Abstract
Background
Immunological stress decreases feed intake, suppresses growth and induces economic losses. However, the underlying molecular mechanism remains unclear. Label-free liquid chromatography and mass spectrometry (LC-MS) proteomics techniques were employed to investigate effects of immune stress on the hepatic proteome changes of Arbor Acres broilers (Gallus Gallus domesticus) challenged with Escherichia coli lipopolysaccharide (LPS).
Results
Proteomic analysis indicated that 111 proteins were differentially expressed in the liver of broiler chickens from the immune stress group. Of these, 28 proteins were down-regulated, and 83 proteins were up-regulated in the immune stress group. Enrichment analysis showed that immune stress upregulated the expression of hepatic proteins involved in defense function, amino acid catabolism, ion transport, wound healing, and hormone secretion. Furthermore, immune stress increased valine, leucine and isoleucine degradation pathways.
Conclusion
The data suggests that growth depression of broiler chickens induced by immune stress is triggered by hepatic proteome alterations, and provides a new insight into the mechanism by which immune challenge impairs poultry production.
Keywords: Broiler chickens, Hepatic proteome, Immune stress, Lipopolysaccharide
Background
Intensive poultry production is conducted in an environment that imposes many stressors on the bird. The stressed bird instigates an integrated response to maintain homeostasis through cross-talk between the central nervous, endocrine and immune systems [1]. Stressors in the bird’s environment, include feeding management, overcrowding, temperature extremes, dust and litter condition, pathogen challenges, vaccination, and psychological factors [2, 3]. All of these stressors can have a cumulative impact on poultry behavior and physiology, thus affecting the immune response and inducing immunologically mediated stress or immune stress [4, 5]. Immune stress is harmful to the bird and can be mitigated by improving the bird’s environment. Studies have shown that stress dysregulates the immune response by increasing the release of inflammatory cytokines and stress hormones [6, 7], reducing NK cell activity, lymphocyte populations, lymphocyte proliferation, antibody production and reactivating latent viral infections [8, 9]. In response to an immune challenge, the appetite and growth performance of the bird will decline [10–12]. Immune stress can also disrupt the balance and composition of the cecal microflora, impair intestinal mucosal immune function, and reduce ileal protein digestibility [13].
A bird or animal’s metabolic priorities are rearranged in response to immune stress, resulting in the redistribution of nutrients away from muscle protein deposition and growth to support upregulation of the immune response [14, 15]. The liver plays a pivotal role in nutrient metabolism, and nutrient repartitioning following an immune challenge when it is enriched with components of the immune system, including macrophages and natural killer T-cells; highlighting the vital role of the liver in immunology [16].
Despite extensive research on the effects of immune stress in broiler chickens, changes in the avian hepatic proteome and the molecular mechanisms induced by an immune insult are not well understood. Proteins as the functional carrier of genes can provide both genomic and functional information [17]. Proteomics represents a new strategy to delineate the molecular basis of the physiological changes in the liver during chicken growth [18, 19]. This approach determines the differential patterns of protein abundance and has been used to demonstrate their functional relationships to external factors [20, 21]. Lipopolysaccharide (LPS) injection is a classical model for inducing immune stress in broiler chickens [7, 10, 22, 23]. In the present experiment, this model was used to investigate the hypothesis that changes in the expression of the hepatic proteome of broilers occur following immune challenge and help explain the response of the bird. Our findings clarify protein expression and biological process changes in the liver of immunologically challenged broilers and provides further information to assist in maintaining the health and productivity of meat or broiler chickens.
Methods
Materials and reagents
All chemicals were purchased from Sigma-Aldrich (St. Louis., Missouri, USA) except modified sequencing grade trypsin that was bought from Promega (Madison, WI, USA). LPS from E. coli (O55:B5) was used in the present experiment.
Bird management
A total of 144 one-day-old, male, Arbor Acres (AA) broiler chickens were purchased from Huadu Chicken Co. (Beijing, China). The chicks were randomly divided into two groups: challenged with saline (control group) or LPS (treatment group). Each group had 6 replicates with 12 birds in each replicate. The distribution of cages was arranged to avoid any location effects within the poultry house. The chickens were reared in two phases and fed a starter diet during d 0–21 and a grower diet during d 22–42. The composition of these corn-soybean based diets are shown in Table 1. All chickens were inoculated and subjected to a photoperiod of 16 h light and 8 h dark in accordance with the AA Broiler Management Guide. The room temperature was maintained at 33–35 °C on d 0–3, at 32–34 °C on d 4–7 and gradually reduced to the maintenance temperature of 20 °C by d 42. The relative humidity was kept at 70% during the first week and thereafter at about 60%.
Table 1.
Ingredient and nutrient composition of experimental broiler diets
| Starter (1–21 d), g/kg | Grower (22–42 d), g/kg | |
|---|---|---|
| Ingredient | ||
| Corn | 593.1 | 604.2 |
| Soybean meal | 298.8 | 288.7 |
| Cotton seed meal | 50.0 | 30.0 |
| Soybean oil | 15.1 | 39.8 |
| L-Lysine | 1.5 | 0.9 |
| DL-Methionine | 1.4 | 1.6 |
| Limestone | 12.7 | 10.2 |
| CaHPO4 | 19.4 | 16.6 |
| NaCl | 3.0 | 3.0 |
| Choline chloride | 2.0 | 2.0 |
| Vitamin premix | 0.3 | 0.3 |
| Mineral premixa | 1.0 | 1.0 |
| Zeolite powder | 1.7 | 1.7 |
| Total | 1000 | 1000 |
| Nutrient concentrationsb | ||
| Metabolic energy, MJ/kg | 12.35 | 13.02 |
| Crude protein | 211.8 | 198.4 |
| Calcium | 10.1 | 8.5 |
| Available phosphorus | 4.5 | 4.0 |
| Total phosphorus | 6.9 | 6.3 |
| Lysine | 11.4 | 10.5 |
| Methionine | 4.9 | 4.8 |
| Methionine + Cysteine | 8.3 | 8.1 |
| Threonine | 7.7 | 2.2 |
aThe premix provided the following per kg diet: vitamin A 10, 000 IU, vitamin D3 2,000 IU, vitamin E 10 IU, vitamin K3 2.5 mg, vitamin B1 1 mg, vitamin B2 6 mg, vitamin B3 10 mg, vitamin B5 40 mg, vitamin B6 3 mg, vitamin B11 0.3 mg, vitamin B12 0.01 mg, biotin 0.12 mg, Cu (as copper sulfate) 8 mg, Fe (as ferrous sulfate) 80 mg, Mn (as manganese sulfate) 60 mg, Zn (as zinc sulfate) 40 mg, Se (as sodium selenite) 0.15 mg, I (as potassium iodide) 0.35 mg
bCalculated values
Experimental treatments and LPS administration
For the first 5 weeks of the study all birds were maintained in a similar manner. On d 36, 38, and 40, all chickens (each group had 6 replicates with 12 birds in each replicate) were injected intravenously with either 1 mL sterile saline (control group) or LPS (treatment group) dissolved in saline at an approximate dose of 5.0 mg/kg body weight (LPS or immune stress group). The injection protocol is the established method used when inducing an immunological challenge with LPS [4, 7, 10, 22, 23]. The protocol commenced at 5 weeks of age to avoid endocrine and physiological changes that occur during the starter phase and to permit additional muscle samples to be collected for meat analysis; results reported separately.
Performance parameters
The body weight of all birds in each replicate was measured on d 36 (before the first injection, W0), d 38 (2 days after the first injection, W38), d 40 (2 days after the second injection, W40) and d 42 (2 days after the third injection, W42). The change in body weight caused by saline or LPS treatment was expressed as body weight gain (W1 = W38- W0, W2 = W40- W0, W3 = W42- W0). W1–3 indicates body weight gain after the first, second or third injection of LPS. Mortality was recorded daily.
Sample collection and parameters determined in blood
On d 42, all birds were weighed after a 12 h-fast. Three birds from each replicate were selected randomly, electrically stunned, and manually slaughtered within 5 min [24]. Blood was collected using vacutainer tubes. The serum, obtained by centrifugation at 1,500 × g for 15 min, was used for the determination of hormones and inflammatory factors. The concentrations of adrenocorticotropic hormone (ACTH), corticosterone (CORT), growth hormone (GH) and insulin-like growth factor-1 (IGF-1), interleukin-1β (IL-1β), interleukin-6 (IL-6), and tumour necrosis factor-α (TNF-α) were determined by quantitative sandwich enzyme immunoassay using commercial kits (Beijing North Institute of Biological Technology, Beijing, China), according to the manufacturer’s instructions.
The middle section of the major or right lobe of the liver was sampled and washed with PBS buffer (NaCl 8 g/L, Na2HPO4 1.44 g/L, KH2PO4 0.24 g/L, KCl 0.2 g/L, pH 7.2) to remove any blood and contaminants on the surface. A liver sample (about 2 g) was taken and put into 5 mL ultra-low temperature freezing tubes (Free Sterile). Samples were immediately frozen in liquid nitrogen and stored at − 80 °C. Likewise, intestinal and muscle samples were also collected and the outcome of their analyses will be published elsewhere.
Protein extraction and digestion
The liver samples of three chickens from each replicate (cage) were combined as a biological replicate, homogenized by pestle in liquid nitrogen. Six biological replicates of each group were analyzed. Protein extraction was performed as previously described [18]. In short, after homogenization the samples were then mixed with a lysis buffer containing 8 mol urea, 2 mol thiourea, 4% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate, 20 mmol Trisbase, 30 mmol dithiothreitol (DTT), and protease inhibitors in ice for 30 min. The sample was then centrifuged at 15,000 × g for 20 min at 10 °C to remove the insoluble fractions. Three volumes of ice-cold acetone were added to the recovered supernatant and allowed to stand at 20 °C for 4 h to precipitate the proteins. Subsequently, the protein pellets were centrifuged at 8,000 × g at 10 °C for 20 min. The supernatant was discarded, followed by extraction of the protein pellet at room temperature. The recovered proteins were re-suspended in 100–150 μL of 5 mol urea, and protein concentration was quantified by the Bradford assay after diluting 50 times. Of each sample, 200 μg of proteins were used by adding four volumes of 40 mmol NH4HCO3, mixing with DTT (final concentration 10 mmol) for 1 h, and then alkylating with iodoacetamide (final concentration 50 mmol) for 1 h in the dark. The surplus iodoacetamide was quenched by DTT (final concentration 30 mmol). To digest protein into peptides, sequencing grade modified trypsin was used (enzyme/protein ratio of 1:100 (W/W)) at 37 °C for 14 h. The enzymatic digestion was stopped by adding 1 μL of formic acid to the solution. The digested peptide samples were desalted using a C18 column (Agilent Technologies Inc., Santa Clara, CA, USA). The eluted peptide solution was collected and extracted using a SpeedVac system (RVC 2–18, Marin Christ, Osterod, Germany) and stored at −80 °C for subsequent LC-MS/MS analysis.
Liquid chromatography and mass spectrometry (LC − MS/MS) analysis
The digested peptide samples were re-dissolved in 50 μL of 0.1% formic acid. Three replicates of each sample were run using a Q-Exactive mass spectrometer (Thermo Fisher Scientific, USA) and coupled to the EASY-nLC 1000 system using a nano electrospray ion source (Thermo Fisher Scientific, USA). To enrich the peptide samples, they were first loaded onto a 2 cm long trap column (75 μm inner diameter fused silica containing 3 μm Aqua C18 beads, Thermo Fisher Scientific, USA) for 2 min in buffer A (0.1% acetic acid) at a flow rate of 10 μL/min. Secondly, the peptides were separated by an analytical column (15 cm long, 50 μm inner diameter fused silica column filing with 2 μm Aqua C18 beads, Thermo Fisher Scientific, USA) using a 120 min gradient. Peptides were gradient eluted for 110 min with a linear gradient from 8% to 30% acetonitrile at a flow rate of 300 nL/min. The eluting peptides from the analytical column were directly infused into a Q-Exactive mass spectrometer via electrospray ionization. The settings for a data-dependent mode to collect the MS and MS/MS data were as follows: one full scan (resolution 70,000 at 400 m/z; 350 to 1,600 m/z) followed by top 20 MS/MS scans using higher-energy collisional dissociation in the linear ion trap mass spectrometer (resolution: 15,000, isolation window: 2 m/z, normalized collision energy: 28) using dynamic exclusion (charge exclusion: unassigned 1, > 8; peptide match: preferred; exclude isotopes: on; dynamic exclusion: 30 s). For identification and abundance level quantification of proteins, the MS/MS data in RAW were retrieved using Xcalibur (version 3.0, Thermo Fisher Scientific, USA) and searched using in-house PEAKS software (version 8.5, Bioinformatics Solutions Inc., CAN).
A database containing protein sequences of Gallus Gallus domesticus including common contaminants was downloaded from NCBI and used, totaling to 76,213 entries (downloaded 25 June, 2020). The parameters of the search database were as follows: trypsin; maximum missed cleavage: 2; precursor ion and MS/MS tolerances: 15 ppm and 0.05 Da; a fixed modification: carbamidomethyl (C, + 57.02); and a variable modification: methionine oxidation (M, + 15.99), asparagine and glutamine deamination (+ 0.984 Da). The fusion-decoy database search strategy with threshold false discovery rate (FDR ≤ 1%) was used to control the FDR at both the protein and peptide levels. A protein was considered as identified only if it had at least one unique peptide. To quantify the relative protein abundance in the livers of broiler chickens both from the control group and immune stress group, three replications of each sample were performed in the quantification module of PEAKS software (version 8.5) via a label-free strategy. Feature detection was performed separately on each sample using the expectation-maximization algorithm. Using the high-performance retention time alignment algorithms, the features of the same peptide from three replicates of each sample were reliably aligned [25]. Normalization was done by dividing each matrix by a factor of the samples obtained as follows: the total ion current (TIC) of the individual sample / the TIC of the reference sample. Quantification of protein abundance in the livers in all samples of broiler chickens was done using the sum of the three highest ion peak intensities of the tryptic peptides.
GO term enrichment analysis
To understand the biological implications of the identified proteins in the liver of broiler chickens, identifiers of protein symbol ID numbers were used as an input for GO term enrichment (functional classes and pathway) using ClueGOv2.3.2, a Cytoscape plug-in (http://www.ici.upmc.fr/cluego/) [26]. The number of proteins identified from the samples was compared with the number of functionally GO annotated proteins in the entire broiler chicken (Gallus Gallus domesticus) genome for enrichment analysis. The significantly enriched GO terms in biological processes and pathways were reported using a right-sided hyper-geometric test and only a P-value < 0.05 was considered. Then, Bonferroni step-down procedure was used to correct the P-value to control FDR. Functional grouping of the terms was based on GO hierarchy. The tree level was ranged from 3 to 8, and kappa score level was 0.4. For comparison purpose, sharing 65% of the terms was considered to be merged.
Protein–protein interaction analysis
A protein–protein interaction network of differential proteins was constructed using the STRING 11.0 (http://string-db.org/) [27]. The network nodes represent proteins, and the edges represent the predicted functional associations.
Statistical analysis
Means of replicate were used as the experimental unit for statistical analysis. The data of blood parameters were analyzed by Independent-Samples T-Test module using SPSS 17.0 software (version 17.0, SPSS Inc., Chicago, IL, USA). Results are presented as the mean ± SE. Differences between means were considered statistically significant at P < 0.05.
Proteins from different samples were considered to be significantly changed in their abundance only when they attained the criteria (P-value < 0.05 and a fold change of > 1.5 or < 0.5).
Results
Growth and wellbeing of all chicks was normal for the first 5 weeks of the study or until the LPS challenge was introduced.
Effects of body weight gain of broilers challenged with LPS
The effects of immune stress on body weight gain of broilers is shown in Table 2. Body weight gain in broilers injected with LPS was significantly lower than in the unchallenged broilers.
Table 2.
Body weight gain of broilers challenged with LPS
| TREATMENT | W0, g | W1, g | W2, g | W3, g |
|---|---|---|---|---|
| Control group | 1966 ± 116 | 182 ± 9.7a | 389 ± 22.9a | 423 ± 27.6a |
| Immune stress group | 1966 ± 107 | −22.7 ± 26.9b | 97.9 ± 46.4b | 112.3 ± 46.7b |
| P-value | 0.49 | 0.0001 | 0.0001 | 0.0001 |
W0, Initial body weight before injection of LPS; W1, Body weight gain 2 days after the first injection of LPS; W2, Body weight gain 2 days after the second injection of LPS; W3, Body weight gain 2 days after the third injection of LPS
a,b In the same column, values with the same or no letter superscripts mean no significant difference (P > 0.05), while with different letter superscripts mean significant difference (P < 0.05)
Changes of serum hormones and cytokines of broilers challenged with LPS
As shown in Table 3, the serum concentrations of ACTH, CORT, IL-1β, TNF-α and IL-6 in broilers injected with LPS were significantly higher than in the unchallenged broilers. However, GH and IGF- І concentrations in serum decreased significantly in the broilers from the immune stress group.
Table 3.
The concentrations of serum hormones and cytokines in broilers challenged with LPS
| TNF-α, fmol/mL | IL-1β, pg/mL | IL-6, ng/mL | GH, ng/mL | CORT, pg/mL | ACTH, pg/mL | IGF-І, ng/mL | |
|---|---|---|---|---|---|---|---|
| Control | 5.88 ± 0.09a | 0.087 ± 0.006a | 60.06 ± 6.87a | 1.37 ± 0.11a | 8.36 ± 0.67a | 5.91 ± 0.63a | 80.46 ± 4.78b |
| LPS | 9.45 ± 0.55b | 0.223 ± 0.041b | 83.93 ± 2.30b | 1.12 ± 0.03b | 10.26 ± 0.35b | 8.24 ± 0.83b | 71.53 ± 3.48a |
| P-value | 0.000 | 0.000 | 0.000 | 0.000 | 0.047 | 0.047 | 0.030 |
a,b In the same column, values with the same or no letter superscripts mean no significant difference (P > 0.05), while with different letter superscripts mean significant difference (P < 0.05)
Qualitative differential analysis of hepatic proteome in broiler chickens between the control and the immune stress group
Protein numbers expressed in the liver of broiler chickens
In the present study, a total of 4,966 proteins were identified in the liver tissues of broiler chickens. In the control group, 4,285 proteins (2,307 groups) were identified and 4,010 proteins (2,182 groups) were identified in the LPS group. As shown in Figs. 1, 3,329 proteins were expressed in both the control and treatment groups.
Fig. 1.
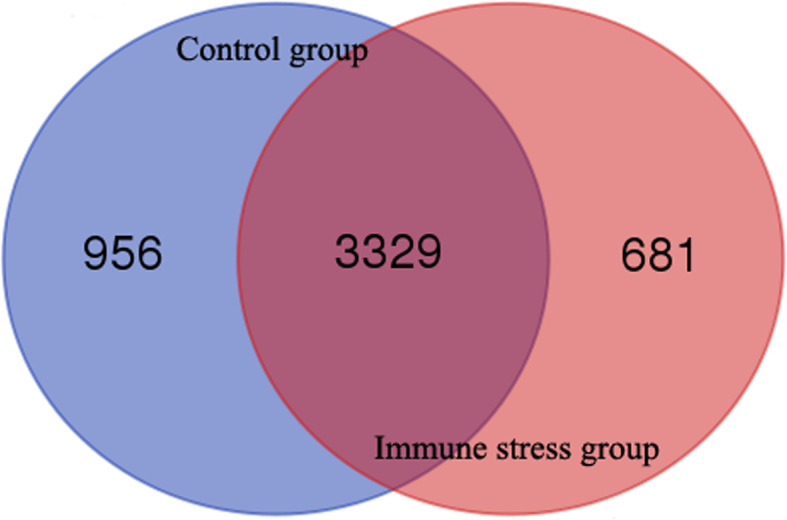
Venn diagram of the number of proteins expressed in the liver of broiler chickens in the control group and immune stress group
GO and KEGG analysis of unique proteins specially expressed in the control group
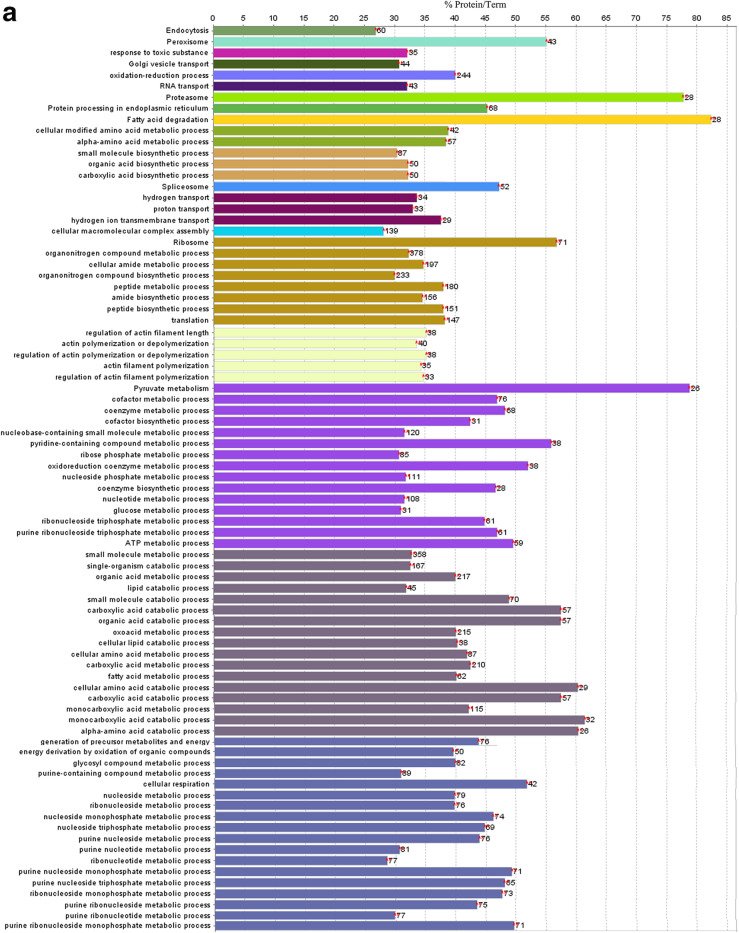
As shown in Fig. 2a, KEGG pathway analysis was performed on specifically expressed proteins in the control group and demonstrated enrichment of endocytosis, peroxisome, Golgi vesicle transport, RNA transport, proteasome, protein processing in endoplasmic reticulum, fatty acid degradation, spliceosome, ribosome and pyruvate metabolism pathways.
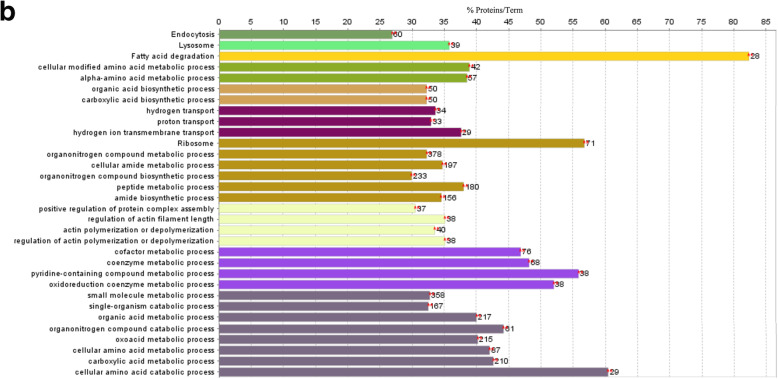
Fig. 2.
Qualitative proteome comparisons of the liver of broiler chickens in the control group and the immune stress group. a and b, GO and KEGG annotation of unique proteins specifically expressed in the control group and the immune stress group, respectively. Terms that begin with an uppercase or lowercase letters are KEGG or GO annotation, respectively. % Proteins/Term stands for the proportion of genes enriched in corresponding functional groups. The bars with the same color represent the same functional groups they belong to. The numbers stand for the genes enriched to the corresponding functional groups
Go analysis showed that the following biological processes were enriched in the control group, including, response to toxic substances, oxidation-reduction, amino acid metabolism, small molecule biosynthesis, transportation (hydrogen or proton transport), proteins biosynthesis (organonitrogen compound metabolic and biosynthetic processes, translation), actin polymerization or depolymerization and its regulation, nucleic acid biosynthesis and metabolism (nucleoside phosphate metabolic process, nucleoside biosynthetic process, nucleoside monophosphate metabolic process etc.), fatty acid metabolism (fatty acid metabolic process and lipid catabolic process, etc.), cofactor and coenzyme biosynthetic and metabolic process, organic acid metabolism (organic acid catabolic or biosynthetic process, carboxylic acid, monocarboxylic acid and glycosyl compound metabolic or biosynthetic process).
GO and KEGG analysis of unique proteins specially expressed in the immune stress group
As shown in Fig. 2b, KEGG pathway analysis was performed on specifically expressed proteins in the immune stress group. Endocytosis, lysosome, fatty acid degradation, ribosome pathways were enriched.
Go analysis showed that the following biological processes were enriched in the LPS group, including, amino acid metabolism, organic acid and carboxylic acid biosynthesis, transportation (hydrogen or proton transport), organonitrogen compound metabolic and biosynthetic processes, positive regulation of protein complex assembly, actin polymerization or depolymerization and its regulation, cofactor and coenzyme metabolism, organic acid metabolism (organic acid, carboxylic acid, oxoacid and amino acid metabolic and catabolic processes).
Quantitative differential analysis of hepatic proteome in broiler chickens between the control and the immune stress group
Label free LC-MS/MS quantitative analysis isolated 111 proteins that were differently expressed in the liver of broilers in the control and immune stress groups. Of the proteins, 83 were up-regulated, but 28 proteins were down-regulated in the immune stress group (Table 4). Down-regulated proteins in the immune stress group were not significantly enriched in GO terms.
Table 4.
Protein information of differential abundance identified in the liver of AA broilers challenged with LPS
| Protein | Accession no. | Symbol ID | Sequence coverage, % | #Unique peptide | Fold change |
|---|---|---|---|---|---|
| 3-hydroxyisobutyryl-CoA hydrolase mitochondrial isoform X1 | gi|971404063 | HIBCH | 13 | 1 | 0.080 |
| Transferase CAF17 mitochondrial | gi|303227895 | IBA57 | 4 | 1 | 0.082 |
| Protein-glutamine gamma-glutamyltransferase 4 | gi|57530757 | TGM4 | 5 | 3 | 0.084 |
| Serine protease inhibitor Kazal-type 2 isoform X2 | gi|971393739 | SPINK2 | 14 | 1 | 0.130 |
| Eukaryotic translation elongation factor 1 epsilon-1 | gi|971382396 | EEF1E1 | 7 | 1 | 0.147 |
| Cathelicidin-3 precursor | gi|906847364 | CATHL3 | 19 | 2 | 0.156 |
| L-amino-acid oxidase precursor | gi|372266150 | IL4I1 | 5 | 2 | 0.157 |
| Chromodomain-helicase-DNA-binding protein 5 isoform X5 | gi|971429121 | CHD5 | 1 | 1 | 0.171 |
| BTB/POZ domain-containing protein KCTD12 | gi|971377767 | KCTD12 | 4 | 1 | 0.193 |
| Cathelicidin-2 precursor | gi|403224971 | CATHL2 | 40 | 4 | 0.194 |
| TBC1 domain family member 10A | gi|971422101 | TBC1D10A | 3 | 1 | 0.196 |
| 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase isoform X2 | gi|971406099 | ACMSD | 6 | 1 | 0.202 |
| Ribonuclease homolog precursor | gi|56118294 | RSFR | 11 | 1 | 0.202 |
| T-cell immunoglobulin and mucin domain-containing protein 4 precursor | gi|57524995 | TIMD4 | 6 | 2 | 0.212 |
| Gallinacin-2 isoform X1 | gi|971390683 | GAL2 | 12 | 1 | 0.219 |
| Lymphocyte antigen 86 precursor | gi|52138689 | LY86 | 9 | 1 | 0.220 |
| 28S ribosomal protein S22 mitochondrial | gi|971410189 | MRPS22 | 3 | 1 | 0.261 |
| Ubiquilin-1 isoform X1 | gi|118104137 | UBQLN1 | 1 | 1 | 0.270 |
| Serine/threonine-protein kinase 4 isoform X1 | gi|971427386 | STK4 | 2 | 1 | 0.272 |
| Myeloid protein 1 precursor | gi|758818508 | MIM1 | 32 | 8 | 0.273 |
| FAS-associated death domain protein | gi|118091445 | FADD | 7 | 1 | 0.280 |
| Protein MRP-126 | gi|760997140 | S100A9 | 25 | 3 | 0.304 |
| Lysozyme g precursor | gi|47825389 | LYG2 | 11 | 2 | 0.313 |
| PREDICTED: Acetyl-CoA carboxylase isoform X2 | gi|971425697 | ACAC | 4 | 6 | 0.322 |
| Phosphomannomutase 2 | gi|71895479 | PMM2 | 4 | 1 | 0.324 |
| Hydroxyacid-oxoacid transhydrogenase mitochondrial isoform X1 | gi|971384192 | ADHFE1 | 8 | 2 | 0.333 |
| Serine/arginine-rich splicing factor 2 | gi|47604918 | SRSF2 | 8 | 1 | 0.337 |
| Tyrosine-protein kinase Lyn | gi|57530388 | LYN | 3 | 1 | 0.347 |
| Trifunctional purine biosynthetic protein adenosine-3 | gi|47825387 | GART | 5 | 1 | 0.351 |
| 59 kDa 2′-5′-oligoadenylate synthase-like protein isoform X1 | gi|971415867 | OASL | 3 | 1 | 0.351 |
| Antigen peptide transporter 2 isoform X1 | gi|971422259 | TAP2 | 2 | 1 | 0.359 |
| Dynein light chain roadblock-type 1 isoform X1 | gi|971427252 | DYNLRB1 | 12 | 1 | 0.372 |
| Gallinacin-7 preproprotein | gi|48976031 | AvBD7 | 15 | 1 | 0.378 |
| Fibrinogen gamma chain precursor | gi|766944255 | FGG | 41 | 13 | 0.386 |
| Splicing factor 3B subunit 6 | gi|50745107 | SF3B6 | 10 | 1 | 0.387 |
| Pre-mRNA-processing factor 39 isoform X1 | gi|363734910 | PRPF39 | 2 | 1 | 0.387 |
| Glutaredoxin-1 | gi|45384038 | GLRX | 10 | 1 | 0.387 |
| Phosphoglucomutase-2 | gi|71897287 | PGM2 | 4 | 1 | 0.389 |
| Chloride intracellular channel protein 2 | gi|71895359 | CLIC2 | 16 | 2 | 0.390 |
| Metallothionein-3 | gi|147901436 | MT3 | 19 | 1 | 0.393 |
| Fructose-bisphosphate aldolase C | gi|330417943 | ALDOC | 23 | 4 | 0.401 |
| GTP cyclohydrolase 1 feedback regulatory protein | gi|313747529 | GCHFR | 20 | 1 | 0.405 |
| Proteasome subunit alpha type-3 | gi|57529899 | PSMA3 | 5 | 1 | 0.415 |
| Heterogeneous nuclear ribonucleoprotein M isoform X2 | gi|971435624 | HNRNPM | 8 | 1 | 0.416 |
| Protein phosphatase 1 regulatory subunit 42 isoform X2 | gi|118087042 | PPP1R42 | 3 | 1 | 0.422 |
| Sorcin | gi|124249424 | SRI | 12 | 2 | 0.425 |
| Gallinacin-1 alpha precursor | gi|45384510 | GAL1 | 12 | 1 | 0.428 |
| Fibrinogen beta chain precursor | gi|267844833 | FGB | 37 | 13 | 0.437 |
| Acid ceramidase precursor | gi|57530079 | ASAH1 | 3 | 1 | 0.438 |
| Cytochrome P450 2D3-like | gi|307078128 | CYP2D6 | 2 | 1 | 0.447 |
| Antigen peptide transporter 1 | gi|209863064 | TAP1 | 2 | 1 | 0.450 |
| Integrin alpha-V-like | gi|971443478 | LOC107056639 | 7 | 1 | 0.458 |
| Protein O-GlcNAcase isoform X1 | gi|971402723 | MGEA5 | 1 | 1 | 0.458 |
| Dihydropteridine reductase | gi|57529509 | QDPR | 16 | 2 | 0.461 |
| RNA-binding protein 39 isoform X2 | gi|513217267 | RBM39 | 2 | 1 | 0.461 |
| Neutral alpha-glucosidase AB-like | gi|971451761 | LOC107051327 | 9 | 1 | 0.464 |
| Complement 4 precursor | gi|116175422 | C4 | 2 | 2 | 0.466 |
| Fibrinogen alpha chain isoform 1 precursor | gi|429484490 | FGA | 13 | 6 | 0.468 |
| Guanylate-binding protein 1-like | gi|971415657 | GBP4L | 2 | 1 | 0.470 |
| F-box only protein 6 | gi|971429573 | FBXO6 | 4 | 1 | 0.471 |
| Erythroblast NAD(P)(+)--arginine ADP-ribosyltransferase isoform X2 | gi|513166631 | ART7C | 3 | 1 | 0.473 |
| Carbonyl reductase [NADPH] 1 isoform X1 | gi|971375185 | CBR1 | 30 | 5 | 0.482 |
| D-2-hydroxyglutarate dehydrogenase mitochondrial | gi|971410344 | D2HGDH | 2 | 1 | 0.485 |
| Protein LOC107050412 | gi|971449891 | LOC107050412 | 4 | 1 | 0.487 |
| Protein GIMAP1 | gi|971379314 | GIMAP1 | 1 | 1 | 0.487 |
| Peroxisomal trans-2-enoyl-CoA reductase | gi|57529732 | PECR | 7 | 1 | 0.495 |
| Filamin-B isoform X1 | gi|971415729 | FLNB | 2 | 2 | 0.505 |
| D-dopachrome decarboxylase | gi|71897241 | DDT | 11 | 1 | 0.508 |
| V-type proton ATPase subunit E 1 | gi|57525423 | ATP6V1E1 | 12 | 2 | 0.511 |
| Choline dehydrogenase mitochondrial isoform X1 | gi|513205065 | CHDH | 1 | 1 | 0.526 |
| ATP-dependent RNA helicase DDX24 | gi|971400168 | DDX24 | 1 | 1 | 0.527 |
| 26S proteasome non-ATPase regulatory subunit 9 | gi|57525182 | PSMD9 | 6 | 1 | 0.534 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 mitochondrial isoform X2 | gi|971398316 | NDUFS3 | 9 | 2 | 0.540 |
| Alanine--glyoxylate aminotransferase 2 mitochondrial | gi|513228840 | AGXT2 | 7 | 2 | 0.553 |
| Ornithine aminotransferase mitochondrial | gi|57529515 | OAT | 4 | 1 | 0.561 |
| Metallothionein | gi|46048711 | MT1 | 43 | 3 | 0.576 |
| 60S ribosomal protein L19 | gi|71896335 | RPL19 | 9 | 1 | 0.589 |
| Epididymal secretory protein E1 precursor | gi|71894903 | NPC2 | 14 | 1 | 0.603 |
| Long-chain fatty acid transport protein 4 | gi|971423093 | SLC27A4 | 2 | 1 | 0.616 |
| Glutaredoxin-3 | gi|475506756 | GLRX3 | 4 | 1 | 0.617 |
| Cytoplasmic FMR1-interacting protein 1 isoform X2 | gi|971376600 | CYFIP1 | 1 | 1 | 0.624 |
| Diamine acetyltransferase 2-like | gi|971451573 | LOC107051219 | 23 | 2 | 0.631 |
| Platelet glycoprotein 4 | gi|71897003 | CD36 | 3 | 1 | 0.646 |
| Ras-related protein Rab-10 | gi|71895051 | RAB10 | 6 | 1 | 1.543 |
| Iron-sulfur cluster assembly enzyme ISCU mitochondrial | gi|971421427 | ISCU | 11 | 1 | 1.574 |
| Nucleophosmin | gi|45383996 | NPM1 | 22 | 5 | 1.601 |
| 40S ribosomal protein S11 | gi|71895103 | RPS11 | 15 | 2 | 1.654 |
| UBX domain-containing protein 4 isoform X4 | gi|513195319 | UBXN4 | 3 | 1 | 1.757 |
| C-factor-like isoform X2 | gi|363738106 | LOC415662 | 38 | 6 | 1.765 |
| Sodium-coupled neutral amino acid transporter 4 isoform X1 | gi|971370947 | SLC38A4 | 2 | 1 | 2.005 |
| UDP-glucose 4-epimerase | gi|363742411 | GALE | 3 | 1 | 2.086 |
| Protein disulfide-isomerase A4 | gi|57530768 | PDIA4 | 7 | 5 | 2.091 |
| Band 4.1-like protein 2 isoform X4 | gi|971387884 | EPB41L2 | 1 | 1 | 2.092 |
| NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 | gi|480540334 | NDUFB1 | 19 | 1 | 2.118 |
| Granulysin precursor | gi|113206146 | GNLY | 30 | 3 | 2.139 |
| Aspartyl aminopeptidase | gi|61098378 | DNPEP | 4 | 2 | 2.149 |
| GTPase IMAP family member 5 | gi|971379352 | GIMAP5 | 4 | 1 | 2.180 |
| Regucalcin isoform X1 | gi|971376431 | RGN | 30 | 7 | 2.180 |
| ATP-citrate synthase isoform X3 | gi|971435352 | ACLY | 6 | 5 | 2.206 |
| Glutathione S-transferase alpha 4 isoform X1 | gi|971389828 | GSTA4L | 10 | 2 | 2.241 |
| Adrenodoxin mitochondrial | gi|310832417 | FDX1 | 5 | 1 | 2.574 |
| Protein syndesmos precursor | gi|45382147 | SDC4 | 8 | 2 | 2.723 |
| Interferon alpha-inducible protein 6 | gi|47777293 | IFI6 | 89 | 6 | 3.222 |
| Regulator of microtubule dynamics protein 1 | gi|475808820 | RMDN1 | 22 | 6 | 3.671 |
| Proteasome subunit beta type-7 | gi|45383366 | PSMB7 | 5 | 1 | 3.850 |
| Tetratricopeptide repeat protein 38-like | gi|363728070 | TTC38L | 3 | 1 | 4.244 |
| Glutathione S-transferase theta-1-like | gi|971421234 | GSTT1L | 11 | 2 | 4.318 |
| ADP/ATP translocase 1 | gi|57530120 | SLC25A4 | 7 | 2 | 5.735 |
| Interferon-induced GTP-binding protein Mx | gi|45382939 | MX | 5 | 2 | 5.960 |
| Cytochrome P450 2H2 precursor | gi|48976111 | CYP2C23b | 28 | 11 | 6.432 |
| Trifunctional purine biosynthetic protein adenosine-3 isoform X1 | gi|971375154 | GART | 5 | 4 | +∞ |
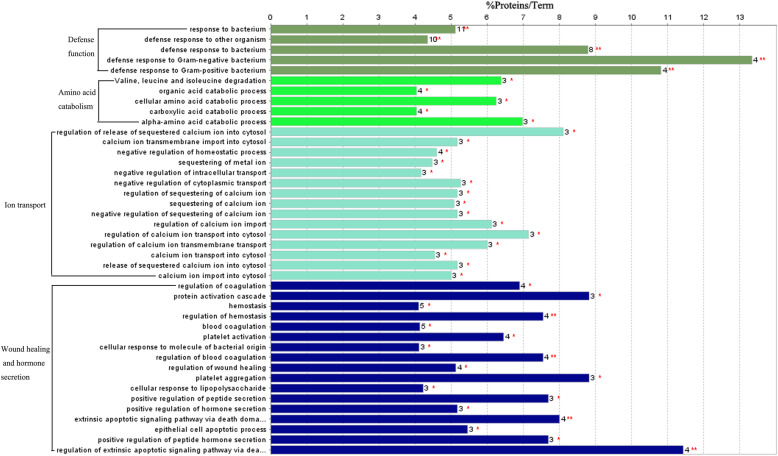
As Fig. 3 and Table 5 show, up-regulated proteins in the immune stress group were significantly enriched in GO terms of defense function, amino acid catabolism, ion transport and regulation, wound healing and hormone secretion and regulation. More specifically, up-regulated proteins in the immune stress group were enriched in valine, leucine and isoleucine degradation pathways. However, there were no GO terms and pathways enriched in down-regulated proteins of the immune stress group.
Fig. 3.
GO and KEGG annotation of up-regulated proteins in the liver of broilers chickens in the immune stress group. Terms that begin with an uppercase or lowercase letters are KEGG or GO annotation. % Proteins/Term stands for the proportion of genes enriched in corresponding functional groups. The bars with the same color represent the same functional groups they belong to. The numbers stand for the genes enriched to the corresponding functional groups
Table 5.
GO annotation of upregulated proteins in broiler chickens in the immune stress group
| GO ID | GO Term | Term P- value | Associated proteins enriched |
|---|---|---|---|
| Biological process | |||
| GO:0009617 | Response to bacterium | 81.0E-9 | AvBD1, AvBD2, AvBD7, CATH2, CATH3, CD36, FGB, LY86, LYN, RSFR, TAP2 |
| GO:0098542 | Defense response to other organism | 1.4E-6 | AvBD1, AvBD2, AvBD7, CATH2, CATH3, CD36, FADD, FGB, OASL, RSFR |
| GO:0042742 | Defense response to bacterium | 87.0E-9 | AvBD1, AvBD2, AvBD7, CATH2, CATH3, CD36, FGB, RSFR |
| GO:0050829 | Defense response to Gram-negative bacterium | 33.0E-6 | AvBD7, CATH2, CATH3, RSFR |
| GO:0050829 | Defense response to Gram-positive bacterium | 79.0E-6 | CATH2, CATH3, CD36, RSFR |
| GO:0016054 | Organic acid catabolic process | 3.3E-3 | AGXT2, HIBCH, OAT, SLC27A4 |
| GO:0009063 | Cellular amino acid catabolic process | 3.3E-3 | AGXT2, HIBCH, OAT |
| GO:0046395 | Carboxylic acid catabolic process | 3.3E-3 | AGXT2, HIBCH, OAT, SLC27A4 |
| GO:1901606 | Alpha-amino acid catabolic process | 2.4E-3 | AGXT2, HIBCH, OAT |
| GO:0051279 | Regulation of release of sequestered calcium ion into cytosol | 1.5E-3 | CLIC2, LYN, SRI |
| GO:0097553 | Calcium ion transmembrane import into cytosol | 5.6E-3 | CLIC2, LYN, SRI |
| GO:0032845 | Negative regulation of homeostatic process | 2.1E-3 | CLIC2, FADD, LYN, SRI |
| GO:0051238 | Sequestering of metal ion | 8.4E-3 | CLIC2, LYN, SRI |
| GO:0032387 | Negative regulation of intracellular transport | 10.0E-3 | CD36, CLIC2, SRI |
| GO:1903650 | Negative regulation of cytoplasmic transport | 5.3E-3 | CD36, CLIC2, SRI |
| GO:0051282 | Regulation of sequestering of calcium ion | 5.6E-3 | CLIC2, LYN, SRI |
| GO:0051208 | Sequestering of calcium ion | 5.9E-3 | CLIC2, LYN, SRI |
| GO:0051283 | Negative regulation of sequestering of calcium ion | 5.6E-3 | CLIC2, LYN, SRI |
| GO:0090279 | Regulation of calcium ion import | 3.5E-3 | CLIC2, LYN, SRI |
| GO:0010522 | Regulation of calcium ion transport into cytosol | 2.2E-3 | CLIC2, LYN, SRI |
| GO:1903169 | Regulation of calcium ion transmembrane transport | 3.7E-3 | CLIC2, LYN, SRI |
| GO:0060402 | Calcium ion transport into cytosol | 8.0E-3 | CLIC2, LYN, SRI |
| GO:0051209 | Release of sequestered calcium ion into cytosol | 5.6E-3 | CLIC2, LYN, SRI |
| GO:1902656 | Calcium ion import into cytosol | 6.2E-3 | CLIC2, LYN, SRI |
| GO:0050818 | Regulation of coagulation | 460.0E-6 | CD36, FGB, FGG, LYN |
| GO:0072376 | Protein activation cascade | 1.2E-3 | C4, FGB, FGG |
| GO:0007599 | Hemostasis | 970.0E-6 | CD36, FGA, FGB, FGG, LYN |
| GO:1900046 | Regulation of hemostasis | 320.0E-6 | CD36, FGB, FGG, LYN |
| GO:0007596 | Blood coagulation | 930.0E-6 | CD36, FGA, FGB, FGG, LYN |
| GO:0030168 | Platelet activation | 590.0E-6 | FGA, FGB, FGG, LYN |
| GO:0071219 | Cellular response to molecule of bacterial origin | 10.0E-3 | CD36, LY86, LYN |
| GO:0030193 | Regulation of blood coagulation | 320.0E-6 | CD36, FGB, FGG, LYN |
| GO:0061041 | Regulation of wound healing | 1.4E-3 | CD36, FGB, FGG, LYN |
| GO:0070527 | Platelet aggregation | 1.2E-3 | FGB, FGG, LYN |
| GO:0071222 | Cellular response to LPS | 9.8E-3 | CD36, LY86, LYN |
| GO:0002793 | Positive regulation of peptide secretion | 1.8E-3 | FGB, FGG, SRI |
| GO:0046887 | Positive regulation of hormone secretion | 5.6E-3 | FGB, FGG, SRI |
| GO:0008625 | Extrinsic apoptotic signaling pathway via death domain receptors | 250.0E-6 | FADD, FGB, FGG, STK4 |
| GO:1904019 | Epithelial cell apoptotic process | 4.8E-3 | FGB, FGG, STK4 |
| GO:0090277 | Positive regulation of peptide hormone secretion | 1.8E-3 | FGB, FGG, SRI |
| GO:1902041 | Regulation of extrinsic apoptotic signaling pathway via death domain receptors | 63.0E-6 | FADD, FGB, FGG, STK4 |
| Pathway | |||
| GO:0000280 | Valine, leucine and isoleucine degradation | 3.1E-3 | AGXT2, HIBCH, IL4I1 |
As shown in Table 6, LPS binding was enriched in up-regulated proteins in the immune stress group using GO annotation based on the molecular function cluster. Moreover, up-regulated proteins in the immune stress group were distributed in the extracellular region, fibrinogen complex, secretory granule, extracellular space and cytoplasm, respectively.
Table 6.
GO annotation of upregulated proteins in broilers chickens in the immune stress group based on molecular function and cellular component
| Molecular function | |||
| GO Term | Description | Count in gene set | False discovery rate |
| GO:0001530 | LPS binding | 2 of 4 | 0.0389 |
| Cellular component | |||
| GO:0005576 | Extracellular region | 12 of 299 | 1.08e-06 |
| GO:0005577 | Fibrinogen complex | 2 of 2 | 0.0021 |
| GO:0030141 | Secretory granule | 3 of 23 | 0.0027 |
| GO:0005615 | Extracellular space | 5 of 167 | 0.0093 |
| GO:0005737 | Cytoplasm | 13 of 1125 | 0.0149 |
Protein and protein interaction (PPI) analysis of differentially expressed proteins in the immune stress group
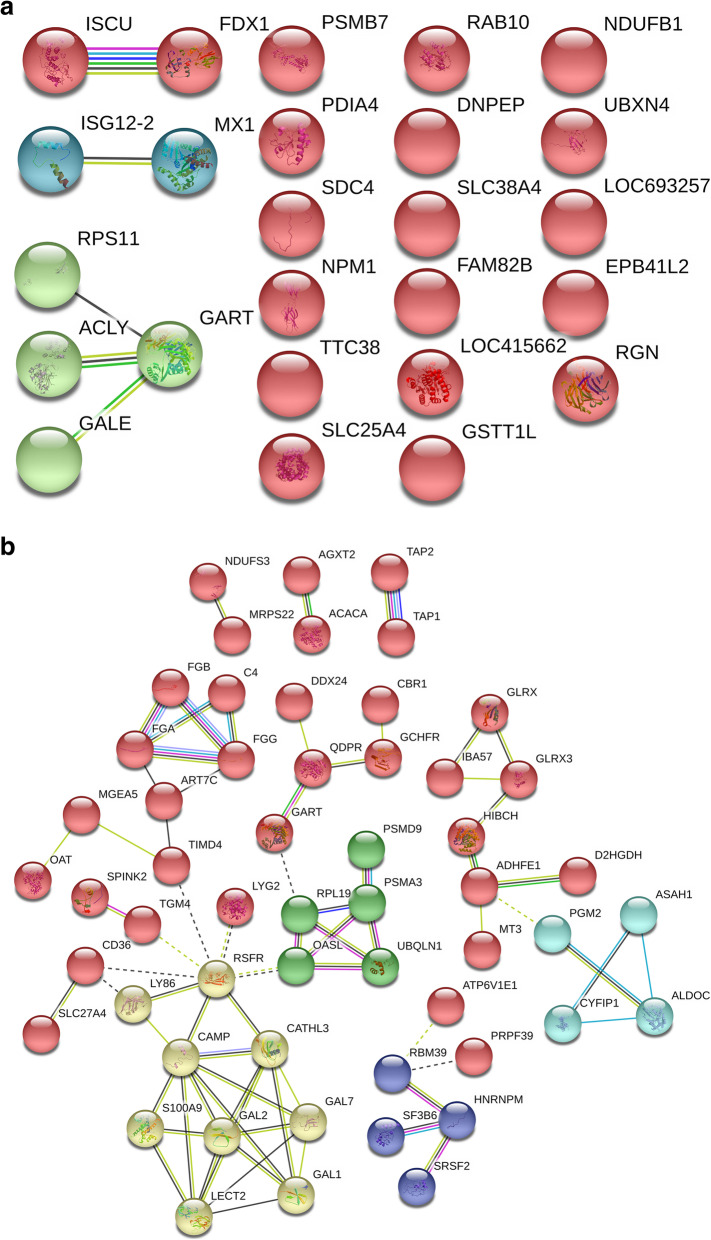
PPI analysis showed that there are only eight proteins connected to the network, including iron-sulfur cluster assembly enzyme (ISCU), adrenodoxin (FDX1), interferon alpha-inducible protein 6 (ISG12–2), interferon-induced GTP-binding protein Mx (MX1), 40S ribosomal protein S11 (RPS11), ATP-citrate synthase isoform X3 (ACLY), UDP-glucose 4-epimerase (GALE), trifunctional purine biosynthetic protein adenosine-3 isoform X1 (GALE). However, there is no significant interaction network (P = 0.248), as Fig. 4a shows.
Fig. 4.
Protein and protein interaction network of differentially expressed proteins in the liver of broilers chickens in the immune stress group. A and B represent interaction network of down-regulated and up-regulated proteins in the liver of broilers chickens in the immune stress group, respectively .Each ball represents node protein, the same color balls represent node proteins clustered in the same sub network. The solid line indicates that the interaction score between the two proteins is more than 0.5 (the dotted line indicates that the score is less than 0.5). Different color solid lines between proteins represent evidence of association. Red lines indicate fusion evidence, green lines indicate neighborhood evidence, blue lines indicate co-occurrence evidence, purple lines indicate experimental evidence, yellow lines indicate text mining evidence, light blue lines indicate database evidence, and black lines indicate co-expression evidence
The results of PPI analysis of upregulated proteins in the immune stress group showed that 77 proteins were connected into the networks with significant interaction between the networks (P = 6.84E-11), as shown in Fig. 4b. Furthermore, cluster analysis showed that the whole network was interconnected by 5 sub-networks, involved in defense function (yellow nodes), protein biosynthesis (Green nodes), RNA splicing and binding (Blue nodes), carboxylic acid metabolism (Cyan) and nutrient metabolism (Red nodes).
Discussion
Immune stress resulting from a LPS challenge inhibited the growth of broilers in this study. The study shows that the concentrations of IL-1β, TNF-α and IL-6 in the serum of broilers injected with LPS was significantly increased. These inflammatory cytokines triggered an up-regulation of the expression of hepatic proteins involved in the immune defense function, amino acid catabolism, ion transport and wound healing and hormone secretion. Moreover, the data revealed that immune stress enhanced the secretion of ACTH and CORT but decreased the secretion of GH and IGF-1. Furthermore, immune stress enhanced hepatic degradation pathways for valine, leucine and isoleucine which would contribute to the growth depression noted by many authors following an immunological challenge [4, 10, 11, 13–15].
Immune stress enhanced the expression of proteins related to defense function
Inflammatory cytokines such as IL-1 and IL-6 can activate B cells and trigger the humoral immune response. Studies have shown that increased humoral response is associated with the inflammatory response [13, 28]. In our study, the serum concentrations of IL-1β and IL-6, following LPS injection, were significantly higher than in unchallenged broilers. Moreover, the present experiment showed that immune stress enhanced the expression of defense function proteins (GO:0009617, GO:0098542, GO:0042742, GO:0050829, GO:0050829), including AvBD1, AvBD2, AvBD7, CATH2, CATH3, CD36, FGB, LY86, LYN, RSFR, TAP2, FADD, and OASL.
These upregulated proteins include effector proteins expressed to directly inactivate pathogens or proteins protecting the chicken’s own tissues against damage. Heterophils are responsible for pathogen inactivation by the release of two classes of antimicrobial peptides, i.e. cathelicidins CATHL1, CATHL2, CATHL3 and gallinacins GAL1, GAL2 and GAL7 (also called avian β-defensins AvBD1, AvBD2 and AvBD7) [29]. These proteins are present in the granules of chicken heterophils associated with response to Salmonella infection [30, 31]. RSFR exhibits multiple enzymatic activities and as a ribonuclease A, it has angiogenic and bactericidal properties [32]. The angiogenic potential of RSFR facilitates the restoration of damaged tissues following inflammation. The bactericidal effects of RFSR protein and its modulatory effect on dendritic cells polarises the immune response towards a Th2 response in chickens [33]. Therefore hepatic upregulation of RSFR, as observed in the immune stress group, suggests that RSFR could contribute to both tissue repair and clearance of residual bacterial pathogens.
Immune stress up-regulated the expression of proteins related to wound healing
Immune stress can lead to delayed wound healing [9]. Up-regulated proteins include those involved in LPS neutralisation and healing of host tissue. In this study, LPS binding (GO:0001530, GO:0071219, GO:0071222) was enriched in GO analysis based on molecular function, including CATHL2, LY86 and complement proteins. Tyrosine-protein kinase Lyn (LYN) plays a role in the LPS-mediated signaling pathway, and in positive regulation of the stress-activated protein kinase signaling cascade. CD36 is involved in the cell surface receptor signaling pathway. Complement 4 precursor is also a defense protein (C4) [34, 35]. Chicken heterophils express lysozyme and two classes of antimicrobial peptides, i.e. cathelicidins and gallinacins. Besides pathogen inactivation, chicken heterophils are also involved in tissue protection and wound healing (GO:0061041) by the expression of RSFR, TGM4, CD36, FGB, FGG and LYN.
Transglutaminases TGM3 and TGM4, are also induced during inflammation [36]. Interestingly, transglutaminase inhibitor cystamine reduced the inflammation induced by 2,4,6-trinitrobenzene sulfonic acid in rats [37]. Transglutaminases catalyse the formation of an isopetide bond between the carboxyamide group of glutamine and the ε amino group of lysine leading to protein cross-linking. TGM3 was induced in the lungs of pigs experimentally infected with Salmonella choleraesuis [38]. TGM3 can cross-link with other proteins during wound healing. In chickens, transglutaminase TGM4 is expressed in B-lymphocytes and to a lesser extent in macrophages [35] and may have a function in wound healing. This would explain up-regulation of TGM4 in the liver of broiler chickens challenged by LPS.
As a consequence of the immune response, blood coagulation is often exploited by pathogens for reason of infective and septic processes. For coagulation, this trigger is usually some form of vascular injury, followed by activation. In the classical waterfall model, each activated protein goes on to activate the next protein in a rapidly expanding cascade of reactions which quickly results in the local formation of a fibrin clot to seal the injury [39]. For example, FG are targeted by bacteria, thus offering a straightforward explanation of positive selection. FG is comprised of the α, β, and γ genes of fibrinogen (FG) (FGA, FGB, and FGG) [40]. In mammals, fibrin (ogen) also serves as a platform for migrating cells, can act as a chemoattractant, and regulates inflammation by activating immune cells, especially macrophages [41]. In the avian thymus, genes encoding fibrinogen subunits (FGA, FGG and FGB) were among the most significantly expressed genes in the broiler after exposure to heat stress and LPS treatments [42]. The present study showed that the biological processes (GO:0050818, GO:0072376, GO:0007599, GO:1900046, GO:0007596, GO:0030168, GO:0030193 and GO:0070527) were enriched, including FGA, FGG and FGB which were up-regulated in the liver of broilers challenged with LPS. This suggests that chickens stimulated by LPS were constantly triggering their body systems to “heal the damage”.
Immune stress enhanced the expression of proteins related to amino acid catabolism
KEGG pathway analysis indicated that the valine, leucine and isoleucine degradation pathway (GO:0000280) was significantly enriched, involving AGXT2, HIBCH, IL4I1. IL4I1 that also play a role in the L-phenylalanine catabolic process. IL4I1 was up-regulated in the spleen [22], the bursa of Fabricius [43] and the thymus gland when birds were exposed to LPS [44]. In this study, IL4I1 was up-regulated in the liver of broilers challenged with LPS. HIBCH is involved in L-valine degradation. AGXT2 plays a role in the glyoxylate catabolic process, L-alanine catabolic process, glycine biosynthetic process and regulation of nitric oxide biosynthesis. Enhancing organic acid catabolism processes (GO:0016054, GO:0009063, GO:0046395, GO:1901606) confirms that body protein and fat anabolism will be reduced by immune stress, resulting in lower feed utilisation and decreased growth performance [14].
OAT has ornithine-oxo-acid transaminase activity and is associated with L-proline biosynthesis. SLC27A4 positively regulates serine/threonine kinase activity and participate in phosphatidylcholine biosynthesis. Up-regulated expression of OAT and SLC27A4 indicates that catabolism will be enhanced in order to meet the nutrients required to synthesize immune effector molecules. This repartitioning of nutrients away from growth and development will reduce bird productivity [15].
Immune stress upregulated the expression of ion transport proteins
Cells of the innate and adaptive immune systems express various ion transporters that allow the influx and efflux of ions across the plasma membrane or their release from intracellular organelles such as the endoplasmic reticulum (ER), mitochondria, and lysosomes [45]. Stimulation of antigen receptors results in a rapid increase in Ca2+ originating from the ER and the extracellular space through PM Ca2+ channels that is required for sustained Ca2+ elevations [46]. SRI is involved in the regulation of high voltage-gated calcium channel activity. CLIC2 is related to the regulation and release of sequestered Ca2+ into the cytosol by sarcoplasmic reticulum. Ca2+ signals also mediate T cell motility. In this study, the up-regulation of proteins associated with ion transport (GO:0051279, GO:0097553, GO:0032845, GO:0051238, GO:0032387, GO:1903650, GO:0051282, GO:0051208, GO:0051283, GO:0090279, GO:0010522, GO:1903169, GO:0060402, GO:0051209 and GO:1902656) suggests that immune stress could trigger the innate and adaptive immune function by inducing the hepatic expression of SRI, CLIC2 and LYN in broilers.
Immune stress increased the expression of proteins related to hormone secretion
When under immune stress, excessive inflammatory cytokines may lead to the activation of the HPA axis, increasing the secretion of ACTH and CORT, and reducing the secretion of the growth promoting hormones such as GH and IGF-1 [47]. In our experiment, function enrichment analysis of up-regulated proteins showed the positive regulation of peptide and hormone secretion (GO:0090277 and GO:0046887) and positive regulation of peptide secretion (GO:0002793) were enriched, including FGB, FGG and SRI.
In the lymphocyte life cycle, T and B cells numbers will be reduced through apoptosis at different stages of ontological development of the immune system to avoid the accumulation and the potential for autoimmunity. However, apoptosis induced by external factors, such as vaccination-induced stress, would cause adverse responses that affect growth performance. It has been shown that stress can trigger the apoptosis of pre-B cells by inducing high concentrations of glucocorticoid, resulting in the reduction of the number of B lymphocytes and suppressed immunity. It has been determined that the infectious bursal disease vaccine can induce apoptotic effects in the bursa of Fabricius [48]. Studies have shown that serum ACTH and CORT concentrations significantly increase due to immune stress induced by LPS [7, 49], and the elevated concentrations of serum ACTH and CORT in these studies are consistent with those observed in the current experiment. High concentrations of ACTH and CORT induces apoptotic effects in spleen lymphocytes [49]. Consistent with these studies, our results show that when apoptosis is induced, enhanced expression of proteins related to the apoptotic signaling pathway (GO:0008625 and GO:1902041) and cell apoptotic process (GO:1904019), involving FADD, FGB, FGG, STK4.
Conclusions
The immune stress induced by LPS triggered alterations in the hepatic proteome of broiler chickens and provides a new insight into the mechanisms by which immune challenge impairs bird growth or productivity. In this regard, impaired growth is secondary to reduced feed intake, which has been well described in the literature [4, 15], and the repartitioning of nutrients. We have demonstrated at a molecular level, that immune stress redirects nutrients which were destined for muscle synthesis and growth to the immune system to support increased functionality. This was evident from increased expression of hepatic proteins involved in defense function, amino acid catabolism, ion transport, wound healing, hormone secretion, and pathogen clearance. The activated immune system can resist immunological challenges but the additional nutritional and metabolic demands imposed on the bird can result in a decline in growth performance.
Acknowledgements
We want to show our appreciation to Professor Li Jianke for his help to design of the present study.
Abbreviations
- LC-MS
Label-free liquid chromatography and mass spectrometry
- LPS
Lipopolysaccharide
- AA
Arbor Acres
- ACTH
Adrenocorticotropic hormone
- CORT
Corticosterone
- GH
Growth hormone
- IGF-1
Insulin-like growth factor-1
- IL-1β
Interleukin-1β
- IL-6
Interleukin-6
- TNF-α
Tumour necrosis factor-α
- DTT
Dithiothreitol
- TIC
Total ion current
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- HIBCH
3-hydroxyisobutyryl-CoA hydrolase mitochondrial isoform X1
- IBA57
Transferase CAF17 mitochondrial
- TGM4
Protein-glutamine gamma-glutamyltransferase 4
- SPINK2
Serine protease inhibitor Kazal-type 2 isoform X2
- EEF1E1
Eukaryotic translation elongation factor 1 epsilon-1
- CATHL3
Cathelicidin-3 precursor
- IL4I1
L-amino-acid oxidase precursor
- CHD5
Chromodomain-helicase-DNA-binding protein 5 isoform X5
- KCTD12
BTB/POZ domain-containing protein KCTD12
- CATHL2
Cathelicidin-2 precursor
- TBC1D10A
TBC1 domain family member 10A
- ACMSD
2-amino-3-carboxymuconate-6-semialdehyde decarboxylase isoform X2
- RSFR
Ribonuclease homolog precursor
- TIMD4
T-cell immunoglobulin and mucin domain-containing protein 4 precursor
- GAL2
Gallinacin-2 isoform X1
- LY86
Lymphocyte antigen 86 precursor
- MRPS22
28S ribosomal protein S22 mitochondrial
- UBQLN1
Ubiquilin-1 isoform X1
- STK4
Serine/threonine-protein kinase 4 isoform X1
- MIM1
Myeloid protein 1 precursor
- FADD
FAS-associated death domain protein
- S100A9
Protein MRP-126
- LYG2
Lysozyme g precursor
- ACAC
PREDICTED: acetyl-CoA carboxylase isoform X2
- PMM2
Phosphomannomutase 2
- ADHFE1
Hydroxyacid-oxoacid transhydrogenase mitochondrial isoform X1
- SRSF2
Serine/arginine-rich splicing factor 2
- LYN
Tyrosine-protein kinase Lyn
- GART
Trifunctional purine biosynthetic protein adenosine-3
- OASL
59 kDa 2'-5'-oligoadenylate synthase-like protein isoform X1
- TAP2
Antigen peptide transporter 2 isoform X1
- DYNLRB1
Dynein light chain roadblock-type 1 isoform X1
- AvBD7
Gallinacin-7 preproprotein
- FGG
Fibrinogen gamma chain precursor
- SF3B6
Splicing factor 3B subunit 6
- PRPF39
Pre-mRNA-processing factor 39 isoform X1
- GLRX
Glutaredoxin-1
- PGM2
Phosphoglucomutase-2
- CLIC2
Chloride intracellular channel protein 2
- MT3
Metallothionein-3
- ALDOC
Fructose-bisphosphate aldolase C
- GCHFR
GTP cyclohydrolase 1 feedback regulatory protein
- PSMA3
Proteasome subunit alpha type-3
- HNRNPM
Heterogeneous nuclear ribonucleoprotein M isoform X2
- PPP1R42
Protein phosphatase 1 regulatory subunit 42 isoform X2
- SRI
Sorcin
- GAL1
Gallinacin-1 alpha precursor
- FGB
Fibrinogen beta chain precursor
- ASAH1
Acid ceramidase precursor
- CYP2D6
Cytochrome P450 2D3-like
- TAP1
Antigen peptide transporter 1
- LOC107056639
Integrin alpha-V-like
- MGEA5
Protein O-GlcNAcase isoform X1
- QDPR
Dihydropteridine reductase
- RBM39
RNA-binding protein 39 isoform X2
- LOC107051327
Neutral alpha-glucosidase AB-like
- C4
Complement 4 precursor
- FGA
Fibrinogen alpha chain isoform 1 precursor
- GBP4L
Guanylate-binding protein 1-like
- FBXO6
F-box only protein 6
- ART7C
Erythroblast NAD(P)(+)--arginine ADP-ribosyltransferase isoform X2
- CBR1
Carbonyl reductase [NADPH] 1 isoform X1
- D2HGDH
D-2-hydroxyglutarate dehydrogenase mitochondrial
- LOC107050412
Protein LOC107050412
- GIMAP1
Protein GIMAP1
- PECR
Peroxisomal trans-2-enoyl-CoA reductase
- FLNB
Filamin-B isoform X1
- DDT
D-dopachrome decarboxylase
- ATP6V1E1
V-type proton ATPase subunit E 1
- CHDH
Choline dehydrogenase mitochondrial isoform X1
- DDX24
ATP-dependent RNA helicase DDX24
- PSMD9
26S proteasome non-ATPase regulatory subunit 9
- NDUFS3
NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 mitochondrial isoform X2
- AGXT2
Alanine--glyoxylate aminotransferase 2 mitochondrial
- OAT
Ornithine aminotransferase mitochondrial
- MT1
Metallothionein
- RPL19
60S ribosomal protein L19
- NPC2
Epididymal secretory protein E1 precursor
- SLC27A4
Long-chain fatty acid transport protein 4
- GLRX3
Glutaredoxin-3
- CYFIP1
Cytoplasmic FMR1-interacting protein 1 isoform X2
- LOC107051219
Diamine acetyltransferase 2-like
- CD36
Platelet glycoprotein 4
- RAB10
Ras-related protein Rab-10
- ISCU
Iron-sulfur cluster assembly enzyme ISCU mitochondrial
- NPM1
Nucleophosmin
- RPS11
40S ribosomal protein S11
- UBXN4
UBX domain-containing protein 4 isoform X4
- LOC415662
C-factor-like isoform X2
- SLC38A4
Sodium-coupled neutral amino acid transporter 4 isoform X1
- GALE
UDP-glucose 4-epimerase
- PDIA4
Protein disulfide-isomerase A4
- EPB41L2
Band 4.1-like protein 2 isoform X4
- NDUFB1
NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1
- GNLY
Granulysin precursor
- DNPEP
Aspartyl aminopeptidase
- GIMAP5
GTPase IMAP family member 5
- RGN
Regucalcin isoform X1
- ACLY
ATP-citrate synthase isoform X3
- GSTA4L
Glutathione S-transferase alpha 4 isoform X1
- FDX1
Adrenodoxin mitochondrial
- SDC4
Protein syndesmos precursor
- IFI6
Interferon alpha-inducible protein 6
- RMDN1
Regulator of microtubule dynamics protein 1
- PSMB7
Proteasome subunit beta type-7
- TTC38L
tetratricopeptide repeat protein 38-like
- GSTT1L
GLUTATHIONE S-transferase theta-1-like
- SLC25A4
ADP/ATP translocase 1
- MX
Interferon-induced GTP-binding protein Mx
- CYP2C23b
Cytochrome P450 2H2 precursor
- GART
Trifunctional purine biosynthetic protein adenosine-3 isoform X1
Authors’ contributions
A. Zheng and W.L.B. contributed to the concept and design of the work. H.C. and G.L. contributed to the design of the work. SAP and A. Zhang executed the experiments. Z.C, W.C and A. Zheng contributed to the analysis and interpretation of the data. A. Zheng drafted the manuscript. W.L.B. and A. Zheng contributed to the final approval of the version for publication. All the authors read and approved the final manuscript.
Funding
Sponsored by National Natural Science Foundation of China (grant no. 31101731) and National Key Research and Development Program of China (No.2018YFD0500600) and The Agricultural Science and Technology Innovation Program (ASTIP).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The feeding trial was conducted according to the guidelines for animal experiments set out by the National Institute of Animal Health. All procedures involving animals such as welfare and ethical issues were approved by the Chinese Academy of Agricultural Sciences (statement no. AEC-CAAS-20191106).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Aijuan Zheng, Email: zhengaijuan@caas.cn.
Anrong Zhang, Email: 15504578605@163.com.
Zhimin Chen, Email: chenzhimin@caas.cn.
Shoaib Ahmed Pirzado, Email: dr.pirzado@gmail.com.
Wenhuan Chang, Email: changwenhuan@caas.cn.
Huiyi Cai, Email: caihuiyi@caas.cn.
Wayne L. Bryden, Email: w.bryden@uq.edu.au
Guohua Liu, Email: liuguohua@caas.cn.
References
- 1.Husband AJ. The immune system and integrated homeostasis. Immunol Cell Biol. 1995;73(4):377–382. doi: 10.1038/icb.1995.58. [DOI] [PubMed] [Google Scholar]
- 2.Lai HT, Nieuwland MG, Kemp B, Aarnink AJ, Parmentier HK. Effects of dust and airborne dust components on antibody responses, body weight gain, and heart morphology of broilers. Poult Sci. 2009;88(9):1838–1849. doi: 10.3382/ps.2009-00129. [DOI] [PubMed] [Google Scholar]
- 3.Star L, Decuypere E, Parmentier HK, Kemp B. Effect of single or combined climatic and hygienic stress in four layer lines: 2. Endocrine and oxidative stress responses. Poult Sci. 2008;87(6):1031–1038. doi: 10.3382/ps.2007-00143. [DOI] [PubMed] [Google Scholar]
- 4.Klasing KC, Laurin DE, Peng RK, Fry DM. Immunologically mediated growth depression in chicks: influence of feed intake, corticosterone and interleukin-1. J Nutr. 1987;117:1629–1637. doi: 10.1093/jn/117.9.1629. [DOI] [PubMed] [Google Scholar]
- 5.Husband AJ. Role of central nervous system and behaviour in the immune response. Vaccine. 1993;11(8):805–816. doi: 10.1016/0264-410X(93)90355-2. [DOI] [PubMed] [Google Scholar]
- 6.Shini S, Kaiser P. Effects of stress, mimicked by administration of corticosterone in drinking water, on the expression of chicken cytokine and chemokine genes in lymphocytes. Stress. 2009;12(5):388–399. doi: 10.1080/10253890802526894. [DOI] [PubMed] [Google Scholar]
- 7.Li K, Zhang P, Shi B, Su J, Yue Y, Tong M, Yan S. Dietary Artemisia ordosica extract alleviating immune stress in broilers exposed to lipopolysaccharide. Ital J Anim Sci. 2017;16(2):301–307. doi: 10.1080/1828051X.2016.1274242. [DOI] [Google Scholar]
- 8.Shini S, Huff GR, Shini A, Kaiser P. Understanding stress-induced immunosuppression: exploration of cytokine and chemokine gene profiles in chicken peripheral leukocytes. Poult Sci. 2010;89(4):841–851. doi: 10.3382/ps.2009-00483. [DOI] [PubMed] [Google Scholar]
- 9.Webster Marketon JI, Glaser R. Stress hormones and immune function. Cell Immunol. 2008;252(1–2):16–26. doi: 10.1016/j.cellimm.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Lai HT, Nieuwland MG, Kemp B, Aarnink AJ, Parmentier HK. Effects of repeated intratracheally administered lipopolysaccharide on primary and secondary specific antibody responses and on body weight gain of broilers. Poult Sci. 2011;90(2):337–351. doi: 10.3382/ps.2010-00997. [DOI] [PubMed] [Google Scholar]
- 11.Marcq C, Cox E, Szalo IM, Thewis A, Beckers Y. Salmonella Typhimurium oral challenge model in mature broilers: bacteriological, immunological, and growth performance aspects. Poult Sci. 2011;90(1):59–67. doi: 10.3382/ps.2010-01017. [DOI] [PubMed] [Google Scholar]
- 12.Star L, Kemp B, van den Anker I, Parmentier HK. Effect of single or combined climatic and hygienic stress in four layer lines: 1. Performance Poultry Sci. 2008;87(6):1022–1030. doi: 10.3382/ps.2007-00142. [DOI] [PubMed] [Google Scholar]
- 13.Yang XJ, Li WL, Feng Y, Yao JH. Effects of immune stress on growth performance, immunity, and cecal microflora in chickens. Poult Sci. 2011;90(12):2740–2746. doi: 10.3382/ps.2011-01591. [DOI] [PubMed] [Google Scholar]
- 14.Husband AJ, Bryden WL. Nutrition, stress and immune activation. Proceed Nutr Soc Australia. 1996;20:60–70. [Google Scholar]
- 15.Klasing KC. Nutrition and the immune system. Br Poult Sci. 2007;48(5):525–537. doi: 10.1080/00071660701671336. [DOI] [PubMed] [Google Scholar]
- 16.Robinson MW, Harmon C, O'Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell Mol Immunol. 2016;13(3):267–276. doi: 10.1038/cmi.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo J, Zheng A, Meng K, Chang W, Bai Y, Li K, Cai H, Liu G, Yao B. Proteome changes in the intestinal mucosa of broiler (Gallus gallus) activated by probiotic enterococcus faecium. J Proteome. 2013;91:226–241. doi: 10.1016/j.jprot.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 18.Zheng A, Luo J, Meng K, Li J, Bryden WL, Chang W, Zhang S, Wang LXN, Liu G, Yao B. Probiotic (enterococcus faecium) induced responses of the hepatic proteome improves metabolic efficiency of broiler chickens (Gallus gallus) BMC Genomics. 2016;17(1):89. doi: 10.1186/s12864-016-2371-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng A, Chang W, Liu G, Yue Y, Li J, Zhang S, et al. Molecular Differences in Hepatic Metabolism between AA Broiler and Big Bone Chickens: a Proteomic Study. PLoS One. 2016;11:e0164702. doi: 10.1371/journal.pone.0164702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng A, Luo J, Meng K, Li J, Zhang S, Li K, Liu G, Cai H, Bryden WL, Yao B. Proteome changes underpin improved meat quality and yield of chickens (Gallus gallus) fed the probiotic enterococcus faecium. BMC Genomics. 2014;15(1):1167. doi: 10.1186/1471-2164-15-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan IM, Cao Z, Liu H, Khan A, Rahman SU, Khan MZ, et al. Impact of cryopreservation on spermatozoa freeze-thawed traits and relevance OMICS to assess sperm Cryo-tolerance in farm animals. Front Vet Sci. 2021;8:609180. doi: 10.3389/fvets.2021.609180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Goor A, Ashwell CM, Persia ME, Rothschild MF, Schmidt CJ, Lamont SJ. Unique genetic responses revealed in RNA-seq of the spleen of chickens stimulated with lipopolysaccharide and short-term heat. PLoS One. 2017;12(2):e0171414. doi: 10.1371/journal.pone.0171414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Zhang H, Chen YP, Yang MX, Zhang LL, Lu ZX, Zhou YM, Wang T. Bacillus amyloliquefaciens supplementation alleviates immunological stress in lipopolysaccharide-challenged broilers at early age. Poult Sci. 2015;94(7):1504–1511. doi: 10.3382/ps/pev124. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Yue HY, Zhang HJ, Xu L, Wu SG, Yan HJ, Gong YS, Qi GH. Transport stress in broilers: I. blood metabolism, glycolytic potential, and meat quality. Poult Sci. 2009;88(10):2033–2041. doi: 10.3382/ps.2009-00128. [DOI] [PubMed] [Google Scholar]
- 25.Lin H, He L, Ma B. A combinatorial approach to the peptide feature matching problem for label-free quantification. Bioinformatics. 2013;29(14):1768–1775. doi: 10.1093/bioinformatics/btt274. [DOI] [PubMed] [Google Scholar]
- 26.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Guo Y, He X, Yuan J, Yang Y, Wang Z. Growth performance and immune responses in chickens after challenge with lipopolysaccharide and modulation by dietary different oils. Animal. 2008;2(2):216–223. doi: 10.1017/S1751731107001188. [DOI] [PubMed] [Google Scholar]
- 29.Li R, Li N, Zhang J, Wang Y, Liu J, Cai Y, et al. Expression of immune-related genes of ducks infected with avian pathogenic Escherichia coli (APEC) Front Microbiol. 2016;7:637. doi: 10.3389/fmicb.2016.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crhanova M, Hradecka H, Faldynova M, Matulova M, Havlickova H, Sisak F, Rychlik I. Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar enteritidis infection. Infect Immun. 2011;79(7):2755–2763. doi: 10.1128/IAI.01375-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuperus T, Coorens M, van Dijk A, Haagsman HP. Avian host defense peptides. Dev Comp Immunol. 2013;41(3):352–369. doi: 10.1016/j.dci.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg HF. RNase a ribonucleases and host defense: an evolving story. J Leukoc Biol. 2008;83(5):1079–1087. doi: 10.1189/jlb.1107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nitto T, Dyer KD, Czapiga M, Rosenberg HF. Evolution and function of leukocyte RNase a ribonucleases of the avian species, Gallus gallus. J Biol Chem. 2006;281(35):25622–25634. doi: 10.1074/jbc.M604313200. [DOI] [PubMed] [Google Scholar]
- 34.Matulova M, Varmuzova K, Sisak F, Havlickova H, Babak V, Stejskal K, Zdrahal Z, Rychlik I. Chicken innate immune response to oral infection with Salmonella enterica serovar Enteritidis. Vet Res. 2013;44(1):37. doi: 10.1186/1297-9716-44-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matulova M, Rajova J, Vlasatikova L, Volf J, Stepanova H, Havlickova H, Sisak F, Rychlik I. Characterization of chicken spleen transcriptome after infection with Salmonella enterica serovar Enteritidis. PLoS One. 2012;7(10):e48101. doi: 10.1371/journal.pone.0048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rychlik I, Elsheimer-Matulova M, Kyrova K. Gene expression in the chicken caecum in response to infections with non-typhoid Salmonella. Vet Res. 2014;45(1):119. doi: 10.1186/s13567-014-0119-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elli L, Ciulla MM, Busca G, Roncoroni L, Maioli C, Ferrero S, Bardella MT, Bonura A, Paliotti R, Terrani C, Braidotti P. Beneficial effects of treatment with transglutaminase inhibitor cystamine on the severity of inflammation in a rat model of inflammatory bowel disease. Lab Investig. 2011;91(3):452–461. doi: 10.1038/labinvest.2010.186. [DOI] [PubMed] [Google Scholar]
- 38.Zhao SH, Kuhar D, Lunney JK, Dawson H, Guidry C, Uthe JJ, Bearson SMD, Recknor J, Nettleton D, Tuggle CK. Gene expression profiling in Salmonella choleraesuis-infected porcine lung using a long oligonucleotide microarray. Mamm Genome. 2006;17(7):777–789. doi: 10.1007/s00335-005-0155-3. [DOI] [PubMed] [Google Scholar]
- 39.Spronk HM, Govers-Riemslag JW, ten Cate H. The blood coagulation system as a molecular machine. BioEssays. 2003;25(12):1220–1228. doi: 10.1002/bies.10360. [DOI] [PubMed] [Google Scholar]
- 40.Rallapalli PM, Orengo CA, Studer RA, Perkins SJ. Positive selection during the evolution of the blood coagulation factors in the context of their disease-causing mutations. Mol Biol Evol. 2014;31(11):3040–3056. doi: 10.1093/molbev/msu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szaba FM, Smiley ST. Roles for thrombin and fibrin(ogen) in cytokine/chemokine production and macrophage adhesion in vivo. Blood. 2002;99(3):1053–1059. doi: 10.1182/blood.V99.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monson MS, Van Goor AG, Persia ME, Rothschild MF, Schmidt CJ, Lamont SJ. Genetic lines respond uniquely within the chicken thymic transcriptome to acute heat stress and low dose lipopolysaccharide. Sci Rep. 2019;9(1):13649. doi: 10.1038/s41598-019-50051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monson MS, Van Goor AG, Ashwell CM, Persia ME, Rothschild MF, Schmidt CJ, et al. Immunomodulatory effects of heat stress and lipopolysaccharide on the bursal transcriptome in two distinct chicken lines. BMC Genomics. 2018;19(1):643. doi: 10.1186/s12864-018-5033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie H, Rath NC, Huff GR, Huff WE, Balog JM. Effects of Salmonella typhimurium lipopolysaccharide on broiler chickens. Poult Sci. 2000;79(1):33–40. doi: 10.1093/ps/79.1.33. [DOI] [PubMed] [Google Scholar]
- 45.Feske S, Wulff H, Skolnik EY. Ion channels in innate and adaptive immunity. Annu Rev Immunol. 2015;33(1):291–353. doi: 10.1146/annurev-immunol-032414-112212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feske S. Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol. 2007;7(9):690–702. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- 47.Dorshkind K, Horseman ND. Anterior pituitary hormones, stress, and immune system homeostasis. BioEssays. 2001;23(3):288–294. doi: 10.1002/1521-1878(200103)23:3<288::AID-BIES1039>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 48.Killian MP, Boviez JD, Gambarotta M, Lombardo DM. Induction of apoptosis in the bursa of Fabricius by vaccination against Gumboro disease. Avian Pathology. 2017;46(5):526–534. doi: 10.1080/03079457.2017.1322684. [DOI] [PubMed] [Google Scholar]
- 49.Li RF, Liu SP, Yuan ZH, Yi JE, Tian YN, Wu J, Wen LX. Effects of induced stress from the live LaSota Newcastle disease vaccination on the growth performance and immune function in broiler chickens. Poult Sci. 2020;99(4):1896–1905. doi: 10.1016/j.psj.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.