmids of flag-tagged human cGAS as indicated were transiently expressed in HeLa cells. The G3BP1–cGAS interaction was analyzed by immunoprecipitation with anti-flag M2 beads (bottom). h, The DNA-binding capacity of the proteins was analyzed by biotin–ISD pull-down in cell lysates of HEK293T that expressed with flag-tagged cGAS and flag-cGAS-C. WCL, whole cell lysates. IP, immunoprecipitation. β-Actin (e,f), GAPDH (h) and α-Tubulin (a,c,d,g), loading controls. α-Tubulin blots in IP samples indicate the purity of IP (a,c,d,g). *P < 0.05, ***P < 0.001, two-tailed t-test (e,f). All data are representative of three experiments, mean ± s.e.m. of triplicate samples in e,f.
IFN-γ-inducible protein 16 (IFI16) regulates both cGAS-mediated cGAMP production and the cGAS downstream factors kinase TBK1 and transcription factor IRF3 in human macrophages23. IFI16 is not required for cGAMP production but is important for STING activation in keratinocytes24. We found that IFI16 interacted with cGAS following DNA transfection in U937 cells (Supplementary Fig. 5e). In addition, using flag-tagged cGAS mutants (carboxy-terminal truncated cGAS (residues 1–160; flag-cGAS-N) and amino-terminal truncated cGAS (residues 161–522; flag-cGAS-C)) we found that flag-cGAS-N was required for cGAS interaction with G3BP1 (Fig. 3g). The N-terminal region of cGAS appears to be dispensable for the activation of cGAS in vitro17 but is required for efficient DNA binding of cGAS25. When full-length cGAS or flag-cGAS-C was expressed ectopically in HEK293T cells, full-length cGAS bound to DNA more efficiently than cGAS-C did (Fig. 3h), suggesting that G3BP1 might regulate the DNA-binding activity of cGAS. Thus, G3BP1 binds to the N terminus of cGAS and may contribute to regulation of the DNA binding of cGAS.
G3BP1 ensures efficient DNA binding and activation of cGAS.
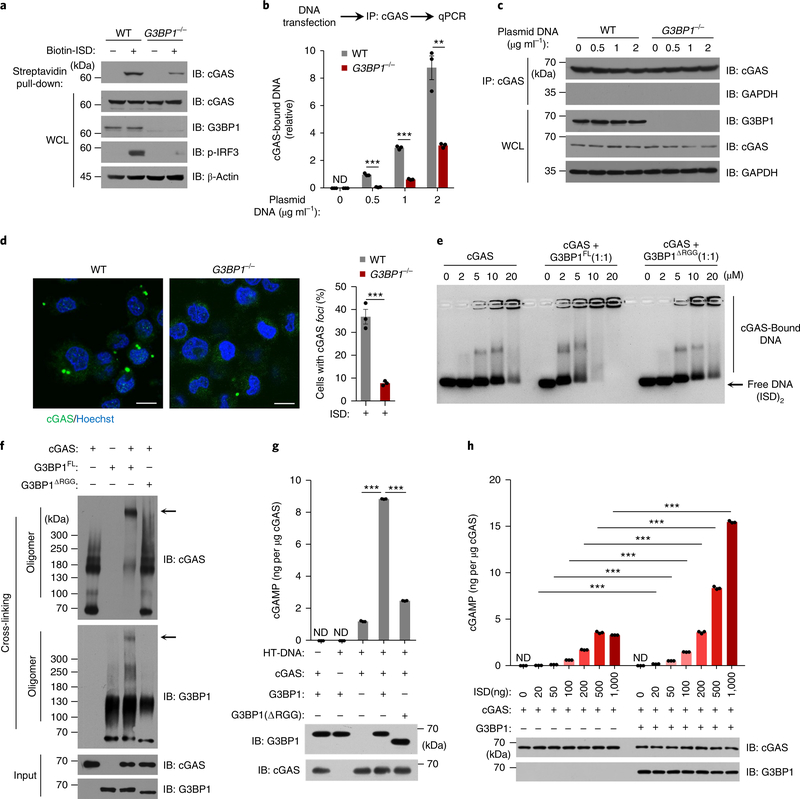
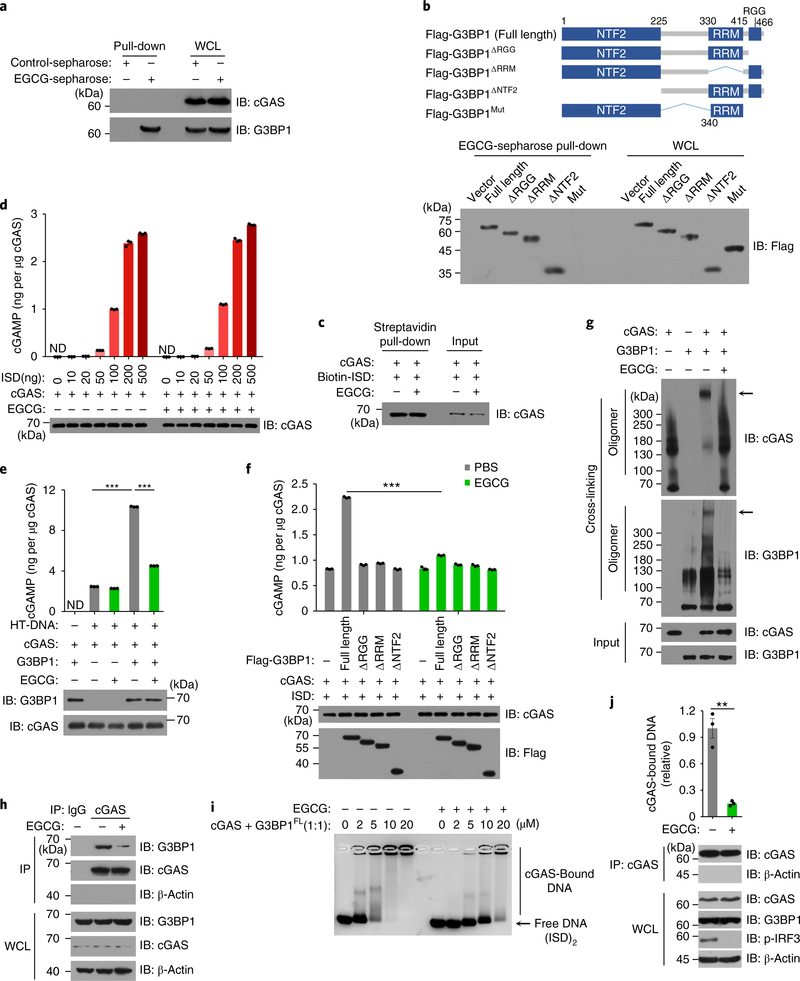
To test whether G3BP1 regulated the DNA-binding activity of cGAS, we transfected biotin-labeled ISD into U937 cells, followed by a streptavidin pull-down to precipitate the DNA-bound endogenous cGAS in both wild-type and G3BP1−/− cells. Absence of G3BP1 led to insufficient DNA binding of cGAS (Fig. 4a–c). Consistently, DNA-triggered cGAS aggregation was significantly reduced in G3BP1−/− U937 cells compared with that of wild-type cells (Fig. 4d). Electrophoretic mobility-shift assay (EMSA) showed that flag-tagged G3BP1, but not the domain-deletion G3BP1 mutants G3BP1ΔRGG, G3BP1ΔNTF2 or G3BP1ΔRRM, markedly enhanced the DNA-binding affinity of cGAS (Fig. 4e and Supplementary Fig. 5f,g), while G3BP1 itself did not bind to DNA (Supplementary Fig. 5h–j). EMSA showed high-molecular-weight complexes that were retained in the wells (Fig. 4e and Supplementary Fig. 5f,g). As the isoelectric point of cGAS is 9.48, we tested whether at high cGAS concentrations the cGAS–DNA complexes would be positively charged and therefore retained in the wells in a standard EMSA electrophoresis buffer (pH ~ 9). When the agarose gel was run at pH 10.5, in which cGAS would be negatively charged, the cGAS–DNA complexes moved into the gel (Supplementary Fig. 6a–d). Importantly, in the presence of G3BP1, the cGAS–DNA complexes were larger than that of cGAS alone, and G3BP1 enhanced the DNA-binding affinity of cGAS (Supplementary Fig. 6a,b). We further found that G3BP1 and cGAS formed a large complex (Fig. 4f). Collectively, these data suggested that G3BP1 increases the DNA-binding affinity of cGAS, probably through changing the structure or oligomerization state of cGAS.
Fig. 4 |. G3BP1 promotes the DNA binding and activation of cGAS.
a, ISD-bound cGAS was analyzed by immunoblotting following biotin–ISD pull-down in U937 cells. b, qPCR analysis of cGAS-bound DNA by immunoprecipitation of cGAS in U937 cells. c, The cGAS immunoprecipitation in b was analyzed by immunoblotting. d, Immunofluorescence analysis of cGAS (green) in U937 cells transfected with ISD (2 μg ml−1) for 3 h (left). The percentage of cells with cGAS foci was quantified (right), at least 100 cells from each group were analyzed. Results are mean ± s.e.m. of n = 3 independent experiments. Scale bars, 10 μm. e, EMSA of cGAS–DNA binding in the presence or absence of flag-tagged G3BP1FL or the mutant as indicated. (ISD)2 (1 μM) were used as DNA probe. Grayscale-inverted image is shown. f, Recombinant cGAS and G3BP1FL or G3BP1ΔRGG were incubated with cross-linker and analyzed with immunoblotting. Arrows, the large cGAS-G3BP1 complexes. g, Recombinant cGAS and G3BP1FL or G3BP1ΔRGG were incubated with HT-DNA. The production of cGAMP was analyzed by LC–MS/MRM. h, The in vitro cGAMP synthesis assay as in g was performed with different amounts of ISD. β-Actin (a) and GAPDH (c), loading controls. GAPDH blot in IP samples indicates the purity of IP (c). **P < 0.01, ***P < 0.001, two-tailed t-test (b,d,g,h). Data are representative of three experiments (a–c,e–h). Data are mean ± s.e.m. of triplicate samples in b,g,h.
We next performed an in vitro cGAMP synthesis assay in which recombinant cGAS protein was incubated with ATP, GTP and DNA, followed by assay of cGAMP production in the presence or absence of G3BP1. Notably, recombinant G3BP1 protein, but not any of the G3BP1 mutants, markedly increased cGAMP production (Fig. 4g and Supplementary Fig. 6e). Further titration of DNA in this assay showed that G3BP1 greatly increased cGAS-mediated cGAMP production (Fig. 4h and Supplementary Fig. 6f). As G3BP1 bound to the N-terminal region of cGAS, G3BP1 failed to enhance the cGAMP synthesis by cGAS-C (Supplementary Fig. 6g). Thus, G3BP1 is critical for efficient DNA binding and activation of cGAS.
EGCG specifically blocks cGAS activation.
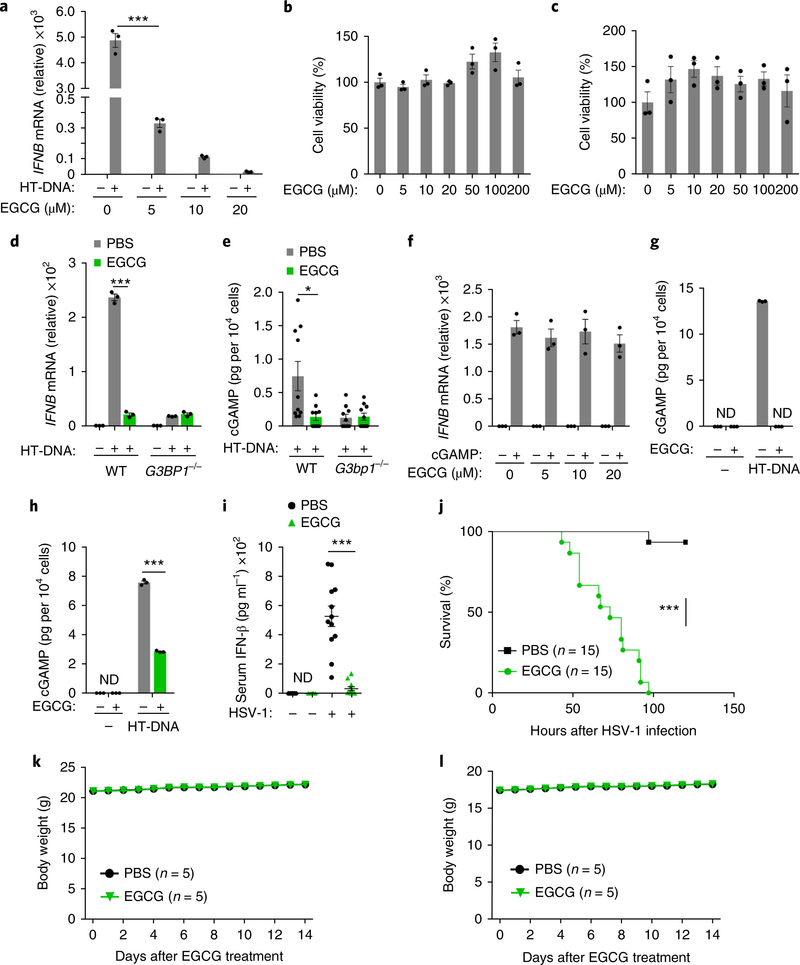
Epigallocatechin gallate (EGCG), a polyphenol isolated from tea leaves, is an inhibitor of G3BP1 (ref. 26). Indeed, EGCG efficiently bound to endogenous G3BP1 in U937 cells (Fig. 5a). The RGG motif and the Ras–GAP binding region (amino acids 225–340) of G3BP1 were reported to be required for the binding of G3BP1 to EGCG26. We expressed a G3BP1 mutant lacking the RGG motif and the Ras–GAP binding region (flag-G3BP1mut) in HEK293T cells. In an EGCG–Sepharose pull-down assay with flag-tagged proteins, EGCG did not bind to flag-G3BP1mut (Fig. 5b). The domain-deletion mutants of G3BP1 (G3BP1ΔRGG, G3BP1ΔNTF2and G3BP1ΔRRM) still bound to EGCG (Fig. 5b), which is consistent with previous reports26. While EGCG did not affect cGAS–DNA binding or cGAMP production mediated by cGAS in the in vitro cGAMP synthesis assay (Fig. 5c–e), it efficiently suppressed cGAMP production in the presence of full-length G3BP1 but not in the presence of G3BP1ΔRGG, G3BP1ΔNTF2 or G3BP1ΔRRM (Fig. 5e,f). Importantly, EGCG disrupted the large G3BP1–cGAS complexes and inhibited the G3BP1-facilitated cGAS–DNA binding in vitro and in U937 cells (Fig. 5g–j). We next tested whether EGCG could inhibit DNA-induced cGAS activation. EGCG robustly suppressed the cGAS-mediated expression of IFNB mRNA and the phosphorylation of IRF3 in U937 and human primary macrophages (Fig. 6a and Supplementary Fig. 7a–g). Notably, EGCG did not show toxicity effects in either bone marrow–derived macrophages (BMDMs) or human primary macrophages, even at a dose ten times higher than that used to inhibit cGAS activation (Fig. 6b,c).
Fig. 5 |. EGCG inhibits G3BP1-promoted cGAS activation.
a, The EGCG–G3BP1 binding was analyzed by EGCG-conjugated Sepharose pull-down in U937 cells. b, Schematic drawing of full-length G3BP1 and G3BP1 mutants (top). EGCG–G3BP1 binding was analyzed by EGCG-conjugated Sepharose pull-down in G3BP1−/− HEK293T cells expressing indicated flag-tagged G3BP1 variants (bottom). c, The in vitro cGAS–DNA binding in the presence or absence of EGCG was analyzed by biotin–ISD pull-down. d, The cGAS alone-mediated cGAMP production, in the presence or absence of EGCG, was analyzed by LC–MS/MRM. e,f, Recombinant cGAS and G3BP1FL or G3BP1 mutants were incubated with ISD in the presence or absence of EGCG (20 μM). The production of cGAMP was analyzed by LC–MS/MRM. g, Recombinant cGAS and G3BP1FL were incubated with cross-linker in the presence or absence of EGCG and analyzed with immunoblotting. h, The cGAS-G3BP1 interaction was analyzed by immunoprecipitation in U937 cells left untreated or treated with EGCG. i, EMSA shows the effect of EGCG on cGAS–DNA binding in the presence of G3BP1, grayscale-inverted image is shown. j, The effect of EGCG on cGAS–DNA binding in U937 cells was analyzed as described in Fig. 4b. β-Actin, loading control. β-Actin blots in IP samples indicate the purity of IP (h,j). PBS was used as control for EGCG. **P < 0.01, ***P < 0.001, two-tailed t-test (e,f,j). Data are representative of three experiments, mean ± s.e.m. of triplicate samples in d–f,j.
Fig. 6 |. EGCG blocks cGAS-mediated interferon production through inhibiting G3BP1.
a,f, qPCR analysis of IFNΒ mRNA expression in human primary macrophages treated with HT-DNA (0.5 μg ml−1) (a) or cGAMP (0.5 μg ml−1) (f) following a 1 hour of pretreatment with EGCG as indicated. b,c, MTS assay shows the cell viability of BMDMs (b) and human primary macrophages (c) incubated with EGCG at indicated concentrations for 48 h. d, The effect of EGCG on HT-DNA-induced IFNB production in U937 cells. e, The cGAMP production in MEFs transfected with HT-DNA following EGCG pretreatment. g,h, EGCG effect on HT-DNA-induced cGAMP production in human primary macrophages (g) and U937 cells (h) was analyzed by LC–MS/MRM, ND, non-detected. i, ELISA of serum IFN-β concentration of WT C57BL/6 mice injected with EGCG (40 mg kg−1) before infection with HSV-1 (1 × 107 pfu per mouse). Uninfected mice, n = 6. j, WT C57BL/6 mice (n = 15 per group) were pretreated with EGCG (40 mg kg−1, i.v.) for 12 h before infection with HSV-1 (i.v., 2 × 106 pfu per mouse). With continuous daily EGCG injection, the survival of mice was monitored for 120 h. k,l, Male (k) and female (l) WT C57BL/6 mice were treated with EGCG (120 mg kg−1) daily for 2 weeks. Body weight was measured every day. *P < 0.05, ***P < 0.001, two-tailed t-test (a,d,e,h,i), two-sided log-rank (Mantel–Cox) test (j). Data are representative of three experiments (a–d,f–h; mean ± s.e.m. of triplicate samples). Results are mean ± s.e.m. of n = 10 mice (e) or n = 12 mice (i) or n = 5 mice (k,l).
We next tested whether the inhibitory effect of EGCG on cGAS-mediated signaling was mediated through inhibition of G3BP1. EGCG did not inhibit the residual cGAS activation in G3BP1-deficient U937 cells or G3BP1-deficient MEFs (Fig. 6d,e). Consistent with our data that G3BP1 deficiency did not inhibit TLR-mediated Tnf expression, EGCG treatment had no inhibitory effect on TLR-mediated TNF expression (Supplementary Fig. 7h). EGCG treatment did not enhance the cGAS–Beclin-1 interaction in HeLa cells (Supplementary Fig. 7i), and EGCG still inhibited the elevated cGAS activation when Becn1 expression was knocked down in mouse L929 cells (Supplementary Fig. 7j), suggesting EGCG is unlikely to inhibit cGAS through Beclin-1. Furthermore, while EGCG had no effect on cGAMP-stimulated IFNB expression in human primary macrophages (Fig. 6f), it substantially inhibited the synthesis of cGAMP in both human primary macrophages and U937 cells after cytoplasmic DNA challenge (Fig. 6g,h). These results indicate that EGCG specifically blocks cGAS activation through inhibiting G3BP1. To test if EGCG was effective in restraining cGAS-mediated IFN production in vivo, we treated mice with EGCG (40 mg kg−1) before HSV-1 infection. Mice were given intravenous (i.v.) injections of EGCG twice, at 12 h and 4 h before infection with HSV-1. The IFN-β secreted in serum from HSV-1-infected mice was analyzed. Treatment with EGCG robustly suppressed the HSV-1-induced concentrations of IFN-β in the serum (Fig. 6i). Moreover, after sublethal infection with HSV-1, the EGCG-treated mice were highly susceptible to HSV-1-induced death compared with PBS-treated mice (Fig. 6j). We next evaluated the potential toxic effect of EGCG in vivo. Mice that received daily intraperitoneal (i.p.) injections with EGCG, at a dose of 120 mg kg−1 (three times higher than the dose used for the viral infection experiments) for 2 weeks, did not show any weight loss (Fig. 6k,l), indicating that EGCG was well tolerated in mice and can be used to block cGAS activation in vivo. Thus, EGCG blocks cGAS activation through inhibiting G3BP1.
EGCG is effective in treating cGAS-mediated autoinflammation.
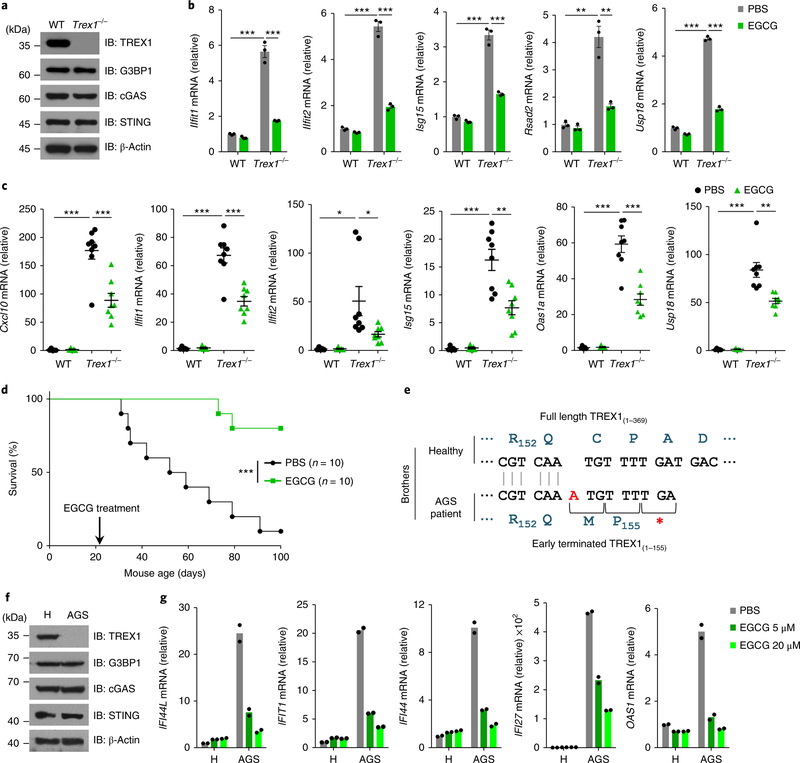
We next explored whether EGCG could be used to treat cGAS-mediated autoimmune diseases. We first isolated bone marrow cells from both wild-type and Trex1−/− mice. The deficiency of TREX1 did not affect the expression of cGAS and G3BP1 (Fig. 7a). Compared with wild-type cells, the Trex1−/− bone marrow cells had highly elevated expression of IFN-stimulated genes (ISGs) (Fig. 7b). Treatment of Trex1−/− bone marrow cells with 20 μM EGCG significantly reduced the abundance of ISG mRNAs (Fig. 7b). To further study the effect of EGCG in vivo, Trex1−/− mice and wild-type littermates received EGCG (40 mg kg−1, i.p.) daily for 14 days. Administration of EGCG strongly inhibited the expression of ISG in the heart of Trex1−/− mice compared with wild-type littermates (Fig. 7c). To determine the pharmacokinetics of EGCG after administration, we monitored the blood concentration of EGCG after i.p. injection. Our data show that although the concentration half-life of EGCG in blood was short, one injection of EGCG led to a concentration higher than 1 μM, which was an effective dose in our cell experiments, for about 9 h in the blood (Supplementary Fig. 7k), suggesting that once-daily injection could be effective in treating Trex1−/− mice. Indeed, daily administration of EGCG significantly prolonged the survival of Trex1−/− mice (Fig. 7d). We further tested the effectiveness of EGCG in human cells. To do so, we isolated peripheral blood mononuclear cells (PBMCs) from a patient with AGS and their healthy sibling27. The previously identified frameshift mutation (c.459_460insA) of TREX1 in the patient with AGS27 was confirmed (Fig. 7e). Immunoblot analysis showed that the PBMCs from the patient had no detectable expression of TREX1 protein (Fig. 7f). The expression of ISGs in the PBMCs from the patient was highly elevated compared with cells from the healthy sibling (Fig. 7g). EGCG treatment strongly inhibited the ISG expression in cells from the patient with AGS (Fig. 7g), suggesting EGCG was effective in treating cGAS-mediated autoinflammatory responses in both the Trex1−/− mouse model and cells from human patients with AGS.
Fig. 7 |. EGCG suppresses cGAS-mediated autoimmune responses.
a, Immunoblot analysis of WT and Trex1−/− bone marrow cells with indicated antibodies. b, qPCR analysis of mRNA expression of indicated ISGs in WT and Trex1−/− bone marrow cells treated with EGCG (20 μM) for 48 h. c, WT mice (n = 5) and Trex1−/− mice (n = 8) were given EGCG (i.p., 40 mg kg−1) for 2 weeks. Relative mRNA expression of indicated ISGs in mouse hearts was measured by qPCR. Results are mean ± s.e.m. d, Trex1−/− mice (n = 10 per group) were given daily injection (i.p.) with EGCG (40 mg kg−1) or PBS for 80 days. The survival of mice was monitored. Statistical analysis was performed with a two-sided log-rank (Mantel–Cox) test. e, Schematic drawing of the TREX1 mutations of the patient with AGS. Blue letters, amino acids. Red ‘A’, insertion. f, Immunoblot analysis of PBMCs from a patient with AGS and his healthy brother (H) with indicated antibodies. g, PBMCs from the siblings in f were treated with EGCG and the ISG expression was measured by qPCR (n = 2 independent experiments). β-Actin, loading control (a,f). *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed t-test (b,c). Data are representative of three experiments (a,b,f). Data are mean ± s.e.m. of triplicate samples in b.
STING-associated vasculopathy with onset in infancy (SAVI) is a recently characterized AGS-like, rare disease28. We ectopically expressed flag-tagged STING mutants found in patients with SAVI, flag-STINGV155M or flag-STINGC206Y in HEK293T cells (Supplementary Fig. 7l). EGCG treatment had no effect on the phosphorylation IRF3 driven by the expression of the mutant STING, whereas BX-795, a known inhibitor of the STING downstream kinase TBK1 (ref. 29), strongly inhibited the STING mutant-driven IRF3 phosphorylation (Supplementary Fig. 7l), further indicating that EGCG acts upstream of STING. Taken together, EGCG is effective in treating cGAS-mediated autoinflammation.
Discussion
In this study we revealed the critical role of G3BP1, a protein known to regulate RNA stress response, in the regulation of the cGAS-mediated DNA-sensing pathway. We found that G3BP1 forms a large complex with cGAS and promotes the oligomerization of cGAS. Our data indicate that G3BP1 primes cGAS for efficient DNA binding and activation. G3BP1 deficiency leads to inefficient DNA binding of cGAS and dramatically inhibits interferon induction by DNA transfection or DNA virus infection. Moreover, we found that EGCG, which is a natural chemical from green tea and is a known inhibitor of G3BP1, potently inhibited DNA-induced cGAS activation and subsequent type I interferon production through specific targeting of G3BP1. Importantly, we further demonstrated that EGCG is effective in treating cGAS-mediated autoinflammatory responses in Trex1−/− mouse model and cells from patients with AGS.
G3BP1 has previously been linked to RNA stress responses, partly through promoting the formation of SG22. However, we found that SG is not involved in G3BP1-mediated cGAS activation and that the function of G3BP1 in cGAS regulation is distinct from its well-established role in SG. Whether cGAS alone is sufficient to respond to cytosolic DNA efficiently remains unclear, although it is known that cGAS binds to DNA directly30,31. A recent study has identified that the protein factors HMGB and TFAM can prearrange DNA structures to facilitate binding of DNA to cGAS32. Our results showed that while G3BP1 itself does not bind to DNA, intact G3BP1 protein is needed for promoting efficient DNA sensing of cGAS. Importantly, considering that the amount of infection-introduced cytosolic DNA would be limited in cells under physiological conditions, G3BP1-dependent priming of cGAS would be very critical for efficient DNA binding when cytosolic DNA is not abundant. Future structural studies are needed to delineate the basis of G3BP1-mediated activation of cGAS.
G3BP1 appears to be critical for the general recognition of different types of DNA by cGAS, regardless of DNA length. Our data also suggest that G3BP1 contributes to cGAS-mediated antiviral responses. A critical coreceptor, PQBP1, has been reported to bind directly to the HIV-1 viral DNA and interact with cGAS to initiate the downstream immune response19. In our study, we found that G3BP1 is also required for the immune response to HIV-1. Thus, PQBP1 is critical for the detection of HIV DNA, and G3BP1 primes cGAS for efficient activation. These two factors seem to work together for sensing and fighting against HIV. IFI16 has been previously shown to cooperate with cGAS to sense nuclear viral DNA33. Additionally, IFI16 has also been reported to regulate cGAS in a cell-type-specific manner23,24. In our study, we found that the IFI16–cGAS interaction is induced by DNA treatment in U937 cells. In contrast, G3BP1 interacts with cGAS in quiescent cells, and DNA treatment led to the dissociation of G3BP1 and cGAS. Thus, G3BP1 and IFI16 probably modulate the different aspects of cGAS activation.
Through defining the role of G3BP1 in cGAS activation, we further demonstrate that the G3BP1 chemical inhibitor EGCG can specifically inhibit inflammation caused by cGAS activation. Because EGCG cannot inhibit the cGAS-mediated immune responses in G3BP1-deficient cells, the inhibitory effect of EGCG on cGAS activity is clearly dependent on G3BP1. Deregulation of cGAS activation has been linked to a number of pathological conditions, including autoimmune diseases10, aging-associated inflammation34,35 and cancers36. Our findings suggest that EGCG can be used as an effective treatment for cGAS-related diseases.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41590-018-0262-4.
Methods
Mice.
G3bp1+/− mice were gifts from J. Tazi (Universitié Montpellier 2). Trex1+/− mice were gifts from D. Barnes and T. Lindahl (Cancer Research UK). All animal experiments were performed in accordance with the NIH guide for the care and use of laboratory animals and with the approval of the Institutional Animal Care and Use Committee of the National Center of Biomedical Analysis.
Wild-type and Trex1−/− mice (3-week-old) were given daily (i.p.) injection of EGCG (40 mg kg−1), PBS was used as control. No randomization was applied since the allocation was based on the genotype of mice.
Reagents.
Anti-G3BP1 (H00010146-M01) was from Abnova; anti-G3BP1 (13057–2-AP) was from Proteintech Group; anti-DDDDK-tag-pAb-HRP-Dire T (PM020–7, anti-flag) was from B&M Biotech; anti-IRF3 (ab68481) and anti-G3BP2 (ab86135) were from Abcam; anti-human cGAS (15102), anti-mouse cGAS (31659), anti-p-IRF3 (4947), anti-STING (13647), anti-TBK1 (3504) and anti-Beclin-1 (3495) were from Cell Signaling; anti-TREX1 (611986) was from BD Transduction Laboratories; anti-human cGAS (HPA031700) and anti-α-Tubulin (T5168) were from Sigma; anti-TIA1 (sc-166247), anti-β-Actin (sc-47778), anti-IFI16 (sc-3428) and anti-HA (sc-7392) were from Santa Cruz Biotechnology; anti-flag M2 affinity gel (a2220), LPS (L2630), EGCG (E4143), PMA (phorbol 12-myristate 13-acetate; 524400), sodium arsenite (S7400) and HT-DNA (D6898) were from Sigma-Aldrich; TBK1 inhibitor BX795 (S1274) was from Selleck; ISD and biotin–ISD were synthesized from Invitrogen; cGAMP (tlrl-cga23), c-di-GMP (tlrl-nacdg), Pam3CSK4 (tlrl-pms), pppRNA (tlrl-3pma) and poly(I:C) (tlrl-pic) were from InvivoGen; rhGM-CSF (215-GM) and rmM-CSF (416-ML) were from R&D Systems. ATP (R0441) and GTP (R0461) were from ThermoFisher Scientific. Anti-human cGAS and anti-human GAPDH were prepared in our laboratory. The cGAS antibody was generated by immunizing rabbits with human cGAS (amino acids 1–160); the GAPDH antibody was generated by immunizing rabbits with human GAPDH protein.
Cell culture and transfection.
HEK293T, HeLa, MEF, BMDM cells were cultured in DMEM containing 10% FBS, 2 mM l-glutamine, 100 U ml−1 penicillin and 100 mg ml−1 streptomycin. U937, L929 and human primary macrophages were cultured in RPMI-1640 medium containing 10% FBS, 2 mM l-glutamine, 100 U ml−1 penicillin and 100 mg ml−1 streptomycin. U937 cells were differentiated with PMA (0.1 μM) from 36 h to approximately 48 h before transfection or other treatment. Human blood samples were obtained following the National Institutes of Health Guide for the use of human samples and with the approval of the Scientific Investigation Board of affiliated hospital of Academy of Military Medical Sciences. Informed consent was obtained from the blood donors by the hospital. For the patient with AGS and their sibling, we isolated PBMCs from their blood after we obtained informed consent. Human primary macrophages were differentiated from PBMCs with rhGM-CSF (10 ng ml−1) for 7 days. PBMCs were isolated from peripheral blood of informed volunteers with HISTOPAQUE (1077, Sigma-Aldrich) according to the manufacturer’s instructions. BMDMs were differentiated from mouse bone marrow cells with rmM-CSF (25 ng ml−1) for 7 days. HeLa, U937 and HEK293T cells were obtained from ATCC. All cell lines were tested to be mycoplasma free by PCR.
Transfection of HT-DNA, ISD or plasmid DNA (peGFP-N1) were performed with Lipofectamine 2000 (Invitrogen) at a final concentration of 2 μg ml−1 for U937 cells and 0.5 μg ml−1 for human primary macrophages and MEFs. cGAMP stimulation was performed as previously described5. Briefly, cells were incubated for 30 min at 37 °C with cGAMP in permeabilization buffer (50 mM HEPES, pH 7; 100 mM KCl; 3 mM MgCl2; 0.1 mM DTT; 85 mM sucrose; 0.2% BSA; 1 mM ATP and 0.1 mM GTP) with 1 μg ml−1 digitonin (Sigma, D141). The permeabilization buffer was replaced with RPMI-1640 medium and cells were cultured for the indicated time.
Generation of gene knockout cell lines via CRISPR/Cas9.
For targeting G3BP1 and G3BP2 with CRISPR/Cas9, we used a LentiCRISPR v2 construct (Addgene, #98290). The guide RNA sequences (G3BP1: 5′-CACCGTACTTGGTCTGGGTCCCCTT-3′; G3BP2: 5′-GGAGTAGTTGTCCAGGTCAT-3′) were designed using the online tool by F. Zhang (http://crispr.mit.edu/). The lentiCRISPR plasmid (with sgRNA cloned) was cotransfected into HEK293T cells with the packaging plasmids pVSVg (Addgene, #8454) and psPAX2 (Addgene, #12260) for 48 h to generate lentivirus. U937 cells were infected with the lentivirus for 24 h, followed by selection with puromycin (2 μg ml−1, Life Technologies) for 5 days. Protein expression was determined by immunoblot analysis.
G3BP1−/− U937 cells or G3bp1−/− MEF cells were infected with lentivirus carrying the guide RNA-resistant wild-type or mutant G3BP1 coding sequences to make rescue cell lines. The positively infected cells were sorted for further analysis.
Plasmids.
cDNA encoding G3BP1 and cGAS was subcloned into pCDH-GFP vector, pcDNA3.0-flag-vector or pXJ40-HA vector for expression in mammalian cells. cDNA encoding G3BP1 and cGAS was subcloned into pCDFDuet-1 vector for expression in bacteria and recombinant protein purification.
RNA isolation and quantitative PCR (qPCR).
Total RNA was extracted with TRI reagent (93289, Sigma). Total RNAs (500 ng) were used to perform the reverse transcription with PrimeScript RT reagent kit (Takara, DRR037A). To determine relative mRNA abundance, qPCR was performed with Powerup SYBR Green Master Mix (Applied Biosystems) on an ABI StepOnePlus system according to the manufacturer’s protocol. Data were analyzed with StepOnePlus software. The primers were synthesized from Invitrogen and sequence information is provided in Supplementary Table 3. Human GAPDH and mouse Hprt were used for normalization.
Immunoprecipitation and pull-down assay.
Cells were lysed with lysis buffer (20 mM Tris–HCl, pH 7.5; 0.5% Nonidet P-40; 10 mM NaCl; 3 mM EDTA and 3 mM EGTA) containing complete protease inhibitor cocktail (Roche, 04693132001), followed by centrifugation at 20,000g for 20 min at 4 °C. The supernatants were immunoprecipitated with indicated antibodies or anti-flag M2 affinity beads. Cell lysates or immunoprecipitates were separated by SDS–PAGE and analyzed with immunoblotting. Proteins were visualized by enhanced chemiluminescence according to the manufacturer’s instruction (Thermo Scientific).
Recombinant flag-tagged G3BP1 and its mutants were incubated at 4 °C with equal amount of recombinant cGAS protein (2 μg) in a buffer (20 mM Tris–HCl, pH 7.5, 0.5% Nonidet P-40, 10 mM NaCl, 3 mM EDTA and 3 mM EGTA containing complete protease inhibitor cocktail (Roche, 04693132001)) at a final volume of 1 ml for 1 h. Interactions were examined with anti-flag M2 affinity beads according to the manufacturer’s instruction.
EGCG pull-down assays were performed as previously described26. Briefly, EGCG was conjugated with cyanogen bromide (CNBr)-activated Sepharose 4B (GE Healthcare). Cells were lysed with lysis buffer (20 mM Tris–HCl, pH 7.5; 0.5% Nonidet P-40; 250 mM NaCl; 3 mM EDTA and 3 mM EGTA) containing complete protease inhibitor cocktail (Roche, 04693132001), followed by centrifugation at 20,000g for 20 min at 4 °C. The supernatants were incubated with EGCG-conjugated Sepharose 4B at 4 °C for 3 h. The beads were then washed three times with lysis buffer. The proteins pulled down were separated by SDS–PAGE and analyzed by immunoblotting.
ELISA.
MEFs or PMA-differentiated U937 cells were seeded into 12-well plates at a density of 4 × 105 cells per well and treated as indicated. Mice were given i.v. injections of EGCG (40 mg kg−1) twice, at 12 h and 4 h respectively, before infection with HSV-1 (1 × 107 pfu per mouse). The secreted interferon in cell culture medium or sera from HSV-1-infected mice was analyzed with ELISA kits (41410, PBL, for human; and 439408, BioLegend, for mouse) according to the manufacturer’s instruction.
Cell viability assay.
BMDMs or human primary macrophages were seeded into 96-well plates at a density of 5 × 104 cells per well and incubated with EGCG at the indicated concentration for 48 h. The cell viability was analyzed with CellTiter One Solution Cell Proliferation Assay kit (G3580, Promega) according to the manufacturer’s instruction.
Immunofluorescence.
Human primary macrophages, HeLa or U937 cells were seeded on coverslips in 24-well plates. After transfection with ISD or poly(I:C), or treatment with arsenite, the cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with Triton X-100 for 10 min and blocked in 3% BSA for 1 h. Cells were then incubated with primary antibodies overnight at 4 °C, including rabbit anti-cGAS (Cell Signaling Technology) and mouse anti-G3BP1 (Abnova). Alexa Fluor 488- and 546-conjugated secondary antibodies (Life Technologies) were incubated for 1 h before images were acquired using a ZEISS LSM 880 (Zeiss) confocal microscope.
Chemical cross-linking of recombinant proteins.
Equal amounts (1 μg) of recombinant cGAS protein and recombinant G3BP1 protein or its mutants were incubated in the cGAMP synthesis buffer (20 mM HEPES, pH 7.5; 5 mM MgCl2; 2 mM ATP and 2 mM GTP) at 25 °C for 15 min before DSP (0.5 mM) was added and incubated for another 10 min. The reactions were stopped by adding loading buffer (2-mercaptoethanol free) and the samples were analyzed with immunoblotting.
Electrophoretic mobility-shift assays.
EMSA was performed as previously described37. Briefly, recombinant cGAS was incubated, in the presence or absence of recombinant G3BP1 (or the mutant), with 1 μM (ISD)2 in the cGAMP synthesis buffer at 37 °C for 90 min. The mixtures were loaded on 1% agarose gel using an electrophoresis buffer (40 mM Tris–HCl at pH 9.2 or 10.5). The gels were then stained with GeneGreen nucleic acid gel stain (Thermo, S7563) and images were acquired using UV Transilluminator (Analytik Jena). The intensity of free DNA bands was analyzed by Image J and the data points were fitted with a binding curve. The Kd was calculated with a binding equilibrium equation using GraphPad Prism.
DNA-binding assay.
Plasmid DNA-binding experiments were performed as previously described19. Briefly, plasmid DNAs (peGFP-N1) were transfected into PMA-differentiated U937 cells for 2 h, followed by formaldehyde cross-linking, which was then quenched with glycine. The cross-linked cells were lysed in an SDS lysis buffer (100 mM Tris–HCl, pH 7.5; 300 mM NaCl; 5 mM MgCl2; 4 mM EDTA; 0.2% SDS; 1% sodium deoxycholate; 1% Trion-X-100; 10% glycerol; 10 mg ml−1 heparin; protease inhibitor cocktail) and centrifuged at 4 °C, 12,000g for 10 min. The cleared cell extracts were diluted 1:1 with H2O and then immunoprecipitated with cGAS antibody. The immunoprecipitants were eluted in elution buffer (100 mM Tris–HCl, pH 7.8; 10 mM EDTA; 1% SDS). After proteinase K treatment and phenol–chloroform extraction, the nucleic acids were precipitated with ethanol and subjected to qPCR analysis.
For the biotin–DNA-binding experiments, the assay was performed according to a previous report17. PMA-differentiated U937 cells were transiently transfected with biotin-labeled ISD for 2 h. The cells were lysed in lysis buffer (20 mM Tris–HCl, pH 7.5; 0.5% Nonidet P-40; 10 mM NaCl; 3 mM EDTA and 3 mM EGTA and protease inhibitor cocktail). The cell lysates were centrifuged at 4 °C, 12,000g for 10 min, and the supernatants were incubated with streptavidin agarose resin (Thermo Scientific) for 60 min. The resin was washed with lysis buffer five times followed by boiling in 1 × SDS-loading buffer and immunoblotting.
In vitro cGAMP synthesis assay.
Plasmids pCDF-His-cGAS and pCDF-His-G3BP1 (full length and mutants) were transformed into Escherichia coli, BL21 (DE3) strain. Cells were collected after an overnight induction with 0.5 mM IPTG at 18 °C and the proteins were purified with HisTrap FF (GE Healthcare, 17–5319-01) according to the manufacturer’s protocol.
Purified recombinant human cGAS protein (1.87 μM, 8 μg) was incubated with DNA in a reaction buffer (20 mM HEPES, pH 7.5; 5 mM MgCl2; 2 mM ATP and 2 mM GTP) at 37 °C for 1.5 h, with or without purified recombinant human G3BP1 protein (1.87 μM). The samples were collected with extraction solvent (40:40:20 (v:v:v) methanol–acetonitrile–sample), and then centrifuged at 4 °C, 12,000g, for 5 min38. Supernatants were collected and lyophilized by a gentle stream of nitrogen gas. The dried extracts were resuspended in ammonium acetate buffer (10 mM ammonium acetate; 0.05% acetate), vortexed and ultrasonicated. After another centrifugation at 4 °C, 12,000g, for 5 min, the supernatants were used for cGAMP quantification by LC–MS/MRM analysis.
To examine the effect of EGCG on cGAMP synthesis, EGCG (20 μM) was incubated with cGAS protein, with or without G3BP1 protein, in reaction buffer at 37 °C for 1 h, then HT-DNA was added and the system was incubated at 37 °C for another 1.5 h. The samples were collected and cGAMP was extracted and quantified as described below.
cGAMP quantitative analysis.
Primary human macrophage, MEFs or PMA-differentiated U937 were transfected with HT-DNA (2 μg ml−1 for U937 cells, 0.5 μg ml−1 for primary human macrophage and MEFs) for the indicated time, followed by cGAMP extraction with extraction solvent (40:40:20 (v:v:v) methanol–acetonitrile–water) as described38.
Quantification of cGAMP was performed on a triple-quadrupole mass spectrometer (Xevo TQ-S, Waters Corp.) equipped with an electrospray ionization source. The nebulizer gas was 99.95% nitrogen, and the collision gas was 99.99% argon with a pressure of 3 × 10−3 mbar in the T-Wave cell. The gas flows of the cone and desolvation were set as 150 and 800 l h−1, respectively. The target compound measurements were performed in the positive mode with a 3.5 kV capillary voltage, 120 °C source temperature and 450 °C desolvation temperature. The optimized ion transitions were: cGAMP m/z 675 → 524; m/z 675 → 136.
RNA interference.
HeLa cells or L929 cells were transfected with siRNAs by RNAiMax transfection reagent for 48 h before stimulation. The siRNAs used were as following: si-cGAS (HSS132955, Sigma-Aldrich), si-Becn1 (5′-CCCTATGGAAATCATTCCT-3′). Protein expression was analyzed by immunoblotting.
G3BP1-specific or TIA1-specific shRNA sequences were designed and cloned into lentiviral pLKO.1 vector (Addgene). U937 cells were infected with lentivirus encoding G3BP1 shRNAs (no. 1: 5′- AGTGCGAGAACAACGAATAAA-3′; no. 2: 5′- TATGGAAAGAACTCTTCTTAT-3′) or TIA1 shRNA (5′-TTTCACCATTTGGACAAATAA-3′), followed by selection with 1 μg ml−1 puromycin.
EGCG pharmacokinetics.
Mice were administrated (i.p.) with EGCG (40 mg kg−1), blood was collected at the indicated time and the EGCG concentration in blood was measured. The quantification of EGCG was performed on a triple-quadrupole mass spectrometer (Xevo TQ-S, Waters Corp.) equipped with an electrospray ionization source. The nebulizer gas was 99.95% nitrogen, and the collision gas was 99.99% argon with a pressure of 3 × 10−3 mbar in the T-Wave cell. The gas flows of the cone and desolvation were set as 150 and 900 l h−1, respectively. The target compound measurements were performed in the negative mode with −2.3 kV capillary voltage, 120 °C source temperature and 450 °C desolvation temperature. The optimized ion transitions were: EGCG m/z 457.0 → 168.9; m/z 457.0 → 125.0.
Identification of cGAS-interacting proteins by mass spectrometry.
Sixty 15-cm dishes of U937 cells were lysed with lysis buffer (20 mM Tris–HCl, pH 7.5; 0.5% Nonidet P-40; 10 mM NaCl; 3 mM EDTA and 3 mM EGTA) containing complete protease inhibitor cocktail (Roche, 04693132001), followed by centrifugation at 20,000g for 20 min at 4 °C. The supernatants were immunoprecipitated with a cGAS antibody. Immunoprecipitates were separated with SDS–PAGE and the gel was stained with Coomassie brilliant blue. The entire lane was cut into 3 mm gel slices and subjected to in-gel digestion before being analyzed with a Q Exactive HF mass spectrometer (ThermoFisher Scientific). The mass spectrometry data were analyzed using the Swissprot database by the MASCOT search engine. Ranking of the identified proteins was based on the peptide coverage of each identification and reproducibility among MS analysis of different samples. The top-ranking proteins were selected and subjected to further validation by immunoprecipitation and immunoblot before detailed study. This study focused on G3BP1.
Statistical analysis.
No statistical methods were used to estimate sample size. A standard two-tailed unpaired Student’s t-test was used for statistical analysis of two groups. Statistically analyzed data are presented as mean ± s.e.m. Statistical analysis of survival curves was performed with a two-sided log-rank (Mantel–Cox) test. P < 0.05 is considered as statistically significant. We performed the statistical analysis using GraphPad Prism. The half-maximum inhibitory concentration was calculated with SPSS (Version 21).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The MS-identified cGAS-interacting protein list is provided in Supplementary Tables 1 and 2. The primer sequences used for qPCR are provided in Supplementary Table 3. The information on antibodies used in our study is provided in Supplementary Table 4. We also provide full scans of all the blots and gels as Supplementary Dataset 1. The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgements
We sincerely thank the patient with AGS and his sibling for donating PBMCs. We thank J. Tazi (Université Montpellier 2) for providing the G3bp1+/− mice, Cancer Research Technology Limited for providing the Trex1+/− mice, C. Widmann (University of Lausanne) for providing HA-G3BP1 plasmid, J. W. Chin (Cambridge) for providing pCDF PylT-1 plasmid, J. U. Jung (University of Southern California) for providing cGAS cDNA construct, H. Shu (Wuhan University) for providing STING cDNA construct, J. Han (Xiamen University) for providing HSV-1, M. Yang (Tsinghua University) for help with protein purification. We thank H. Yu (UT Southwestern Medical Center), Y. Zheng (Carnegie Institution for Science) and H. Qi (Tsinghua University) for helpful discussion and critical reading of the manuscript. This work was supported by grants from the China National Natural Science Foundation (No. 81771708 to T.L., No. 81521064 to X.-M.Z.).
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41590-018-0262-4.
References
- 1.Takeuchi O & Akira S Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Wu J & Chen ZJ Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Gurtler C & Bowie AG Innate immune detection of microbial nucleic acids. Trends Microbiol. 21, 413–420 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao P et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao P et al. Structure-function analysis of STING activation by c[G(2′,5′) pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154, 748–762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita M et al. Gene-targeted mice lacking the Trex1 (DNase III) 3′ –> 5′ DNA exonuclease develop inflammatory myocarditis. Mol. Cell. Biol. 24, 6719–6727 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crow YJ & Manel N Aicardi–Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15, 429–440 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Aicardi J & Goutières F A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann. Neurol. 15, 49–54 (1984). [DOI] [PubMed] [Google Scholar]
- 10.Gao D et al. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl Acad. Sci. USA 112, E5699–E5705 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray EE, Treuting PM, Woodward JJ & Stetson DB Cutting Edge: cGAS is required for lethal autoimmune disease in the Trex1-deficient mouse model of Aicardi–Goutières syndrome. J. Immunol. 195, 1939–1943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Protter DS & Parker R Principles and properties of stress granules. Trends Cell Biol. 26, 668–679 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onomoto K, Yoneyama M, Fung G, Kato H & Fujita T Antiviral innate immunity and stress granule responses. Trends Immunol. 35, 420–428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luecke S et al. cGAS is activated by DNA in a length-dependent manner. EMBO Rep. 18, 1707–1715 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zekri L et al. Control of fetal growth and neonatal survival by the RasGAP-associated endoribonuclease G3BP. Mol. Cell. Biol. 25, 8703–8716 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burdette DL et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun L, Wu J, Du F, Chen X & Chen ZJ Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao D et al. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoh SM et al. PQBP1 is a proximal sensor of the cGAS-dependent innate response to HIV-1. Cell 161, 1293–1305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilks N et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 15, 5383–5398 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Q et al. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 15, 228–238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tourrière H et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell. Biol. 160, 823–831 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Jonsson KL et al. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 8, 14391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almine JF et al. IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Commun. 8, 14392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao J et al. Nonspecific DNA binding of cGAS N terminus promotes cGAS activation. J. Immunol. 198, 3627–3636 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Shim JH et al. Epigallocatechin gallate suppresses lung cancer cell growth through Ras-GTPase-activating protein SH3 domain-binding protein 1. Cancer Prev. Res 3, 670–679 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Guo H et al. Clinicopathological and genetic analysis of Aicardi-Goutières syndrome. Chinese J. Neurol. 47, 96–100 (2014). [Google Scholar]
- 28.Liu Y et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371, 507–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark K, Plater L, Peggie M & Cohen P Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation. J. Biol. Chem. 284, 14136–14146 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Civril F et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou W et al. Structure of the human cGAS-DNA complex reveals enhanced control of immune surveillance. Cell 174, 300–311.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreeva L et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 549, 394–398 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Orzalli MH et al. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl Acad. Sci. USA 112, E1773–E1781 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackenzie KJ et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harding SM et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl. Acad. Sci. USA 114, 1637–1642 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 1019–1031 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu C et al. Oxygen promotes biofilm formation of Shewanella putrefaciens CN32 through a diguanylate cyclase and an adhesin. Sci. Rep 3, 1945 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS-identified cGAS-interacting protein list is provided in Supplementary Tables 1 and 2. The primer sequences used for qPCR are provided in Supplementary Table 3. The information on antibodies used in our study is provided in Supplementary Table 4. We also provide full scans of all the blots and gels as Supplementary Dataset 1. The data that support the findings of this study are available from the corresponding authors upon reasonable request.