Abstract
Background
Although social cognitive dysfunction is a major feature of frontotemporal dementia (FTD), it has been poorly studied in familial forms. A key goal of studies is to detect early cognitive impairment using validated measures in large patient cohorts.
Methods
We used the Revised Self-Monitoring Scale (RSMS) as a measure of socioemotional sensitivity in 730 participants from the genetic FTD initiative (GENFI) observational study: 269 mutation-negative healthy controls, 193 C9orf72 expansion carriers, 193 GRN mutation carriers and 75 MAPT mutation carriers. All participants underwent the standardised GENFI clinical assessment including the ‘CDR® plus NACC FTLD’ scale and RSMS. The RSMS total score and its two subscores, socioemotional expressiveness (EX score) and modification of self-presentation (SP score) were measured. Volumetric T1-weighted magnetic resonance imaging was available from 377 mutation carriers for voxel-based morphometry (VBM) analysis.
Results
The RSMS was decreased in symptomatic mutation carriers in all genetic groups but at a prodromal stage only in the C9orf72 (for the total score and both subscores) and GRN (for the modification of self-presentation subscore) groups. RSMS score correlated with disease severity in all groups. The VBM analysis implicated an overlapping network of regions including the orbitofrontal cortex, insula, temporal pole, medial temporal lobe and striatum.
Conclusions
The RSMS indexes socioemotional impairment at an early stage of genetic FTD and may be a suitable outcome measure in forthcoming trials.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13195-021-00865-w.
Keywords: Frontotemporal dementia, Familial, C9orf72, GRN, MAPT, RSMS, CDR® plus NACC FTLD, VBM
Background
Frontotemporal dementia (FTD) is a complex and heterogeneous neurodegenerative disease, manifesting itself as a diverse spectrum of clinical syndromes. However, despite differences in presentation, many people with FTD develop impaired social cognition [1], a set of psychological processes which includes the ability to evaluate social and emotional cues from others and then select an appropriate behavioural response, a phenomenon often referred to as ‘socioemotional sensitivity’ or ‘self-monitoring’. In both healthy and clinical populations, the Revised Self-Monitoring Scale (RSMS) [2] has often been used to study socioemotional sensitivity and responsiveness as well as the neural networks that underlie them [3, 4].
Unlike many neurodegenerative diseases, FTD is highly heritable with approximately a third of patients having a causative autosomal dominant genetic mutation [5]. Mutations are most commonly found in one of three genes, chromosome 9 open reading frame 72 (C9orf72), progranulin (GRN) and microtubule-associated protein tau (MAPT) [6], with the most common clinical presentation being behavioural variant FTD (bvFTD) [7]. However, whilst social cognitive dysfunction has been studied extensively in sporadic FTD, few investigations have looked at genetic cohorts exclusively.
The Genetic FTD Initiative (GENFI) is a multicentre natural history study aimed at investigating early biomarkers in a large genetic FTD cohort, including measures of cognition [5]. This study sought to assess whether the RSMS could detect early changes in social cognition and what the underlying neural correlates of the RSMS were in people with mutations in C9orf72, GRN and MAPT.
Methods
Participants
Participants were recruited from the fifth data freeze of GENFI, incorporating data from 24 sites. Of the 849 participants enrolled in the second phase of the study, cross-sectional data on the RSMS was available from 730 participants, consisting of 269 healthy controls (family members who tested negative for the mutation carried within the family), 193 C9orf72 expansion carriers, 193 GRN mutation carriers and 75 MAPT mutation carriers (Table 1). All participants provided written informed consent.
Table 1.
Demographics and the RSMS total, EX and SP scores for each genetic group, split by global CDR® plus NACC FTLD score (0, 0.5, 1+). N represents number of participants, mean (standard deviation) shown for age, education and cognitive test scores. In the symptomatic (1+) groups, MMSE scores were significantly lower in GRN mutation carriers than in the C9orf72 expansion carrier group but no other differences were seen, whilst no differences were seen in the CDR® plus NACC FTLD-SB
| N | Sex | Age (years) | Education (years) | MMSE (/30) | CDR plus NACC FTLD-SB | RSMS total (/65) | RSMS EX (/30) | RSMS SP (/35) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| % male | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | |||
| Controls | 269 | 42 | 46.2 (13.0) | 14.4 (3.4) | 29.3 (1.1) | 0.2 (0.4) | 47.8 (8.4) | 23.3 (4.2) | 24.5 (5.3) | |
| C9orf72 | 0 | 93 | 41 | 43.9 (11.6) | 14.3 (3.0) | 29.1 (1.2) | 0.0 (0.0) | 47.1 (10.5) | 22.8 (5.4) | 24.3 (6.0) |
| 0.5 | 34 | 44 | 49.7 (11.2) | 14.0 (2.6) | 28.4 (2.2) | 1.1 (0.7) | 41.9 (11.4) | 19.8 (6.2) | 22.1 (6.3) | |
| 1+ | 66 | 65 | 62.7 (9.5) | 13.0 (3.8) | 23.3 (6.8) | 11.1 (5.6) | 23.5 (12.3) | 9.6 (7.0) | 14.0 (6.6) | |
| GRN | 0 | 122 | 34 | 45.6 (12.2) | 14.7 (3.5) | 29.5 (0.8) | 0.0 (0.0) | 47.9 (8.9) | 23.6 (4.0) | 24.3 (5.9) |
| 0.5 | 24 | 46 | 51.3 (13.8) | 14.0 (4.3) | 28.6 (2.3) | 0.9 (0.8) | 43.8 (10.7) | 21.6 (6.3) | 22.2 (5.6) | |
| 1+ | 47 | 47 | 63.0 (7.4) | 11.7 (3.4) | 20.1 (7.7) | 9.8 (6.2) | 28.6 (12.1) | 12.9 (6.7) | 15.6 (6.1) | |
| MAPT | 0 | 41 | 41 | 38.3 (11.0) | 14.3 (3.3) | 29.5 (0.8) | 0.0 (0.0) | 50.7 (9.7) | 24.0 (4.5) | 26.7 (6.0) |
| 0.5 | 13 | 31 | 46.4 (12.8) | 13.6 (2.5) | 28.1 (2.3) | 1.1 (0.8) | 50.1 (14.2) | 23.8 (7.5) | 26.3 (7.1) | |
| 1+ | 21 | 57 | 58.9 (9.4) | 13.6 (4.0) | 21.9 (8.1) | 10.3 (6.0) | 22.8 (18.9) | 9.4 (9.5) | 13.4 (9.8) | |
Assessments
All participants were given the standardised GENFI clinical assessment battery including a medical history, physical examination, the Mini-Mental State Examination, and the CDR® Dementia Staging Instrument with the National Alzheimer Coordinating Centre Frontotemporal Lobar Degeneration component (CDR® plus NACC FTLD) (Table 1). The CDR® plus NACC FTLD is a clinical measure of disease severity in FTD, consisting of a core six cognitive/functional domains with a further 2 domains addressing behaviour and language [8]. Each domain is rated on a five-point scale ranging from 0 (normal), 0.5 (questionably or minimally impaired), 1 (mildly but definitely impaired), 2 (moderately impaired), to 3 (severely impaired). The sum of ratings across all eight domains is used to generate the CDR® plus NACC FTLD sum of boxes (CDR® plus NACC FTLD-SB) (Table 1). A second measure, a global CDR® plus NACC FTLD score can also be generated, using a specific algorithm [9]. We used this global score to classify each of the genetic groups cross-sectionally into those who scored 0 (i.e. were asymptomatic), 0.5 (possibly or mildly symptomatic i.e. prodromal), and 1 or more (fully symptomatic mutation carriers). A neuropsychological assessment was also performed including the Trail Making Test Parts A and B, the WAIS-R Digit Symbol test, the D-KEFS Color-Word Interference Test Ink Naming, category fluency (animals), the Faux Pas recognition test, and the Facial Emotion Recognition Test.
Demographics
Demographics are shown in Table 1. There was a significant difference in sex between these groups: symptomatic C9orf72 carriers had a significantly higher percentage of males than in the mildly symptomatic and asymptomatic C9orf72 carrier groups and in the controls (X2(1) = 4.08, p = 0.044, X2(1) = 9.12, p = 0.003 and X2(1) = 11.79, p = 0.001, respectively). There was also a significant difference in age between groups (F(9,720)) = 27.5, p < 0.001): asymptomatic MAPT mutation carriers were significantly younger and mildly symptomatic GRN mutation carriers were significantly older than controls (p < 0.001 and p = 0.043 respectively). All symptomatic mutation carriers were significantly older than controls (p < 0.001). Analysis of differences in years spent in education (F(9,720)) = 4.09, p < 0.001) showed that symptomatic C9orf72 and GRN mutation carriers spent significantly fewer years when compared to controls (p = 0.003 and p < 0.001, respectively). All analyses were therefore adjusted for sex, age and education.
Revised Self-Monitoring Scale (RSMS)
The RSMS is a widely used questionnaire made up of 13 items designed to measure an individual’s awareness of social behaviour and sensitivity to subtle emotional expressions during face-to-face interaction [10]. Items include ‘In conversations, the subject is sensitive to even the slightest change in the facial expression of the person he/she is conversing with’ and ‘If someone is lying to the subject, he/she usually knows it at once from that person’s manner or expression’. Each item is rated by a participant’s informant on a 6-point scale, ranging from ‘certainly, always false’ (0 points) to ‘certainly, always true’ (6 points). As well as a total score, two subscores of the RSMS can also be calculated: socioemotional expressiveness i.e. the ability to understand subtle social cues in others (EX score, out of 30), and modification of self-presentation i.e. the ability to change one’s behaviour when it is not appropriate for the current social situation (SP score, out of 35).
Statistical analysis
Statistical analyses were performed using StataCorp. 2019. Stata Statistical Software: Release 16. College Station, TX: StataCorp LLC. In the healthy control group, Spearman rank correlations were performed to assess the relationship between the RSMS total score, age, sex and education. Cross-sectional RSMS total, EX and SP scores were compared between groups (healthy controls, and 0, 0.5 and 1+ in each genetic group) using a linear regression model adjusting for age, sex and education, with 95% bias-corrected bootstrapped confidence intervals with 1000 repetitions (to correct for non-normally distributed data). Spearman rank correlations were performed in each genetic group to investigate the association between RSMS total score and disease severity (as measured by CDR® plus NACC FTLD-SB). Finally, non-parametric partial correlations adjusting for age, sex, education and disease severity were also performed to assess the relationship between RSMS total score and cognition.
Image acquisition and processing
Participants underwent volumetric T1-weighted magnetic resonance imaging according to the harmonized GENFI protocol on a 3T scanner. All images underwent quality control and any scans with movement or artefacts were eliminated from analysis. In addition, any scans displaying moderate to severe vascular disease or any lesion presentation were also excluded. 377 scans were included in the analysis: 151 C9orf72 expansion carriers, 162 GRN mutation carriers and 64 MAPT mutation carriers. Voxel-based morphometry (VBM) was subsequently performed using Statistical Parametric Mapping (SPM) 12(www.fil.ion.ucl.ac.uk/spm), running under Matlab R2014a (Mathworks, USA). T1-weighted images were normalised and segmented into grey matter (GM), white matter (WM) and cerebrospinal fluid (CSF) probability maps using standard procedures and a fast-diffeomorphic image registration algorithm (DARTEL) [11]. Prior to analysis, GM segmentations were then transformed into Montreal Neurological Institute (MNI) space, modulated and smoothed using a Gaussian kernel with 6-mm fill-width at half maximum, before applying a mask image as reported in Ridgway et al. 2009 [12]. In order to investigate the neural correlates of socioemotional sensitivity in each genetic group, multiple regression models were performed to explore the relationship of RSMS total score and GM density in mutation carriers in each genetic group. Age, sex, scanner type and total intracranial volume (TIV, calculated using SPM [13]) were included as nuisance covariates. The Family-Wise Error (FWE) correction for multiple comparisons was set at 0.05. However, if no findings were observed at that strict level of correction, results were reviewed at an uncorrected p value of 0.001.
Results
Healthy control performance on the RSMS
Mean (standard deviation) RSMS total score was 47.8 (8.4) in controls (Tables S1 and S2). Overall, there was no significant difference between performance in females (n = 157: 48.5 (8.0)) and males (n = 112: 46.8 (9.0) (p = 0.21). No significant correlations between RSMS total score and age (rho = 0.01, p = 0.87) or education (rho = 0.12, p = 0.06) were observed.
Cross-sectional analysis of mutation carriers
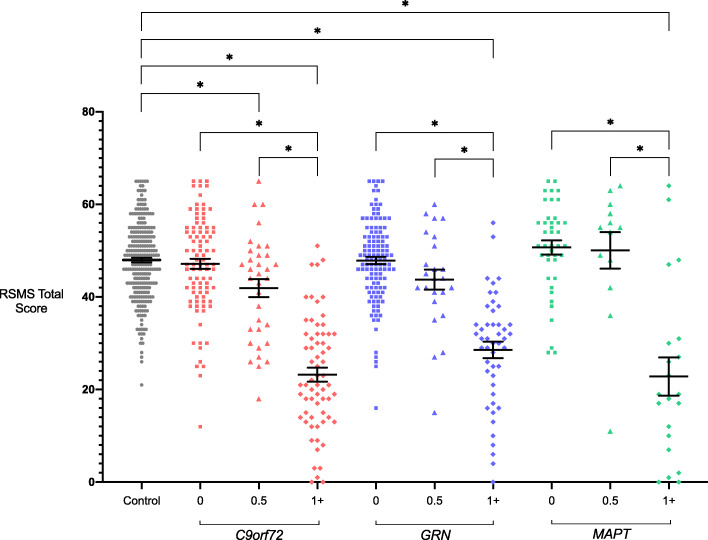
Mean RSMS total scores in all symptomatic (CDR 1+) mutation carriers were significantly lower than in healthy controls (Tables 1 and 2, Fig. 1): C9orf72 23.5 (12.3), GRN 28.6 (12.1) and MAPT 22.8 (18.9). In the CDR 0.5 groups, the C9orf72 group also scored significantly lower than controls with a trend for a lower score in the GRN group and no difference in the MAPT group: C9orf72 41.9 (11.4), GRN 43.8 (12.1) and MAPT 50.1 (14.2). No significant differences were observed between the asymptomatic (CDR 0) mutation carrier groups and controls.
Table 2.
Adjusted mean differences in RSMS total scores between the genetic groups stratified by global CDR® plus NACC FTLD scores, with 95% bias-corrected confidence intervals. Significant values are shown in bold
| C9ORF72 | GRN | MAPT | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.5 | 1+ | 0 | 0.5 | 1+ | 0 | 0.5 | 1+ | |||||||||||
| Controls | −0.91 | −5.34 | −22.39 | −0.35 | −4.05 | −17.41 | 1.99 | 2.07 | −24.28 | ||||||||||
| −3.41 | 1.60 | −9.36 | -1.32 | −25.72 | −19.05 | −2.17 | 1.47 | −8.53 | 0.42 | −20.94 | −13.88 | −1.21 | 5.19 | −5.72 | 9.86 | −32.48 | −16.08 | ||
| C9ORF72 | 0 | −4.43 | −21.48 | 0.55 | −3.15 | −16.50 | 2.90 | 2.97 | −23.37 | ||||||||||
| −9.10 | 0.24 | −25.68 | −17.29 | −2.29 | 3.39 | −8.19 | 1.90 | −20.84 | −12.17 | −0.91 | 6.71 | −5.02 | 10.97 | −31.89 | −14.86 | ||||
| 0.5 | −17.05 | 4.99 | 1.29 | −12.07 | 7.33 | 7.41 | −18.94 | ||||||||||||
| −21.93 | −12.16 | 0.61 | 9.36 | −4.54 | 7.11 | −17.17 | −6.98 | 2.19 | 12.48 | −1.18 | 15.99 | −27.88 | −10.01 | ||||||
| 1+ | 22.04 | 18.34 | 4.98 | 24.38 | 24.45 | −1.89 | |||||||||||||
| 18.41 | 25.66 | 12.93 | 23.74 | 0.40 | 9.55 | 19.79 | 28.97 | 16.28 | 32.63 | −10.40 | 6.61 | ||||||||
| GRN | 0 | −3.70 | −17.06 | 2.35 | 2.42 | −23.93 | |||||||||||||
| −8.60 | 1.20 | −20.77 | −13.35 | −1.31 | 6.00 | −5.61 | 10.44 | −32.26 | −15.60 | ||||||||||
| 0.5 | −13.36 | 6.05 | 6.12 | −20.23 | |||||||||||||||
| −18.76 | −7.96 | 0.72 | 11.38 | −2.59 | 14.83 | −29.17 | −11.29 | ||||||||||||
| 1+ | 19.41 | 19.48 | −6.87 | ||||||||||||||||
| 14.54 | 24.27 | 11.10 | 27.85 | −15.59 | 1.85 | ||||||||||||||
| MAPT | 0 | 0.07 | −26.27 | ||||||||||||||||
| −7.46 | 7.60 | −35.02 | −17.53 | ||||||||||||||||
| 0.5 | −26.35 | ||||||||||||||||||
| −36.96 | −15.74 | ||||||||||||||||||
| 1+ | |||||||||||||||||||
Fig. 1.
RSMS total scores in each genetic group, stratified by global CDR® plus NACC FTLD scores. Bars represent the mean score and standard error of the mean in each group. Significant differences from controls and within each genetic group are starred. Differences between different genetic groups are not shown
Within each genetic group, there was a significantly lower RSMS total score in the symptomatic group compared with the CDR 0.5 and CDR 0 groups (Tables 1 and 2, Fig. 1).
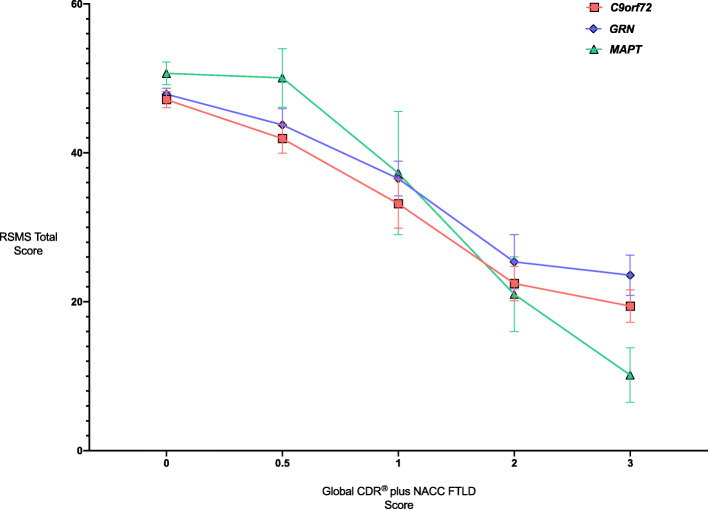
Stratifying by individual global CDR® plus NACC FTLD score (0, 0.5, 1, 2 and 3), all genetic groups show decreasing RSMS total score with increasing CDR (Fig. 2).
Fig. 2.
Mean RSMS total scores in each genetic group by individual global CDR® plus NACC FTLD score. Error bars represent standard error of the mean
RSMS EX and SP scores followed a similar pattern as for RSMS total performance (Table 1, Tables S3 and S4, Figures S1 and S2): the mean scores in all symptomatic (CDR 1+) mutation carriers and the C9orf72 CDR 0.5 group were significantly lower than in healthy controls for both EX and SP scores. However, additionally, the GRN CDR 0.5 group had significantly lower mean SP score than controls. Within each genetic group, there was a significantly lower RSMS EX and SP score in the symptomatic groups compared with the CDR 0.5 and CDR 0 groups, with EX score also lower in the C9orf72 CDR 0.5 group compared with the CDR 0 group (Tables S3 and S4, Figures S1 and S2).
Relationship between RSMS and CDR® plus NACC FTLD-SB
A strong negative correlation between RSMS total score and CDR® plus NACC FTLD-SB scores was observed for all genetic groups (Figure S3): C9orf72 (r = −0.67, p < 0.001), GRN (r = −0.59, p < 0.001) and MAPT (r = −0.53, p < 0.001).
Relationship between RSMS and cognition
A weak positive correlation was found between RSMS total score and one test of social cognition, the Facial Emotion Recognition test, in the C9orf72 group only (r = 0.18, p = 0.018; Table S5). However, no significant correlations were found on other tests of cognition except for category fluency where there was a weak positive correlation in both the C9orf72 (r=0.15, p = 0.047) and GRN (r=0.15, p = 0.047) groups.
Neural correlates of RSMS in each genetic group
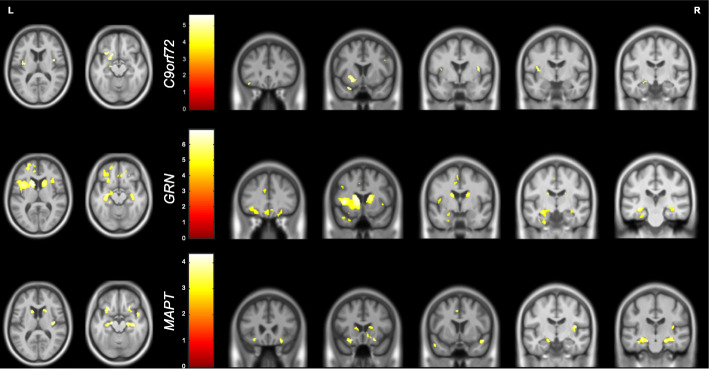
The VBM analysis revealed positive associations of the RSMS total score with grey matter volume corrected for multiple comparisons in the C9orf72 and GRN groups, but only at an uncorrected p value of <0.001 for the MAPT group. Overlapping neural correlates were seen in each of the genetic groups, with an association of decreased score with lower grey matter volume in the orbitofrontal lobe, insula, temporal pole, medial temporal lobe and both caudate and putamen (Fig. 3, Table S6).
Fig. 3.
Neural correlates of RSMS total score. Results for C9orf72 and GRN groups are shown corrected at p < 0.05, with results for the MAPT group shown at p < 0.001 uncorrected. Results are shown on a study-specific T1-weighted MRI template in MNI space
Discussion
In this study, we have shown that the RSMS detects social cognitive impairment in genetic FTD, including early difficulties within the CDR 0.5 group of C9orf72 mutation carriers for the total score and for both C9orf72 and GRN mutation carriers for the modification of self-presentation (SP) subscore. RSMS total score is highly correlated with ‘CDR® plus NACC FTLD’ score and with an overlapping ‘social cognitive’ network of regions including orbitofrontal, anteromedial temporal, insula and striatal areas.
The results here show that the RSMS score decreases with increasing disease severity as measured by the CDR® plus NACC FTLD score, with a significant negative correlation between both scores in each genetic group i.e. RSMS decreases as CDR® plus NACC FTLD increases. This relationship has also been described in a recent study [14], although that study did not separate mutation carriers into separate genetic groups.
Carriers of C9orf72 repeat expansions at CDR 0.5 (i.e. possibly or mildly symptomatic) perform significantly worse on the total RSMS score and both subscores than controls, whilst GRN mutation carriers have a significantly lower SP subscore and a trend to lower total and EX scores than controls. These results highlight the potential for the RSMS to detect early deficits in social cognition in these genetic cohorts of FTD, prior to phenoconversion to being fully symptomatic.
The profiles of RSMS performance in MAPT mutation carriers seem to be somewhat unique. Symptomatic MAPT mutation carriers scored much lower at baseline than the other MAPT mutation carriers, a result that is consistent with findings from other cross-sectional [5, 15–17] and longitudinal [18] familial FTD studies. This could imply that self-monitoring in MAPT mutation carriers is relatively stable in early disease stages until soon before or at the point of conversion when there is a rapid decline in social cognitive function, as opposed to a more gradual (and earlier) deterioration in GRN and C9orf72 mutation carriers [19–22].
Analysis of the relationship of the RSMS with cognitive test scores reveals only very weak correlations, suggesting that the RSMS is likely to be measuring a different aspect of behaviour than the current psychometric tests. In C9orf72 mutation carriers, there was a weak correlation with the Facial Emotion Recognition Test i.e. the ability to detect the emotions of others in their faces, suggesting some overlap in social cognitive abilities in performing these tasks within this group. However, the term social cognition encompasses a number of different skills which are dissociable [23], and socioemotional sensitivity (as measured by the RSMS) is likely to be represent a distinct (if nonetheless overlapping) domain. Weak correlations were also seen with category fluency in the C9orf72 and GRN groups. The reason for this relationship is unclear but interestingly, prior studies have shown an association between verbal fluency and both social relationships and activity [24, 25], hypothesizing that fluency is better when social interaction can be maintained.
Previous studies in sporadic FTD have described links between deficits in empathic perspective taking and a ‘social cognition network’ comprising bifrontal (particularly orbitofrontal), anterior and inferior temporal and insula cortical regions [26–28]. Subcortical structures such as the amygdala and caudate have also been implicated in driving such dysfunction [28]. Results of the VBM analysis in this study highlighted frontal involvement across all mutation carrier groups, in particular the orbitofrontal cortex, a region known to be involved in decision-making and coordinating complex social and emotional behaviours [29–31] with its atrophy and circuitry disruption having been previously described in patients with behavioural variant FTD [32]. Previous studies specifically utilising the RSMS as a tool to measure social cognition have identified a positive association between socioemotional sensitivity and functional connectivity within the brain’s salience network, largely between the right anterior insula and both cortical and subcortical nodes [10], as well as between right supramarginal and angular gyri, and right frontal pole [33, 34]. Here, we demonstrate widespread insula involvement, anteriorly in C9orf72 and GRN mutation carriers and posteriorly in MAPT mutation carriers, in addition to anterior cingulate cortex involvement in GRN mutation carriers exclusively, another crucial element of the salience network [35].
Other brain regions associated with such behavioural deficits in FTD include the inferior and medial temporal gyri [4], areas particularly involved in emotion perception and recognition. Grey matter volume of the temporal pole was positively correlated with RSMS score in all mutation groups, with C9orf72 carriers also exhibiting an association with superior temporal gyrus and GRN and MAPT carriers showing a correlation with inferior temporal gyri specifically. Our results also show an association of the basal ganglia, particularly the caudate and putamen, in all genetic groups. These subcortical regions are also known to be implicated in emotion recognition [36–38], an integral factor in an individual’s performance on the RSMS.
Overall, there appears to be a network of brain regions associated with impairment of socioemotional sensitivity in FTD that includes frontal, temporal, insula and striatal areas, including significant crossover with areas involved in the salience network, thus supporting the established role of aberrant saliency detection in FTD-related social cognitive dysfunction.
Limitations
These data should be interpreted in light of some limitations. Despite the large nature of GENFI in comparison to other FTD studies, one limitation lies in the relatively small numbers in some of the groups once stratified. Future studies should aim to replicate these findings in larger cohorts, as well as investigate longitudinal changes in socioemotional sensitivity over time.
Another limitation lies in the design of the RSMS, due to the inclusion of reverse scoring. While every effort is taken to ensure the informant understands how to answer correctly, we cannot eliminate the chance of misinterpretation.
Although the RSMS has been examined in a number of studies previously, and the data presented here suggests it could potentially be included as an outcome measure in genetic FTD trials, there has been limited validation of the questionnaire so far and more work will be necessary e.g. investigation of test-retest reliability.
Lastly, while global CDR® plus NACC FTLD scoring is a validated and robust tool used to measure disease severity in FTD, the assessment of motor and neuropsychiatric symptoms is not included. With FTD representing a diverse spectrum of symptomatic profiles, a limitation of this study lies in possible mis-categorisation of individuals who might be at a more advanced stage of their disease but present with symptoms that are not specifically addressed by this scale.
Conclusions
In summary, this study describes the ability of the RSMS to detect early changes in socioemotional behaviour in distinct genetic cohorts of FTD and illustrates the neural correlates of self-monitoring in these populations. Whilst further studies will be needed to validate the RSMS and explore how it changes over time, the present data suggests it may well serve as a useful outcome measure in future clinical trials.
Supplementary Information
Additional file 1: Figure S1. RSMS EX scores in each genetic carrier group, stratified by Global CDR® plus NACC FTLD scores. Significant differences from controls and within each carrier group are starred. Differences between carrier groups are not shown. Figure S2. RSMS SP scores in each genetic carrier group, stratified by Global CDR® plus NACC FTLD scores. Significant differences from controls and within each carrier group are starred. Differences between carrier groups are not shown. Figure S3. Negative correlations between RSMS total and CDR® plus FTLD NACC SOB scores were observed across all mutation carrier groups: C9orf72 (r = -0.67, p < 0.001), GRN (r = -0.59, p < 0.001), MAPT (r = -0.53, p < 0.001). Each dot represents one mutation carrier. Table S1. RSMS total test scores (mean and SD) in healthy controls split by age group. Table S2. Cumulative frequency of RSMS total test scores in healthy controls. Table S3. Adjusted mean differences in RSMS EX scores between the genetic groups stratified by Global CDR® plus NACC FTLD scores with 95% bias-corrected confidence intervals (significant values in bold). Table S4. Adjusted mean differences in RSMS SP scores between the genetic groups stratified by Global CDR® plus NACC FTLD scores with 95% bias-corrected confidence intervals (significant values in bold). Table S5. Correlation of RSMS total test score with cognitive tests. Significant results are in bold. Table S6. Positive neuroanatomical correlates of grey matter volume with the RSMS total score in each genetic group.
Acknowledgements
We thank all participants and their family members for taking part in the GENFI study.
GENFI Consortium Authors
• Sónia Afonso - Instituto Ciencias Nucleares Aplicadas a Saude, Universidade de Coimbra, Coimbra, Portugal;
• Maria Rosario Almeida - Faculty of Medicine, University of Coimbra, Coimbra, Portugal;
• Sarah Anderl-Straub – Department of Neurology, University of Ulm, Ulm, Germany;
• Christin Andersson - Department of Clinical Neuroscience, Karolinska Institutet, Stockholm, Sweden;
• Anna Antonell - Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain;
• Silvana Archetti - Biotechnology Laboratory, Department of Diagnostics, ASST Brescia Hospital, Brescia, Italy;
• Andrea Arighi - Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Neurodegenerative Diseases Unit, Milan, Italy; University of Milan, Centro Dino Ferrari, Milan, Italy;
• Mircea Balasa - Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain;
• Myriam Barandiaran - Cognitive Disorders Unit, Department of Neurology, Donostia University Hospital, San Sebastian, Gipuzkoa, Spain; Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain;
• Nuria Bargalló - Imaging Diagnostic Center, Hospital Clínic, Barcelona, Spain;
• Robart Bartha - Department of Medical Biophysics, The University of Western Ontario, London, Ontario, Canada; Centre for Functional and Metabolic Mapping, Robarts Research Institute, The University of Western Ontario, London, Ontario, Canada;
• Benjamin Bender - Department of Diagnostic and Interventional Neuroradiology, University of Tübingen, Tübingen, Germany;
• Alberto Benussi - Centre for Neurodegenerative Disorders, Department of Clinical and Experimental Sciences, University of Brescia, Italy;
• Maxime Bertoux – Inserm 1172, Lille, France; CHU, CNR-MAJ, Labex Distalz, LiCEND Lille, France;
• Anne Bertrand - Sorbonne Université, Paris Brain Institute – Institut du Cerveau – ICM, Inserm U1127, CNRS UMR 7225, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France; Inria, Aramis project-team, F-75013, Paris, France; Centre pour l'Acquisition et le Traitement des Images, Institut du Cerveau et la Moelle, Paris, France
• Valentina Bessi - Department of Neuroscience, Psychology, Drug Research, and Child Health, University of Florence, Florence, Italy;
• Sandra Black - Sunnybrook Health Sciences Centre, Sunnybrook Research Institute, University of Toronto, Toronto, Canada;
• Sergi Borrego-Ecija - Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain;
• Jose Bras - Center for Neurodegenerative Science, Van Andel Institute, Grand Rapids, Michigan, MI 49503, USA;
• Alexis Brice - Sorbonne Université, Paris Brain Institute – Institut du Cerveau – ICM, Inserm U1127, CNRS UMR 7225, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France; Reference Network for Rare Neurological Diseases (ERN-RND);
• Rose Bruffaerts - Laboratory for Cognitive Neurology, Department of Neurosciences, KU Leuven, Leuven, Belgium;
• Agnès Camuzat - Sorbonne Université, Paris Brain Institute – Institut du Cerveau – ICM, Inserm U1127, CNRS UMR 7225, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France;
• Marta Cañada - CITA Alzheimer, San Sebastian, Gipuzkoa, Spain
• Valentina Cantoni - Centre for Neurodegenerative Disorders, Neurology Unit, Department of Clinical and Experimental Sciences, University of Brescia, Brescia, Italy;
• Paola Caroppo - Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy;
• Miguel Castelo-Branco - Faculty of Medicine, University of Coimbra, Coimbra, Portugal;
• Olivier Colliot - Sorbonne Université, Paris Brain Institute – Institut du Cerveau – ICM, Inserm U1127, CNRS UMR 7225, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France; Inria, Aramis project-team, F-75013, Paris, France; Centre pour l'Acquisition et le Traitement des Images, Institut du Cerveau et la Moelle, Paris, France;
• Thomas Cope – Department of Clinical Neuroscience, University of Cambridge, Cambridge, UK;
• Vincent Deramecourt - Univ Lille, France; Inserm 1172, Lille, France; CHU, CNR-MAJ, Labex Distalz, LiCEND Lille, France;
• María de Arriba - Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain;
• Giuseppe Di Fede - Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy;
• Alina Díez - Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain
• Diana Duro - Faculty of Medicine, University of Coimbra, Coimbra, Portugal;
• Chiara Fenoglio - Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Neurodegenerative Diseases Unit, Milan, Italy; University of Milan, Centro Dino Ferrari, Milan, Italy;
• Camilla Ferrari - Department of Neuroscience, Psychology, Drug Research, and Child Health, University of Florence, Florence, Italy;
• Catarina B. Ferreira -Laboratory of Neurosciences, Institute of Molecular Medicine, Faculty of Medicine, University of Lisbon, Lisbon, Portugal;
• Nick Fox – Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Morris Freedman - Baycrest Health Sciences, Rotman Research Institute, University of Toronto, Toronto, Canada;
• Giorgio Fumagalli - Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Neurodegenerative Diseases Unit, Milan, Italy; University of Milan, Centro Dino Ferrari, Milan, Italy;
• Aurélie Funkiewiez - Centre de référence des démences rares ou précoces, IM2A, Département de Neurologie, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France; Sorbonne Université, Paris Brain Institute – Institut du Cerveau – ICM, Inserm U1127, CNRS UMR 7225, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France;
• Alazne Gabilondo -Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain;
• Roberto Gasparotti - Neuroradiology Unit, University of Brescia, Brescia, Italy
• Serge Gauthier - Alzheimer Disease Research Unit, McGill Centre for Studies in Aging, Department of Neurology & Neurosurgery, McGill University, Montreal, Québec, Canada;
• Stefano Gazzina - Neurology, ASST Brescia Hospital, Brescia, Italy
• Giorgio Giaccone - Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy;
• Ana Gorostidi - Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain;
• Caroline Greaves – Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Rita Guerreiro - Center for Neurodegenerative Science, Van Andel Institute, Grand Rapids, Michigan, MI 49503, USA;
• Carolin Heller – Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Tobias Hoegen - Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany;
• Begoña Indakoetxea - Cognitive Disorders Unit, Department of Neurology, Donostia University Hospital, San Sebastian, Gipuzkoa, Spain; Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain;
• Vesna Jelic - Division of Clinical Geriatrics, Karolinska Institutet, Stockholm, Sweden;
• Hans-Otto Karnath - Division of Neuropsychology, Hertie-Institute for Clinical Brain Research and Center of Neurology, University of Tübingen, Tübingen, Germany;
• Ron Keren -The University Health Network, Toronto Rehabilitation Institute, Toronto, Canada;
• Gregory Kuchcinski - Univ Lille, France; Inserm 1172, Lille, France; CHU, CNR-MAJ, Labex Distalz, LiCEND Lille, France;
• Tobias Langheinrich - Division of Neuroscience and Experimental Psychology, Wolfson Molecular Imaging Centre, University of Manchester, Manchester, UK; Manchester Centre for Clinical Neurosciences, Department of Neurology, Salford Royal NHS Foundation Trust, Manchester, UK;
• Thibaud Lebouvier - Univ Lille, France; Inserm 1172, Lille, France; CHU, CNR-MAJ, Labex Distalz, LiCEND Lille, France;
• Maria João Leitão - Centre of Neurosciences and Cell Biology, Universidade de Coimbra, Coimbra, Portugal;
• Albert Lladó - Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain;
• Gemma Lombardi - Department of Neuroscience, Psychology, Drug Research and Child Health, University of Florence, Florence, Italy;
• Sandra Loosli -Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany;
• Carolina Maruta - Laboratory of Language Research, Centro de Estudos Egas Moniz, Faculty of Medicine, University of Lisbon, Lisbon, Portugal;
• Simon Mead - MRC Prion Unit, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Lieke Meeter - Department of Neurology, Erasmus Medical Center, Rotterdam, Netherlands;
• Gabriel Miltenberger - Faculty of Medicine, University of Lisbon, Lisbon, Portugal;
• Rick van Minkelen - Department of Clinical Genetics, Erasmus Medical Center, Rotterdam, Netherlands;
• Sara Mitchell - Sunnybrook Health Sciences Centre, Sunnybrook Research Institute, University of Toronto, Toronto, Canada;
• Katrina Moore – Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London UK;
• Benedetta Nacmias - Department of Neuroscience, Psychology, Drug Research and Child Health, University of Florence, Florence, Italy;
• Annabel Nelson - Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Linn Öijerstedt - Center for Alzheimer Research, Division of Neurogeriatrics, Department of Neurobiology, Care Sciences and Society, Bioclinicum, Karolinska Institutet, Solna, Sweden; Unit for Hereditary Dementias, Theme Aging, Karolinska University Hospital, Solna, Sweden;
• Jaume Olives - Alzheimer’s disease and Other Cognitive Disorders Unit, Neurology Service, Hospital Clínic, Barcelona, Spain;
• Sebastien Ourselin - School of Biomedical Engineering & Imaging Sciences, King's College London, London, UK;
• Alessandro Padovani - Centre for Neurodegenerative Disorders, Department of Clinical and Experimental Sciences, University of Brescia, Italy
• Jessica Panman – Department of Neurology, Erasmus Medical Center, Rotterdam, Netherlands;
• Janne M. Papma - Department of Neurology, Erasmus Medical Center, Rotterdam;
• Yolande Pijnenburg - Amsterdam University Medical Centre, Amsterdam VUmc, Amsterdam, Netherlands;
• Cristina Polito - Department of Biomedical, Experimental and Clinical Sciences “Mario Serio”, Nuclear Medicine Unit, University of Florence, Florence, Italy
• Enrico Premi - Stroke Unit, ASST Brescia Hospital, Brescia, Italy
• Sara Prioni - Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy;
• Catharina Prix -Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany;
• Rosa Rademakers [as London Ontario geneticist] - Department of Neurosciences, Mayo Clinic, Jacksonville, Florida, USA;
• Veronica Redaelli -Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy;
• Daisy Rinaldi - Centre de référence des démences rares ou précoces, IM2A, Département de Neurologie, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France; Sorbonne Université, Paris Brain Institute – Institut du Cerveau – ICM, Inserm U1127, CNRS UMR 7225, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France; Département de Neurologie, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France; Reference Network for Rare Neurological Diseases (ERN-RND);
• Tim Rittman – Department of Clinical Neurosciences, University of Cambridge, Cambridge, UK;
• Ekaterina Rogaeva -Tanz Centre for Research in Neurodegenerative Diseases, University of Toronto, Toronto, Canada;
• Adeline Rollin - CHU, CNR-MAJ, Labex Distalz, LiCEND Lille, France;
• Pedro Rosa-Neto -Translational Neuroimaging Laboratory, McGill Centre for Studies in Aging, McGill University, Montreal, Québec, Canada;
• Giacomina Rossi - Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy;
• Martin Rossor – Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Beatriz Santiago - Neurology Department, Centro Hospitalar e Universitario de Coimbra, Coimbra, Portugal;
• Dario Saracino – Sorbonne Université, Paris Brain Institute – Institut du Cerveau – ICM, Inserm U1127, CNRS UMR 7225, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France; Inria, Aramis project-team, F-75013, Paris, France; Centre de référence des démences rares ou précoces, IM2A, Département de Neurologie, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France;
• Sabrina Sayah - Sorbonne Université, Paris Brain Institute – Institut du Cerveau – ICM, Inserm U1127, CNRS UMR 7225, AP-HP - Hôpital Pitié-Salpêtrière, Paris, France;
• Elio Scarpini - Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Neurodegenerative Diseases Unit, Milan, Italy; University of Milan, Centro Dino Ferrari, Milan, Italy;
• Sonja Schönecker - Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany;
• Harro Seelaar - Department of Neurology, Erasmus Medical Centre, Rotterdam, Netherlands;
• Elisa Semler -Department of Neurology, University of Ulm, Ulm;
• Rachelle Shafei – Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Christen Shoesmith - Department of Clinical Neurological Sciences, University of Western Ontario, London, Ontario, Canada;
• Imogen Swift - Department of Neurodegenerative Disease, Dementia Research Centre, UCL Institute of Neurology, Queen Square, London, UK;
• Miguel Tábuas-Pereira - Neurology Department, Centro Hospitalar e Universitario de Coimbra, Coimbra, Portugal;
• Mikel Tainta - Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain;
• Ricardo Taipa - Neuropathology Unit and Department of Neurology, Centro Hospitalar do Porto - Hospital de Santo António, Oporto, Portugal;
• David Tang-Wai -The University Health Network, Krembil Research Institute, Toronto, Canada;
• David L. Thomas - Neuroimaging Analysis Centre, Department of Brain Repair and Rehabilitation, UCL Institute of Neurology, Queen Square, London, UK;
• Paul Thompson - Division of Neuroscience and Experimental Psychology, Wolfson Molecular Imaging Centre, University of Manchester, Manchester, UK;
• Hakan Thonberg - Center for Alzheimer Research, Division of Neurogeriatrics, Karolinska Institutet, Stockholm, Sweden;
• Carolyn Timberlake – Department of Clinical Neurosciences, University of Cambridge, Cambridge, UK;
• Pietro Tiraboschi - Fondazione IRCCS Istituto Neurologico Carlo Besta, Milano, Italy;
• Emily Todd – Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Philip Van Damme - Neurology Service, University Hospitals Leuven, Belgium; Laboratory for Neurobiology, VIB-KU Leuven Centre for Brain Research, Leuven, Belgium;
• Mathieu Vandenbulcke - Geriatric Psychiatry Service, University Hospitals Leuven, Belgium; Neuropsychiatry, Department of Neurosciences, KU Leuven, Leuven, Belgium;
• Michele Veldsman - Nuffield Department of Clinical Neurosciences, Medical Sciences Division, University of Oxford, Oxford, UK;
• Ana Verdelho - Department of Neurosciences and Mental Health, Centro Hospitalar Lisboa Norte - Hospital de Santa Maria & Faculty of Medicine, University of Lisbon, Lisbon, Portugal;
• Jorge Villanua - OSATEK, University of Donostia, San Sebastian, Gipuzkoa, Spain;
• Jason Warren – Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Carlo Wilke - Department of Neurodegenerative Diseases, Hertie-Institute for Clinical Brain Research and Center of Neurology, University of Tübingen, Tübingen, Germany; Center for Neurodegenerative Diseases (DZNE), Tübingen, Germany;
• Ione Woollacott – Dementia Research Centre, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Elisabeth Wlasich - Neurologische Klinik, Ludwig-Maximilians-Universität München, Munich, Germany;
• Henrik Zetterberg - Dementia Research Institute, Department of Neurodegenerative Disease, UCL Institute of Neurology, Queen Square, London, UK;
• Miren Zulaica - Neuroscience Area, Biodonostia Health Research Insitute, San Sebastian, Gipuzkoa, Spain.
Abbreviations
- FTD
Frontotemporal dementia
- RSMS
Revised Self-Monitoring Scale
- GENFI
Genetic FTD initiative
- C9orf72
Chromosome 9 open-reading frame 72
- GRN
Progranulin
- MAPT
Microtubule-associated protein tau
- CDR® plus NACC FTLD
CDR® Dementia Staging Instrument with National Alzheimer Coordinating Centre Frontotemporal Lobar Degeneration component
- VBM
Voxel-based morphometry
- bvFTD
Behavioural variant FTD
- CDR® plus NACC FTLD-SB
CDR® plus NACC FTLD sum of boxes
- EX
RSMS socio-emotional expressiveness subscore
- SP
RSMS modification of self-presentation subscore
- SPM
Statistical parametric mapping
- GM
Grey matter
- WM
White matter
- CSF
Cerebrospinal fluid
- DARTEL
Fast-diffeomorphic image registration algorithm
- MNI
Montreal Neurological Institute
- TIV
Total intracranial volume
- FWE
Family-Wise Error
Authors’ contributions
HDF, LLR and JDR designed the study, processed the data, performed analyses and wrote the first draft of the manuscript. All other authors have collected and helped to analyse data and contributed to the writing and critical revision of the manuscript. The author(s) read and approved the final manuscript.
Funding
The Dementia Research Centre is supported by Alzheimer’s Research UK, Brain Research Trust and The Wolfson Foundation. This work was supported by the NIHR Queen Square Dementia Biomedical Research Unit, the NIHR UCL/H Biomedical Research Centre and the Leonard Wolfson Experimental Neurology Centre (LWENC) Clinical Research Facility as well as an Alzheimer’s Society grant [AS-PG-16-007]. This work was also supported by the MRC UK GENFI grant [MR/M023664/1], the Italian Ministry of Health (CoEN015 and Ricerca Corrente) and the Canadian Institutes of Health Research as part of a Centres of Excellence in Neurodegeneration grant, a Canadian Institutes of Health Research operating grant, The Bluefield Project and the JPND GENFI-PROX grant [2019-02248]. JDR is supported by an MRC Clinician Scientist Fellowship [MR/M008525/1] and has received funding from the NIHR Rare Disease Translational Research Collaboration [BRC149/NS/MH], the Bluefield Project and the Association for Frontotemporal Degeneration. MB is supported by a Fellowship award from the Alzheimer’s Society, UK [AS-JF-19a-004-517]. JBR is supported by the Wellcome Trust [103838], the Medical Research Council and NIHR Cambridge Biomedical Research Centre. This work was also funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology [EXC 2145 SyNergy – ID 390857198]. Several authors of this publication are members of the European Reference Network for Rare Neurological Diseases - Project ID No 739510.
Availability of data and materials
Data are available upon reasonable request. The raw data of this project are part of GENFI and are not publicly available in accordance with the ethical approval. Data can be accessed upon reasonable request to JDR (j.rohrer@ucl.ac.uk).
Declarations
Ethics approval and consent to participate
Data was collected at GENFI sites under individual ethics approval approved by the local ethics board. Written consent was obtained from all participants before entering the study.
Consent for publication
Consent for publication of data is included within the written consent when entering into the study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jonathan D. Rohrer, Email: j.rohrer@ucl.ac.uk
on behalf of the Genetic FTD Initiative, GENFI:
Sónia Afonso, Maria Rosario Almeida, Sarah Anderl-Straub, Christin Andersson, Anna Antonell, Silvana Archetti, Andrea Arighi, Mircea Balasa, Myriam Barandiaran, Nuria Bargalló, Robart Bartha, Benjamin Bender, Alberto Benussi, Maxime Bertoux, Anne Bertrand, Valentina Bessi, Sandra Black, Sergi Borrego-Ecija, Jose Bras, Alexis Brice, Rose Bruffaerts, Agnès Camuzat, Marta Cañada, Valentina Cantoni, Paola Caroppo, Miguel Castelo-Branco, Olivier Colliot, Thomas Cope, Vincent Deramecourt, María de Arriba, Giuseppe Di Fede, Alina Díez, Diana Duro, Chiara Fenoglio, Camilla Ferrari, Catarina B. Ferreira, Nick Fox, Morris Freedman, Giorgio Fumagalli, Aurélie Funkiewiez, Alazne Gabilondo, Roberto Gasparotti, Serge Gauthier, Stefano Gazzina, Giorgio Giaccone, Ana Gorostidi, Caroline Greaves, Rita Guerreiro, Carolin Heller, Tobias Hoegen, Begoña Indakoetxea, Vesna Jelic, Hans-Otto Karnath, Ron Keren, Gregory Kuchcinski, Tobias Langheinrich, Thibaud Lebouvier, Maria João Leitão, Albert Lladó, Gemma Lombardi, Sandra Loosli, Carolina Maruta, Simon Mead, Lieke Meeter, Gabriel Miltenberger, Rick van Minkelen, Sara Mitchell, Katrina Moore, Benedetta Nacmias, Annabel Nelson, Linn Öijerstedt, Jaume Olives, Sebastien Ourselin, Alessandro Padovani, Jessica Panman, Janne M. Papma, Yolande Pijnenburg, Cristina Polito, Enrico Premi, Sara Prioni, Catharina Prix, Rosa Rademakers, Veronica Redaelli, Daisy Rinaldi, Tim Rittman, Ekaterina Rogaeva, Adeline Rollin, Pedro Rosa-Neto, Giacomina Rossi, Martin Rossor, Beatriz Santiago, Dario Saracino, Sabrina Sayah, Elio Scarpini, Sonja Schönecker, Harro Seelaar, Elisa Semler, Rachelle Shafei, Christen Shoesmith, Imogen Swift, Miguel Tábuas-Pereira, Mikel Tainta, Ricardo Taipa, David Tang-Wai, David L. Thomas, Paul Thompson, Hakan Thonberg, Carolyn Timberlake, Pietro Tiraboschi, Emily Todd, Philip Van Damme, Mathieu Vandenbulcke, Michele Veldsman, Ana Verdelho, Jorge Villanua, Jason Warren, Carlo Wilke, Ione Woollacott, Elisabeth Wlasich, Henrik Zetterberg, and Miren Zulaica
References
- 1.Desmarais P, Lanctôt KL, Masellis M, Black SE, Herrmann N. Social inappropriateness in neurodegenerative disorders. Int Psychoger. 2018;30(2):197–207. doi: 10.1017/S1041610217001260. [DOI] [PubMed] [Google Scholar]
- 2.Lennox RD, Wolfe RN. Revision of the self-monitoring scale. J Pers Soc Psychol. 1984;46(6):1349–1364. doi: 10.1037/0022-3514.46.6.1349. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann SG. The emotional consequences of social pragmatism: the psychophysiological correlates of self-monitoring. Biol Psychol. 2006;73(2):169–174. doi: 10.1016/j.biopsycho.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Shdo SM, Ranasinghe KG, Gola KA, Mielke CJ, Sukhanov PV, Miller BL, Rankin KP. Deconstructing empathy: neuroanatomical dissociations between affect sharing and prosocial motivation using a patient lesion model. Neuropsychologia. 2018;116(Pt A):126–135. doi: 10.1016/j.neuropsychologia.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rohrer JD, Nicholas JM, Cash DM, van Swieten J, Dopper E, Jiskoot L, van Minkelen R, Rombouts SA, Cardoso MJ, Clegg S, Espak M, Mead S, Thomas DL, de Vita E, Masellis M, Black SE, Freedman M, Keren R, MacIntosh BJ, Rogaeva E, Tang-Wai D, Tartaglia MC, Laforce R, Jr, Tagliavini F, Tiraboschi P, Redaelli V, Prioni S, Grisoli M, Borroni B, Padovani A, Galimberti D, Scarpini E, Arighi A, Fumagalli G, Rowe JB, Coyle-Gilchrist I, Graff C, Fallström M, Jelic V, Ståhlbom AK, Andersson C, Thonberg H, Lilius L, Frisoni GB, Binetti G, Pievani M, Bocchetta M, Benussi L, Ghidoni R, Finger E, Sorbi S, Nacmias B, Lombardi G, Polito C, Warren JD, Ourselin S, Fox NC, Rossor MN. Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the Genetic Frontotemporal dementia Initiative (GENFI) study: a cross-sectional analysis. Lancet Neurol. 2015;14(3):253–262. doi: 10.1016/S1474-4422(14)70324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warren JD, Rohrer JD, Rossor MN. Frontotemporal dementia. BMJ (Online). 2013;347(aug12 3). 10.1136/bmj.f4827. [DOI] [PMC free article] [PubMed]
- 7.Greaves CV, Rohrer JD. An update on genetic frontotemporal dementia. J Neurol. 2019;266(8):2075–2086. doi: 10.1007/s00415-019-09363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyagawa T, Brushaber D, Syrjanen J, Kremers W, Fields J, Forsberg LK, Heuer HW, Knopman D, Kornak J, Boxer A, Rosen H, Boeve B, on Behalf of the ARTFL/LEFFTDS Consortium Use of the CDR® plus NACC FTLD in mild FTLD: data from the ARTFL/LEFFTDS consortium. Alzheimer’s Dement. 2020;16(1):79–90. doi: 10.1016/j.jalz.2019.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyagawa T, Brushaber D, Syrjanen J, Kremers W, Fields J, Forsberg LK, Heuer HW, Knopman D, Kornak J, Boxer A, Rosen HJ, Boeve BF, Appleby B, Bordelon Y, Bove J, Brannelly P, Caso C, Coppola G, Dever R, Dheel C, Dickerson B, Dickinson S, Dominguez S, Domoto-Reilly K, Faber K, Ferrell J, Fishman A, Fong J, Foroud T, Gavrilova R, Gearhart D, Ghazanfari B, Ghoshal N, Goldman JS, Graff-Radford J, Graff-Radford N, Grant I, Grossman M, Haley D, Hsiung R, Huey E, Irwin D, Jones D, Jones L, Kantarci K, Karydas A, Kaufer D, Kerwin D, Kraft R, Kramer J, Kukull W, Litvan I, Lucente D, Lungu C, Mackenzie I, Maldonado M, Manoochehri M, McGinnis S, McKinley E, Mendez MF, Miller B, Multani N, Onyike C, Padmanabhan J, Pantelyat A, Pearlman R, Petrucelli L, Potter M, Rademakers R, Ramos EM, Rankin K, Rascovsky K, Roberson ED, Rogalski E, Sengdy P, Shaw L, Tartaglia MC, Tatton N, Taylor J, Toga A, Trojanowski JQ, Wang P, Weintraub S, Wong B, Wszolek Z. Utility of the global CDR® plus NACC FTLD rating and development of scoring rules: data from the ARTFL/LEFFTDS Consortium. Alzheimer’s Dement. 2020;16(1):106–117. doi: 10.1002/alz.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toller G, Brown J, Sollberger M, Shdo SM, Bouvet L, Sukhanov P, Seeley WW, Miller BL, Rankin KP. Individual differences in socioemotional sensitivity are an index of salience network function. Cortex. 2018;103:211–223. doi: 10.1016/j.cortex.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 12.Ridgway GR, et al. Issues with threshold masking in voxel-based morphometry of atrophied brains. Neuroimage. 2009;44(1):99–111. doi: 10.1016/j.neuroimage.2008.08.045. [DOI] [PubMed] [Google Scholar]
- 13.Malone IB, Leung KK, Clegg S, Barnes J, Whitwell JL, Ashburner J, Fox NC, Ridgway GR. Accurate automatic estimation of total intracranial volume: a nuisance variable with less nuisance. Neuroimage. 2015;104:366–372. doi: 10.1016/j.neuroimage.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toller G, et al. Revised Self-Monitoring Scale. Neurology. 2020. 10.1212/wnl.0000000000009451.
- 15.Dopper EGP, Rombouts SARB, Jiskoot LC, Heijer T, Graaf JRA, Koning I, Hammerschlag AR, Seelaar H, Seeley WW, Veer IM, van Buchem MA, Rizzu P, van Swieten JC. Structural and functional brain connectivity in presymptomatic familial frontotemporal dementia. Neurology. 2013;80(9):814–823. doi: 10.1212/WNL.0b013e31828407bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geschwind DH, Robidoux J, Alarcón M, Miller BL, Wilhelmsen KC, Cummings JL, Nasreddine ZS. Dementia and neurodevelopmental predisposition: cognitive dysfunction in presymptomatic subjects precedes dementia by decades in frontotemporal dementia. Ann Neurol. 2001;50(6):741–746. doi: 10.1002/ana.10024. [DOI] [PubMed] [Google Scholar]
- 17.Barandiaran M, Estanga A, Moreno F, Indakoetxea B, Alzualde A, Balluerka N, Martí Massó JF, de Munain AL. Neuropsychological features of asymptomatic c.709-1G>A progranulin mutation carriers. J Int Neuropsychol Soc. 2012;18(6):1086–1090. doi: 10.1017/S1355617712000823. [DOI] [PubMed] [Google Scholar]
- 18.Jiskoot LC, Dopper EGP, Heijer T, Timman R, van Minkelen R, van Swieten JC, Papma JM. Presymptomatic cognitive decline in familial frontotemporal dementia: a longitudinal study. Neurology. 2016;87(4):384–391. doi: 10.1212/WNL.0000000000002895. [DOI] [PubMed] [Google Scholar]
- 19.Janssen JC, Schott JM, Cipolotti L, Fox NC, Scahill RI, Josephs KA, Stevens JM, Rossor MN. Mapping the onset and progression of atrophy in familial frontotemporal lobar degeneration. J Neurol Neurosurg Psychiatry. 2005;76(2):162–168. doi: 10.1136/jnnp.2003.032201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferman TJ, McRae CA, Arvanitakis Z, Tsuboi Y, Vo A, Wszolek ZK. Early and pre-symptomatic neuropsychological dysfunction in the PPND family with the N279K tau mutation. Park Relat Disord. 2003;9(5):265–270. doi: 10.1016/S1353-8020(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 21.Rohrer JD, Warren JD, Barnes J, Mead S, Beck J, Pepple T, Boyes R, Omar R, Collinge J, Stevens JM, Warrington EK, Rossor MN, Fox NC. Mapping the progression of progranulin-associated frontotemporal lobar degeneration. Nat Clin Pract Neurol. 2008;4(8):455–460. doi: 10.1038/ncpneuro0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiskoot LC, Panman JL, van Asseldonk L, Franzen S, Meeter LHH, Donker Kaat L, van der Ende EL, Dopper EGP, Timman R, van Minkelen R, van Swieten JC, van den Berg E, Papma JM. Longitudinal cognitive biomarkers predicting symptom onset in presymptomatic frontotemporal dementia. J Neurol. 2018;265(6):1381–1392. doi: 10.1007/s00415-018-8850-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adolphs R. The social brain: Neural basis of social knowledge. Annu Rev Psychol. 2009;60(1):693–716. doi: 10.1146/annurev.psych.60.110707.163514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly ME, Duff H, Kelly S, McHugh Power JE, Brennan S, Lawlor BA, Loughrey DG. The impact of social activities, social networks, social support and social relationships on the cognitive functioning of healthy older adults: a systematic review. Syst Rev. 2017;6(1):259. doi: 10.1186/s13643-017-0632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown CL, Gibbons LE, Kennison RF, Robitaille A, Lindwall M, Mitchell MB, Shirk SD, Atri A, Cimino CR, Benitez A, MacDonald SWS, Zelinski EM, Willis SL, Schaie KW, Johansson B, Dixon RA, Mungas DM, Hofer SM, Piccinin AM. Social activity and cognitive functioning over time: a coordinated analysis of four longitudinal studies. J Aging Res. 2012;2012:1–12. doi: 10.1155/2012/287438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumfor F, Piguet O. Disturbance of emotion processing in frontotemporal dementia: a synthesis of cognitive and neuroimaging findings. Neuropsychol Rev. 2012;22(3):280–297. doi: 10.1007/s11065-012-9201-6. [DOI] [PubMed] [Google Scholar]
- 27.Couto B, Manes F, Montañés P, Matallana D, Reyes P, Velasquez M, et al. Structural neuroimaging of social cognition in progressive non-fluent aphasia and behavioral variant of frontotemporal dementia. Front Hum Neurosci. 2013;7. 10.3389/fnhum.2013.00467. [DOI] [PMC free article] [PubMed]
- 28.Eslinger PJ, Moore P, Anderson C, Grossman M. Social cognition, executive functioning, and neuroimaging correlates of empathic deficits in frontotemporal dementia. J Neuropsychiatry Clin Neurosci. 2011;23(1):74–82. doi: 10.1176/appi.neuropsych.23.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beer JS, John OP, Scabini D, Knight RT. Orbitofrontal cortex and social behavior: integrating self-monitoring and emotion-cognition interactions. J Cogn Neurosci. 2006;18(6):871–879. doi: 10.1162/jocn.2006.18.6.871. [DOI] [PubMed] [Google Scholar]
- 30.Rolls ET. The functions of the orbitofrontal cortex. Brain Cogn. 2004;55(1):11–29. doi: 10.1016/S0278-2626(03)00277-X. [DOI] [PubMed] [Google Scholar]
- 31.Kringelbach ML, Rolls ET. The functional neuroanatomy of the human orbitofrontal cortex: evidence from neuroimaging and neuropsychology. Prog Neurobiol. 2004;72(5):341–372. doi: 10.1016/j.pneurobio.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62(1):42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Multani N, Taghdiri F, Anor CJ, Varriano B, Misquitta K, Tang-Wai DF, et al. Association between social cognition changes and resting state functional connectivity in frontotemporal dementia, Alzheimer’s disease, Parkinson’s disease, and healthy controls. Front Neurosci. 2019;13. 10.3389/fnins.2019.01259. [DOI] [PMC free article] [PubMed]
- 34.Parthimos TP, Karavasilis E, Rankin KP, Seimenis I, Leftherioti K, Papanicolaou AC, Miller B, Papageorgiou SG, Papatriantafyllou JD. The neural correlates of impaired self-monitoring among individuals with neurodegenerative dementias. J Neuropsychiatry Clin Neurosci. 2019;31(3):201–209. doi: 10.1176/appi.neuropsych.17120349. [DOI] [PubMed] [Google Scholar]
- 35.Menon V. Brain Mapping: An Encyclopedic Reference. 2015. Salience network. [Google Scholar]
- 36.Calder AJ, Keane J, Lawrence AD, Manes F. Impaired recognition of anger following damage to the ventral striatum. Brain. 2004;127(9):1958–1969. doi: 10.1093/brain/awh214. [DOI] [PubMed] [Google Scholar]
- 37.Kemp J, Berthel MC, Dufour A, Després O, Henry A, Namer IJ, Musacchio M, Sellal F. Caudate nucleus and social cognition: neuropsychological and SPECT evidence from a patient with focal caudate lesion. Cortex. 2013;49(2):559–571. doi: 10.1016/j.cortex.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Peirce JE, Péron J. The basal ganglia and the cerebellum in human emotion. Soc Cogn Affect Neurosci. 2020;15(5):599–613. doi: 10.1093/scan/nsaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. RSMS EX scores in each genetic carrier group, stratified by Global CDR® plus NACC FTLD scores. Significant differences from controls and within each carrier group are starred. Differences between carrier groups are not shown. Figure S2. RSMS SP scores in each genetic carrier group, stratified by Global CDR® plus NACC FTLD scores. Significant differences from controls and within each carrier group are starred. Differences between carrier groups are not shown. Figure S3. Negative correlations between RSMS total and CDR® plus FTLD NACC SOB scores were observed across all mutation carrier groups: C9orf72 (r = -0.67, p < 0.001), GRN (r = -0.59, p < 0.001), MAPT (r = -0.53, p < 0.001). Each dot represents one mutation carrier. Table S1. RSMS total test scores (mean and SD) in healthy controls split by age group. Table S2. Cumulative frequency of RSMS total test scores in healthy controls. Table S3. Adjusted mean differences in RSMS EX scores between the genetic groups stratified by Global CDR® plus NACC FTLD scores with 95% bias-corrected confidence intervals (significant values in bold). Table S4. Adjusted mean differences in RSMS SP scores between the genetic groups stratified by Global CDR® plus NACC FTLD scores with 95% bias-corrected confidence intervals (significant values in bold). Table S5. Correlation of RSMS total test score with cognitive tests. Significant results are in bold. Table S6. Positive neuroanatomical correlates of grey matter volume with the RSMS total score in each genetic group.
Data Availability Statement
Data are available upon reasonable request. The raw data of this project are part of GENFI and are not publicly available in accordance with the ethical approval. Data can be accessed upon reasonable request to JDR (j.rohrer@ucl.ac.uk).