SUMMARY
RNA helicases play critical roles in various biological processes, including serving as viral RNA sensors in innate immunity. Here, we find that RNA helicase DEAH-box helicase 15 (DHX15) is essential for type I interferon (IFN-I, IFN-β), type III IFN (IFN-λ3), and inflammasome-derived cytokine IL-18 production by intestinal epithelial cells (IECs) in response to poly I:C and RNA viruses with preference of enteric RNA viruses, but not DNA virus. Importantly, we generate IEC-specific Dhx15-knockout mice and demonstrate that DHX15 is required for controlling intestinal inflammation induced by enteric RNA virus rotavirus in suckling mice and reovirus in adult mice in vivo, which owes to impaired IFN-β, IFN-λ3, and IL-18 production in IECs from Dhx15-deficient mice. Mechanistically, DHX15 interacts with NLRP6 to trigger NLRP6 inflammasome assembly and activation for inducing IL-18 secretion in IECs. Collectively, our report reveals critical roles for DHX15 in sensing enteric RNA viruses in IECs and controlling intestinal inflammation.
In brief
Xing et al. show that DHX15 functions as an RNA virus sensor to produce IFN-β, IFN-λ3, and IL-18 in IECs and is required to control RNA virus-induced intestinal inflammation in vivo. They further demonstrate that DHX15 interacts with NLRP6 to trigger NLRP6 inflammasome activation for IL-18 secretion in IECs.
Graphical Abstract
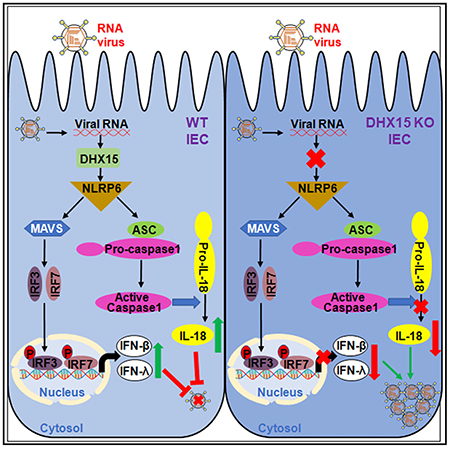
INTRODUCTION
The innate immune system is the first line of defense against virus infection. The innate immune cells have developed the ability to recognize viruses through sensing viral nucleic acids, either in the endosomes or in the cytosol (Ablasser and Hur, 2020). Up to now, a number of cytosolic DNA virus sensors (Ablasser and Hur, 2020), including cyclic guanosine monophosphate (GMP)-AMP synthase (cGAS) (Ablasser et al., 2013; Gao et al., 2013; Sun et al., 2013) and DDX41 (Zhang et al., 2011b), have been identified. These DNA sensors use adaptors such as stimulator of interferon genes (STING) (Ishikawa and Barber, 2008; Ishikawa et al., 2009; Zhong et al., 2008) to induce the type I interferon (IFN-I, IFN-α/β) response and activate the inflammasome response (Swanson et al., 2017; Wang et al., 2020). In parallel, a number of RNA virus sensors have been identified (Ablasser and Hur, 2020), including Toll-like receptors (TLRs) recognizing endosomal viral RNA and RNA helicases recognizing cytosolic viral RNA. Many helicase family members can sense cytosolic viral RNA. RNA helicases RIG-I (Myong et al., 2009; Yoneyama et al., 2004), MDA-5 (Yoneyama et al., 2005), DDX3 (Oshiumi et al., 2010), DEAH-box helicase 9 (DHX9) (Zhang et al., 2011c), DHX15 (Lu et al., 2014; Mosallanejad et al., 2014), DHX29 (Sugimoto et al., 2014), DDX60 (Oshiumi et al., 2015), DDX1/DDX21/DHX36 (Zhang et al., 2011a), and DHX33 (Mitoma et al., 2013) use adaptors such as mitochondrial antiviral-signaling protein (MAVS) and Toll/interleukin-1 (IL-1) receptor (TIR)-domain-containing adaptor-inducing interferon-β (TRIF) to induce the IFN-I response (Kawai et al., 2005; Seth et al., 2005; Xu et al., 2005) and nucleotide-binding oligomerization domain (NOD)-like receptor family pyrin-domain-containing 3 (NLRP3) to activate the inflammasome response and subsequent release of both IL-1β and IL-18 (Mitoma et al., 2013).
Enteric viruses enter the host through the mucosal surface of the intestinal tract to cause inflammational diseases in the intestinal tract. Intestinal epithelial cells (IECs) lining the intestinal tract serve as a first line of defense against invading enteric viruses. IECs are equipped with different kinds of DNA and RNA virus sensors that recognize the invading enteric viruses and initiate the antiviral innate immune response by producing IFN-I and type III IFN (IFN lambdas [IFNλs]) (Durbin et al., 2013; Lazear et al., 2015; Sen et al., 2012). These IFN-I and IFNλs invoke innate antiviral mechanisms within virus-infected and uninfected bystander cells and coordinately regulate the development of adaptive immune responses against enteric viruses (Deal et al., 2013; Wack et al., 2015). In addition, inflammasome serves an important role in host defense by recognizing viral infection and triggering responses from the innate immune system (Kanneganti, 2010; Muruve et al., 2008; Wang et al., 2014). IECs are also equipped with different kinds of DNA and RNA virus inflammasome receptors that recognize the invading enteric viruses and initiate inflammasome activation by producing inflammasome-derived cytokine IL-18 (Lei-Leston et al., 2017; Li and Zhu, 2020). Recently, NLRP9b inflammmasome was reported to restrict rotavirus infection in IECs (Zhu et al., 2017), suggesting the critical role of inflammasome activation and IL-18 from IECs in controlling enteric virus infection.
DEAH-box helicase 15 (DHX15) is an outstanding member of the DEAD-box RNA helicase subfamily in the DExD/H helicase family (Linder, 2006). DHX15 has been shown to function in multiple biological processes, including pre-mRNA splicing (Yoshimoto et al., 2009), ribosome assembly, and biogenesis (Chen et al., 2014; Memet et al., 2017; Studer et al., 2020). A few studies suggest that DHX15 contributes to carcinogenesis in leukemia (Chen et al., 2018), breast cancer (Lin et al., 2009), prostate cancer (Jing et al., 2018), and hepatocellular carcinoma (Xie et al., 2019) and acts as a tumor suppressor gene in glioma (Ito et al., 2017) and gastric cancer (Xiao et al., 2016). Importantly, we and other groups have shown that DHX15 is an RNA virus sensor through binding double-stranded RNA (dsRNA) from RNA virus and induces the production of IFN-I and proinflammatory cytokines in dendritic cells (DCs) in response to dsRNA and RNA viruses in vitro (Lu et al., 2014; Mosallanejad et al., 2014; Pattabhi et al., 2019). A recent study indicates that NLRP6 interacts with DHX15, and both are required in sensing enteric viruses, including encephalomyocarditis virus (EMCV) and norovirus in the murine intestine to produce both IFN-I and IFN-λs (Wang et al., 2015), suggesting that DHX15 may function as an RNA virus sensor to induce IFNs in IECs. We previously identified DHX15 as an RNA virus sensor in DCs in vitro (Lu et al., 2014). However, the in vivo roles of DHX15 as an RNA virus sensor in IECs and intestinal inflammation induced by enteric viruses are still unknown. In this study, we generate IEC-specific Dhx15-knockout (KO) mice. We find that DHX15 is essential for IFN-β, IFN-λ3, and inflammasome-derived cytokine IL-18 production by human and mouse IECs in response to poly I:C and enteric RNA viruses, but not DNA and DNA virus. DHX15 is also shown to be required for controlling intestinal inflammation induced by enteric RNA viruses in vivo. Mechanistically, DHX15 recruits NLRP6 to trigger NLRP6 inflammasome assembly and activation, to result in IL-18 secretion in IECs. Thus, our studies reveal critical roles of DHX15 in sensing enteric RNA viruses in IECs and controlling intestinal inflammation induced by enteric RNA viruses.
RESULTS
DHX15 is essential for IFN-β, IFN-λ3, and IL-18 production by human HT-29 IECs in response to poly I:C
Previously, we identified DHX15 as an RNA virus sensor, which activates and induces MAVS-dependent IFN-I production in DCs in response to dsRNA poly I:C and RNA viruses in vitro (Lu et al., 2014). To further investigate the biological function of DHX15 in human IECs, we established the stable knockdown of DHX15 human HT-29 IECs through use of short hairpin RNA (shRNA). The DHX15-targeting shRNA produced an efficient knockdown of DHX15 expression (Figure 1A). In addition, the shRNA targeting MAVS, STING, retinoic acid-inducible gene I (RIG-I), or melanoma differentiation-associated gene 5 (MDA5) efficiently knocked down the expression of MAVS, STING, RIG-I, or MDA5 (Figures 1A and S1A). These cells were then stimulated by poly I:C and the production of IFN-I IFN-β and type III IFN IFN-λ3, inflammasome cytokine IL-18, and proinflammatory cytokines IL-6 and tumor necrosis factor-α (TNF-α) by the cultured HT-29 IECs was measured by ELISA. The control (sh-Ctrl) HT-29 IECs produced high levels of IFN-β (Figures 1B and S1B), IFN-λ3 (Figures 1C and S1C), IL-18 (Figures 1D and S1D), IL-6 (Figure S2A), and TNF-α (Figure S2B) following poly I:C stimulation. Production of these cytokines was strongly attenuated in DHX15- and MAVS-knockdown HT-29 IECs in response to poly I:C (Figures 1B–1D and S2), which further confirmed our previous data that DHX15 positively regulates the production of IFN-β, IL-6, and TNF-α in myeloid DCs in response to poly I:C and RNA virus (Lu et al., 2014). However, the production of these cytokines was not affected or was only slightly affected in STING-knockdown HT-29 IECs (Figures 1B–1D and S2), which confirmed a previous report showing that STING plays a critical role in DNA sensing but no role in poly I:C sensing (Ishikawa et al., 2009). Furthermore, the production of IFN-β, IFN-λ3, and IL-18 was not affected or was only slightly affected in HT-29 IECs with the knockdown of RIG-I or MDA5 (Figures S1B–S1D).
Figure 1. DHX15 is essential for producing IFN-β, IFN-λ3, and IL-18 by human HT-29 IECs in response to poly I:C.
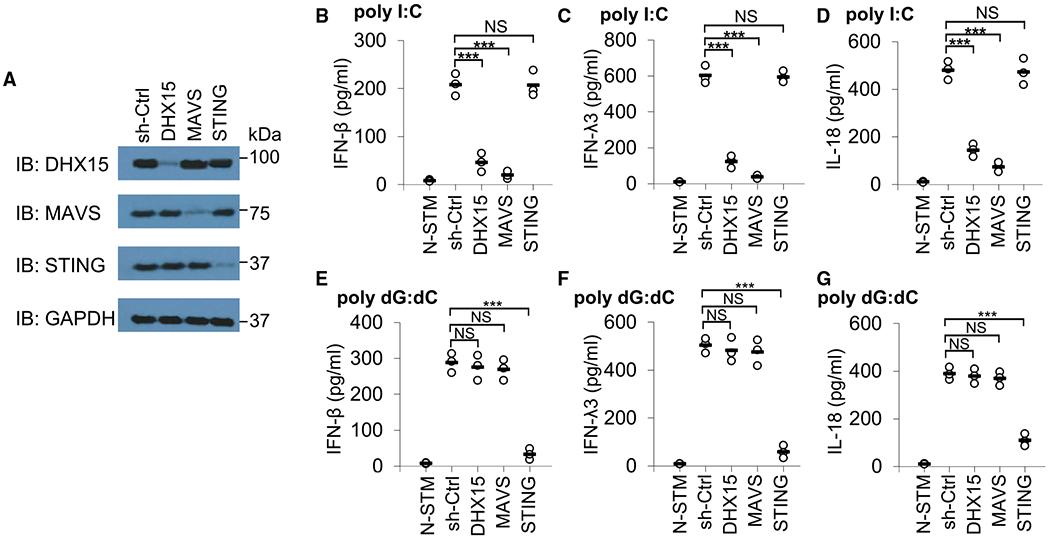
(A) Immunoblot (IB) showing the knockdown efficiency of shRNAs targeting the indicated genes in HT-29 IECs. Nontargeting shRNA served as a control (sh-Ctrl). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) blots are shown as loading controls. The position of protein markers (shown in kDa) is indicated at right. (B–G) ELISA of IFN-β (B and E), IFN-λ3 (C and F), and IL-18 (D and G) production from human HT-29 IECs with the indicated shRNA after a 20-h stimulation with 5 μg/mL poly I:C (B–D) or 2.5 μg/mL poly dG:dC (E–G) delivered by Lipofectamine 3000. N-STM, scrambled shRNA-treated HT-29 IECs without stimulation. Each circle represents an individual independent experiment, and small solid black lines indicate the average of triplicates. NS, not significant; p > 0.05, ***p < 0.001 (unpaired t test).
We next determined whether DHX15 was essential for sensing DNA in HT-29 IECs. Similarly, the HT-29 IECs were treated with sh-Ctrl and knockdown shRNAs targeting DHX15, MAVS, STING, RIG-I, or MDA5 followed by poly dG:dC stimulation, and the production of IFN-β, IFN-λ3, and IL-18 by the cultured HT-29 IECs was then measured by ELISA. As a result, the knockdown of DHX15, MAVS (Figures 1E–1G), RIG-I, or MDA5 (Figures S1B–S1D) in HT-29 IECs had little effect on IFN-β, IFN-λ3, and IL-18 production in response to poly dG:dC, which confirmed a previous report showing that the RNA-sensing adaptor molecule MAVS is not required for cytokine production in response to cytosolic DNA (Sun et al., 2006). However, STING knockdown led to a significant reduction in those cytokine productions in response to poly dG:dC (Figures 1E–1G). Collectively, these data suggest that DHX15 functions independent of RIG-I and MDA5 and is essential for IFN-β, IFN-λ3, and IL-18 production by human HT-29 IECs in response to dsRNA poly I:C.
DHX15 is required for producing IFN-β, IFN-λ3, and IL-18 in human HT-29 IECs upon RNA virus infection
Enteric viruses such as rotavirus and reovirus could replicate in IECs, and the impact of enteric viruses on intestine homeostasis and inflammation is just beginning to be unraveled (Bányai et al., 2018; Metzger et al., 2018). To examine the function of DHX15 in sensing RNA virus infection in human IECs, cytokine production was measured after culturing sh-Ctrl and knockdown shRNA-treated human HT-29 IECs followed by infection with two enteric RNA viruses (simian rotavirus SA-11 strain or reovirus T3D strain) and two non-enteric RNA viruses (vesicular stomatitis virus [VSV] Indiana strain and influenza A virus [Flu] PR8 strain). DHX15-knockdown HT-29 IECs had 50%–70% reduction in IFN-β, IFN-λ3, and IL-18 production in response to enteric RNA virus rotavirus or reovirus (Figures 2A–2F), while there was 20%–30% reduction in IFN-β, IFN-λ3, and IL-18 production from DHX15-knockdown HT-29 IECs after infection with non-enteric RNA virus VSV and Flu virus (Figures S3A–S3C), suggesting that DHX15 was essential for sensing RNA viruses, especially enteric RNA viruses, to produce IFN-β, IFN-λ3, and IL-18 in human HT-29 IECs. Although MAVS-knockdown HT-29 IECs had significant reduction in IFN-ββ, IFN-λ3, and IL-18 levels, STING-knockdown HT-29 IECs had little effect on cytokine production in response to these enteric RNA viruses (Figures 2A–2F). To further determine whether DHX15 senses DNA virus in human IECs, cytokine levels were measured after culturing sh-Ctrl and knockdown shRNA-treated human HT-29 IECs with HSV-1 DNA virus. DHX15 and MAVS knockdown in HT-29 IECs had little effect on IFN-β, IFN-λ3, and IL-18 production by HT-29 IECs in response to HSV-1 infection (Figures 2G–2I). In contrast, STING-knockdown HT-29 IECs had significant reduction in the production of those cytokines (Figures 2G–2I). Importantly, knockdown of DHX15 or MAVS in HT-29 IECs enhanced the viral replication of enteric RNA virus rotavirus or reovirus, but not DNA virus HSV-1 (Figures 2J–2L). Therefore, these data indicate that DHX15 plays an important role in sensing RNA virus infection with preference of enteric RNA virus in human IECs.
Figure 2. DHX15 is required for IFN-β, IFN-λ3, and IL-18 production in human HT-29 IECs upon enteric RNA virus infection.
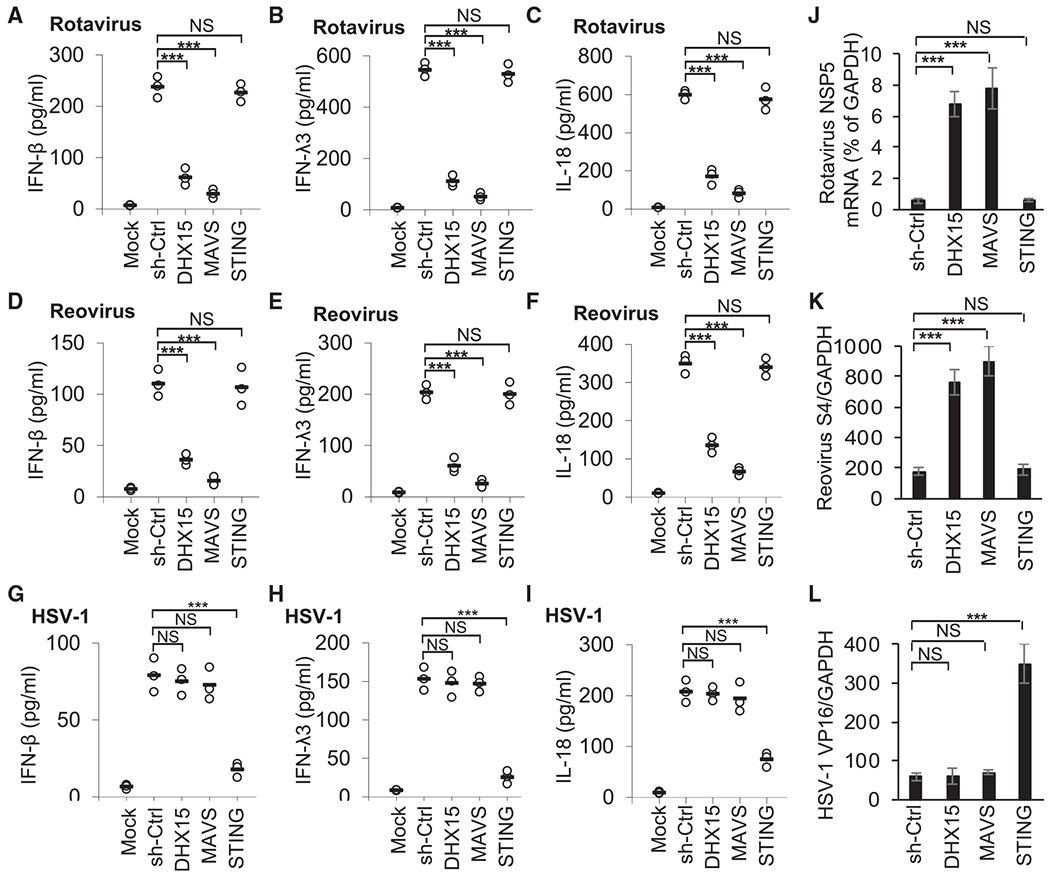
(A–I) ELISA of IFN-β (A, D, and G), IFN-λ3 (B, E, and H), and IL-18 (C, F, and I) production from human HT-29 IECs with the indicated shRNA after a 20-h infection with enteric RNA viruses, including simian rotavirus SA-11 strain (A–C) and reovirus T3D strain (D–F), or DNA virus HSV-1 KOS strain (G–I) at a multiplicity of infection (MOI) of 10. Mock, scrambled shRNA (sh-Ctrl)-treated human HT-29 IECs without virus infection. Each circle represents an individual independent experiment, and small solid black lines indicate the average of triplicates.
(J–L) Quantification of expression of rotavirus NSP5 gene (J), reovirus S4 gene (K), and HSV-1 VP16 gene (L) relative to GAPDH in human HT-29 IECs infected by rotavirus (J), reovirus (K), or HSV-1(L) as in (A)–(I). Data are represented as means ± SEMs. NS, p > 0.05, ***p < 0.001 (unpaired t test).
DHX15 positively regulates production of IFN-β, IFN-λ3, and IL-18 in mouse IECs upon RNA virus infection
We have found that DHX15 is essential for IFN-β, IFN-λ3, and IL-18 production in human IECs in response to dsRNA and RNA viruses, especially enteric RNA viruses. This finding prompted us to examine the roles of DHX15 in mouse IECs and intestinal inflammation induced by enteric RNA virus in vivo. We crossed the Dhx15-targetd mice with FRT deleter (Rosa26-FLPe) mice to generate Dhx15-flox mice (Dhx15fl/fl), which were further crossed with Villin-Cre transgenic mice to generate IEC-specific Dhx15-KO mice, Dhx15fl/fl; Villin-Cre (Dhx15IEC-KO) (Figure S4A). In addition, the deletion of Dhx15 was confirmed by the PCR analysis of genomic DNA (Figure S4B). Furthermore, mouse primary IECs were isolated from wild-type (WT) Dhx15fl/fl and Dhx15IEC-KO mice, and immunoblot (IB) analysis confirmed that DHX15 was deleted in mouse IECs from Dhx15IEC-KO mice (Figure S4C). In addition, flow cytometry analysis showed that the purity of primary IECs isolated from WT Dhx15fl/fl and Dhx15IEC-KO mice was >94% (Figure S5) and KO of DHX15 in IECs did not change the expression of differentiation markers, including epithelial cell adhesion molecule (EpCAM) and E-cadherin (Figure S5), indicating that DHX15 does not affect the expression of differentiation markers in mouse IECs. The epithelial barrier function of IECs plays a crucial role in regulating intestinal homeostasis and inflammation (Schulzke et al., 2009). However, the mouse IECs from WT Dhx15fl/fl and Dhx15IEC-KO mice had a comparable level of expression of several major tight junction proteins, including E-cadherin, claudin-2, occludin, and zonula occludens-1 (ZO-1) (Figure S6), suggesting that DHX15 is dispensable for the expression of epithelial tight junction proteins. Next, we assessed the immune cell subset frequencies in spleen and mesenteric lymph node (mLN) from WT Dhx15fl/fl and Dhx15IEC-KO mice by flow cytometry (Figure S7). Flow cytometry analysis revealed comparable frequencies (Figures S7A and S7B) and absolute numbers (Figures S7C and S7D) of CD4+ T cells, CD8+ T cells, B cells, and natural killer (NK) cells in spleen (Figures S7A and S7C) and mLN (Figures S7B and S7D) from WT Dhx15fl/fl and Dhx15IEC-KO mice. These data show that the IEC-specific Dhx15-KO mice is successfully generated for subsequent in vivo studies.
To further confirm the function of DHX15 in sensing enteric RNA virus infection in mouse IECs, mouse primary IECs were isolated from WT Dhx15fl/fl and Dhx15IEC-KO mice and were infected with two enteric RNA viruses (rotavirus EW strain and reovirus) and two non-enteric RNA viruses (VSV and Flu virus) for detecting cytokine production by ELISA. Mouse IECs from Dhx15IEC-KO mice produced much smaller levels of IFN-β, IFN-λ3, and IL-18 than those from WT Dhx15fl/fl mice in response to enteric RNA virus rotavirus (Figures 3A–3C) or reovirus (Figures 3D–3F), while reduction folds of IFN-β, IFN-λ3, and IL-18 in mouse IECs infected by non-enteric RNA viruses VSV and Flu virus were less than those IECs infected by enteric RNA viruses rotavirus and reovirus (Figures S8A–S8C). To further determine whether DHX15 senses DNA virus in mouse IECs, the cytokine levels were measured after culturing mouse primary IECs from WT Dhx15fl/fl and Dhx15IEC-KO mice with herpes simplex virus-1 (HSV-1) DNA virus infection. DHX15 KO had little effect on IFN-β, IFN-λ3, and IL-18 production by mouse IECs in response to HSV-1 infection (Figures 3G–3I). Furthermore, DHX15 deficiency in mouse IECs enhanced the viral replication of enteric RNA virus rotavirus or reovirus, but not DNA virus HSV-1 (Figures 3J–3L). These results indicate that DHX15 positively regulates IFN-β, IFN-λ3, and IL-18 production in mouse primary IECs upon infection with RNA viruses, especially enteric RNA viruses.
Figure 3. DHX15 positively regulates production of IFN-β, IFN-λ3, and IL-18 in mouse primary IECs upon enteric RNA virus infection.
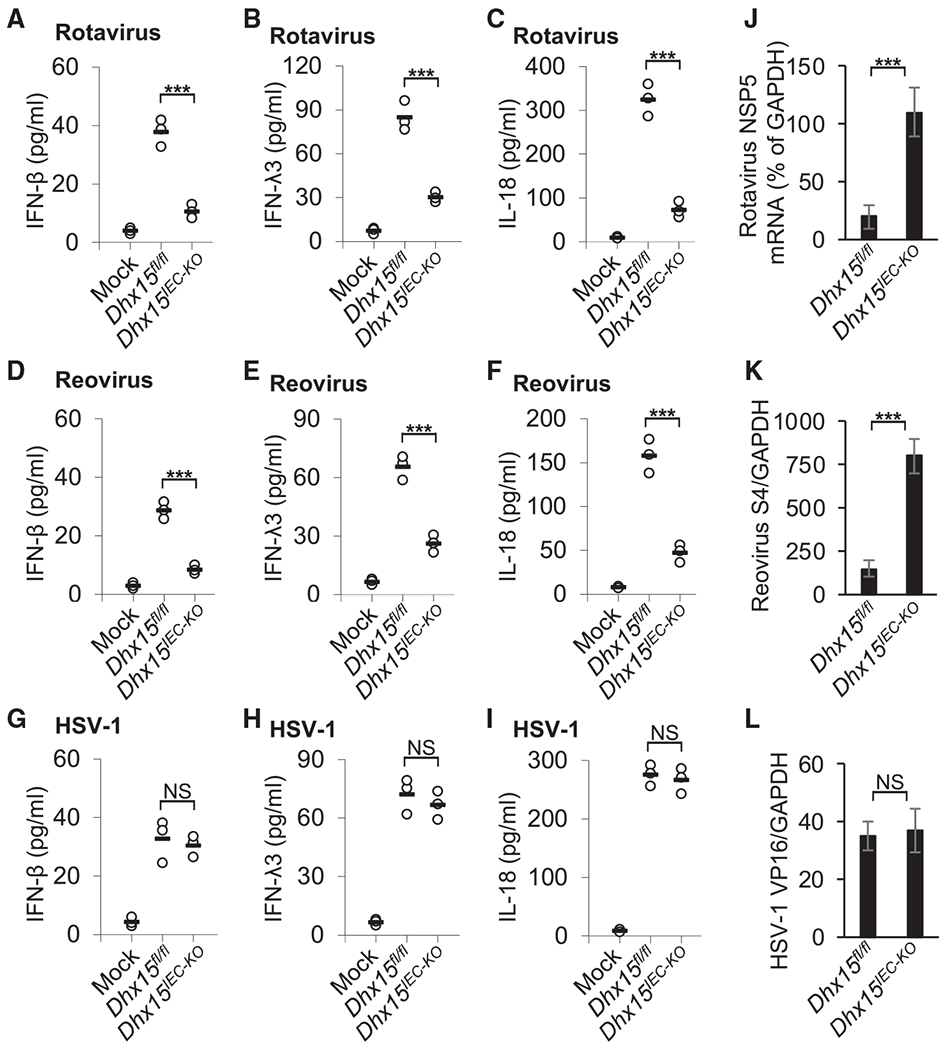
(A–I) ELISA of IFN-β (A, D, and G), IFN-λ3 (B, E, and H) and IL-18 (C, F, and I) production in mouse primary IECs from wild-type Dhx15fl/fl and Dhx15IEC-KO mice after a 20-h infection with enteric RNA viruses, including rotavirus EW strain (A–C) and reovirus T3D strain (D–F), or DNA virus HSV-1 KOS strain (G–I) at a MOI of 10. Mock, cells without virus infection. Each circle represents an individual independent experiment, and small solid black lines indicate the average of triplicates.
(J–L) Quantification of expression of rotavirus NSP5 gene (J), reovirus S4 gene (K), and HSV-1 VP16 gene (L) relative to GAPDH in mouse primary IECs from wild-type Dhx15fl/fl and Dhx15IEC-KO mice infected by rotavirus (J), reovirus (K), or HSV-1(L) as in (A)–(I). Data are represented as means ± SEMs. NS, p > 0.05, ***p < 0.001 (unpaired t test).
DHX15 is essential for controlling intestinal inflammation induced by enteric rotavirus infection in vivo
Next, we evaluated the importance of DHX15 in controlling intestinal inflammation triggered by enteric rotavirus infection in suckling mice in vivo. We first challenged WT Dhx15fl/fl and Dhx15IEC-KO suckling mice orally with mouse rotavirus and monitored diarrhea over time. Dhx15IEC-KO suckling mice exhibited more frequent incidences of diarrhea compared to their WT Dhx15fl/fl littermates (Figure 4A). In addition, we infected WT Dhx15fl/fl and Dhx15IEC-KO suckling mice orally with rotavirus for 1 day and then measured IFN-β, IFN-λ3, and IL-18 in intestine homogenates from infected mice. The qRT-PCR analysis showed that the expression of Ifnb1, Ifnl2/3, and Il18 in the intestines of Dhx15IEC-KO suckling mice was significantly lower than that in intestines from WT Dhx15fl/fl littermates (Figures 4B–4D). In addition, Dhx15IEC-KO suckling mice produced 2- to 3-fold less IFN-λ3 and IL-18 than did WT Dhx15fl/fl littermates, in response to rotavirus (Figures 4E and 4F), whereas the IFN-β protein level was not detectable in intestines from suckling mice. Furthermore, we harvested intestine and fecal samples at day 5 post-infection, and determined viral titers of rotavirus by qRT-PCR. We detected higher viral loads in the intestine and more increased fecal shedding of rotavirus in Dhx15IEC-KO suckling mice than in WT Dhx15fl/fl littermates (Figures 4G and 4H). These data demonstrate that DHX15 is essential for controlling intestinal inflammation induced by enteric rotavirus infection in suckling mice in vivo.
Figure 4. DHX15 is essential to control intestinal inflammation induced by enteric rotavirus infection in suckling mice in vivo.
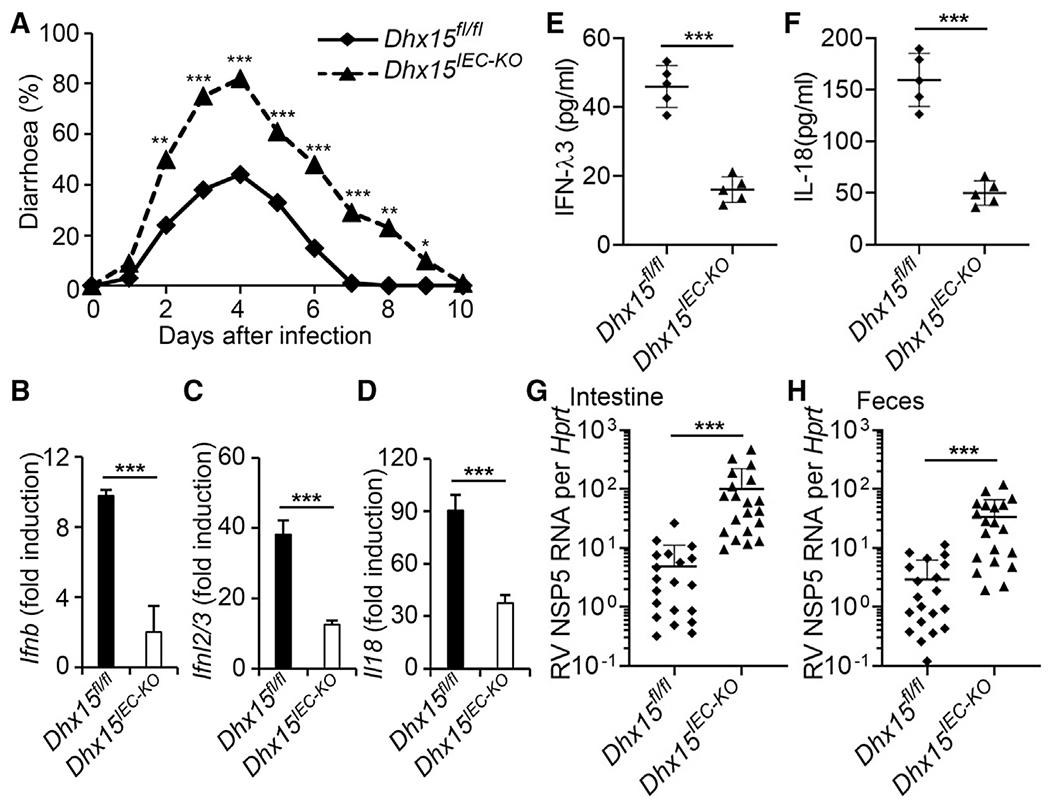
(A) Diarrhea duration and percentage of mice with diarrhea (score ≥ 2) from 8-day-old wild-type Dhx15fl/fl and Dhx15IEC-KO suckling mice (n = 20 per strain) orally inoculated by gavage with 1 DD50 rotavirus EW strain.
(B–F) The wild-type Dhx15fl/fl and Dhx15IEC-KO suckling mice (n = 5 per strain) were orally inoculated by gavage with 1 DD50 rotavirus EW strain. At day 1 post-inoculation, mice were euthanized, and intestine tissues were excised for qRT-PCR detection of Ifnb (B), Ifnl2/3 (C), and Il18 (D) expression. In addition, the excised intestine was homogenized in PBS for the detection of IFN-λ3 (E) and IL-18 (F) in intestine homogenates by ELISA. Data are represented as means ± SEMs.
(G and H) The wild-type Dhx15fl/fl and Dhx15IEC-KO suckling mice (n = 20 per strain) were orally inoculated by gavage with 1 DD50 rotavirus EW strain. At day 5 post-inoculation, mice were euthanized, and intestine tissues (G) and feces (H) were collected for qRT-PCR detection of rotavirus levels. Mock, mouse without rotavirus infection. *p < 0.05, **p < 0.01, and ***p < 0.001 (unpaired t test).
DHX15 is required for control of intestinal inflammation induced by enteric reovirus infection in vivo
We next evaluated whether DHX15 is required for the control of intestinal inflammation induced by enteric reovirus in adult mice in vivo. The 5-week-old WT Dhx15fl/fl and Dhx15IEC-KO adult mice were inoculated intragastrically with enteric reovirus and mice survival was monitored over time. The challenge of Dhx15IEC-KO mice with reovirus led to lethal infection (Figure 5A). In contrast, all of the WT Dhx15fl/fl mice survived from reovirus infection (Figure 5A). In addition, we infected WT Dhx15fl/fl and Dhx15IEC-KO mice intragastrically with enteric reovirus for 1 day and then measured IFN-β, IFN-λ3, and IL-18 in intestine homogenates from infected mice. As expected, Dhx15IEC-KO mice produced 2- to 3-fold less IFN-β, IFN-λ3, and IL-18 than did WT Dhx15fl/fl mice, in response to reovirus (Figures 5B–5D). In addition, we harvested intestine tissues and feces at day 4 post-infection, and determined viral titers of reovirus in those organs by plaque assay. We detected significantly higher viral loads in the intestine and more increased fecal shedding of reovirus in Dhx15IEC-KO mice than in WT Dhx15fl/fl mice (Figures 5E and 5F). Next, we assessed the impact of DHX15 in leukocyte homing to the intestine and mLN from WT Dhx15fl/fl and Dhx15IEC-KO mice with reovirus infection for 2 days by flow cytometry. Flow cytometry analysis revealed more frequencies (Figure 5G) and more numbers (Figure 5H) of CD4+ T cells and CD8+ T cells homing to intestine and mLN (Figures 5G and 5H) from Dhx15IEC-KO mice compared to those from WT Dhx15fl/fl mice, while there were more frequencies and numbers of B cells and NK cells homing to intestine and mLN (Figures 5G and 5H) from WT Dhx15fl/fl mice than those from Dhx15IEC-KO mice. These data suggested that T cells were involved in promoting intestinal inflammation induced by reovirus infection, while B cells and NK cells were inclined to protecting from intestinal inflammation after reovirus infection. Furthermore, intestine histopathology and histology score showed that the intestine from Dhx15IEC-KO mice exhibited more severe inflammatory lesions and inflammation compared to WT Dhx15fl/fl mice at day 4 after reovirus infection (Figures 5I and 5J). These collective findings indicate that DHX15 is required for the control of intestinal inflammation induced by enteric reovirus infection in adult mice in vivo.
Figure 5. DHX15 is required for control of intestinal inflammation induced by enteric reovirus infection in adult mice in vivo.
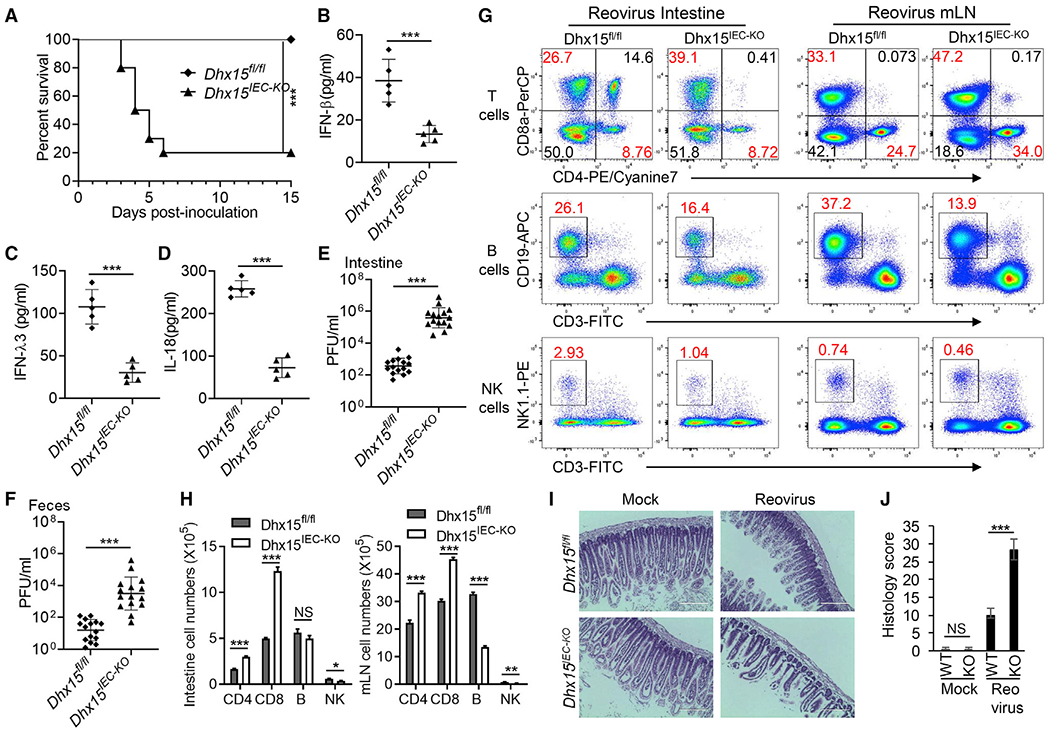
(A) Survival of 5-week-old wild-type Dhx15fl/fl and Dhx15IEC-KO adult mice (n = 10 per strain) after intragastric injection of reovirus T3D strain (1 × 108 plaque-forming units [PFUs] per mouse).
(B–D) The wild-type Dhx15fl/fl and Dhx15IEC-KO mice (n = 5 per strain) were inoculated intragastrically with 1 × 108 PFUs reovirus T3D strain. At day 1 post-inoculation, mice were euthanized, and intestine tissues were excised and homogenized in PBS. Levels of IFN-β (B), IFN-λ3 (C), and IL-18 (D) in intestine homogenates were quantified by ELISA.
(E and F) The wild-type Dhx15fl/fl and Dhx15IEC-KO mice (n = 15 per strain) were inoculated intragastrically with 1 × 108 PFU of reovirus T3D strain. At day 4 post-inoculation, mice were euthanized, feces were collected, and intestinal tissues were excised. The viral titers in intestine homogenates (E) and shedding in feces (F) were determined by plaque assay. Results are expressed as mean viral titers for 15 animals for each time point. Error bars indicate SEMs.
(G) Flow cytometry analysis of CD4+ T cells, CD8+ T cells, B cells, and NK cells of intestine lamina propria lymphocytes (left panel) and mesenteric lymph nodes (mLN) (right panel) from wild-type Dhx15fl/fl and Dhx15IEC-KO mice infected with reovirus for 2 days using CD3-FITC, CD4-PE/cyanine7, CD8a-PerCP/cyanine5.5, CD19-APC, and NK1.1-PE antibodies.
(H) The absolute cell numbers in intestine (left) and mLN (right) from wild-type Dhx15fl/fl and Dhx15IEC-KO mice (n = 3 mice) for representative flow cytometry data in (G).
(I) Hematoxylin and eosin (H&E) staining of intestine sections from wild-type Dhx15fl/fl and Dhx15IEC-KO mice as in (E). Scale bars represent 200 μm.
(J) Graph depicting histology scores for inflammation and tissue damage of intestine sections in (I). Data are represented as means ± SEMs. NS, p > 0.05, *p < 0.05, **p < 0.01, and ***p < 0.001 (unpaired t test).
DHX15 recruits NLRP6 to promote inflammasome assembly and activation
We previously identified DHX15 as an RNA virus sensor to interact with MAVS for producing IFN-β in DCs in vitro (Lu et al., 2014). Recently, NLRP6 has been shown to regulate intestinal antiviral innate immunity through binding viral RNA via DHX15 and interacting with MAVS to induce the production of IFN-β and IFN-λ (Wang et al., 2015), further confirming our data that DHX15 is required for producing IFN-β and IFN-λ in IECs in response to enteric RNA viruses. However, the mechanisms by which DHX15 regulates the inflammasome activation in IECs are unknown. It is reported that NLRP6 inflammasome signaling regulates epithelial IL-18 secretion (Levy et al., 2015). We next evaluated whether DHX15 interacts with NLRP6 to trigger NLRP6 inflammasome assembly and activation to result in IL-18 secretion in IECs. We detected the interaction between DHX15 and NLRP6 at an endogenous protein level in mouse wild-type IECs, but not in Dhx15-deficient IECs (Figure 6A). In addition, the interaction of DHX15 and NLRP6 was significantly enhanced by enteric reovirus infection (Figures 6A and S9A). To further map the binding sites between DHX15 and NLRP6, we analyzed the interactions among FLAG-tagged recombinant NLRP6 and hemagglutinin (HA)-tagged recombinant full-length DHX15 and truncation mutants of DHX15 (Figure 6B). Both recombinant full-length DHX15 and its truncation mutants N (amino acids [aa] 1–433 of DHX15) and DEXDc (aa 1–398 of DHX15), except for mutant C (aa 661–794 of DHX15), interacted with NLRP6, suggesting that the DEXDc domain of DHX15 bound NLRP6 (Figure 6C). In contrast, the full length of NLRP6 is required for interaction with DHX15 (data not shown), which further confirmed a previous report (Wang et al., 2015). The assembly and activation of NLRP6 inflammasome are required for IL-18 production (Levy et al., 2015). We then investigated whether DHX15 promotes the assembly and activation of NLRP6 inflammasome. Coimmunoprecipitation assay showed that ASC could pull down NLRP6 to assemble the NLRP6 inflammasome in HEK293T cells. Importantly, the overexpression of full-length DHX15 could promote the interaction of ASC and NLRP6; however, the DHX15 mutant C could not due to its lost interaction with NLRP6 (Figure 6D), indicating that DHX15 could promote the assembly of NLRP6 inflammasome. To further validate whether DHX15 promotes the NLRP6 inflammasome activation for IL-18 maturation, we reconstituted NLRP6 inflammasome components in HEK293T cells. In the reconstituted system, NLRP6 inflammasome activation resulted in IL-18 cleavage and maturation (Figures 6E and S9B). The overexpression of full-length DHX15, but not for vector control, significantly increased NRLP6 inflammasome activation by inducing more IL-18 cleavage and maturation (Figures 6E and S9B), suggesting that DHX15 promotes NLRP6 inflammasome activation. In addition, IEC-specific deficiency of DHX15 did not affect the expression of RNA sensors, including RIG-I, MDA5, and DHX9, and their adaptor MAVS that are responsible for the production of IFN-β and IFN-λ in IECs, neither for the expression of inflammasome components NRLP6 and NLRP9 in mouse primary IECs (Figure S9C). However, reovirus infection induced more expression of DHX15, MAVS, and NLRP6 in mouse IECs from wild-type Dhx15fl/fl mice than those from Dhx15IEC-KO mice (Figure S9C). Furthermore, the coimmunoprecipitation assay showed that NRLP6 could recruit and interact with DHX15 and MAVS, but not RIG-I and MDA5, to form the interaction complex in mouse IECs after reovirus infection (Figure S9D). This suggested that DHX15 operated independently of RIG-I and MDA5 for inducing NLRP6 inflammasome activation in mouse IECs after RNA virus infection. Collectively, our results demonstrate that DHX15 interacts with NLRP6 to trigger NLRP6 inflammasome assembly and activation to result in IL-18 secretion in IECs.
Figure 6. DHX15 recruits NLRP6 to promote the inflammasome assembly and activation.
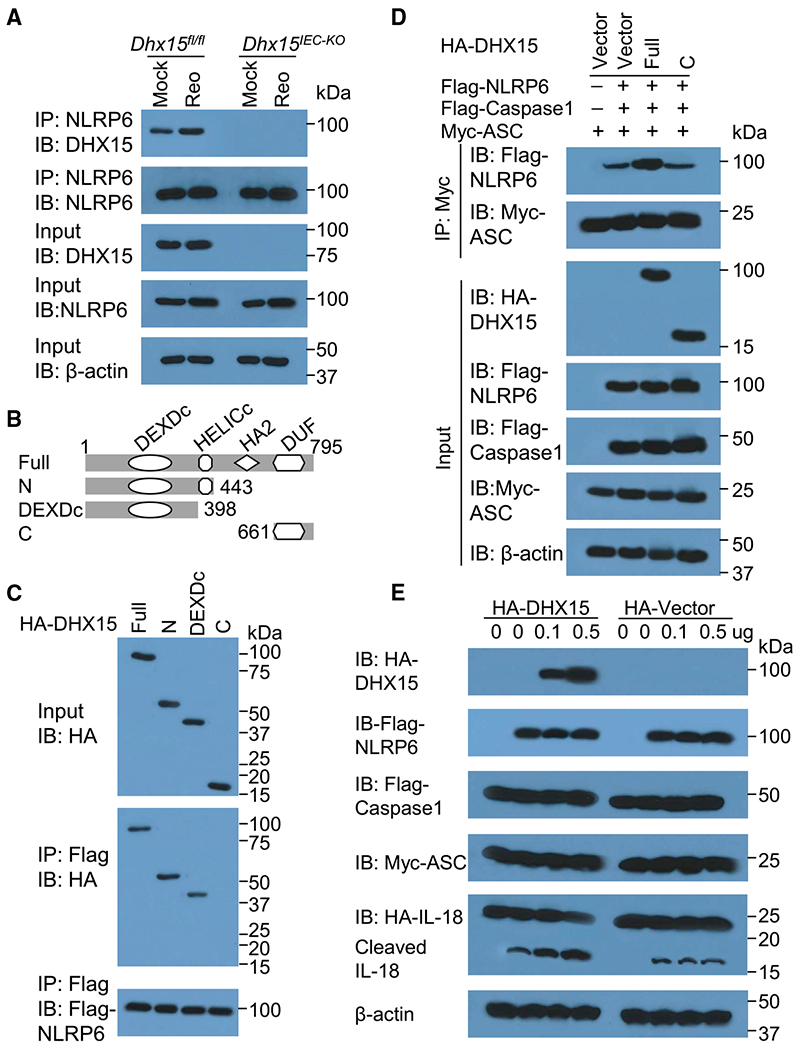
(A) IB analysis of endogenous proteins of DHX15 and NLRP6 precipitated with anti-NLRP6 from whole-cell lysates of IECs from wild-type Dhx15fl/fl and Dhx15IEC-KO mice infected without (Mock) or with reovirus (Reo) T3D strain at MOI of 5.
(B) Schematic diagram showing full-length (Full) DHX15 and its truncations with deletion of various domains (left margin); numbers at ends indicate amino acid positions (top). DEXDc, DEAD-like helicases superfamily domain; HELICc, helicase superfamily c-terminal domain; HA2, helicase-associated domain 2; DUF, domain of unknown function.
(C) IB analysis of purified FLAG-tagged NLRP6 with anti-FLAG (bottom blot), and IB analysis (with anti-HA) of purified HA-tagged full-length DHX15 (Full) and DHX15 truncation mutants alone (top blot) including N-terminal domain (N), DEAD-like helicases superfamily domain (DEXDc), and C-terminal domain (C), or after incubation with FLAG-tagged NLRP6 and immunoprecipitation with anti-FLAG (center blot).
(D) IB analysis of DHX15, NLRP6, caspase-1, and ASC in HEK293T cells transfected with Myc-ASC and co-transfected with or without FLAG-caspase-1, FLAG-NLRP6, HA-DHX15 (Full for full-length, or C for its truncate containing C-terminal domain), or HA-vector control followed by immunoprecipitation with anti-Myc antibody.
(E) IB analysis of DHX15, NLRP6, caspase-1, ASC, full-length IL-18, and its cleaved IL-18 in HEK293T cells transfected with HA-IL-18, Myc-ASC, and FLAG-caspase-1, and co-transfected with or without FLAG-NLRP6, HA-DHX15, or HA-vector control with different doses as indicated. The position of protein markers (shown in kDa) is indicated at right.
DISCUSSION
RNA helicases represent a large family of proteins that have been detected in almost all of the biological systems in which RNA plays a central role (Jankowsky, 2011). The increasing number of in vitro studies show that RNA helicases are involved in immune responses toward viruses by acting as viral RNA sensors or immune signaling adaptors (Ranji and Boris-Lawrie, 2010). However, there is a lack of in vivo studies involved in the tissue- or cell-specific functions of RNA helicases due to the lethality of mice with global KO of RNA helicases. Previously, we have shown that DHX15, a member of RNA helicases, functions as dsRNA and an RNA virus sensor in DCs for producing IFN-β in vitro. In this study, we found that DHX15 was essential to produce IFN-β, IFN-λ3, and IL-18 in both human and mouse IECs in response to dsRNA poly I:C and RNA viruses, especially enteric RNA viruses, including rotavirus and reovirus in vitro. This suggested that DHX15 is also an RNA sensor for RNA viruses with the preference of enteric RNA viruses in IECs to produce not only IFN-β but also IFN-λ3 and inflammasome-derived cytokine IL-18. Our previous data show that DHX15 senses and binds viral dsRNA (Lu et al., 2014). We believe that enteric RNA viruses, including rotavirus and reovirus, may replicate much better and produce more viral dsRNA for DHX15 sensing in the cytosol of IECs than non-enteric RNA viruses VSV and Flu. To further investigate the in vivo roles of DHX15 in intestinal antiviral response and enteric virus-induced intestinal inflammation, we generated the Dhx15 floxed mice (Dhx15fl/fl) and IEC-specific Dhx15-KO mice (Dhx15IEC-KO). Importantly, IEC-specific Dhx15-KO mice are susceptible to infection with enteric rotavirus in suckling mice and reovirus in adult mice. The increased intestinal inflammation induced by enteric viruses in IEC-specific Dhx15-deficient mice was due to the impaired production of IFN-β, IFN-λ3, and IL-18 from IECs. Our flow cytometry data suggested that T cells were involved in promoting the intestinal inflammation induced by reovirus infection, while B cells and NK cells were inclined to protect from intestinal inflammation after reovirus infection. We have previously shown that DHX15 functions as an RNA virus sensor to interact with MAVS to produce IFN-β in DCs in vitro (Lu et al., 2014). Recently, NLRP6 has been shown to regulate intestinal antiviral innate immunity through binding viral RNA via DHX15 and interacting with MAVS to induce the production of IFN-β and IFN-λ in IECs (Wang et al., 2015), further confirming our data that DHX15 is a RNA sensor of enteric RNA viruses and is required for producing IFN-β and IFN-λ in IECs after viral infection. It has been reported that NLRP6 inflammasome signaling regulates the epithelial IL-18 secretion (Levy etal., 2015). Mechanistically, we demonstrate that DHX15 interacts with NLRP6 to trigger NLRP6 inflammasome assembly and activation for inducing IL-18 secretion in IECs. Therefore, we demonstrate that DHX15 is required for intestinal inflammation induced by enteric RNA viruses in vivo. Our findings indicate promise for potential therapeutic applications involving targeting RNA helicase DHX15 to reduce intestinal inflammation to treat intestinal diseases such as inflammatory bowel disease (IBD).
The importance of the type I IFN system for controlling enteric viral infections varies greatly depending on the challenge virus. For example, IFN-α/β plays an important role in restricting virus-induced disease after oral inoculation of mice with poliovirus or reovirus (Johansson et al., 2007; Ohka et al., 2007; Teijaro et al., 2013), but it is of moderate importance in restricting rotavirus, which exhibits a high tropism for gut epithelial cells (Angel et al., 1999; Feng et al., 2008; Pott et al., 2011). We found that IFN-β in intestine from IEC-specific Dhx15-deficient mice with reovirus infection was significantly reduced, which confirmed the previous observation that IFN-β plays a critical role in restricting reovirus-induced disease in mice (Johansson et al., 2007). We have previously shown that DHX15 senses the dsRNA of RNA viruses and activates MAVS-dependent signaling to produce IFN-β in DCs in vitro (Lu et al., 2014). Here, we found that DHX15 also served as an RNA virus sensor and was essential to the IFN-β production in both human and mouse IECs in response to enteric RNA viruses.
Type I IFN receptors are expressed ubiquitously and are expected to evoke antiviral defenses in all tissues and cell types, including IECs, whereas the type III IFN receptor is largely expressed in epithelial cells (Sommereyns et al., 2008). Therefore, IFN-λs are largely produced by epithelial cells including IECs, and the IFN-λ system is essential for the efficient control of rotavirus replication in IECs (Pott et al., 2011). In the present study, IEC-specific Dhx15-KO mice are susceptible to intestinal inflammation induced by either enteric rotavirus in suckling mice or reovirus in adult mice in vivo, which owes to the significantly dampened IFN-λ3 production of IECs from Dhx15-deficient mice. Our findings further confirmed the important role of IFN-λ in controlling enteric virus infection and virus-induced intestinal inflammation. It was reported in 2015 that NLRP6 is required for host defense against infection by enteric RNA viruses, including EMCV and norovirus, through interacting with DHX15, which senses dsRNA from those enteric RNA viruses and mediates the production of both IFN-I and IFN-λ in the mouse intestine (Wang et al., 2015). Here, we found that DHX15 was required for IFN-β and IFN-λ production in both human and mouse IECs by sensing enteric RNA viruses, including rotavirus and reovirus.
The inflammasome is a caspase-1-containing complex that activates the proinflammatory cytokines IL-1β and IL-18 and results in the proinflammatory cell death known as pyroptosis (Lupfer et al., 2015). Increasing evidence has highlighted the importance of inflammasome activation in the control of virus infection (Lupfer et al., 2015; Zhu et al., 2017). We found that the inflammasome-derived cytokine IL-18 in the intestines of IEC-specific Dhx15-deficient mice with enteric RNA virus infection was dramatically reduced in either suckling mice or adult mice, indicating that inflammasome activation in IECs is important to control enteric RNA virus infection and virus-induced intestinal inflammation. We found that DHX15 is essential for IL-18 production in both human and mouse IECs in response to enteric RNA viruses, including rotavirus and reovirus in vitro and in vivo. Mechanistically, we demonstrate that DHX15 interacts with NLRP6 to trigger NLRP6 inflammasome assembly and activation for inducing IL-18 secretion in IECs.
In conclusion, we generate the IEC-specific Dhx15-KO mice and demonstrate that DHX15 is required for intestinal inflammation induced by enteric RNA viruses in vivo and that DHX15 recruits NLRP6 inflammasome signaling to induce IL-18 secretion in IECs. Most important, severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) is an RNA virus that causes the ongoing coronavirus disease 2019 (COVID-19) pandemic (Guan et al., 2020; Zhu et al., 2020). Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA are frequently observed in COVID-19 patients, and SARS-CoV-2 could efficiently infect human IECs (Lamers et al., 2020; Zang et al., 2020). Thus, our findings on DHX15 in controlling enteric RNA virus-induced intestinal inflammation may provide potential therapeutic applications involving targeting RNA helicase DHX15 to control SARS-CoV-2 and the intestinal inflammation induced by SARS-CoV-2.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Zhiqiang Zhang (zzhang@houstonmethodist.org).
Materials availability
All plasmids generated in this study are available upon request.
Data and code availability
This study did not generate/analyze datasets or code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Dhx15fl/fl conditional knockout mice were generated at Taconic-Artemis (Cologne, Germany) in close consultation with our lab as follows: Mouse genomic fragments of the Dhx15 locus were subcloned using RPCIB-731 BAC library via ET recombination and recloned into a basic targeting vector placing an F3-site flanked neomycin resistance cassette in intron 3 and a thymidine kinase cassette downstream of the 3′ UTR. LoxP sites flanked exon 4. The targeting vector was sequenced to confirm correctness. The linearized DNA vector was electroporated into C57BL/6N embryonic stem cells, neomycin selection (500 μg/ml) started on day 2 and counter selection with ganciclovir (2 μM) started on day 5 after electroporation. Embryonic stem cell clones were isolated on day 8 and analyzed by Southern blotting according to standard procedure. Blastocysts were isolated from the uterus of BALB/c females at day 3.5 post coitum and 10-15 targeted C57BL/6NTac embryonic stem cells were injected into each blastocyst. After recovery, 6 injected blastocysts were transferred to each uterine horn of 2.5 days post coitum, pseudopregnant females. Chimerism of offspring was measured by coat color contribution of embryonic stem cells to the BALB/c host (black/white). Highly chimeric mice were bred to strain C57BL/6 females transgenic for the Flp recombinase gene to remove the neomycin resistance cassette in mice carrying the conditional knockout allele (Dhx159fl/WT), which were further self-crossed to generate Dhx15fl/fl mice. Germline transmission was identified by the presence of black strain C57BL/6 offspring. The Dhx15fl/fl mice were backcrossed to the C57BL/6 background strain over at least 6 generations before use in subsequent experiments.
The Dhx15fl/fl mice were crossed with the Villin-Cre transgenic mice (Madison et al., 2002) (Stock No: 021504, The Jackson Laboratory) that express Cre recombinase in villus and crypt epithelial cells of the small and large intestines to generate IEC-specific Dhx15-knockout mice, Dhx15fl/fl; Villin-Cre (Dhx15IEC-KO). All animals were on the C57BL/6 genetic background and maintained in the specific pathogen-free facility at Houston Methodist Research Institute in Houston, Texas. Animal use and care were approved by the Houston Methodist Animal Care Committee, in accordance with institutional animal care and use committee guidelines.
In vivo rotavirus infection
Rotavirus EW is a non-cell culture-adapted wild-type murine rotavirus strain. The virus titration of rotavirus was expressed as 50% diarrhea dose (DD50) defined as the highest dilution that causes diarrhea in 50% of suckling C57BL/6 mice (Burns et al., 1995). For rotavirus infection in mice, 8-day-old wild-type Dhx15fl/fl and Dhx15IEC-KO mice were orally inoculated by gavage with 1 DD50 rotavirus in 50 μL PBS. The appearance of diarrhea was monitored over time by changes in color and consistency of feces. At day 5 after infection, mice were euthanized and intestine were collected. Rotavirus titer in intestinal tissues was detected by quantitative RT-PCR (qRT-PCR) based on rotavirus gene 11 (NSP5) sequences. At day 1 after infection, mice were euthanized and intestines were collected. The cytokines Ifnb, Ifnl2/3, and Il18 in intestine tissues were determined by qRT-PCR. The cytokines IFN-λ and IL-18production in intestine homogenate was measured by ELISA.
For diarrhea experiment of rotavirus infection, diarrhea was documented, and tissue samples were collected and measured in a double-blinded manner. The percentage and severity of diarrhea among the littermates during the course of infection was recorded as previously described (Ball et al., 1996). In brief, diarrhea was scored on the basis of color, consistency and amount, and numbered as follows: 0 = normal; 1 = pasty; 2 = semi-liquid; 3 = liquid, and consider score ≥ 2 as diarrhea.
In vivo reovirus infection
For reovirus infection in mice, five-week-old wild-type Dhx15fl/fl and Dhx15IEC-KO mice were inoculated intragastrically with 1 × 108 plaque-forming units (PFU) of Reovirus (Reovirus type 3 strain Dearing, T3D) in PBS. Reovirus T3D strain was purchased from ATCC (ATCC® VR-824™). For the survival experiments, mice were monitored daily for survival after reovirus infection. At day 1 post-infection, mice were euthanized and the intestine tissue was excised and homogenized in PBS (1ml PBS per 1g tissue). The cytokine production in intestine homogenate was measured by ELISA. At day 2 post-infection, mice were euthanized and the intestine and mesenteric lymph nodes were excised for flow cytometry analysis. At day 4 post-infection, mice were euthanized and the intestine tissue was excised for determining reovirus titer by standard plaque assays.
METHOD DETAILS
Cells culture and lentiviral infection
Human intestinal epithelial cells (IECs) line HT-29 was obtained from ATCC (ATCC HTB-38) and cultured in complete advanced DMEM/F12 medium. The HT-29 IECs were infected with a pLKO.1 lentiviral vector carrying a scrambled shRNA (RHS6848, Horizon Discovery) or target gene sequences (Horizon Discovery) as described in our previous studies (Xing et al., 2016, 2017; Zhang et al., 2011b, 2013). After 24 h of culture, cells were selected by the addition of puromycin (2 ng/ml) to the medium. Cells were stimulated for 16 h with poly I:C (20 μg/ml) delivered by Lipofectamine 3000. The knockdown efficiency was detected with immunoblot analysis.
Isolation of mouse intestinal epithelial cells
Isolation of mouse primary intestinal epithelial cells (IECs) was performed as described previously (Atarashi et al., 2011). Briefly, 6-week-old C57BL/6 mouse intestines were opened longitudinally, washed in phosphate-buffered saline (PBS) and cut into 5-mm fragments. The epithelial integrity was disrupted by treatment with 1 mM dithiothreitol (DTT) on a shaker. Liberated IECs were collected and separated by Percoll gradient (Sigma Aldrich). Interface cells were collected and used as IECs. Purified IECs were cultured in high-glucose-formulated DMEM, supplemented with 10% FBS, 4 mM glutamine, 20 mM HEPES, 1 mM sodium pyruvate, and 100 U/mL penicillin/streptomycin. The purity of isolated IEC was confirmed using FACS analysis with antibodies against IEC markers, PE-anti-E-Cadherin antibody (Catalog: 147304, BioLegend) and FITC-anti-Cytokeratin 18 antibody (Catalog: MA1-10326, ThermoFisher Scientific). Isolated IEC purity and survival rate were both > 94%.
Isolation of lamina propria lymphocytes from intestine
Small intestines were cut into1-cm pieces and treated with RPMI 1640 containing 0.5 mmol/L EDTA for 20 minutes at 37°C to remove epithelial cells and mucus. After washing 4 or 5 times with prewarmed PBS, tissues were minced and dissociated in RPMI 1640 containing 10% fetal bovine serum, 0.5 mg/mL collagenase D and 100 μg/mL DNase I twice for 30 minutes at 37°C with shaking. Lamina propria lymphocytes were enriched by using 40% and 75% Percoll gradients with no brake centrifugation (2200 rpm for 20 minutes at room temperature).
Virus plaque titration
For reovirus infection in mice, viral titers in intestine from infected mice were determined by plaque assay on L929 cells (Virgin et al., 1991). Weights of organs were measured before the assay, and PFU were calculated per mg of tissue. Briefly, tissue was homogenized in 800 μL of PBS. The homogenates were treated with chloroform (10% final concentration), centrifuged briefly and serial dilutions of the aqueous supernatants were incubated on L929 cells at room temperature. After 1 h, the inoculum was removed and cells were covered with 2% agar solution with amphotericin-B. After six days, 2% agar solution containing 2% neutral red solution was added and plaques were visualized with neutral red on the second day (Xing et al., 2011, 2012, 2013, 2017).
Histology
Intestines were removed from wild-type Dhx15fl/fl and Dhx15IEC-KO mice infected by reovirus. These removed intestines were washed using PBS before being fixed with 10% formaldehyde for 24 h at room temperature. The tissues were embedded in paraffin and processed by standard techniques. Longitudinal 5-μm sections were stained with Haematoxylin & Eosin (H&E) and viewed with a digital inverted light microscope (EVOS, Thermo Fisher Scientific, Waltham, MA) as previously described (Xing et al., 2016, 2017; Zhang et al., 2020). Histology score was assessed based on intestinal inflammation and tissue damage. Inflammation was assessed by the presence of infiltrating mononuclear cells, polymorphonuclear cells and lymphocytic cells (scores from 0 to 3, with 0 absent, 1 mild, 2 moderate, 3 severe). For the evaluation of tissue damage four scores were ascribed to crypt hyperplasia, epithelial injury and death of epithelial cells (0 absent, 1 mild, 2 moderate, 3 severe).
In vitro immunoprecipitation and immunoblot analysis
For the endogenous immunoprecipitation interaction assay, mouse primary IECs were infected with or without reovirus at MOI of 5 for 6 hours and then lysed with lysis buffer (50 mM Tris-Cl [pH7.5], 1 mM EDTA, 150 mM NaCl, 1.0% NP-40) containing protease inhibitor cocktail (ThermoFisher Scientific). The cell lysates were incubated with anti-NLRP6 antibody and protein A/G agarose beads for immunoblot analysis. For the preparation of purified DHX15 and NLRP6, HEK293T cells were transfected with expression plasmids encoding full-length or truncated versions of HA- tagged DHX15 including N-terminal domain (N, amino acids 1 to 443 of DHX15), DEAD-like helicases superfamily domain (DEXDc, amino acids 1 to 398 of DHX15) and C-terminal domain (C, amino acids 661 to 794 of DHX15) constructed in pCMV-HA vector (Catalog: 631604, Clontech) as described in our previous study (Lu et al., 2014), or full-length Flag-NLRP6 (Hara et al., 2018). Lysates were prepared from the transfected cells, followed by incubation with anti-HA or anti-Flag beads. Proteins were eluted from the beads after beads were washed six times with PBS. For precipitation with anti-Flag beads, purified HA-tagged full-length DHX15 or truncations of DHX15 were incubated for 2 h with purified Flag-tagged NLRP6. Beads were added; after 2 h of incubation, the bound complexes were pelleted by centrifugation. Proteins and beads were analyzed by immunoblot analysis. HT-29 IECs or mouse primary IECs were washed twice with phosphate-buffered saline (PBS) on ice and lysed in NP-40 lysis buffer with complete protease inhibitor for immunoblot analysis. For immunoblot analysis, all protein samples were dissolved in SDS sample buffer and resolved by 10%–15% SDS-PAGE. After electrophoresis, separated proteins were transferred onto polyvinylidenedifluoride (PVDF) membrane. The membrane was then blocked with 5% nonfat milk. After incubation with specific primary antibody, horseradish peroxidase-conjugated secondary antibody was applied. The positive immune reactive signal was detected by an enhanced chemiluminescence system (ThermoFisher Scientific) as previously described (Xing et al., 2011,2012, 2013, 2015a, 2015b, 2016).
NLRP6 inflammasome reconstitution in HEK293T cells
HEK293T cells were plated in six-well microplates and incubated overnight. The cells were transfected with plasmids including HA-pro-IL-18 (MG50073-CY, SinoBiological, 1000ng/well), Myc-ASC (Cat: 73952, Addgene, 200ng/well), Flag-Caspase-1 (Cat: 21142, Addgene, 200ng/well), with or without Flag-NLRP6 (500ng/well), HA-DHX15 or HA-Vector (100 or 500ng/well) using Lipofectamine 3000. Cells were collected 24 h after transfection and lysed in NP-40 buffer with complete protease inhibitors. IL-18 maturation was assessed by immunoblot analysis.
Flow cytometry
Mouse primary IECs were isolated from wild-type Dhx15fl/fl and Dhx15IEC-KO mice. The cells were then fixed and stained with APC/Cyanine7 anti-mouse CD45 antibody (30-F11, Biolegend), PE anti-mouse CD324 (E-Cadherin) antibody (DECMA-1, Biolegend), PE/Cyanine7 anti-mouse CD326 (EpCAM) antibody (G8.8, Biolegend) and their isotype matched control antibodies for their differentiation. Cells isolated from spleen, mesenteric lymph node (LN), and lamina propria lymphocytes were stained using live/dead Zombie Aqua Fixable viability Kit (Biolegend) for 10 minutes followed by staining with fluorochrome-conjugated antibodies on ice for 20 minutes, washed twice in PBS/BSA, and fixed in 1% paraformaldehyde prior to flow cytometry analysis. Flow cytometry data were acquired on a LSR-II flow cytometer (Beckton Dickinson) and analyzed using FlowJo v10 software (Tree Start) as previously described (Xing et al., 2016).
Quantitative RT-PCR
RNA was isolated using the RNeasy Kit (QIAGEN) according to the manufacturer’s instructions. The isolated RNA was used to synthesize cDNA with the iScript cDNA Synthesis Kit (Bio-Rad). iTaq SYBR Green Supermix with ROX (Bio-Rad) was used for quantitative RT-PCR (qRT-PCR) (Xing et al., 2010; Xing et al., 2018). PCRs were performed in triplicate. Primer sequences used for qRT-PCR are shown in Table S1.
QUANTIFICATION AND STATISTICAL ANALYSIS
A two-tailed unpaired Student’s t test was used for statistical analysis with Microsoft Excel and GraphPad Prism Software. P values of less than 0.05 were considered significant unless specifically indicated.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-DHX15 | Abcam | Cat#ab254591; RRID: AB_2892059 |
| anti-MAVS | Cell Signaling Technology | Cat#3993S; RRID:AB_823565 |
| anti-STING | Cell Signaling Technology | Cat#13647S; RRID:AB_2732796 |
| anti-DHX9 | Abcam | Cat#ab26271; RRID:AB_777725 |
| anti-RIG-I | Cell Signaling Technology | Cat#3743S; RRID:AB_2269233 |
| anti-MDA5 | Cell Signaling Technology | Cat#5321S; RRID:AB_10694490 |
| anti-NLRP6 | Sigma-Aldrich | Cat#SAB1302240; RRID:AB_2750643 |
| anti-NLRP9 | Novus Biologicals | Cat#NBP2-24661; RRID:AB_2892061 |
| anti-GAPDH | Sigma-Aldrich | Cat#G9295; RRID:AB_1078992 |
| anti-β-actin | Sigma-Aldrich | Cat#A3854; RRID:AB_262011 |
| APC/Cyanine7 anti-mouse CD45 | BioLegend | Cat#103116; RRID:AB_312981 |
| PE anti-mouse/human CD324 (E-Cadherin) | BioLegend | Cat#147304; RRID:AB_2563040 |
| PE/Cyanine7 anti-mouse CD326 (Ep-CAM) | BioLegend | Cat#118216; RRID:AB_1236471 |
| FITC anti-mouse CD3 | BioLegend | Cat#100204; RRID:AB_312661 |
| PE/Cyanine7 anti-mouse CD4 | BioLegend | Cat#100528; RRID:AB_312729 |
| PerCP/Cyanine5.5 anti-mouse CD8a | BioLegend | Cat#100734; RRID:AB_2075238 |
| APC anti-mouse CD19 | BioLegend | Cat#152410; RRID:AB_2629839 |
| PE anti-mouse NK-1.1 | BioLegend | Cat#108708; RRID:AB_313395 |
| Bacterial and virus strains | ||
| Reovirus T3D strain | ATCC | Cat#VR-824 |
| Rotavirus EW strain | Dr. Harry B. Greenberg (Stanford University) | Dr. Harry B. Greenberg (Stanford University) |
| Rotavirus SA-11 strain | ATCC | Cat#VR-1565 |
| Herpes simplex virus 1(HSV-1) KOS strain | ATCC | Cat#VR-1493 |
| Vesicular stomatitis virus (VSV) Indiana strain | ATCC | Cat#VR-1238 |
| Influenza A virus A/PR/8/34 strain | ATCC | Cat#VR-95 |
| Biological samples | ||
| Collagenase D | Sigma-Aldrich | Cat#11088882001 |
| DNase I | Sigma-Aldrich | Cat#10104159001 |
| Chemicals, peptides, and recombinant proteins | ||
| UltraPure 0.5M EDTA, pH 8.0 | Invitrogen | Cat#15575020 |
| Percoll | GE Healthcare | Cat#17089101 |
| Critical commercial assays | ||
| Human IFN-β ELISA kit | PBL InterferonSource | Cat#41415-1 |
| Mouse IFN-β ELISA kit | PBL InterferonSource | Cat#42410-1 |
| Human IFN-lambda 3 (IFN-λ3) ELISA kit | R&D systems | Cat#D28B00 |
| Mouse IFN-lambda 3 (IFN-λ3) ELISA kit | R&D systems | Cat#DIY1789B-05 |
| Human IL-18 ELISA kit | R&D systems | Cat#DY318-05 |
| Mouse IL-18 ELISA kit | R&D systems | Cat#DY7625-05 |
| Human IL-6 ELISA kit | R&D systems | Cat#DY206-05 |
| Human TNF-α ELISA kit | R&D systems | Cat#DY210-05 |
| RNeasy Mini Kit (250) | QIAGEN | Cat#74106 |
| iScript cDNA Synthesis Kit | Bio-Rad | Cat#1708891 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat#1725125 |
| Zombie Aqua Fixable Viability Kit | BioLegend | Cat#423102 |
| Poly(I:C) HMW | InvivoGen | Cat#tlrl-pic-5 |
| Poly(dG:dC) | InvivoGen | Cat#tlrl-pgcn |
| Experimental models: cell lines | ||
| Human intestinal epithelial cells (IECs) line HT-29 | ATCC | Cat#HTB-38 |
| HEK293T cells | ATCC | Cat#CRL-3216 |
| L929 cells | ATCC | Cat#CCL-1 |
| Experimental models: organisms/strains | ||
| Dhx15fl/fl conditional knockout mice | This paper | Taconic-Artemis |
| Villin-Cre transgenic mice | The Jackson Laboratory | Stock No: 021504 |
| IEC-specific Dhx15-knockout
mice,
Dhx15fl/fl; Villin-Cre (Dhx15IEC-KO) |
This paper | This paper |
| Oligonucleotides | ||
| shRNA for human DHX15 | Horizon Discovery | clone ID: TRCN0000000009 |
| shRNA for human MAVS | Horizon Discovery | clone ID: TRCN0000146651 |
| shRNA for human STING | Horizon Discovery | clone ID: TRCN0000161052 |
| shRNA for human RIG-I | Horizon Discovery | clone ID: TRCN0000151446 |
| shRNA for human MDA5 | Horizon Discovery | clone ID: TRCN0000050852 |
| See Table S1 for primer sequences | This study | N/A |
| Recombinant DNA | ||
| HA-DHX15 | This paper | This paper |
| HA-pro-IL-18 | SinoBiological | Cat#MG50073-CY |
| Myc-ASC | Addgene | Cat#73952 |
| Flag-Caspase-1 | Addgene | Cat#21142 |
| Flag-NLRP6 | Dr. Gabriel Núñez (University of Michigan) | Dr. Gabriel Núñez (University of Michigan) |
| Software and algorithms | ||
| Prism (version 8) | GraphPad Software | https://www.graphpad.com; RRID:SCR_002798 |
| FlowJo (version 10) | BD Biosciences | https://www.flowjo.com/ |
| Office 365 | Microsoft | https://www.office.com/ |
Highlights.
DHX15 senses RNA viruses to produce IFN-β, IFN-λ3, and IL-18 in IECs
IEC-specific Dhx15-knockout mice are generated
DHX15 is required for controlling RNA virus-induced intestinal inflammation in vivo
DHX15 interacts with NLRP6 to trigger inflammasome activation in IECs
ACKNOWLEDGMENTS
We thank Dr. Harry B. Greenberg (Stanford University) and Dr. Gabriel Núñez (University of Michigan) for providing the kind gifts of the Rotavirus EW strain and plasmid Flag-NLRP6, respectively. This work was supported by Lupus Research Alliance grant 519418 (to Z.Z.), National Institutes of Health grant R56AI148215 and R01AI155488 (to Z.Z.), and American Heart Association Career Development Award 20CDA35260116 (to J.X.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109205.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ablasser A, and Hur S (2020). Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat. Immunol 21, 17–29. [DOI] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Röhl I, Hopfner KP, Ludwig J, and Hornung V (2013). cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel J, Franco MA, Greenberg HB, and Bass D (1999). Lack of a role for type I and type II interferons in the resolution of rotavirus-induced diarrhea and infection in mice. J. Interferon Cytokine Res 19, 655–659. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball JM, Tian P, Zeng CQ, Morris AP, and Estes MK (1996). Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272, 101–104. [DOI] [PubMed] [Google Scholar]
- Bányai K, Estes MK, Martella V, and Parashar UD (2018). Viral gastroen-teritis. Lancet 392, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JW, Krishnaney AA, Vo PT, Rouse RV, Anderson LJ, and Greenberg HB (1995). Analyses of homologous rotavirus infection in the mouse model. Virology 207, 143–153. [DOI] [PubMed] [Google Scholar]
- Chen YL, Capeyrou R, Humbert O, Mouffok S, Kadri YA, Lebaron S, Henras AK, and Henry Y (2014). The telomerase inhibitor Gno1p/PINX1 activates the helicase Prp43p during ribosome biogenesis. Nucleic Acids Res. 42, 7330–7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XL, Cai YH, Liu Q, Pan LL, Shi SL, Liu XL, Chen Y, Li JG, Wang J, Li Y, et al. (2018). ETS1 and SP1 drive DHX15 expression in acute lymphoblastic leukaemia. J. Cell. Mol. Med 22, 2612–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal EM, Lahl K, Narváez CF, Butcher EC, and Greenberg HB (2013). Plasmacytoid dendritic cells promote rotavirus-induced human and murine B cell responses. J. Clin. Invest 123, 2464–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RK, Kotenko SV, and Durbin JE (2013). Interferon induction and function at the mucosal surface. Immunol. Rev 255, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng N, Kim B, Fenaux M, Nguyen H, Vo P, Omary MB, and Greenberg HB (2008). Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J. Virol 82, 7578–7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu YT, Du F, Aroh C, Yan N, Sun L, and Chen ZJ (2013). Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341, 903–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, et al. ; China Medical Treatment Expert Group for Covid-19 (2020). Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med 382, 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Seregin SS, Yang D, Fukase K, Chamaillard M, Alnemri ES, Inohara N, Chen GY, and Núñez G (2018). The NLRP6 Inflammasome Recognizes Lipoteichoic Acid and Regulates Gram-Positive Pathogen Infection. Cell 175, 1651–1664.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, and Barber GN (2008). STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Ma Z, and Barber GN (2009). STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Koso H, Sakamoto K, and Watanabe S (2017). RNA helicase DHX15 acts as a tumour suppressor in glioma. Br. J. Cancer 117, 1349–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E (2011). RNA helicases at work: binding and rearranging. Trends Biochem. Sci 36, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y, Nguyen MM, Wang D, Pascal LE, Guo W, Xu Y, Ai J, Deng FM, Masoodi KZ, Yu X, et al. (2018). DHX15 promotes prostate cancer progression by stimulating Siah2-mediated ubiquitination of androgen receptor. Oncogene 37, 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C, Wetzel JD, He J, Mikacenic C, Dermody TS, and Kelsall BL (2007). Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J. Exp. Med 204, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD (2010). Central roles of NLRs and inflammasomes in viral infection. Nat. Rev. Immunol 10, 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, and Akira S (2005). IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol 6, 981–988. [DOI] [PubMed] [Google Scholar]
- Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, Ravelli RBG, Paul van Schayck J, Mykytyn AZ, Duimel HQ, et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear HM, Nice TJ, and Diamond MS (2015). Interferon-λ: Immune Functions at Barrier Surfaces and Beyond. Immunity 43, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei-Leston AC, Murphy AG, and Maloy KJ (2017). Epithelial Cell Inflammasomes in Intestinal Immunity and Inflammation. Front. Immunol 8, 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, et al. (2015). Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 163, 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, and Zhu S (2020). NLRP6 inflammasome. Mol. Aspects Med 76, 100859. [DOI] [PubMed] [Google Scholar]
- Lin ML, Fukukawa C, Park JH, Naito K, Kijima K, Shimo A, Ajiro M, Nishidate T, Nakamura Y, and Katagiri T (2009). Involvement of G-patch domain containing 2 over expression in breast carcinogenesis. Cancer Sci 100, 1443–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P (2006). Dead-box proteins: a family affair–active and passive players in RNP-remodeling. Nucleic Acids Res. 34, 4168–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Lu N, Weng L, Yuan B, Liu YJ, and Zhang Z (2014). DHX15 senses double-stranded RNA in myeloid dendritic cells. J. Immunol 193, 1364–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer C, Malik A, and Kanneganti TD (2015). Inflammasome control of viral infection. Curr. Opin. Virol 12, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, and Gumucio DL (2002). Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem 277, 33275–33283. [DOI] [PubMed] [Google Scholar]
- Memet I, Doebele C, Sloan KE, and Bohnsack MT (2017). The G-patch protein NF-κB-repressing factor mediates the recruitment of the exonuclease XRN2 and activation of the RNA helicase DHX15 in human ribosome biogenesis. Nucleic Acids Res. 45, 5359–5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger RN, Krug AB, and Eisenächer K (2018). Enteric Virome Sensing-Its Role in Intestinal Homeostasis and Immunity. Viruses 10, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitoma H, Hanabuchi S, Kim T, Bao M, Zhang Z, Sugimoto N, and Liu YJ (2013). The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity 39, 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosallanejad K, Sekine Y, Ishikura-Kinoshita S, Kumagai K, Nagano T, Matsuzawa A, Takeda K, Naguro I, and Ichijo H (2014). The DEAH-box RNA helicase DHX15 activates NF-κB and MAPK signaling downstream of MAVS during antiviral responses. Sci. Signal 7, ra40. [DOI] [PubMed] [Google Scholar]
- Muruve DA, Pétrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, and Tschopp J (2008). The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452, 103–107. [DOI] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner KP, and Ha T (2009). Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323, 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohka S, Igarashi H, Nagata N, Sakai M, Koike S, Nochi T, Kiyono H, and Nomoto A (2007). Establishment of a poliovirus oral infection system in human poliovirus receptor-expressing transgenic mice that are deficient in alpha/beta interferon receptor. J. Virol 81, 7902–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Sakai K, Matsumoto M, and Seya T (2010). DEAD/H BOX 3 (DDX3) helicase binds the RIG-I adaptor IPS-1 to up-regulate IFN-beta-inducing potential. Eur. J. Immunol 40, 940–948. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Miyashita M, Okamoto M, Morioka Y, Okabe M, Matsumoto M, and Seya T (2015). DDX60 Is Involved in RIG-I-Dependent and Independent Antiviral Responses, and Its Function Is Attenuated by Virus-Induced EGFR Activation. Cell Rep. 11, 1193–1207. [DOI] [PubMed] [Google Scholar]
- Pattabhi S, Knoll ML, Gale M Jr., and Loo YM (2019). DHX15 Is a Coreceptor for RLR Signaling That Promotes Antiviral Defense Against RNA Virus Infection. J. Interferon Cytokine Res 39, 331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pott J, Mahlakõiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, and Hornef MW (2011). IFN-lambda determines the intestinal epithelial antiviral host defense. Proc. Natl. Acad. Sci. USA 108, 7944–7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranji A, and Boris-Lawrie K (2010). RNA helicases: emerging roles in viral replication and the host innate response. RNA Biol. 7, 775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulzke JD, Ploeger S, Amasheh M, Fromm A, Zeissig S, Troeger H, Richter J, Bojarski C, Schumann M, and Fromm M (2009). Epithelial tight junctions in intestinal inflammation. Ann. N Y Acad. Sci 1165, 294–300. [DOI] [PubMed] [Google Scholar]
- Sen A, Rothenberg ME, Mukherjee G, Feng N, Kalisky T, Nair N, Johnstone IM, Clarke MF, and Greenberg HB (2012). Innate immune response to homologous rotavirus infection in the small intestinal villous epithelium at single-cell resolution. Proc. Natl. Acad. Sci. USA 109, 20667–20672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, and Chen ZJ (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122, 669–682. [DOI] [PubMed] [Google Scholar]
- Sommereyns C, Paul S, Staeheli P, and Michiels T (2008). IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS Pathog 4, e1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer MK, Ivanović L, Weber ME, Marti S, and Jonas S (2020). Structural basis for DEAH-helicase activation by G-patch proteins. Proc. Natl. Acad. Sci. USA 117, 7159–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Mitoma H, Kim T, Hanabuchi S, and Liu YJ (2014). Helicase proteins DHX29 and RIG-I cosense cytosolic nucleic acids in the human airway system. Proc. Natl. Acad. Sci. USA 111, 7747–7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, and Chen ZJ (2006). The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24, 633–642. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, and Chen ZJ (2013). Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson KV, Junkins RD, Kurkjian CJ, Holley-Guthrie E, Pendse AA, El Morabiti R, Petrucelli A, Barber GN, Benedict CA, and Ting JP (2017). A noncanonical function of cGAMP in inflammasome priming and activation. J. Exp. Med 214, 3611–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro JR, Ng C, Lee AM, Sullivan BM, Sheehan KC, Welch M, Schreiber RD, de la Torre JC, and Oldstone MB (2013). Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW 4th, Mann MA, Fields BN, and Tyler KL (1991). Monoclonal antibodies to reovirus reveal structure/function relationships between capsid proteins and genetics of susceptibility to antibody action. J. Virol 65, 6772–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wack A, Terczyńska-Dyla E, and Hartmann R (2015). Guarding the frontiers: the biology of type III interferons. Nat. Immunol 16, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jiang W, Yan Y, Gong T, Han J, Tian Z, and Zhou R (2014). RNA viruses promote activation of the NLRP3 inflammasome through a RIP1-RIP3-DRP1 signaling pathway. Nat. Immunol 15, 1126–1133. [DOI] [PubMed] [Google Scholar]
- Wang P, Zhu S, Yang L, Cui S, Pan W, Jackson R, Zheng Y, Rongvaux A, Sun Q, Yang G, et al. (2015). Nlrp6 regulates intestinal antiviral innate immunity. Science 350, 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hu D, Wu C, Feng Y, Li A, Liu W, Wang Y, Chen K, Tian M, Xiao F, et al. (2020). STING promotes NLRP3 localization in ER and facilitates NLRP3 deubiquitination to activate the inflammasome upon HSV-1 infection. PLoS Pathog. 16, e1008335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YF, Li JM, Wang SM, Yong X, Tang B, Jie MM, Dong H, Yang XC, and Yang SM (2016). Cerium oxide nanoparticles inhibit the migration and proliferation of gastric cancer by increasing DHX15 expression. Int. J. Nanomedicine 11, 3023–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Liao H, Zhang C, and Zhang S (2019). Overexpression and clinical relevance of the RNA helicase DHX15 in hepatocellular carcinoma. Hum. Pathol 84, 213–220. [DOI] [PubMed] [Google Scholar]
- Xing J, Wu F, Wang S, Krensky AM, Mody CH, and Zheng C (2010). Granulysin production and anticryptococcal activity is dependent upon a far upstream enhancer that binds STAT5 in human peripheral blood CD4+ T cells. J. Immunol 185, 5074–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Wang S, Lin F, Pan W, Hu CD, and Zheng C (2011). Comprehensive characterization of interaction complexes of herpes simplex virus type 1 ICP22, UL3, UL4, and UL20.5. J. Virol 85, 1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Wang S, Lin R, Mossman KL, and Zheng C (2012). Herpes simplex virus 1 tegument protein US11 downmodulates the RLR signaling pathway via direct interaction with RIG-I and MDA-5. J. Virol 86, 3528–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Ni L, Wang S, Wang K, Lin R, and Zheng C (2013). Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-κB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J. Virol 87, 9788–9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Chai Z, Ly H, and Liang Y (2015a). Differential Inhibition of Macrophage Activation by Lymphocytic Choriomeningitis Virus and Pichinde Virus Is Mediated by the Z Protein N-Terminal Domain. J. Virol 89, 12513–12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Ly H, and Liang Y (2015b). The Z proteins of pathogenic but not nonpathogenic arenaviruses inhibit RIG-I-like receptor-dependent interferon production. J. Virol 89, 2944–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Weng L, Yuan B, Wang Z, Jia L, Jin R, Lu H, Li XC, Liu YJ, and Zhang Z (2016). Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat. Immunol 17, 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhang A, Zhang H, Wang J, Li XC, Zeng MS, and Zhang Z (2017). TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat. Commun 8, 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhang A, Minze LJ, Li XC, and Zhang Z (2018). TRIM29 Negatively Regulates the Type I IFN Production in Response to RNA Virus. J. Immunol 201, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, and Shu HB (2005). VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19, 727–740. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, and Fujita T (2004). The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol 5, 730–737. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M Jr., Akira S, et al. (2005). Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol 175, 2851–2858. [DOI] [PubMed] [Google Scholar]
- Yoshimoto R, Kataoka N, Okawa K, and Ohno M (2009). Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res. 37, 891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, et al. (2020). TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol 5, eabc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, and Liu YJ (2011a). DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity 34, 866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, and Liu YJ (2011b). The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol 12, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Lu N, Facchinetti V, and Liu YJ (2011c). DHX9 pairs with IPS-1 to sense double-stranded RNA in myeloid dendritic cells. J. Immunol 187, 4501–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Bao M, Lu N, Weng L, Yuan B, and Liu YJ (2013). The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol 14, 172–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Xing J, Xia T, Zhang H, Fang M, Li S, Du Y, Li XC, Zhang Z, and Zeng MS (2020). EphA2 phosphorylates NLRP3 and inhibits inflammasomes in airway epithelial cells. EMBO Rep 21, e49666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, and Shu HB (2008). The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550. [DOI] [PubMed] [Google Scholar]
- Zhu S, Ding S, Wang P, Wei Z, Pan W, Palm NW, Yang Y, Yu H, Li HB, Wang G, et al. (2017). Nlrp9b inflammasome restricts rotavirus infection in intestinal epithelial cells. Nature 546, 667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. ; China Novel Coronavirus Investigating and Research Team (2020). A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med 382, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets or code.


