Abstract
Complex microbial communities in environmental systems play a key role in the detoxification of chemical contaminants by transforming them into less active metabolites or by complete mineralization. Biotransformation, i.e., transformation by microbes, is well understood for a number of priority pollutants, but a similar level of understanding is lacking for many emerging contaminants encountered at low concentrations and in complex mixtures across natural and engineered systems. Any advanced approaches aiming to reduce environmental exposure to such contaminants (e.g., novel engineered biological water treatment systems, design of readily degradable chemicals, or improved regulatory assessment strategies to determine contaminant persistence a priori) will depend on understanding the causal links among contaminant removal, the key driving agents of biotransformation at low concentrations (i.e., relevant microbes and their metabolic activities), and how their presence and activity depend on environmental conditions. In this Perspective, we present the current understanding and recent methodological advances that can help to identify such links, even in complex environmental microbiomes and for contaminants present at low concentrations in complex chemical mixtures. We discuss the ensuing insights into contaminant biotransformation across varying environments and conditions and ask how much closer we have come to designing improved approaches to reducing environmental exposure to contaminants.
1. Drivers behind Biotransformation: Changes at Low Concentrations
Microbes, including bacteria, archaea, fungi, and other small eukaryotes, harbor a tremendous diversity of catabolic capacities and are nearly ubiquitous. They therefore play a key role in reducing environmental exposure to chemical contaminants by transforming them into less bioactive compounds, which can continue all the way to complete mineralization.1,2 Compared to other types of environmental transformations such as photolysis or abiotic redox reactions, microbial transformation is generally considered to be the most important degradation process in terms of mass balance.3 However, there is a growing body of evidence that increasing levels of chemical contamination in the environment directly impair human and environmental health4 (see, e.g., loss of invertebrate biodiversity in pesticide-polluted streams5 or compromised reproduction in humans due to exposure to endocrine-disrupting chemicals6). Given the need to manage increasingly closed local and regional water cycles7 and the ever-increasing production and consumption of chemicals,8 we can no longer just passively rely on microbes to reduce contaminant levels. Rather, we should aim to actively take advantage of their capabilities to decontaminate water resources by developing optimized engineered systems at critical control points of the water cycle (e.g., at wastewater and drinking water treatment facilities) or by designing new chemicals in a way that conserves their intended activity while rendering them readily transformable to innocuous substances in the environment. Designing systems and/or chemicals optimized for microbial transformation requires understanding (i) the causal links between contaminant removal and the specific agents of biotransformation (i.e., relevant microbes and catabolic pathways down to the enzyme level) and (ii) the conditions under which biotransformation can occur.
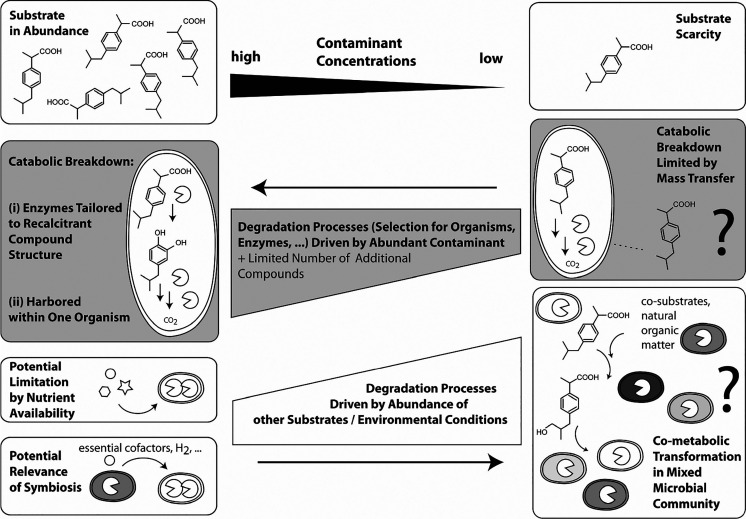
In general, any observed contaminant biotransformation outcome (i.e., whether a given contaminant is removed, at what rate, and yielding what kind of products) is the result of a complex interplay of several factors (Figure 1). These factors are (i) the bioavailable concentration of the contaminants in question, (ii) the composition and capabilities of the microbial community (e.g., expressed metabolic pathways or single enzymes), and (iii) the presence of the proper substrates and electron donors or acceptors.9,10 Different environmental conditions (e.g., redox conditions, temperature, humidity, nutrient status, chemical exposure, microbial residence time, etc.) determine these factors and hence exert control on contaminant biotransformation. For each individual contaminant, the extent to which it is biotransformed is in great part determined by those factors and by its chemical structure, which determines its intrinsic recalcitrance, i.e., its ability to interact with specific enzymes.
Figure 1.
Biodegradation as driven by an interplay among environmental conditions, compound structure, compound concentration, and potential active agents of biotransformation. The selective pressure imposed by the substrate decreases at lower concentrations, so that environmental conditions become more important. Consequently, drivers of biodegradation may change fundamentally when moving from high (left) to low (right) concentrations.
Over the past 30–40 years, a large body of biotransformation knowledge has accrued for a number of major legacy contaminants, mostly in a bioremediation context, i.e., for highly concentrated mixtures (e.g., chlorinated solvents,11−14 BTEX,9,15 or explosives/munitions chemicals16). In such a context, contaminants are typically metabolically degraded; i.e., they can be used as sources of carbon or other nutrients, and/or as electron donors or acceptors by microbes, and, therefore, drive the general biodegradation setting (i.e., selection for organisms, evolution of pathways, etc.) (Figure 1, left side). Consequently, to explore optimal conditions for bioremediation, research has focused on identifying specific degraders or consortia that become dominant under such thermodynamically favorable conditions, involving experimentation with enriched or pure cultures, characterization of metabolic pathways involving growth assays, and enzyme purification and characterization. More advanced work has focused on the intricate metabolic interactions in contaminant-degrading consortia, thus introducing a community level perspective.17,18 This research has taught us the importance of mutualistic and symbiotic interactions within complex microbiota, where partner organisms either generate necessary electron donors (e.g., hydrogen19) or supply cofactors (e.g., corrinoids20) essential for efficient pollutant removal (Figure 1, left side).
More recently, awareness of the widespread presence of highly complex mixtures of hundreds of anthropogenic chemicals at low concentrations (i.e., in the low microgram per liter to nanogram per liter range) throughout aquatic and terrestrial environments has increased.21 Biodegradation of these chemicals of emerging concern at low concentrations is far less understood but is key to developing and deploying strategies to reduce environmental exposure to such contaminants (Figure 1, right side). While the importance of substrate availability, i.e., the concentration of the contaminant in question, is recognized in principle,22 bioavailability limitations have primarily been expected from diffusion through microbial biofilms,23 or when compounds are sorbed to the solid matrix in heterogeneous environments such as soils.24 In contrast, for suspended microbes in water, dissolved contaminant concentrations next to the cell have commonly been considered to be directly available. Yet with decreasing pollutant concentrations, it can be expected that metabolic degradation, i.e., biodegradation by abundant, specialized degraders with targeted, efficient enzymes, will become less competitive, even for fully dissolved and hence available contaminants, because energy gained from degrading the available contaminant may no longer be sufficient to fulfill the degraders’ energy requirements. In that case, mixed substrate usage and co-metabolic (“fortuitous”) transformation by promiscuous enzymes expressed for different purposes may potentially become more prominent. With this, exertion of selective pressure by the contaminant decreases, while the influence of environmental conditions, e.g., the presence of a co-substrate such as natural organic matter and the composition and activity of microbial communities (which itself is a product of the environmental conditions), on contaminant degradation increases (Figure 1, right side). The following questions specifically arise in this context. (1) Is there a concentration below which contaminant availability becomes so limiting that a niche for catabolic breakdown by degraders no longer exists? (2) If so, are there examples and circumstances under which chemicals are efficiently transformed despite low concentrations? (3) What are the underlying degradation mechanisms in those cases? Are those compounds still metabolically degraded, meaning that the complete enzymatic toolbox is hosted within one degrader or small degrader consortium that fulfill their overall energy and nutrient requirements from mixed substrate usage, or are those compounds co-metabolically (“fortuitously”) transformed and hence do not serve as growth substrates for a specific degrader or degrader consortium? In the latter case, enzymes, the active agents, may no longer be harbored by a single organism only, but may be distributed over a microbial community (“metaorganism”) (Figure 1, right side). An important consequence of co-metabolic degradation is that degradation might not be complete but stop at any given stage, raising questions about the fate and effect of recalcitrant transformation products thus formed.
The major challenge in acquiring mechanistic insights into biodegradation under low concentrations is that well-established direct approaches of our disciplines may no longer be adequate. Enrichment cultivations, requiring high contaminant concentrations and reducing highly diverse communities to simple, few-membered communities, are not likely to inform on complex environmental settings relevant at low concentrations. The limitations are even more pronounced if the responsible enzymes and catabolic fluxes are distributed over multiple organisms. Advanced methodological approaches are thus essential for probing and understanding contaminant biotransformation in complex mixtures with other contaminants and in complex microbial communities. In this Perspective, we therefore first survey recent methodological advances that open new windows of opportunity for gaining mechanistic insights into contaminant biotransformation at relevant environmental concentrations and at the level of complex microbial communities (section 2). We then present concrete examples of successful application of these methods to gain mechanistic insights into low-concentration contaminant transformation across varying environments and conditions (section 3). In section 4, we discuss the extent to which these emerging insights may eventually inform the design of improved chemical and engineering approaches to reduce environmental exposure to contaminants. Finally, in section 5, we put everything into context and highlight approaches that seem to have the greatest potential for future progress in contaminant biotransformation research.
2. Novel Methods for Elucidating Biotransformation at Trace Levels
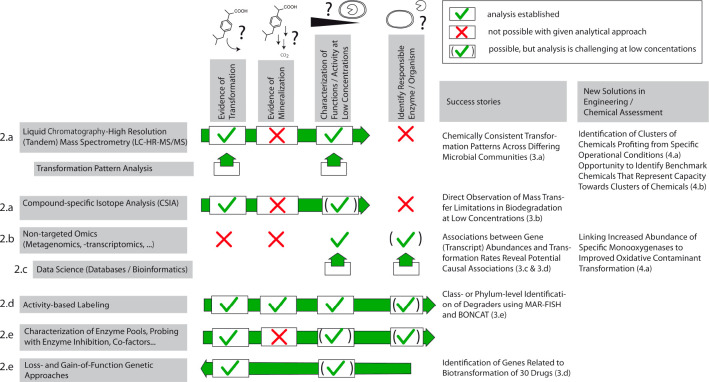
In the following, we focus on recent methodological advances that are, or have the potential to be, instrumental in providing mechanistic insights into contaminant biotransformation at low, environmentally relevant concentrations and at the level of complex microbial communities. These methods can support us in answering two major questions in this context. (a) Is the contaminant transformed or even fully degraded? (b) Can the observed degradation activity be linked to specific microbial community functions (i.e., responsible organisms or enzymes)? Figure 2 summarizes the extent to which the different methods presented in the following contribute to answering these questions. One specific feature of this type of research is its need for being highly interdisciplinary, bringing together methods from analytical chemistry, molecular biology, high-throughput experimentation, and data sciences.
Figure 2.
Realized and potential contributions of novel analytical and experimental methods for elucidating the mechanisms and agents of biotransformation at trace levels and for conceiving new solutions for environmental engineering and chemical assessment.
2a. High-Resolution and High-Sensitivity Chemical Analytics for Detecting Chemical Contaminant Transformation
The detection and characterization of low-level contaminant transformation in the environment has been revolutionized by two major technological advancements in analytical chemistry: high-resolution mass spectrometry (HRMS) and compound-specific isotope analysis (CSIA). These are complemented by different labeling-based approaches that can be applied in the case of metabolic degradation and will be presented separately further below.
Toward the turn of the century, the advent of softer ionization techniques, readily coupled to both gas and liquid chromatography, allowed for the detection of intact molecular ions. Robust and sensitive HRMS instruments now make the detection and quantification of low concentrations (i.e., down to a few nanograms per liter) of individual contaminants possible, also in complex contaminant mixtures and without the need for spiked reference standards.25 For transformation research, this has opened two unprecedented possibilities. First, transformation products can be sought26,27 and identified with reasonable certainty and accuracy28 using suspect and nontarget workflows both in batch experiments and in full-scale treatment systems.29 This allows characterization of contaminant degradation with respect to not only parent compound half-lives but also the actual enzymatic transformation reaction(s) that has taken place. Second, the possibility of resolving individual compound signals now allows us to study transformation processes in complex substance mixtures, generating information about biotransformation outcomes (e.g., rates and products) for many, structurally diverse chemicals under fully consistent conditions (see section 3a for an example). The sensitivity of these methods even allows the observation of biotransformation at ambient contaminant concentrations present in the environments of interest (e.g., impacted rivers). This allowed the demonstration, for instance, that biotransformation kinetics for some compounds differed between experiments that were run with the chemical residue levels present in the environment (down to50 ng/L) compared to those spiked to a specific, higher starting concentration of, e.g., 50 μg/L. This study highlighted the importance of understanding how community pre-exposure to contaminants and relative spike levels influence observed biotransformation kinetics.30
CSIA, the second analytical innovation, accesses a previously untapped source of information, naturally occurring stable isotopes in organic contaminants. Analysis by gas or liquid chromatography, coupled to online conversion to CO2, N2, or H2, and subsequent isotope ratio mass spectrometry (IRMS) (ref (31) and references cited therein) can measure contaminant isotopic signatures (13C/12C, 15N/14N, etc.) and deliver two different types of information. First, isotopic fingerprinting can elucidate different origins of the same chemical compound. Second, gradual changes in contaminant isotope ratios (over time or with transport distance) can detect natural contaminant degradation, because enzyme reactions usually prefer molecules with light isotopes (kinetic isotope effect). This leaves molecules with heavy isotopes behind, resulting in characteristic changes in contaminant isotope ratios that allow the detection of biodegradation. CSIA of multiple elements (C, Cl, or N) within the same contaminant can even reveal reaction-specific characteristic isotope effect patterns. This made it possible to discriminate underlying transformation mechanisms, such as different atrazine degradation pathways and diverging reaction mechanisms in reductive dehalogenases.32,33 In section 3b, we give an example of how CSIA can further be used to discriminate between diffusion and enzymatic biotransformation as rate-limiting steps in contaminant degradation. For studies at low contaminant concentrations, CSIA poses the challenge that rare isotopes (13C and 15N) need to be analyzed with the same precision as their more abundant counterparts. To this end, optimized and validated extraction methods need to be developed that push respective quantification limits into the submicrogram per liter range.34
2b. Nontargeted Omics Approaches for Resolving Microbial Community Functions in Relation to Trace Contaminant Degradation
To explore the role of microorganisms in complex communities as active agents in low-level contaminant transformation (Figure 1, right side), modern high-throughput DNA sequencing technologies (Illumina, Nanopore, MinION, etc.) are becoming an essential tool. Analysis of specific marker genes, such as that of rRNA, provides information about the phylogenetic placement of populations and on community composition, today with a resolution at the genus or even species level. Sequencing of the entire DNA (metagenome) or mRNA (metatranscriptome) pool of a complex microbial community, termed metagenomic or metatranscriptomic sequencing, respectively, additionally allows us to access the metabolic and biochemical capacities of individual populations and entire communities and can thus point toward potentially active biotransformation functions in the analyzed microbiome. These technologies have enabled the generation of ample knowledge about microbiota from diverse habitats, such as oceans,35 soil,36 wastewater,37 and human and animal hosts.38 Wu et al.,37 for instance, analyzed 16S rRNA gene sequences from more than 1200 wastewater treatment plants (WWTPs) on all six continents and used the data to elucidate which operational and environmental factors most strongly influence community composition and functioning in terms of BOD removal.
However, such sequencing approaches come with two major limitations in elucidating the main drivers of contaminant transformation. First, the identification of genes of interest in such large data sets may be all but trivial if the relevant catabolic pathways and reactions for a certain contaminant transformation have not been resolved a priori. Second, even if pathways are known and thus can be tracked, metagenomic or metatranscriptomic detection does not provide direct evidence that the respective pathway is actively involved in a given contaminant degradation process. In that context, it has been argued that (meta)proteomic data, i.e., the nontarget enumeration of enzymes in a given microbial community, describe the pool of enzymes potentially involved in contaminant transformation most directly39 and hence, particularly in combination with separation of the enzyme pools by compartment (intra- and extracellular), have the greatest potential of the different omics approaches to provide new mechanistic insights. As a prerequisite, however, protein identification and annotation require the availability of sample-specific metagenome data, which need to be interpreted and annotated with due caution.40 Also, wherever a certain pollutant induces the expression of a specific catabolic pathway, a variety of omics approaches (metagenomics, -transcriptomics, or -proteomics) may be successfully applied to identify functionally relevant microbiome components.41 Yet at low contaminant levels, the expected potential lack of metabolic pathway induction, particularly for co-metabolic transformation, questions the applicability of this approach for identifying agents of transformation.
Alternatively, omics-based profiling of different microbial communities may be combined with data-driven analysis pipelines to generate correlation-based hypotheses about the link between functions of interest, e.g., contaminant biotransformation and specific microbes or genes (see section 3c for an example).42 However, despite the first success stories, three major challenges are associated with correlational approaches. First, they often suffer from poor statistical power due to the high number of taxonomic and genetic features in a (meta)transcriptome/proteome compared to the typically limited number of observations and biological replicates. Second, the lack of standardized procedures for sample preparation, data treatment, and analysis may hamper quantitative analyses and cross-comparisons between studies. In particular, the normalization of transcript abundances across diverse communities and sequencing libraries is the subject of ongoing debate43,44 yet may strongly affect the outcome of quantitative or correlative interpretations. Third, the reliance on gene annotation and available databases to interpret microbial functions is often compromised by the fact that new annotations build on existing annotations in databases. Error rates in functional assignment for large, uncurated sequence databases have been documented to be as high as 80%, a compounding problem that has been termed “annotation rot”.45 In the following, we therefore discuss how pertinent databases and bioinformatics tools currently being developed may aid in alleviating these annotation challenges.
2c. Databases and Bioinformatics Approaches to Link Observed Transformations to Enzyme Functions and Underlying Gene Sequences
Protein and genome databases are essential for any workflow trying to link observed biotransformation reactions to genes and enzymes potentially catalyzing them. While there are a large number of freely available resources covering enzymatic reactions and pathway maps (e.g., KEGG, BRENDA, Rhea, and MetaCyc), protein sequences including functional annotations (e.g., SwissProt, UniProt, Pfam, and EggNOG), and genomes of individual bacterial strains (e.g., proGenomes2, RefSeq, Ensembl, and PATRIC), these are mostly focused on central and secondary metabolism and contain scarce information about contaminant biotransformation. For our purpose, databases that specifically collate relevant and well-curated information about contaminant biotransformation (i.e., pathways, enzymes, metabolites, and kinetics) therefore play a crucial role. Prominent examples include the Eawag-BBD/PPS (http://eawag-bbd.ethz.ch),46 originally developed at the University of Minnesota, and its successor, enviPath (http://envipath.org),47 or the plastics microbial biodegradation database PMBD (http://pmbd.genome-mining.cn/home/).48 Relative to Eawag-BBD/PPS, enviPath provides opportunities to store information about biotransformation kinetics besides pathway information and to supply both pathways and kinetics with metadata on environmental and operational conditions. These should allow the exploration of the dependence of contaminant removal on environmental conditions to learn about the causes underlying the observed variability in microbial biotransformation and to learn how to shape conditions to foster effective biotransformation (see section 3a for further discussion).
On the basis of these and other databases containing pathway and enzyme information, reaction mining and machine learning methods have been applied to develop a number of derivative, online tools that predict likely microbial contaminant biotransformation reactions and pathways based on chemical structure, such as Eawag-PPS,49 enviPath,47 BNICE,50 and Catalogic.51 Information contained therein can also be used to link observed or predicted transformation reactions to enzyme classes that can likely catalyze the respective transformation reaction52−55 and hence support a more targeted mining of omics data for associations between genes or enzymes and specific biotransformation reactions.42
One general limitation when trying to associate genes with observed biotransformation reactions is that only a limited number of biodegradation genes fall into families with distinct and limited enzymatic activities such as s-triazine ring opening56 or fumarate addition.57 Instead, most biodegradation reactions are catalyzed by enzymes in large protein families that are multifunctional. This requires more sophisticated sequence analysis to find signatures of specific reaction subclasses. In a biosynthesis context, for instance, AdenylPred was developed as an online tool (http://z.umn.edu/adenylpred) that can predict substrate type for adenylate-forming enzymes on the basis of their sequence.58 More toward biodegradation, a similar machine learning approach has been developed that predicts substrates of bacterial nitrilases on the basis of sequence.59 These and similar approaches rely on classifying multifunctional families into subfamilies based on their sequence similarity. By choosing appropriate similarity cutoff criteria, the user can observe clusters of highly related sequences that generally correlate with specific functions. These tools have delineated numerous biodegradation pathways and their enzymes and as such are contributing to combating “annotation rot”.60,61 A parallel approach is to directly predict the binding and reactivity of specific pollutants by known enzymes with X-ray structures via docking and molecular dynamics simulations.62,63 This is particularly useful with pervasive microbial biodegradative enzymes, for example, with aromatic hydrocarbon oxygenases, for which many X-ray structures of divergent members of the class are available. Once enzymes with predicted reactivity toward specific substrates are identified, sequence signatures derived from BLAST-type alignments can be used to identify closely related genes and enzymes in other microorganisms.
2d. Labeling-Based, Targeted Molecular or Cellular Approaches for Tracing Transformation Pathways and Responsible Organisms
Labeling represents a further paramount approach for linking process-relevant microbial populations to specific pollutant-degrading functions within complex microbiota.64 Process-based labeling typically involves addition of a labeled substrate, along with the detection of label incorporation (mostly stable isotopes or fluorogenic substrates) into degradation products, cellular biomarkers, enzymes, or entire cells. While labeled degradation products enable us to distinguish between co-metabolic transformation and catabolic breakdown to CO2, incorporation in biomolecules provides a unique angle for linking a process to responsible microorganisms. However, while some of the respective labeling tools have already found their way into bioremediation practice,65 their application to pollutant degradation at low concentrations or even to co-metabolic biotransformation remains a major challenge. The most important limitation is the substantial amount of label (e.g., 13C) that needs to be assimilated from the pollutant into biomass to allow for canonical process-based labeling strategies, such as stable isotope probing (SIP) of nucleic acids.66
Therefore, a major research need lies with the invention and application of labeling strategies capable of tracing low-level or co-metabolic activities. Here, recently emerging indirect and combinatorial labeling strategies can possibly lead the way. First, if hypotheses about the “main” metabolism of co-metabolic degraders are available, parallel labeling strategies can be designed to substantiate co-metabolic degradation. For instance, the monooxygenase systems of methanotrophs are known to be capable of co-metabolic micropollutant degradation67 and methylotrophs have been reported as labeled in experiments with 13C-labeled micropollutants.68 Experimental settings with switched labeled methylotrophic substrates and micropollutants could directly link such metabolic activities for the same complex inocula.68 Next, SIP studies with labeled contaminants have been conducted in parallel across wide ranges of contaminant concentrations (e.g., for endocrine disruptors69), thus attempting to extrapolate the involvement of identified populations at low concentrations in situ.
Recent advances in the detection of labeled gene products (transcriptome-SIP and protein-SIP70,71), albeit accomplished only for catabolic degradation to date, suggest that such methods may also be useful for dissecting functional adaptations during the co-metabolic degradation of contaminants within complex microbiota, if assumptions about the main catabolic activities of degraders are at hand. However, technical thresholds of labeling and detection thresholds may again limit actual applications at low concentrations. Here, advanced mass spectrometric approaches (e.g., NanoSIMS72) or high-sensitivity approaches for detecting cellular labeling such as Raman spectroscopy-based cell sorting, which can recognize and sort out labeled cells at the single-cell level,73 may offer important advantages. These approaches may be particularly promising when combined with general activity-based labeling strategies using 18O- or deuterium-labeled water.73,74 In that case, rigorous comparative experiments with and without added contaminants can potentially identify cells specifically involved in contaminant degradation and thus provide access for downstream single-cell genomics or even strain cultivation.75 Indirect labeling can also alleviate notorious limitations in acquiring 13C-labeled substrates for organic contaminants, which either can be very costly or are not commercially available. Such “next-generation physiology” approaches75 using cellular labeling with isotopic or fluorogenic substrates will be indispensable to truly link specific microbial populations to contaminant degradation in the environment.
2e. From Targeted to High-Throughput Screening Approaches for Uncovering Enzyme Activity and Specificity in Contaminant Transformation
A number of techniques have been developed and used regularly in different contexts to confirm enzymatic activity on specific substrates. These should also be highly instrumental as a second confirmatory tier to the workflows described above in cases in which there are strong hypotheses about the activity of a limited range of enzymes on a number of potential substrates. Such techniques include enzyme inhibition or addition of cofactors (e.g., refs (76) and (77)) and heterologous cloning and expression of target genes in host cells to confirm activity on specific substrates in whole cell assays or with purified enzymes. More recently, the latter approach has been developed further into more of a screening approach by using prior bioinformatics analysis to identify clusters of enzyme homologues that are suspected to catalyze certain types of transformation reactions, followed by cloning and experimental testing of the gene products on a range of potential substrates. These screening approaches typically rely on the availability or synthesis of substrates that produce fast readouts upon enzymatic conversion (e.g., fluorogenic probes78 or products with specific absorbances such as p-nitrophenolate79,80).
There are also a number of techniques emerging that allow the discovery of enzymatic activity on contaminants of interest in a more untargeted manner. They typically work with cell-free lysates81 and often involve some splitting of the enzyme pool into smaller pools, which facilitates downstream (meta)proteomic analysis if they show activity on specific contaminants of interest. Splitting might be achieved by different techniques such as gel electrophoresis and activity testing on gel slices,82 or a number of physical and physicochemical treatment steps, e.g., separating the activated sludge enzyme pool into extracellular, EPS-bound, and intracellular fractions.83 Another proteomics-based approach to identifying contaminant-degrading enzymes employs contaminant-specific chemical probes designed to pull down enzymes that convey a biotransformation of interest, as elegantly applied for the identification of β-glucuronidases.84 The currently probably most untargeted and high-throughput tools for uncovering microbiome-encoded genes involved in contaminant biotransformation take advantage of loss-of-function and gain-of-function genetic approaches, which combine genetic screening of whole genomes with phenotypic (e.g., antibiotic resistance) or high-throughput mass spectrometry-based readouts (see section 3d for an example).85,86
3. Emerging Insights from Application of Novel Methods
Using a combination of existing and emerging technologies as introduced above, a number of novel insights into low-concentration contaminant transformation in complex microbial communities were recently achieved.
3a. HRMS-Based Analysis Reveals Transformation-Specific Patterns
Emerging sets of consistent information about both biotransformation kinetics and reactions across large numbers of diverse chemicals, now accessible through HRMS/MS analysis, allow us to study patterns of contaminant biotransformation as a function of environmental and operational conditions and across different microbial communities. Kinetic analysis of biotransformation experiments with differently conditioned activated sludge and riverine biofilm communities revealed characteristic trends for groups of substances undergoing similar types of initial transformation reactions,87−89 suggesting that shared enzymes or enzyme systems that are conjointly regulated catalyze biotransformation reactions within such groups. In a number of recent studies exploring the influence of environmental or operational conditions on contaminant biotransformation across large sets of literature-retrieved data, global models embracing all compounds were therefore compared to class-specific models for groups of contaminants hypothesized to undergo similar initial biotransformation reactions. The latter generally showed improved performance in explaining observed variability in biotransformation kinetics.90,91 However, given that substrate specificities of enzymes are strong and difficult to predict,59 more such analyses across wider collections of compounds and different microbial systems are warranted to better understand the robustness of such reaction-specific groupings. Further explorations should also provide additional insights into whether the observation of reaction-specific grouping is exclusive to contaminants being transformed co-metabolically by the same unspecific enzyme system(s) or whether such patterns also emerge in the case of metabolic turnover.
3b. Mass Transfer Limitations to Metabolic Degradation as Uncovered by CSIA
Recently, compound-specific isotope fractionation unequivocally demonstrated a largely overlooked role of mass transfer limitation in contaminant degradation at low concentrations. Isotope fractionation is fully expressed if enzymatic turnover is limiting but becomes masked if mass transfer into bacterial cells becomes rate-determining. With bacteria cultivated on atrazine in a chemostat, a drastic decrease in the level of isotope fractionation was observed at a threshold concentration of ∼50 μg/L atrazine,73,74 and a similar decrease in isotope fractionation at lower concentrations was observed for the herbicide metabolite 2,6-dichlorobenzamide (BAM) in an inoculated sediment tank.92 First, this direct observation of mass transfer limitation demonstrated that diffusion into the cells through the cell membrane can become rate-limiting when concentrations decrease to levels close to the Monod/Michaelis–Menten constant of enzymatic breakdown93−95 (Figure 1, right side). Second, it revealed that the onset of mass transfer limitation occurs at residual concentrations that are orders of magnitude higher than the nanogram per liter concentrations at which products of biotransformation are observed by LC-HRMS in the environment. Third, mass transfer limitation appeared to trigger adaptation at the cellular level so that growth and turnover rates of catabolic degradation became significantly smaller once this threshold was reached.94,96 Because substrate uptake is a general prerequisite for metabolic degradation, mass transfer through the cell membrane is expected to become limiting at low concentrations eventually in all bacteria. Further studies may help us to understand how such mass transfer thresholds vary among organisms, in the presence of other substrates and as a function of molecular properties and enzymatic transformation rates.
3c. Association Mining Links Gene Transcripts to Biotransformation Reactions
The observation of reaction-specific variability in contaminant biotransformation across different microbial communities suggests the possibility of identifying genes for certain prevalent biotransformation reactions that would allow characterization of a given microbial community for its capacity to perform the respective reaction on a range of structurally related compounds. Under the assumption of co-metabolic transformation, Achermann et al.97 have therefore searched for correlations between biotransformation rate constants and gene transcript abundances across a series of activated sludge communities grown at different retention times. They limited the risk of identifying noncausal relationships by restricting the gene search space to those that encode enzymes potentially catalyzing the observed biotransformation reactions. In doing so, they were able to demonstrate linear and proportional relationships between biotransformation rate constants and relevant gene transcripts both for nitrification as a major community function and, more importantly, for biotransformation of two nitrile-containing contaminants (i.e., bromoxynil and acetamiprid) present at trace concentrations. While conversion of bromoxynil by nitrile hydratase had been known before, evidence of activity of nitrile hydratase-type enzymes on acetamiprid was only demonstrated later.97,98 Similar correlational approaches between kinetic and gene (transcript) abundance data have also been used by others to highlight enzyme classes potentially involved in pharmaceutical biotransformation during wastewater treatment99 and in contaminant biotransformation in soil columns representing managed aquifer systems.100,101
3d. Gain-of-Function Genetic Libraries Highlight Relevant Enzyme Functions in Gut Microbiota
A recent study illustrated the use of high-throughput mass spectrometry screening and genetic gain-of-function screens to identify microbial agents (i.e., strains and enzymes) that are responsible for the microbial metabolism of medical drugs in gut microbiota.85 Using a combinatorial pooling strategy, 271 drugs of maximal structural diversity were tested for potential biotransformation by 76 sequenced bacterial isolates from the human gut. Almost two-thirds of the tested drugs were found to be biotransformed by at least one gut isolate, and each tested bacterial strain transformed between 12 and 76 different pharmaceuticals. Next, a gain-of-function approach was employed to experimentally test all gene-encoded enzymes of the bacterial strains to be found most versatile in their ability to catalyze contaminant transformations. Concretely, the genomic DNA of a given strain was sheared into 2–8 kb fragments, which were cloned in an Escherichia coli expression vector, resulting in tens of thousands of E. coli clones. The biotransformation capacity of these clones was then tested against the original mixture of drugs to identify actively biotransforming clones and enzymes. With this approach, four of the most competent previously tested bacterial strains were profiled, and 30 genes that collectively biotransformed 20 different drugs were identified. Finally, the identified microbial agents were assessed as potential bioindicators for the biotransforming capacity of complex microbial communities for specific drugs. For this purpose, metagenomic sequencing of fecal communities was combined with the communities’ in vitro biotransformation rates, which demonstrated significant association between the abundance of the biotransforming enzymes/bacterial strains and the experimentally determined biotransformation rates. Altogether, this study illustrated how the combination of high-throughput mass spectrometry, bacterial culturing, and genetics can provide mechanistic insight into biotransformation processes at the molecular level and how this information could be exploited to identify bioindicators that explain and potentially predict the behavior of complex microbial communities.
3e. Activity-Based Labeling Pinpoints Bacterial Classes Involved in Contaminant Degradation
As mentioned above, a process-based, specific labeling of degrader biomarkers or cells with (stable) isotopes or fluorogenic substrates is not yet routinely applied to low-level or co-metabolic pollutant breakdown. Still, a few promising demonstrations exist, such as a recent phylum level assignment of distinct pharmaceutical degradation capacities at microgram per liter concentrations in wastewater biofilms via microautoradiography FISH (MAR-FISH).102 Although no further taxonomic or catabolic detail was provided, labeled populations all belonged to minor phyla within the sludge microbiota, and the study is an important proof of principle that degraders of low-concentration contaminants can be successfully labeled via substrate queries. In a commendably comprehensive study, the desulfurization of perfluorooctanesulfonate (PFOS) at low to high nanogram per liter concentrations was recently linked to distinct microbial clades within Antarctic coastal waters.103 Here, indirect cellular labeling with bio-orthogonal noncanonical amino acid tagging (BONCAT)75 revealed that distinct members of the Gammaproteobacteria, Roseobacter, and SAR11 groups were most responsive to PFOS amendment. Flanked by clearly apparent community level transcriptome adaptations within the microbial sulfur metabolism, this study shows how even the incomplete degradation of perfluorinated compounds at low concentrations can be traced by cellular labeling approaches.
4. Approaching New Solutions in Engineering and Chemical Assessment by Taking Advantage of Advances in Contaminant Biotransformation Research
4a. Design of Novel Engineered Treatment Systems
Processes based on biodegradation principles are an intriguing alternative for removal of chemicals from water due to lower energy requirements as compared to those of physiochemical processes, no generation of residuals, little or no need for auxiliary chemicals, and the possibility of tailoring them to transform specific chemicals or structures. The advances in analytical chemistry and biomolecular microbiology can now provide insights into the presence and nature of contaminants before and after treatment, and we can use all of the metadata to understand what it takes in terms of physical, chemical, and biological environments to bring about effective biotransformation.
These advantages have already been well recognized in the fields of chemical engineering and industrial biotechnology where contemporary concepts of modifying complex microbial communities for targeted biotransformation of well-defined chemicals are established, known as synthetic biology (SynBio) or metabolic engineering.104,105 However, while these concepts hold promise, they have not yet delivered beyond the laboratory scale (with some notable exceptions, e.g., PETase106) and applying these concepts to engineered applications for water treatment is still challenging. While scalability could likely be resolved, efficiency is a determining factor for adoption of a new treatment process. For the design of a compact, efficient, and competitive process, short hydraulic retention times are desired, requiring concentrated amounts of active biomass with enhanced biotransformation rates. Given the mostly low contaminant concentrations, neither of these conditions is likely to occur naturally, thus requiring a number of engineering interventions to provide a robust and efficient process. These might include a combination of enhanced biofilm processes, physical partitioning, and tailored chemical reactions in bulk solution and on surfaces. For instance, competent degrader strains may be introduced into the biological treatment system, which, however, need to be reliably sustained over extended time periods to minimize restart periods and process down times.107,108 Other operational elements that may be considered in the design of novel biological processes include targeting, enriching, and delivering proper degraders or consortia, maintaining suitable and stable redox conditions, preconditioning (i.e., preacclimation in side streams; bioaugmentation), and maximizing exposure (i.e., by improving bioavailability and attachment of biofilms).
Addition of suitable degraders has been successfully used in drinking water treatment settings under ambient conditions, for instance, for the pesticide metabolite 2,6-dichlorobenzamide (BAM), which is very persistent and one of the most frequently detected contaminants in groundwater. Instead of application of advanced post-treatment, augmenting conventional sand filters with the previously identified degrader strain Aminobacter sp. MSH1 resulted in efficient biodegradation of BAM at environmentally relevant concentrations.109 While performance was limited by a loss of degraders, it was later improved by immobilization of the Aminobacter using carriers.110
Extending solid retention times (SRTs) and applying higher dissolved oxygen levels during conventional activated sludge (CAS) processes can increase microbial diversity and the number of opportunities for co-metabolic biotransformation of chemicals.91,111 In particular, employing biofilm-based systems after conventional secondary treatment can offer a high degree of biodiversity within the biofilm structure and operational conditions that are easier to adjust.112 A tight control of more favorable operational conditions creates niches for establishment of consortia that are capable of degrading chemicals that persist under normal conditions. This was demonstrated recently at lab and full scale by establishing sequential biofilters characterized by carbon-starving and oxic conditions resulting in significantly improved removal of mixtures of chemicals at ambient concentrations occurring in impaired surface water and wastewater effluents.113,114 Functional gene presence and expression profiles derived from metagenomic and metatranscriptomic analyses of these systems suggest higher ratios of genes associated with the biodegradation and metabolism of diverse contaminants, including vanillate monooxygenase (EC 1.14.13.82), 2-hydroxyhexa-2,4-dienoate hydratase (EC 4.2.1.132), and 2-oxopent-4-enoate hydratase (EC 4.2.1.80).101
4b. Development and Authorization of New Chemical Substances
New capabilities brought about by modern sensitive analytical techniques for generating biotransformation data for multiple chemicals in different environments under realistic conditions also create new opportunities to strengthen chemical regulation. Environmental persistence, a keystone parameter in chemical regulation, is assessed with laboratory tests that provide quantitative outcomes that are difficult to reproduce and have an unknown relationship to biodegradation rates in the environment. The emerging data will make it possible to explore the relationship between laboratory test outcome and environmental biodegradation rates and how this relationship depends on chemical structure and environmental conditions.
The new analytical capabilities are also strengthening the basis for the application of benchmarking techniques that build on measuring the relative behavior of the test chemical with respect to benchmark chemicals rather than quantifying absolute biodegradation rates. Wisely chosen benchmark chemicals can provide information about test system properties and correct for sources of variability in biodegradation rates.115 Benchmarking can now be applied on large sets of experimental biotransformation data to explore the potential for read-across of the biotransformation rate of a given chemical between different environments. For a data set of 27 chemicals with widely ranging properties and biodegradation rates from 35 different laboratory reactors, normalization against the biodegradation rate of one of the chemicals reduced the mean between-reactor variability from 0.65 to 0.35 log unit (unpublished data). For a number of groups of chemicals that underwent similar initial biotransformation reactions, variability was even further reduced when the benchmark was chosen from that group.
In addition, after correction for bioavailability, good correlations were recently reported between biotransformation half-lives in soil obtained through regulatory simulation studies and short-term experiments with activated sludge for a set of ∼40 chemicals. This not only provides evidence that the relative biotransformation behavior of chemicals might even be conserved to some extent between environments but also highlights the potential to read-across biodegradation rates between environmental systems.116 Using efficient, short-term tests (e.g., with activated sludge and mixtures of test chemicals) for estimating outcomes of regulatory tests might pave the way toward considering biotransformation potential at an early stage of compound development and hence support the development of environmentally benign new chemicals.116
In line with the new EU Green Deal strategy toward a toxic-free environment, these developments promise more precise regulatory test protocols with greater environmental relevance and eventually also experimental strategies or accurate predictive tools to include consideration of degradability in chemical development. However, in a regulatory context, uptake of new testing strategies or data evaluation protocols is slow because of the need to agree on legally defensible instruments that afford long-term planning safety to chemical companies. Here, low-key innovations on the data evaluation side, such as benchmarking, might support weight-of-evidence evaluations without the need to modify regulations.
5. Lessons Learned
Despite all of the advances in gaining deeper mechanistic insights into contaminant biotransformation at relevant environmental concentrations (Figure 2), we conclude that we are just at the beginning of a system’s understanding that could ultimately result in improved approaches to reduce environmental exposure to contaminants. In our opinion, the following aspects are key to reaching this goal.
First, while there is evidence suggesting that contaminant transformation at trace level concentrations and in complex mixtures by environmental microbial communities involves both metabolic and co-metabolic degradation, experimental and analytical methods for discerning the two mechanisms under relevant conditions need to be further improved. This seems essential to further understand controls of one or the other and, in a next step, being able to apply these controls to promote contaminant biotransformation in treatment systems. Specific questions of interest include the following.
There are threshold concentrations for metabolic degradation, but why are they, at least for some compounds, lower than previously assumed? What role does mixed substrate usage play in those cases? Or may low-level degradation be partly driven by other selection criteria, e.g., that some contaminants are not primarily used as an energy or carbon source, but as a source of other essential elements (N, S, P etc.)?
For co-metabolic degradation, are there a number of low-specificity enzymes that are behind those transformation-specific patterns seen across different microbial communities? If so, what are they and what conditions lead to their expression and/or high abundance?
Second, approaches for linking the presence of enzymes or microbes to activity and thus for helping to identify the key players of biotransformation need to be further developed and tested for different contaminants and environmental microbial communities. From Figure 2, we can identify approaches that have the greatest potential to fully identify responsible enzymes and organisms as highlighted by the green arrows. Overall, we see the most promise in techniques that are untargeted and allow for discovery, yet also believe that there is potential in improving more data-driven approaches. More specifically, these include the following.
Next-generation, activity-based labeling approaches such as MAR-FISH, BONCAT, or Raman-based sorting of labeled cells can potentially pick out degraders in complex, realistic settings. However, this approach will most likely remain limited to primarily elucidating “metabolic degradation” (Figure 2, 2d).
High-throughput screening libraries (“loss- and gain-of-function genetic approaches”) for bacterial strains of specific interest (i.e., because of their known broad contaminant biotransformation potential) are an untargeted way of discovering abundant functions, e.g., unspecific enzymes that can co-metabolically transform a wide range of contaminants (Figure 2, 2e). While they are currently still experimentally and analytically very expensive, they do not rely on any predictions of reactions, transforming enzymes, etc., and hence help to circumvent limitations of databases such as annotation issues.
Open repository databases that are specifically focused on contaminant biotransformation will be essential to further our understanding of specific conditions that foster biotransformation and to expand our knowledge of enzymes and strains involved in contaminant degradation. The latter should eventually also help data-driven approaches (i.e., correlation analysis between chemical and omics data) to be used to their full potential (Figure 2, 2c). In this context, it might also be of interest to include information about metabolism in gut microbiota, because gut microbes not only are exposed to many of the same chemicals we also encounter in wastewater but also might be released to WWTPs and receiving environments.117
Third, considering the potential highlighted by some of the success stories featured in sections 3 and 4, new solutions in engineering and chemical assessments that take full advantage of novel biological approaches are conceivable, including the introduction of key degraders or co-substrates, creating and controlling operational conditions that foster co-metabolic degradation, or induction of factors that result in upregulation of key functional genes. However, the success of such endeavors will critically rely on a better integration of disciplinary knowledge, particularly among wastewater engineers, microbiologists, and (bio)chemists. In that sense and in light of the recent strong push toward green and sustainable chemistry in Europe and worldwide,118,119 we invite all peers to take advantage of the ideas and techniques presented here to work together toward using the full potential of microbes to provide for toxic-free environments.
Acknowledgments
M.E., K.F., and T.L. acknowledge support of the European Research Council (ERC) under the European Union’s Seventh Framework Program (FP7/2007-2013), in the frame of Grant Agreement 616644 (POLLOX), Grant Agreement 614768 (PROduCTS), and Grant Agreement 616861 (MicroDegrade), respectively, for funding of research discussed here. The authors also thank all participants of the TransCon2019 conference (TransCon2019: Understanding and managing microbial biotransformation of environmental contaminants, Centro Stefano Franscini, Monte Verità, Ascona, Switzerland, 2019) for sharing their ideas and inspiring workshop discussions, which ultimately led to the conception of this Perspective.
The authors declare no competing financial interest.
References
- de Lorenzo V. Systems biology approaches to bioremediation. Curr. Opin. Biotechnol. 2008, 19 (6), 579–589. 10.1016/j.copbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Ufarté L.; Laville É.; Duquesne S.; Potocki-Veronese G. Metagenomics for the discovery of pollutant degrading enzymes. Biotechnol. Adv. 2015, 33 (8), 1845–1854. 10.1016/j.biotechadv.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Fenner K.; Canonica S.; Wackett L. P.; Elsner M. Evaluating Pesticide Degradation in the Environment: Blind Spots and Emerging Opportunities. Science 2013, 341 (6147), 752–758. 10.1126/science.1236281. [DOI] [PubMed] [Google Scholar]
- Diamond M. L.; de Wit C. A.; Molander S.; Scheringer M.; Backhaus T.; Lohmann R.; Arvidsson R.; Bergman Å.; Hauschild M.; Holoubek I.; Persson L.; Suzuki N.; Vighi M.; Zetzsch C. Exploring the planetary boundary for chemical pollution. Environ. Int. 2015, 78, 8–15. 10.1016/j.envint.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Beketov M. A.; Kefford B. J.; Schafer R. B.; Liess M. Pesticides reduce regional biodiversity of stream invertebrates. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (27), 11039–11043. 10.1073/pnas.1305618110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwacka A.; Zamkowska D.; Radwan M.; Jurewicz J. Exposure to modern, widespread environmental endocrine disrupting chemicals and their effect on the reproductive potential of women: an overview of current epidemiological evidence. Hum. Fertil. 2019, 22 (1), 2–25. 10.1080/14647273.2017.1358828. [DOI] [PubMed] [Google Scholar]
- Harris-Lovett S. R.; Binz C.; Sedlak D. L.; Kiparsky M.; Truffer B. Beyond User Acceptance: A Legitimacy Framework for Potable Water Reuse in California. Environ. Sci. Technol. 2015, 49 (13), 7552–7561. 10.1021/acs.est.5b00504. [DOI] [PubMed] [Google Scholar]
- Bernhardt E. S.; Rosi E. J.; Gessner M. O. Synthetic chemicals as agents of global change. Frontiers in Ecology and the Environment 2017, 15 (2), 84–90. 10.1002/fee.1450. [DOI] [Google Scholar]
- Meckenstock R. U.; Elsner M.; Griebler C.; Lueders T.; Stumpp C.; Aamand J.; Agathos S. N.; Albrechtsen H.-J.; Bastiaens L.; Bjerg P. L.; Boon N.; Dejonghe W.; Huang W. E.; Schmidt S. I.; Smolders E.; Sørensen S. R.; Springael D.; van Breukelen B. M. Biodegradation: Updating the Concepts of Control for Microbial Cleanup in Contaminated Aquifers. Environ. Sci. Technol. 2015, 49 (12), 7073–7081. 10.1021/acs.est.5b00715. [DOI] [PubMed] [Google Scholar]
- Poursat B. A. J.; van Spanning R. J. M.; de Voogt P.; Parsons J. R. Implications of microbial adaptation for the assessment of environmental persistence of chemicals. Crit. Rev. Environ. Sci. Technol. 2019, 49 (23), 2220–2255. 10.1080/10643389.2019.1607687. [DOI] [Google Scholar]
- Tratnyek P. G.; Edwards E.; Carpenter L.; Blossom S. Environmental occurrence, fate, effects, and remediation of halogenated (semi)volatile organic compounds. Environmental Science: Processes & Impacts 2020, 22 (3), 465–471. 10.1039/D0EM90008G. [DOI] [PubMed] [Google Scholar]
- Hug L. A.; Maphosa F.; Leys D.; Löffler F. E.; Smidt H.; Edwards E. A.; Adrian L. Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philos. Trans. R. Soc., B 2013, 368 (1616), 20120322. 10.1098/rstb.2012.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins G. D.; McCarty P. L. Field Evaluation of in Situ Aerobic Cometabolism of Trichloroethylene and Three Dichloroethylene Isomers Using Phenol and Toluene as the Primary Substrates. Environ. Sci. Technol. 1995, 29 (6), 1628–1637. 10.1021/es00006a029. [DOI] [PubMed] [Google Scholar]
- Duhamel M.; Wehr S. D.; Yu L.; Rizvi H.; Seepersad D.; Dworatzek S.; Cox E. E.; Edwards E. A. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-dichloroethene and vinyl chloride. Water Res. 2002, 36 (17), 4193–4202. 10.1016/S0043-1354(02)00151-3. [DOI] [PubMed] [Google Scholar]
- Lueders T. The ecology of anaerobic degraders of BTEX hydrocarbons in aquifers. FEMS Microbiol. Ecol. 2017, 93 (1), fiw220. 10.1093/femsec/fiw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biological Remediation of Explosive Residues; Springer International Publishing: Cham, Switzerland, 2014. [Google Scholar]
- Truskewycz A.; Gundry T. D.; Khudur L. S.; Kolobaric A.; Taha M.; Aburto-Medina A.; Ball A. S.; Shahsavari E. Petroleum Hydrocarbon Contamination in Terrestrial Ecosystems-Fate and Microbial Responses. Molecules 2019, 24 (18), 3400. 10.3390/molecules24183400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug L. A.; Beiko R. G.; Rowe A. R.; Richardson R. E.; Edwards E. A. Comparative metagenomics of three Dehalococcoides-containing enrichment cultures: the role of the non-dechlorinating community. BMC Genomics 2012, 13, 327. 10.1186/1471-2164-13-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhamel M.; Edwards E. A. Growth and Yields of Dechlorinators, Acetogens, and Methanogens during Reductive Dechlorination of Chlorinated Ethenes and Dihaloelimination of 1,2-Dichloroethane. Environ. Sci. Technol. 2007, 41 (7), 2303–2310. 10.1021/es062010r. [DOI] [PubMed] [Google Scholar]
- Yan J.; Şimşir B.; Farmer A. T.; Bi M.; Yang Y.; Campagna S. R.; Löffler F. E. The corrinoid cofactor of reductive dehalogenases affects dechlorination rates and extents in organohalide-respiring Dehalococcoides mccartyi. ISME J. 2016, 10 (5), 1092–1101. 10.1038/ismej.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S. D.; Ternes T. A. Water Analysis: Emerging Contaminants and Current Issues. Anal. Chem. 2014, 86 (6), 2813–2848. 10.1021/ac500508t. [DOI] [PubMed] [Google Scholar]
- Bosma T. N. P.; Middeldorp P. J. M.; Schraa G.; Zehnder A. J. B. Mass transfer limitation of biotransformation: Quantifying bioavailability. Environ. Sci. Technol. 1997, 31 (1), 248–252. 10.1021/es960383u. [DOI] [Google Scholar]
- Flemming H.-C.; Wingender J.; Szewzyk U.; Steinberg P.; Rice S. A.; Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14 (9), 563–575. 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- Semple K. T.; Doick K. J.; Wick L. Y.; Harms H. Microbial interactions with organic contaminants in soil: Definitions, processes and measurement. Environ. Pollut. 2007, 150 (1), 166–176. 10.1016/j.envpol.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Hollender J.; Schymanski E. L.; Singer H. P.; Ferguson P. L. Nontarget Screening with High Resolution Mass Spectrometry in the Environment: Ready to Go?. Environ. Sci. Technol. 2017, 51 (20), 11505–11512. 10.1021/acs.est.7b02184. [DOI] [PubMed] [Google Scholar]
- Helbling D. E.; Hollender J.; Kohler H.-P. E.; Singer H.; Fenner K. High-throughput identification of microbial transformation products of organic micropollutants. Environ. Sci. Technol. 2010, 44 (17), 6621–6627. 10.1021/es100970m. [DOI] [PubMed] [Google Scholar]
- Perez S.; Barcelo D. Application of advanced MS techniques to analysis and identification of human and microbial metabolites of pharmaceuticals in the aquatic environment. TrAC, Trends Anal. Chem. 2007, 26 (6), 494–514. 10.1016/j.trac.2007.05.004. [DOI] [Google Scholar]
- Schymanski E.; Jeon J.; Gulde R.; Fenner K.; Ruff M.; Singer H.; Hollender J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. 10.1021/es5002105. [DOI] [PubMed] [Google Scholar]
- Schollée J. E.; Schymanski E. L.; Avak S. E.; Loos M.; Hollender J. Prioritizing Unknown Transformation Products from Biologically-Treated Wastewater Using High-Resolution Mass Spectrometry, Multivariate Statistics, and Metabolic Logic. Anal. Chem. 2015, 87 (24), 12121–12129. 10.1021/acs.analchem.5b02905. [DOI] [PubMed] [Google Scholar]
- Li Z.; McLachlan M. S. Biodegradation of Chemicals in Unspiked Surface Waters Downstream of Wastewater Treatment Plants. Environ. Sci. Technol. 2019, 53 (4), 1884–1892. 10.1021/acs.est.8b05191. [DOI] [PubMed] [Google Scholar]
- Elsner M.; Jochmann M. A.; Hofstetter T. B.; Hunkeler D.; Bernstein A.; Schmidt T. C.; Schimmelmann A. Current challenges in compound-specific stable isotope analysis of environmental organic contaminants. Anal. Bioanal. Chem. 2012, 403 (9), 2471–2491. 10.1007/s00216-011-5683-y. [DOI] [PubMed] [Google Scholar]
- Elsner M. Stable isotope fractionation to investigate natural transformation mechanisms of organic contaminants: principles, prospects and limitations. J. Environ. Monit. 2010, 12 (11), 2005–2031. 10.1039/c0em00277a. [DOI] [PubMed] [Google Scholar]
- Lihl C.; Douglas L. M.; Franke S.; Pérez-de-Mora A.; Meyer A. H.; Daubmeier M.; Edwards E. A.; Nijenhuis I.; Sherwood Lollar B.; Elsner M. Mechanistic Dichotomy in Bacterial Trichloroethene Dechlorination Revealed by Carbon and Chlorine Isotope Effects. Environ. Sci. Technol. 2019, 53 (8), 4245–4254. 10.1021/acs.est.8b06643. [DOI] [PubMed] [Google Scholar]
- Torrentó C.; Bakkour R.; Glauser G.; Melsbach A.; Ponsin V.; Hofstetter T. B.; Elsner M.; Hunkeler D. Solid-phase extraction method for stable isotope analysis of pesticides from large volume environmental water samples. Analyst 2019, 144 (9), 2898–2908. 10.1039/C9AN00160C. [DOI] [PubMed] [Google Scholar]
- Sunagawa S.; Coelho L. P.; Chaffron S.; Kultima J. R.; Labadie K.; Salazar G.; Djahanschiri B.; Zeller G.; Mende D. R.; Alberti A.; Cornejo-Castillo F. M.; Costea P. I.; Cruaud C.; d'Ovidio F.; Engelen S.; Ferrera I.; Gasol J. M.; Guidi L.; Hildebrand F.; Kokoszka F.; Lepoivre C.; Lima-Mendez G.; Poulain J.; Poulos B. T.; Royo-Llonch M.; Sarmento H.; Vieira-Silva S.; Dimier C.; Picheral M.; Searson S.; Kandels-Lewis S.; Bowler C.; de Vargas C.; Gorsky G.; Grimsley N.; Hingamp P.; Iudicone D.; Jaillon O.; Not F.; Ogata H.; Pesant S.; Speich S.; Stemmann L.; Sullivan M. B.; Weissenbach J.; Wincker P.; Karsenti E.; Raes J.; Acinas S. G.; Bork P.; Boss E.; Bowler C.; Follows M.; Karp-Boss L.; Krzic U.; Reynaud E. G.; Sardet C.; Sieracki M.; Velayoudon D. Structure and function of the global ocean microbiome. Science 2015, 348 (6237), 1261359. 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- Bahram M.; Hildebrand F.; Forslund S. K.; Anderson J. L.; Soudzilovskaia N. A.; Bodegom P. M.; Bengtsson-Palme J.; Anslan S.; Coelho L. P.; Harend H.; Huerta-Cepas J.; Medema M. H.; Maltz M. R.; Mundra S.; Olsson P. A.; Pent M.; Põlme S.; Sunagawa S.; Ryberg M.; Tedersoo L.; Bork P. Structure and function of the global topsoil microbiome. Nature 2018, 560 (7717), 233–237. 10.1038/s41586-018-0386-6. [DOI] [PubMed] [Google Scholar]
- Wu L.; Ning D.; Zhang B.; Li Y.; Zhang P.; Shan X.; Zhang Q.; Brown M.; Li Z.; Van Nostrand J. D.; Ling F.; Xiao N.; Zhang Y.; Vierheilig J.; Wells G. F.; Yang Y.; Deng Y.; Tu Q.; Wang A.; Acevedo D.; Agullo-Barcelo M.; Alvarez P. J. J.; Alvarez-Cohen L.; Andersen G. L.; de Araujo J. C.; Boehnke K.; Bond P.; Bott C. B.; Bovio P.; Brewster R. K.; Bux F.; Cabezas A.; Cabrol L.; Chen S.; Criddle C. S.; Deng Y.; Etchebehere C.; Ford A.; Frigon D.; Gómez J. S.; Griffin J. S.; Gu A. Z.; Habagil M.; Hale L.; Hardeman S. D.; Harmon M.; Horn H.; Hu Z.; Jauffur S.; Johnson D. R.; Keller J.; Keucken A.; Kumari S.; Leal C. D.; Lebrun L. A.; Lee J.; Lee M.; Lee Z. M. P.; Li Y.; Li Z.; Li M.; Li X.; Ling F.; Liu Y.; Luthy R. G.; Mendonça-Hagler L. C.; de Menezes F. G. R.; Meyers A. J.; Mohebbi A.; Nielsen P. H.; Ning D.; Oehmen A.; Palmer A.; Parameswaran P.; Park J.; Patsch D.; Reginatto V.; de los Reyes F. L.; Rittmann B. E.; Robles A. N.; Rossetti S.; Shan X.; Sidhu J.; Sloan W. T.; Smith K.; de Sousa O. V.; Stahl D. A.; Stephens K.; Tian R.; Tiedje J. M.; Tooker N. B.; Tu Q.; Van Nostrand J. D.; De los Cobos Vasconcelos D.; Vierheilig J.; Wagner M.; Wakelin S.; Wang A.; Wang B.; Weaver J. E.; Wells G. F.; West S.; Wilmes P.; Woo S.-G.; Wu L.; Wu J.-H.; Wu L.; Xi C.; Xiao N.; Xu M.; Yan T.; Yang Y.; Yang M.; Young M.; Yue H.; Zhang B.; Zhang P.; Zhang Q.; Zhang Y.; Zhang T.; Zhang Q.; Zhang W.; Zhang Y.; Zhou H.; Zhou J.; Wen X.; Curtis T. P.; He Q.; He Z.; Brown M.; Zhang T.; He Z.; Keller J.; Nielsen P. H.; Alvarez P. J. J.; Criddle C. S.; Wagner M.; Tiedje J. M.; He Q.; Curtis T. P.; Stahl D. A.; Alvarez-Cohen L.; Rittmann B. E.; Wen X.; Zhou J. Global diversity and biogeography of bacterial communities in wastewater treatment plants. Nat. Microbiology 2019, 4 (7), 1183–1195. 10.1038/s41564-019-0426-5. [DOI] [PubMed] [Google Scholar]
- Gilbert J. A.; Blaser M. J.; Caporaso J. G.; Jansson J. K.; Lynch S. V.; Knight R. Current understanding of the human microbiome. Nat. Med. 2018, 24 (4), 392–400. 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner M. Metaproteomics: Much More than Measuring Gene Expression in Microbial Communities. mSystems 2019, 4 (3), e00115-19. 10.1128/mSystems.00115-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins-Schiffman E.; May D. H.; Mikan M.; Riffle M.; Frazar C.; Harvey H. R.; Noble W. S.; Nunn B. L. Critical decisions in metaproteomics: achieving high confidence protein annotations in a sea of unknowns. ISME J. 2017, 11 (2), 309–314. 10.1038/ismej.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taş N.; Brandt B. W.; Braster M.; van Breukelen B. M.; Röling W. F. M. Subsurface landfill leachate contamination affects microbial metabolic potential and gene expression in the Banisveld aquifer. FEMS Microbiol. Ecol. 2018, 94 (10), 1–12. 10.1093/femsec/fiy156. [DOI] [PubMed] [Google Scholar]
- Johnson D. R.; Helbling D. E.; Men Y.; Fenner K. Can meta-omics help to establish causality between contaminant biotransformations and genes or gene products?. Environmental Science: Water Research & Technology 2015, 1 (3), 272–278. 10.1039/C5EW00016E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P.; Kin K.; Lynch V. J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012, 131 (4), 281–285. 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- Soneson C.; Love M.; Robinson M. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences [version 2; peer review: 2 approved]. F1000Research 2015, 4 (4), 1521. 10.12688/f1000research.7563.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnoes A. M.; Brown S. D.; Dodevski I.; Babbitt P. C. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput. Biol. 2009, 5 (12), e1000605. 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J.; Ellis L. B. M.; Wackett L. P. The University of Minnesota Biocatalysis/Biodegradation Database: Improving Public Access. Nucleic Acids Res. 2010, 38, D488–D491. 10.1093/nar/gkp771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker J.; Lorsbach T.; Gütlein M.; Schmid E.; Latino D.; Kramer S.; Fenner K. enviPath – The environmental contaminant biotransformation pathway resource. Nucleic Acids Res. 2016, 44 (Database Issue), D502–D508. 10.1093/nar/gkv1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Z.; Zhang H. PMBD: a Comprehensive Plastics Microbial Biodegradation Database. Database 2019, 2019, baz119. 10.1093/database/baz119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. F.; Ellis L. B. M.; Wackett L. P. The University of Minnesota Pathway Prediction System: multi-level prediction and visualization. Nucleic Acids Res. 2011, 39, W406–W411. 10.1093/nar/gkr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley S. D.; Broadbelt L. J.; Hatzimanikatis V. Computational Framework for Predictive Biodegradation. Biotechnol. Bioeng. 2009, 104 (6), 1086–1097. 10.1002/bit.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S.; Pavlov T.; Dimitrova N.; Georgieva D.; Nedelcheva D.; Kesova A.; Vasilev R.; Mekenyan O. Simulation of chemical metabolism for fate and hazard assessment. II CATALOGIC simulation of abiotic and microbial degradation. SAR QSAR Environ. Res. 2011, 22 (7–8), 719–755. 10.1080/1062936X.2011.623322. [DOI] [PubMed] [Google Scholar]
- Hadadi N.; MohammadiPeyhani H.; Miskovic L.; Seijo M.; Hatzimanikatis V. Enzyme annotation for orphan and novel reactions using knowledge of substrate reactive sites. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (15), 7298. 10.1073/pnas.1818877116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid E.; Fenner K. enviLink: A database linking contaminant biotransformation rules to enzyme classes in support of functional association mining. bioRxiv 2021, 10.1101/2021.05.20.442588. [DOI] [Google Scholar]
- Yamanishi Y.; Hattori M.; Kotera M.; Goto S.; Kanehisa M. E-zyme: predicting potential EC numbers from the chemical transformation pattern of substrate-product pairs. Bioinformatics 2009, 25 (12), I179–I186. 10.1093/bioinformatics/btp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S. A.; Cuesta S. M.; Furnham N.; Holliday G. L.; Thornton J. M. EC-BLAST: a tool to automatically search and compare enzyme reactions. Nat. Methods 2014, 11 (2), 171–174. 10.1038/nmeth.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukema K. G.; Tassoulas L. J.; Robinson S. L.; Konopatski J. F.; Bygd M. D.; Wackett L. P. Cyanuric Acid Biodegradation via Biuret: Physiology, Taxonomy, and Geospatial Distribution. Appl. Environ. Microbiol. 2020, 86 (2), e01964-19. 10.1128/AEM.01964-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Netzer F.; Kuntze K.; Vogt C.; Richnow H. H.; Boll M.; Lueders T. Functional Gene Markers for Fumarate-Adding and Dearomatizing Key Enzymes in Anaerobic Aromatic Hydrocarbon Degradation in Terrestrial Environments. Journal of Molecular Microbiology and Biotechnology 2016, 26 (1–3), 180–194. 10.1159/000441946. [DOI] [PubMed] [Google Scholar]
- Robinson S. L.; Terlouw B. R.; Smith M. D.; Pidot S. J.; Stinear T. P.; Medema M. H.; Wackett L. P. Global analysis of adenylate-forming enzymes reveals beta-lactone biosynthesis pathway in pathogenic Nocardia. J. Biol. Chem. 2020, 295 (44), 14826–14839. 10.1074/jbc.RA120.013528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z.; Eakes J.; Cooper C. J.; Foster C. M.; Standaert R. F.; Podar M.; Doktycz M. J.; Parks J. M. Machine learning-based prediction of enzyme substrate scope: Application to bacterial nitrilases. Proteins: Struct., Funct., Genet. 2021, 89 (3), 336–347. 10.1002/prot.26019. [DOI] [PubMed] [Google Scholar]
- Zhao S.; Kumar R.; Sakai A.; Vetting M. W.; Wood B. M.; Brown S.; Bonanno J. B.; Hillerich B. S.; Seidel R. D.; Babbitt P. C.; Almo S. C.; Sweedler J. V.; Gerlt J. A.; Cronan J. E.; Jacobson M. P. Discovery of new enzymes and metabolic pathways by using structure and genome context. Nature 2013, 502 (7473), 698–702. 10.1038/nature12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun S.; Korczynska M.; Wichelecki D. J.; San Francisco B.; Zhao S. W.; Rodionov D. A.; Vetting M. W.; Al-Obaidi N. F.; Lin H.; O’Meara M. J.; Scott D. A.; Morris J. H.; Russel D.; Almo S. C.; Osterman A. L.; Gerlt J. A.; Jacobson M. P.; Shoichet B. K.; Sali A. Prediction of enzymatic pathways by integrative pathway mapping. eLife 2018, 7, 27. 10.7554/eLife.31097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukema K. G.; Escalante D. E.; Maltby M. M.; Bera A. K.; Aksan A.; Wackett L. P. In Silico Identification of Bioremediation Potential: Carbamazepine and Other Recalcitrant Personal Care Products. Environ. Sci. Technol. 2017, 51 (2), 880–888. 10.1021/acs.est.6b04345. [DOI] [PubMed] [Google Scholar]
- Escalante D. E.; Aukema K. G.; Wackett L. P.; Aksan A. Simulation of the Bottleneck Controlling Access into a Rieske Active Site: Predicting Substrates of Naphthalene 1,2-Dioxygenase. J. Chem. Inf. Model. 2017, 57 (3), 550–561. 10.1021/acs.jcim.6b00469. [DOI] [PubMed] [Google Scholar]
- Cupples A. M. Contaminant-Degrading Microorganisms Identified Using Stable Isotope Probing. Chem. Eng. Technol. 2016, 39 (9), 1593–1603. 10.1002/ceat.201500479. [DOI] [Google Scholar]
- Fischer A.; Manefield M.; Bombach P. Application of stable isotope tools for evaluating natural and stimulated biodegradation of organic pollutants in field studies. Curr. Opin. Biotechnol. 2016, 41, 99–107. 10.1016/j.copbio.2016.04.026. [DOI] [PubMed] [Google Scholar]
- Sims G. K.; Gomez A. M.; Kanissery R.. Stable DNA Isotope Probing to Examine Organisms Involved in Biodegradation. In Microbial Metabolism of Xenobiotic Compounds; Arora P. K., Ed.; Springer Singapore: Singapore, 2019; pp 55–77. [Google Scholar]
- Benner J.; De Smet D.; Ho A.; Kerckhof F.-M.; Vanhaecke L.; Heylen K.; Boon N. Exploring methane-oxidizing communities for the co-metabolic degradation of organic micropollutants. Appl. Microbiol. Biotechnol. 2015, 99 (8), 3609–3618. 10.1007/s00253-014-6226-1. [DOI] [PubMed] [Google Scholar]
- Dai H.; Gao J.; Wang S.; Li D.; Wang Z. The key active degrader, metabolic pathway and microbial ecology of triclosan biodegradation in an anoxic/oxic system. Bioresour. Technol. 2020, 317, 124014. 10.1016/j.biortech.2020.124014. [DOI] [PubMed] [Google Scholar]
- Sathyamoorthy S.; Hoar C.; Chandran K. Identification of Bisphenol A-Assimilating Microorganisms in Mixed Microbial Communities Using 13C-DNA Stable Isotope Probing. Environ. Sci. Technol. 2018, 52 (16), 9128–9135. 10.1021/acs.est.8b01976. [DOI] [PubMed] [Google Scholar]
- Bradford L. M.; Vestergaard G.; Táncsics A.; Zhu B.; Schloter M.; Lueders T. Transcriptome-Stable Isotope Probing Provides Targeted Functional and Taxonomic Insights Into Microaerobic Pollutant-Degrading Aquifer Microbiota. Front. Microbiol. 2018, 9, 2696. 10.3389/fmicb.2018.02696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W.-Y.; Su J.-Q.; Richnow H. H.; Adrian L. Identification of dominant sulfamethoxazole-degraders in pig farm-impacted soil by DNA and protein stable isotope probing. Environ. Int. 2019, 126, 118–126. 10.1016/j.envint.2019.02.001. [DOI] [PubMed] [Google Scholar]
- Mayali X. NanoSIMS: Microscale Quantification of Biogeochemical Activity with Large-Scale Impacts. Annual Review of Marine Science 2020, 12 (1), 449–467. 10.1146/annurev-marine-010419-010714. [DOI] [PubMed] [Google Scholar]
- Lee K. S.; Palatinszky M.; Pereira F. C.; Nguyen J.; Fernandez V. I.; Mueller A. J.; Menolascina F.; Daims H.; Berry D.; Wagner M.; Stocker R. An automated Raman-based platform for the sorting of live cells by functional properties. Nature Microbiology 2019, 4 (6), 1035–1048. 10.1038/s41564-019-0394-9. [DOI] [PubMed] [Google Scholar]
- Schwartz E.; Hayer M.; Hungate B. A.; Mau R. L.. Stable Isotope Probing of Microorganisms in Environmental Samples with H218O. In Stable Isotope Probing: Methods and Protocols; Dumont M. G., Hernández García M., Eds.; Springer: New York, 2019; pp 129–136. [DOI] [PubMed] [Google Scholar]
- Hatzenpichler R.; Krukenberg V.; Spietz R. L.; Jay Z. J. Next-generation physiology approaches to study microbiome function at single cell level. Nat. Rev. Microbiol. 2020, 18 (4), 241–256. 10.1038/s41579-020-0323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gil L.; Krah D.; Ghattas A. K.; Carballa M.; Wick A.; Helmholz L.; Lema J. M.; Ternes T. A. Biotransformation of organic micropollutants by anaerobic sludge enzymes. Water Res. 2019, 152, 202–214. 10.1016/j.watres.2018.12.064. [DOI] [PubMed] [Google Scholar]
- Men Y.; Achermann S.; Helbling D. E.; Johnson D. R.; Fenner K. Relative contribution of ammonia oxidizing bacteria and other members of nitrifying activated sludge communities to micropollutant biotransformation. Water Res. 2017, 109, 217–226. 10.1016/j.watres.2016.11.048. [DOI] [PubMed] [Google Scholar]
- Zumstein M. T.; Kohler H. P. E.; McNeill K.; Sander M. High-Throughput Analysis of Enzymatic Hydrolysis of Biodegradable Polyesters by Monitoring Cohydrolysis of a Polyester-Embedded Fluorogenic Probe. Environ. Sci. Technol. 2017, 51 (8), 4358–4367. 10.1021/acs.est.6b06060. [DOI] [PubMed] [Google Scholar]
- Hajighasemi M.; Tchigvintsev A.; Nocek B.; Flick R.; Popovic A.; Hai T.; Khusnutdinova A. N.; Brown G.; Xu X.; Cui H.; Anstett J.; Chernikova T. N.; Brüls T.; Le Paslier D.; Yakimov M. M.; Joachimiak A.; Golyshina O. V.; Savchenko A.; Golyshin P. N.; Edwards E. A.; Yakunin A. F. Screening and Characterization of Novel Polyesterases from Environmental Metagenomes with High Hydrolytic Activity against Synthetic Polyesters. Environ. Sci. Technol. 2018, 52 (21), 12388–12401. 10.1021/acs.est.8b04252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S. L.; Smith M. D.; Richman J. E.; Aukema K. G.; Wackett L. P. Machine learning-based prediction of activity and substrate specificity for OleA enzymes in the thiolase superfamily. Synth. Biol. 2020, 5 (1), ysaa004. 10.1093/synbio/ysaa004. [DOI] [Google Scholar]
- Krah D.; Ghattas A.-K.; Wick A.; Bröder K.; Ternes T. A. Micropollutant degradation via extracted native enzymes from activated sludge. Water Res. 2016, 95, 348–360. 10.1016/j.watres.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S. Q.; Chan W. W. M.; Fletcher K. E.; Seifert J.; Liang X. M.; Loffler F. E.; Edwards E. A.; Adrian L. Functional Characterization of Reductive Dehalogenases by Using Blue Native Polyacrylamide Gel Electrophoresis. Appl. Environ. Microbiol. 2013, 79 (3), 974–981. 10.1128/AEM.01873-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumstein M. T.; Helbling D. E. Biotransformation of antibiotics: Exploring the activity of extracellular and intracellular enzymes derived from wastewater microbial communities. Water Res. 2019, 155, 115–123. 10.1016/j.watres.2019.02.024. [DOI] [PubMed] [Google Scholar]
- Jariwala P. B.; Pellock S. J.; Goldfarb D.; Cloer E. W.; Artola M.; Simpson J. B.; Bhatt A. P.; Walton W. G.; Roberts L. R.; Major M. B.; Davies G. J.; Overkleeft H. S.; Redinbo M. R. Discovering the Microbial Enzymes Driving Drug Toxicity with Activity-Based Protein Profiling. ACS Chem. Biol. 2020, 15 (1), 217–225. 10.1021/acschembio.9b00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M.; Zimmermann-Kogadeeva M.; Wegmann R.; Goodman A. L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019, 570 (7762), 462–467. 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg K. J.; Reyes A.; Wang B.; Selleck E. M.; Sommer M. O. A.; Dantas G. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 2012, 337 (6098), 1107. 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achermann S.; Falås P.; Joss A.; Mansfeldt C. B.; Men Y.; Vogler B.; Fenner K. Trends in Micropollutant Biotransformation along a Solids Retention Time Gradient. Environ. Sci. Technol. 2018, 52 (20), 11601–11611. 10.1021/acs.est.8b02763. [DOI] [PubMed] [Google Scholar]
- Desiante W. L.; Minas N.; Fenner K. Micropollutant biotransformation and bioaccumulation in natural stream biofilms. Water Res. 2021, 193, 116846. 10.1016/j.watres.2021.116846. [DOI] [PubMed] [Google Scholar]
- Meynet P.; Davenport R. J.; Fenner K. Understanding the Dependence of Micropollutant Biotransformation Rates on Short-Term Temperature Shifts. Environ. Sci. Technol. 2020, 54 (19), 12214–12225. 10.1021/acs.est.0c04017. [DOI] [PubMed] [Google Scholar]
- Nolte T. M.; Pinto-Gil K.; Hendriks A. J.; Ragas A. M. J.; Pastor M. Quantitative structure–activity relationships for primary aerobic biodegradation of organic chemicals in pristine surface waters: starting points for predicting biodegradation under acclimatization. Environmental Science: Processes & Impacts 2018, 20 (1), 157–170. 10.1039/C7EM00375G. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Fenner K.; Helbling D. E. Clustering micropollutants based on initial biotransformations for improved prediction of micropollutant removal during conventional activated sludge treatment. Environmental Science: Water Research & Technology 2020, 6 (3), 554–565. 10.1039/C9EW00838A. [DOI] [Google Scholar]
- Sun F.; Mellage A.; Gharasoo M.; Melsbach A.; Cao X.; Zimmermann R.; Griebler C.; Thullner M.; Cirpka O. A.; Elsner M. Mass-Transfer-Limited Biodegradation at Low Concentrations—Evidence from Reactive Transport Modeling of Isotope Profiles in a Bench-Scale Aquifer. Environ. Sci. Technol. 2021, 55 (11), 7386–7397. 10.1021/acs.est.0c08566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrl B. N.; Kundu K.; Gharasoo M.; Marozava S.; Elsner M. Rate-Limiting Mass Transfer in Micropollutant Degradation Revealed by Isotope Fractionation in Chemostat. Environ. Sci. Technol. 2019, 53 (3), 1197–1205. 10.1021/acs.est.8b05175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu K.; Marozava S.; Ehrl B.; Merl-Pham J.; Griebler C.; Elsner M. Defining lower limits of biodegradation: atrazine degradation regulated by mass transfer and maintenance demand in Arthrobacter aurescens TC1. ISME J. 2019, 13 (9), 2236–2251. 10.1038/s41396-019-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampara M.; Thullner M.; Richnow H. H.; Harms H.; Wick L. Y. Impact of Bioavailability Restrictions on Microbially Induced Stable Isotope Fractionation. 2. Experimental Evidence. Environ. Sci. Technol. 2008, 42 (17), 6552–6558. 10.1021/es702781x. [DOI] [PubMed] [Google Scholar]
- Kundu K.; Weber N.; Griebler C.; Elsner M. Phenotypic heterogeneity as key factor for growth and survival under oligotrophic conditions. Environ. Microbiol. 2020, 22 (8), 3339–3356. 10.1111/1462-2920.15106. [DOI] [PubMed] [Google Scholar]
- Achermann S.; Mansfeldt C. B.; Müller M.; Johnson D. R.; Fenner K. Relating Metatranscriptomic Profiles to the Micropollutant Biotransformation Potential of Complex Microbial Communities. Environ. Sci. Technol. 2020, 54 (1), 235–244. 10.1021/acs.est.9b05421. [DOI] [PubMed] [Google Scholar]
- Guo L.; Fang W.-W.; Guo L.-L.; Yao C.-F.; Zhao Y.-X.; Ge F.; Dai Y.-J. Biodegradation of the Neonicotinoid Insecticide Acetamiprid by Actinomycetes Streptomyces canus CGMCC 13662 and Characterization of the Novel Nitrile Hydratase Involved. J. Agric. Food Chem. 2019, 67 (21), 5922–5931. 10.1021/acs.jafc.8b06513. [DOI] [PubMed] [Google Scholar]
- Stadler L. B.; Love N. G. Oxygen Half-Saturation Constants for Pharmaceuticals in Activated Sludge and Microbial Community Activity under Varied Oxygen Levels. Environ. Sci. Technol. 2019, 53 (4), 1918–1927. 10.1021/acs.est.8b06051. [DOI] [PubMed] [Google Scholar]
- Li D.; Alidina M.; Drewes J. E. Role of primary substrate composition on microbial community structure and function and trace organic chemical attenuation in managed aquifer recharge systems. Appl. Microbiol. Biotechnol. 2014, 98 (12), 5747–5756. 10.1007/s00253-014-5677-8. [DOI] [PubMed] [Google Scholar]
- Li D.; Sharp J. O.; Drewes J. E. Microbial genetic potential for xenobiotic metabolism increases with depth during biofiltration. Environmental Science: Processes & Impacts 2020, 22 (10), 2058–2069. 10.1039/D0EM00254B. [DOI] [PubMed] [Google Scholar]
- Falås P.; Jewell K. S.; Hermes N.; Wick A.; Ternes T. A.; Joss A.; Nielsen J. L. Transformation, CO2 formation and uptake of four organic micropollutants by carrier-attached microorganisms. Water Res. 2018, 141, 405–416. 10.1016/j.watres.2018.03.040. [DOI] [PubMed] [Google Scholar]
- Cerro-Gálvez E.; Roscales J. L.; Jiménez B.; Sala M. M.; Dachs J.; Vila-Costa M. Microbial responses to perfluoroalkyl substances and perfluorooctanesulfonate (PFOS) desulfurization in the Antarctic marine environment. Water Res. 2020, 171, 115434. 10.1016/j.watres.2019.115434. [DOI] [PubMed] [Google Scholar]
- Swartz J. R. Expanding biological applications using cell-free metabolic engineering: An overview. Metab. Eng. 2018, 50, 156–172. 10.1016/j.ymben.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Beal J.; Goñi-Moreno A.; Myers C.; Hecht A.; de Vicente M. d. C.; Parco M.; Schmidt M.; Timmis K.; Baldwin G.; Friedrichs S.; Freemont P.; Kiga D.; Ordozgoiti E.; Rennig M.; Rios L.; Tanner K.; de Lorenzo V.; Porcar M. The long journey towards standards for engineering biosystems. EMBO Rep. 2020, 21 (5), e50521. 10.15252/embr.202050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier V.; Topham C. M.; Gilles A.; David B.; Folgoas C.; Moya-Leclair E.; Kamionka E.; Desrousseaux M. L.; Texier H.; Gavalda S.; Cot M.; Guémard E.; Dalibey M.; Nomme J.; Cioci G.; Barbe S.; Chateau M.; André I.; Duquesne S.; Marty A. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580 (7802), 216–219. 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- Benner J.; Helbling D. E.; Kohler H. P.; Wittebol J.; Kaiser E.; Prasse C.; Ternes T. A.; Albers C. N.; Aamand J.; Horemans B.; Springael D.; Walravens E.; Boon N. Is biological treatment a viable alternative for micropollutant removal in drinking water treatment processes?. Water Res. 2013, 47 (16), 5955–76. 10.1016/j.watres.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Ellegaard-Jensen L.; Horemans B.; Raes B.; Aamand J.; Hansen L. H. Groundwater contamination with 2,6-dichlorobenzamide (BAM) and perspectives for its microbial removal. Appl. Microbiol. Biotechnol. 2017, 101 (13), 5235–5245. 10.1007/s00253-017-8362-x. [DOI] [PubMed] [Google Scholar]
- Albers C.; Feld L.; Ellegaard-Jensen L.; Aamand J. Degradation of trace concentrations of the persistent groundwater pollutant. Water Res. 2015, 83, 61–70. 10.1016/j.watres.2015.06.023. [DOI] [PubMed] [Google Scholar]
- Horemans B.; Raes B.; Vandermaesen J.; Simanjuntak Y.; Brocatus H.; T’Syen J.; Degryse J.; Boonen J.; Wittebol J.; Lapanje A.; Sørensen S. R.; Springael D. Biocarriers Improve Bioaugmentation Efficiency of a Rapid Sand Filter for the Treatment of 2,6-Dichlorobenzamide-Contaminated Drinking Water. Environ. Sci. Technol. 2017, 51 (3), 1616–1625. 10.1021/acs.est.6b05027. [DOI] [PubMed] [Google Scholar]
- Vuono D. C.; Regnery J.; Li D.; Jones Z. L.; Holloway R. W.; Drewes J. E. rRNA Gene Expression of Abundant and Rare Activated-Sludge Microorganisms and Growth Rate Induced Micropollutant Removal. Environ. Sci. Technol. 2016, 50 (12), 6299–6309. 10.1021/acs.est.6b00247. [DOI] [PubMed] [Google Scholar]
- Torresi E.; Polesel F.; Bester K.; Christensson M.; Smets B. F.; Trapp S.; Andersen H. R.; Plósz B. G. Diffusion and sorption of organic micropollutants in biofilms with varying thicknesses. Water Res. 2017, 123, 388–400. 10.1016/j.watres.2017.06.027. [DOI] [PubMed] [Google Scholar]
- Hellauer K.; Karakurt S.; Sperlich A.; Burke V.; Massmann G.; Hübner U.; Drewes J. E. Establishing sequential managed aquifer recharge technology (SMART) for enhanced removal of trace organic chemicals: Experiences from field studies in Berlin, Germany. J. Hydrol. 2018, 563, 1161–1168. 10.1016/j.jhydrol.2017.09.044. [DOI] [Google Scholar]
- Regnery J.; Wing A. D.; Kautz J.; Drewes J. E. Introducing sequential managed aquifer recharge technology (SMART) – From laboratory to full-scale application. Chemosphere 2016, 154, 8–16. 10.1016/j.chemosphere.2016.03.097. [DOI] [PubMed] [Google Scholar]
- McLachlan M. S.; Zou H.; Gouin T. Using Benchmarking To Strengthen the Assessment of Persistence. Environ. Sci. Technol. 2017, 51 (1), 4–11. 10.1021/acs.est.6b03786. [DOI] [PubMed] [Google Scholar]
- Fenner K.; Screpanti C.; Renold P.; Rouchdi M.; Vogler B.; Rich S. Comparison of Small Molecule Biotransformation Half-Lives between Activated Sludge and Soil: Opportunities for Read-Across?. Environ. Sci. Technol. 2020, 54 (6), 3148–3158. 10.1021/acs.est.9b05104. [DOI] [PubMed] [Google Scholar]
- Quintela-Baluja M.; Abouelnaga M.; Romalde J.; Su J.-Q.; Yu Y.; Gomez-Lopez M.; Smets B.; Zhu Y.-G.; Graham D. W. Spatial ecology of a wastewater network defines the antibiotic resistance genes in downstream receiving waters. Water Res. 2019, 162, 347–357. 10.1016/j.watres.2019.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Communication from the Commission to the European Parliament, The Council, The European Economic and Social Committee and the Committee of the Regions - Chemicals Strategy for Sustainability Towards a Toxic-Free Environment. 2020; p 25.
- Communication from the Commission to the European Parliament, the Council and the European Economic and Social Committee - European Union Strategic Approach to Pharmaceuticals in the Environment. 2019; p 13.