Kumar et al. identify a multiprotein complex called DISCO (distal centriole complex) required to nucleate distal appendages and restrain centriole elongation, which are essential for the initiation of cilium assembly. Without DISCO, cells fail to ciliate and transduce Hedgehog signals, which are critical for mammalian development.
Abstract
Cells inherit two centrioles, the older of which is uniquely capable of generating a cilium. Using proteomics and superresolved imaging, we identify a module that we term DISCO (distal centriole complex). The DISCO components CEP90, MNR, and OFD1 underlie human ciliopathies. This complex localizes to both distal centrioles and centriolar satellites, proteinaceous granules surrounding centrioles. Cells and mice lacking CEP90 or MNR do not generate cilia, fail to assemble distal appendages, and do not transduce Hedgehog signals. Disrupting the satellite pools does not affect distal appendage assembly, indicating that it is the centriolar populations of MNR and CEP90 that are critical for ciliogenesis. CEP90 recruits the most proximal known distal appendage component, CEP83, to root distal appendage formation, an early step in ciliogenesis. In addition, MNR, but not CEP90, restricts centriolar length by recruiting OFD1. We conclude that DISCO acts at the distal centriole to support ciliogenesis by restraining centriole length and assembling distal appendages, defects in which cause human ciliopathies.
Introduction
Centrioles are ancient, microtubule-based structures with two main functions: (1) they are core components of the centrosome, the primary microtubule organizing center; and (2) they are the foundations for cilia, cellular antennae specialized for signaling. While the main barrel of each centriole is composed of nine tradially arranged microtubule triplets, many proteins apart from tubulin comprise the centrioles (Winey and O’Toole, 2014; Keller et al., 2005, 2009; Jakobsen et al., 2011; Andersen et al., 2003). To date, the parts list of centrioles is incomplete.
Centriole structure can vary between species, but within a particular cell type, centriole length is nearly uniform, as are centriolar number and timing of assembly (Goehring and Hyman, 2012; Kong et al., 2020). Coordination with the cell cycle entrains this uniformity; centrioles duplicate during interphase, and at mitosis, each daughter cell inherits an older (mother) centriole and a younger (daughter) centriole (Nigg and Holland, 2018; Breslow and Holland, 2019).
The differences in centriolar age dictate different structures and functions. Only the mother centriole can assemble a cilium, and only it possesses subdistal and distal appendages. Through the distal appendages, the mother centriole attaches to preciliary vesicles, an early step in ciliogenesis (Schmidt et al., 2012; Sillibourne et al., 2013; Tanos et al., 2013). In contrast, subdistal appendages are dispensable for ciliogenesis (Tanos et al., 2013; Mazo et al., 2016; Chong et al., 2020). Although components of appendages have been identified (Bowler et al., 2019; Yang et al., 2018; Chong et al., 2020), how they are assembled remains enigmatic.
Related to their roles in fundamental cellular processes such as cell division and intercellular communication, centriole dysfunction causes diverse human developmental disorders, including ciliopathies and microcephaly (Nigg and Holland, 2018; Reiter and Leroux, 2017). For example, mutations in CEP90 (centrosomal protein of 90 kD; also known as PIBF1), MNR (moonraker; also known as KIAA0753 or OFIP), or OFD1 cause Joubert syndrome (JBTS), a ciliopathy characterized by brainstem and cerebellar malformations (Wheway et al., 2015; Shen et al., 2020; Hebbar et al., 2018; Stephen et al., 2017; Hammarsjö et al., 2017; Coene et al., 2009). Similarly, in many cancers, the number or structure of centrioles is dysregulated (Gönczy, 2015; Marteil et al., 2018).
CEP90 has been identified as a component of centriolar satellites, protein assemblies that orbit the centrosome (Kim et al., 2012; Kodani et al., 2015). Knockdown of CEP90 alters the localization of centriolar satellites, which has been proposed to compromise ciliogenesis (Kim and Rhee, 2011; Kim et al., 2012).
Using a combination of label-retention expansion microscopy and structured illumination microscopy (LR-ExSIM; Shi et al., 2019), we found that CEP90, in addition to being a component of centriolar satellites, is a component of the distal end of centrioles. Proteomic analyses revealed that CEP90 is part of an evolutionarily conserved complex, which we term DISCO (distal centriole complex), that includes MNR and OFD1. Mouse genetic studies showed that CEP90 and MNR are key regulators of mother centriole function, essential for normal vertebrate development, Hedgehog signaling, and ciliogenesis. In investigating how CEP90 and MNR function in ciliogenesis, we found that both are required for the assembly of distal appendages. Mutant analysis demonstrated that the components of DISCO are recruited to the centriole in a hierarchical manner, culminating in CEP90, which recruits the most proximal distal appendage protein, CEP83, to initiate distal appendage formation. Thus, our work identifies an evolutionarily conserved complex that functions at the distal mother centriole to build distal appendages, an early and critical step in ciliogenesis.
Results
CEP90 localizes to centriolar satellites and a ring at the distal ends of centrioles
CEP90 has previously been identified as a component of centriolar satellites (Kim and Rhee, 2011). We confirmed that CEP90 localizes to centriolar satellites by immunostaining human retinal pigment epithelial (RPE1) cells with antibodies to CEP90 and PCM1, a marker of centriolar satellites (Fig. 1 a). In addition, a centrosomal pool of CEP90 did not colocalize with PCM1 (Fig. 1 a). To more precisely identify where centrosomal CEP90 localized, we dispersed centriolar satellites with nocodazole and found that CEP90 localized with γ-tubulin at centrioles (Fig. 1 b). 3D structured illumination microscopy (3D-SIM) revealed that CEP90 forms rings at centrioles (Fig. 1 b).
Figure 1.
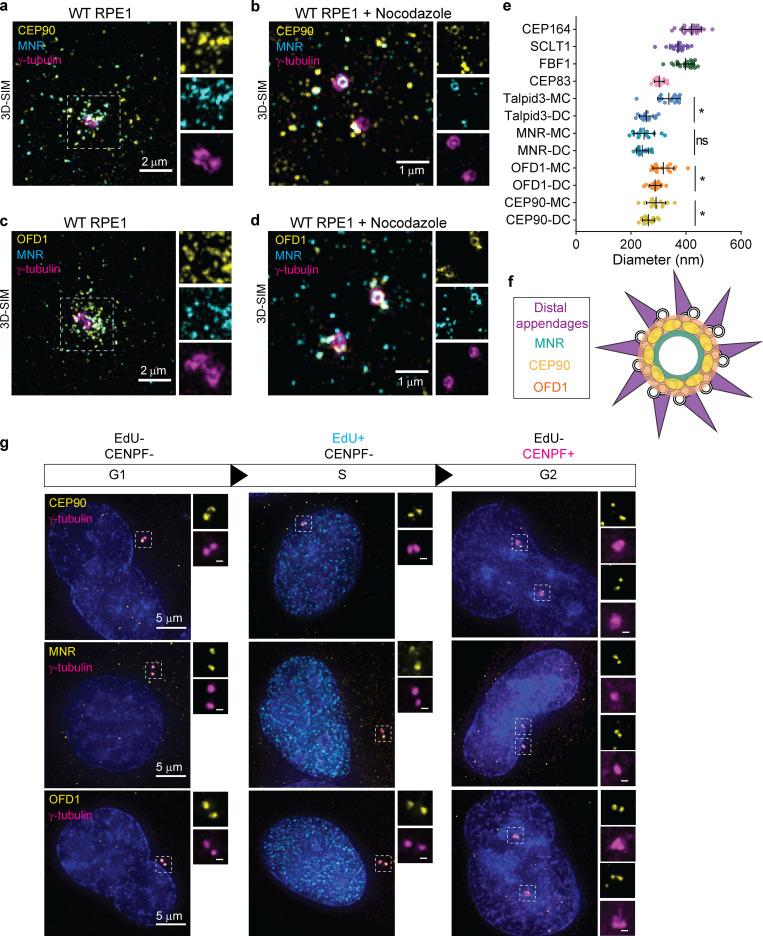
CEP90 localizes to centriolar satellites and the distal centriole. (a) 3D-SIM of immunostained RPE1 cells reveals localization of CEP90 (yellow) at centriolar satellites (PCM1, cyan) and centrioles (TubulinAc, magenta) in WT, cycling RPE1 cells. Scale bar = 1 µm. (b) Treatment of serum-starved RPE1 cells with nocodazole disperses the centriolar satellites, highlighting 3D-SIM of CEP90 (yellow) rings at centrioles (γ-tubulin, magenta). Distal appendage component CEP164 (cyan) indicates the mother centriole in c and d. Scale bar = 1 µm. (c) 3D-SIM of serum-starved PCM1−/− RPE1 cells shows that CEP90 (yellow) localizes to centrioles (γ-tubulin, magenta) independent of centriolar satellites. Scale bar of main panel and insets = 1 µm and 0.5 µm respectively. (d) 3D-SIM confirms localization of CEP90 (yellow) at centrioles (γ-tubulin, magenta) in nocodazole-treated, serum-starved eYFP-CEP90–expressing PCM1−/− RPE1 cells. Scale bar = 1 µm. (e) Quantification of eYFP-CEP90, Centrobin, and CEP164 fluorescence intensity at daughter (DC) and mother (MC) centrioles from 3D-SIM images, n = 10–20 cells. Horizontal lines indicate means ± SEM. ***, P < 0.0005, unpaired t test. AU, arbitrary units. (f) LR-ExSIM of RPE1 cells immunostained for CEP90 (yellow) and CEP164 (magenta) reveals that rings of CEP90 are composed of discrete puncta. CEP90 rings are smaller and more proximal to CEP164 rings. Scale bar = 1 µm. (g) Example of an LR-ExSIM image of a radially oriented centriole used to quantify the number and angle between adjacent puncta of CEP90. Scale bar = 0.5 µm. (h) Histogram of number of discrete puncta of CEP90 and CEP164 observed per centriole in LR-ExSIM images. n = 12–17 measurements. (i) Histogram of the angular spacing between adjoining centriolar CEP90 puncta observed by LR-ExSIM. n = 66 measurements. (j) Schematic of the ring of CEP90 punctae (yellow), distal appendages (magenta), and subdistal appendages (blue) at the distal centriole. CEP90 decorates the distal end of mother and daughter centrioles.
Independent of nocodazole, we disrupted centriolar satellites in RPE1 cells by deleting PCM1, a scaffold for centriolar satellites, using CRISPR-Cas9 (Fig. S1). As described previously, centriolar satellites were not detected in PCM1−/− RPE1 cells (Odabasi et al., 2019). Consistent with the loss of satellites, localization of CEP90 to puncta around the centrosome was absent in PCM1−/− RPE1 cells (Fig. 1 c). As in nocodazole-treated cells, the rings of CEP90 at centrioles remained in PCM1−/− RPE1 cells (Fig. 1 c), indicating that CEP90 localizes to centrioles independently of PCM1 and centriolar satellites. To confirm antibody staining specificity, we immunostained exogenously expressed eYFP-tagged CEP90 in nocodazole-treated PCM1−/− RPE1 cells (Fig. 1 d). Localization of eYFP-CEP90 resembled endogenous CEP90 staining and suggested that CEP90, unlike Centrobin and CEP164, localized to both mother and daughter centrioles with similar fluorescence intensity (Fig. 1 e).
Figure S1.
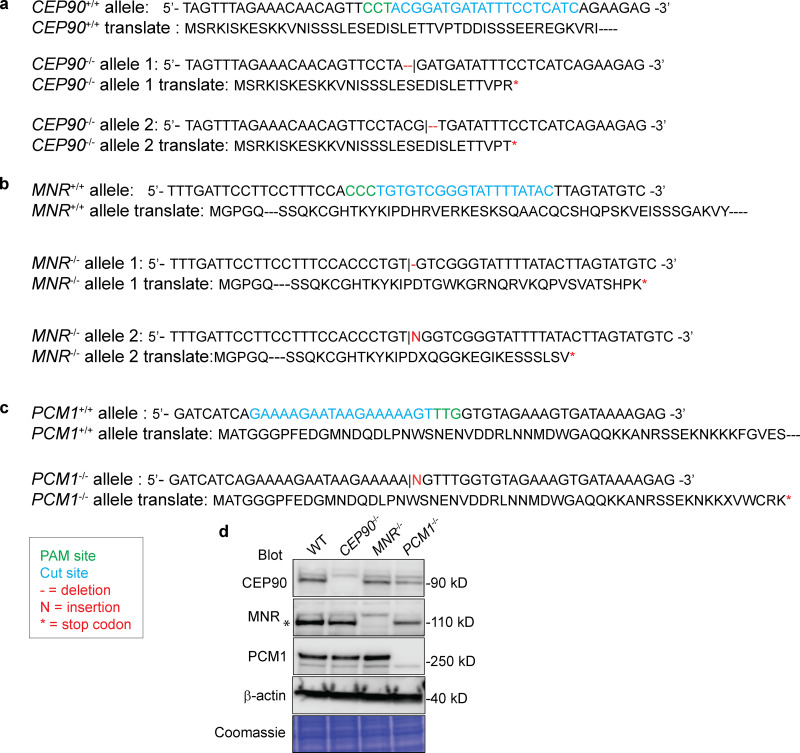
Generation of CEP90−/−, MNR−/−, and PCM1−/− cell lines.(a–c) Sequence analysis of genomic DNA isolated from control and CEP90−/− (a), MNR−/− (b), and PCM1−/− (c) RPE1 cell lines generated using CRISPR-Cas9. Insertions and deletions, and translation products resulting from genome editing are indicated. The protospacer adjacent motif (PAM) is indicated in green. (d) Immunoblot analysis of whole-cell lysates from control, CEP90−/−, MNR−/−, and PCM1−/− RPE1 cell lines confirms loss of protein in mutant cell lines. The specific MNR band is indicated with an asterisk, and the top band is nonspecific.
Expansion microscopy involves physically expanding samples embedded in a hydrogel (Wassie et al., 2019). To precisely map the localization of CEP90 at centrioles, we combined expansion microscopy together with multicolor LR-ExSIM to minimize signal loss during expansion and provide ∼30-nm lateral resolution (Shi et al., 2019). The distal appendage component CEP164 imaged by LR-ExSIM was comparable to data previously obtained using stochastic optical reconstruction microscopy (STORM); CEP164 formed a discontinuous ring at the mother centriole (Shi et al., 2019, 2017). Intriguingly, LR-ExSIM resolved that the ring of CEP90 is composed of discrete puncta with a mode of nine and separated by a mean angle of ∼36 nm (Fig. 1, f–i). In agreement with 3D-SIM microscopy, the CEP90 ring at the mother centriole as observed by LR-ExSIM had a smaller diameter (228 ± 23 nm) and was proximal to the distal appendages. Thus, CEP90 comprises a ninefold ring at the distal centriole (Fig. 1 j).
CEP90, OFD1, and MNR form DISCO
To gain insight into the function of CEP90 at the distal centriole, we proteomically identified CEP90 interactors. More specifically, we generated RPE1 cell lines stably expressing GFP (control) or YFP-CEP90. To discriminate proteins interacting with CEP90 at centriolar satellites and those interacting with CEP90 at centrioles, we also generated PCM1−/− RPE1 cell lines stably expressing GFP or YFP-CEP90 in which centriolar satellites are disrupted (Fig. 2 a). We immunoprecipitated GFP from these cell lines and detected coimmunoprecipitating proteins by liquid chromatography tandem mass spectrometry. Significance analysis of interactome (SAINTexpress; Teo et al., 2014) analysis with Bayesian false discovery rate < 0.05 indicated high-confidence interactors (Fig. 2, b and c). Some CEP90 interactors, such as PCM1, CEP131, and BBS4, were identified in WT cells, but not in PCM1−/− cells, suggesting that these interactions are dependent on centriolar satellites. Consistent with this conclusion, many of these PCM1-dependent interactors, including CEP131 and BBS4, are components of centriolar satellites (Hori and Toda, 2017).
Figure 2.
CEP90 forms a complex with OFD1 and MNR. (a) Schematic depicting workflow used to identify CEP90 interactors at centriolar satellites and centrioles using WT and PCM1−/− RPE1 cells. PCM1−/− cells localize CEP90 to the distal centriole, and WT cells localize CEP90 to the distal centriole and centriolar satellites. (b) Venn diagram comparing high-confidence interactors of CEP90 in WT and PCM1−/− RPE1 cells. (c) Interactome dot representation of selected CEP90 interactors identified in the proteomic screen. Hits were grouped based on their cellular localization. Average number of peptide spectra (AvgSpec) is represented by dot shade. Abundance of a peptide spectrum produced in relation to the most abundant spectrum is depicted by dot size. Bayesian false discovery rate (BFDR) is represented by rim color. (d) Immunoblot of a subset of CEP90 interactions identified by proteomics were validated by coimmunoprecipitation (IP). CEP90 interacts with PCM1, OFD1 and MNR, but not α-tubulin or GAPDH. FT, flow through. Specific MNR band is indicated with an asterisk. The top band is nonspecific, as it is undiminished in the MNR-knockout cell lysates. (e) Coulson plot showing the phylogenetic distribution of a subset of centriolar proteins in select ciliated metazoan species. Orthologues identified with high confidence are indicated with a filled circle, and a subset of CEP90 interactors further explored in this study are highlighted in blue. The dendrogram on top (made using interactive tree of life; Ciccarelli et al., 2006) shows the evolutionary relationship between species.
To identify centriolar interactors of CEP90, we assessed which interactors were detected in both WT and PCM1−/− RPE1 cells, as these interactions are predicted to be independent of centriolar satellites. Notably, OFD1 and MNR, previously described components of centriolar satellites and centrosomes that interact with each other (Chevrier et al., 2016), were detected as CEP90 interactors (Fig. 2, b and c). Using coimmunoprecipitation, we confirmed that CEP90 interacts with PCM1, MNR, and OFD1 (Fig. 2 d). Phylogenetic analysis revealed that CEP90, MNR, and OFD1 are largely coconserved in metazoans but absent in ecdysozoa such as Drosophila melanogaster and Caenorhabditis elegans (Fig. 2 e).
To assess whether MNR and OFD1 colocalize with CEP90, we immunostained RPE1 cells. CEP90 colocalized with MNR at centriolar satellites (Fig. 3 a) and, in cells treated with nocodazole to disperse centriolar satellites, at centrioles (Fig. 3 b). Similarly, MNR and OFD1 colocalized at centriolar satellites (Fig. 3 c), and centrioles in nocodazole-treated cells (Fig. 3 d). Thus, like CEP90 and OFD1, MNR is a component of both centriolar satellites and centrioles.
Figure 3.
CEP90 colocalizes with OFD1 and MNR. (a) Immunostaining of RPE1 cells for CEP90 (yellow), MNR (cyan), and γ-tubulin (magenta) demonstrating that CEP90 colocalizes with MNR at centriolar satellites. Scale bar = 2 µm. (b) 3D-SIM of RPE1 cells treated with nocodazole to disperse centriolar satellites highlights colocalization of CEP90 (yellow) and MNR (cyan) at centrioles (γ-tubulin, magenta). Scale bar = 1 µm. (c) Immunostaining of OFD1 (yellow), MNR (cyan), and γ-tubulin (magenta) reveals CEP90 and MNR colocalization at centriolar satellites. Scale bar = 2 µm. (d) 3D-SIM of immunostained cells treated with nocodazole reveals a ring of OFD1 (yellow) colocalizing with MNR (cyan) at centrioles (γ-tubulin, magenta). Scale bar = 1 µm. (e) Measurements of ring diameters measured in 3D-SIM images. For distal centriole proteins, ring diameters were measured at the mother (MC) and daughter centriole (DC). Scatter dot plot shows mean ± SD. *, P < 0.05, unpaired t test. n = 13–22 centrioles. (f) Schematic representation of a radial view of a mother centriole with distal appendages (purple), OFD1 (orange), CEP90 (yellow), and MNR (green). (g) Localization of distal centriole proteins (yellow) to centrioles (γ-tubulin, magenta) is cell cycle dependent. Cells in G1 phase were identified as lacking both EdU and CENPF staining, cells in S phase as being positive for EdU but lacking CENPF, and cells in G2 phase as being positive for CENPF but lacking EdU staining. Two puncta of CEP90, OFD1, and MNR were observed at one centrosome during G1 and S phases and four puncta at two centrosomes after centrosome duplication during G2 phase. Scale bars represent 5 µm in main panels and 0.5 µm in insets.
To gain insight into the spatial organization of distal centriole proteins with respect to distal appendages, we analyzed radially oriented centrioles with 3D-SIM (Fig. 3 e). Consistent with previous observations, distal appendage components CEP164 and CEP83 were organized into respectively larger and smaller rings at the mother centriole (Yang et al., 2018; Bowler et al., 2019). Of the distal centriole proteins, MNR was organized into the smallest ring (244 ± 29 nm; Fig. 3, e and f). Talpid3, OFD1, and CEP90 also were organized into rings that, intriguingly, were of different diameters at the mother and daughter centrioles. At the mother centriole, rings (from largest to smallest) were composed of Talpid3 (345 ± 27 nm), OFD1 (317 ± 37 nm), and CEP90 (291 ± 35 nm). At the daughter centriole, these rings were smaller and ordered differently from largest to smallest as OFD1 (288 ± 21 nm), CEP90 (263 ± 21 nm), and Talpid3 (255 ± 24 nm; Fig. 3, e and f), indicating that the distal centriole is reorganized when the daughter centriole matures into a mother.
Centriole biogenesis is coordinated with the cell cycle. Procentrioles form during S phase and elongate in G2. To ascertain when distal centriole proteins are recruited to centrioles, we used 5-ethynyl-2′-deoxyuridine (EdU) incorporation and Centromere protein F (CENPF) staining to distinguish between cells in different stages of the cell cycle (Viol et al., 2020). CEP90, OFD1, and MNR localized to mother and daughter centrioles in cells in G1 (EdU negative, CENPF negative) and S phase (EdU positive, CENPF negative; Fig. 3 g). In G2 (EdU negative, CENPF positive), both centrosomes (marked by γ-tubulin) each possessed two puncta of CEP90, OFD1, and MNR (Fig. 3 g). Therefore, CEP90, OFD1, and MNR are recruited to the distal end of elongating procentrioles in G2.
CEP90 and MNR are critical for vertebrate development, ciliogenesis, and Hedgehog signaling
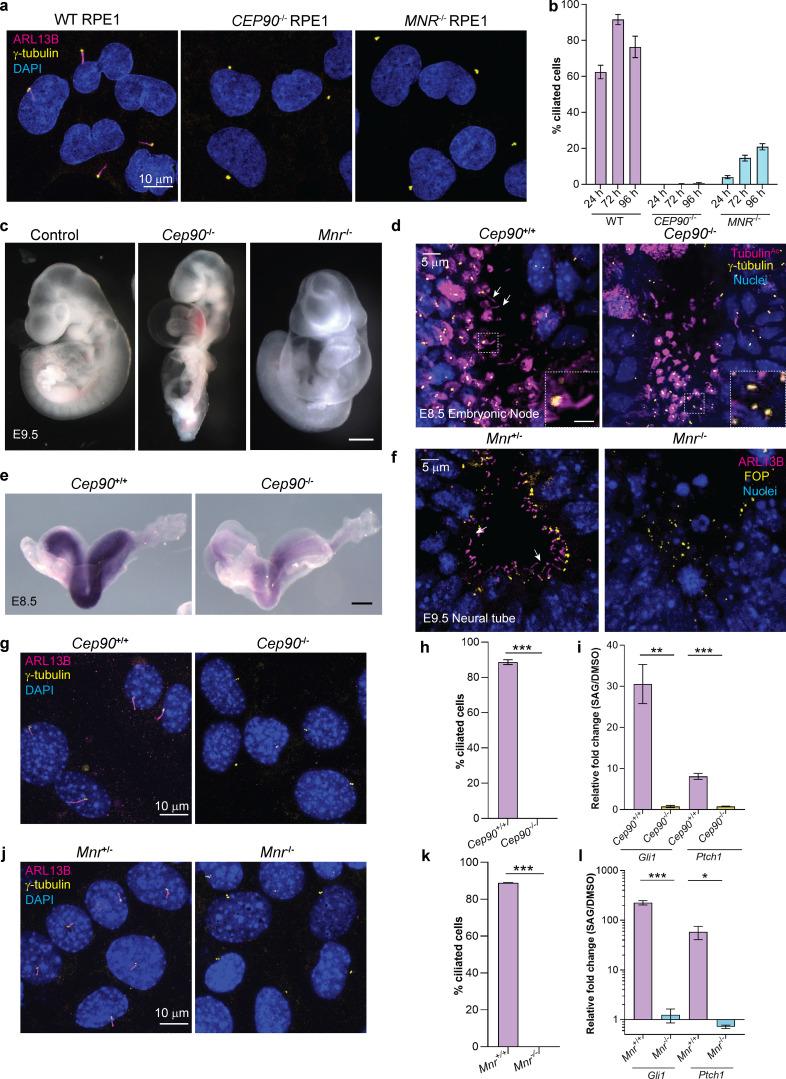
To assess the function of CEP90 and MNR, we generated RPE1 cell lines lacking CEP90 and MNR using CRISPR-Cas9. Immunoblot analyses confirmed loss of protein in the mutant cell lines (Fig. S1, a–d). Notably, CEP90−/− and MNR−/− RPE1 cells possessed centrioles but both lacked cilia (Fig. 4, a and b). As we previously identified a role for OFD1 in cilium assembly (Singla et al., 2010; Hunkapiller et al., 2011), we conclude that the DISCO complex comprised of CEP90, MNR, and OFD1 is essential for ciliogenesis.
Figure 4.
CEP90 and MNR are essential for ciliogenesis. (a) WT, CEP90−/−, and MNR−/− serum-starved RPE1 cells were immunostained for cilia (ARL13B, magenta), centrosomes (γ-tubulin, yellow), and nuclei (Hoechst, blue). Scale bar = 10 µm. (b) Quantification of ciliation frequency of WT, CEP90−/−, and MNR−/− RPE1 cells serum starved for times indicated. n > 100 cells from two biological replicates. Bar graph shows mean ± SEM. (c) Images of control, Cep90−/−, and Mnr−/− embryos at E9.5. Scale bar = 100 µm. (d) Whole-mount immunostaining of nodes of littermate control and Cep90−/− embryos at E8.5 for centrosomes (γ-tubulin, yellow), cilia (TubulinAc, magenta), and nuclei (Hoechst, blue). Arrows in the control image point to cilia projecting into the node lumen, which are absent in Cep90−/− embryos. Scale bars represent 5 µm in the main panels and 2 µm in insets. (e) In situ hybridization for Gli1 in E8.5 littermate control and Cep90−/− embryos revealing decreased expression in the absence of CEP90 indicative of disrupted Hedgehog signaling. Scale bar = 300 µm. (f) Immunostaining of E9.5 neural tube sections of littermate control and Mnr−/− embryos for centrosomes (FOP, yellow), cilia (ARL13B, magenta), and nuclei (Hoechst, blue), indicating that MNR is required for ciliogenesis in vivo. Scale bar = 5 µm. (g) MEFs derived from Cep90+/+ and Cep90−/− embryos were serum starved for 24 h and immunostained for cilia (ARL13B, magenta), centrosomes (γ-tubulin, yellow), and nuclei (Hoechst, blue). Scale bar = 10 µm. (h) Quantification of ciliation frequency shows loss of cilia in Cep90−/− MEFs. Bar graph shows mean ± SEM. Asterisks indicate P < 0.0005 determined using unpaired t test. n > 100 cells from two biological replicates. (i) qRT-PCR of HH target genes Gli1 and Ptch1 in serum-starved Cep90+/+ and Cep90−/− MEFs stimulated with 200nM SAG for 24h relative to DMSO-treated controls. Bar graph shows mean ± SEM. Asterisks indicate P < 0.05 determined using unpaired t test (**, P < 0.005; ***, P < 0.0005). n = 3 biological replicates. (j) MEFs derived from Mnr+/+ and Mnr−/− embryos were serum starved for 24 h and immunostained for cilia (ARL13B, magenta), centrosomes (γ-tubulin, yellow) and nuclei (Hoechst, blue). Scale bar = 10 µm. (k) Quantification of ciliation frequency shows loss of cilia in Mnr−/− MEFs. Bar graph shows mean ± SEM. Asterisks indicate P < 0.0005 determined using an unpaired t test. n > 100 cells from two biological replicates. (l) qRT-PCR of HH target genes Gli1 and Ptch1 in serum-starved Mnr+/+ and Mnr−/− MEFs stimulated with 200 nM SAG for 24 h relative to DMSO-treated controls. Bar graph shows mean ± SEM. Asterisks indicate P < 0.05 determined using unpaired t test (***, P < 0.0005; *, P < 0.05). n = 3 biological replicates.
To query the function of CEP90 and MNR in vertebrates, we obtained Cep90 and Mnr mutant mice from IMPC. The Cep90 mutant mice (Pibf1tm1.1 (KOMP)Vlcg) contain a deletion of seven exons, and the Mnr mutant mice (4933427D14Riktm1 (KOMP)Vlcg) contain a deletion of 10 exons.
Heterozygous Cep90 and Mnr mice were viable and fertile, with no obvious phenotypes. Homozygous Cep90 and Mnr mice did not survive beyond embryonic day 9.5 (E9.5). At E9.5, Cep90−/− and Mnr−/− embryos displayed pericardial edema (Fig. 4 c) and unlooped, midline hearts. One key role for primary cilia is in transducing extracellular signals including Hedgehog proteins, secreted morphogens required for vertebrate development. As lack of Smoothened (Smo), a central component of the Hedgehog signal transduction pathway, or lack of cilia also produces unlooped, midline hearts (Corbit et al., 2005; Huangfu et al., 2003), we examined the presence of cilia in Cep90−/− and Mnr−/− embryos. γ-Tubulin–positive centrioles were detected in both WT and Cep90−/− embryonic nodes at E8.5, but cilia were absent from Cep90−/− nodes (Fig. 4 d). Consistent with a defect in ciliogenesis, Cep90−/− embryos also displayed attenuated expression of the Hedgehog target gene Gli1, indicating defects in Hedgehog signal transduction (Fig. 4 e). Similarly, cilia detected by ARL13B staining were absent in neural tube sections from E9.5 Mnr−/− embryos (Fig. 4 f). Mouse embryonic fibroblasts (MEFs) derived from Cep90−/− and Mnr−/− embryos recapitulated the ciliogenesis phenotype observed in CEP90−/− and MNR−/− RPE1 cells (Fig. 4, g, h, j, and k). Furthermore, qRT-PCR analysis confirmed that induction of the Hedgehog target genes Gli1 and Ptch1 in response to the Hedgehog pathway activator Smoothened agonist (SAG) was abrogated in Cep90−/− or Mnr−/− MEFs (Fig. 4, i and l). Thus, CEP90 and MNR are both required for vertebrate development, ciliogenesis, and Hedgehog signaling.
CEP90 ciliopathy mutations affect ciliogenesis and centriolar satellite morphology
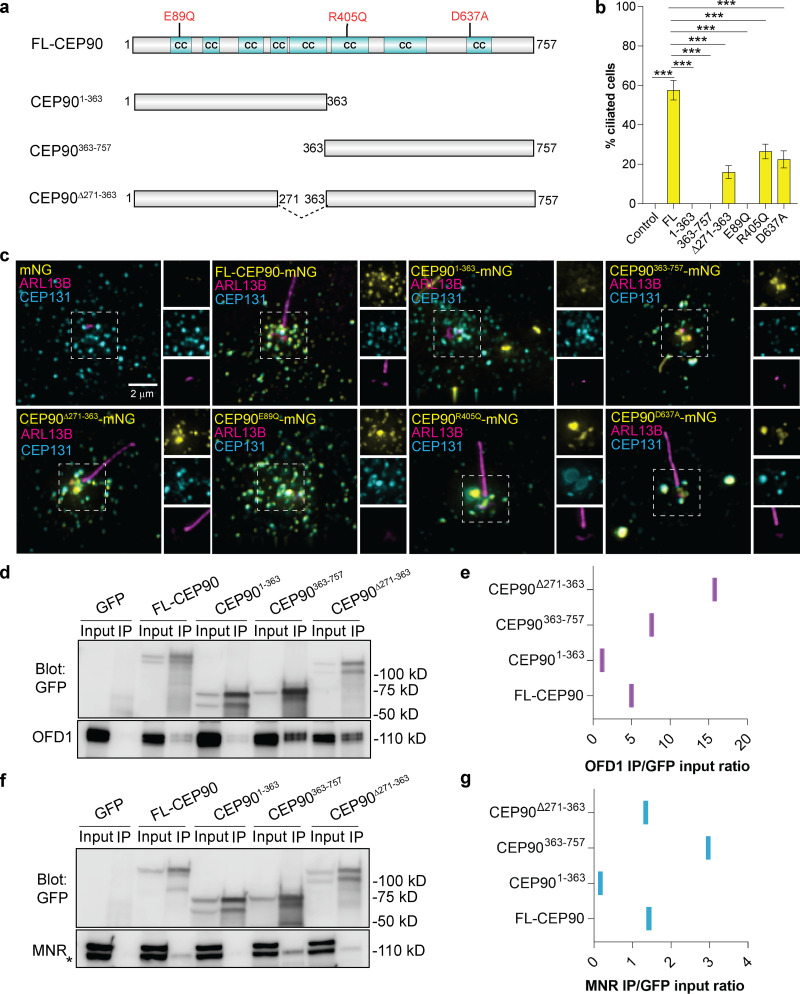
To identify regions in CEP90 required for cilium assembly, we tested whether CEP90 lacking the N terminus, C terminus, or central domain (Kim et al., 2012) could support ciliogenesis in CEP90−/− RPE1 cells (Fig. 5 a). Unlike expression of the full-length CEP90, CEP90 lacking the N terminal (CEP90363–757) or the C-terminal (CEP901–363) regions failed to rescue ciliogenesis defects in CEP90−/− RPE1 cells (Fig. 5, b and c). A construct of CEP90 lacking residues required for interaction with PCM1 (CEP90Δ271–363) localized to the centrosomal region and partially rescued ciliogenesis defects in CEP90−/− RPE1 cells (Fig. 5, a–c). These data support a role for both N- and C-terminal regions of CEP90 in ciliogenesis, further highlighting its function as a critical centriolar scaffold.
Figure 5.
CEP90 ciliopathy mutations affect ciliogenesis and centriolar satellite morphology. (a) Schematic representation of full-length (FL) human CEP90, truncation constructs, and disease-associated mutations. Coiled-coil (CC) domains identified by MARCOIL (Zimmermann et al., 2018) at 90% threshold are indicated in cyan. (b) Quantification of ciliation frequency of CEP90−/− RPE1 (control) and CEP90−/− cells expressing mNeonGreen-tagged disease variants and truncations of CEP90. Graph shows mean ± SEM. ***, P < 0.05, one-way ANOVA. n > 100 cells from two biological replicates. (c) CEP90−/− RPE1 cells and CEP90−/− cells expressing the denoted mNeonGreen (mNG)–tagged versions of CEP90 were serum starved for 24 h and immunostained for cilia (ARL13B), centriolar satellites (CEP131), and mNeonGreen. Scale bar = 2 µm. (d) Immunoprecipitation of GFP-tagged full-length and truncation constructs of CEP90 blotted for GFP and OFD1. (e) Quantification of OFD1 band intensities relative to corresponding GFP input band intensities. (f) Immunoprecipitation of GFP-tagged full-length and truncation constructs of CEP90 blotted for GFP and MNR. (g) Quantification of MNR band intensities relative to corresponding GFP input band intensities. Asterisk represents the MNR band; the top band is nonspecific.
We also examined whether disease-associated variants of CEP90 compromise its ability to support cilia assembly or centriolar satellite localization. Previously, we identified a rare variant in CEP90 (CEP90E89Q) associated with microcephaly (Kodani et al., 2015). Surprisingly, this variant of CEP90 displayed normal localization to centrioles and centriolar satellites (Fig. 5 c) but failed to support ciliogenesis in CEP90−/− RPE1 cells (Fig. 5, b and c). We also assessed two mutant forms of CEP90 (CEP90R405Q and CEP90D637A) implicated in JBTS (Wheway et al., 2015; Fig. 5, b and c). Interestingly, both Joubert-associated variants of CEP90 perturbed centriolar satellite morphology (Fig. 5 c) and showed reduced ability to support ciliogenesis in CEP90−/− RPE1 (Fig. 5 b). Thus, our data suggest that distinct human disease-associated mutations in CEP90 differentially compromise centriolar satellite morphogenesis and ciliogenesis.
To identify regions of CEP90 required for interaction with the DISCO components OFD1 and MNR, we performed coimmunoprecipitation experiments with CEP90 truncation constructs. Both OFD1 and MNR preferentially interacted with the C-terminal region of CEP90 (CEP90363–757), and CEP90 lacking the C-terminal region (CEP901–363) failed to immunoprecipitate OFD1 and MNR (Fig. 5, d–g). Therefore, the C-terminal region of CEP90 (in which mutations cause JBTS) is critical for interaction with the DISCO subunits OFD1 and MNR (Wheway et al., 2015).
MNR, but not CEP90, restricts centriole length
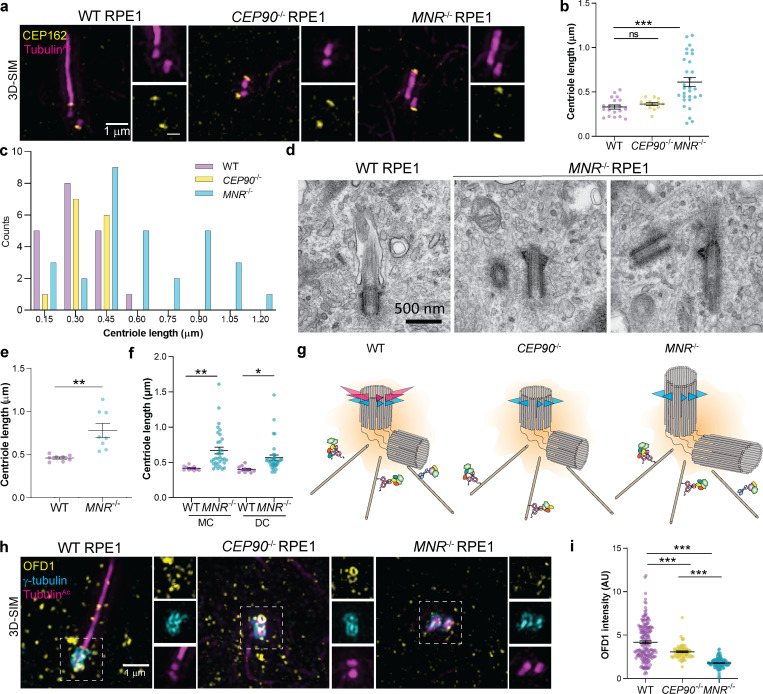
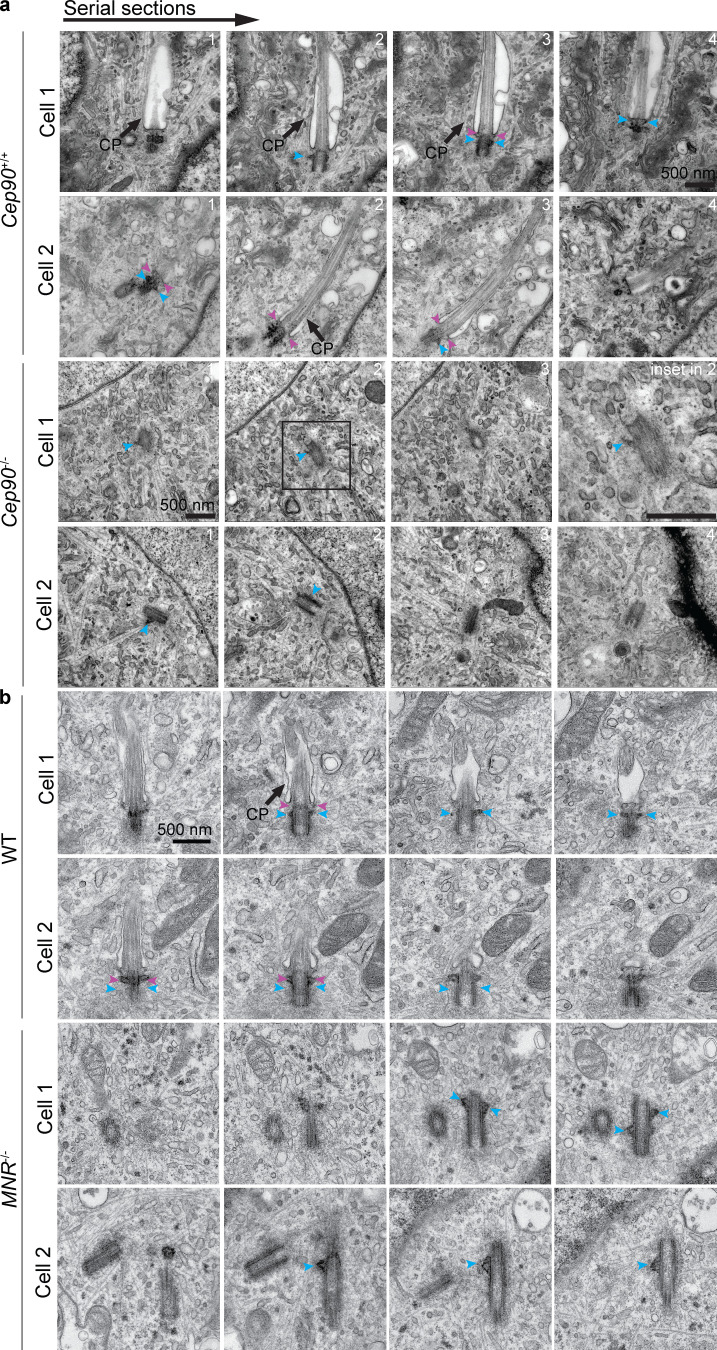
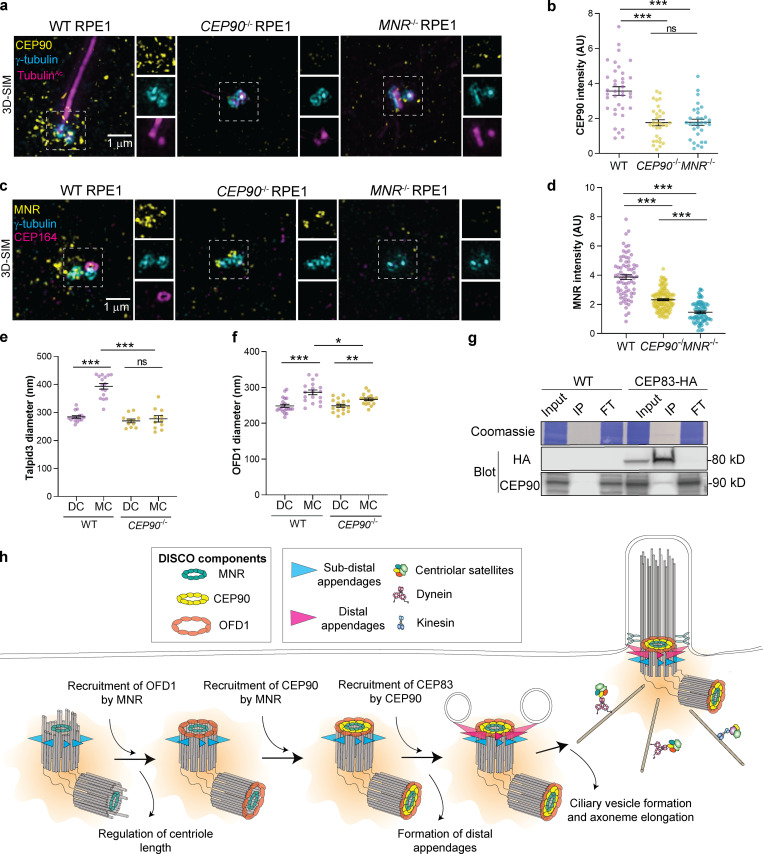
Since the distal centriole proteins OFD1, Talpid3, and C2CD3 control centriole length, we analyzed CEP90−/− and MNR−/− centrioles labeled with TubulinAc and distal centriole protein CEP162 by 3D-SIM. Compared with control centrioles, centriole length was not altered in CEP90−/− cells. However, centriole length was highly variable in MNR−/− cells, with ∼30% of MNR−/− cells containing hyperelongated centrioles (some of which were over a micron long; Fig. 6, a–c). The hyperelongated centrioles in MNR−/− cells possessed the distal centriole component CEP162 at one end, suggesting that proximal distal polarity is maintained despite the elongation (Fig. 6 a). Serial-section transmission EM (TEM) analysis confirmed the presence of elongated centrioles in MNR−/− RPE1 cells (Fig. 6, d and e). Both mother and daughter centrioles (distinguished by localization of Ninein to the mother centriole-specific subdistal appendage) were hyperelongated in MNR−/− cells (Fig. 6 f). Therefore, MNR restrains centriole lengthening of both mother and daughter centrioles (Fig. 6, f and g).
Figure 6.
MNR, but not CEP90, restricts centriole length. (a) 3D-SIM images of WT, CEP90−/−, and MNR−/− RPE1 cells immunostained for cilia/centrioles (TubulinAc, magenta) and CEP162 (yellow), a distal centriolar protein. Scale bars represent 1 µm in the main panels and 0.5 µm in insets. (b) Graph of centriolar lengths measured using 3D-SIM images. Centriole lengths are longer and have a wider distribution in MNR−/− cells than in WT or CEP90−/− cells. Error bars indicate mean ± SEM. ***, P < 0.05, one-way ANOVA. n = 14–30 centrioles. (c) Histogram of centriole lengths observed in WT, MNR−/−, and CEP90−/− cells. n = 14–30 centrioles. (d) Serial-section TEM confirms the presence of elongated centrioles in MNR−/− RPE1 cells. Scale bar = 500 nm. (e) Centriole lengths of WT and MNR−/− RPE1 cells measured using TEM images. Horizontal lines indicate means ± SEM. Asterisks indicate P < 0.005 determined using unpaired t test. n = 8 centrioles per condition. (f) WT and MNR−/− RPE1 cells were serum-starved and immunostained with antibodies to TubulinAc and subdistal appendage component Ninein to distinguish mother (MC) and daughter centrioles (DC). Graph of centriolar lengths measured using 3D-SIM. MNR restrains centriole lengthening of both mother and daughter centrioles. Error bars indicate mean ± SEM. Asterisks indicate P < 0.05 determined using unpaired t test (*, P < 0.05; **, P < 0.005). n = 10–35 measurements. (g) Schematic depicting distinct roles of CEP90 and MNR in regulating centriole length. (h) 3D-SIM imaging of serum-starved WT, CEP90−/−, and MNR−/− RPE1 cells immunostained for OFD1 (yellow), centrioles (γ-tubulin, cyan), and cilia (TubulinAc, magenta). Boxed regions are depicted in insets throughout. OFD1 localizes to centrioles in WT and CEP90−/− cells, but not MNR−/− cells. Scale bar = 1 µm. (i) Quantification of OFD1 fluorescence intensity at centrioles in WT, CEP90−/−, and MNR−/− cells. Horizontal lines indicate means ± SEM. ***, P < 0.05, one-way ANOVA. n = 64–187 cells.
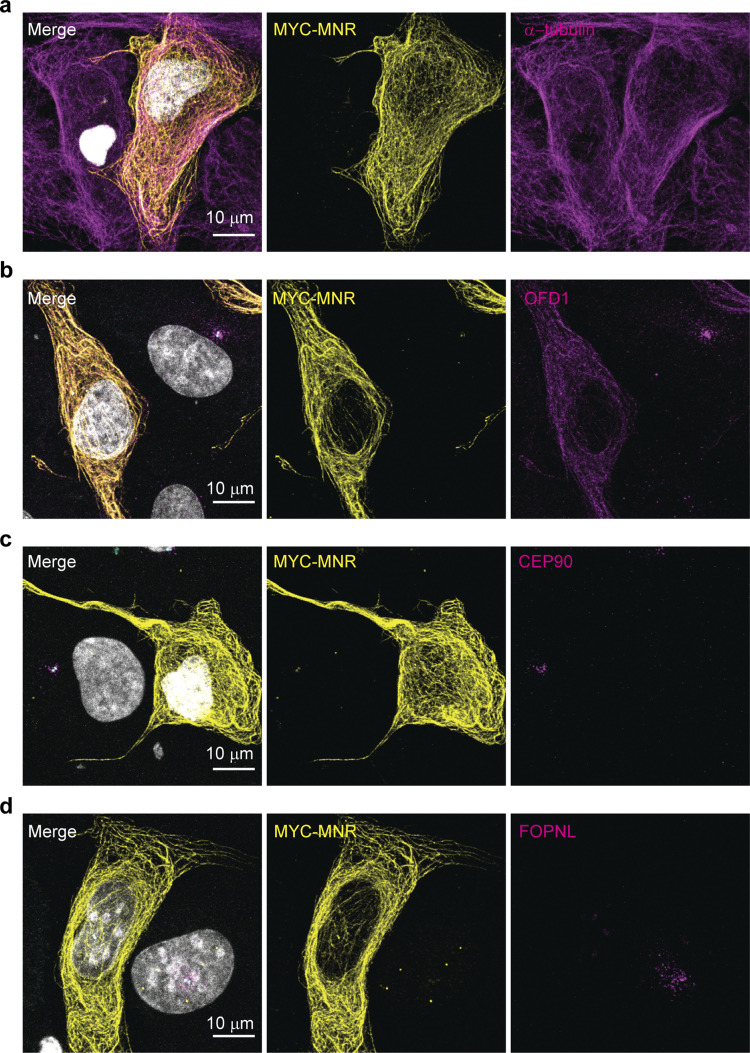
The abnormal centriole morphology in MNR−/− cells is reminiscent of the phenotype we previously observed in OFD1 mutant cells (Singla et al., 2010). Therefore, we examined the localization of OFD1 in MNR−/− RPE1 cells. OFD1 failed to localize to the centrioles in the absence of MNR, but not CEP90 (Fig. 6, h and i). Overexpressed MNR localizes to microtubules (Chevrier et al., 2016), and microtubule-associated MNR can sequester endogenous OFD1 (Fig. S2). Therefore, MNR is necessary and sufficient to recruit OFD1 (Fig. 6, h and i), and our data support a model in which MNR recruits OFD1 to restrict centriole length (Feng et al., 2017; Srivastava and Panda, 2017), with CEP90 being dispensable for centriole length control and dedicated to ciliogenesis.
Figure S2.
Overexpressed MNR localizes to microtubules and recruits endogenous OFD1. (a) RPE1 cells transiently transfected with MYC-tagged MNR and immunostained with antibodies to MYC-tag and α-tubulin. Scale bar = 10 µm. (b) RPE1 cells transiently transfected with MYC-tagged MNR and immunostained with antibodies to MYC-tag and OFD1. Scale bar = 10 µm. (c) RPE1 cells transiently transfected with MYC-tagged MNR and immunostained with antibodies to MYC-tag and CEP90. Scale bar = 10 µm. (d) RPE1 cells transiently transfected with MYC-tagged MNR and immunostained with antibodies to MYC-tag and FOPNL. Scale bar = 10 µm.
CEP90 and MNR are required for the removal of CP110 and CEP97 from the mother centriole at the initiation of ciliogenesis
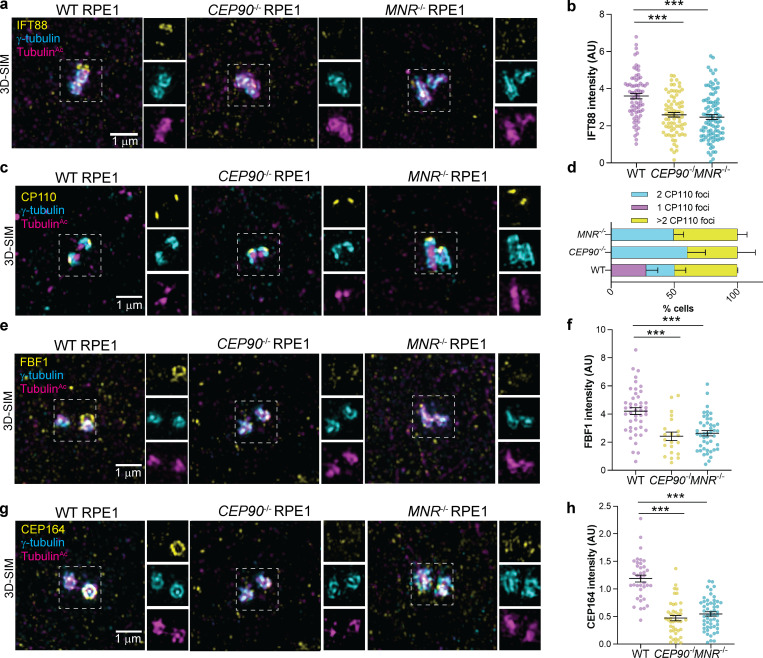
Ciliogenesis depends on a specialized protein transport machinery called intraflagellar transport (IFT). We examined whether IFT88 localization to basal bodies depends on CEP90 or MNR, and found that IFT88 was decreased at the mother centriole of both CEP90−/− and MNR−/− RPE1 cells in both serum-starved (Fig. 7, a and b) and cycling cells (Fig. S3, a and b).
Figure 7.
CEP90 and MNR are required for early ciliogenesis. (a) WT, CEP90−/−, and MNR−/− RPE1 cells immunostained for IFT88 (yellow), centrioles (γ-tubulin, cyan), and cilia (TubulinAc, magenta. IFT88 is not recruited to the centrosome of CEP90−/− or MNR−/− cells. (b) Quantification of IFT88 fluorescence intensity at WT, CEP90−/−, and MNR−/− centrosomes. Error bars indicate mean ± SEM. ***, P < 0.05, unpaired t test. n = 38–63 measurements. (c) WT, CEP90−/−, and MNR−/− serum-starved RPE1 cells immunostained for CP110 (yellow), centrioles (γ-tubulin, cyan), and cilia (TubulinAc, magenta). Scale bar = 1 µm. (d) Quantification of whether CP110 localizes to one or two centrioles. In the absence of CEP90 or MNR, CP110 continues to localize to the distal mother centriole. n > 50 cells from two independent experiments. Error bars indicate mean ± SEM. (e) WT, CEP90−/−, and MNR−/− serum-starved RPE1 cells immunostained for CEP97 (yellow), γ-tubulin (cyan), and TubulinAc (magenta). Scale bar = 1 µm. (f) Quantification of whether CEP97 localizes to one or two centrioles. As with CP110, CEP90 and MNR are required to remove CEP97 from the distal mother centriole. n > 50 cells from two independent experiments. (g) 3D-SIM images of RPE1 cells immunostained for Myosin Va (Myo-Va, yellow) and centrioles (γ-tubulin, magenta). Myo-Va cannot localize near centrosomes (left), can localize to preciliary vesicles (denoted centrosomal Myo-Va, middle), or can localize to the ciliary pocket (denoted ciliary Myo-Va, right). Scale bar = 1 µm. (h) Quantification of three distinct Myo-Va staining patterns in WT, CEP90−/− and MNR−/− cells. n > 50 cells from two independent experiments. (i) Serial-section TEM images of serum-starved Cep90+/+ and Cep90−/− MEFs confirms the absence of preciliary vesicle docking at the Cep90−/− mother centriole. Scale bar = 200 nm. n = 9 cells for both genotypes. (j) Serial-section TEM images of WT and MNR−/− RPE1 cells confirms the absence of preciliary vesicle docking at the MNR−/− mother centriole. Scale bar = 250 nm. n = 10 cells for WT and n = 6 for MNR−/− cells. (k) Model based on our data highlighting the role of CEP90 and MNR in maturation of the mother centriole.
Figure S3.
CEP90 and MNR regulate distal appendage assembly irrespective of ciliation status. (a) Cycling WT, CEP90−/−, and MNR−/− RPE1 cells stained with antibodies to IFT88, γ-tubulin (centrosome marker), and TubulinAc (cilia and centriole marker). 3D-SIM imaging reveals IFT88 at the centrosome in WT, but not CEP90−/− and MNR−/−, cells. Scale bars for main panels and insets represent 1 µm. (b) Quantification of IFT88 fluorescence intensity at centrioles. Scatter dot plots show mean ± SEM. ***, P < 0.001, one-way ANOVA. n = 73–96 measurements. (c) Cycling WT, CEP90−/−, and MNR−/− RPE1 cells immunostained for CP110 (yellow), centrioles (FOP, cyan), and TubulinAc (cilia and centriole marker, magenta). Scale bar = 1 µm. (d) Quantification of CP110 foci. (e) Cycling WT, CEP90−/−, and MNR−/− RPE1 cells stained with antibodies to FBF1, γ-tubulin (centrosome marker), and TubulinAc (cilia and centriole marker). 3D-SIM imaging reveals FBF1 at the mother centriole in WT, but not CEP90−/− and MNR−/−, cells. Scale bars for main panels and insets represent 1 µm. (f) Quantification of FBF1 fluorescence intensity at centrioles. Scatter dot plots show mean ± SEM. ***, P < 0.001, one-way ANOVA. n = 21–45 measurements. (g) Cycling WT, CEP90−/−, and MNR−/− RPE1 cells stained with antibodies to CEP164, γ-tubulin (centrosome marker), and TubulinAc (cilia and centriole marker). 3D-SIM imaging reveals CEP164 at one of the two centrioles in WT, but not CEP90−/− and MNR−/−, cells. Scale bars for main panels and insets represent 1 µm. (h) Quantification of CEP164 fluorescence intensity at centrioles. Scatter dot plots show mean ± SEM. ***, P < 0.001, one-way ANOVA. n = 37–53 measurements.
A critical early step in ciliogenesis is the removal from the distal mother centriole of two proteins that can inhibit ciliogenesis, CP110 and CEP97. We tested whether CEP90 or MNR function removal of CP110 and CEP97 by examining CEP90−/− and MNR−/− RPE1 cells. We found that cells lacking either CEP90 or MNR fail to remove CP110 and CEP97 (Fig. 7, c–f). Taken together, these results reveal that CEP90 and MNR are required for early steps of ciliogenesis.
CEP90 and MNR are required for ciliary vesicle docking and distal appendage assembly
As ciliary vesicle formation contributes to removing CP110 and CEP97 from the mother centriole (Westlake et al., 2011; Lu et al., 2015), we investigated whether persistence of CP110 and CEP97 in CEP90−/− and MNR−/− cells was due to defective recruitment of preciliary vesicles.
To assess whether CEP90 and MNR affect preciliary vesicle recruitment, we examined the localization of Myosin-Va–positive preciliary vesicles (Wu et al., 2018) to centrioles in WT, CEP90−/−, and MNR−/− RPE1 cells. Consistent with the defect in distal appendage formation, cells lacking either CEP90 or MNR showed reduced Myosin-Va at the mother centriole (Fig. 7, g and h). To confirm the requirement for both CEP90 and MNR in preciliary vesicle recruitment, we examined WT and knockout cells by serial-section TEM, which confirmed that centrioles in cells lacking CEP90 or MNR fail to dock to preciliary vesicles (Fig. 7, i and j; and Fig. S4). Thus, CEP90 and MNR are essential for ciliary vesicle recruitment to the mother centriole, a key early step of ciliogenesis (Fig. 7 k).
Figure S4.
Serial-section TEM confirms loss of distal appendages in cells lacking CEP90 or MNR. (a) TEM images ofCep90+/+ and Cep90−/− MEF cells confirms the absence of preciliary vesicle docking and distal appendages at the Cep90−/− mother centriole. Blue arrowheads indicate subdistal appendages, and pink arrowheads indicate distal appendages. (b) TEM images of WT and MNR−/− RPE1 cells confirms the absence of preciliary vesicle docking and distal appendages at the MNR−/− mother centriole. Centrioles are hyperelongated in the absence of MNR. Blue arrowheads indicate subdistal appendages, and pink arrowheads indicate distal appendages. Scale bars = 500 nm. CP, ciliary pocket.
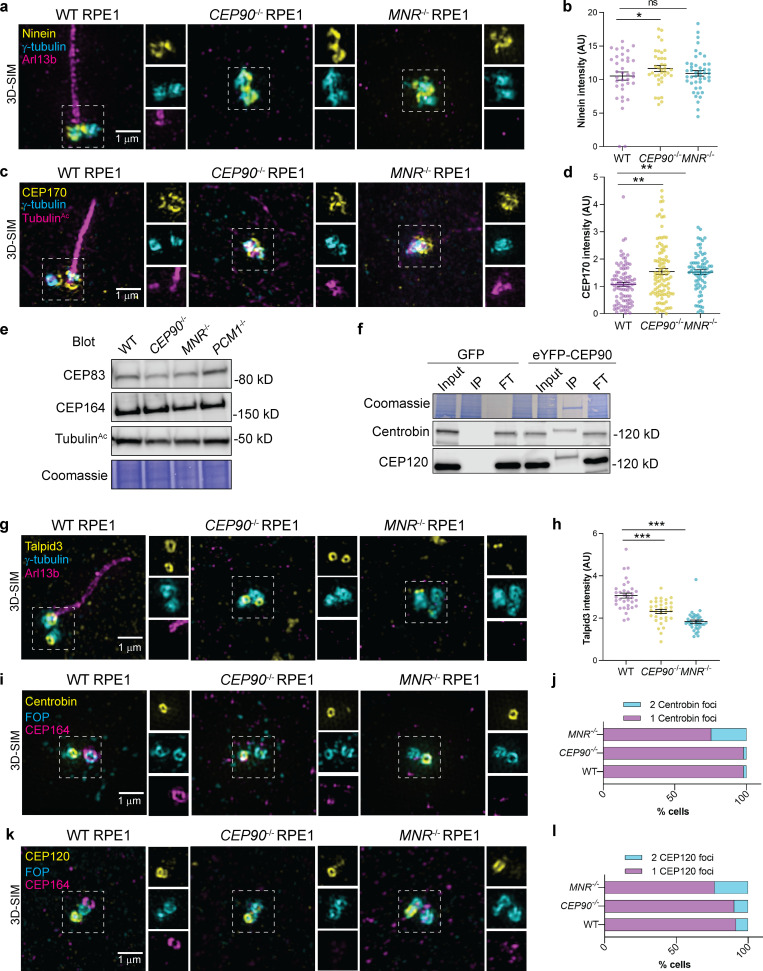
An early step of ciliogenesis is the acquisition of appendages by the mother centriole, defining its maturation into a basal body. Subdistal appendages mediate the anchoring of the basal body to microtubules and regulate the spatial positioning of the cilium in the cell (Mazo et al., 2016). In CEP90−/− and MNR−/− RPE1 cells, the localization of components of subdistal appendages, such as Ninein and CEP170, were unaffected (Fig. S5, a–d), indicating that CEP90 and MNR are dispensable for subdistal appendage formation.
Figure S5.
CEP90 regulates distal appendage assembly independent of Talpid3 recruitment and removal of Centrobin.(a) 3D-SIM imaging of serum-starved WT, CEP90−/−, and MNR−/− RPE1 cells immunostained for Ninein (yellow), centrioles (γ-tubulin, cyan), and cilia (ARL13B, magenta). Boxed regions are depicted in insets throughout. Ninein localizes to centrioles in WT, CEP90−/−, and MNR−/− cells. Scale bar = 1 µm. (b) Quantification of Ninein fluorescence intensity at centrioles in WT, CEP90−/−, and MNR−/− cells. Horizontal lines indicate means ± SEM. Asterisks indicate P < 0.05 determined using one-way ANOVA. n = 36–45 measurements. (c) 3D-SIM imaging of serum-starved WT, CEP90−/− and MNR−/− RPE1 cells immunostained for CEP170 (yellow), centrioles (γ-tubulin, cyan) and cilia (TubulinAc, magenta). Boxed regions are depicted in insets throughout. CEP170 localizes to centrioles in WT, CEP90−/−, and MNR−/− cells. Scale bar = 1 µm. (d) Quantification of CEP170 fluorescence intensity at centrioles in WT, CEP90−/−, and MNR−/− cells. Horizontal lines indicate means ± SEM. Asterisks indicate P < 0.005 determined using one-way ANOVA. n = 73–99 measurements. (e) Immunoblot of distal appendage proteins CEP83 and CEP164 in WT, CEP90−/−, MNR−/−, and PCM1−/− RPE1 cells. (f) Coimmunoprecipitation of daughter centriole proteins Centrobin and CEP120 with CEP90. IP indicates eluate, and FT indicates flow-through. (g) WT, CEP90−/−, and MNR−/− RPE1 cells were serum starved for 24 h and stained with antibodies to Talpid3, γ-tubulin (centrosome marker), and ARL13B (cilia marker). 3D-SIM imaging reveals ring of Talpid3 at mother and daughter centrioles. Scale bars represent 1 µm for main panels and insets. (h) Quantification of Talpid3 fluorescence intensity at centrioles. Scatter dot plots show mean ± SEM. Asterisks indicate P < 0.0005, determined using one-way ANOVA. n = 33–35 measurements. (i) WT, CEP90−/−, and MNR−/− serum-starved RPE1 cells immunostained for Centrobin (yellow), centrioles (FOP, cyan), and distal appendages (CEP164, magenta). Scale bar = 1 µm. (j) Quantification of whether Centrobin localizes to one or two centrioles. n > 50 cells from two biological replicates. CEP90 and MNR are not required to remove Centrobin from the distal mother centriole. (k) WT, CEP90−/−, and MNR−/− serum-starved RPE1 cells immunostained for CEP120 (yellow), centrioles (FOP, cyan), and distal appendages (CEP164, magenta). Scale bar = 1 µm. (l) Quantification of whether CEP120 localizes to one or two centrioles. n > 50 cells from two biological replicates. CEP90 and MNR are not required to remove Centrobin or CEP120 from the distal mother centriole.
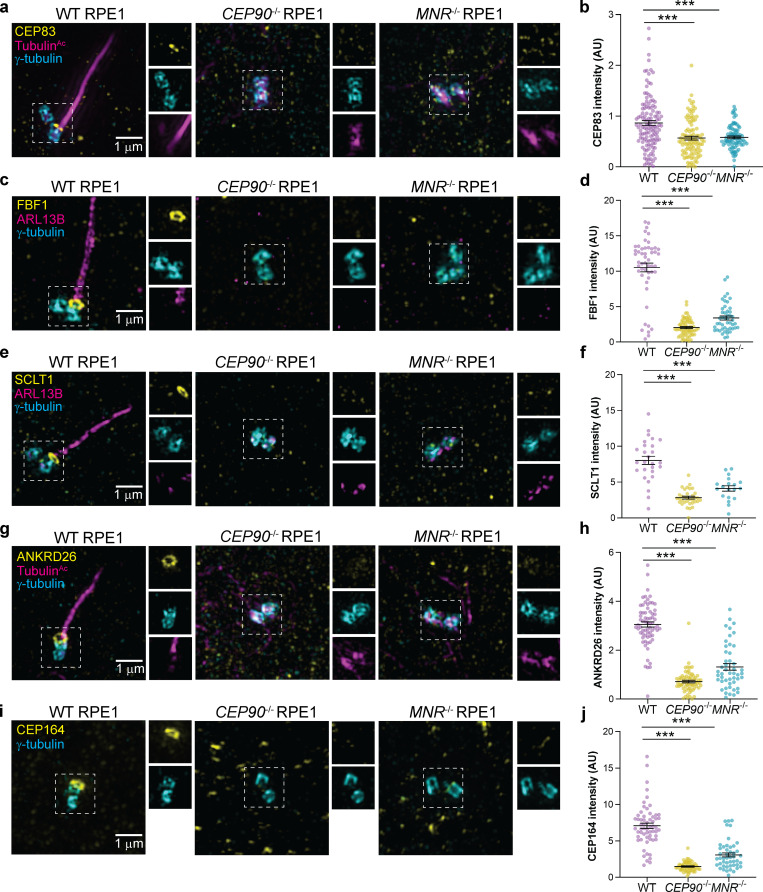
A function of distal appendages is the recruitment of small preciliary vesicles to the mother centriole that fuse and give rise to the ciliary membrane (Tanos et al., 2013; Schmidt et al., 2012). CEP90−/− and MNR−/− centrioles fail to recruit distal appendage proteins CEP83, FBF1, SCLT1, ANKRD26, and CEP164, indicating that they are essential for distal appendage formation (Fig. 8, c–g). FBF1 and CEP164 failed to localize to the mother centriole in cycling CEP90−/− and MNR−/− cells, indicating that CEP90 and MNR organize the distal appendage irrespective of whether serum starvation initiates ciliogenesis (Fig. S3, e–h). CEP90 and MNR are not critical for distal appendage protein abundance (Fig. S5 e), suggesting that they are not required for distal appendage protein stability but rather are essential for distal appendage assembly at the mother centriole.
Figure 8.
CEP90 and MNR recruit distal appendage components to the mother centriole.(a–j) 3D-SIM images and quantification of centrosomal intensity of WT, CEP90−/−, and MNR−/− RPE1 cells immunostained for γ-tubulin (cyan), TubulinAc (magenta), and distal centriole components (yellow) CEP83 (a and b), FBF1 (c and d), SCLT1 (e and f), ANKRD26 (g and h), and CEP164 (i and j). Scale bars = 1 µm. Horizontal lines in scatter dot plots indicate means ± SEM. ***, P < 0.001, one-way ANOVA. n ≥ 40 measurements per condition.
Recent studies show that daughter centriole proteins are removed from the mother centriole to enable distal appendage assembly (Wang et al., 2018; Mahjoub et al., 2010). Since CEP90 localizes to the distal daughter centriole as well as the distal mother centriole, we hypothesized that CEP90 may interact with distal daughter centriole proteins. Coimmunoprecipitation revealed that CEP90 did interact with daughter centriole components Centrobin and CEP120 (Fig. S5 f). As removing daughter centriole proteins requires distal centriole proteins, such as Talpid3, we examined the localization of Talpid3 and found that Talpid3 is recruited to the distal centriole in WT, CEP90−/−, and MNR−/− RPE1 cells (Fig. S5, g and h). Moreover, daughter centriole components Centrobin and CEP120 localized to only one centriole in WT, CEP90−/−, and MNR−/− RPE1 cells (Fig. S5, i–l). Therefore, CEP90 and MNR regulate distal appendage formation through a mechanism independent of Talpid3 recruitment or daughter centriole protein removal.
CEP90 functions at the mother centriole to support distal appendage formation
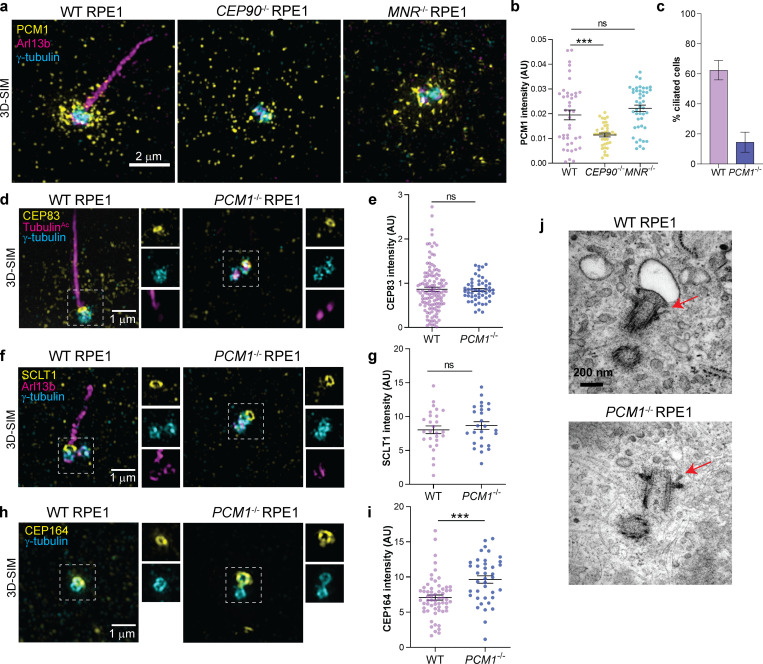
Using siRNA-mediated gene knockdown, a previous study showed that CEP90 promotes centriolar satellite accumulation in the vicinity of centrosomes (Kim et al., 2012). We confirmed that, in CEP90−/− RPE1 cells, PCM1-positive centriolar satellites failed to accumulate around centrosomes (Fig. 9, a and b). Interestingly, loss of MNR did not affect targeting of PCM1-positive centriolar satellites to the centrosome (Fig. 9, a and b), indicating that CEP90 and MNR have some distinct functions in centriolar satellite distribution. These observations raised the interesting possibility that CEP90-dependent targeting of centriolar satellites to the centrosomal area promotes distal appendage formation and ciliogenesis.
Figure 9.
Centriolar satellites are dispensable for distal appendage assembly at the mother centriole. (a) 3D-SIM images of WT, CEP90−/−, and MNR−/− RPE1 cells immunostained with antibodies to PCM1, a centriolar satellite marker, ARL13B and γ-tubulin. Scale bar = 2 µm. (b) Quantification of PCM1 intensity in the pericentrosomal area. PCM1-positive centriolar satellites fail to accumulate around centrosomes in CEP90−/−, but not MNR−/−, cells. ***, P < 0.05, ordinary one-way ANOVA. n = 37–44 measurements. (c) Ciliogenesis is disrupted in PCM1−/− RPE1 cells 24 h after serum starvation. n > 100 cells from two biological replicates. Bar graph shows mean ± SEM. (d) WT and PCM1−/− RPE1 cells were serum starved for 24 h and stained with antibodies to CEP83, γ-tubulin (centrosome marker), and TubulinAc (cilia marker). 3D-SIM imaging reveals ring of CEP83 at the mother centrioles in PCM1−/− RPE1 cells. Scale bar = 1 µm. (e) Quantification of CEP83 fluorescence intensity at centrioles. Scatter dot plots show mean ± SEM. ns, unpaired t test. n = 53–124 measurements. (f) WT and PCM1−/− RPE1 cells were serum starved for 24 h and stained with antibodies to SCLT1, γ-tubulin (centrosome marker), and ARL13B (cilia marker). 3D-SIM imaging reveals ring of SCLT1 at the mother centrioles in WT and PCM1−/− RPE1 cells. Scale bar = 1 µm. (g) Quantification of SCLT1 fluorescence intensity at centrioles. Scatter dot plots show mean ± SEM. ns, unpaired t test. n = 26–28 measurements. (h) WT and PCM1−/− RPE1 cells were serum starved for 24 h and stained with antibodies to CEP164 and γ-tubulin (centrosome marker). 3D-SIM imaging reveals ring of CEP164 at the mother centrioles in PCM1−/− RPE1 cells. Scale bar = 1 µm. (i) Quantification of CEP164 fluorescence intensity at centrioles. Scatter dot plots show mean ± SEM. ***, P < 0.001, unpaired t test. n = 40–60 measurements. (j) Representative TEM images of WT and PCM1−/− RPE1 cells serum starved for 1 h. Distal appendages are marked with arrows. Scale bar = 200 nm.
As PCM1−/− cells lack centriolar satellites but retain CEP90 at centrioles (Fig. 1 c), these cells can help disentangle the functions of CEP90 and MNR at centriolar satellites and centrioles. In accordance with previous observations (Wang et al., 2016; Odabasi et al., 2019), RPE1 cells lacking PCM1 displayed compromised ciliogenesis (Fig. 9 c). In stark contrast to CEP90−/− cells, distal appendage components CEP83, SCLT1, and CEP164 localized equivalently to the mother centrioles of PCM1−/− and WT RPE1 cells (Fig. 9, d–i), indicating that distal appendage assembly is independent of PCM1. Furthermore, serial-section TEM confirmed the presence of distal appendages in PCM1−/− RPE1 cells (Fig. 9 j). As the critical centriolar satellite scaffold PCM1 is not required for either CEP90 localization to centrioles or distal appendage formation, we conclude that the centriolar satellite population of CEP90 is dispensable for distal appendage assembly.
If the centriolar satellite population is dispensable, we predicted that MNR recruitment of CEP90 to the distal centriole would be critical for distal appendage assembly and ciliogenesis. Indeed, we found that although CEP90 protein levels were unchanged in MNR−/− RPE1 cells (Fig. S1 d), CEP90 failed to localize to MNR−/− centrioles, while MNR localized to the centrioles in the absence of CEP90 (Fig. 10, a–d).
Figure 10.
MNR recruits CEP90, which recruits CEP83 to build distal appendages. (a) 3D-SIM imaging of WT, CEP90−/−, and MNR−/− serum-starved RPE1 cells immunostained for CEP90 (yellow), γ-tubulin (cyan), and TubulinAc (magenta). Scale bar = 1 µm. (b) Quantification of CEP90 fluorescence intensity at centrioles. Horizontal lines in scatter dot plots indicate means ± SEM. ***, P < 0.0001, one-way ANOVA. n = 31–37 measurements. CEP90 fails to localize to MNR−/− and CEP90−/− centrioles, although protein levels of CEP90 remained unchanged in MNR−/− RPE1 cells (Fig. S1 d). (c) 3D-SIM imaging of WT, CEP90−/−, and MNR−/− serum-starved RPE1 cells immunostained for MNR (yellow), γ-tubulin (cyan), and CEP164 (magenta). Scale bar = 1 µm. (d) Quantification of MNR fluorescence intensity at centrioles. Horizontal lines in scatter dot plots indicate means ± SEM. ***, P < 0.0001, one-way ANOVA. n = 73–122 measurements. MNR localization is reduced but present at CEP90−/− centrioles. (e) Quantification of Talpid3 diameter at mother (MC) and daughter centrioles (DC) in serum-starved WT and CEP90−/− cells. Horizontal lines in scatter dot plots indicate means ± SEM. ***, P < 0.05, one-way ANOVA. n = 11–16 measurements. The Talpid3 ring diameter is increased at WT mother centrioles, but not CEP90−/− mother centrioles. (f) Quantification of OFD1 diameter at mother (MC) and daughter centrioles (DC) in serum-starved WT and CEP90−/− cells. Horizontal lines in scatter dot plots indicate means ± SEM. Asterisks indicate P < 0.05 (*, P < 0.05; **, P < 0.005; ***, P < 0.0005), one-way ANOVA. n = 17–20 measurements. In the absence of CEP90, the mother centriole ring of OFD1 does not expand as in WT cells. (g) WT or RPE1 cells expressing CEP83-HA were lysed, immunoprecipitated with anti-HA antibody–bound beads, and immunoblotted with antibodies to HA and CEP90. (h) Model of the hierarchical recruitment of the DISCO complex. MNR recruitment of OFD1 restrains centriole elongation and MNR recruits CEP90, which, in turn, recruits CEP83, the base of the distal appendage.
To investigate how CEP90 can be specifically required for the formation distal appendages, we investigated whether CEP90 reorganizes distal centriole proteins at the mother centriole to promote distal appendage assembly. 3D-SIM analysis revealed that Talpid3 and OFD1 diameters were smaller in mother centrioles of CEP90−/− cells compared with WT (Fig. 10, e and f). These results support a model where CEP90 functions in the transformation of a daughter to a mother centriole by reorganizing the distal centriole.
To investigate how CEP90 participates in distal appendage formation, we examined whether CEP90 interacts with the most proximal component of the distal appendage known, CEP83. Coimmunoprecipitation analysis revealed an interaction between CEP90 and CEP83 (Fig. 10 g). We propose that MNR recruits CEP90 to the distal centriole, and CEP90, in turn, recruits CEP83 to initiate distal appendage assembly (Fig. 10 h). Therefore, distal centriolar proteins are recruited in a hierarchical manner, with MNR recruiting OFD1 to restrict centriole length and CEP90 being dedicated to distal appendage assembly.
Discussion
CEP90 and MNR are critical for distal appendage assembly and ciliogenesis
Through the process of mitosis, vertebrate daughter cells each inherit two centrioles. Yet, only the older, mother centriole templates a cilium. In no small part, this unique function of the mother centriole depends on its distal appendages, acquired during the previous G2/M phase. In this study, we identify a multiprotein complex, which we name DISCO, that is composed of CEP90, OFD1, and MNR and is critical for distal appendage formation and ciliogenesis.
Consequent to their role in distal appendage formation, CEP90 and MNR are also required for subsequent events in ciliogenesis, including recruitment of the preciliary vesicles that give rise to the ciliary membrane, and loading of IFT88 (Sillibourne et al., 2013; Tanos et al., 2013). Unlike some other centriolar proteins, such as CEP120 (Tsai et al., 2019), CEP90 and MNR are specifically required for distal appendage formation and are dispensable for subdistal appendage formation, revealing that discrete mother centriolar complexes support distal and subdistal appendage assembly.
In mice, we found that CEP90 and MNR are essential for embryonic development, particularly for Hedgehog signaling. Primary cilia are required for Hedgehog signal transduction in vertebrates. A central component of the Hedgehog pathway is the seven-pass transmembrane protein Smo. Interestingly, Cep90, Mnr, and Smo mutants arrested at similar points in embryonic development and displayed similar phenotypes, emphasizing the importance of CEP90 and MNR in ciliary Hedgehog signaling.
MNR recruits OFD1 to regulate centriole length
CEP90 and MNR colocalize with their interactor OFD1 at the distal ends of centrioles. We found that distal centriole proteins are recruited to the centriole in a hierarchical manner. MNR forms the innermost ring at the distal centriole and is essential for the recruitment of OFD1 and CEP90, whereas CEP90 is dispensable for the recruitment of the other complex members. Overexpressed MNR localizes to microtubules (Chevrier et al., 2016) bringing along OFD1. Therefore, we propose that MNR binds to the distal centriolar microtubules to recruit OFD1, culminating in CEP90 recruitment (Fig. 10).
We previously demonstrated that OFD1 was essential to restrain centriolar length (Singla et al., 2010). We have found that one of its partners, MNR, is also critical to restrict centriolar elongation. However, its other partner, CEP90, does not control centriolar length. Thus, within the complex composed of MNR, OFD1, and CEP90, the subunits have distinct functions, with CEP90 dedicated to founding distal appendages. Consequently, the requirement for MNR and OFD1 in distal appendage assembly may be secondary to their roles in recruiting CEP90 to the distal centriole. The most parsimonious model is that MNR (and perhaps OFD1) at the distal centriole recruits CEP90 to build distal appendages and that MNR recruits OFD1 to restrict centriole elongation.
CEP90 recruits CEP83 to initiate distal appendage assembly at the mother centriole
The functions of centriolar satellites, although intimately associated with centrosomes, remain unclear (Odabasi et al., 2019; Prosser and Pelletier, 2020). CEP90 is a component of centriolar satellites, where it interacts with PCM1, the major scaffolding protein of centriolar satellites (Kim et al., 2012). Like CEP90, its interactors MNR and OFD1 are centriolar satellite proteins. As loss of CEP90 disrupts the pericentrosomal localization of centriolar satellites, we initially hypothesized that centriolar satellites transport CEP90 to centrioles or that CEP90 acts at centriolar satellites to transport distal appendage components to the mother centriole. In PCM1−/− RPE1 cells, centriolar satellites are disrupted but CEP90 remains at the distal centriole, indicating that satellites are dispensable for CEP90 localization at centrioles. Moreover, to our surprise, we found that PCM1−/− RPE1 cells assemble distal appendages. Thus, centriolar satellites (and CEP90 at centriolar satellites) are dispensable for distal appendage assembly, indicating that CEP90 functions at the distal centriole to build distal appendages.
A combination of expansion and structured illumination superresolution microscopy revealed that CEP90 decorates the distal centriole in a discontinuous ring-like pattern with ninefold symmetry; this ring of CEP90 is proximal to and smaller than the ring of CEP164. While antibodies to endogenous CEP90 label the daughter centriole more heavily than the mother centriole, epitope-tagged CEP90 localizes equivalently to both the mother and daughter centrioles. We speculate that reduced accessibility of the CEP90 antibody once distal appendages are assembled at the mother centriole may limit endogenous CEP90 immunofluorescence.
At the centriole, CEP90 interacts with and is critical for the recruitment of the distal appendage component CEP83. As CEP83 is at or near the root of the distal appendage (Tanos et al., 2013), CEP90 is required to initiate distal appendage formation. In support of this conclusion, no distal appendage-like structures were observable in serial-section transmission electron micrographs of cells lacking CEP90. These data raise the tantalizing possibility that the ninefold ring of CEP90 at the distal centriole templates the assembly of the distal appendages.
Superresolved microscopy also revealed that OFD1 and Talpid3 are structurally different in the mother and daughter centrioles, with the diameter of both rings increasing upon transition from daughter to mother. CEP90 is essential for OFD1 and Talpid3 rings to dilate to the mother centriole–specific diameter. We propose that this CEP90-dependent reorganization of distal centriole directs adoption of mother centriole–specific properties, including distal appendage assembly and ciliogenesis. Future work will focus on establishing how CEP90 reorganizes OFD1 and Talpid3 at the distal mother centriole to initiate distal appendage assembly.
In addition to MNR, OFD1, and CEP90, Talpid3 and C2CD3 localize to the distal centriole and are required for distal appendage formation (Singla et al., 2010; Thauvin-Robinet et al., 2014; Ye et al., 2014; Wang et al., 2018). We did not identify C2CD3 or Talpid3 in our proteomic analysis, suggesting that they might be members of a distinct DISCO (Tsai et al., 2019). Although CEP90 interacts with daughter centriole proteins CEP120 and Centrobin, Talpid3 recruitment, and the subsequent removal of Centrobin and CEP120 from the distal mother centriole, occur normally in CEP90−/− RPE1 cells. Therefore, CEP90 acts independently or downstream of Talpid3 recruitment and daughter centriole protein removal to regulate distal appendage assembly.
Mutations in CEP90 cause JBTS (Kodani et al., 2015; Wheway et al., 2015; Hebbar et al., 2018), while mutations in OFD1 and MNR cause orofaciodigital syndrome and JBTS (Singla et al., 2010; Stephen et al., 2017; Chevrier et al., 2016; Coene et al., 2009). JBTS and orofaciodigital syndrome have partially overlapping clinical features. Ultrastructural studies have identified a cogwheel-like structure at the distal domain of human centrioles (Paintrand et al., 1992; Ibrahim et al., 2009), raising the possibility that DISCO may comprise part of this structure. We propose that DISCO both controls centriolar length and builds distal appendages and that inherited defects in this distal centriolar cogwheel attenuate ciliogenesis, Hedgehog signaling, and embryonic patterning, resulting in JBTS and orofaciodigital syndrome.
Materials and methods
Mouse lines
Cep90 (Pibf1tm1.1(KOMP)Vlcg) and Mnr (4933427D14Riktm1.1(KOMP)Vlcg) mice, generated in a C57BL/6NJ background, were obtained from the International Mouse Phenotyping Consortium. Mice were housed in a barrier facility with veterinary supervision and given food and water ad libitum. All mouse protocols were approved by the Institutional Animal Care and Use Committee at the University of California, San Francisco and the University of Alabama, Birmingham.
Cell lines and cell culture
Human retinal epithelial (RPE1-hTERT) cells were cultured in DMEM/F12 (Thermo Fisher Scientific; catalog 10565042) supplemented with 10% FBS at 37°C in 5% CO2. To induce ciliation, cells were serum starved in Opti-MEM reduced serum media for indicated times. 293T cells were cultured in DMEM supplemented with 10% FBS and 1X GlutaMAX at 37°C in 5% CO2.
RPE1 cell lines stably expressing eYFP-CEP90 were generated using lentiviruses containing the human CEP90 cDNA (a gift from Kunsoo Rhee, Seoul National University, Seoul, South Korea; Kim and Rhee, 2011; Kim et al., 2012) in the pLVX-IRES-Puro (Clontech) plasmid background. CEP90−/− RPE1 cells expressing mNeonGreen-tagged full-length, truncation, and disease-associated mutation variants of CEP90 were generated using lentiviruses containing human CEP90 cDNA in a pLVX-EF1αΔ-mNeonGreen plasmid background. Lentiviruses were generated using the Lenti-X Packaging Single Shot system (Takara Bio) according to the manufacturer’s guidelines. Infected cells were plated in a glass-bottom 96-well plate (Cellvis; catalog P96-1.5H-N) using limiting dilution, and monoclonal cell lines expressing eYFP-CEP90 were manually selected based on fluorescence.
RPE1 cells stably expressing HA-CEP83 under the control of tetracycline-inducible promoter were a gift from Barbara Tanos (Brunel University, London, UK) and Meng-Fu Bryan Tsou (Sloan Kettering Institute, New York, NY; Tanos et al., 2013). CEP83 expression was induced with 1 µg/ml doxycycline (Fischer Chemical; catalog BP26535) for 48 h.
Cep90−/− and Mnr−/− MEFs were derived from E8.5 embryos along with littermate WT control MEFs. Cells were cultured in DMEM supplemented with 10% FBS (Invitrogen) and Glutamax-I (Invitrogen) and subsequently immortalized by transduction with SV40 large T antigen.
All cell lines were routinely tested for mycoplasma contamination and found negative.
Generation of CEP90−/−, MNR−/−, and PCM1−/− RPE1 cells by CRISPR-Cas9 gene targeting
RNA guided targeting of CEP90 and MNR was achieved by coexpression of Cas9 along with gRNAs. The pSpCas9 (BB)-2A-GFP (PX458) was a gift from Feng Zhang (Massachusetts Institute of Technology, Cambridge, MA; Addgene; plasmid #48138; http://n2t.net/addgene:48138; RRID:Addgene_48138). The gRNA sequences used for CEP90 and MNR were 5′-GATGAGGAAATATCATCCGT-3′ and 5′-GTATAAAATACCCGACCACA-3′, respectively.
PCM1−/− RPE1 cells were generated by electroporating recombinant Cas9 along with single guide RNAs (sgRNAs; Synthego; CRISPRevolution sgRNA EZ kit). The sgRNA targeting exon 4 of human PCM1: 5′-GAAAAGAAUAAGAAAAAGUU-3′ was used. 1.5 nmol sgRNA was resuspended in 15 µl nuclease-free Tris/EDTA buffer for a final concentration of 100 µM (100 pmol/µl). The RNP mixture containing 1.8 µl of sgRNA with 3 µl (90 pmol) Truecut Cas9 v2 (Thermo Fisher Scientific; catalog A36498) in a total volume of 5 µl was incubated at room temperature for 15 min. The RNP mixture was electroporated into hTERT-RPE1 cells using the Neon transfection system (Thermo Fisher Scientific) according to the manufacturer’s instructions using the following parameters: 1,350 V pulse voltage, 20 ms pulse width, and two pulses. All electroporated cells were serially diluted, and single colonies were screened by Western blotting, immunofluorescence, and PCR analyses. For genotyping, the following PCR primers were used: 5′-TGGGATGCACTAAATTGCCTA-3′ and 5′-TTACCTGCCGTTTGAAGACA-3′ for CEP90 alleles, 5′-TCCAGTGAACCAACTCACAGA-3′ and 5′-TAGGAGCGTGGCTGTGTCTAT-3′ for MNR alleles, and 5′-ACAGGCCATGTTAATTTTTGCT-3′ and 5′-CCATCCCCAGTGATTAAAATTC-3′ for PCM1 alleles. PCR products were cloned and sequenced.
Plasmids and transfections
MYC-DDK–tagged Mnr (4933427D14Rik) cDNA in cloned in a pCMV6 plasmid was obtained from OriGene (MR211309). hTERT-RPE1 cells were transfected using TransIT-LT1 transfection reagent (Mirus Bio) according to manufacturer guidelines.
For coimmunoprecipitation experiments in Fig. 5, C-terminal GFP-2x Strep-tagged full-length and truncation versions of CEP90 were cloned into pLVX-EF1α-IRES-Puro backbone and transfected into 293T cells using TransIT-293 transfection reagent (Mirus Bio) according to manufacturer guidelines.
Antibodies
Table S1 lists primary antibodies and the dilution at which they were used. Secondary antibodies conjugated to Alexa Fluor 488, 568, and 647 were purchased from Thermo Fisher Scientific and used at a 1:500 dilution.
Immunofluorescence
For immunostaining, cells were fixed with either 100% cold methanol for 3 min or 4% paraformaldehyde in Dulbecco's PBS for 15 min at room temperature and then incubated in blocking buffer (2.5% BSA and 0.1% Triton X-100 in PBS) for 1 h at room temperature. Paraformaldehyde fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min at room temperature before addition of blocking buffer. Coverslips were incubated with primary antibodies in blocking buffer overnight at 4°C, washed three times with PBS and incubated in secondary antibodies in blocking buffer for 1–2 h at room temperature. Nuclei were counterstained with Hoechst 33352 (Thermo Fisher Scientific; catalog H3570). Coverslips were washed three times with PBS and mounted with Prolong Diamond (Thermo Fisher Scientific; catalog P36961).
In experiments requiring disruption of the microtubule cytoskeleton, cells were treated with 20 µM nocodazole (Sigma-Aldrich; catalog SML1665) for 2 h at 37°C before fixation.
Identification of cell cycle stages was performed as described previously (Viol et al., 2020) using the Click-iT EdU Alexa Fluor 555 imaging kit (Life Technologies). Briefly, PCM1−/− RPE1 cells were treated with 10 µM EdU for 30 min and fixed in 100% cold methanol for 3 min. The Click-IT reaction was performed according to manufacturer guidelines, and samples were subsequently processed for indirect immunofluorescence. Cells lacking CENPF and EdU in the nucleus were categorized as G1. EdU-positive, CENPF-negative cells were categorized as being in S phase, while cells with nuclear CENPF staining and without EdU staining were classified as being in G2 phase.
Superresolution microscopy
3D-SIM and 2D-SIM was performed using the DeltaVision OMX-SR microscope (GE Healthcare) using the 60×/1.42 NA oil-immersion objective and three scientific complementary metal–oxide–semiconductor cameras. Immersion oil with refractive index of 1.518 was used for most experiments. Z stacks of 5–6 µm were collected using a 0.125 µm step size. Raw images were reconstructed using SoftWorx 6.5.2 (GE Healthcare) using default parameters.
Label-retention expansion microscopy was performed as described previously (Shi et al., 2019). Briefly, RPE1 cells were cultured on 16-well chambered slides (Sigma-Aldrich; catalog GBL112358-8EA) coated with 0.1% gelatin (Sigma-Aldrich; catalog G1393). Cells were serum starved in Opti-MEM to induce ciliation for 24 h before fixation. After incubation with primary antibodies, cells were incubated with appropriate secondary antibodies conjugated to N-hydroxysuccinimido–methacrylic acid–biotin or N-hydroxysuccinimido–methacrylic acid–digitonin. After gel polymerization and proteinase K digestion, gels were stained with fluorescently labeled streptavidin or digitonin antibodies and images were acquired on the DeltaVision OMX-SR.
Fluorescence intensity measurements and statistical analyses
For fluorescence intensity measurements, z stacks were acquired on the DeltaVision OMX-SR using widefield settings. Identical laser power and exposure settings were used for control and experimental samples. Average-intensity projections were generated using ImageJ (Schneider et al., 2012), and images were transferred to CellProfiler image analysis software (McQuin et al., 2018). Cilia number was quantified using Hoechst-stained nuclei to count the total number of cells and a cilia marker (ARL13B/TubulinAc) to identify cilia using the object identification module in CellProfiler using difference in signal intensity and size to segment cilia. For quantification of centrosomal intensity, a mask around the centrosomal area was generated using a centrosomal marker (e.g., γ-tubulin or FGFR1 Oncogene Partner [FOP]) to identify the centrosome in CellProfiler. This centrosomal mask was used to determine fluorescence intensity (integrated intensity) and area (in pixels) for the channel of interest. Fluorescence intensity values in a pericentrosomal area were used to measure background. Cells were serum starved for 24 h before fixing to synchronize cells in G0/G1 and eliminate cell cycle–dependent alterations in centrosomal proteins. Data were exported to Microsoft Excel, and graphs were generated in GraphPad Prism 8.
Statistical analyses were performed using GraphPad Prism 8. Results represented are mean ± SD. Statistical differences between datasets were analyzed using one-way ANOVA with Tukey’s multiple comparison tests or a two-tailed unpaired Student’s t test. Data distribution was assumed to be normal, but this was not formally tested. P value < 0.05 was considered significant and is indicated with an asterisk.
TEM
For EM, cells were plated on eight-well Permanox slides (Nunc; catalog177445), gently washed in 0.1 M phosphate buffer (PB), and fixed in 3.5% EM-grade glutaraldehyde (Electron Microscopy Sciences; catalog 16210) in 0.1 M PB for 10 min at 37°C. Fixative was removed and replaced with fresh fixative, and samples were incubated at 4°C for 1 h. Slides were washed three times with 0.1 M PB and processed for TEM as described previously (Singla et al., 2010).
Coimmunoprecipitation and Western blotting
Cells grown in 10-cm Petri dishes were washed twice with ice-cold Dulbecco's PBS and lysed for 30 min on ice with frequent pipetting. Lysis buffer used was 10 mM Tris/Cl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 0.5% Nonidet P40 substitute supplemented with 1x protease inhibitor (Roche; cOmplete mini, EDTA free, catalog 4693159001) and 1x phosphatase inhibitor (Thermo Fisher Scientific; PhosStop, catalog 4906845001) cocktails. Lysates were cleared by centrifugation at 17,000 g for 10 min at 4°C. GFP-trap magnetic agarose (Chromotek; catalog gtma-10) was used for eYFP-CEP90 coimmunoprecipitation assays and HA-magnetic beads (Pierce; catalog 88837) for CEP83-HA coimmunoprecipitation assays. Coimmunoprecipitation assays were performed according to manufacturer guidelines. Samples were separated on 4–15% Criterion TGX Precast gels (Bio-Rad) and transferred to Immobilon polyvinylidene fluoride membranes (0.45 µm pore size) for chemiluminescence detection.
Mass spectrometry and data analysis
Eluates obtained after coimmunoprecipitation were reduced by the addition of 1 mM DTT at 60°C for 15 min, cooled to room temperature, and alkylated by the addition of 3 mM iodoacetamide for 45 min in the dark. Alkylation was quenched by the addition of 3 mM DTT, and proteins were digested overnight at 37°C with 1 µg trypsin (0.5 µg/µl; Promega). Following digestion, peptides were acidified with trifluoroacetic acid (0.5% final, pH < 2), desalted using UltraMicroSpin Columns (The NEST Group; PROTO 300 C18 300 Å) according to the manufacturer’s specifications, and dried under vacuum centrifugation. Samples were resuspended in 4% formic acid, 4% acetonitrile solution and separated by a reversed-phase gradient over a nanoflow column (360-µm outer diameter × 75-µm inner diameter) packed with 25 cm of 1.8 µm Reprosil C18 particles (Dr. Maisch HPLC GmbH). The HPLC buffers were 0.1% formic acid and 100% acetonitrile on 0.1% formic acid for buffer A and B, respectively. The gradient was operated at 400 nl/min from 0 to 28% buffer B over 40 min, followed by a column wash at 95% B, with a total acquisition time of 50 min. Eluting peptides were analyzed in on a Bruke timsTOF Pro mass spectrometry system equipped with a Bruker nanoElute high-pressure liquid chromatography system interfaced via a captiveSpray source. A data-dependent parallel accumulation–serial fragmentation (PASEF) acquisition (Meier et al., 2018) method was used for data acquisition using the following parameters: 100–1,700 m/z range, 0.85–1.30 V*s/cm2 trapped ion mobility range, 1,600 V spray voltage, intensities of 200,000 were repeated for in PASEF 0 times, intensities of 100,000 to 200,000 were repeated for in PASEF five times, intensities of less than 100,000 were repeated for in PASEF 10 times, four PASEF tandem mass spectrometry scans with a total cycle time of 0.53 s, and active exclusion for 0.4 min. Data were searched against the human proteome database (canonical sequences downloaded from UniProt March 21, 2018) using MaxQuant (Cox and Mann, 2008; Prianichnikov et al., 2020). Peptide and protein identifications were filtered to a 1% false discovery rate at the peptide and protein level, and protein–protein interaction analysis was performed using SAINTexpress (Teo et al., 2014).
Online supplemental material
Fig. S1 depicts CRISPR-Cas9–mediated mutations of CEP90, MNR, and PCM1 and effects on corresponding proteins as assessed by immunoblotting. Fig. S2 shows that overexpressed MNR localizes to microtubules and sequesters endogenous OFD1. Fig. S3 shows that CEP90 and MNR regulate distal appendage assembly irrespective of whether the cell possesses a cilium. Fig. S4 includes serial-section TEMs of WT, Cep90−/−, and MNR−/− cells. Fig. S5 shows that CEP90 and MNR regulate distal appendage assembly independent of Talpid3 recruitment and removal of daughter centriole proteins. Table S1 lists all primary antibodies used in this study.
Supplementary Material
lists primary antibodies used in this study for immunofluorescence and Western blotting.
Acknowledgments
We thank E. Yu for help with animal husbandry and genotyping of mice used in this study. We thank members of the Reiter and Yoder laboratories for helpful discussions.
This work was supported by the National Institutes of Health (grant R01HD089918 to J.F. Reiter and B. Yoder, grants R01R01DE029454 and R01AR054396 to J.F. Reiter, Pathway to Independence Award K99GM126136/R00GM126136 to X. Shi, grant R01GM124334 to B. Huang); the Valencian Council for Innovation, Universities, Science and Digital Society (grant PROMETEO/2019/075 to J.M. Garcia-Verdugo); the Spanish Ministry of Science, Innovation and Universities (grant PCI2018-093062 to V. Herranz-Pérez); and the University of California, San Francisco Mary Anne Koda-Kimble Seed Award for Innovation (X. Shi). D. Kumar is supported a Jane Coffin Childs Memorial Fund for Medical Research postdoctoral fellowship and the Program for Breakthrough Biomedical Research Award, which is partially funded by the Sandler Foundation. B. Huang and J.F. Reiter are Chan Zuckerberg Initiative Biohub Investigators.
The authors declare no competing financial interests.
Author contributions: conceptualization, D. Kumar, B. Yoder, and J.F. Reiter; data acquisition, D. Kumar, A. Rains, V. Herranz-Pérez, Q. Lu, X. Shi, D.L. Swaney, and E. Stevenson; funding acquisition, D. Kumar, N.J. Krogan, C. Westlake, J.M. Garcia-Verdugo, B. Huang, B. Yoder, and J.F. Reiter; writing – original draft, D. Kumar; writing – review & editing, D. Kumar, A. Rains, V. Herranz-Pérez, B. Yoder, and J.F. Reiter.
Data availability
The mass spectrometry data files (raw and search results) have been deposited to the ProteomeXchange Consortium (Deutsch et al., 2017; Perez-Riverol et al., 2019; http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with dataset identifier PXD022372.
References
- Andersen, J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., and Mann M.. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 426:570–574. 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- Bowler, M., Kong D., Sun S., Nanjundappa R., Evans L., Farmer V., Holland A., Mahjoub M.R., Sui H., and Loncarek J.. 2019. High-resolution characterization of centriole distal appendage morphology and dynamics by correlative STORM and electron microscopy. Nat. Commun. 10:993. 10.1038/s41467-018-08216-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow, D.K., and Holland A.J.. 2019. Mechanism and Regulation of Centriole and Cilium Biogenesis. Annu. Rev. Biochem. 88:691–724. 10.1146/annurev-biochem-013118-111153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier, V., Bruel A.-L., Van Dam T.J.P., Franco B., Lo Scalzo M., Lembo F., Audebert S., Baudelet E., Isnardon D., Bole A., et al. 2016. OFIP/KIAA0753 forms a complex with OFD1 and FOR20 at pericentriolar satellites and centrosomes and is mutated in one individual with oral-facial-digital syndrome. Hum. Mol. Genet. 25:497–513. 10.1093/hmg/ddv488 [DOI] [PubMed] [Google Scholar]
- Chong, W.M., Wang W.-J., Lo C.-H., Chiu T.-Y., Chang T.-J., Liu Y.-P., Tanos B., Mazo G., Tsou M.B., Jane W.-N., et al. 2020. Super-resolution microscopy reveals coupling between mammalian centriole subdistal appendages and distal appendages. eLife. 9:e53580. 10.7554/eLife.53580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli, F.D., Doerks T., Von Mering C., Creevey C.J., Snel B., and Bork P.. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science. 311:1283–1287. [DOI] [PubMed] [Google Scholar]
- Coene, K.L.M., Roepman R., Doherty D., Afroze B., Kroes H.Y., Letteboer S.J.F., Ngu L.H., Budny B., van Wijk E., Gorden N.T., et al. 2009. OFD1 is mutated in X-linked Joubert syndrome and interacts with LCA5-encoded lebercilin. Am. J. Hum. Genet. 85:465–481. 10.1016/j.ajhg.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit, K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y.R., and Reiter J.F.. 2005. Vertebrate Smoothened functions at the primary cilium. Nature. 437:1018–1021. 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Cox, J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Deutsch, E.W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D.S., Bernal-Llinares M., Okuda S., Kawano S., et al. 2017. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45(D1):D1100–D1106. 10.1093/nar/gkw936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., Song Y., Shen M., Xie S., Li W., Lu Y., Yang Y., Ou G., Zhou J., Wang F., et al. 2017. Microtubule-binding protein FOR20 promotes microtubule depolymerization and cell migration. Cell Discov. 3:17032. 10.1038/celldisc.2017.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring, N.W., and Hyman A.A.. 2012. Organelle growth control through limiting pools of cytoplasmic components. Curr. Biol. 22:R330–R339. 10.1016/j.cub.2012.03.046 [DOI] [PubMed] [Google Scholar]
- Gönczy, P.2015. Centrosomes and cancer: revisiting a long-standing relationship. Nat. Rev. Cancer. 15:639–652. 10.1038/nrc3995 [DOI] [PubMed] [Google Scholar]
- Hammarsjö, A., Wang Z., Vaz R., Taylan F., Sedghi M., Girisha K.M., Chitayat D., Neethukrishna K., Shannon P., Godoy R., et al. 2017. Novel KIAA0753 mutations extend the phenotype of skeletal ciliopathies. Sci. Rep. 7:15585. 10.1038/s41598-017-15442-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbar, M., Kanthi A., Shukla A., Bielas S., and Girisha K.M.. 2018. A biallelic 36-bp insertion in PIBF1 is associated with Joubert syndrome. J. Hum. Genet. 63:935–939. 10.1038/s10038-018-0462-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori, A., and Toda T.. 2017. Regulation of centriolar satellite integrity and its physiology. Cell. Mol. Life Sci. 74:213–229. 10.1007/s00018-016-2315-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu, D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., and Anderson K.V.. 2003. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 426:83–87. 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Hunkapiller, J., Singla V., Seol A., and Reiter J.F.. 2011. The ciliogenic protein Oral-Facial-Digital 1 regulates the neuronal differentiation of embryonic stem cells. Stem Cells Dev. 20:831–841. 10.1089/scd.2010.0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, R., Messaoudi C., Chichon F.J., Celati C., and Marco S.. 2009. Electron tomography study of isolated human centrioles. Microsc. Res. Tech. 72:42–48. 10.1002/jemt.20637 [DOI] [PubMed] [Google Scholar]
- Jakobsen, L., Vanselow K., Skogs M., Toyoda Y., Lundberg E., Poser I., Falkenby L.G., Bennetzen M., Westendorf J., Nigg E.A., et al. 2011. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 30:1520–1535. 10.1038/emboj.2011.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, L.C., Romijn E.P., Zamora I., Yates J.R. III, and Marshall W.F.. 2005. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15:1090–1098. 10.1016/j.cub.2005.05.024 [DOI] [PubMed] [Google Scholar]
- Keller, L.C., Geimer S., Romijn E., Yates J. III, Zamora I., and Marshall W.F.. 2009. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell. 20:1150–1166. 10.1091/mbc.e08-06-0619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K., and Rhee K.. 2011. The pericentriolar satellite protein CEP90 is crucial for integrity of the mitotic spindle pole. J. Cell Sci. 124:338–347. 10.1242/jcs.078329 [DOI] [PubMed] [Google Scholar]
- Kim, K., Lee K., and Rhee K.. 2012. CEP90 is required for the assembly and centrosomal accumulation of centriolar satellites, which is essential for primary cilia formation. PLoS One. 7:e48196. 10.1371/journal.pone.0048196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani, A., Yu T.W., Johnson J.R., Jayaraman D., Johnson T.L., Al-Gazali L., Sztriha L., Partlow J.N., Kim H., Krup A.L., et al. 2015. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. eLife. 4:e07519. 10.7554/eLife.07519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, D., Sahabandu N., Sullenberger C., Vásquez-Limeta A., Luvsanjav D., Lukasik K., and Loncarek J.. 2020. Prolonged mitosis results in structurally aberrant and over-elongated centrioles. J. Cell Biol. 219:e201910019. 10.1083/jcb.201910019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Q., Insinna C., Ott C., Stauffer J., Pintado P.A., Rahajeng J., Baxa U., Walia V., Cuenca A., Hwang Y.S., et al. 2015. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 17:228–240. 10.1038/ncb3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub, M.R., Xie Z., and Stearns T.. 2010. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J. Cell Biol. 191:331–346. 10.1083/jcb.201003009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteil, G., Guerrero A., Vieira A.F., de Almeida B.P., Machado P., Mendonça S., Mesquita M., Villarreal B., Fonseca I., Francia M.E., et al. 2018. Over-elongation of centrioles in cancer promotes centriole amplification and chromosome missegregation. Nat. Commun. 9:1258. 10.1038/s41467-018-03641-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazo, G., Soplop N., Wang W.-J., Uryu K., and Tsou M.F.. 2016. Spatial Control of Primary Ciliogenesis by Subdistal Appendages Alters Sensation-Associated Properties of Cilia. Dev. Cell. 39:424–437. 10.1016/j.devcel.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin, C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D., et al. 2018. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 16:e2005970. 10.1371/journal.pbio.2005970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier, F., Brunner A.D., Koch S., Koch H., Lubeck M., Krause M., Goedecke N., Decker J., Kosinski T., Park M.A., et al. 2018. Online parallel accumulation–serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics. 17:2534–2545. 10.1074/mcp.TIR118.000900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg, E.A., and Holland A.J.. 2018. Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19:297–312. 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odabasi, E., Gul S., Kavakli I.H., and Firat-Karalar E.N.. 2019. Centriolar satellites are required for efficient ciliogenesis and ciliary content regulation. EMBO Rep. 20:e47723. 10.15252/embr.201947723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintrand, M., Moudjou M., Delacroix H., and Bornens M.. 1992. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 108:107–128. 10.1016/1047-8477(92)90011-X [DOI] [PubMed] [Google Scholar]
- Perez-Riverol, Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47(D1):D442–D450. 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prianichnikov, N., Koch H., Koch S., Lubeck M., Heilig R., Brehmer S., Fischer R., and Cox J.. 2020. Maxquant software for ion mobility enhanced shotgun proteomics. Mol. Cell. Proteomics. 19:1058–1069. 10.1074/mcp.TIR119.001720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser, S.L., and Pelletier L.. 2020. Centriolar satellite biogenesis and function in vertebrate cells. J. Cell Sci. 133:jcs239566. 10.1242/jcs.239566 [DOI] [PubMed] [Google Scholar]
- Reiter, J.F., and Leroux M.R.. 2017. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 18:533–547. 10.1038/nrm.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, K.N., Kuhns S., Neuner A., Hub B., Zentgraf H., and Pereira G.. 2012. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J. Cell Biol. 199:1083–1101. 10.1083/jcb.201202126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y., Wang H., Liu Z., Luo M., Ma S., Lu C., Cao Z., Yu Y., Cai R., Chen C., et al. 2020. Identification of two novel pathogenic variants of PIBF1 by whole exome sequencing in a 2-year-old boy with Joubert syndrome. BMC Med. Genet. 21:192. 10.1186/s12881-020-01130-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X., Garcia G. III, Van De Weghe J.C., McGorty R., Pazour G.J., Doherty D., Huang B., and Reiter J.F.. 2017. Super-resolution microscopy reveals that disruption of ciliary transition-zone architecture causes Joubert syndrome. Nat. Cell Biol. 19:1178–1188. 10.1038/ncb3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, X., Li Q., Dai Z., Tran A., Feng S., Ramirez A.D., Lin Z., Wang X., Chow T.T., Seiple I.B., and Huang B.. 2019. Label-retention expansion microscopy. bioRxiv. 687954. doi:. 10.1101/687954 [DOI] [PMC free article] [PubMed]
- Sillibourne, J.E., Hurbain I., Grand-Perret T., Goud B., Tran P., and Bornens M.. 2013. Primary ciliogenesis requires the distal appendage component Cep123. Biol. Open. 2:535–545. 10.1242/bio.20134457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla, V., Romaguera-Ros M., Garcia-Verdugo J.M., and Reiter J.F.. 2010. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell. 18:410–424. 10.1016/j.devcel.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, S., and Panda D.. 2017. A centrosomal protein FOR20 regulates microtubule assembly dynamics and plays a role in cell migration. Biochem. J. 474:2841–2859. 10.1042/BCJ20170303 [DOI] [PubMed] [Google Scholar]
- Stephen, J., Vilboux T., Mian L., Kuptanon C., Sinclair C.M., Yildirimli D., Maynard D.M., Bryant J., Fischer R., Vemulapalli M., et al. NISC Comparative Sequencing Program . 2017. Mutations in KIAA0753 cause Joubert syndrome associated with growth hormone deficiency. Hum. Genet. 136:399–408. 10.1007/s00439-017-1765-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos, B.E., Yang H.J., Soni R., Wang W.J., Macaluso F.P., Asara J.M., and Tsou M.F.B.. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27:163–168. 10.1101/gad.207043.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo, G., Liu G., Zhang J., Nesvizhskii A.I., Gingras A.C., and Choi H.. 2014. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics. 100:37–43. 10.1016/j.jprot.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauvin-Robinet, C., Lee J.S., Lopez E., Herranz-Pérez V., Shida T., Franco B., Jego L., Ye F., Pasquier L., Loget P., et al. 2014. The oral-facial-digital syndrome gene C2CD3 encodes a positive regulator of centriole elongation. Nat. Genet. 46:905–911. 10.1038/ng.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, J.J., Hsu W.B., Liu J.H., Chang C.W., and Tang T.K.. 2019. CEP120 interacts with C2CD3 and Talpid3 and is required for centriole appendage assembly and ciliogenesis. Sci. Rep. 9:6037. 10.1038/s41598-019-42577-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viol, L., Hata S., Pastor-Peidro A., Neuner A., Murke F., Wuchter P., Ho A.D., Giebel B., and Pereira G.. 2020. Nek2 kinase displaces distal appendages from the mother centriole prior to mitosis. J. Cell Biol. 219:e201907136. 10.1083/jcb.201907136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Lee K., Malonis R., Sanchez I., and Dynlacht B.D.. 2016. Tethering of an E3 ligase by PCM1 regulates the abundance of centrosomal KIAA0586/Talpid3 and promotes ciliogenesis. eLife. 5:e12950. 10.7554/eLife.12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Failler M., Fu W., and Dynlacht B.D.. 2018. A distal centriolar protein network controls organelle maturation and asymmetry. Nat. Commun. 9:3938. 10.1038/s41467-018-06286-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassie, A.T., Zhao Y., and Boyden E.S.. 2019. Expansion microscopy: principles and uses in biological research. Nat. Methods. 16:33–41. 10.1038/s41592-018-0219-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlake, C.J., Baye L.M., Nachury M.V., Wright K.J., Ervin K.E., Phu L., Chalouni C., Beck J.S., Kirkpatrick D.S., Slusarski D.C., et al. 2011. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc. Natl. Acad. Sci. USA. 108:2759–2764. 10.1073/pnas.1018823108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheway, G., Schmidts M., Mans D.A., Szymanska K., Nguyen T.T., Racher H., Phelps I.G., Toedt G., Kennedy J., Wunderlich K.A., et al. University of Washington Center for Mendelian Genomics . 2015. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat. Cell Biol. 17:1074–1087. 10.1038/ncb3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey, M., and O’Toole E.. 2014. Centriole structure. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130457. 10.1098/rstb.2013.0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.T., Chen H.Y., and Tang T.K.. 2018. Myosin-Va is required for preciliary vesicle transportation to the mother centriole during ciliogenesis. Nat. Cell Biol. 20:175–185. 10.1038/s41556-017-0018-7 [DOI] [PubMed] [Google Scholar]
- Yang, T.T., Chong W.M., Wang W.J., Mazo G., Tanos B., Chen Z., Tran T.M.N., Chen Y.D., Weng R.R., Huang C.E., et al. 2018. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 9:2023. 10.1038/s41467-018-04469-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, X., Zeng H., Ning G., Reiter J.F., and Liu A.. 2014. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc. Natl. Acad. Sci. USA. 111:2164–2169. 10.1073/pnas.1318737111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, L., Stephens A., Nam S.Z., Rau D., Kübler J., Lozajic M., Gabler F., Söding J., Lupas A.N., and Alva V.. 2018. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 430:2237–2243. 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists primary antibodies used in this study for immunofluorescence and Western blotting.
Data Availability Statement
The mass spectrometry data files (raw and search results) have been deposited to the ProteomeXchange Consortium (Deutsch et al., 2017; Perez-Riverol et al., 2019; http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with dataset identifier PXD022372.