Bourke et al. present a new method for using light to trigger protein trafficking through the secretory network. They use the method to characterize how synaptic proteins traffic from different subcellular domains in neurons.
Abstract
Here we introduce zapalog-mediated endoplasmic reticulum trap (zapERtrap), which allows one to use light to precisely trigger forward trafficking of diverse integral membrane proteins from internal secretory organelles to the cell surface with single cell and subcellular spatial resolution. To demonstrate its utility, we use zapERtrap in neurons to dissect where synaptic proteins emerge at the cell surface when processed through central (cell body) or remote (dendrites) secretory pathways. We reveal rapid and direct long-range trafficking of centrally processed proteins deep into the dendritic arbor to synaptic sites. Select proteins were also trafficked to the plasma membrane of the axon initial segment, revealing a novel surface trafficking hotspot. Proteins locally processed through dendritic secretory networks were widely dispersed before surface insertion, challenging assumptions for precise trafficking at remote sites. These experiments provide new insights into compartmentalized secretory trafficking and showcase the tunability and spatiotemporal control of zapERtrap, which will have broad applications for regulating cell signaling and function.
Introduction
Integral membrane and secreted proteins are synthesized, processed, and delivered to the appropriate subcellular location through a complex set of cellular organelles collectively termed the secretory network. In eukaryotes, the early secretory network is principally defined by the ER, the ER–Golgi intermediate compartment (ERGIC), and the Golgi apparatus (GA). In most cells, the physical arrangement of these organelles favors centripetal movement of proteins toward perinuclear GA after they leave the ER. Following GA processing, proteins are sorted and distributed to their appropriate subcellular addresses in distinct classes of mobile vesicle carriers (for reviews, see Barlowe and Miller, 2013; Lee et al., 2004; Lippincott-Schwartz et al., 2000).
A powerful approach for investigating secretory trafficking has been to sequester a protein of interest in the ER and trigger its release while simultaneously visualizing it en route to its functional destination (Boncompain et al., 2012; Chen et al., 2013; Horton and Ehlers, 2003; Presley et al., 1997, Rivera et al., 2000; Toomre et al., 2000). However, previously developed “trap and release” strategies generally lack spatial control, which would allow precision control of protein trafficking in individual cells or even subcellular domains. Such a tool would open the door to many new applications, including tuning receptor levels on user-defined cell populations; releasing specific growth factors, morphogens, or hormones from targeted cells; and dissecting subcellular trafficking pathways in large and complex cells of the nervous system, whose secretory organelles are broadly distributed and poorly understood (Bourke et al., 2018; Kennedy and Hanus, 2019). Here we describe the development of zapalog-mediated ER trap (zapERtrap), which uses a brief pulse of visible light to initiate forward trafficking of diverse cargo proteins from the ER. Because light can be precisely steered over user-defined cell populations or even subcellular domains, protein trafficking can be triggered with exceptional spatial and temporal control. Furthermore, zapERtrap overcomes significant limitations of a previous light-triggered secretion tool we developed, including bulky protein tags and UVB excitation light (less than ∼310 nm), which is challenging to focus for local activation with conventional optics (Chen et al., 2013).
We demonstrate the utility of zapERtrap by using it to investigate compartmentalized trafficking in neurons, which face the challenge of regulating the abundance and localization of channel, receptor, and adhesion proteins throughout elaborate cellular processes that can project long distances (Bourke et al., 2018; Kennedy and Hanus, 2019; Ramírez and Couve, 2011). In the neuronal cell body, or soma, the general architecture and organization of secretory organelles resemble conventional cells where secretory trafficking from the somatic ER to the perinuclear GA occurs (Horton and Ehlers, 2003; Horton et al., 2005; Torre and Steward, 1996). However, whether post-GA transport vesicles are directly delivered over long distances into dendrites via previously described active transport mechanisms remains unclear (Kapitein et al., 2010; Lipka et al., 2016). Alternatively, cargo could enter dendrites following insertion into the somatic membrane followed by lateral diffusion and/or recapture into distinct long-range trafficking organelles. In either case, computational modeling suggests that long-range delivery of somatically processed cargoes to dendrites may be too slow, taking many hours to days, to efficiently accommodate protein demand in remote regions (Williams et al., 2016). Indeed, evidence is accumulating for a distinct, local secretory processing network that could satisfy protein demand in neuronal dendrites. For example, ER-bound ribosomes and mRNAs encoding integral membrane and secreted proteins have been detected in dendrites, implying ongoing local translation of proteins at the dendritic ER (Cajigas et al., 2012; Cui-Wang et al., 2012; Wu et al., 2017). Experiments directly visualizing ER exit and accumulation in the ERGIC established that early trafficking events can occur in dendrites (Bowen et al., 2017; Hanus et al., 2014; Horton and Ehlers, 2003). However, whether subsequent trafficking to the cell surface is restricted to the same dendritic region as cargoes progress through sparsely distributed and highly mobile organelle networks has remained largely overlooked (Bowen et al., 2017; Cui-Wang et al., 2012).
To address these issues, we used zapERtrap to characterize where and when two archetypal synaptic proteins, the cell adhesion molecule neuroligin 1 (NL1) and GluA1-containing AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid)-type glutamate receptors, first appear at the cell surface when released from the ER in different subcellular domains. We observe that NL1 and GluA1 locally released from the somatic ER are directly transported deep into the dendritic arbor before surface delivery at or near synaptic sites. Neural activity differentially affected the timing and extent of surface trafficking of the two cargoes, consistent with distinct secretory trafficking routes originating from the cell body. Locally releasing these cargoes from dendritic ER resulted in surprisingly diffuse surface delivery, with no enrichment of total surface protein near the site of ER release, challenging current assumptions. Finally, select cargoes were prevented from entering axons through robust plasma membrane insertion at the axon initial segment (AIS), revealing a novel surface trafficking hotspot in neurons. Together these data provide the first look at the spatiotemporal dynamics of secretory trafficking from distinct subcellular domains in any cell type. More broadly, zapERtrap opens the door to previously unapproachable questions concerning how proteins are processed, trafficked, and secreted in space and time in complex cellular environments.
Results
Developing and validating an approach for light-triggered protein secretion
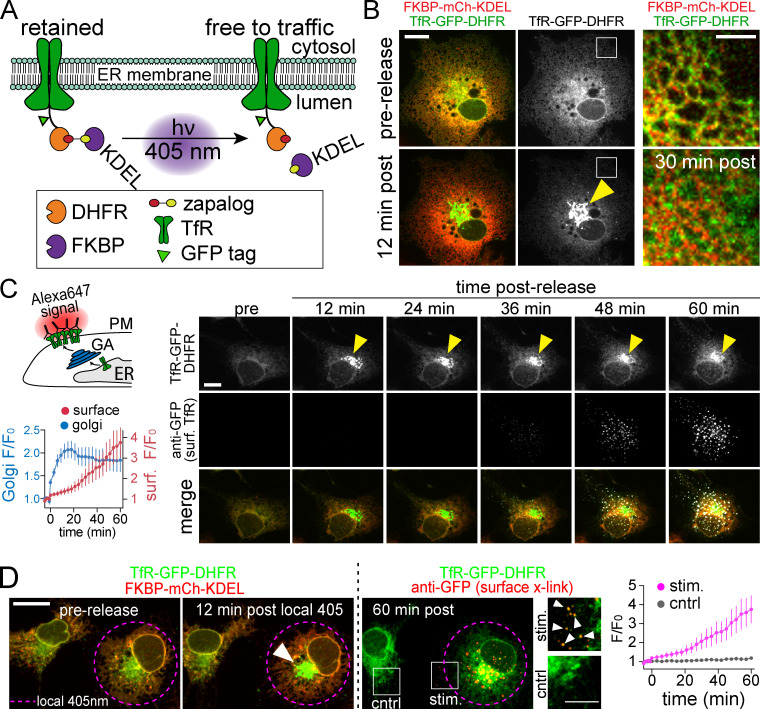
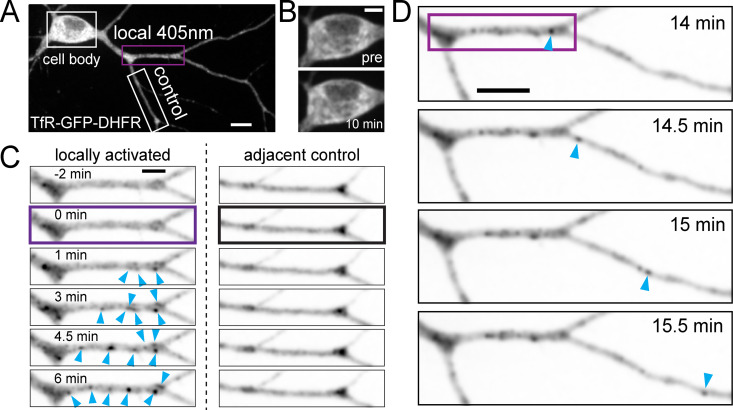
zapERtrap relies on a small-molecule protein dimerizer “zapalog,” which consists of the antibiotic trimethoprim (TMP) tethered to a synthetic ligand of FK506-binding protein (SLF) through a photocleavable linker (Gutnick et al., 2019). Zapalog dimerizes FK506-binding protein (FKBP), which binds to SLF, and dihydrofolate reductase (DHFR), which binds to TMP. Illumination with low-intensity 405-nm light disrupts the linker bridging SLF and TMP to rapidly dissociate FKBP and DHFR. To use zapalog for light-inducible ER release, we fused DHFR to the lumenal domain of several different integral membrane cargo molecules (Fig. 1 A). To sequester these molecules in the ER, we targeted FKBP-XFP (where XFP is either EGFP or mCh) to the ER and appended a C-terminal “KDEL” ER retention motif. In the presence of zapalog, FKBP-XFP-KDEL dimerizes with DHFR-fused cargo molecules, trapping them in the ER. Photocleavage of zapalog with 405-nm light liberates the cargo molecule from the ER retention module (FKBP-XFP-KDEL), allowing forward trafficking to proceed (Fig. 1 A).
Figure 1.
Developing zapERtrap for light-triggered protein secretion. (A) Schematic of zapERtrap strategy. DHFR is fused to the lumenal domain of a membrane protein cargo (TfR shown here). An ER-retained version of FKBP (C-terminal KDEL tag) is expressed in the lumen of the ER. The zapalog compound bridges these domains, retaining the DHFR-fused protein in the ER. 405-nm light disrupts the zapalog tether between DHFR and FKBP-KDEL, allowing forward trafficking to proceed. (B) A pulse of 405-nm light triggers redistribution of TfR-GFP-DHFR (green) from the ER to the GA (arrowhead). Magnified images (taken from the region marked by the white boxes) before (top) and 12 min following (bottom) ER release are shown on the right. Scale bar, 10 µm. Inset scale bar, 5 µm. (C) Live cell antibody surface labeling following ER release. Top left: Schematic of strategy visualizing surface accumulation using extracellular Alexa647-anti-GFP. Right: Image time series of TfR-GFP-DHFR accumulation in the GA (top; arrowhead) and on the cell surface (middle) following ER release. Scale bar, 7 µm. Bottom left: Kinetics of TfR-GFP-DHFR accumulation in the GA (blue) and on the surface (red) following release; mean ± SEM (n = 6 cells from two independent experiments). (D) Focal 405-nm illumination (left; pink dashed line) triggers TfR-GFP-DHFR (green) trafficking to the GA (arrowhead) and the surface (right, Alexa647-anti-GFP puncta shown in red) only in the photoactivated cell. Magnified images show TfR-GFP-DHFR surface label (arrowheads) in the photoactivated cell (stim.; top) and control cell (bottom). Scale bar, 10 µm. Inset, 5 µm. The average time courses of TfR-GFP-DHFR surface accumulation for photoactivated cells (pink line) and neighboring control cells (gray line) are plotted; mean ± SEM (n = 6–8 cells from two independent experiments). cntrl, control; surf, surface.
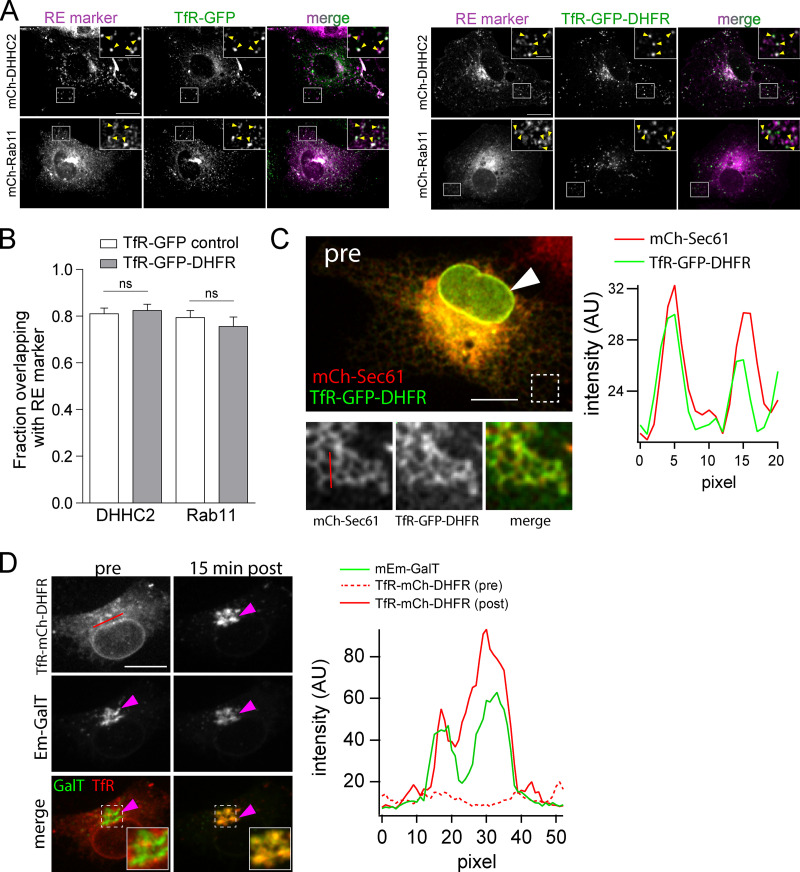
Due to its robust trafficking properties, we initially validated the zapERtrap system using transferrin receptor (TfR) as a model cargo in COS-7 cells. In the absence of zapalog, TfR-GFP-DHFR strongly colocalizes with recycling endosome (RE) markers Rab11 and DHHC2, confirming that the DHFR tag does not disrupt normal subcellular targeting (Fig. S1, A and B). However, in the presence of 500 nM zapalog and coexpressed mCh-FKBP-KDEL, TfR-GFP-DHFR was strongly retained in the ER (Fig. 1 B and Fig. S1 C). Following brief (10–50 ms) full-field 405-nm illumination, we observed robust redistribution of TfR-GFP-DHFR from the ER to the GA, with kinetics similar to previously reported ER to GA trafficking (Fig. 1, B and C; Fig. S1 D; and Video 1; Boncompain et al., 2012; Chen et al., 2013; Horton and Ehlers, 2003; Presley et al., 1997). To measure subsequent plasma membrane insertion, we included an Alexa Fluor 647–conjugated antibody against GFP (Alexa647-anti-GFP) in the extracellular solution (Fig. 1 C). As the GFP-tagged cargo is presented on the cell surface, rapid antibody binding results in signal amplification as well as restriction of lateral mobility through cross-linking, allowing us to monitor TfR-GFP-DHFR surface delivery in real time with high sensitivity (Fig. 1 C and Video 1). Following release of ER-retained TfR-GFP-DHFR, surface-bound Alexa647-anti-GFP could be detected within 30 min, similar to previous reports for the latency between ER release and surface appearance (Fig. 1 C; Chen et al., 2013; Presley et al., 1997).
Figure S1.
The DHFR tag does not perturb subcellular localization.(A) Subcellular localization of TfR-GFP (left) versus TfR-GFP-DHFR (right) in COS-7 cells (in the absence of zapalog) relative to the RE markers mCh-DHHC2 (top) and mCh-Rab11 (bottom). Insets show magnified images of the regions marked by the white box. Yellow arrowheads in insets denote colocalized puncta. Scale bar, 20 µm. Inset scale bar, 5 µm. (B) Quantification of the data shown in A. Colocalization between TfR-GFP or TfR-GFP-DHFR and the RE markers mCh-DHHC2 and mCh-Rab11 was assessed by calculating the proportion of either TfR-GFP (white bars) or TfR-GFP-DHFR (gray bars) that overlap with the respective RE marker; mean ± SEM (unpaired t test; P = 0.7346 [DHHC2], P = 0.4558 [Rab11]; n = 10 cells/condition). (C) TfR-GFP-DHFR (green) colocalizes with ER marker mCh-Sec61 (red) in COS-7 cells expressing FKBP-KDEL in the presence of zapalog. The white arrowhead denotes the nuclear envelope, which is contiguous with the ER and contains both red and green signals. Bottom: Enlarged images of mCh-Sec61 and TfR-GFP-DHFR at the cell periphery (denoted by dashed box in top panel) where ER morphology is distinct. A plot of pixel intensities along the red line (in blow-up panel of mCh-Sec61) is plotted to the right. Scale bar, 10 µm; magnified images are 6 × 6 µm. (D) Shown is a COS-7 cell expressing TfR-mCh-DHFR, the GA marker mEmerald-GalT, and FKBP-KDEL in the presence of zapalog before (left) and 15 min following 405-nm light exposure (right). The magenta arrowheads denote the GA. The insets show colocalization between mEmerald-GalT and TfR-mCh-DHFR. Note the display settings were adjusted for the TfR-mCh-DHFR 15 min after image to avoid apparent saturation of the signal as it concentrated in the GA. Pixel intensities along the red line shown in the upper left are plotted for mEmerald-GalT (green line) and TfR-mCh-DHFR before ER release (red dashed line) and 15 min after ER release (solid red line). Scale bar, 10 µm; magnified images are 5 × 5 µm.
Video 1.
Spatially restricted ER release of TfR-GFP-DHFR from an individual COS-7 cell. Single cell ER release in COS-7 cells expressing TfR-GFP-DHFR (green) and FKBP-mCh-KDEL (channel not displayed) in the presence of Alexa647-anti-GFP (white) before and after focal 405-nm illumination. The photoactivated cell is marked with a purple circle the first frame after photoactivation. Note the accumulation of TfR-GFP-DHFR at the GA and then the surface only in the targeted cell. The duration of the video is 50 min with an acquisition rate of 1 frame/2 min. Scale bar, 20 µm. Playback speed, 2 min between frames.
One of the major potential applications of this approach is local control of secretory trafficking from user-defined cell populations. To test if this was possible with zapERtrap, we initiated ER release in only one of the COS-7 cells within the imaging field with focally directed 405-nm excitation (Fig. 1 D). We only observed accumulation of TfR-GFP-DHFR in the GA of photoactivated cells (Fig. 1 D and Video 1). Accordingly, only photoactivated cells accumulated surface TfR-GFP-DHFR (Fig. 1 D).
Spatial and temporal properties of synaptic protein trafficking in hippocampal neurons
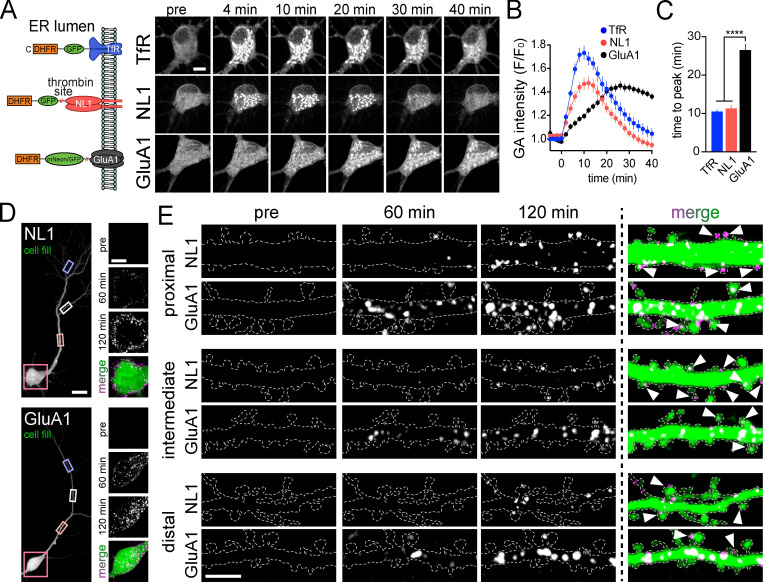
We next used zapERtrap to investigate synaptic protein trafficking in neurons. We engineered DHFR fusions with the AMPA-type glutamate receptor GluA1 (DHFR-mNeon/GFP-GluA1) and the synaptic cell adhesion molecule neuroligin 1 (DHFR-GFP-NL1; Fig. 2 A). We first confirmed that the DHFR tag does not perturb their normal subcellular localization in primary hippocampal neurons (Fig. S2 A). All cargoes were efficiently retained in the ER with zapalog and released with a single 50-ms 405-nm light pulse (Fig. 2, A–C; Video 2; and Video 3).
Figure 2.
Spatial and temporal properties of synaptic protein trafficking in hippocampal neurons following global ER release. (A) Schematic of constructs used for ER release experiments in hippocampal neurons. Right: Image series of TfR-GFP-DHFR (top), DHFR-GFP-NL1 (middle), and DHFR-mNeon-GluA1 (bottom) accumulation in the GA of hippocampal neurons following light exposure at time 0. Scale bar, 10 µm. (B) Time course of TfR-GFP-DHFR (blue), DHFR-GFP-NL1 (red), and DHFR-mNeon-GluA1 (black) trafficking to the GA following global (whole cell) light-triggered ER release; mean ± SEM (n = 11–27 cells from two or three independent experiments). (C) Comparison of the time to peak accumulation for TfR-GFP-DHFR (blue), DHFR-GFP-NL1 (red), and DHFR-mNeon-GluA1 (black) following ER release. ****, P < 0.0001 (one-way ANOVA, Tukey’s multiple comparisons test). (D) Surface expression of DHFR-GFP-NL1 (top) and DHFR-GFP-GluA1 (bottom) was detected by including Alexa647-anti-GFP in the extracellular solution. Somatic regions before and 60 min and 120 min following ER release are shown (right). Scale bar, 20 µm. Inset scale bar, 10 µm. (E) Appearance of DHFR-GFP-NL1 and DHFR-GFP-GluA1 at the surface of proximal (top), intermediate (middle), and distal (bottom) dendrites. Examples show surface signal before and 60 min and 120 min following ER release. Arrowheads denote spines with surface label. Scale bar, 5 µm.
Figure S2.
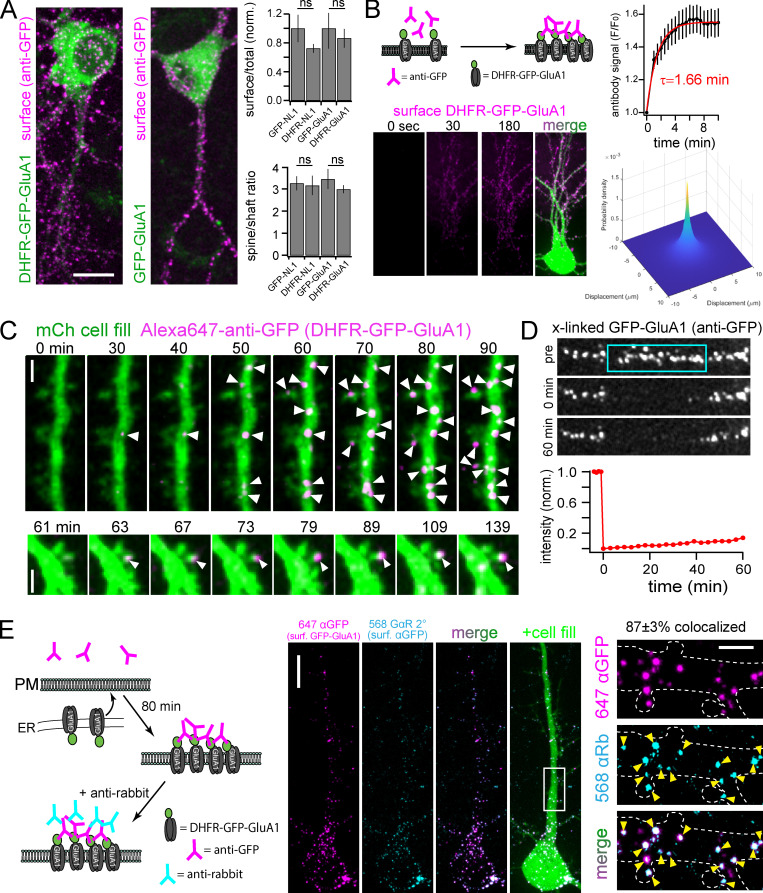
Localization of DHFR fused synaptic proteins and validation of antibody cross-linking for immobilizing surface proteins. (A) DHFR does not disrupt basal trafficking. Hippocampal neurons expressing the indicated constructs for 18 h (in the absence of zapalog) were labeled with Alexa647-anti-GFP under nonpermeabilizing conditions. The image panels show representative surface labeling (magenta) for DHFR-GFP-GluA1 (left) compared with GFP-GluA1 (right). The ratios of total surface (Alexa647-anti-GFP) to total (GFP) signal (top) and spine to shaft surface signal (bottom) are plotted for DHFR-GFP-NL1 and DHFR-GFP-GluA1 and compared with constructs without DHFR. The surface/total ratio signal is normalized to control (non-DHFR fused constructs); mean ± SEM, n = at least 8 neurons/condition from two separate cultures (Student’s t test). Scale bar, 10 µm. (B) Kinetics of Alexa647-anti-GFP binding to surface cargoes. Schematic of antibody cross-linking/labeling strategy is shown (top left). Bottom right: Antibody binding/cross-linking signal (magenta) measured in live neurons expressing DHFR-GFP-GluA1 immediately before and 30 s and 180 s following antibody addition. Kinetic data (top right) were fit with a single exponential (solid red line, τ = 1.66 min, n = 6 neurons). The two-dimensional plot (bottom right) models the probability of receptor localization following surface insertion and antibody cross-linking as a function of displacement from the membrane insertion site (see Materials and methods for details). Scale bar, 10 µm. (C) Representative examples of stable antibody binding to DHFR-GFP-GluA1 as it appears at the cell surface. A neuron expressing DHFR-GFP-GluA1 and cell fill (green) was imaged at the times indicated after ER release in the continuous presence of extracellular Alexa647-anti-GFP (magenta). Note the stable, sequential appearance of new surface puncta (arrowheads) on dendrites and select spines. Two representative examples are shown from different cells. Scale bar, 2 µm. (D) Alexa647-anti-GFP labeled DHFR-GFP-GluA1 is shown before, immediately following, and 60 min after photobleaching a small region (designated by blue box). Quantification of Alexa647 signal within the photobleached region is shown below; mean ± SEM (n = 10 dendritic regions from seven neurons). Scale bar, 5 µm. (E) The majority of cross-linked/labeled DHFR-GFP-GluA1 remains on the cell surface. DHFR-GFP-GluA1 was released from the ER and allowed to traffic to the surface for 80 min in the continuous presence of Alexa647- anti-GFP (magenta, generated in rabbit). Alexa647-anti-GFP was washed off, and Alexa568-anti-rabbit (teal) was added to label surface anti-GFP and confirm its localization at the cell surface. Magnified images of the highlighted region (white box) are shown to the right. Yellow arrowheads denote colocalized puncta. 87 ± 3% (mean ± SEM) of the Alexa647-anti-GFP puncta overlapped with Alexa568-anti-rabbit puncta. Data are averaged from five neurons. Scale bar, 10 µm (left); 2 µm (right). norm., normalized.
Video 2.
Effective retention and light-triggered release from the ER using zapERtrap in a hippocampal neuron. Shown is a hippocampal neuron expressing TfR-GFP-DHFR (green) along with ER-targeted FKBP-mCh-KDEL (red) in the presence of 500 nM zapalog before and after a single pulse of full-field 405-nm illumination (indicated by the white dot) delivered at time 0. Note the rapid light-triggered redistribution of the retained cargo (green) to mobile intracellular dendritic organelles and the stationary GA in the soma. Dimensions of the video are 82 µm × 82 µm. Playback speed, 1 min between frames.
Video 3.
Kinetics of somatic GA accumulation for different cargoes. Shown are hippocampal neurons expressing FKBP-mCh-KDEL (channel not displayed) and either TfR-GFP-DHFR (left), DHFR-GFP-NL1 (center), or DHFR-mNeon-GluA1 (right) before and after full-field 405-nm illumination (indicated by the white dot) delivered at time 0. The duration of the videos is 67 min with a baseline acquisition rate of 1 frame/min (first 5 frames) and a post-release acquisition rate of 1 frame/2 min. Time stamp displays the time elapsed following full-field 405-nm illumination. Scale bar, 10 µm. Playback speed, 2 min between frames.
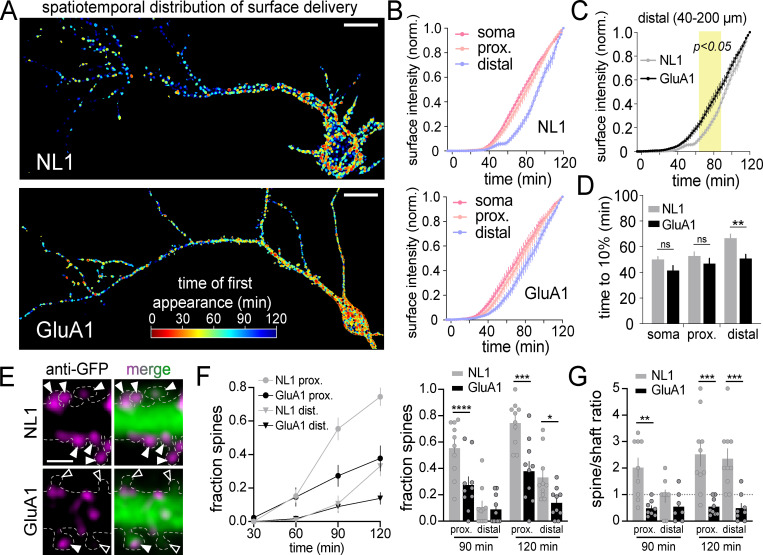
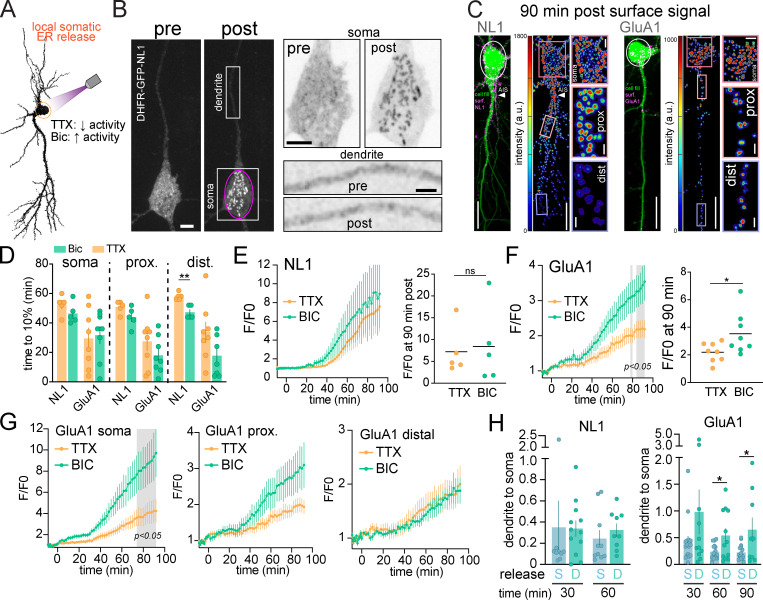
We next quantified where and when GluA1 and NL1 appear at the neuronal surface following global (i.e., the entire cell was exposed to 405-nm light) ER release. To isolate the contribution of secretory trafficking from lateral diffusion and subsequent recycling mechanisms, we included Alexa647-anti-GFP in the extracellular solution to continuously cross-link, immobilize, and label cargoes in real time, as they are presented (Fig. S2, B–E; Heine et al., 2008; Penn et al., 2017). At 120 min after release, both cargoes had accumulated at the surface of the soma and proximal regions of the dendritic arbor (Fig. 2, D and E; and Video 4). However, their dendritic distribution appeared markedly different. For GluA1, signal was easily discernable in distal dendrites (>40 µm from the soma) compared with NL1, which appeared more enriched on the cell body (Fig. 2, D and E). To quantify this, we segmented discrete surface signal puncta, measuring when they occurred, and classifying whether they occurred in the soma, proximal (0–40 µm from the soma), or distal (40–200 µm from the soma) dendritic domains (Fig. 3, A–D; and Video 5). Surprisingly, even though GA accumulation was nearly 2.5-fold slower for GluA1 compared with NL1 (Fig. 2 C), detectable GluA1 surface insertion within the somatic region was not delayed (time to 10% of total surface accumulation: 50.0 ± 2.29 min [NL1], 41.5 ± 3.81 min [GluA1]; Fig. 3, B–D). For proximal dendritic regions, the rates of surface accumulation were also similar between the two cargoes (time to 10% surface accumulation: 52.8 ± 3.07 min [NL1], 46.9 ± 4.03 min [GluA1]; Fig. 2 E; and Fig. 3, B and D). However, in distal dendrites, GluA1 was detected significantly earlier following ER release compared with NL1 (time to 10% surface accumulation: 66.6 ± 3.18 min [NL1], 50.9 ± 3.14 min [GluA1]; Fig. 2 E; and Fig. 3, B–D). Thus, GluA1 traffics to the surface more uniformly across the dendrites and soma, while NL1 is inserted in the soma and proximal dendrites followed by delayed insertion at distal sites, suggesting distinct trafficking pathways for each cargo (Fig. 3 D and Video 5).
Video 4.
Surface accumulation of cargoes following whole-cell illumination in neurons. Shown are hippocampal neurons expressing FKBP-mCh-KDEL (channel not displayed) and either DHFR-GFP-NL1 (top) or DHFR-GFP-GluA1 (bottom) before and after full-field. 405-nm illumination in the presence of extracellular Alexa647-anti-GFP (magenta). The first frame after photoexcitation is denoted by the appearance of the white circle (upper left corner). Left: The surface signal. Right: The merge between the cell fill (green) and surface signal (magenta). Time stamp displays the time elapsed following full-field 405-nm illumination. Scale bar, 20 µm. Playback speed, 2 min between frames.
Figure 3.
Subcellular distribution of NL1 and GluA1 surface presentation following global ER release. (A) The timing and location of surface trafficking for DHFR-GFP-NL1 (top) and DHFR-GFP-GluA1 (bottom) are shown following global light-triggered ER release. Surface signal with a shorter latency of appearance is rendered in warmer colors. Scale bars, 20 µm. (B) Time courses of DHFR-GFP-NL1 (top) and DHFR-GFP-GluA1 (bottom) surface delivery at different cellular domains following global ER release. Shown are the normalized intensities for surface signal at the soma (pink line) and proximal (peach line; up to 40 µm from the soma) or distal regions of the dendritic arbor (lavender line; 40–200 µm from the soma); mean ± SEM (NL1: n = 10 neurons/time point from four independent experiments; GluA1: n = 11 neurons/time point from two independent experiments). (C) Comparison of DHFR-GFP-NL1 (gray) and DHFR-GFP-GluA1 (black) surface accumulation (from B) at distal dendrites (40–200 µm from the soma) following ER release. The yellow shaded region denotes P < 0.05 (two-way ANOVA, Bonferroni’s multiple comparisons test). (D) Time to 10% of total surface accumulation is plotted for DHFR-GFP-NL1 (gray) and DHFR-GFP-GluA1 (black) in different cellular domains; mean ± SEM; **, P < 0.01 (Student's t test). (E) Representative images of DHFR-GFP-NL1 (top) and DHFR-GFP-GluA1 (bottom) surface signal (magenta, Alexa647-anti-GFP) in spines 90 min following ER release. Solid arrowheads denote cargo-positive spines. Open arrowheads mark spines that lack detectable surface cargo. The outline of the cell (dashed line) was drawn based on the cell fill (green signal in merge). Scale bar, 2 µm. (F) Time course of the fraction of spines in proximal (circles) and distal (triangles) dendrites with detectable DHFR-GFP-NL1 (gray) or DHFR-GFP-GluA1 (black) signal following ER release. A comparison of the fraction of DHFR-GFP-NL1– and DHFR-GFP-GluA1–positive spines at 90 and 120 min is shown on the right; mean ± SEM; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (Student's t test; n = 10 neurons/time point for NL1 and GluA1). (G) Plotted is the ratio of the total spine surface signal to the total dendritic shaft surface signal for DHFR-GFP-NL1 (gray) or DHFR-GFP-GluA1 (black) in proximal and distal dendrites, 90 and 120 min following ER release; mean ± SEM; **, P < 0.01; ***, P < 0.001 (Student's t test; n = 8–10 neurons/time point [NL1], n = 7–9 neurons/time point [GluA1]). dist., distal; norm., normalized; prox., proximal.
Video 5.
Segmented surface signal displaying where and when NL1 and GluA1 appear on the cell surface following global release. Newly appearing surface signal (anti-GFP binding to DHFR-GFP-NL1 or DHFR-GFP-GluA1) was masked, segmented, and displayed only during the first frame of appearance and for three subsequent frames (even though the signal was persistent) to visualize where and when surface signal appeared (rendered in green; see Materials and methods). The cells were exposed to full-field 405-nm illumination at time 0. The cell outlines (purple) were drawn based on a cell fill mask. The first frame after photoexcitation is denoted by the appearance of the white circle (upper right corner). The duration of the videos is 130 min with an acquisition rate of 1 frame/2 min. Scale bar, 20 µm. Playback speed, 2 min between frames.
We next quantified GluA1 and NL1 accumulation at the surface of dendritic spines, the primary postsynaptic compartments of excitatory synapses. While the surface labeling method cannot unambiguously distinguish between direct insertion into spines and lateral diffusion into spines before antibody cross-linking and detection (see Materials and methods), we observed large differences in the fraction of spines with accumulated signal. NL1 was targeted to a larger fraction of spines and more of the total dendritic signal resided in spines, compared with GluA1 (Fig. 3, E–G). Thus, new proteins can be rapidly incorporated into spines, although the extent of spine delivery appears to be cargo dependent.
The AIS is a surface trafficking hotspot for select cargoes
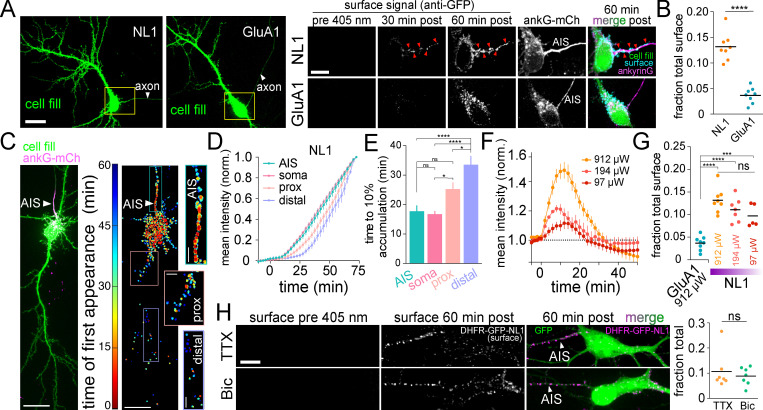
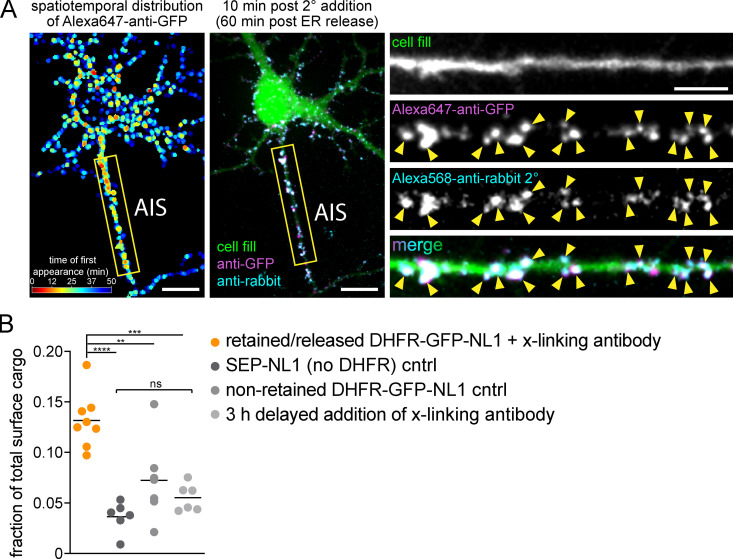
Following global ER release, we observed robust surface insertion of NL1, but not GluA1, at the AIS (Fig. 4, A–E; and Video 6). To ensure AIS signal did not arise from intracellular vesicles loaded with NL1 that had surfaced and bound Alexa647-anti-GFP elsewhere in the cell, we added an Alexa568 anti-rabbit antibody (which labels Alexa647-anti-GFP) to cells that had accumulated Alexa647-anti-GFP signal for 50 min following ER release. We observed robust Alexa647-anti-GFP puncta colocalization with Alexa568-anti-rabbit puncta, supporting direct delivery of DHFR-GFP-NL1 to the AIS surface (Fig. S3 A). We next tested the possibility that AIS insertion was due to overloading the secretory pathway, causing NL1 to “spill over” into a nonrelevant, AIS-directed pathway. Here we leveraged the ability to precisely titrate the amount of NL1 released from the ER simply by decreasing the intensity of the excitation light. We calibrated the amount of NL1 release by quantifying its accumulation in the GA following excitation with different light intensities (Fig. 4 F). Importantly, we observed strong AIS delivery, even at 405-nm excitation intensities near the threshold for reliably detecting surface trafficking (Fig. 4, F and G). Surprisingly, we observed very little DHFR-GFP-GluA1 signal at the AIS even at saturating light intensities that released the maximum level of GluA1 from the ER (Fig. 4, A, B, and G). We also tested whether NL1 trafficking at the AIS is regulated by neuronal firing. We observed no difference in NL1 AIS accumulation in the presence of tetrodotoxin (TTX), which suppresses action potentials, or the γ-aminobutyric acid type A receptor antagonist bicuculline (Bic), which causes increased action potentials (Fig. 4 H). Combined, these data reveal a novel trafficking route to the surface of the AIS that is independent of neural activity, but highly cargo selective. We note that we were able to detect AIS enrichment in our experiments because our detection strategy effectively traps and immobilizes cargo soon after it reaches the cell surface, thus integrating the accumulated AIS signal. If we delayed addition of our immobilizing/detection antibody to 3 h after ER release, we did not observe any enrichment at the AIS (Fig. S3 B). Likewise, no AIS enrichment could be detected in cells constitutively expressing superecliptic pHluorin (SEP)-tagged NL1 or DHFR-GFP-NL1 that had not been ER retained (Fig. S3 B). Thus, under normal steady-state conditions, NL1 that is initially presented at the AIS must quickly exit the AIS via lateral diffusion or be rapidly recaptured by endocytosis and trafficked to postsynaptic sites.
Figure 4.
The AIS is a surface trafficking hotspot for specific cargoes. (A) Representative images of DHFR-GFP-NL1 (left) and DHFR-GFP-GluA1 (right) expressing neurons expressing the AIS marker ankyrinG-mCh (not shown) and a cell fill (green). The axon is indicated by the white arrowhead. Magnified images to the right (taken from the yellow boxes) display surface signal at the AIS for DHFR-GFP-NL1 (top) and DHFR-GFP-GluA1 (bottom) before and 60 min after ER release. Note that Alexa647-anti-GFP was present in the extracellular solution following ER release to continuously trap and label proteins as they surfaced. Red arrowheads denote accumulated cargo at the surface of the AIS. Scale bars, 20 µm; inset, 10 µm. (B) AIS surface signal (expressed as a fraction of total surface signal throughout the entire cell) for DHFR-GFP-NL1 (orange) or DHFR-GFP-GluA1 (blue) 60 min after ER release; mean ± SEM; ****, P < 0.0001 (Student's t test, n = 8 from two independent experiments for NL1 and GluA1). (C) Surface trafficking at the AIS occurs early following ER release. Confocal image (left) and heatmap displaying the timing and location of DHFR-GFP-NL1 surface appearance (right). Insets show the AIS and proximal and distal dendrites. Scale bars, 20 µm. Inset scale bars, 5 µm. (D) Shown are mean NL1 surface intensities (normalized to their maximum values) at the AIS (teal line), soma (pink line), proximal dendrites (5–40 µm from the soma; peach line) or distal dendrites (40–200 µm from the soma; lavender line); mean ± SEM (n = 10 neurons/time point from two independent experiments). (E) Time to reach 10% of maximum DHFR-GFP-NL1 surface signal is plotted for each subcellular compartment; mean ± SEM; *, P < 0.05; ****, P < 0.0001 (one-way ANOVA, Tukey’s multiple comparisons test; n = 10 neurons from two independent experiments). (F) ER release can be titrated by decreasing photoexcitation power. DHFR-GFP-NL1 accumulation in the GA is plotted following illumination with decreasing 405-nm light intensities (912 µW, 194 µW, and 97 µW); mean ± SEM (n = 7–14 neurons/condition from at least two independent experiments). (G) DHFR-GFP-NL1 appears at the surface of the AIS even when decreasing amounts are released from the ER. GluA1 surface signal at the AIS following exposure to a saturating light intensity (912 µW) is shown for comparison; mean ± SEM; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA, Tukey’s multiple comparisons test; n = 5–8 neurons/condition from two independent experiments). (H) DHFR-GFP-NL1 traffics to the surface of the AIS when network activity is elevated (Bic) or suppressed (TTX). Student's t test; n = 7 neurons/condition from three independent experiments. norm., normalized; prox., proximal.
Video 6.
NL1 is inserted into the plasma membrane of the AIS. Shown is a hippocampal neuron expressing DHFR-GFP-NL1 along with unlabeled FKBP-KDEL, ankyrinG-mCh (red), and a GFP cell fill (green) before and after full-field 405-nm illumination in the presence of Alexa647-anti-GFP (surface signal is shown in grayscale). The first frame after photoexcitation is denoted by the appearance of the white circle (upper right corner). The duration of the video is 50 min with a baseline acquisition rate of 1 frame/2 min and a post-release acquisition rate of 1 frame/2.5 min. Time stamp displays the time elapsed following full-field 405-nm illumination at time 0. Scale bar, 10 µm. Playback speed, 2.5 min between frames.
Figure S3.
Control experiments for surface NL1 accumulation at the AIS. (A) Antibody-labeled DHFR-GFP-NL1 signal is localized to the cell surface. DHFR-GFP-NL1 was released from the ER and continuously cross-linked and labeled with Alexa647-anti-GFP as it appeared at the cell surface for 50 min after ER release. Left: The timing and distribution of accumulated Alexa647-anti-GFP (generated in rabbit) immediately before addition of Alexa568-anti-rabbit secondary to label cell surface Alexa647-anti-GFP. The center panel shows the same neuron 10 min following addition of Alexa568-anti-rabbit (cyan). Insets to the right show the AIS, taken from the yellow box in the image to the left. The robust colocalization of Alexa647-anti-GFP and Alexa568-anti-rabbit (arrowheads) confirms accumulated DHFR-GFP-NL1 is on the cell surface. Scale bars, 10 µm. Inset scale bar, 5 µm. (B) Comparison of the fraction of total surface cargo at the AIS for retained/released DHFR-GFP-NL1 in the continuous presence of cross-linking antibody (orange) versus three trafficking controls: SEP-GluA1 (dark gray), nonretained (no zapalog) DHFR-GFP-NL1 (gray), and DHFR-GFP-NL1 that was released and allowed to traffic for 3 h before addition of cross-linking antibody (light gray). The retained/released DHFR-GFP-NL1 is the same data shown in Fig. 4 B with Alexa647-anti-GFP present for the duration of the experiment; mean ± SEM; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA, Tukey’s multiple comparisons test; n = 8 [retained/released DHFR-GFP-NL1]; n = 6 [SEP-NL1]; n = 7 [nonretained DHFR-GFP-NL1]; n = 6 [3 h delayed addition cntrl]; n = number of neurons). cntrl, control.
Proteins originating in the dendritic secretory pathway are broadly dispersed as they traffic to the cell surface
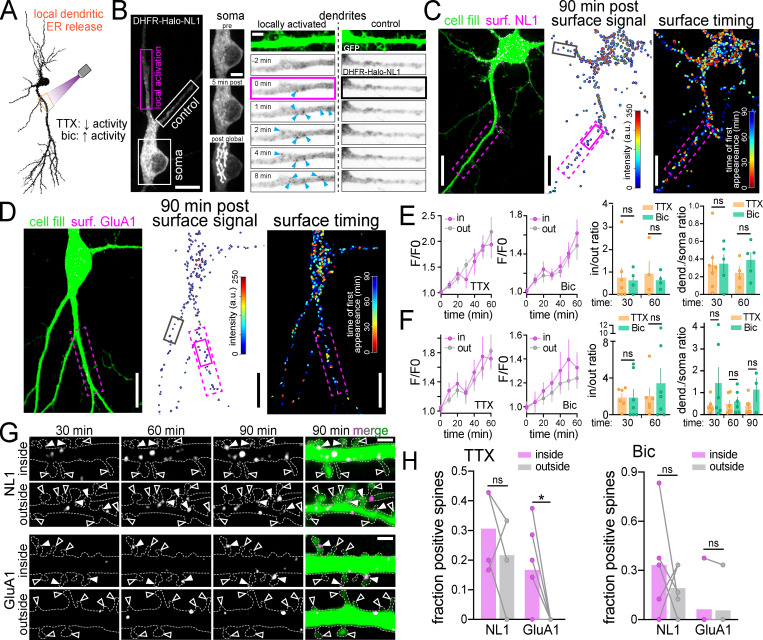
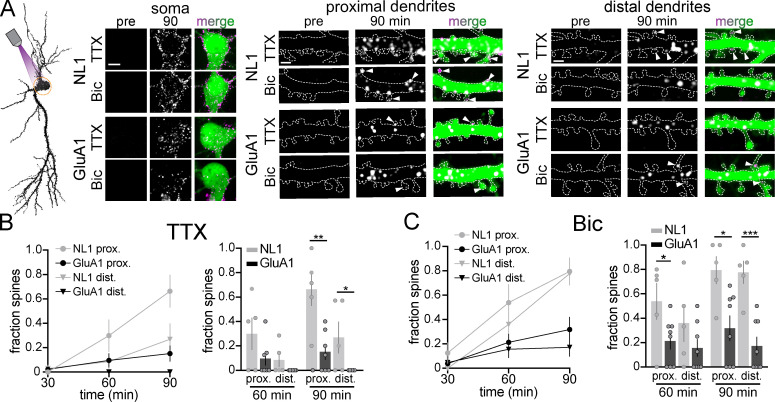
It is widely assumed that synaptic receptor proteins generated in dendrites are precisely delivered to nearby synaptic sites. Thus, we used zapERtrap to test whether protein cargoes exiting the ER within a specific dendritic branch reach the surface of the same branch, or are dispersed as they move through mobile downstream trafficking organelles. We also asked whether neural activity influences the spatial pattern of surface expression by applying TTX or Bic before ER release (Fig. 5 A). We locally released DHFR-GFP-GluA1 and DHFR-GFP-NL1 from dendritic segments ∼30–40 µm in length and 30–50 µm from the soma. Following focal light exposure, we observed the intracellular GFP signal coalesce into punctate intracellular structures within tens of seconds in the illuminated dendrite but not in neighboring dendrites or the soma, consistent with spatially restricted ER exit and ERGIC accumulation in dendrites (Fig. 5 B; and Fig. S4, A–C; Bowen et al., 2017). These structures were initially stable before abruptly moving away from their sites of appearance, often leaving the photo-targeted region (Fig. S4 D and Video 7). Accordingly, we observed widespread surface delivery for both NL1 and GluA1 not only at the targeted dendrite but also at untargeted control dendrites and even the cell body, especially for NL1 (Fig. 5, C and D). We could not detect enrichment of total surface signal within or near the illuminated region (Fig. 5, E and F). Despite no enrichment in overall surface GluA1 at the targeted dendrite, we found that spines within the illuminated region were significantly more enriched with GluA1 compared with similarly sized regions in neighboring control dendrites, but only when action potentials were suppressed with TTX (Fig. 5, G and H). These data are consistent with previous reports of local synthesis and GluA1 trafficking to local synapses following suppression of synaptic transmission (Sutton et al., 2006). In contrast, NL1 was delivered to a similar fraction of spines inside and outside of the ER release zone, with no activity dependence, suggesting a distinct trafficking pathway from GluA1 (Fig. 5 H).
Figure 5.
Local ER release from dendrites reveals the rate, spatial dynamics, and activity dependence of remote secretory trafficking. (A) Schematic of experimental strategy. NL1 and GluA1 were locally released from a user-defined dendritic branch in the presence of either TTX to suppress or Bic to elevate neuronal activity. (B) Representative image (left) and time series of intracellular DHFR-HaloTag-NL1 (labeled with JF646) signal at the soma (center) and dendrites (right) shortly following local dendritic ER release (pink rectangle). Blue arrowheads mark the appearance of vesicular structures following illumination. The neighboring unstimulated control branch (white rectangle) is shown for comparison. At the end of the experiment, the cell was exposed to global full-field illumination and imaged 10 min later. Note the robust DHFR-GFP-NL1 accumulation in the somatic GA following global but not local dendritic release (middle). Scale bar, 10 µm; magnified panels, 5 µm. (C) Images showing cellular morphology (cell fill, left), DHFR-GFP-NL1 surface signal (center), and timing/location of surface trafficking (right) following local dendritic ER release. The photoactivated region is denoted by the dashed pink rectangle. The black and pink (solid lines) boxes denote the dendritic regions shown in G. Scale bars, 10 µm. (D) Images showing cellular morphology (cell fill, left), surface DHFR-GFP-GluA1 signal (center), and timing/location of surface trafficking (right) following local dendritic ER release (pink dashed rectangle). The black and pink (solid lines) boxes denote the dendritic regions shown in G. Scale bars, 10 µm. (E) Time course of DHFR-GFP-NL1 surface signal inside (in) and outside (out) the ER release zone in the presence of TTX (left) or Bic (right); mean ± SEM. No significant differences were found at any time point (two-way ANOVA, Bonferroni’s multiple comparisons test). The plots to the right show the ratio of signal in versus out of the release zone and the ratio of total dendritic to somatic signal 30 and 60 min following dendritic ER release in the presence of TTX (orange) or Bic (green); mean ± SEM (Student's t test; n = 4–7 from at least three independent experiments). (F) Time course of DHFR-GFP-GluA1 surface signal inside (in) and outside (out) the ER release zone in the presence of TTX (left) or Bic (right). No significant differences were found at any time point (two-way ANOVA, Bonferroni’s multiple comparisons test); mean ± SEM. The plots to the right show the ratio of signal in versus out of the release zone and the ratio of total dendritic to somatic signal 30 and 60 min following dendritic ER release in the presence of TTX (orange) or Bic (green); mean ± SEM (Student's t test; n = 3–7 from at least two independent experiments). (G) Image series of surface-labeled DHFR-GFP-NL1 (top) and DHFR-GFP-GluA1 (bottom) inside and outside the photoactivated region depicted in C and D. Solid and open arrowheads denote cargo-positive and -negative spines, respectively. Scale bars, 2 µm. (H) Comparison of the fraction of spines with DHFR-GFP-GluA1 or DHFR-GFP-NL1 within the release zone (purple) versus randomly selected regions of the same size in separate control dendrites (gray) 60 min following local dendritic ER release in the presence of TTX (left) or Bic (right); mean ± SEM; *, P < 0.05 (paired t test; n = 4–6 from at least three independent experiments). dend., dendritic; surf., surface.
Figure S4.
zapERtrap can be used to locally release cargoes from the ER within defined regions of the dendritic arbor. (A) TfR-GFP-DHFR was released from the dendritic segment under the purple box by local illumination with 405-nm light. The adjacent dendrite (white rectangle) serves as a nonphotoactivated control. Scale bar, 10 µm. (B) Somatic TfR-GFP-DHFR signal showed no detectable difference before and 10 min following local dendritic release. Scale bar, 5 µm. (C) Time-lapse of intracellular TfR signal following local dendritic release in the targeted branch and an adjacent control branch (white box in A). Note the rapid appearance of punctate trafficking organelles (arrowheads) in the released branch but not the control branch. Scale bar, 5 µm. (D) Zoomed-out images of the dendrite shown in C, starting 14 min after local dendritic photorelease (purple box). An example of a mobile intracellular cargo vesicle (blue arrowhead) exiting the photoactivated region is shown. Scale bar, 10 µm.
Video 7.
Following local dendritic ER release, cargo can be transported out of the release zone. Shown are hippocampal neurons expressing either TfR-GFP-DHFR (top) or DHFR-Halo-NL1 (bottom, labeled with JaneliaFluor 646) along with FKBP-mCh-KDEL (signal not shown). ER-retained cargo was released at time 0 using focally directed 405-nm excitation. The timing and location of ER release are shown by purple rectangles. Arrows denote examples of mobile carriers exiting the release zone. The dimensions of the top panel (TfR) are 13 µm × 45 µm; bottom panel (NL1), 20 µm × 45 µm. Playback speed, 15 sec between frames.
The cell body secretory pathway directs cargoes over long distances to dendrites and spines
To address how far somatically derived proteins can directly traffic into dendrites and the effects of neural activity on the spatiotemporal dynamics of long-range transport, we used focal illumination to release target proteins from the somatic ER in the presence of TTX or Bic. Once more, we included Alexa647-anti-GFP in the extracellular solution to continuously label and cross-link cargoes as they appeared at the surface (Fig. 6 A). Following local somatic release, cargo rapidly accumulated in the cell body GA with no detectable accumulation occurring in ERGIC or GA-like structures in either proximal or distal dendrites (Fig. 6 B). Thus, cargoes released from the soma primarily use the cell body secretory network rather than laterally diffusing in the ER membrane to dendritic exit sites (Cui-Wang et al., 2012; Aridor et al., 2004). In the minutes following somatic ER release, both DHFR-GFP-NL1 and DHFR-GFP-GluA1 appeared at the surface of the soma as well as at the surface of dendrites tens to hundreds of micrometers into the dendritic arbor (Figs. 6 C and Fig. S5 A). NL1 also rapidly appeared at the surface of the AIS, demonstrating that AIS-directed NL1 originates from the cell body (Fig. 6 C). Elevating neural activity with Bic increased the total amount of GluA1 delivered to the cell surface but did not have a significant effect on the timing of surface delivery at the soma or dendrites (Fig. 6, D, F, and G). Intriguingly, activity accentuated GluA1 surface delivery primarily near the site of ER release (soma) and proximal dendrites, consistent with previous studies demonstrating activity-dependent stalling and fusion of transport vesicles, perhaps before they could be transported deeper into the dendritic arbor (Fig. 6 G; Hangen et al., 2018). Conversely, elevating activity did not impact the total amount of NL1 surface delivery but significantly decreased the time to achieve detectable surface signal at distal dendrites (Fig. 6, D and E). Surprisingly, we also observed surface accumulation of somatically released cargoes in dendritic spines, indicating that somatically derived proteins undergo long-range trafficking to postsynaptic compartments within tens of minutes. Spine targeting of GluA1 and NL1 occurred in a proximal (higher fraction) to distal (lower fraction) gradient and was elevated at distal synapses when neural activity was elevated (Fig. S5, B and C). While NL1 and GluA1 originating from either the somatic or dendritic secretory networks could be delivered to dendrites and spines, we noted that the dendrite/soma ratio of total surface signal for GluA1 (but not NL1) was significantly higher when GluA1 was released from dendrites, supporting a higher degree of compartmentalized dendritic trafficking for GluA1 (Fig. 6 H).
Figure 6.
Local release from the cell body ER reveals direct, long range trafficking to dendrites. (A) Schematic of experimental strategy. DHFR-GFP-NL1 and DHFR-GFP-GluA1 were locally released from the soma in the presence of TTX to suppress or Bic to elevate neuronal activity. (B) Example of DHFR-GFP-NL1 intracellular localization before (left) and 14 min after (right) somatic ER release (pink circle). The magnified images to the right show the soma (top) and a section of dendrite (bottom) before and after ER release. Note the absence of vesicular structures appearing in the dendrites shortly following somatic release. Scale bar, 6 µm. Top inset scale bar, 6 µm. Bottom inset scale bar, 3 µm. (C) Merged confocal images showing cell fill (green) and surface signal (Alexa647-anti-GFP, magenta) for DHFR-GFP-NL1 (left) and DHFR-GFP-GluA1 (right) 90 min following local ER release (white circles). Surface signal is shown in the heatmap images. Insets show the soma, proximal, and distal dendrites 90 min after release. Scale bars, 20 µm. Soma inset scale bars, 5 µm. Dendrite inset scale bars, 2 µm. (D) Time to 10% surface accumulation following somatic ER release in the presence of TTX (orange bars) or Bic (green bars) for NL1 and GluA1. Mean ± SEM; **, P < 0.01 (Student's t test; n = 5–8 neurons from at least three independent experiments). (E) Time course of DHFR-GFP-NL1 surface trafficking (over the entire cell) following somatic ER release in the presence of TTX (orange line) or Bic (green line). Surface signal at 90 min is shown to the right; mean ± SEM (Student's t test; n = 5 neurons/condition from three independent experiments). (F) Time course of DHFR-GFP-GluA1 surface trafficking (over the entire cell) following somatic ER release in the presence of TTX (orange line) or Bic (green line). The gray shaded regions designate P < 0.05, two-way ANOVA, Bonferroni’s multiple comparisons test; n = 8 neurons/condition from two independent experiments. Surface signal at 90 min is shown on the right; mean ± SEM; *, P < 0.05 (Student's t test). (G) Time course of DHFR-GFP-GluA1 surface trafficking at the soma (left), proximal dendrites (<40 µm from the soma; center), and distal dendrites (40–200 µm from the soma; right) in the presence of TTX or Bic; mean ± SEM. The gray shaded region designates P < 0.05, two-way ANOVA, Bonferroni’s multiple comparisons test. (H) The ratio of total dendritic to somatic signal is plotted at different time points following local ER release from the soma (blue) or from dendrites (teal) for DHFR-GFP-NL1 (left) and DHFR-GFP-GluA1 (right); mean ± SEM; *, P < 0.05 (Student's t test; n = 9–16 neurons from at least two independent experiments). dist., distal; prox., proximal.
Figure S5.
Subcellular distribution of NL1 and GluA1 surface accumulation following somatic ER release. (A) Images of DHFR-GFP-NL1 (top) and DHFR-GFP-GluA1 (bottom) surface accumulation at the soma (left), proximal dendrites (middle), and distal dendrites (right) before and 90 min following somatic ER release in the presence of TTX or Bic. Merged images show the Alexa647-anti-GFP signal (magenta) and the cell fill (green). White arrowheads indicate spines with detectable levels of surface signal. Scale bars, 5 µm (somatic images); 2 µm (dendritic images). (B) Time course of the fraction of spines in proximal (circles) and distal (triangles) dendrites that contain surface NL1 (gray) or GluA1 (black) following somatic ER release in the presence of TTX. A comparison of the fraction of NL1- and GluA1-positive spines at 90 min and 120 min is shown on the right; mean ± SEM; *, P < 0.05; **, P < 0.01 (unpaired t test; n = 5 neurons [NL1], n = 7 neurons [GluA1] from at least two independent experiments). (C) Time course of the fraction of spines in proximal (circles) and distal (triangles) dendrites that contain surface NL1 (gray) or GluA1 (black) following somatic ER release in the presence of Bic. A comparison of the fraction of NL1- and GluA1-positive spines at 90 min and 120 min is shown on the right; mean ± SEM; *, P < 0.05; ***, P < 0.001 (unpaired t test; n = 5 neurons [NL1], n = 8 neurons [GluA1] from at least two independent experiments). dist., distal; prox., proximal.
Contribution of lateral diffusion/recycling to distal localization of new surface proteins
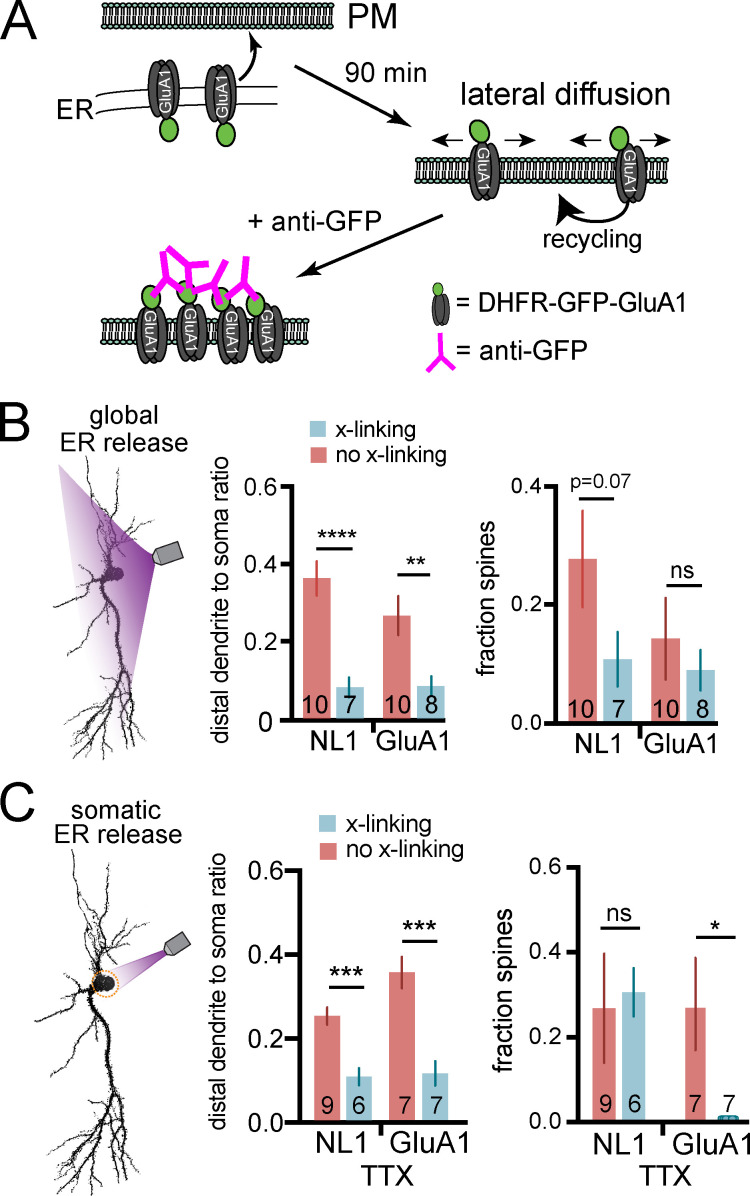
All experiments to this point were performed under antibody cross-linking conditions, where cargo was continuously immobilized and labeled as it appeared at the surface. Under these conditions, we observed modest amounts of surface NL1 and GluA1 in the most distal regions of dendrites, suggesting the secretory network may not be sufficient for direct delivery to remote sites (Figs. 3 and 6). To test whether subsequent trafficking via lateral diffusion significantly contributes to protein targeting to distal dendrites and spines, we repeated global and somatic ER release experiments under conditions where we allowed surface trafficking and lateral diffusion to occur for 90 min before we labeled and visualized the distribution of inserted proteins (Fig. 7 A). When we allowed lateral diffusion to occur following global ER release, we observed significantly more NL1 and GluA1 in distal dendrites compared with the same experiment performed under cross-linking conditions (Fig. 7 B). Thus, lateral diffusion/recycling plays a significant role in fulfilling protein demand at distal dendrites.
Figure 7.
Contribution of lateral diffusion/recycling to protein localization in distal dendrites and spines following global and somatic ER release. (A) Experimental schematic. Cargoes are released from the ER in the absence of cross-linking antibody and allowed to traffic for 90 min before adding Alexa647-anti-GFP to label surface protein. (B) Distribution of surface DHFR-GFP-NL1 and DHFR-GFP-GluA1 following global ER release under cross-linking (anti-GFP present throughout the experiment) and non–cross-linking (anti-GFP added 90 min post-ER release) conditions. The ratio of total surface signal in dendrites (>40 µm from the soma) versus soma is shown in the middle panel. The fraction of spines with each cargo is plotted to the right. n values correspond to the number of cells, from at least two independent experiments (**, P < 0.01; ****, P < 0.0001; Student's t test). (C) Same experiment as in B, except cargoes were locally released from the somatic ER. n values correspond to the number of cells, from at least two independent experiments (*, P < 0.05; ***, P < 0.001; Student's t test).
We also performed local somatic release experiments under non–cross-linking conditions to test how lateral diffusion/recycling refines protein localization of somatically derived proteins. We performed these experiments in the presence of TTX for comparison with our cross-linking experiments where we observed only modest trafficking to distal dendritic regions and negligible accumulation of GluA1 in spines (Fig. 6 and Fig. S5 B). In contrast, under non–cross-linking conditions, we observed significantly more surface signal for both cargoes at distal dendrites and significantly more spine accumulation of GluA1 (Fig. 7 C). This experiment demonstrates that the somatic secretory network can target proteins deep into the dendritic arbor, but subsequent lateral diffusion and recycling mechanisms play a key role in refining subcellular distribution and spine targeting, especially at remote sites.
Discussion
Allowing precise spatial, temporal, and gain control, zapERtrap adds new capabilities to previously developed “trap and release” strategies for controlling secretory trafficking (Boncompain et al., 2012; Chen et al., 2013; Hangen et al., 2018; Presley et al., 1997; Rivera et al., 2000; Toomre et al., 2000). We expect the approach will be easily adapted for local control of secreted substances or for regulating the abundance of diverse surface proteins with single-cell (and subcellular) resolution in different cell types. In addition to applications for precision control of cellular properties and signaling, zapERtrap will be valuable for investigating secretory trafficking pathways, especially in large and complex cells like neurons and glia, where long-range and decentralized secretory networks are poorly understood (Kennedy and Hanus, 2019).
Distinct trafficking networks for different cargoes originating in the same subcellular domain
Previous studies support distinct secretory trafficking pathways for diverse neuronal proteins (Bowen et al., 2017; Gu et al., 2016; Hanus et al., 2016; Jeyifous et al., 2009). For example, a significant fraction of AMPA receptors appears to use a nonconventional trafficking route that bypasses the GA, while NL1 appears to use a conventional, GA-dependent route (Bowen et al., 2017; Hanus et al., 2016). However, it remained unclear if the trafficking route is dictated by the identity of the cargo molecule or the subcellular location of entry into the secretory network. Our data indicate that molecular identity rather than subcellular location dictates the trafficking route since we observed major differences between NL1 and GluA1 even when both were targeted for ER release from the same subcellular domain. Differences include rates of GA accumulation, distinct effects of activity on the distribution and abundance of surface trafficking, and insertion at the AIS. What could account for these differences? Slower GluA1 trafficking to the GA could be explained by a fraction of GluA1 bypassing the GA through more dispersed ERGIC organelles and the relatively slow ER exit previously observed in imaging and biochemical studies (Bowen et al., 2017; Greger et al., 2002). We speculate that following transport to the GA and/or ERGIC, NL1 and GluA1 are packaged into distinct classes of transport vesicles that are differentially impacted by neural activity. For example, increased network activity mobilized NL1 deeper into the dendritic arbor, consistent with previous studies demonstrating activity dependence of kinesin-directed vesicle trafficking in dendrites and spines (Ghiretti et al., 2016; Hoerndli et al., 2015; McVicker et al., 2016). On the other hand, increased activity promoted more GluA1 surface insertion with less impact on spatial distribution, consistent with activity-dependent stalling and fusion of GluA1 transport vesicles (Hangen et al., 2018). This also agrees with previous studies demonstrating forward trafficking AMPA receptors localize to REs, whose fusion with the dendritic plasma membrane (PM) is activity dependent (Bowen et al., 2017; Hiester et al., 2017; Kennedy et al., 2010; Park et al., 2004).
The secretory network targets select proteins to the AIS
One of our most striking observations was rapid and selective NL1 trafficking to the surface of the AIS. While previous studies demonstrate that intracellular trafficking vesicles harboring somatodendritic proteins are prevented from axonal entry immediately proximal to or within the AIS, no study to our knowledge has demonstrated direct insertion into the PM of the AIS (Al-Bassam et al., 2012; Farías et al., 2015; Song et al., 2009). This has escaped previous detection because under normal conditions, proteins using this route must only be transiently presented at the AIS, with little steady-state accumulation. We could only detect NL1 at the AIS surface when we continuously cross-linked and trapped it as it appeared there. Since NL1 normally localizes to excitatory synapses in dendrites, why would it first traffic to the AIS? We speculate that fusion of NL1-containing transport vesicles at the AIS is an intermediate sorting checkpoint for separating comingled dendritic and axonal proteins that left the GA in the same transport vesicle. Future experiments will elucidate a more complete catalog of proteins trafficked to the AIS as well as their fate following AIS insertion.
The dendritic secretory network is not organized for precise local trafficking under basal conditions
While accumulating evidence supports local synthesis of diverse integral membrane proteins and secreted factors at remote dendritic sites, an untested assumption is that the dendritic secretory network is organized for their precise, local delivery to nearby sites on the cell surface. Often overlooked is how spatially precise surface targeting could be, given that proteins must first traffic through multiple organelle networks, some of which are highly mobile (Bowen et al., 2017; Horton and Ehlers, 2003; Mikhaylova et al., 2016). Indeed, we observed that proteins released from the ER within a defined dendritic segment often exited the targeted area in mobile transport vesicles and appeared at the cell surface over a much broader area than the targeted ER exit zone. It is possible that additional synaptic signaling mechanisms could modify secretory organelles or their associated motor proteins to limit their mobility and spatially constrain trafficking to nearby synapses. Future experiments using more sophisticated activity manipulations at the level of individual synapses will test whether local synaptic secretory cargoes can be precisely directed to synapses undergoing plasticity.
Materials and methods
Experimental model and subject details
All hippocampal neurons were derived from both male and female neonatal Sprague–Dawley rat pups. Timed pregnant dams (typically embryonic day 16) were obtained from Charles River Laboratories and housed under standard conditions (12-h light/dark cycle, food and water ad libitum) until the litter was born. All animal procedures were performed in accordance with a protocol approved by the University of Colorado Denver Institutional Animal Care and Use Committee.
Primary cell culture and transfection
Hippocampi were dissected from postnatal day 0–1 rats and dissociated with 20 U/ml papain (Worthington) in dissociation medium (0.14 M NaCl, 5.4 mM KCl, 0.17 mM Na2HPO4 · 7 H2O, 0.22 mM KH2PO3, 9.9 mM Hepes, 44 mM sucrose, 33 mM glucose, 1.5 mM CaCl2, 0.5 mM EDTA, 1 mM NaOH, and 0.2 mg/ml cysteine) for 1 h at room temperature and triturated in MEM (GIBCO). Cells were plated on poly-D-lysine–coated 18-mm glass coverslips at 25,000 cells/cm2 in MEM supplemented with 10% fetal bovine serum (HyClone), 50 U/ml penicillin, and 50 µg/ml streptomycin. After 1 d, the medium was replaced with Neurobasal-A (GIBCO) supplemented with B27 (Invitrogen) and GlutaMAX (GIBCO). Neurons were fed with Neurobasal-A containing B27, GlutaMAX, and mitotic inhibitors (uridine + 5-fluoro-2′deoxyridine) by replacing half of the medium on day 6 or day 7 and then weekly. Neurons were maintained at 37°C in a humidified incubator at 5% CO2. Neurons were transfected between 14 and 18 d in vitro with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations and allowed to express for 16–24 h in the presence of 0.5–1 µM zapalog.
Cell line maintenance and transfection
COS-7 cells were maintained and propagated in DMEM (GIBCO) supplemented with 10% fetal bovine serum (HyClone), 50 U/ml penicillin and 50 µg/ml streptomycin at 37°C with 5% CO2. COS-7 cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations and allowed to express for 24 h in the presence of 0.5–1 µM zapalog. COS-7 cell lines were obtained from American Type Culture Collection, expanded, and frozen. Parent cell lines were freshly thawed and validated by cellular morphology and growth characteristics and confirmed to be mycoplasma negative.
Methods details
Molecular cloning
Full sequences of constructs and oligonucleotides used in this study are available upon request. To generate DHFR-tagged cargo molecules, DHFR was inserted following the signal peptide at the N terminus of FP-NL1 and FP-GluA1 and at the C terminus of TfR-FP (where FP is EGFP, mCh, HaloTag, or mNeon) using standard restriction digest cloning or Gibson assembly. We included a thrombin cleavage site between the FP and the open reading frames of the cargo proteins so that any accumulated background surface signal could be eliminated. All constructs were verified by sequencing at the Barbara Davis Sequencing Core at the University of Colorado School of Medicine. The DHHC2-mCh plasmid was a gift from Dr. Mark Dell’Acqua (University of Colorado Anschutz Medical Campus, Aurora, CO); the mEmerald-GalT (aa 1–82 Golgi targeting sequence of human GalT) plasmid was a gift from Dr. Michael Davidson (Addgene plasmid #54108); the mCh-Sec61 plasmid was a gift from Dr. Jennifer Lippincott-Schwartz (Janelia Farm Research Campus, Ashburn, VA; Addgene plasmid #90994); the AnkyrinG-mCherry plasmid was a gift from Dr. Katharine R. Smith (University of Colorado Anschutz Medical Campus, Aurora, CO); the cDNAs for GluA1 and TfR were gifts from Dr. Michael Ehlers (Duke University, Durham, NC); the NL1 open reading frame was a gift from Dr. Peter Scheiffele (University of Basel, Basel, Switzerland; Addgene clone #15262); and the Rab11a reading frame was a gift from Dr. Richard Pagano’s laboratory (Addgene clone #12674).
Imaging
Live cell imaging was performed at 32°C on an Olympus IX71 equipped with a spinning-disk scan head (Yokogawa). Excitation illumination was delivered from an acousto-optic tunable filter controlled laser launch (Andor). Images were acquired using a 60× Plan Apochromat 1.4 NA objective and collected on a 1,024 × 1,024–pixel Andor iXon EMCCD camera using Metamorph (Molecular Devices) data acquisition software. For most experiments, a 4.8-µm z-stack (0.4-µm step size) was acquired at each time point. For global release experiments, full-field illumination was performed using a 10–50 ms pulse of light at 80% laser power (912 µW measured from the objective) using a 100-mW 405-nm laser. For local release experiments, we focally stimulated the preparation using galvanometric mirrors (FRAPPA; Andor) to steer a diffraction-limited 405-nm spot with a 500-µs dwell time. For all local release experiments (except for laser intensity titration experiments), zapalog photolysis was triggered with 23.0 µW/µm2 405-nm illumination (6% total laser power from a 100-mW fiber-coupled laser).
Live cell surface labeling and immunocytochemistry
Primary antibodies used in this study include Alexa Fluor 555–conjugated anti-GFP (Invitrogen; cat no. A31851; Research Resource Identifier [RRID]: 2536188; 4 µg/ml; 1:500 dilution) and Alexa Fluor 647–conjugated anti-GFP (Invitrogen; cat no. A31852; RRID: 162553; 2.67 µg/ml; 1:750 dilution). Goat anti-rabbit 568 secondary antibody (Invitrogen; A-11036; RRID: 10563566) was used at 1:750 dilution. For experiments visualizing intracellular GluA1 trafficking to the GA, we used mNeon as a fluorescent probe due to its increased brightness and slightly better pH resistance compared with EGFP. For experiments imaging intracellular vesicle formation and movement in dendrites, we used halo-tagged NL1 labeled with JaneliaFluor 646 HaloTag ligand due to its brightness, photostability, and resistance to the low pH in the lumen of trafficking vesicles. JaneliaFluor 646 HaloTag ligand was added to the cell culture media at a final concentration of 100 nM, 30 min before imaging. The JaneliaFluor 646 halotag dye was a gift from Dr. Luke Lavis at the Howard Hughes Medical Institute Janelia Farm Research Campus, Ashburn, VA.
Primary hippocampal neurons were transfected with DHFR-tagged cargoes and FKBP-XFP-KDEL (where XFP is mCh or GFP) using a standard Lipofectamine 2000 (Invitrogen)–based transfection protocol according to the manufacturer’s instructions. Zapalog compound (500 nM to 1 µM) was added immediately following transfection and present throughout the experiment. Synthesis of zapalog was performed by Medicilon Inc. with details described in Gutnick et al. (2019). Note that care must be taken to prevent exposure of the samples to room light to avoid photolysis of the zapalog compound. All cell culture manipulations were performed under a red safe light. Neurons were imaged in artificial cerebrospinal fluid (ACSF) containing 130 mM NaCl, 5 mM KCl, 10 mM Hepes, 30 mM glucose, 2 mM CaCl2, 1 mM MgCl2, and 0.5–1 µM zapalog, pH 7.4. Prior to baseline image acquisition, Alexa647-anti-GFP (Invitrogen; 2.67 µg/ml; 1:750 dilution) was added to the imaging ACSF for subsequent real-time detection of surface presentation of released cargoes. We eliminated minor background signal arising from cargo that escaped the ER by treatment with thrombin (Sigma-Aldrich; cat no. T6884) at 1 U/ml 10–30 min before imaging. ER retention efficiency was quantified by comparing surface signal measured after a 10-min exposure to Alexa647-anti-GFP (Invitrogen/Molecular Probes, cat no. A31852; RRID: AB_162553) in hippocampal neurons maintained in the dark expressing either DHFR-GFP-NL1 or DHFR-GFP-GluA1 (along with FKBP-mCh-KDEL) in the presence and absence of zapalog compound. NL1 and GluA1 are retained at 85 ± 7% and 80 ± 6% efficiency, respectively. These efficiencies likely underestimate the efficiency of retention for cells used in this study since we confirmed the absence of background surface signal arising from recycling or surface receptors that may have escaped the ER. This was done by incubating the cells with labeled anti-GFP antibody for a prolonged baseline period (∼20 min) in the presence of extracellular Alexa647-anti-GFP before ER release. Cells displaying detectable surface signal before 405-nm light exposure were excluded from analysis. Following 405-nm light exposure, the kinetics of surface presentation for DHFR-GFP-NL1 and DHFR-GFP-GluA1 (measured by accumulated Alexa647-anti-GFP signal) were well fit by a single exponential function with τ values = 4.1 ± 0.3 h (GluA1) and 2.1 ± 0.2 h (NL1; n = 6 neurons for GluA1 and 5 neurons for NL1 from two independent cultures), similar to previous values using a different ER retention/release approach (Bowen et al., 2017). For activity dependence experiments, TTX (2 µM final concentration) was added to live cells 30 min before imaging and included in the imaging ACSF. Bic (30 µM final concentration) was added to imaging ACSF immediately before ER release. For all experiments, we confined our analysis to neurons with a pyramidal-shaped cell body, large apical dendrite, and presence of dendritic spines.
For live cell surface insertion experiments using cross-linking anti-GFP antibodies, note that the appearance of surface-bound anti-GFP signal does not represent the precise site of secretory cargo membrane insertion since antibody binding/cross-linking takes tens of seconds, during which time receptors can diffuse from the insertion site. Receptors inserted into the cell membrane become antibody X-linked after a period Δt that is described by an exponential function with τ = 100 s (Fig. S2 B). During this time, the particles are free to diffuse throughout the membrane according to a 2D isotropic diffusion model with D = 0.06 µm2/s (empirically determined value for extrasynaptic AMPA receptors; see Borgdorff and Choquet, 2002). We modeled this process by convolving the exponential distribution (to generate immobilization times Δt), with a Gaussian distribution (to describe the surface diffusion). Each dimension was simulated independently, and the Gaussian distribution was described by σ2 = 2D Δt. We used a stochastic simulation to numerically estimate the probability density function for receptor displacement in two dimensions. This simulation is shown in the graph in Fig. S2 B, which plots localization probability versus distance from the origin (insertion site). The Rayleigh distribution, with a σ2 = 2D Δt, describes the absolute distance a protein will immobilize relative to its insertion location on a 2D surface given that the protein diffuses for a time Δt. We used a similar stochastic simulation to estimate the convolved effect of exponentially distributed immobilization times (Δt) with this Rayleigh distribution. This model estimates 50% of the receptors will be immobilized within 3.8 µm of the insertion site. Thus, while our approach does not pinpoint the exact location of membrane delivery, it reports the cellular domain of insertion within several micrometers.
Quantification and statistical analysis
All quantification was performed on raw fluorescent images using MATLAB or ImageJ to measure pixel intensities. Background intensity values were estimated either in ImageJ by measuring pixel intensities in image regions with no detectable signal or in MATLAB by interpolating the background intensity within the cell based on the background intensity values outside the cell. Images were expanded in ImageJ for display only.
GraphPad Prism 8 was used for performing statistical analyses and plotting data. Heatmaps were generated using MATLAB. For all statistical tests, a P value of <0.05 was considered significant. Asterisks denote the following significance levels: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Quantification of surface accumulation
Quantification of surface accumulation was performed using custom analysis software written in MATLAB with the freely available MATLAB Toolbox DipImage (Delft University). For each neuron, two masks were generated. The first mask was used to identify the entire cell and used the fluorescent signal of the nonspecific cell-fill marker. The second mask identified locations of protein surface accumulation after ER release and was generated using the fluorescent signal of the Alexa647-labeled anti-GFP antibody. For mask generation, images were first filtered using the Laplacian of the Gaussian image transform (implemented by taking only the positive terms from the "dxx"+ "dyy" operation in DipImage) followed by a user-selected threshold, identified by selecting the maximum intensity within a background region. To quantify the distance of each labeled object along the cell from the cell soma, two approaches were used. Both approaches required a user-selected soma region that was drawn as a polygon over the entire somatic body. The first approach quantifies the intensity density of cell surface labeling as a function of distance from the cell soma. A geometric distance transform was applied using the "bwdistgeodesic" function in MATLAB, with the whole cell mask and identified soma polygon as input. This distance transform was then used to generate a series of masks at variable distances, whose intensity density was used to calculate intensity as a function of distance. Due to nonuniformity of the sample illumination, a custom background was calculated by interpolating the background signal within the cell using the background signal from all regions outside of the cell. Using the MATLAB function "regionfill," we interpolated a background within the region of the whole cell mask, using background signal. The second approach quantifies the number of aggregated surface receptor puncta as a function of time and distance from the cell soma. To uniquely identify individual puncta, each spatially isolated puncta within the mask was labeled using the DipImage "label" function. The distance between each label and the cell soma was calculated by identifying the minimum value after subtracting the geometric distance transform of the soma mask and the geometric distance transform of the individual labeled puncta (found using the MATLAB function "bwdistgeodesic" as described above), the minimum value representing the minimum path length from the puncta to the soma, along the whole cell mask. The fraction of cargo-positive spines was calculated within 10-µm regions inside the ER release zone versus a region of the same size at a comparable distance from the soma and made from a separate dendrite with similar spine density/morphology.
Online supplemental material
Fig. S1 shows that the DHFR tag does not perturb subcellular localization. Fig. S2 shows localization of DHFR-fused synaptic proteins and validation of antibody cross-linking for immobilizing surface proteins. Fig. S3 shows control experiments for surface NL1 accumulation at the AIS. Fig. S4 shows that zapERtrap can be used to locally release cargoes from the ER within defined regions of the dendritic arbor. Fig. S5 shows subcellular distribution of NL1 and GluA1 surface accumulation following somatic ER release. Video 1 shows spatially restricted ER release of TfR-GFP-DHFR from an individual COS-7 cell. Video 2 shows effective retention and light-triggered release from the ER using zapERtrap in a hippocampal neuron. Video 3 shows kinetics of somatic GA accumulation for different cargoes. Video 4 shows surface accumulation of cargoes following whole-cell illumination in neurons. Video 5 shows segmented surface signal displaying where and when NL1 and GluA1 appear on the cell surface following global release. Video 6 shows that NL1 is inserted into the plasma membrane of the AIS. Video 7 shows that following local dendritic ER release, cargo can be transported out of the release zone.
Acknowledgments
We thank Mark Dell’Acqua and Chandra Tucker for critical discussions.
A.M. Bourke is supported by the National Science Foundation (DGE-1553798) and the Howard Hughes Medical Institute Gilliam Fellowship. M.J. Kennedy is supported by grants from the National Institute of Neurological Disorders and Stroke (R01NS082271, R01NS10755, R35NS116879, and UF1NS107710). This work was also supported by National Institute of General Medical Sciences grant R01GM069808 awarded to T.S. Schwarz.
The authors declare no competing financial interests.
Author contributions: Conceptualization: M.J. Kennedy; Methodology: M.J. Kennedy, A.M. Bourke, A.B. Bowen, T.L. Schwarz, and A. Gutnick; Software: S.L. Schwartz and A.M. Bourke; Formal Analysis: A.M. Bourke; Investigation: M.J. Kennedy, A.M. Bourke, C.S. Winborn, M.S. Kleinjan, D.J. Kareemo, and A.B. Bowen; Writing – Original Draft: M.J. Kennedy and A.M. Bourke; Funding: M.J. Kennedy and A.M. Bourke; Resources: T.L. Schwarz and A. Gutnick; Supervision: M.J. Kennedy.
Data and software availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthew J. Kennedy (matthew.kennedy@cuanschutz.edu). Plasmids generated in this study will be made available through Addgene. The code generated during this study is available at https://github.com/mjkennedylab/zapERtrap.
References
- Al-Bassam, S., Xu M., Wandless T.J., and Arnold D.B.. 2012. Differential trafficking of transport vesicles contributes to the localization of dendritic proteins. Cell Rep. 2:89–100. 10.1016/j.celrep.2012.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor, M., Guzik A.K., Bielli A., and Fish K.N.. 2004. Endoplasmic reticulum export site formation and function in dendrites. J. Neurosci. 24:3770–3776. 10.1523/JNEUROSCI.4775-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe, C.K., and Miller E.A.. 2013. Secretory protein biogenesis and traffic in the early secretory pathway. Genetics. 193:383–410. 10.1534/genetics.112.142810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain, G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., and Perez F.. 2012. Synchronization of secretory protein traffic in populations of cells. Nat. Methods. 9:493–498. 10.1038/nmeth.1928 [DOI] [PubMed] [Google Scholar]
- Borgdorff, A.J., and Choquet D.. 2002. Regulation of AMPA receptor lateral movements. Nature. 417:649–653. 10.1038/nature00780 [DOI] [PubMed] [Google Scholar]
- Bourke, A.M., Bowen A.B., and Kennedy M.J.. 2018. New approaches for solving old problems in neuronal protein trafficking. Mol. Cell. Neurosci. 91:48–66. 10.1016/j.mcn.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen, A.B., Bourke A.M., Hiester B.G., Hanus C., and Kennedy M.J.. 2017. Golgi-independent secretory trafficking through recycling endosomes in neuronal dendrites and spines. eLife. 6:e27362. 10.7554/eLife.27362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas, I.J., Tushev G., Will T.J., tom Dieck S., Fuerst N., and Schuman E.M.. 2012. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 74:453–466. 10.1016/j.neuron.2012.02.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Gibson E.S., and Kennedy M.J.. 2013. A light-triggered protein secretion system. J. Cell Biol. 201:631–640. 10.1083/jcb.201210119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui-Wang, T., Hanus C., Cui T., Helton T., Bourne J., Watson D., Harris K.M., and Ehlers M.D.. 2012. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 148:309–321. 10.1016/j.cell.2011.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farías, G.G., Guardia C.M., Britt D.J., Guo X., and Bonifacino J.S.. 2015. Sorting of Dendritic and Axonal Vesicles at the Pre-axonal Exclusion Zone. Cell Rep. 13:1221–1232. 10.1016/j.celrep.2015.09.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiretti, A.E., Thies E., Tokito M.K., Lin T., Ostap E.M., Kneussel M., and Holzbaur E.L.F.. 2016. Activity-Dependent Regulation of Distinct Transport and Cytoskeletal Remodeling Functions of the Dendritic Kinesin KIF21B. Neuron. 92:857–872. 10.1016/j.neuron.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger, I.H., Khatri L., and Ziff E.B.. 2002. RNA editing at arg607 controls AMPA receptor exit from the endoplasmic reticulum. Neuron. 34:759–772. 10.1016/S0896-6273(02)00693-1 [DOI] [PubMed] [Google Scholar]
- Gu, Y., Chiu S.-L., Liu B., Wu P.-H., Delannoy M., Lin D.-T., Wirtz D., and Huganir R.L.. 2016. Differential vesicular sorting of AMPA and GABAA receptors. Proc. Natl. Acad. Sci. USA. 113:E922–E931. 10.1073/pnas.1525726113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutnick, A., Banghart M.R., West E.R., and Schwarz T.L.. 2019. The light-sensitive dimerizer zapalog reveals distinct modes of immobilization for axonal mitochondria. Nat. Cell Biol. 21:768–777. 10.1038/s41556-019-0317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangen, E., Cordelières F.P., Petersen J.D., Choquet D., and Coussen F.. 2018. Neuronal Activity and Intracellular Calcium Levels Regulate Intracellular Transport of Newly Synthesized AMPAR. Cell Rep. 24:1001–1012.e3. 10.1016/j.celrep.2018.06.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanus, C., Kochen L., Tom Dieck S., Racine V., Sibarita J.-B., Schuman E.M., and Ehlers M.D.. 2014. Synaptic control of secretory trafficking in dendrites. Cell Rep. 7:1771–1778. 10.1016/j.celrep.2014.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanus, C., Geptin H., Tushev G., Garg S., Alvarez-Castelao B., Sambandan S., Kochen L., Hafner A.-S., Langer J.D., and Schuman E.M.. 2016. Unconventional secretory processing diversifies neuronal ion channel properties. eLife. 5:e20609. 10.7554/eLife.20609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine, M., Groc L., Frischknecht R., Béïque J.C., Lounis B., Rumbaugh G., Huganir R.L., Cognet L., and Choquet D.. 2008. Surface mobility of postsynaptic AMPARs tunes synaptic transmission. Science. 320:201–205. 10.1126/science.1152089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiester, B.G., Bourke A.M., Sinnen B.L., Cook S.G., Gibson E.S., Smith K.R., and Kennedy M.J.. 2017. L-Type Voltage-Gated Ca2+ Channels Regulate Synaptic-Activity-Triggered Recycling Endosome Fusion in Neuronal Dendrites. Cell Rep. 21:2134–2146. 10.1016/j.celrep.2017.10.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerndli, F.J., Wang R., Mellem J.E., Kallarackal A., Brockie P.J., Thacker C., Madsen D.M., and Maricq A.V.. 2015. Neuronal Activity and CaMKII Regulate Kinesin-Mediated Transport of Synaptic AMPARs. Neuron. 86:457–474. 10.1016/j.neuron.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, A.C., and Ehlers M.D.. 2003. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J. Neurosci. 23:6188–6199. 10.1523/JNEUROSCI.23-15-06188.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, A.C., Rácz B., Monson E.E., Lin A.L., Weinberg R.J., and Ehlers M.D.. 2005. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 48:757–771. 10.1016/j.neuron.2005.11.005 [DOI] [PubMed] [Google Scholar]
- Jeyifous, O., Waites C.L., Specht C.G., Fujisawa S., Schubert M., Lin E.I., Marshall J., Aoki C., de Silva T., Montgomery J.M., et al. 2009. SAP97 and CASK mediate sorting of NMDA receptors through a previously unknown secretory pathway. Nat. Neurosci. 12:1011–1019. 10.1038/nn.2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein, L.C., Schlager M.A., Kuijpers M., Wulf P.S., van Spronsen M., MacKintosh F.C., and Hoogenraad C.C.. 2010. Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr. Biol. 20:290–299. 10.1016/j.cub.2009.12.052 [DOI] [PubMed] [Google Scholar]
- Kennedy, M.J., and Hanus C.. 2019. Architecture and Dynamics of the Neuronal Secretory Network. Annu. Rev. Cell Dev. Biol. 35:543–566. 10.1146/annurev-cellbio-100818-125418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, M.J., Davison I.G., Robinson C.G., and Ehlers M.D.. 2010. Syntaxin-4 defines a domain for activity-dependent exocytosis in dendritic spines. Cell. 141:524–535. 10.1016/j.cell.2010.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.C.S., Miller E.A., Goldberg J., Orci L., and Schekman R.. 2004. Bi-directional protein transport between the ER and Golgi. Annu. Rev. Cell Dev. Biol. 20:87–123. 10.1146/annurev.cellbio.20.010403.105307 [DOI] [PubMed] [Google Scholar]
- Lipka, J., Kapitein L.C., Jaworski J., and Hoogenraad C.C.. 2016. Microtubule-binding protein doublecortin-like kinase 1 (DCLK1) guides kinesin-3-mediated cargo transport to dendrites. EMBO J. 35:302–318. 10.15252/embj.201592929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Roberts T.H., and Hirschberg K.. 2000. Secretory protein trafficking and organelle dynamics in living cells. Annu. Rev. Cell Dev. Biol. 16:557–589. 10.1146/annurev.cellbio.16.1.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVicker, D.P., Awe A.M., Richters K.E., Wilson R.L., Cowdrey D.A., Hu X., Chapman E.R., and Dent E.W.. 2016. Transport of a kinesin-cargo pair along microtubules into dendritic spines undergoing synaptic plasticity. Nat. Commun. 7:12741. 10.1038/ncomms12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova, M., Bera S., Kobler O., Frischknecht R., and Kreutz M.R.. 2016. A Dendritic Golgi Satellite between ERGIC and Retromer. Cell Rep. 14:189–199. 10.1016/j.celrep.2015.12.024 [DOI] [PubMed] [Google Scholar]
- Park, M., Penick E.C., Edwards J.G., Kauer J.A., and Ehlers M.D.. 2004. Recycling endosomes supply AMPA receptors for LTP. Science. 305:1972–1975. 10.1126/science.1102026 [DOI] [PubMed] [Google Scholar]
- Penn, A.C., Zhang C.L., Georges F., Royer L., Breillat C., Hosy E., Petersen J.D., Humeau Y., and Choquet D.. 2017. Hippocampal LTP and contextual learning require surface diffusion of AMPA receptors. Nature. 549:384–388. 10.1038/nature23658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley, J.F., Cole N.B., Schroer T.A., Hirschberg K., Zaal K.J., and Lippincott-Schwartz J.. 1997. ER-to-Golgi transport visualized in living cells. Nature. 389:81–85. 10.1038/38001 [DOI] [PubMed] [Google Scholar]
- Ramírez, O.A., and Couve A.. 2011. The endoplasmic reticulum and protein trafficking in dendrites and axons. Trends Cell Biol. 21:219–227. 10.1016/j.tcb.2010.12.003 [DOI] [PubMed] [Google Scholar]
- Rivera, V.M., Wang X., Wardwell S., Courage N.L., Volchuk A., Keenan T., Holt D.A., Gilman M., Orci L., Cerasoli F. Jr., et al. 2000. Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science. 287:826–830. 10.1126/science.287.5454.826 [DOI] [PubMed] [Google Scholar]
- Song, A.-H., Wang D., Chen G., Li Y., Luo J., Duan S., and Poo M.-M.. 2009. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 136:1148–1160. 10.1016/j.cell.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Sutton, M.A., Ito H.T., Cressy P., Kempf C., Woo J.C., and Schuman E.M.. 2006. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 125:785–799. 10.1016/j.cell.2006.03.040 [DOI] [PubMed] [Google Scholar]
- Toomre, D., Steyer J.A., Keller P., Almers W., and Simons K.. 2000. Fusion of constitutive membrane traffic with the cell surface observed by evanescent wave microscopy. J. Cell Biol. 149:33–40. 10.1083/jcb.149.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre, E.R., and Steward O.. 1996. Protein synthesis within dendrites: glycosylation of newly synthesized proteins in dendrites of hippocampal neurons in culture. J. Neurosci. 16:5967–5978. 10.1523/JNEUROSCI.16-19-05967.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, A.H., O’Donnell C., Sejnowski T.J., and O’Leary T.. 2016. Dendritic trafficking faces physiologically critical speed-precision tradeoffs. eLife. 5:e20556. 10.7554/eLife.20556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., Whiteus C., Xu C.S., Hayworth K.J., Weinberg R.J., Hess H.F., and De Camilli P.. 2017. Contacts between the endoplasmic reticulum and other membranes in neurons. Proc. Natl. Acad. Sci. USA. 114:E4859–E4867. 10.1073/pnas.1701078114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthew J. Kennedy (matthew.kennedy@cuanschutz.edu). Plasmids generated in this study will be made available through Addgene. The code generated during this study is available at https://github.com/mjkennedylab/zapERtrap.