Abstract
Studying the dynamic interaction between host cells and pathogen is vital but remains technically challenging. We describe herein a time-resolved chemical proteomics strategy enabling host and pathogen temporal interaction profiling (HAPTIP) for tracking the entry of a pathogen into the host cell. A novel multifunctional chemical proteomics probe was introduced to label living bacteria followed by in vivo crosslinking of bacteria proteins to their interacting host-cell proteins at different time points initiated by UV for label-free quantitative proteomics analysis. We observed over 400 specific interacting proteins crosslinked with the probe during the formation of Salmonella-containing vacuole (SCV). This novel chemical proteomics approach provides a temporal interaction profile of host and pathogen in high throughput and would facilitate better understanding of the infection process at the molecular level.
Keywords: chemical proteomics, in vivo labeling, pathogen–host interactions, photo crosslinking, quantitative proteomics
Uncovering specific interactions between host cells and pathogens and measuring the proteome changes in host cells after pathogen infection is critical for understanding the biology of pathogen infection.[1] The study of infectious diseases has greatly benefited from the contribution of proteomic approaches, which can provide a global view of host–pathogen interactions.[2] However, there are still questions to be addressed. Most of the existing strategies for mapping of the host–pathogen interaction or the changes in the proteome of the host only focus on certain infection time points,[3–5] and thereby fall short in providing a temporal interaction profile that can cover a more complete infection cycle. Moreover, a lot of these interactions may be weak and transient, and thus are difficult to capture by traditional approaches.
Recently, several chemical proteomic approaches were introduced to identify receptors for ligands.[6,7] These studies also suggest their application in finding new putative pathogen–host interactions. As these approaches mainly aimed at capturing receptor through chemical reactions occurring on the host-cell surface, they are not suitable for revealing the interactome after the pathogen enters the host cells. On the other hand, the use of photoactivatable probes offers a key advantage of the light-controlled conversion of transient noncovalent interactions into covalent isolable complexes, thus enabling large-scale identification of ligand (e.g., drug or lipid)-binding proteins or substrates of enzyme within the cells by mass spectrometry (MS).[8–11]
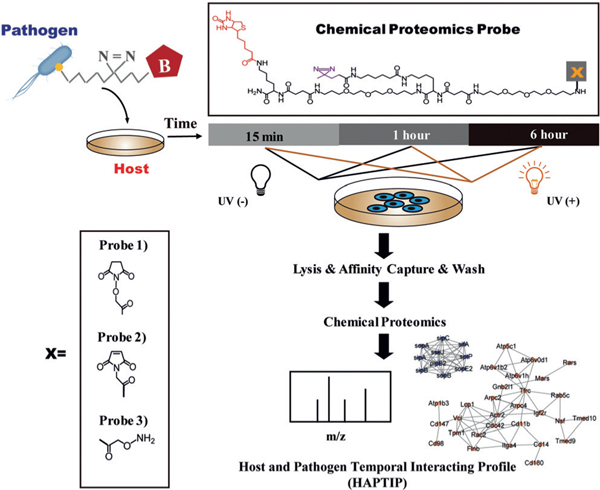
Herein, we report the development of a novel chemical proteomics strategy, termed host and pathogen temporal interaction profiling (HAPTIP), to globally map the host–pathogen interactome and demonstrate its application to the Salmonella infection process. Salmonella are Gram-negative bacterial pathogens that can infect a wide range of animals including humans, farm animals, and even plants.[12] Upon infection, Salmonella replicates within host cells in a membrane-bound compartment, called the Salmonella-containing vacuole (SCV).[13] Intravacuolar bacterial replication depends on tightly controlled interactions with host-cell vesicular compartments. Therefore, it is crucial, albeit technically challenging, to characterize the interactions involved in the Salmonella infection process. The core of the HAPTIP method is a new multifunctional chemical proteomics probe bearing a labeling group that conjugates the probe to Salmonella surface, a photo-reactive diazirine group that allows for covalent crosslinking of Salmonella proteins to their interacting host-cell proteins thereby facilitating the discovery of transient or weak interactions, and an isolation group for purifying the interacting proteins for quantitative proteomics analysis. We apply the HAPTIP to study the host–pathogen interactome in a time-resolved manner throughout the course of SCV formation. To the best of our knowledge, this is the first attempt to chemically label living bacteria for proteome profiling of the host-cell response.
Labeling living bacteria chemically requires careful considerations; the labeling reaction and resulting covalent attachment should have minimal impact on the function and activity of bacteria, and the labeling should have good efficiency for the follow-up identification of low abundance, specific interacting proteins in the host cells. Accordingly, we prepared three probes with different types of labeling group (Figure 1): 1) N-hydroxysuccinimide (NHS) group that reacts with the amine group on the cell surface proteins (Supporting Information, Figure S1), 2) maleimide (MAL) group that reacts with the thiol group on the cell surface proteins (Supporting Information, Figure S2), and 3) aminooxy (ONH2) group that conjugates the reagent to the glycans on bacteria surface through an oxime click reaction (Supporting Information, Figure S3). The three probes were chosen based on the availability of corresponding surface reactive groups (amine, thiol, and glycan groups) on the pathogen and mild reaction conditions. Labeling on primary amine groups is expected to be most efficient, leading to potentially significant impact on bacteria activities.[14] On the other hand, sulfhydryl groups (thiols) are present in most proteins but are not as abundant as primary amines, and we expected labeling through thiols would have a minimum effect on Salmonella activity. For the ONH2-probe, the oxime click reaction was chosen because it conjugates the aldehyde group generated after mildly oxidizing glycans on the Salmonella surface under physiological condition.[15,16] As the oxidation conditions we utilized only oxidize the terminal sialic acid on the cell surface and the probe was designed with a cell impermeant polyethylene glycol chain, the labeling is expected to attach the probe on the outermost surface, thus facilitating the capture of interacting proteins with minimum steric hindrance. Moreover, because lipopolysaccharides (LPS) present on the outer membrane of Gram-negative bacteria can recognize and bind cell surface receptors during infection, we assumed that labeling the glycan chain would help to identify LPS-interacting proteins as well.
Figure 1.
Experimental workflow for tracking the entry of Salmonella using the HAPTIP method.
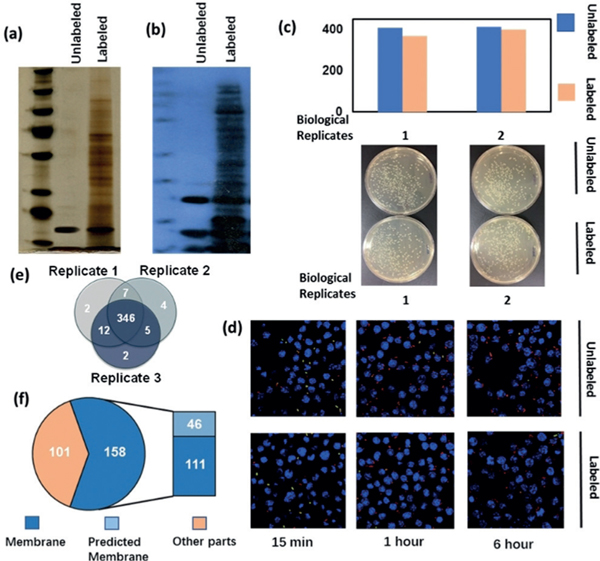
Salmonella was labeled with three distinctive probes, and we examined the labeling efficiency and its effects on Salmonella growth, infection efficiency, and intracellular survival. Results showed that the NHS-probe labeling affected bacterial growth (Supporting Information, Figure S1 c) although the labeling efficiency was high (Supporting Information, Figure S1 b). The labeling efficiency of Malprobe was relatively low owing to the limited number of free thiol groups on the cell surface (Supporting Information, Figure S2 b) and the bacterial growth was not affected (Supporting Information, Figure S2 c). The ONH2-probe showed good labeling efficiency as revealed by SDS-PAGE (Figure 2 a) and western blot detected by anti-biotin antibody (Figure 2 b). Importantly, the labeling had little influence on Salmonella growth (Figure 2 c). Subsequently, we compared the ability of Salmonella to infect host cells with or without ONH2-functionalized probe labeling. Encouragingly, the infection rate and intracellular survival in RAW264.7 macrophage cells with ONH2-probe-labeled Salmonella proved to be almost identical to the unlabeled Salmonella (Figure 2d and Supporting Information, Figure S4). We further carried out mass spectrometric analysis on ONH2-labeled bacteria proteins enriched by NeutrAvidin beads. Bacteria without labeling but captured by the NeutrAvidin beads was also analyzed by MS as the control. After subtracting the control hits, a total of 378 proteins were identified from three replicates (Figure 2e and Supporting Information, Table S1). Among them, subcellular location of 259 proteins were known or predicted. Gene ontology cellular component (GOCC) analysis showed that more than 60 % of these proteins were membrane proteins (Figure 2 f). Cell-invasion proteins including SipB, SipD, and secreted effector protein SifA, SopD, SopE2, SseC, and SseJ, which are known to be important in Salmonella infection were identified, indicating that the labeling was primarily on the surface of Salmonella. Therefore, we finally chose the ONH2-functionalized probe for all following studies. The synthesis and characterization of the multifunctional probe are outlined in Supporting Information, Figures S5–8.
Figure 2.
a) SDS-PAGE (silver stain) of Salmonella unlabeled and labeled with probe, lysed, and captured by NeutrAvidin beads. b) Western blot (against biotin antibody) of Salmonella unlabeled and labeled with probe lysed, and captured by NeutrAvidin beads. c) Enumeration of colony forming units (cfu) for labeled and unlabeled bacteria.d) Differential inside/outside staining showing the infection and survival rates of unlabeled and labeled Salmonella to macrophage cells. e) Proteins identified from labeled Salmonella and captured by NeutrAvidin beads (After removing control). f) GO cellular component of proteins identified from labeled Salmonella and captured by NeutrAvidin beads (after removing control).
As illustrated in Figure 1, to temporally profile the interaction between Salmonella and host cells, labeled Salmonella were incubated with macrophage cells at the multiplicity of infection (MOI) of 50 for set intervals ranging from 15 min to 6 h. Three time points were chosen based on the critical formation of SCV.[13] After washing away the externally attached Salmonella, macrophage samples with different infection times were irradiated with 365 nm UV light for 10 min to covalently crosslink proteins within close proximity to Salmonella. We reasoned that the probe with a photoreactive diazirine group, which is known to be specific in crosslinking molecules in close proximity, only react with proteins that are in direct contact with Salmonella, thereby minimizing crosslinking of other non-specific interacting proteins.[17] The irradiated cells were then harvested and lysed with lysis buffer, and NeutrAvidin beads were used to isolate the putative targets. The beads were subsequently washed with vigorous washing conditions. Control isolation (without UV radiation) was also performed in the same biological context tested. The proteins enriched by NeutrAvidin beads either from samples and controls were digested on-beads sequentially with Lys-C and trypsin, and the resulting peptides were characterized and quantified by label-free LC-MS/MS.
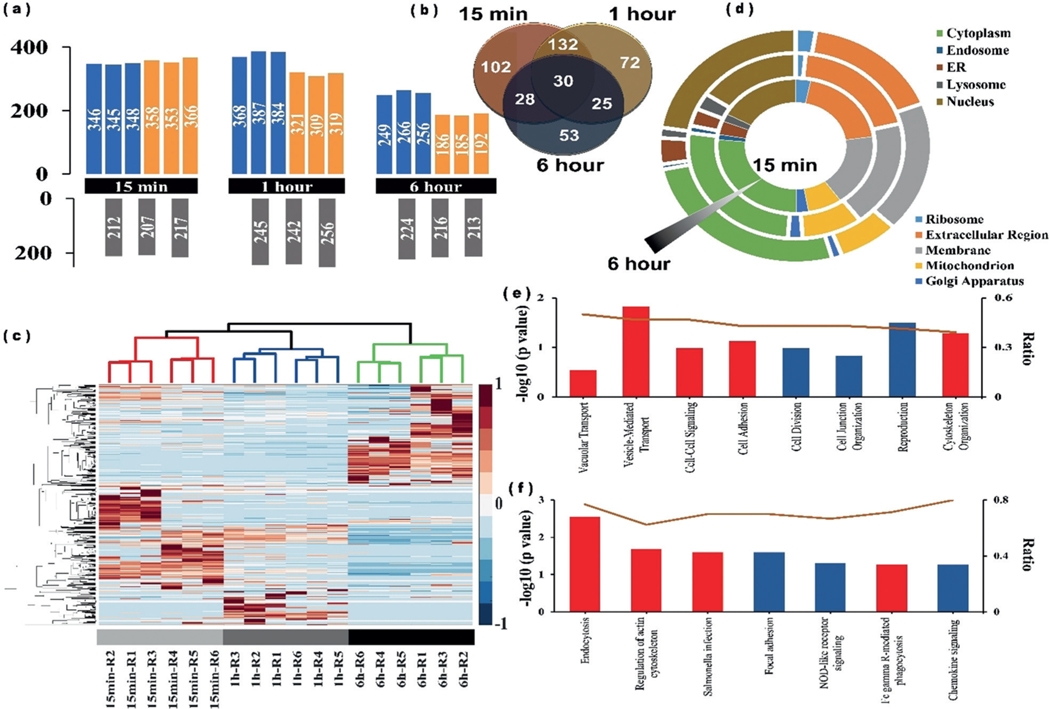
In total, we identified 442 crosslinked proteins in two biological replicates over the three time points during Salmonella internalization into macrophage cells (Supporting Information, Table S2). Proteins identified from samples and controls in each time point is shown in Figure 3 a. Among them, 292 (66 %), 232 (52 %), and 136 (31 %) proteins were exclusively captured at 15 min, 1 h, and 6 h, respectively (Figure 3 b). High technical and biological reproducibility were achieved, as two independent biological replicates and three technical replicates correlated well in each time point (Figure 3c and Supporting Information, Figure S9). The correlation analysis also revealed that the 15 min and 1 h results were more closely related, as both of these two time points are within the relatively early endocytosis process. The three time points clearly separated in the principal component analysis (PCA) plot, also with the 15 min and 1 h sharing more similarity (Supporting Information, Figure S10). We then analyzed the proteomics data to see whether these proteins can fall into distinct biological processes or pathways. GOCC analysis showed the proteins with the highest levels of enrichment originated from extracellular regions (including vesicles, organelles, and exosomes) or from membrane-bound vesicles or organelles, especially at 0.5 and 1 h (Figure 3 d). The GOCC results agree with the infection process, as adhesion to the host-cell surface is important for many bacteria to initiate infection, and further fuse with the cellular membrane to form SCV. We identified more nuclear proteins in the 6 h samples than the other two time points. The reason for this observation may be that the SCV membrane gets damaged in the late endocytosis process and the pathogen may escape into the host-cell cytosol and become associated with nuclear proteins. The functional analysis indicates many proteins identified in these three time points are related to biological processes implicated in endocytosis, Salmonella infection, regulation of actin cytoskeleton as well as focal adhesion (Figure 3 e) and pathway analysis shows that these proteins are involved in a number of biological processes linked to vacuolar transport, cell–cell signaling, and vesicle mediated transport (Figure 3 f).
Figure 3.
a) Number of proteins identified from each time point in two biological replicates. Upper panel shows proteins identified from UV + sample, lower panel shows proteins identified from UV sample as control, each bar represents the identified protein number from a single MS run. b) Venn diagram of the crosslinked proteins at the three time points. c) Heatmap analysis of the identified proteins from each time points. Rows in heatmap denote quantified proteins, and columns denote sample. Red color denotes highly expressed protein, whereas blue denotes lowly expressed protein. d) GO cellular component of the identified proteins from each time points. e) GO biological process enrichment analysis of the identified proteins from each time points. f) KEGG pathway analysis of the identified proteins from each time points. The ratio value is defined as the protein number of identified for each functional category normalized by protein number of genome background. Among them, several important biological process and pathway were particularly highlighted with red.
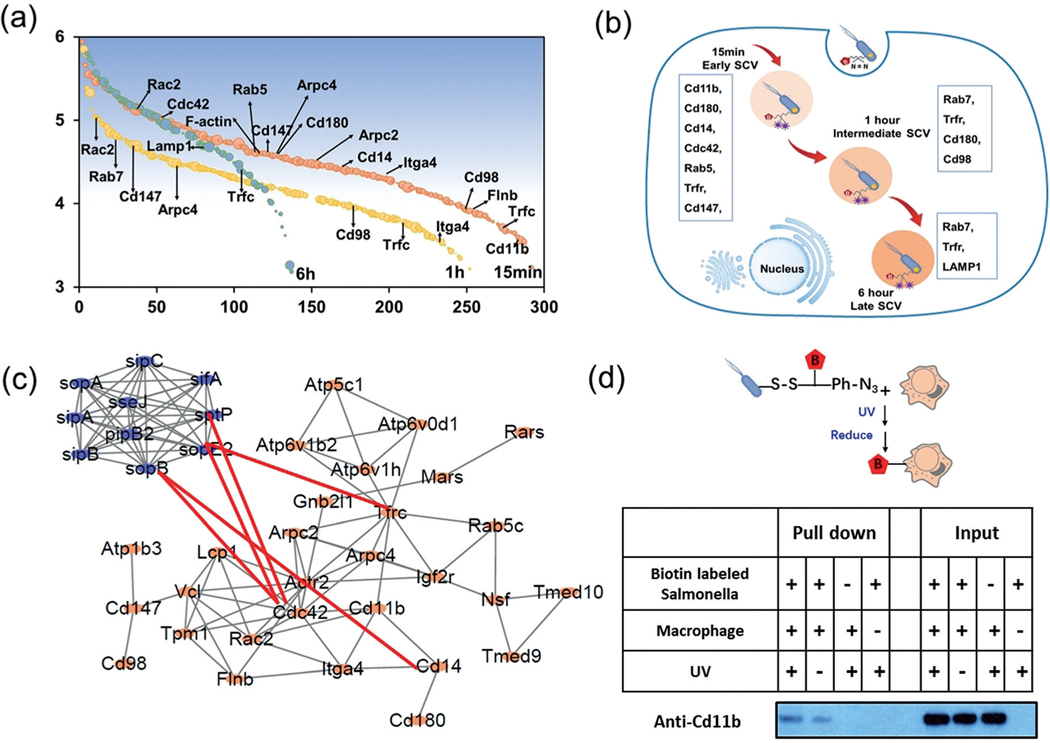
Among specific proteins identified in each time point (Figure 4 a), at the very early time point of 15 min infection, Salmonella was internalized by phagocytic cells using the actin polymerization pathway, and proteins involved in this process are expected to crosslink with Salmonella. Indeed, Arpc 2, Arpc 4, and F-actin capping proteins were identified, which agrees with the fact that actin polymerization is stimulated in the initial steps of the internalization.[18] Other known interactions were also identified, for example Cdc 42, which is known to be activated by Salmonella effector SopB/SigD;[19] and Cd14, which is reported as the receptor for Salmonella.[20] Early SCV markers, Rab 5 and Trfc are also present in the 15 min sample. In the 1 h sample, several proteins identified are overlapped with the 15 min samples, including Rab5, Rab7, Trfc, Cd14, Cd147, and Cd180, while Cdc42 disappeared. In the 6 h sample, early and intermediate SCV matures into late SCV by loss of early endosomal proteins and simultaneous acquisition of selective late endosomal and lysosomal proteins including LAMP1,[21] while proteins that participate in the early SCV formation all disappeared, including the above-mentioned Cd family proteins and early SCV markers, Rab5. Overall, these identified proteins include not only known SCV markers but also known receptors and adaptors for Salmonella, like Cd14. Cd14 has been recognized as a co-receptor (along with the Toll-like receptor TLR4 and MD-2) for the recognition of lipopolysaccharide from Salmonella surface.[22] We identified Cd14 in the early infection stage but not in the stage of intermediate and late SCV, demonstrating the direct interaction between Salmonella and Cd14 during the infection. Meanwhile, proteins from Salmonella that are known to interact with the host cells and help the uptake of Salmonella including SifA, SipB, SipC, SopA, SopD, SopE, and SopE2 were identified (Supporting Information, Table S3),[13,18] further indicating the capability of this method to identify interaction between pathogen and host. Together, these results demonstrate that the HAPTIP strategy enabled us to trace the entry of Salmonella into host cells and identify the specific interacting proteins at different SCV formation processes (Figure 4 b). Pathway analyses revealed the known interaction between the identified proteins from the host cell, and our results further provided the new information on the network among host-cell and pathogen proteins (Figure 4 c).
Figure 4.
a) Proteins identified from each time point in two biological replicates after removing the control. Y-axis denotes log 10 transformed iBAQ value of identified proteins (Shaded backgroud denotes the relative iBAQ intensity), and x-axis denotes proteins. b) Illustration of Salmonella–host interacting proteins involved in different SCV process. c) IPA Interaction network of identified proteins from macrophages and from Samonella, known interaction were connected with red line. d) Validation of interaction between Salmonella and macrophage. Host proteins were pulled down by biotin-labeled Salmonella using NeutrAvidin beads and analyzed by western blotting using antibodies against the protein. Before pull-down, samples were treated with or without 365 nm UV irradiation.
In addition to proteins that have been previously reported to interact with Salmonella, we also discovered several macrophage proteins that were not previously known to interact with Salmonella during early stages of infection, such as Cd98, Cd180, Cd147, and Cd11b. Some of these were reported as receptor or ligands for LPS in other bacteria but have not been reported in the Salmonella–macrophage system. For example, the lipid raft-associated protein Cd98 is required for vaccinia virus endocytosis,[23] while the immunoglobulin superfamily member Cd147 is a critical host receptor for the meningococcal pilus components PilE and PilV.[24] Among them, we chose an interacting protein, Cd11b, to further verify its interaction with Salmonella. Cd11b was previously reported to bind LPS and promote TLR4 signaling and, importantly, may participate in LPS uptake into cells.[25] However, its role has not been clearly described in the process of Salmonella entry. To verify the interaction between Cd11b and macrophages in vitro, we applied a biotin switch method using a commercially available crosslinking reagent. Salmonella was first labeled by sulfosuccinimidyl-2-[6-(biotinamido)-2-(p-azidobenzamido) hexanoamido]ethyl-1,3′-dithiopropionate (Sulfo-SBED), and the whole bacteria extract then was incubated with the lysate of macrophage. The mixture was irradiated with the same UVas described before and the crosslinked proteins were purified by NeutrAvidin beads. The recovered proteins were treated with the gel-loading buffer that contains the reduction reagent dithiothreitol to reduce the disulfide bond on the crosslinking reagent, which resulted in the transfer of the biotin group to Salmonella-interacting proteins in host cells to facilitate their pull-down by NeutrAvidin beads. The western blot result confirmed the crosslinked Cd11b after UV crosslinking, while a much weaker signal was observed without UV crosslinking (Figure 4 d). The interaction between Cd11b and Salmonella was further confirmed by a standard pull-down experiment (Supporting Information, Figure S11). Together, the HAPTIP strategy allowed us to identify novel interactions between Salmonella and macrophages, which provided leads for future exploration on understanding the molecular mechanism of the bacterial infection process.
The interaction between host cells and pathogens are highly dynamic and complex with many questions to be answered. It is extremely valuable to provide a dynamic picture of such interactions during the infection process. Towards this goal, we developed a novel time-resolved chemical proteomics strategy. It is conceivable that the general strategy of HAPTIP can be applicable to many bacteria or virus, thus contributing to the discovery and understanding of host–pathogen interactions in multiple infection systems.
Supplementary Material
Acknowledgements
This project has been funded by NIH grants 1R01GM111788, S10 RR025044 and NSF grant 1506752 to W. A. Tao and 2016YFA0501300 to H. Lu and Y. Zhang.
Footnotes
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: https://doi.org/10.1002/anie.201911078.
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Ying Zhang, Department of Biochemistry, Department of Chemistry, Center for Cancer Research, Purdue University West Lafayette, IN 47907 (USA); Minghang Hospital and Institutes of Biomedical Sciences Fudan University 131 Dong’an Road, Shanghai 200032 (China).
Der-Shyang Kao, Department of Biochemistry, Department of Chemistry, Center for Cancer Research, Purdue University West Lafayette, IN 47907 (USA).
Bing Gu, Department of Biological Sciences, Purdue University West Lafayette, IN 47907 (USA).
Rajdeep Bomjan, Department of Biological Sciences, Purdue University West Lafayette, IN 47907 (USA).
Mayank Srivastava, Department of Biochemistry, Department of Chemistry, Center for Cancer Research, Purdue University West Lafayette, IN 47907 (USA).
Haojie Lu, Minghang Hospital and Institutes of Biomedical Sciences Fudan University 131 Dong’an Road, Shanghai 200032 (China).
Daoguo Zhou, Department of Biological Sciences, Purdue University West Lafayette, IN 47907 (USA).
W. Andy Tao, Department of Biochemistry, Department of Chemistry, Center for Cancer Research, Purdue University West Lafayette, IN 47907 (USA).
References
- [1].Garrett WS, Science 2015, 348, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Beltran PMJ, Federspiel JD, Sheng X, Cristea IM, Mol. Syst. Biol 2017, 13, 922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Weekes MP, Tomasec P, Huttlin EL, Fielding CA, Nusinow D, Stanton RJ, Wang EC, Aicheler R, Murrell I, Wilkinson GW, Lehner PJ, Gygi SP, Cell 2014, 157, 1460–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Scaturro P, Stukalov A, Haas DA, Cortese M, Draganova K, Płaszczyca A, Bartenschlager R, Götz M, Pichlmair A, Nature 2018, 561, 253–257. [DOI] [PubMed] [Google Scholar]
- [5].Yang YF, Hu M, Yu KW, Zeng XM, Liu XY, Protein Cell 2015, 6, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Frei AP, Jeon OY, Kilcher S, Moest H, Henning LM, Jost C, Plückthun A, Mercer J, Aebersold R, Carreira EM, Wollscheid B, Nat. Biotechnol 2012, 30, 997–1001. [DOI] [PubMed] [Google Scholar]
- [7].Sobotzki N, Schafroth MA, Rudnicka A, Koetemann A, Marty F, Goetze S, Yamauchi Y, Carreira EM, Wollscheid B, Nat. Commun 2018, 9, 1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bar-Peled L, Kemper EK, Suciu RM, Vinogradova EV, Backus KM, Horning BD, Paul TA, Ichu TA, Svensson RU, Olucha J, Chang MW, Kok BP, Zhu Z, Ihle NT, Dix MM, Jiang P, Hayward MM, Saez E, Shaw RJ, Cravatt BF, Cell 2017, 171, 696–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].He D, Xie X, Yang F, Zhang H, Su HM, Ge Y, Song HP, Chen PR, Angew. Chem. Int. Ed 2017, 56, 14521–14525; Angew. Chem. 2017, 129, 14713 – 14717. [DOI] [PubMed] [Google Scholar]
- [10].Broncel M, Serwa RA, Ciepla P, Krause E, Dallman MJ, Magee AI, Tate EW, Angew. Chem. Int. Ed 2015, 54, 5948–5951; Angew. Chem. 2015, 127, 6046 – 6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang LN, Yang L, Pan L, Kadasala NR, Xue L, Schuster RJ, Parker LL, Wei A, Tao WA, J. Am. Chem. Soc 2015, 137, 12772–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Haraga A, Ohlson MB, Miller SI, Nat. Rev. Microbiol 2008, 6, 53–66. [DOI] [PubMed] [Google Scholar]
- [13].Steele-Mortimer O, Curr. Opin. Microbiol 2008, 11, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Greg TH, Bioconjugate Techniques, 2nd ed., Academic Press, New York, 2008, pp. 171 and 183. [Google Scholar]
- [15].Zeng Y, Ramya TNC, Dirksen A, Dawson PE, Paulson JC, Nat. Methods 2009, 6, 207–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Xiao HP, Suttapitugsakul S, Sun FX, Wu RH, Acc. Chem. Res 2018, 51, 1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li ZQ, Hao PL, Li L, Tan CYJ, Cheng XM, Chen GYJ, Sze SK, Shen HM, Yao SQ, Angew. Chem. Int. Ed 2013, 52, 8551–8556; Angew. Chem. 2013, 125, 8713 – 8718. [DOI] [PubMed] [Google Scholar]
- [18].Schleker S, Sun JC, Raghavan B, Srnec M, Muller N, Koepfinger M, Murthy L, Zhao ZM, Klein-Seetharaman J, Proteomics Clin. Appl 2012, 6, 117–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rogers LD, Kristensen AR, Boyle EC, Robinson DP, Ly RT, Finlay BB, Foster LJ, J. Proteomics 2008, 71, 97–108. [DOI] [PubMed] [Google Scholar]
- [20].Rapsinski GJ, Newman TN, Oppong GO, van Putten JP, Tükel Ç, J. Biol. Chem 2013, 288, 14178–14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Knodler LA, Steele-Mortimer O, Traffic 2003, 4, 587–599. [DOI] [PubMed] [Google Scholar]
- [22].Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, Granucci F, Kagan JC, Cell 2011, 147, 868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schroeder N, Chung CS, Chen CH, Liao CL, Chang W, J. Virol 2012, 86, 4868–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bernard SC, Simpson N, Join-Lambert O, Federici C, Laran-Chich MP, Maissa N, Bouzinba-Segard H, Morand PC, Chretien F, Taouji S, Chevet E, Janel S, Lafont F, Coureuil M, Segura A, Niedergang F, Marullo S, Couraud PO, Nassif X, Bourdoulous S, Nat. Med 2014, 20, 725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kagan JC, Trends Immunol. 2017, 38, 696–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.