Abstract
The bacterial glycocalyx is a quintessential drug target comprised of structurally distinct glycans. Bacterial glycans bear unusual monosaccharide building blocks whose proper construction is critical for bacterial fitness, survival, and colonization in the human host. Despite their appeal as therapeutic targets, bacterial glycans are difficult to study due to the presence of rare bacterial monosaccharides that are linked and modified in atypical manners. Their structural complexity ultimately hampers their analytical characterization. This review highlights recent advances in bacterial chemical glycobiology and focuses on the development of chemical tools to probe, perturb, and image bacterial glycans and their biosynthesis. Current technologies have enabled the study of bacterial glycosylation machinery even in the absence of detailed structural information.
Keywords: bacteria, glycans, chemical tools, bioorthogonal chemistry, lectin, inhibit
Graphical Abstract
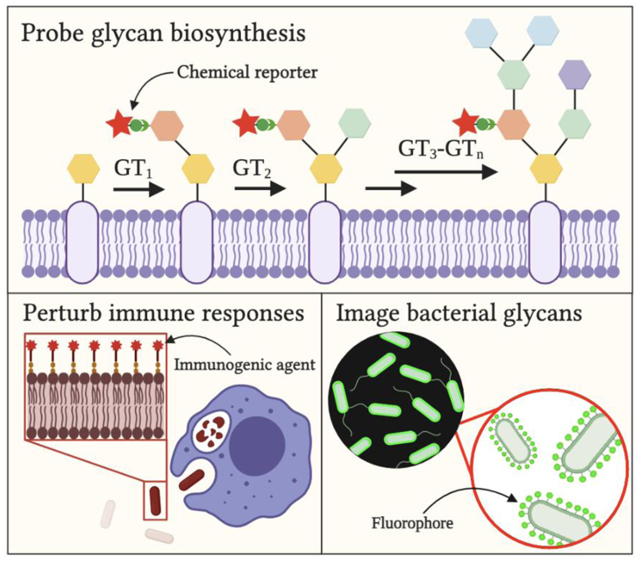
1. Introduction
Bacterial glycans are attractive therapeutic targets. These macromolecules cover the cell wall of bacteria to form a sugar coat, termed the glycocalyx, that modulates host-pathogen interactions.1, 2 The glycans found in the bacterial glycocalyx range from lipid-linked structures such as lipopolysaccharide (LPS), to polysaccharides including capsular polysaccharide, to peptide cross-linked hybrids like peptidoglycan, to glycosylated proteins (Fig. 1). Critically, the component glycans of these structures bear rare bacterial monosaccharide building blocks which vary between bacterial species and are absent from eukaryotic cells.1, 2 Bacterial glycans are comprised of over 100 monosaccharides,3 as opposed to just nine monosaccharides commonly used in mammalian glycans. Exclusively bacterial monosaccharides include muramic acid, heptose, rhamnose, pseudaminic acid, bacillosamine, and countless others (Fig. 1). Beyond their structural diversity, glycans are compelling targets due to their evident association with virulence in priority pathogens. For example, lipopolysaccharide modification is key to immune system evasion in Pseudomonas aeruginosa4 and Salmonella enterica5; capsular polysaccharides contribute to biofilm formation and host cell adhesion in Streptococcus pneumoniae6 and Klebsiella pneumoniae7; and protein glycosylation enables flagellation and motility in Camplyobacter jejuni8 and Helicobacter pylori.9 Therefore, probing bacterial glycans and their biosynthesis can potentially reveal attractive drug targets for the development of pathogen-specific antibiotics.
Figure 1. Structures of common bacterial glycans.
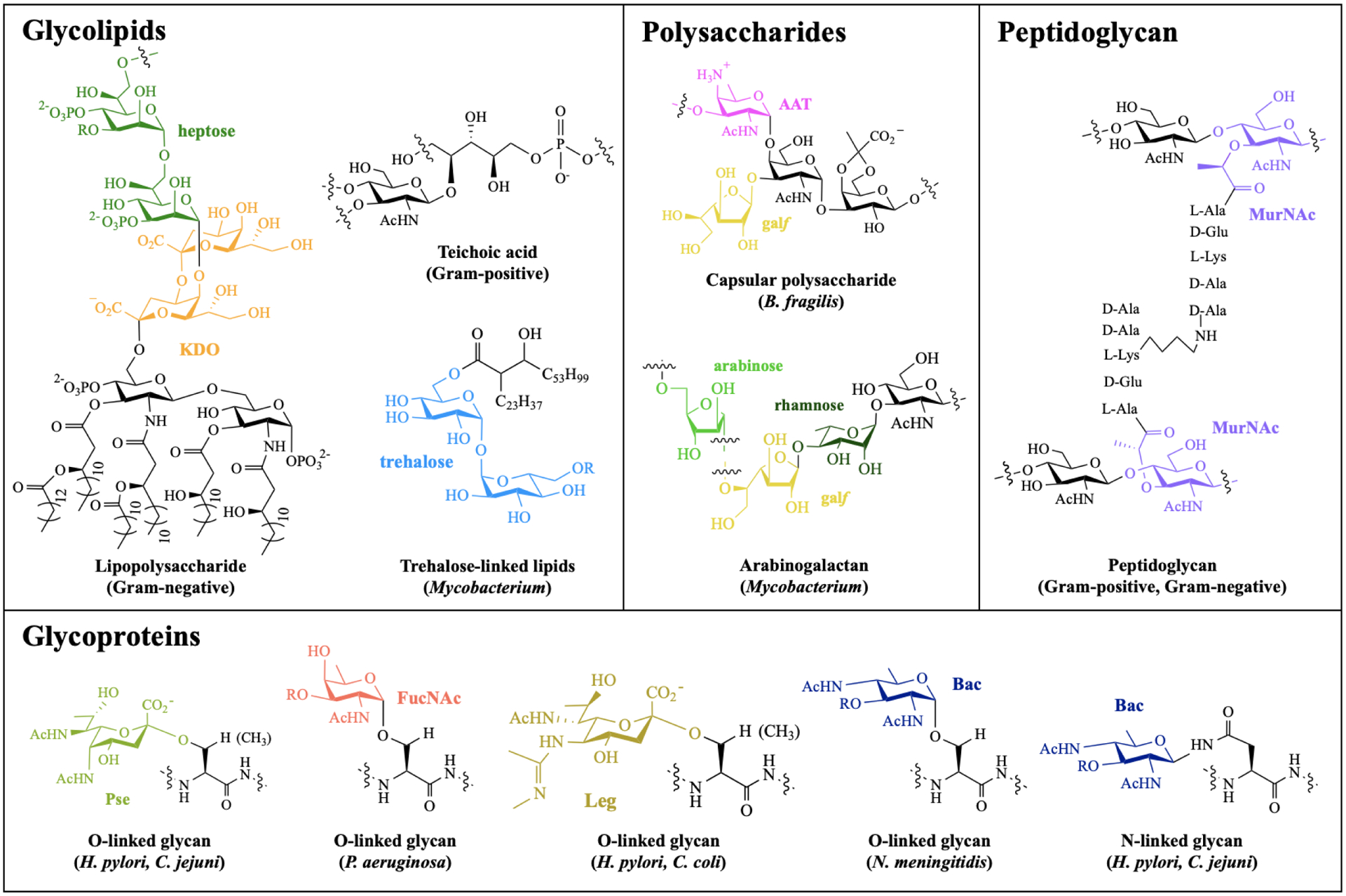
Exclusively bacterial monosaccharides are highlighted in color, with known species distribution in parentheses. Abbreviations: heptose = L-glycero-d-mannoheptose; KDO = 3-deoxy-d-manno-oct-2-ulosonic acid; AAT = 2-acetamido-4-amino-2,4,6-trideoxyhexose; galf = galactofuranose; MurNAc = muramic acid; Pse = pseudaminic acid; FucNAc = N-acetylfucosamine; Leg = legionaminic acid; Bac = bacillosamine.
Despite their critical roles in host-pathogen interactions and their intrigue as molecular targets, the systematic study of bacterial glycans lags behind that of their eukaryotic counterparts. Since traditional approaches to study glycans were designed for eukaryotic building blocks, bacterial glycans are often refractory to analysis with established tools due to the presence of unusual monosaccharides.10 For example, in many instances, glycan-degrading enzymes (e.g. glycosidases) do not recognize bacterial glycans as substrates,11 and automated glycan analysis software (e.g. mass spectrometry) does not detect prokaryotic building blocks.12 While there are rigorous biochemical and analytical approaches to characterize bacterial glycan structures and their biosynthetic pathways,13–19 these studies are time-consuming and not readily generalizable. These approaches are inherently limited by analytical challenges associated with isolation and structural assignment of complex, branched, heterogeneous glycans composed of myriad building blocks, many of which are isomers with identical masses. Further, the diversity of glycan structures across different bacterial isolates and strains makes information gleaned in one species often not translatable to another, and even from one strain to another in the same species. As a result, there is a need for accessible chemical tools to accelerate the study and inhibition of bacterial glycan biosynthesis.
In this review, we present current chemical probes to study bacterial glycobiology. We begin with a brief overview of the evolution of the toolkit since the discovery of protein glycosylation in bacteria. Then, we highlight recently developed technologies, categorizing them into chemical tools that (1) probe, (2) perturb, and (3) image bacterial glycans. Finally, we discuss current gaps in the toolkit and potential future directions, with inspiration from successful methods used to study glycans in mammalian systems.
2. Evolution of the chemical toolkit to study bacterial glycans
The study of bacterial glycans was pioneered with rigorous analytical approaches to determine their molecular structures and unveil their biosynthetic pathways. Though these approaches have been highly successful, they are time-consuming and the results are not readily transferable as the structures elucidated differ among classes of bacterial glycans and across bacterial strains. Recently, these traditional approaches have been complemented with new technologies that uncover important functional information about bacterial glycans and their biosynthesis.
2.1. Traditional approaches
Traditionally, intensive biochemical and genetic tools have been deployed to probe bacterial glycans. An early study by Barreteau and colleagues20 deconstructed peptidoglycan biosynthesis in Escherichia coli into cytoplasmic steps using extensive gel filtration and reverse-phase high pressure liquid chromatography techniques. Similarly, Hartley et al.19 characterized the O-linked glycosylation pathway in Neisseria gonorrhoeae via a multi-step strategy that involved the preparation of genetic constructs, protein purification, and enzymatic assays. Kido and Kobayashi21 revealed the genes required in structure determination of E. coli polysaccharides by performing site-directed mutagenesis and immunoblot analyses. Additional examples have been thoroughly reviewed by Cooper,22 Silhavy, Kahn, Walker and others23. These efforts form the bedrock of our current understanding of glycans in the bacterial cell envelope.
In addition, conventional approaches have relied heavily on chemical tools that illuminate the molecular fingerprints of bacterial glycans. For example, mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy have permitted detailed structural characterization of diverse bacterial glycans.24–26 MS approaches rely on fragmentation strategies to partition complex, unusual glycan structures into smaller analyzable components. For these studies, the use of enzymatic degradation with glycosidases3 and chemical degradation (e.g. partial acid hydrolysis, beta elimination, and Smith degradation27) to cleave complex glycoconjugates into simpler fragments has been critical to deconvolute and assign structures. Meanwhile, NMR techniques involve heavy computation to decipher carbohydrate couplings in complex glycans.28
While these studies have led to robust characterization of glycans in model bacterial species, specific insights are not necessarily translatable among bacteria. For example, in seminal work by Koomey, Imperiali, and coworkers, specific enzyme activity assays were employed to analyze and assign the biochemical function of glycosyltransferases encoded by protein glycosylation genes in Neisseria spp.19–21 This strategy required prior knowledge of target glycans’ structures and thus is nontrivial to apply to species whose glycan structures remain unknown. Likewise, analysis of LPS structures via MS and NMR studies yielded critical information about LPS biosynthesis in multiple Gram-negative bacteria,26, 30 yet this approach involved time- and labor-intensive purification and enrichment of target glycans. Structural variability of the LPS O-antigen among Gram-negative bacteria makes information gleaned about LPS biosynthesis in E. coli29 not readily transferable to LPS biosynthesis in other Gram-negative bacteria, for example, or even across bacterial isolates of the same species. Taken together, these strategies yield invaluable insights yet are not always feasible for timely probing of unknown structures in priority and emerging antibiotic-resistant bacteria.
2.2. Emerging approaches
Over the last few decades, chemical tools have been developed to probe bacterial glycan biosynthesis via complementary approaches to those described above. Metabolic oligosaccharide engineering (MOE) is a chemical approach pioneered by Bertozzi,30, 31 Reutter,32, 33 and coworkers to study glycans in eukaryotic systems34 and more recently to study bacterial glycans (Fig. 2). In this two-step chemical approach, cells are treated with an unnatural sugar adorned with a bioorthogonal functional group, termed a “chemical handle” (e.g. azide), that is processed by permissive glycosyltransferases to yield cellular glycans bearing the modified sugar. Then, cells are incubated with an exquisitely selective reaction partner (e.g. alkyne) that reacts with the chemical handle-containing glycan via a bioorthogonal reaction35, 36 to form a covalent adduct. Using this two-step approach, metabolically labeled cellular glycans can be elaborated with a reporter of choice (e.g. fluorophore, biochemical epitope tag) to ease their detection and study.
Figure 2. General schematic of metabolic oligosaccharide engineering (MOE).
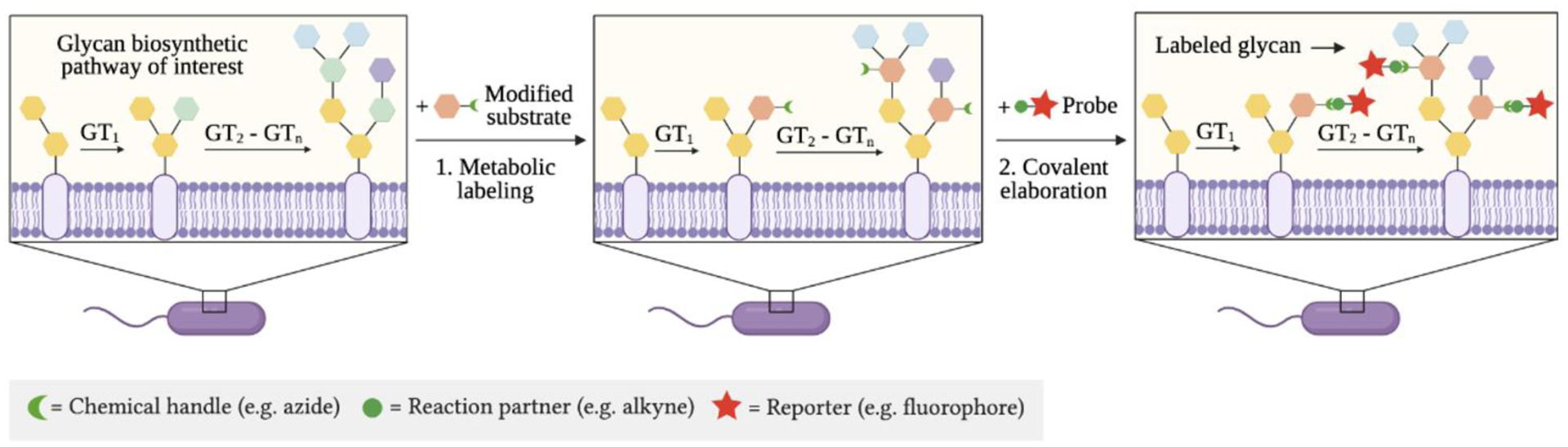
First, a sugar modified with a chemical handle is metabolically incorporated via permissive glycosyltransferases (GTs) into cellular glycans. Second, an exquisitely selective partner undergoes bioorthogonal reaction with the chemical handle to yield a detectable covalent adduct bearing a reporter.
A suite of bioorthogonal reactions have been developed to probe glycans in living systems.37 For example, “click chemistry” between azides and alkynes,38 Staudinger ligation between azides and triarylphosphines,39 and inverse electron demand Diels-Alder between cyclopropenes and tetrazines40 are three popular bioorthogonal reactions that have been deployed to study bacterial glycans. As described in more detail below, this approach enables the delivery of reagents which facilitate the visualization and perturbation of bacterial glycans and their biosynthesis. While initial reports of MOE in bacteria focused on unnatural versions of ubiquitous monosaccharides, the chemical toolkit for the study of bacterial glycosylation has expanded considerably in recent years. A variety of metabolic reporters, crafted from analogs of monosaccharides, D-amino acids, and fatty acids, has emerged to probe, perturb and image bacterial glycan biosynthesis. Critically, because all that is required in MOE is an unnatural substrate bearing a chemical handle, this approach offers the opportunity to study bacterial glycoconjugates even without full knowledge of their structures.
Lectin-based assays and microarrays have also emerged as complementary approaches to study bacterial glycans. Lectins make up a class of proteins that bind chiefly to sugars. Early precedents in bacterial glycobiology relied on plant-derived lectins to discover protein glycosylation in model bacteria such as C. jejuni.12 These initial successes helped to spur important developments in this arena by Mahal,41 Gildersleeve,42 Paulson,43 and others,44 who pioneered and popularized the use of glycan-binding agents to examine the bacterial glycocalyx. As described in more detail below, lectin-based approaches enable the inspection of glycan interactions on the surface of bacterial cells to illuminate key host-pathogen dynamics. Similar to MOE, lectin-based studies and microarray platforms offer high-throughput parallel approaches to yield important insights into bacterial glycans and their biosynthesis.
In the following sections, we describe contemporary tools in categories based on their objectives: (1) probing, (2) perturbing, and (3) imaging bacterial glycans. These technologies reveal important functions of glycans that, in combination with traditional structural analysis, accelerate the construction of glycosylation pathways and determination of key drug targets in medically significant pathogens.
3. Probing bacterial glycans
Recent advances in chemical biology have introduced tools to yield insights into glycans and their biosynthesis in the absence of detailed structural information. Here we highlight the development and application of novel metabolic labeling methods, lectins, and microarray technologies to probe bacterial glycans.
3.1. Metabolic labeling
Bacteria containing suitably promiscuous metabolic pathways can uptake unnatural substrates bearing chemical reporters and incorporate them into their cellular glycans. This observation was first exploited by Nishimura and coworkers45 when they incorporated a ketone-bearing substrate into E. coli’s peptidoglycan. Since that foundational demonstration, metabolic labeling of bacterial glycans has been deployed to understand the steps and key players involved in bacterial glycan biosynthesis. A wide range of metabolic precursors bearing chemical reporters has been developed to study bacterial glycans.46 In this section, we describe representative metabolic labeling strategies and key information these probes have yielded about bacterial glycosylation.
Bacterial monosaccharide analogs have emerged as excellent metabolic reporters of bacterial glycans. Initially, unnatural azide-containing sugar analogs of common monosaccharides such as N-acetylgalactosamine (GalNAc),47 N-acetylglucosamine (GlcNAc),48, 49 fucose (Fuc),50 and sialic acid (Sia)51 facilitated the detection of glycans in bacterial systems. However, these substrates have a relatively narrow incorporation efficiency across bacterial species. For example, Clark et al.52 demonstrated that an azide-containing variant of GlcNAc, peracetylated N-azidoacetylglucosamine (GlcNAz), was robustly incorporated into glycoproteins in the gastric pathogen H. pylori, yet was not appreciably incorporated into glycoproteins produced by C. jejuni, M. smegmatis, B. fragilis, Pseudomonas aeruginosa, nor Burkholderia thailandensis. In response to this observation, Dube, Kulkarni and coworkers53 developed azide-containing analogs of the rare bacterial deoxy amino d-sugars bacillosamine, DATDG, and FucNAc to label glycoproteins in a larger array of Gram-negative species (Fig. 3A). The authors reported that several pathogenic bacteria robustly incorporated rare monosaccharide analogs into their glycans, while symbiotic bacteria and mammalian cells did not incorporate a detectable amount of these analogs onto their cell surfaces. This observation indicates the potential of atypical bacterial sugars to be pathogen-selective probes in complex microbial communities such as the gut microbiome.
Figure 3. Metabolic labeling of bacterial glycans with a variety of unnatural substrates.
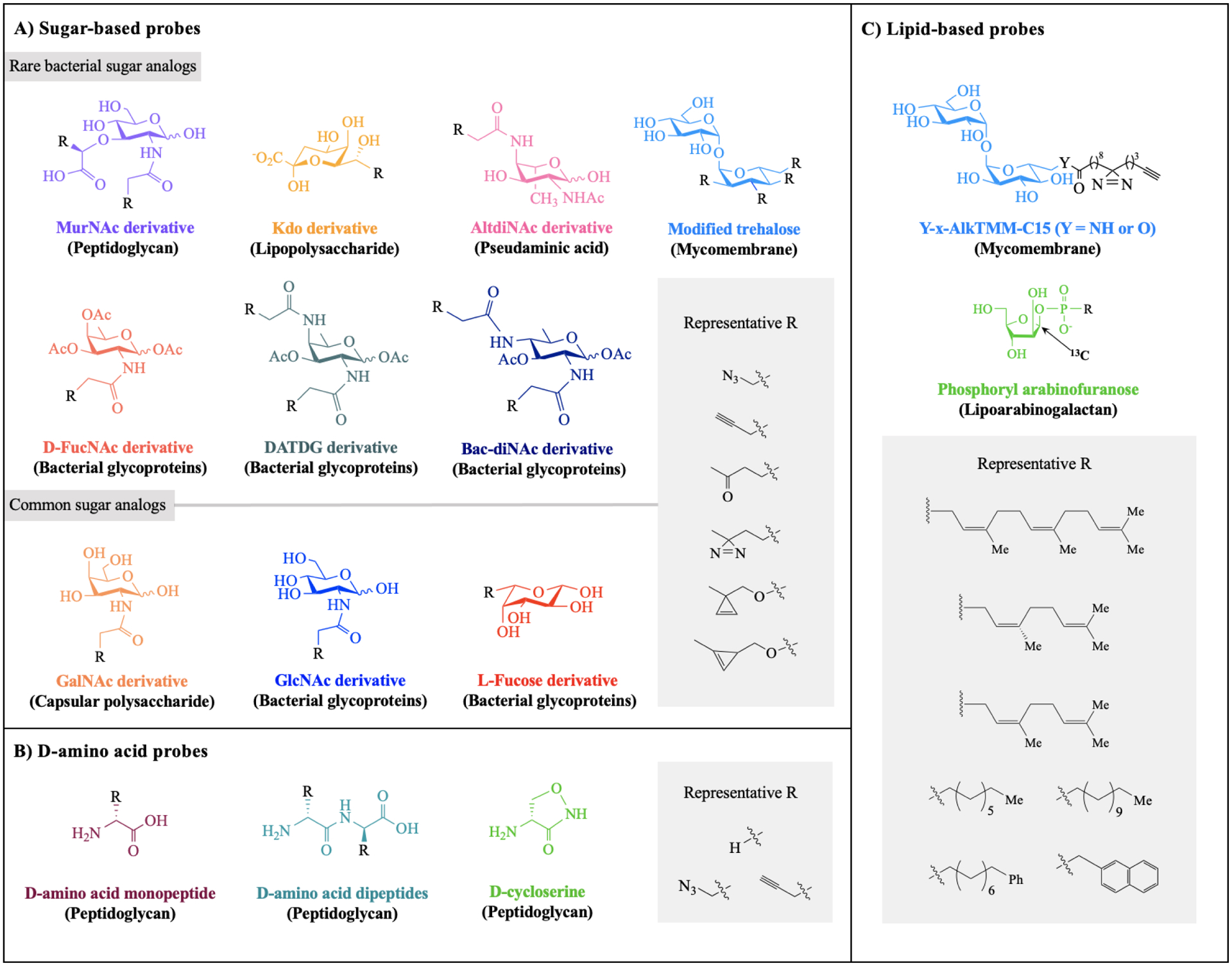
Shown here is a panel of substrate analogs accessed from A) rare and common sugar building blocks, B) D-amino acids, and C) glycolipids. For each scaffold, chemical modification is permissible at positions with R groups and known metabolic fate indicated in parentheses. Abbreviations: MurNAc = muramic acid; KDO = 3-deoxy-d-manno-oct-2-ulosonic acid; AltdiNAc = 2,4-diacetamido-2,4,6-trideoxy-l-altrose; D-FucNAc = N-acetyl-d-fucosamine; DATDG = 2,4-diacetamido-2,4,6-trideoxyhexose; Bac-diNAc = N,N’-diacetylbacillosamine; GalNAc = N-acetylgalactosamine; GlcNAc = N-acetylglucosamine; AlkTMM = alkylated trehalose monomycolate.
These findings set the stage for deploying a broad array of rare monosaccharide analogs, including MurNAc, AltdiNAc, and trehalose derivatives, to probe bacterial glycosylation (Fig. 3A). These analogs have yielded incisive insights into bacterial glycan biosynthesis. For example, Andolina and coworkers54 reported that the modified hexose probe 2-acetamido-4-azidoacetamido-2,4,6-trideoxy-l-altrose was metabolically converted in situ to pseudaminic acid (Pse) and led to installation of an azide in pseudaminic acid-containing glycans. Using this strategy, the authors validated the presence of Pse biosynthesis in a series of pathogenic bacteria that appeared, based on bioinformatics studies, to have Pse biosynthesis enzymes. Given the role of Pse in host-pathogen interactions of Gram-negative bacteria,55 access to this information could underpin glycan interference strategies. In another example, Moulton and colleagues56 utilized GlcNAz in an MOE-based screen to identify H. pylori’s O-linked protein glycosylation genes. By tracking glycoprotein biosynthesis in wildtype H. pylori versus glycosylation mutants, the authors were able to develop a model of glycoprotein biosynthesis and gain insights into the functional consequences of disrupting glycoprotein biosynthesis in this system.
In addition to monosaccharide-based probes, D-amino acids (DAAs) are versatile probes for metabolic labeling of bacterial peptidoglycan (PG) (Fig. 3B). Previously, Pires,57 VanNieuwenhze,58–61 Walker,62 Bertozzi63 and others64–68 established the uptake of unnatural DAAs into bacteria with PG. More recently, VanNieuwenhze and coworkers69 explored the mechanism of DAA incorporation into bacterial PG. The authors revealed that bacteria processed synthetic DAAs via two extracytoplasmic pathways, through D,D-transpeptidases and L,D-transpeptidases, rather than via cytoplasmic incorporation. This mechanistic finding prompted the development of a new fluorescent DAA probe for metabolic labeling of subcellular muropeptides,69 enabling the study of PG biosynthesis from not only the cell surface but also the cytoplasm. In another example, the Siegrist group70 reported the use of monopeptide DAA probes as PG precursors in mycobacterial cell wall synthesis (Fig. 3B). Through this strategy, Siegrist and colleagues tracked the remodelling of M. tuberculosis’s sidewall. They discovered the pathogen’s ability to relocate peptidoglycan assembly to repair the cell wall in response to damage caused by antibiotics. Thus, DAA probes have produced medically significant insights that could inform PG interference strategies.
Mycolate and trehalose analogs can be harnessed for selective metabolic labeling of mycobacterial glycans (Fig. 3C). Mycobacteria are unique for their mycomembrane, an outer membrane comprised primarily of trehalose-linked lipids, namely mycolic acids.71 Although many anti-tuberculosis drugs target the mycomembrane, its biosynthesis and host-interaction mechanisms are not fully understood.72 For example, mycolate-interacting proteins are important biomolecules involved in mycomembrane metabolism and host interactions, yet are currently underexplored. To fill this gap, the Swarts group recently reported the application of photoactivatable trehalose monomycolate analogs bearing diazirines and alkynes for in vivo targeting and analysis of mycolate-interacting proteins in M. smegmatis (Fig. 3C).73 Once metabolically incorporated into the mycomembrane, the glycolipid probes underwent photo-crosslinking with target proteins and subsequent click-chemistry-mediated purification and characterization. This approach enriched over 100 mycolate-interacting proteins involved in mycobacterial modulation of host interactions and enabled their structural and functional characterization. The strategy featured here is an example of proximity labeling, a method pioneered by Ting,74 Burke,75 and others to enrich neighboring proteins with biotin using engineered enzymes.76 Proximity labeling has been extended for in vivo tagging using a heterobifunctional probe with a photoactivatable moiety and biotin.77 Since this technology is well-established in proteomic studies, its recent application in mycobacterial glycobiology by the Swarts group is a novel advance. Moreover, this strategy should be generalizable to any unnatural substrates that are processed into bacterial glycans, thus setting the stage for studies beyond mycobacterial species.
Unnatural glycolipid-based substrates are powerful probes to label glycans that are recalcitrant to metabolic labeling with unnatural monosaccharides. Recent work by Calabretta and colleagues78 featured the use of synthetic arabinofuranosyl phospholipids to probe cell wall glycans in Corynebacterium glutamicum and M. smegmatis, models of M. tuberculosis (Fig. 3C). In the study, a glycolipid probe based on a late-stage biosynthetic intermediate was developed. This lipid-linked glycan restored cell wall arabinan in a C. glutamicum arabinan mutant and rescued wild-type cells treated with a cell wall synthesis inhibitor, leading to the full recovery of the cell envelope in both cases. Moreover, the incorporation of a 13C label into the arabinofuranosyl moiety on the probes facilitated NMR characterization of cell wall glycans. These findings highlight the ability of unnatural glycolipids to serve as trackable biosynthetic intermediates of cell wall synthesis in mycobacteria. Further, they offer an important alternative if analogs based on early biosynthetic intermediates (e.g. monosaccharides) fail. Taken together, these recent reports provide illustrative examples of how metabolic labeling can be used to probe bacterial glycan biosynthesis and function.
3.2. Lectins
Over the last decade, new applications of lectins in probing bacterial glycans have emerged. Early examples in bacterial glycobiology relied on plant-derived lectins, which continue to advance the field. Beyond the important role plant lectins have played in detecting and facilitating the study of different classes of bacterial glycans, novel applications are still unfolding. For example, Kalograiaki et al.79 recently demonstrated the use of plant lectins for bacteria typing. In their report, the group determined that two galactose-specific plant lectins, Ricinus communis (RCA) and Viscum album (VAA) lectins, bound differentially to non-capsulated Haemophilus influenzae in clinical samples. The specific display of cell surface glycans had differential effects on RCA and VAA binding to H. influenzae. In particular, RCA bound to H. influenzae strains displaying truncated lipooligosaccharides, whereas VAA only recognized fully elaborated lipooligosaccharides. This study affirms the utility of bacteria-binding plant lectins and reinforces plants as important sources of biological probes crucial to the advancement of bacterial glycobiology. Future studies can examine the potential of using RCA and VAA lectins to detect and differentiate strains of H. influenzae in infection models.
Beyond plant lectins, advances in the field have also relied on mammalian lectins produced by the innate immune system to interrogate cell surface glycans that distinguish host cells from foreign pathogens.80 The three main classes of human innate immune-associated lectins are S-type lectins (galectins), siglecs, and C-type lectins.81 However, since these lectins recognize and bind to pathogenic glycan epitopes in a density-dependent manner,82 they have had limited utility for profiling isolated bacterial glycans. Specifically, innate immune lectin binding depends on the density and number of glycan epitopes present on the cell surface rather than just the affinity of single epitopes. As a result of multivalent binding, these lectins bind with low affinity and high avidity to abundant glycans rather than rare epitopes of interest. In response to these limitations, current efforts in this area seek to identify new lectins capable of specific recognition to facilitate precise bacterial glycan analyses.
Bacteria-focused lectins allow the inspection of bacterial glycans and their dynamics. One novel class of lectins that binds specifically to microbes is X-type lectins, or intelectins.83 Intelectins were recently demonstrated to play an important role in the human immune system.84 Lee and coworkers85 identified two types of intelectins, human intelectin-1 (hIntL-1) and human intelectin-2 (hIntL-2), encoded by the human genome. Subsequently, Kiessling and coworkers86 discovered that hIntL-1 bound exclusively to a diverse array of microbial glycan epitopes, including d-Galf- and KDO-presenting epitopes, not present on human cells. Later studies by Wangkanont87 and Chen et al.88 revealed that the mechanism of intelectins in recognizing bacterial glycans is conserved from Xenopus laevis to zebrafish to humans. This observation presents intelectins as naturally occurring bacteria-focused lectins that can act as selective probes for scrutinizing the bacterial glycocalyx. Thus, intelectins are potentially useful for analyzing the bacterial glycoform, unveiling key glycan-mediated mechanisms of infection, and tracking glycan-binding interactions crucial to pathogenesis. For more information, Wesenser and colleagues86 published an excellent review on soluble human lectins that have the potential to be harnessed for detection and characterization of bacterial glycans.
Additionally, microbial lectins are complementary probes for identifying epitopes on the host cell surface responsible for bacterial adhesion and colonization. In a recent example, Sykorova and colleagues89 discovered two novel lectins, BP39L and CV39L, from the pathogens Burkholderia pseudomallei and Chromobacterium violaceum, respectively. Functional assays verified the binding preference of BP39L for mannosylated saccharides and CV39L for more complex β-l-fucose-presenting polysaccharides. These data imply that host cells presenting identified epitopes are likely vulnerable to B. pseudomallei and C. violaceum infections. Moreover, determining potential binding partners of bacterial lectins facilitates the development of therapeutics that prevent infection by blocking bacterial adhesion to host cells. As a proof-of-concept study, Le and coworkers90 recently examined the binding preferences of fucose-specific lectins, namely LecB, BC2L-C, and AFL, produced by cystic fibrosis-causing pathogens. The authors designed a panel of multivalent α-l-fucoside-containing glycoclusters and screened for potential inhibitors of fucose-specific lectins. Notably, they demonstrated that a tetravalent, α-l-fucose-presenting glycocluster inhibited P. aeruginosa adhesion to human epithelial bronchial cells in vitro. This study sets the stage for a class of anti-adhesive inhibitors, highlighted in section 4.2, which impedes key glycan-binding interactions to block bacterial entry to the host and reduces infection as a result.
3.3. Microarrays
Microarrays allow the high-throughput analysis of multiple biomolecular interactions simultaneously using small and concentrated amounts of sample.91 Microarrays are assembled by tethering the probes of interest (e.g. lectins, glycans, bacteria) to a surface (e.g. glass slides). Once assembled, the binding of targets to the arrays is assessed, typically via a fluorescent mechanism. The immobilized probes range from DNA, proteins, and more recently, to glycans. Respectively, these probes enable massive parallel analyses to query the presence of genes, proteins, and glycans of interest in samples, as well as their binding profiles. Although carbohydrate microarrays were originally developed to study glycan-mediated interactions in mammalian models,92 this technology has extended its reach to bacterial systems. In this section, we highlight the use of different microarrays to probe glycan expression and binding preferences in bacteria.
Lectin microarrays are effective in exposing differential bacterial glycosignatures (Fig. 4A). Recent work by the Drickamer group93 employed mammalian lectin arrays to screen for bacteria-specific receptors. Using cow C-type lectins as probes, they discovered distinct binding preferences across pathogens including E. coli, K. pneumoniae, Staphylococcus aureus, and Mycobacterium bovis. In addition to providing an unbiased search for lectins that bound to a particular microbe, these microarrays revealed previously unknown binding events, indicating novel glycan compositions on some of the studied bacteria. This assay can be adapted for myriad purposes: probing glycan structure by assaying the binding of different lectins to one bacterial species, comparing binding specificity of potential receptors across bacterial species, assessing binding to different host lectins across pathogens, and more. In all applications, lectin microarrays have the potential to shed light on the diversity of bacterial glycoforms. Elucidating these glycopatterns can facilitate the development of functional assays that explore the role of distinctive bacterial glycan coatings in modulating the dynamics between bacteria, host cells, and the environment.
Figure 4. Microarray assays probe glycan-binding events pertinent to host-pathogen interactions.
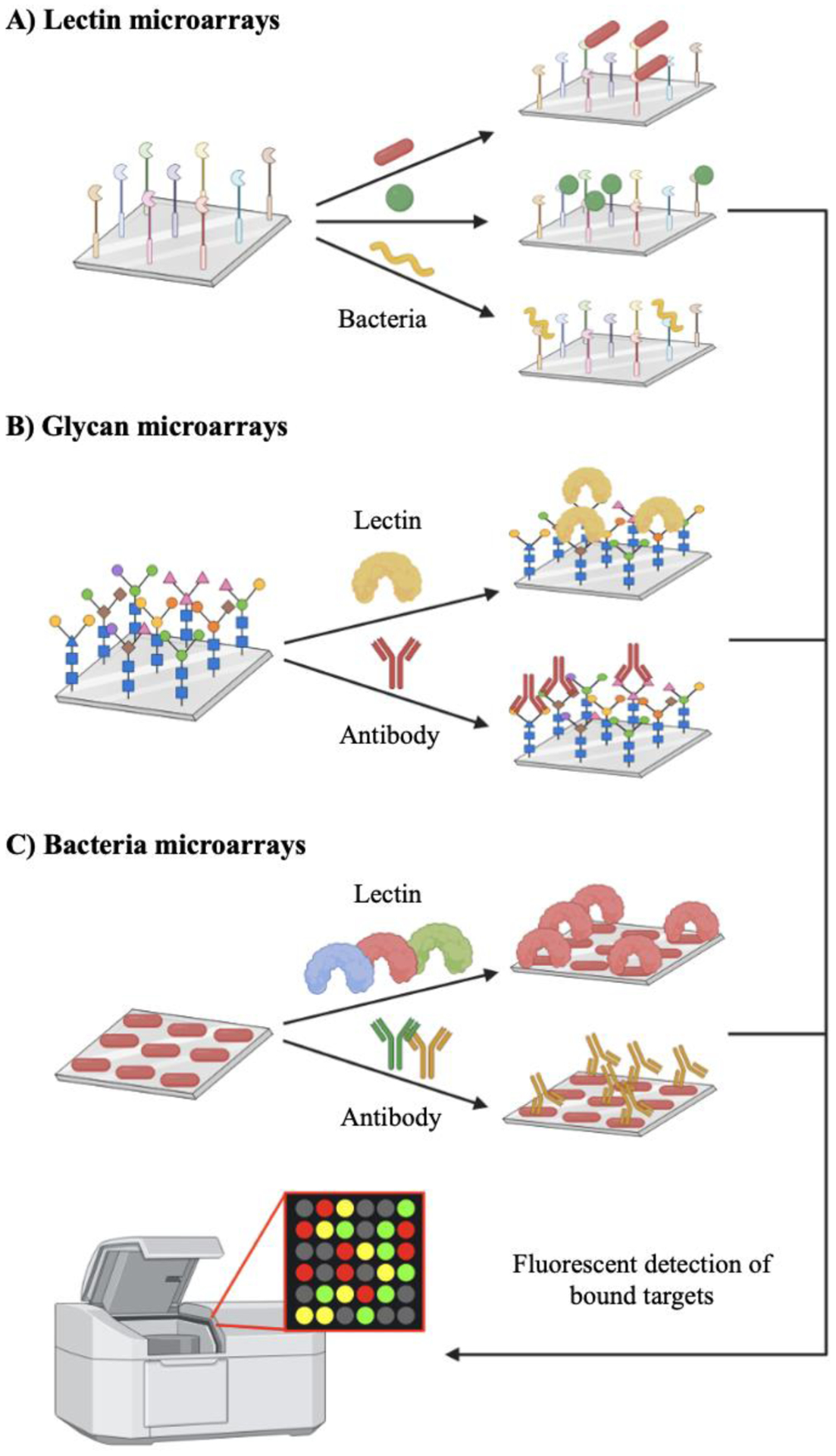
A) Lectin microarrays differentiate bacteria based on distinct binding patterns. B) Glycan microarrays elucidate glycan epitopes key to host adhesion and identify novel immunogenic targets. C) Bacteria microarrays reveal intact glycan-binding interactions on bacterial surfaces.
Microarrays incorporating mammalian and bacterial carbohydrates have been recently applied to characterize bacterial glycans and their binding interactions (Fig. 4B). One use of these microarrays is determining structures of the bacterial glycome responsible for key host-pathogen interactions. For example, Dupin and coworkers94 used a human glycocluster microarray to identify binding partners of LecB, a lectin produced by P. aeruginosa to facilitate adhesion to host cells. The microarray consisted of oligosaccharides and polysaccharides collected from bacterial fermentation and immobilized on a DNA chip, where their interaction with LecB was monitored. The authors ultimately uncovered carbohydrate residues responsible for multivalent binding to LecB. In another study, Wang and colleagues95 demonstrated the use of a PG microarray for the high-throughput screening of peptidoglycan binding proteins. The authors discovered PG fragments that served as ligands for human peptidoglycan recognition proteins.
Additionally, glycan microarrays can unveil bacterial glycan epitopes that are potential vaccine candidates.96–98 In one seminal example, the Seeberger group99 developed a glycoconjugate vaccine against S. pneumoniae using a combination of automated glycan assembly and glycan microarray-based screening. First, the authors generated synthetic glycans from S. pneumoniae capsular polysaccharides via automated glycan assembly100, 101 and identified immunogenic glycan epitopes using a glycanmicroarray platform. With iterative output from this assay, they crafted and refined a glycoconjugate vaccine that elicited a robust antibacterial immune response in rabbits. This study is an excellent example of applying glycan microarrays to discover and enhance glycoconjugate therapeutics against priority pathogens.
As an alternative to probing pure glycans and their fragments, whole bacteria microarrays can facilitate the characterization of intact bacterial glycans in context (Fig. 4C). In a pioneering study, Solis and coworkers102 printed K. pneumoniae on a nitrocellulose-coated glass slide. After validating this approach by detecting the binding of bacterial cells to anti-K. pneumoniae antibodies, the authors examined bacterial glycosylation patterns and screened for inhibitors of host-pathogens interactions. Glycosylation patterns were derived from the binding behavior of bacteria microarrays to a panel of plant lectins with and without the presence of competing carbohydrates. The authors observed that specific carbohydrates reduced the ability of lectins to bind to bacterial cells via competitive interference. This result confirms these carbohydrates as potential inhibitors of host-pathogen interactions. Thus, future studies can couple this assay with recently developed sugar-based inhibitors,53, 103, 104 highlighted in section 4.1., to screen for potential leads that impede bacterial attachment and colonization of the host. In more recent work, Solis and colleagues105 used bacterial microarrays to detect the presence of target glycan structures on the surface of nontypeable H. influenzae. In addition to detecting H. influenzae glycans, they observed strain-selective binding of innate immune lectins to nontypeable clinical isolates. Thus, bacterial microarrays enable typing and pattern recognition of bacterial glycans without relying on intensive glycan synthesis and purification.
3.4. Summary
Above, we featured a series of complementary tools for probing bacterial glycans in the absence of complete structural information. Metabolic labeling is one of the most versatile techniques available to probe glycosylation in all classes of bacteria, and is accessible through a wide range of precursors, from monosaccharides, to D-amino acids, to lipid substrate analogs. While metabolic labeling allows the tailoring of substrates to study glycans of interest, lectins offer a complementary approach to study bacterial glycans. Recent discoveries have revealed several lectins that bind exclusively to bacterial glycans, allowing further scrutiny of the bacterial glycocalyx. In addition, microbial lectins elucidate host glycan epitopes key to bacterial adhesion and infection. Finally, microarrays extend on the use of lectins to allow high-throughput screening of glycan-binding interactions. This technology has been adapted to directly utilize bacterial glycans or whole bacterial cells for capturing native glycosignatures and binding events. Taken together, these approaches comprise an enabling toolkit that can uncover medically important glycosylation pathways and targets in priority pathogens.
4. Perturbing bacterial glycans
New tools to study bacterial glycosylation are capable of manipulating glycans and their biosynthesis in real-time. Here we feature the advancements in methods to perturb bacterial glycans, including glycosylation inhibitors and molecules that block glycan binding interactions, and the consequences of these glycan-perturbing agents.
4.1. Perturbation approach: Inhibit glycan biosynthesis
Bacterial glycans are crucial to bacterial fitness and pathogenesis. Indeed, genetic and small molecule inhibition of glycocalyx construction leads to bacteria with reduced viability.106–111 Additionally, pathogenic bacteria coat themselves with unique glycans key to host cell binding and colonization.4 These characteristics, coupled to the long history of using antibiotics to disrupt glycan biosynthesis, suggest bacterial glycans are attractive targets for therapeutic intervention.
Traditional small molecule inhibitors are potent in obstructing bacterial glycan biosynthesis (Fig. 5A). Recent work by Naclerio et al.112 identified novel compounds that suppressed lipoteichoic acid biosynthesis in methicillin-resistant S. aureus (MRSA). Lipoteichoic acid (LTA) is crucial to survival and pathogenicity in Gram-positive bacteria. In the study, the authors discovered small molecule inhibitors containing the 1,3,4-oxadiazolyl core that interrupted S. aureus LTA biosynthesis and caused dramatic bactericidal effects in vivo. These compounds exhibited inhibition potency from 4 to 8-fold greater than blockbuster antibiotics against MRSA such as vancomycin and linezolid. Similarly, a study by Imperiali and coworkers113 employed a fragment-based screening approach for the high-throughput identification of small molecule inhibitors of PgID, a key enzyme in C. jejuni bacillosamine biosynthesis (Fig. 5B). Optimized compounds effectively inhibited the target enzyme in the nanomolar range, yet they did not impact in vivo glycosylation of cell surface proteins. Thus, further structural optimization will be required to enhance the antibacterial potency of these compounds. Though these efforts are at different stages, they are incredibly important steps in the process of teasing out the impacts of small molecule inhibitors on glycan biosynthesis and validating different targets.
Figure 5. Mechanisms of action of bacterial glycosylation inhibitors and representative structures of each class of inhibitors.
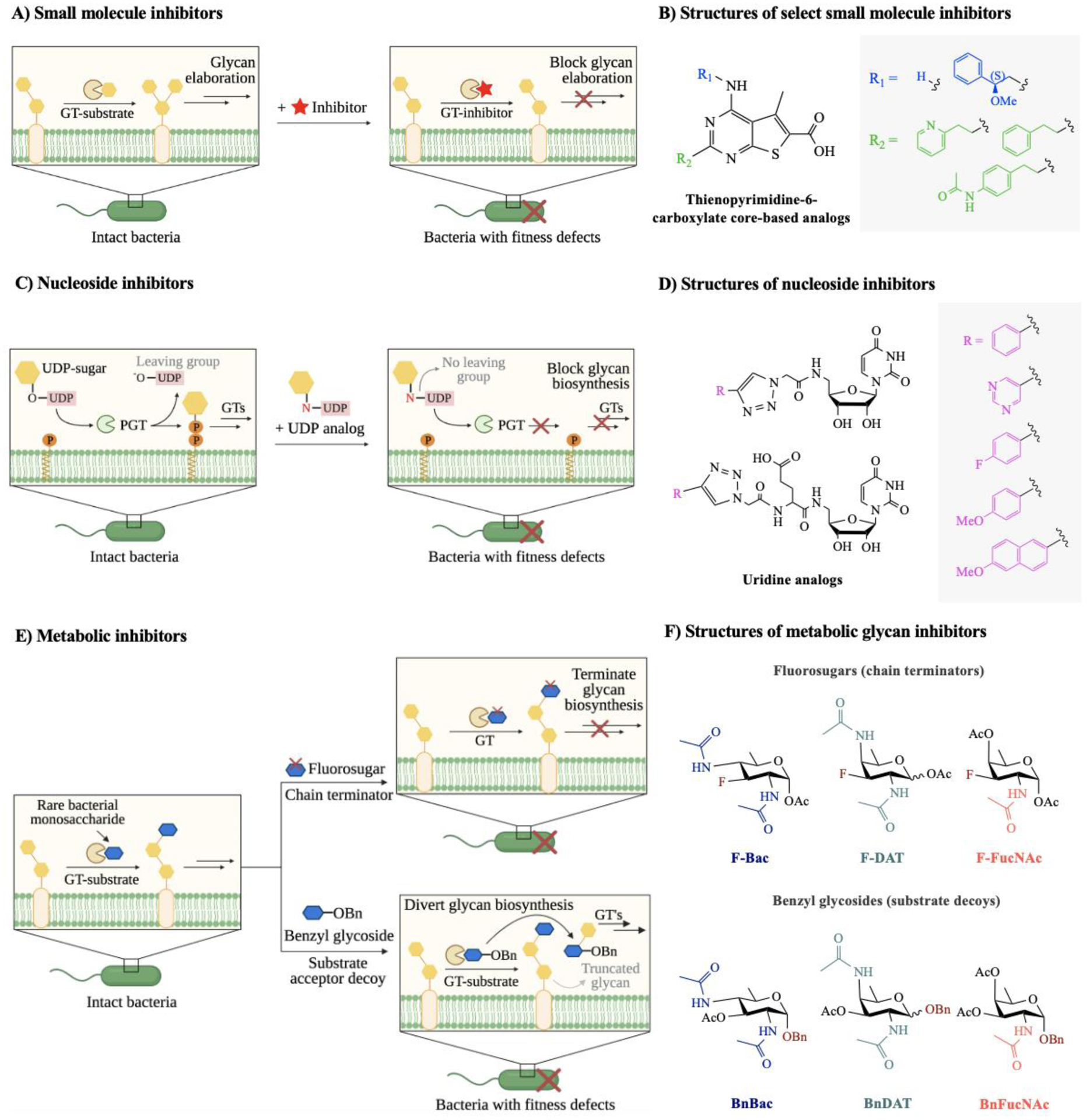
A) Small molecule inhibitors compete with natural substrates in the active site of the target glycosyltransferase (GT), leading to the reduction of glycan production in the associated glycan biosynthetic pathway. B) Structures of representative small molecule inhibitors accessed from thienopyrimidine-6-carboxylate core-based analogs.113 C) Phosphoglycosyl transferases (PGTs) initiate glycan biosynthesis on a lipid carrier by catalyzing the transfer of a nucleoside sugar to a polyprenyl phosphate. Uridine analogs inhibit PGT activity and block addition of monosaccharides onto the lipid carrier. D) Structures of representative nucleoside inhibitors accessed from uridine analogs.115 E) Metabolic inhibitors consist of two classes: fluorosugars (chain terminators) and benzyl glycosides (substrate decoys). Fluorosugars terminate glycan elongation, while benzyl glycosides mimic endogenous glycan acceptors and divert glycan biosynthesis onto decoy substrates. F) Structures of representative metabolic inhibitors accessed from rare deoxy amino sugars, featuring chain terminators and substrate decoys.53 Abbreviations: Bac = bacillosamine (blue hexagon); DAT = 2,4-diacetamido-2,4,6-trideoxygalactose; FucNAc = N-acetylfucosamine.
Nucleoside inhibitors can also impede key players in target bacterial glycosylation pathways (Fig. 5C). Recent work by Imperiali and coworkers114 explored a modular design based on the scaffold of potent nucleoside antibiotics to yield inhibitors of bacterial phosphoglycosyltransferases (PGTs). PGTs are essential enzymes for bacterial glycan assembly; they add the first phosphorylated monosaccharide onto a lipid-carrier to create a lipid-linked oligosaccharide, which is further elaborated by the sequential action of glycosyltransferases to yield higher-order glycans for glycoprotein, glycolipid, and polysaccharide biosynthesis. Their strategy yielded potent inhibitors that disrupted PGT activity in the micromolar range, suggesting nucleoside-based small molecules are novel anti-PGT agents that can impede early-stage glycan biosynthesis. In another study exploring nucleoside-based inhibitors, Madec and colleagues115 reported a library of uridinyl nucleoside analogs that exhibited inhibition of bacterial sugar-modifying enzymes (Fig. 5D). Then, they screened the inhibitors for their efficacy in interrupting the functions of glycosyltransferases from representative Gram-negative and -positive bacteria. Uridine analogs inhibited UDP-sugar processing enzymes in an enzyme-specific fashion in the micromolar range. These studies establish nucleoside inhibitors as potential leads that target bacterial glycosyltransferases of therapeutic interest.
Recently, novel metabolic inhibitors were developed to interrupt bacterial glycan biosynthesis via different mechanisms (Fig. 5E). Dube, Kulkarni, and coworkers53 designed and synthesized two classes of metabolic inhibitors based on the rare bacterial monosaccharides bacillosamine, DATDG, and FucNAc (Fig. 5F). One class of inhibitors, termed chain terminators, relied on fluorosugars that were thought to be incorporated into the growing bacterial glycan but terminated further elongation, ultimately yielding truncated cellular glycans by serving as dead-end substrates. In the other class of inhibitors, termed substrate decoys, bacterial glycan biosynthesis was diverted onto benzyl glycosides to outcompete endogenous substrates and yield truncated cellular glycans. The authors demonstrated that both classes of metabolic inhibitors led to glycoprotein biosynthesis defects in H. pylori. Moreover, disrupting H. pylori’s glycocalyx precipitated downstream functional defects in growth, motility, and biofilm formation. Not only did the inhibitors alter H. pylori’s protein glycosylation, but they acted in a bacteria-selective fashion. These inhibitors pave the way for the systematic study and targeting of pathogen-specific glycosylation machinery.
Metabolic inhibitors have also been developed to selectively interfere with the mycobacterial glycocalyx. Sucheck and colleagues103 utilized non-hydrolyzable derivatives of trehalose to inhibit mycobacterial trehalose biosynthesis. The modified trehalose derivatives inhibited trehalose 6-phosphate phosphatase in the micromolar range. However, further investigation of the functional effects of these inhibitors in cells is necessary to validate this strategy. In conceptually related work, Wolber and coworkers104 inhibited biofilm formation in M. smegmatis using 2-, 5-, and 6-position-substituted trehalose analogs. In addition to impacting biofilm formation, these compounds moderately inhibited in vitro growth in the micromolar range. Hence, metabolic inhibitors offer a generalizable approach to disarming the glycocalyx in a variety of pathogenic bacteria.
4.2. Perturbation approach: Block glycan binding
One key function of bacterial glycans is modulating host-pathogen interactions via specific glycan-binding events. Bacteria adhere to host cells by presenting surface glycans that are exquisitely selective to target molecules on the host cell surface.116 Further, bacterial adhesins can bind to host surface glycans in a lock and key fashion, facilitating bacterial entry and colonization of the host. Blocking host-pathogen glycan-binding interactions poses one approach to interfere with the infection cycle (Fig. 6A). Toward this end, anti-adhesive agents that bind and block glycans or glycan-binding proteins offer great promise for preventing infections.
Figure 6. Blocking glycan binding events key to pathogenesis.
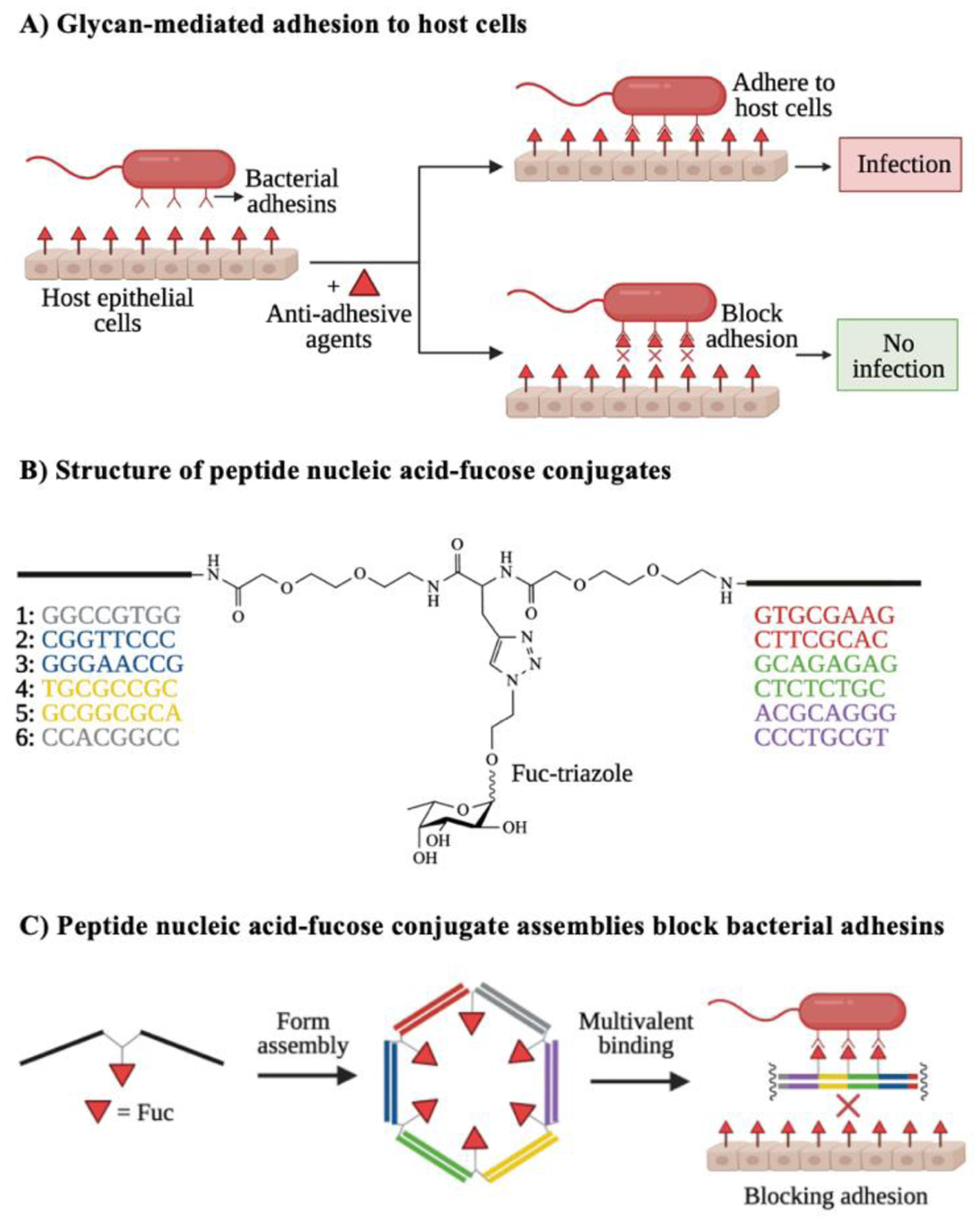
A) Bacterial adhesion to host cells often depends on host-pathogen interactions. Anti-adhesive agents that bind to either bacterial adhesins or host receptors can block these glycan-mediated binding events, resulting in reduced infection. B) Structure of representative synthetic anti-adhesive compounds, featuring peptide nucleic acid-fucose conjugates. C) Peptide nucleic acid-fucose conjugates can form assemblies that bind to bacterial adhesins in a multivalent fashion and block bacterial entry to the host. Images B and C are inspired by Figure 2 from Machida et al.117
New advances in chemical biology have produced novel anti-adhesives of host-pathogen glycan-binding events. Machida and coworkers117 recently reported the use of nucleic acid-based oligomeric assemblies with peptide nucleic acid-fucose conjugates to disrupt the binding of Burkholderia ambifaria’s lectins to host epithelial cells (Fig. 6B). The peptide nucleic acid-fucose conjugates bound with high affinity to the fucose-binding lectin BambL of B. ambifaria, forming unstable assemblies that reduced bacterial adhesion to host cells (Fig. 6C). These assemblies fully blocked the pathogen’s host cellular entry, demonstrating synthetic agents have the potential to interfere with bacterial glycan-binding interactions.
Natural products are an important source of bioactive compounds that can exhibit anti-adhesive properties. Fu and colleagues118 discovered a rhamnose binding protein that inhibited the biofilm of P. aeruginosa. The authors identified a hemolymph plasma lectin cloned from a horseshoe crab that exhibited selective binding to rhamnose-containing moieties on P. aeruginosa’s biofilm. Subsequent experiments revealed the lectin’s ability to impede biofilm formation and disperse mature biofilms, leading to the reduction of P. aeruginosa cytotoxicity to human lung cells and zebrafish embryos in vivo. These results suggest that horseshoe lectins are potent anti-adhesives of host-pathogen interactions. In another study, Gottesman and colleagues119 targeted the BabA receptor of H. pylori using rhamnogalacturonans from the plant Abelmoschus esculentus. The authors observed that highly branched and esterified rhamnogalacturonans effectively blocked the BabA-Lewis B interaction responsible for H. pylori adhesion to human gastric epithelial cells. Moreover, they discovered pectins from apple fruits could similarly hinder H. pylori binding to human galectin-3. Since designing anti-adhesive agents is inherently challenging, primarily due to the ability of any cell to rearrange its glycan coat via phase variation,120 natural products bearing anti-adhesive properties significantly ease the development of anti-adhesive drugs against pathogenic bacteria.
4.3. Consequence of perturbation: Potentiate antibiotics
Bacterial glycans are both intriguing targets of antibiotics and the cell’s first line of defense against antibiotics. Altering the bacterial cell wall thus has the potential to hypersensitize bacteria to antibiotics by removing their first line of defense. Indeed, genetic knockouts of glycosylation genes in antibiotic-resistant bacteria have potentiated existing antibiotics in Streptomyces coelicolor,121 Mycobacterium abscessus,122 and Enterococcus faecium.123 Similarly, small molecule-induced remodelling of the bacterial cell wall has rendered bacteria more vulnerable to antibiotics,124, 125 as described in more detail in this section.
Small molecules that impede biofilm formation can sensitize bacteria to antibiotics (Fig. 7A). The bacterial biofilm is an extracellular matrix composed of polysaccharides, proteins, and nucleic acids that serves as a physical barrier for bacterial cells against environmental insults.126 Biofilm formation is a key mechanism of antibiotic resistance in pathogenic bacteria, suggesting that biofilm inhibition can recover bacterial susceptibility to antibiotics.127 Recent work by Yoshii and colleagues128 demonstrated the recovery of methicillin-resistant S. aureus (MRSA) sensitivity to β-lactam antibiotics using norgestimate as a biofilm inhibitor. The authors revealed that norgestimate, an acetylated progestin, disrupted staphylococcal biofilm formation by supressing production of polysaccharide intercellular adhesins and extracellular proteins. Moreover, norgestimate induced abnormal features in cell wall morphology, including increased thickness and rippled septa. These characteristics were correlated with MRSA-heightened sensitivity to β-lactams and point to the promise of biofilm disruption strategies based on glycan biosynthesis. In another study, Su and coworkers129 explored the ability of synthetic antibiofilm agents to resensitize MRSA and Acinetobacter baumannii to the antibiotic oxacillin. The authors synthesized a library of 4,5-disubstituted-2-aminoimidazole-triazole conjugates and confirmed their potency in dispersing MRSA and A. baumannii biofilms at low-micromolar concentrations. Lead compounds were reported to increase bacteria sensitivity to oxacillin by 2–4 fold.
Figure 7. Approaches to potentiate antibiotics against resistant bacteria.
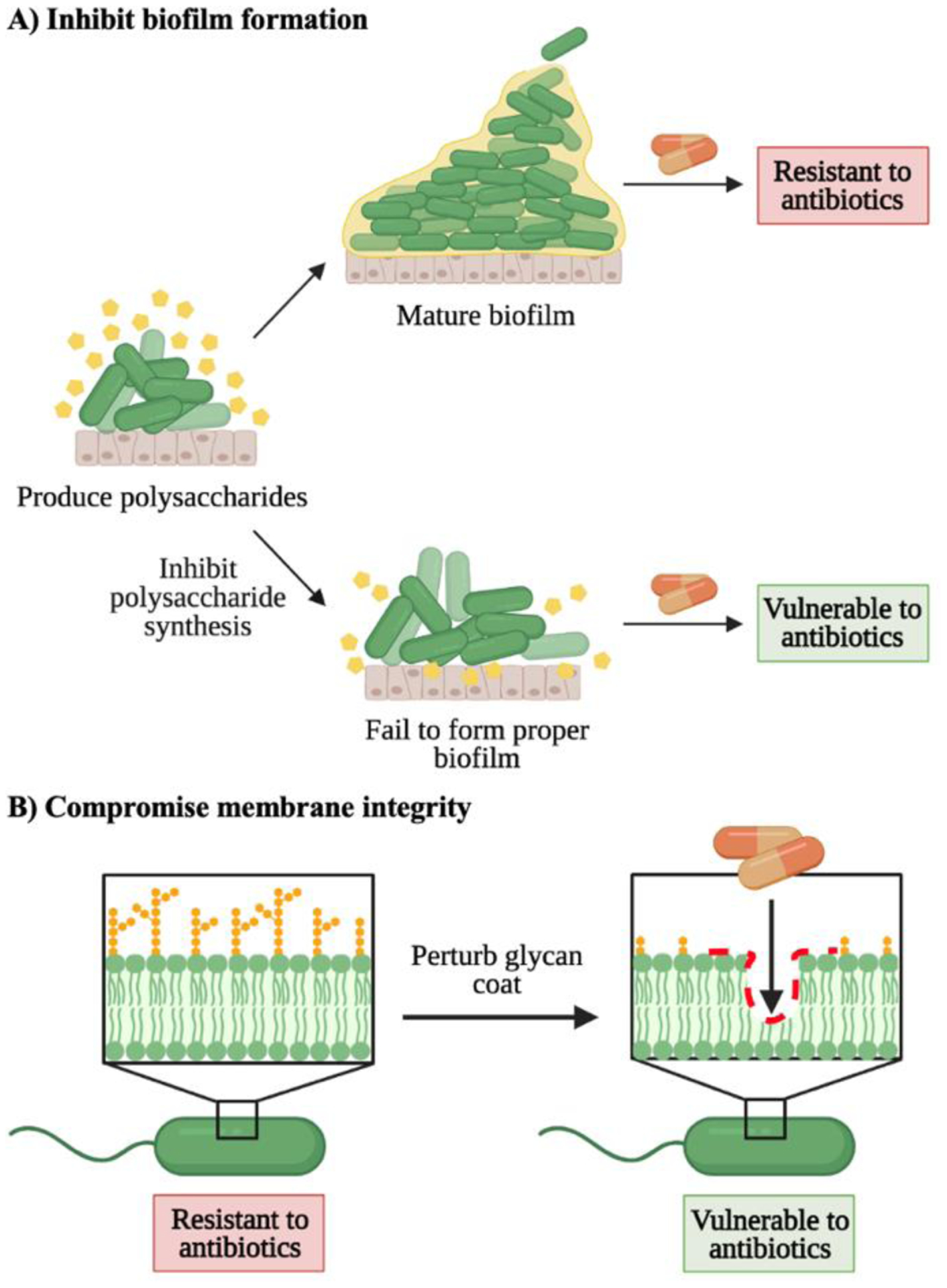
A) Inhibiting polysaccharide production during biofilm formation sensitizes bacteria to antibiotics by removing their protective barrier. B) Membrane-altering agents weaken the bacterial glycan coat and render bacteria vulnerable to antibiotics.
Chemical agents that compromise the bacterial membrane can also potentiate antibiotics (Fig. 7B). Si et al.130 developed a glycosylated cationic block β-peptide that reversed antibiotic resistance in Gram-negative bacteria. The peptide inhibited the integrity of the bacterial outer membrane and dissipated the transmembrane electrochemical potential. These effects led to the resensitization of carbapenem- and colistin-resistant Gram-negative bacteria to antibiotics in vitro and in vivo. Likewise, Lyu and colleagues131 recently reported using amphiphilic tobramycin-lysine conjugates to potentiate antibiotics in multidrug-resistant Gram-negative bacteria. The authors revealed that the novel tobramycin-lysine conjugates penetrated the outer membrane and synergized rifampicin and minocycline against multidrug- and extensively drug-resistant P. aeruginosa isolates. Thus, perturbing the outer-membrane glycan coating can render bacteria more vulnerable to antibiotics.
4.4. Consequence of perturbation: Engage immune system
Bacterial pathogens are notorious for their ability to evade attack by the host’s immune system. To escape immune detection, bacteria employ tactics including modifying cell surfaces, expressing proteins that block host immune factors, and mimicking host antigens.132 Glycoimmunotherapy approaches engage the host immune system to treat cancer, yet these strategies are less developed for treatment of bacterial disease. The tremendous success of carbohydrate-based vaccines to prevent bacterial infection suggests that immune-based strategies to treat bacterial disease hold great promise. Here we highlight the recent development of glycoconjugates that flag bacterial cells for immune-mediated clearance (Fig. 8A).
Figure 8. Perturbing the bacterial glycocalyx to recruit immune responses.
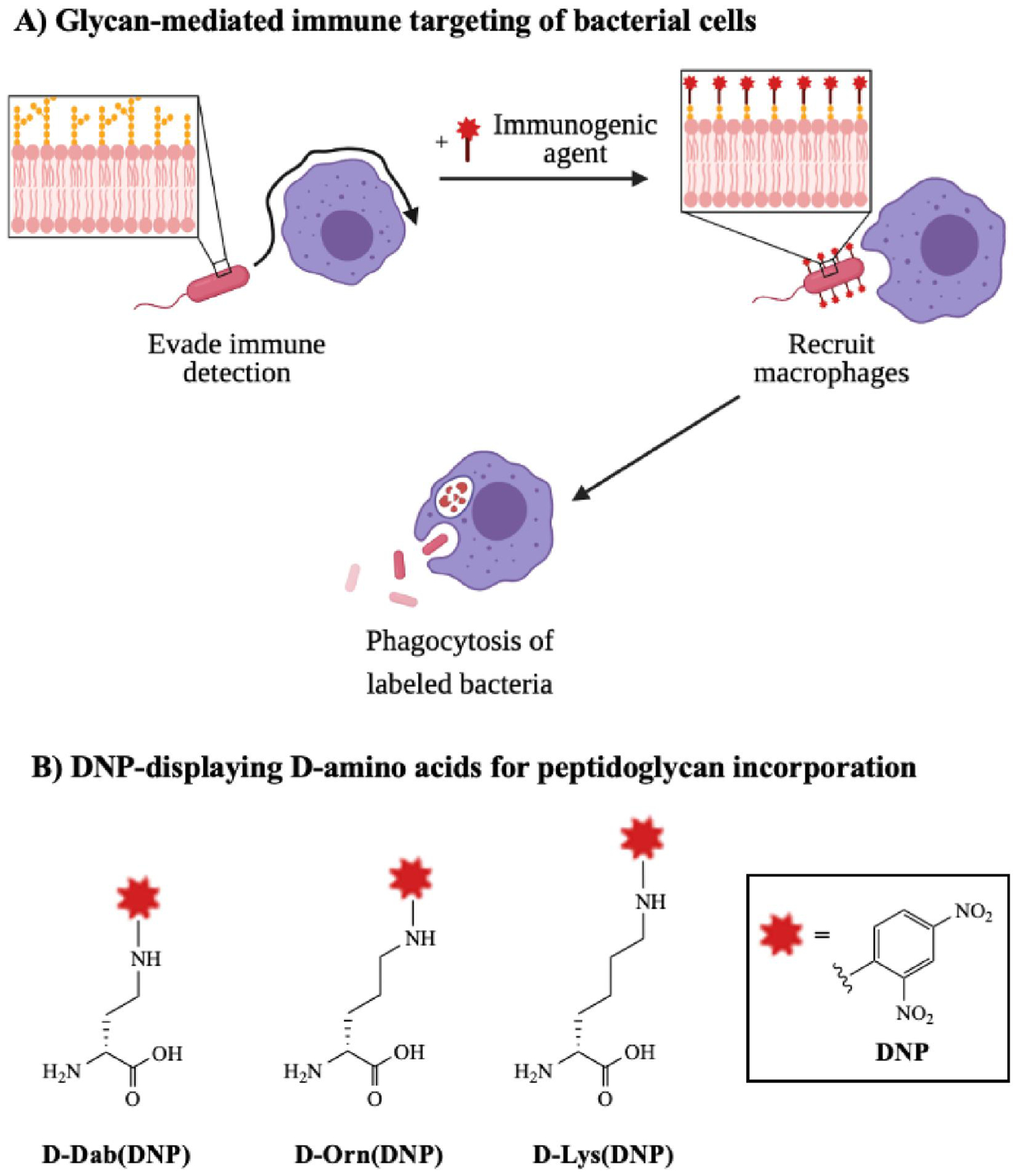
A) Bacteria typically evade host immune detection by modifying surface glycans to block immune factors or mimic host antigens. Unnatural substrates bearing immune stimulants can exploit permissive metabolic pathways to become expressed on bacterial surfaces, ultimately recruiting macrophages to seek and destroy pathogens. B) Structures of representative immunogenic agents accessed from unnatural d-amino acids bearing 2.4-dinitrophenyl (DNP), which can be incorporated into the peptidoglycan and recruit anti-DNP antibodies.133
Pires and coworkers pioneered a glycoimmunotherapy approach against bacterial pathogens. In one study, Fura et al.133 established a D-amino acid-based antibody recruitment therapy to promote clearance of planktonic Gram-positive bacteria by host immune cells. This strategy exploited substrate promiscuity in biosynthetic enzymes to incorporate unnatural D-amino acids bearing immunogenic 2,4-dinitrophenyl (DNP) into cell surface peptidoglycan (Fig. 8B). As a result, bacteria synthesized peptidoglycan displaying DNP, which recruited anti-DNP antibodies and elicited a host immune response. In a subsequent study, Feigman et al.134 designed a class of polymyxin B molecules conjugated to antigenic epitopes. These conjugates facilitated binding to surface glycans on Gram-negative bacteria and subsequent detection by human serum antibodies. Conjugates elicited specific killing of bacteria by immune cells in the presence of human serum, demonstrating the potency of immunotherapeutic agents in aiding the elimination of foreign pathogens.
4.5. Summary
Glycan biosynthesis inhibitors derived from antibacterial scaffolds, nucleoside-based analogs, and rare bacterial monosaccharides represent three complementary approaches to inactivate or divert bacterial glycosylation enzymes, in some cases in a bacteria-selective manner. Perturbation of bacterial glycan biosynthesis precipitates important functional consequences. Altering the bacterial glycan coat is one approach to block host-pathogen glycan-binding interactions and stop bacterial entry into host cells. Further, perturbed glycan biosynthesis provides a mechanism to hypersensitive or resensitize bacteria to antibiotics. Finally, alterations in glycan architecture offer a means to engage the immune system with unmasked or novel immune-recruiting epitopes. Newly invented probes allow the directed manipulation of the bacterial glycocalyx and yield important insights into possible therapeutic targets in bacterial pathogens.
5. Imaging bacterial glycans
The emergence of novel imaging probes has facilitated the tracking of bacterial glycans in physiologically relevant contexts. Here we describe the development and application of these tools, highlighting the bioorthogonal imaging of unnatural sugars, the development of direct fluorescent metabolic probes and fluorescent antibiotics, and the application of tagged isoprenoids to track bacterial glycan biosynthesis in vitro and in vivo.
5.1. Bioorthogonal imaging to track unnatural sugars
As discussed in section 3.1., chemically modified sugars are effective metabolic reporters of bacterial glycans and their biosynthesis. By attaching a bioorthogonal handle to an unnatural sugar, the handle can selectively recruit an agent of choice, such as a fluorescent tag, to illuminate labeled glycans. This approach enables rapid access to functional information without requisite structural details.
In studies of specific bacterial species, sugar analogs serve as excellent bioorthogonal imaging agents (Fig. 9A). For example, DeMeester et al.135 developed a library of functionalized uridine diphosphate N-acetyl muramic acid (UDP-NAM) analogs. Click chemistry with azido UDP-NAM derivatives allowed the fluorescent imaging of the peptidoglycan in both Gram-negative and Gram-positive bacteria, specifically E. coli and Lactobacillus acidophilus (Fig. 9B). Using a similar approach, Taylor et al.136 revealed key proteins defining cell wall growth in H. pylori via imaging with a bioorthogonal NAM analog. After verifying that alkyne-containing NAM was incorporated into the cell wall, the authors conjugated the sugar with an azido fluorescent dye via click chemistry. Bacterial cells were imaged in 3D and analyzed to construct cell wall growth patterns. A protein crucial to forming H. pylori’s helical shape was identified, uncovering a potentially compelling drug target.
Figure 9. Imaging strategies using bioorthogonal probes.
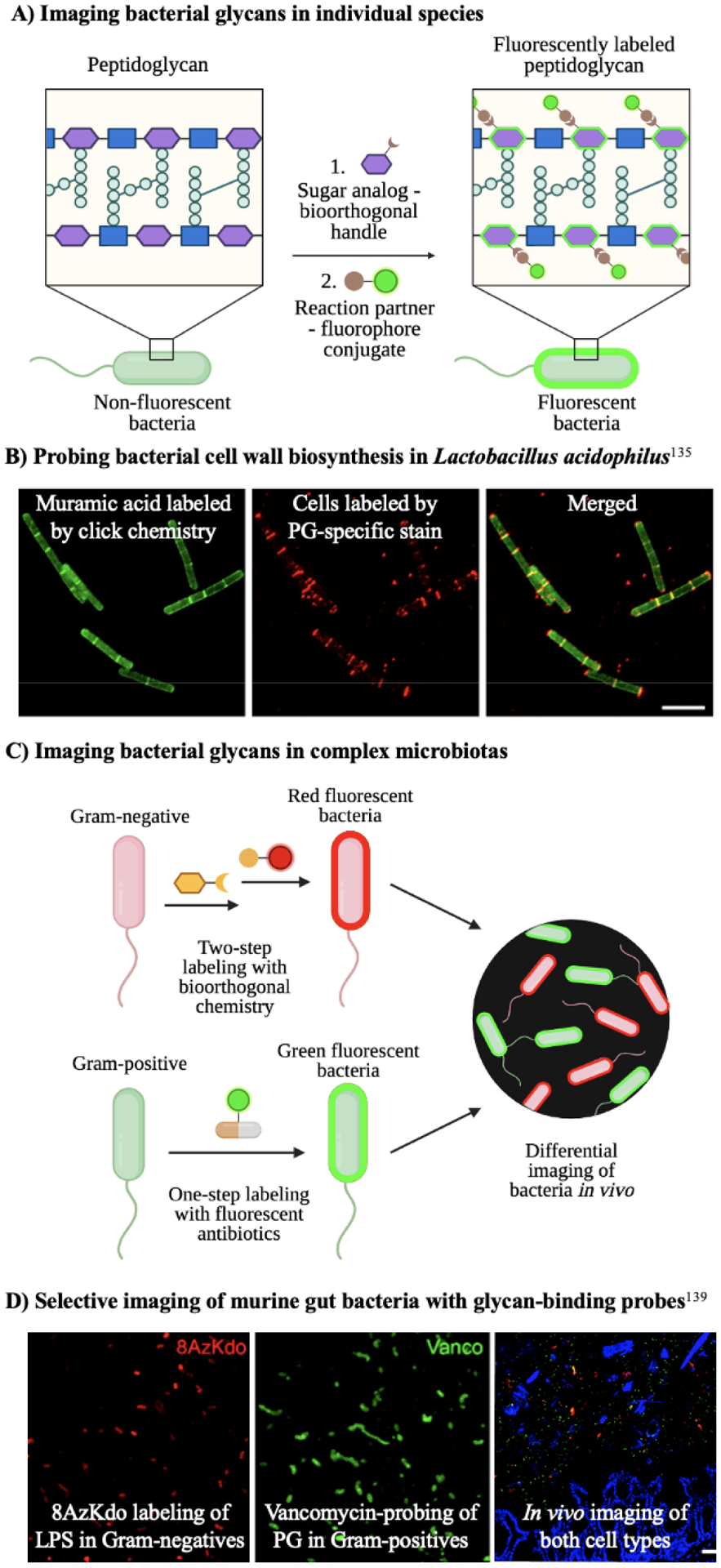
A) Sugar analogs bearing bioorthogonal groups illuminate bacterial glycans on target species. The sugars GlcNAc and MurNAc are depicted by blue squares and purple hexagons, respectively. B) Clickable muramic acid derivatives fluorescently labeled the peptidoglycan in Lactobacillus acidophilus. Reproduced with permission from DeMeester et al.135 C) Labeling bacteria via bioorthogonal probes alongside fluorescent antibiotics allows the differential imaging of Gram-negative and Gram-positive bacteria in vivo. D) Selective labeling of Gram-negative bacteria by 8-azido-8-deoxy-Kdo (8AzKdo, yellow hexagon) and Gram-positive bacteria by fluorescent vancomycin in the gut microbiome. Reproduced with permission from Wang et al.139
Using a different unnatural sugar in a separate model system, Swarts and colleagues137 employed azide-containing trehalose (TreAz) analogs to label glycans in mycobacterial trehalose metabolic pathways. TreAz was incorporated into cell surface glycolipids and subsequently underwent bioorthogonal ligation with alkyne-modified probes to illuminate the mycobacterial glycoform. The authors also demonstrated the diversity in the labeling routes of TreAz analogs, suggesting this approach could uncover multiple pathways important to mycobacterial trehalome biosynthesis. Similarly, Foley et al.138 used trehalose monomycolates bearing terminal alkynes to tag arabinogalactan mycolates and trehalose dimycolates, major constituents of the mycomembrane. Alkyne-labeled mycolates were visualized in situ via click chemistry with azido fluorophores. This technique allowed the imaging of intact mycomembrane components and facilitated the investigation of mycobacterial physiology and pathogenicity.
In addition to viewing bacterial glycans in monocultures of bacteria, sugar analogs enable live imaging of bacterial glycopatterns in complex microflora. Wang and coworkers139 presented an approach to selectively classify Gram-negative and Gram-positive bacteria in the mouse gut based on metabolic labeling and subsequent bioorthogonal chemistry (Fig. 9C). The authors utilized the azide-modified monosaccharide 8-azido-8-deoxy-Kdo (8AzKdo) to label and visualize lipopolysaccharides in Gram-negative species. On the other hand, Gram-positive bacteria were tagged by fluorophore-conjugated vancomycin. This strategy allowed multicomponent imaging of both types of bacteria in complex gut microbiotas (Fig. 9D). Additionally, Geva-Zatorsky and colleagues48 reported the in vivo imaging of commensal bacteria in the intestine using metabolic oligosaccharide engineering. Azido analogs of the naturally abundant sugar N-acetyl-galactosamine were incorporated into capsular polysaccharide A of B. fragilis and nine other anaerobic bacteria. Click chemistry enabled visualization of gut host-microbe interactions and elucidation of the fate of labeled glycans in a murine infection model. Since not all gut anaerobes are amenable to cultivation or genetic manipulation in vitro, in vivo labeling is critical to study the microbiome.
In a similar study, Hudak and coworkers140 demonstrated the live visualization of distinctive glycan components of endogenous anaerobic bacteria in the intestine. The authors confirmed the unnatural sugars GalNAz, KdoAz, and D-amino acids were metabolically incorporated into capsular polysaccharide (CPS), lipopolysaccharide (LPS), and peptidoglycan (PG) in intestinal microbes, respectively. Coupled to click chemistry with fluorescent reporters, this approach enabled the simultaneous tracking of CPS, LPS, and PG on the surface of live bacteria as they moved into specific intestinal immune cells in the living host. The localization of these bacterial glycans shed light on the progression of intestinal diseases, particularly by revealing how, when and which cell types encounter them, and whether differences in their accumulation correlate with disease. Thus, these illustrative examples of multicomponent imaging in mixed bacterial populations will open the door to understanding interspecies and glycan dynamics in complex systems. This information ultimately facilitates the development of therapeutics targeting key glycan-mediated host-pathogen interactions.
5.2. Fluorescent metabolic probes
One recent advance in imaging the bacterial glycocalyx involves the use of small molecule fluorophores that are metabolically incorporated into target glycans (Fig. 10). By exploiting promiscuous bacterial metabolic pathways, modified glycoconjugate building blocks, such as sugars and D-amino acids, are accepted. Fluorophore-containing probes enable the real-time visualization of native glycans with minimal disruptions. This efficient method has been widely implemented to reveal important functions and mechanistic information about bacterial glycosylation.
Figure 10. Mechanisms of action of metabolic imaging probes.
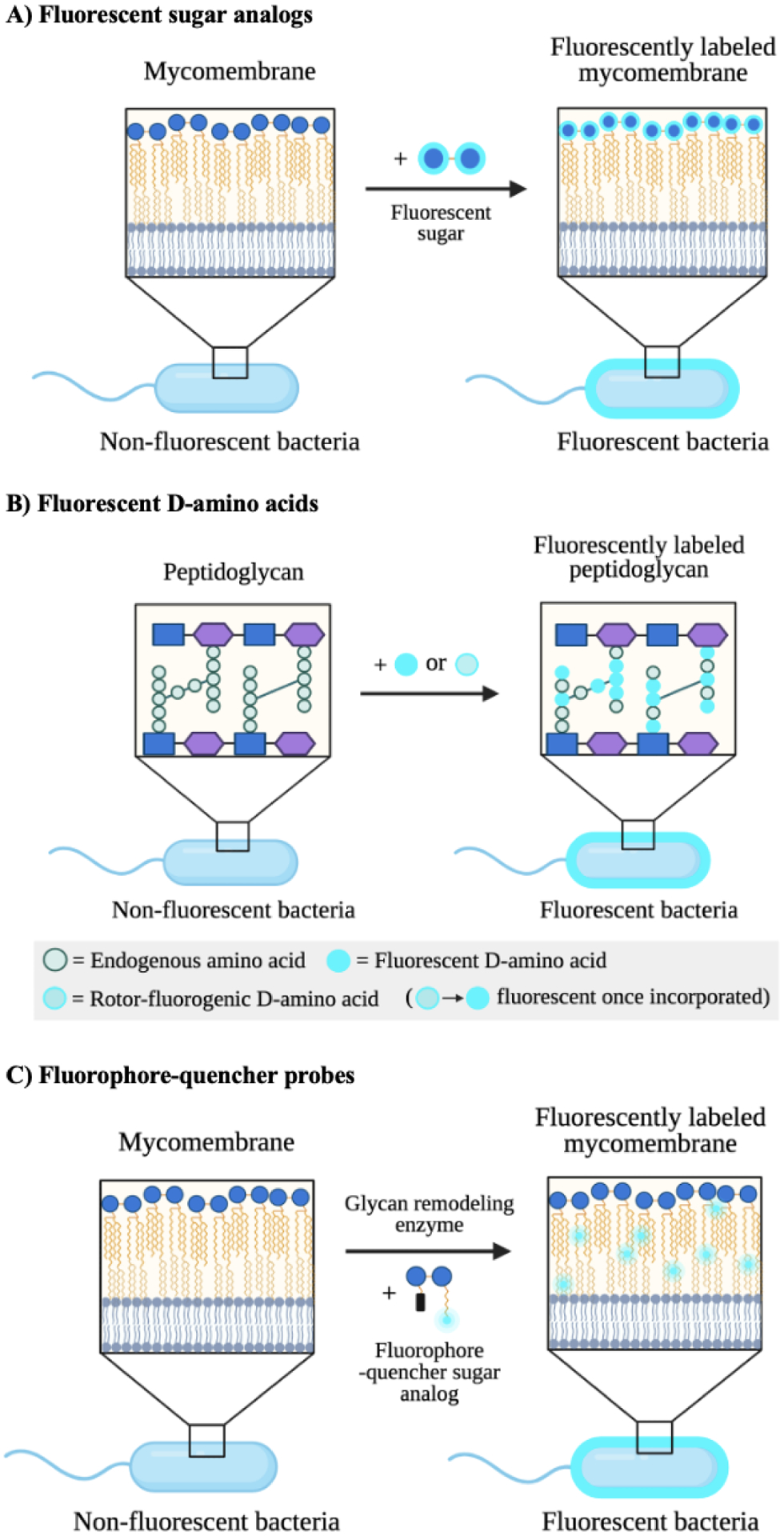
A) Fluorescent sugar analogs are metabolically incorporated into endogenous glycans to enable their detection and tracking via fluorescence. Trehalose is depicted by two linked blue circles. B) Fluorescent and rotor-fluorogenic D-amino acids are metabolically incorporated into peptidoglycan and facilitate the real-time imaging of peptidoglycan biosynthesis. Rotor-fluorogenic D-amino acids turn fluorescent once incorporated into cellular peptidoglycan. C) Fluorophore-quencher probes accessed from sugar analogs become fluorescent upon specific enzyme-catalyzed reactions and allow the direct visualization of enzyme activity during cell wall biosynthesis.
Fluorescent sugar analogs represent a useful class of metabolic imaging agents. Zlitni et al.141 developed a fluorescent derivative of the dietary sugar maltotriose to monitor bacterial infections. The probe was taken up by Gram-negative and Gram-positive bacteria in vivo in a wound murine model, allowing the detection of infection, analysis of infection burden, and illumination of antibiotic effectiveness. In addition, the maltotriose scaffold exhibited stable pharmacokinetics, demonstrating its suitability for imaging in live models. Thus, fluorescent maltotriose analogs can facilitate the real-time visualization of bacterial sugar uptake. This example illustrates the potential to track uptake of fluorescent sugars that are required for the construction of higher order bacterial glycans.
Recent work by Rodriguez-Rivera and colleagues142 reported the use of fluorescein-trehalose analogs to study mycomembrane dynamics in live cells (Fig. 10A). Trehalose-fluorophore molecules were metabolically incorporated into trehalose mycolates of mycobacterial species, enabling the live observation of labeled glycolipid mobility. Through this technique, the authors revealed the diversity in mycomembrane fluidity across species and the effect of anti-tuberculosis drugs on mycomembrane dynamics. In a related study, Swarts and coworkers143 developed a two-step chemoenzymatic synthesis of trehalosamine, an aminoglycoside antibiotic against mycobacteria and an essential intermediate in syntheses of trehalose-based imaging probes. The authors observed that trehalose-fluorophore conjugates elaborated from trehalosamine labeled distinctive trehalose metabolism pathways. Thus, future investigations can employ fluorescent trehalosamine derivatives to explore the antimicrobial activities of trehalosamine-based antibiotics in a pathway-specific manner. In more recent work, the Swarts group144 demonstrated that the 18F-modified trehalose analogue 2-deoxy-2-[18F]fluoro-D-trehalose was metabolized by M. smegmatis but not by various mammalian cell lines. This observation suggests that 18F-modified trehalose analogues can be coupled with positron emission tomography (PET) for in vivo selective imaging of infections caused by trehalose-processing pathogens such as M. tuberculosis. Since PET imaging using 18F-labeled sugar probes is a clinically established method,145, 146 future studies can explore the application of 18F analogs of metabolic reporters, such as those featured in section 3.1., in illuminating the bacterial glycocalyx.
Quencher-fluorophore probes based on sugars have been developed as “smart” imaging agents of bacterial cell wall biosynthesis (Fig. 10C). A study by Hodges and coworkers147 demonstrated the visualization of mycolic acid membrane synthesis using a quencher-trehalose-fluorophore as a mycolyltransferase substrate analog. The mock substrate turned fluorescent upon mycolyltransferase-mediated hydrolysis and allowed the real-time tracking of mycolyltransferase activity during cell wall biosynthesis. In a similar study, Holmes and colleagues148 designed a fluorescence-quenched analog of trehalose dimycolate. The compound became fluorescent as a result of hydrolysis by trehalose dimycolate hydrolase and mycomembrane trehalose degrading enzymes. These studies indicate the potency of quencher fluorophore probes in tracking the activity of cell wall synthesis enzymes and elucidating the glycoassembly machinery.
Fluorescent D-amino acids are versatile imaging agents of bacterial peptidoglycan biosynthesis (Fig. 10B). Recently, Hsu et al.149 introduced a family of fluorescent D-amino acids used for the non-invasive imaging of bacterial peptidoglycan. These probes were accepted by endogenous bacterial transpeptidases and covalently appended onto PG, overcoming toxicity and permeability issues. Their high bioorthogonality and PG specificity enabled the real-time visualization of PG synthesis in live cells. Similarly, Boersma and coworkers150 reported the fluorescent D-amino acid-aided imaging of PG dynamics in wildtype versus PG hydrolase mutants of S. pneumoniae. The authors were able to detect regions of new PG turnover during live cell division and revealed the bacteria’s mechanism of PG remodelling during pathogenesis. In an important advance, VanNieuwenhze and colleagues68 presented rotor-fluorogenic D-amino acids for the continuous imaging of transpeptidation reactions in vitro and in vivo. Rotor-fluorogenic D-amino acids are “smart” probes that turn fluorescent only when incorporated into bacterial PG, where bond rotation is restricted, allowing the temporal screening of PG biosynthesis in live cells. Ultimately, these studies highlight fluorescent D-amino acids as powerful tools to uncover important functions and antibiotic targets of the bacterial peptidoglycan in biologically relevant contexts.
5.3. Fluorescent glycan-binding antibiotics
Antibiotics are key to eradicating bacterial infections and, as demonstrated recently, yield incisive information about bacterial pathogenesis. To this end, fluorescent antibiotics have been developed to track drug-bacteria interactions within organelles, whole cells, and live animals. Common cell wall-targeting antibiotics provide a rich source of fluorescent probes for imaging bacterial glycans.151–155 In this section, we feature a number of recent advancements in applying fluorescent antibiotics to visualize bacterial glycosylation. For a more extensive description of fluorescent antibiotics, we refer readers to a comprehensive review by Stone and colleagues.156
Fluorescent antibiotics illuminate important functions and pathways within bacterial glycan biosynthesis. Recent work by McInerney and colleagues157 utilized a fluorescent polymyxin analog to label lipopolysaccharides (LPS) in Gram-negative bacteria. The probe enabled the high-throughput quantitation of polymyxin-LPS affinities and revealed novel drug targets in the outer-membrane of Gram-negative pathogens. Likewise, Akram et al.158 accomplished real-time imaging of Gram-negative bacteria in distal human lungs using a fluorescent antimicrobial peptide that targeted lipid A, an essential component of LPS. The authors validated the probe’s ability to generate safe and rapid pulmonary molecular detection of Gram-negative pathogens in humans. In a similar study, Mao and coworkers159 developed a fluorescent β-lactam analog to selectively visualize β-lactamase activity in extended-spectrum antibiotic-resistant bacteria. This probe bears a ratiometric fluorescent sensor, which allowed the measurement of fluorescent signals independent of their intensities and environmental conditions. As a result, the probe sensitively detected β-lactamase activity in bacterial pathogens and rapidly distinguished extended-spectrum antibiotic-resistant bacteria from other pathogens. These studies exemplify the utility of fluorescent antibiotics in yielding key information about bacterial pathogenesis and uncovering novel therapeutic targets.
5.4. Polyisoprenoids to track glycan biosynthesis
Polysaccharides are major building blocks of the bacterial cell envelope that facilitate bacterial infection and colonization of host cells. Bacterial polysaccharides are often assembled on a lipid polyisoprenoid carrier. Briefly, glycosyltransferases act in succession to transfer monosaccharides onto a lipid carrier to yield a polysaccharide (Fig. 11A). Fluorescent polyisoprenoids are versatile scaffolds for characterizing polysaccharide biosynthesis, tracking their assembly in real time, and understanding of their role as it relates to bacterial pathogenesis (Fig. 11).
Figure 11. Imaging bacterial glycans via fluorescent polyisoprenoids.
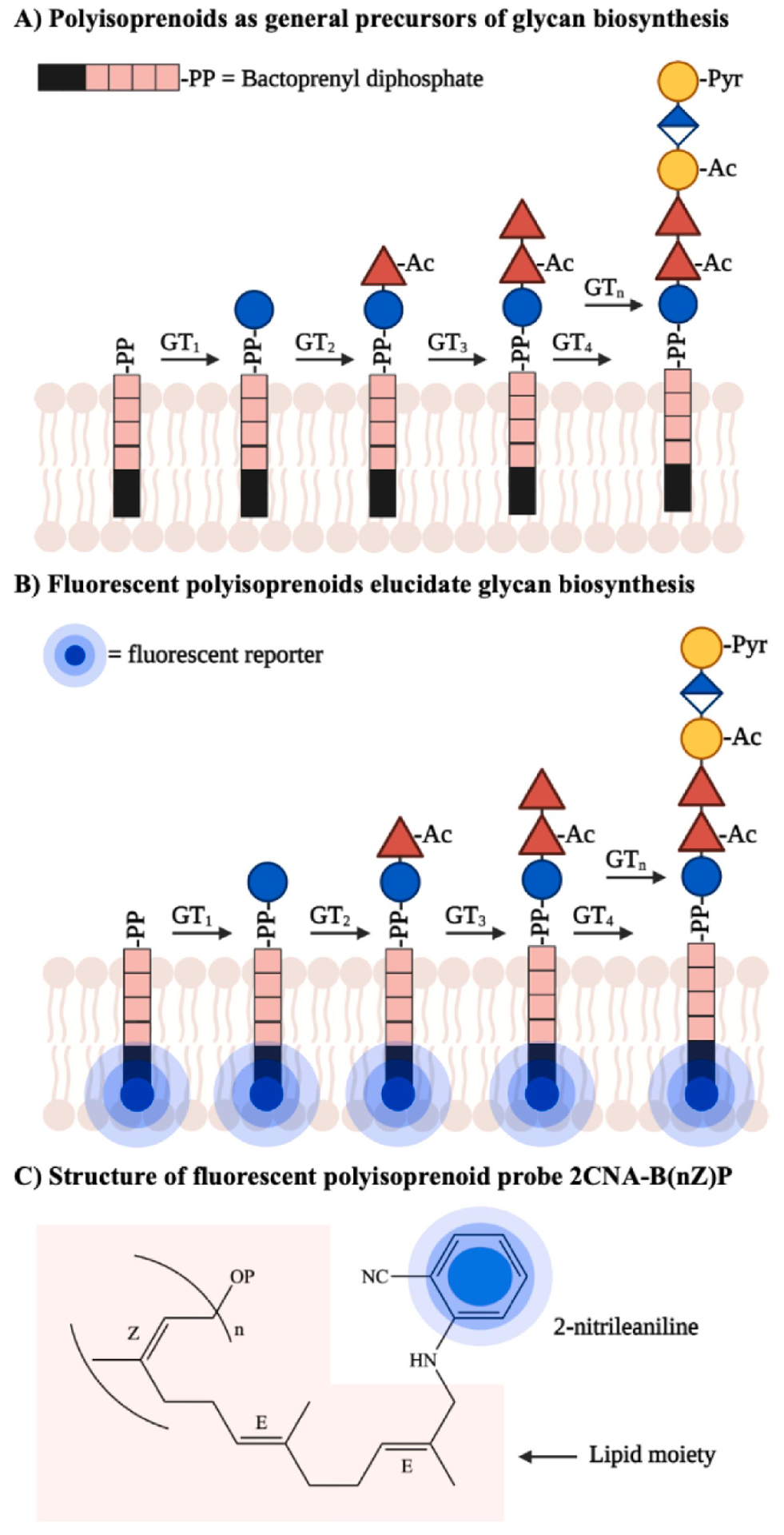
A) Bacterial polyisoprenoids act as lipid carriers upon which glycosyltransferases (GTs) construct higher order glycans, including polysaccharides. They are critical for biosynthesis of a number of glycans. B) A fluorescent reporter can be metabolically incorporated into bacterial polyisoprenoids to monitor glycan assembly steps and characterize glycosyltransferase activities. Glycans at different stages during biosynthesis can be imaged. C) Example of an imaging probe which consists of the fluorescent reporter 2-nitrileaniline appended to bactoprenyl phosphate. Figure adapted from Scott et al.161 Abbreviations: Ac = acetyl; Pyr = pyruvyl; 2CNA-B(nZ)P = 2-nitrileanilinobactoprenyl phosphate.
Recent advancements in the development of fluorescent polyisoprenoids have yielded important insights into bacterial glycosylation machinery. Troutman and colleagues160 took advantage of undecaprepyl pyrophosphate synthase’s substrate promiscuity to incorporate a fluorescent isoprenoid unit into bacterial polyisoprenoids in vitro. Tagged isoprenoids allowed substrate and product tracking and quantification by reverse-phase high performance liquid chromatography (HPLC). Using this general strategy, they traced the enzymatic biosynthesis of B. fragilis capsular polysaccharide A tetrasaccharide repeating unit onto a fluorescent polyisoprenoid in a sequential, one-pot reaction. The authors uncovered the first evidence of a pyruvate acetal modification of galactose in the polysaccharide biosynthesis pathway. This process revealed the pyruvyltransferase WcfO to be a novel drug target due to its crucial role in producing functional repeating tetrasaccharide units of the capsular polysaccharide.
Similarly, Scott et al.161 employed a fluorescent polyisoprenoid to identify six key proteins involved in the biosynthesis of colanic acid, a polysaccharide secreted by E. coli to protect it from harsh conditions (Fig. 11B). As a starting substrate, the polyisoprenoid probe (Fig. 11C) enabled the coupled fluorescence HPLC-mass spectrometry analyses of subsequent monosaccharide additions onto colanic acid repeating units by each of the six enzymes in this pathway. This approach illuminated the enzymes’ activities in colanic acid production in vitro. This knowledge is significant for understanding bacterial glycan biosynthesis, assigning functional roles to glycosyltransferases, and ultimately developing effective therapeutic interventions for targeting bacterial pathogens that utilize polyisoprenoids. Future in vivo applications of these probes will validate the ability of this strategy to shed light on bacterial glycoassembly pathways in the context of live cells.
5.5. Summary
We have highlighted a suite of emergent chemical tools to image bacterial glycans and examples of their most recent applications. One novel approach entails the application of bioorthogonal chemistry for two-step labeling and tracking of target glycans. In addition, single-step labeling with fluorescent-metabolic probes derived from widely available glycoconjugate building blocks, namely sugars and amino acids, are powerful imaging agents that allow the real-time visualization of bacterial glycosylation activities in vitro and in vivo. Similarly, fluorescent glycan-related antibiotics are efficient reporters of the bacterial surface glycocalyx, particularly its interactions with drugs and host cells. Finally, fluorescent polyisoprenoids represent a versatile class of molecular imaging tools to track and characterize polysaccharide biosynthesis. Ultimately, these probes yield valuable insights into the bacterial glycosylation apparatus and uncover new drug targets key to fighting antibiotic resistance. For further reviews on small molecule imaging reporters of the bacterial glycoform, we recommend two excellent reads by Kocaoglu and Carlson162 and Parker et al.163
6. Current gaps and future directions
Recent advances in bacterial chemical glycobiology have shed light on the roles and functions of bacterial glycans and indicated potential therapeutic targets. Nevertheless, before identifying leads to ultimately develop new narrow-spectrum antibiotics, paramount challenges remain unaddressed. In this section, we highlight the present knowledge gaps in studies of bacterial glycans and the need for novel tools to answer fundamental questions about these biomolecules.
6.1. Characterizing the structures of bacterial glycans
While the current toolkit has yielded high-level information about the importance of bacterial glycans to function and pathogenesis, molecular details of these glycans in many instances are still unknown. One common approach to elucidating glycan structure employs mass spectrometry. However, this strategy has several major limitations: (1) low instrumental sensitivity to heterogeneous glycoforms, (2) lack of database search algorithms to identify the components of bacterial glycans, (3) poor differentiation of monosaccharides that are structural isomers with the exact same mass, and (4) challenges with purification of these complex biomolecules.164 The first limitation requires the enrichment of specific glycan subtypes, which has been achieved by several chemical tagging methods.165–168 On the other hand, the second limitation requires the truncation of glycans to produce sample pools with predictable masses, which complicates the analysis of whole glycan structures. The third limitation is unavoidable. Finally, efforts have been made to facilitate in line chromatographic separation prior to mass spectrometry analysis.169
To overcome some of these constraints, Woo and colleaeges164 developed a mass-independent methodology termed Isotope-Targeted Glycoproteomics (IsoTaG). In brief, this approach consists of metabolic labeling of cellular glycans, enrichment of labeled species using an isotopic recoding affinity probe, and analysis of enriched glycans via tandem mass spectrometry. Since the initial development, IsoTaG has been used to identify over 1375 N- and 2159 O-linked glycoproteins in human cells.170 This novel method is effective for characterizing whole glycan structures, yet it has only been utilized to study glycoproteomes in mammalian systems.171, 172 Given the key components of this strategy are not species-specific, IsoTaG is potentially generalizable to studies of glycans in bacterial systems.
6.2. Metabolic labeling of glycans in complex microbial communities
We have featured several applications of metabolic probes for the study of bacterial glycan biosynthesis (section 3.1). However, these probes have only been employed to investigate a small number of bacterial species rather than complex microbial communities. A few recent studies offer notable exceptions. In one study, Wang and coworkers139 explored the ability of murine gut microbes to incorporate 8-azido-8-deoxy-Kdo into lipopolysaccharides. The authors elaborated azide-labeled glycans in these complex communities with a fluorescent probe, then enriched and identified fluorescently-labeled Gram-negative bacteria via 16S rRNA sequencing. This study indicates the feasibility of labeling glycans on select bacteria in complex microbial communities. As another proof-of-concept study, more recent work by Wolan et al.173 reported the metabolic labeling of sialic acid-expressing microbes in a human fecal microbiome. By culturing the fecal microbiome with an azide-containing sialic acid analog, the authors were able to label sialic acid-presenting microbes and identify a new strain of E. coli that incorporated sialic acids onto its surface to evade host immune detection. Moreover, they found that levels of sialic acids present on gut bacteria was regulated by commensal microbes present within the samples. These findings illustrate that the bacterial glycocalyx is tailored by the microbial community. Thus, studying bacterial glycans in context is of paramount importance.
Taken together, these novel findings demonstrate the necessity of examining bacterial glycopatterns in complex microbial communities. In particular, this strategy allows the characterization of bacterial glycan biosynthesis in a high-throughput manner, since a variety of sugar analogs can be introduced and assessed for patterns of uptake. Ultimately, this approach provides an unbiased means to explore the role of glycans in modulating microbiome dynamics as it relates to human health and disease.
6.3. Expanding the panel of metabolic probes
Although bacteria utilize over 100 monosaccharides in glycan biosynthesis, only a small percentage of these sugars have been chemically modified into metabolic probes (Fig. 3A). Thus, extending the panel of bacterial sugar analogs would facilitate the identification of elusive glycan structures and distinctive glycan coats across bacterial species. To this end, the Kulkarni group174 presented an expedient synthetic route to access rare deoxy amino L-sugar building blocks. This work, and many others,175 set the stage for the determination of key building blocks that differentiate glycans from one bacterial species to another. Beyond monosaccharide building blocks, unusual oligosaccharides amenable to chemical modifications are crucial to decoding the bacterial glycocalyx. While expeditious and high-throughput syntheses of oligosaccharides have been demonstrated,176, 177 these strategies are biased toward accessing mammalian and structurally defined molecules. Thus, future studies can extend on precedented work to produce atypical, bacterial oligosaccharides as probes for specific epitope labeling. Ultimately, the expanded set of metabolic probes can pave the way to understanding bacterial glycan biosynthesis and identifying potential therapeutic targets.
7. Conclusion
The growing problem of antibiotic resistance suggests the ineffectiveness of our current antibiotic arsenal and the need for new antibiotic targets. Bacterial glycans are compelling molecular targets that can be harnessed to develop both broad-spectrum and narrow-spectrum antibiotics. Despite having critical roles in bacterial function and pathogenesis, glycans are difficult to study due to the sheer diversity of building blocks present in their complex structures. Fortunately, recent advances in chemical biology have expedited the study of bacterial glycans despite these challenges. By exploiting promiscuous metabolic pathways in bacteria, chemical reporters can be incorporated into cellular glycans to facilitate the study of biosynthetic intermediates and their end products, along with their critical binding interactions, in physiologically relevant contexts. In particular, we have highlighted a suite of chemical tools to probe, perturb, and image bacterial glycans. These probes have been applied to investigate the role of glycans in a wide range of priority pathogens, as well as commensal species, in both in vitro and in vivo studies. Thus, these tools set the stage for understanding the fundamental functions of these biomolecules and potentially revealing novel pathogen-specific drug targets.
Additional steps are still needed to fill the gaps in the current toolkit to study bacterial glycans. One persistent shortcoming of the toolkit is the lack of rapid and accessible methods to characterize the full structures of bacterial glycans. This knowledge is critical for identifying druggable molecular targets. In addition, less attention has been given to understanding the role of glycans in modulating mixed microbial dynamics. Applying chemical tools to probe complex microbial glycopatterns in a physiologically relevant context is thus an important next step. Finally, there is a need for more available sugar analogs of bacterial monosaccharides and unusual oligosaccharides representative of bacterial rare glycan epitopes. Given these atypical molecules are major constituents of bacterial glycans, an extended panel of probes crafted from these scaffolds will enable a wider search for unknown glycans as well as species-specific incorporation of different sugars. Ultimately, these steps will reveal crucial information about bacterial glycosylation as it relates to human health and disease.
8. Acknowledgement
We would like to thank Karen Moulton, Owen Tuck, Adedunmola Adewale, Andrew Mulholland, Chiamaka Okoye, Jacob Kassama, and Missy Demczak for thoughtful comments on this manuscript. We gratefully acknowledge the National Institutes of Health (award R15GM109397 to DHD) for supporting this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Dube DH; Champasa K; Wang B, Chemical tools to discover and target bacterial glycoproteins. Chem Commun 2011, 47 (1), 87–101. [DOI] [PubMed] [Google Scholar]
- 2.Tra VN; Dube DH, Glycans in pathogenic bacteria--potential for targeted covalent therapeutics and imaging agents. Chem Commun 2014, 50 (36), 4659–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edens RE, Polysaccharides: structural diversity and functional versatility; Dumitriu S, Ed.; Marcel Dekker: New York. 2005. [Google Scholar]
- 4.Cigana C; Curcurù L; Leone MR; Ieranò T; Lorè NI; Bianconi I; Silipo A; Cozzolino F; Lanzetta R; Molinaro A; Bernardini ML; Bragonzi A, Pseudomonas aeruginosa exploits lipid A and muropeptides modification as a strategy to lower innate immunity during cystic fibrosis lung infection. PLoS One 2009, 4 (12), e8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray GL; Attridge SR; Morona R, Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J Bacteriol 2006, 188 (7), 2735–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paton JC; Trappetti C, Streptococcus pneumoniae capsular polysaccharide. Microbiol Spectr 2019, 7 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H; Wilksch JJ; Strugnell RA; Gee ML, Role of capsular polysaccharides in biofilm formation: an AFM nanomechanics study. ACS Appl Mater Interfaces 2015, 7 (23), 13007–13. [DOI] [PubMed] [Google Scholar]
- 8.Goon S; Kelly JF; Logan SM; Ewing CP; Guerry P, Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol Microbiol 2003, 50 (2), 659–71. [DOI] [PubMed] [Google Scholar]
- 9.Schirm M; Soo EC; Aubry AJ; Austin J; Thibault P; Logan SM, Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol Microbiol 2003, 48 (6), 1579–92. [DOI] [PubMed] [Google Scholar]
- 10.Balonova L; Hernychova L; Bilkova Z, Bioanalytical tools for the discovery of eukaryotic glycoproteins applied to the analysis of bacterial glycoproteins. Expert Rev Proteomics 2009, 6 (1), 75–85. [DOI] [PubMed] [Google Scholar]
- 11.Walsh I; Zhao S; Campbell M; Taron CH; Rudd PM, Quantitative profiling of glycans and glycopeptides: an informatics’ perspective. Curr Opin Struct Biol 2016, 40, 70–80. [DOI] [PubMed] [Google Scholar]
- 12.Linton D; Allan E; Karlyshev AV; Cronshaw AD; Wren BW, Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol Microbiol 2002, 43 (2), 497–508. [DOI] [PubMed] [Google Scholar]
- 13.Kelly J; Jarrell H; Millar L; Tessier L; Fiori LM; Lau PC; Allan B; Szymanski CM, Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J Bacteriol 2006, 188 (7), 2427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linton D; Dorrell N; Hitchen PG; Amber S; Karlyshev AV; Morris HR; Dell A; Valvano MA; Aebi M; Wren BW, Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol Microbiol 2005, 55 (6), 1695–703. [DOI] [PubMed] [Google Scholar]
- 15.Wacker M; Linton D; Hitchen PG; Nita-Lazar M; Haslam SM; North SJ; Panico M; Morris HR; Dell A; Wren BW; Aebi M, N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 2002, 298 (5599), 1790–3. [DOI] [PubMed] [Google Scholar]
- 16.Young NM; Brisson JR; Kelly J; Watson DC; Tessier L; Lanthier PH; Jarrell HC; Cadotte N; St Michael F; Aberg E; Szymanski CM, Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J Biol Chem 2002, 277 (45), 42530–9. [DOI] [PubMed] [Google Scholar]
- 17.Hartley MD; Morrison MJ; Aas FE; Børud B; Koomey M; Imperiali B, Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N’-diacetylbacillosamine. Biochemistry 2011, 50 (22), 4936–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Power PM; Roddam LF; Rutter K; Fitzpatrick SZ; Srikhanta YN; Jennings MP, Genetic characterization of pilin glycosylation and phase variation in Neisseria meningitidis. Mol Microbiol 2003, 49 (3), 833–47. [DOI] [PubMed] [Google Scholar]
- 19.Power PM; Seib KL; Jennings MP, Pilin glycosylation in Neisseria meningitidis occurs by a similar pathway to wzy-dependent O-antigen biosynthesis in Escherichia coli. Biochem Biophys Res Commun 2006, 347 (4), 904–8. [DOI] [PubMed] [Google Scholar]
- 20.Barreteau H; Kovac A; Boniface A; Sova M; Gobec S; Blanot D, Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev 2008, 32 (2), 168–207. [DOI] [PubMed] [Google Scholar]
- 21.Kido N; Kobayashi H, A single amino acid substitution in a mannosyltransferase, WbdA, converts the Escherichia coli O9 polysaccharide into O9a: generation of a new O-serotype group. J Bacteriol 2000, 182 (9), 2567–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper CA; Mainprize IL; Nickerson NN, Genetic, biochemical, and structural analyses of bacterial surface polysaccharides. Adv Exp Med Biol 2015, 883, 295–315. [DOI] [PubMed] [Google Scholar]
- 23.Silhavy TJ; Kahne D; Walker S, The bacterial cell envelope. Cold Spring Harb Perspect Biol 2010, 2 (5), a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caroff M; Karibian D, Structure of bacterial lipopolysaccharides. Carbohydr Res 2003, 338 (23), 2431–47. [DOI] [PubMed] [Google Scholar]
- 25.Hitchen PG; Dell A, Bacterial glycoproteomics. Microbiology 2006, 152 (Pt 6), 1575–1580. [DOI] [PubMed] [Google Scholar]
- 26.Kay E; Lesk VI; Tamaddoni-Nezhad A; Hitchen PG; Dell A; Sternberg MJ; Muggleton S; Wren BW, Systems analysis of bacterial glycomes. Biochem Soc Trans 2010, 38 (5), 1290–3. [DOI] [PubMed] [Google Scholar]
- 27.Banoub JH; El Aneed A; Cohen AM; Joly N, Structural investigation of bacterial lipopolysaccharides by mass spectrometry and tandem mass spectrometry. Mass Spectrom Rev 2010, 29 (4), 606–50. [DOI] [PubMed] [Google Scholar]
- 28.Battistel MD; Azurmendi HF; Yu B; Freedberg DI, NMR of glycans: shedding new light on old problems. Prog Nucl Magn Reson Spectrosc 2014, 79, 48–68. [DOI] [PubMed] [Google Scholar]
- 29.Liu B; Furevi A; Perepelov AV; Guo X; Cao H; Wang Q; Reeves PR; Knirel YA; Wang L; Widmalm G, Structure and genetics of Escherichia coli O antigens. FEMS Microbiol Rev 2020, 44 (6), 655–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahal LK; Yarema KJ; Bertozzi CR, Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science 1997, 276 (5315), 1125–8. [DOI] [PubMed] [Google Scholar]
- 31.Saxon E; Bertozzi CR, Cell surface engineering by a modified Staudinger reaction. Science 2000, 287 (5460), 2007–10. [DOI] [PubMed] [Google Scholar]
- 32.Keppler OT; Horstkorte R; Pawlita M; Schmidt C; Reutter W, Biochemical engineering of the N-acyl side chain of sialic acid: biological implications. Glycobiology 2001, 11 (2), 11r–18r. [DOI] [PubMed] [Google Scholar]
- 33.Keppler OT; Stehling P; Herrmann M; Kayser H; Grunow D; Reutter W; Pawlita M, Biosynthetic modulation of sialic acid-dependent virus-receptor interactions of two primate polyoma viruses. J Biol Chem 1995, 270 (3), 1308–14. [DOI] [PubMed] [Google Scholar]
- 34.Sminia TJ; Zuilhof H; Wennekes T, Getting a grip on glycans: A current overview of the metabolic oligosaccharide engineering toolbox. Carbohydr Res 2016, 435, 121–141. [DOI] [PubMed] [Google Scholar]
- 35.Bertozzi CR, A decade of bioorthogonal chemistry. Acc Chem Res 2011, 44 (9), 651–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sletten EM; Bertozzi CR, Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl 2009, 48 (38), 6974–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prescher JA; Dube DH; Bertozzi CR, Chemical remodelling of cell surfaces in living animals. Nature 2004, 430 (7002), 873–7. [DOI] [PubMed] [Google Scholar]
- 38.Chang PV; Prescher JA; Sletten EM; Baskin JM; Miller IA; Agard NJ; Lo A; Bertozzi CR, Copper-free click chemistry in living animals. Proc Natl Acad Sci U S A 2010, 107 (5), 1821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laughlin ST; Bertozzi CR, Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat Protoc 2007, 2 (11), 2930–44. [DOI] [PubMed] [Google Scholar]
- 40.Blackman ML; Royzen M; Fox JM, Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J Am Chem Soc 2008, 130 (41), 13518–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu KL; Mahal LK, A lectin microarray approach for the rapid analysis of bacterial glycans. Nat Protoc 2006, 1 (2), 543–9. [DOI] [PubMed] [Google Scholar]
- 42.Hsu KL; Gildersleeve JC; Mahal LK, A simple strategy for the creation of a recombinant lectin microarray. Mol Biosyst 2008, 4 (6), 654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Reilly MK; Paulson JC, Siglecs as targets for therapy in immune-cell-mediated disease. Trends Pharmacol Sci 2009, 30 (5), 240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karmakar S; Stowell SR; Cummings RD; McEver RP, Galectin-1 signaling in leukocytes requires expression of complex-type N-glycans. Glycobiology 2008, 18 (10), 770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadamoto R; Niikura K; Sears PS; Liu H; Wong CH; Suksomcheep A; Tomita F; Monde K; Nishimura S, Cell-wall engineering of living bacteria. J Am Chem Soc 2002, 124 (31), 9018–9. [DOI] [PubMed] [Google Scholar]
- 46.Prescher JA; Bertozzi CR, Chemistry in living systems. Nat Chem Biol 2005, 1 (1), 13–21. [DOI] [PubMed] [Google Scholar]
- 47.Hang HC; Yu C; Kato DL; Bertozzi CR, A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc Natl Acad Sci U S A 2003, 100 (25), 14846–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geva-Zatorsky N; Alvarez D; Hudak JE; Reading NC; Erturk-Hasdemir D; Dasgupta S; von Andrian UH; Kasper DL, In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat Med 2015, 21 (9), 1091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vocadlo DJ; Hang HC; Kim EJ; Hanover JA; Bertozzi CR, A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci U S A 2003, 100 (16), 9116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Besanceney-Webler C; Jiang H; Wang W; Baughn AD; Wu P, Metabolic labeling of fucosylated glycoproteins in Bacteroidales species. Bioorg Med Chem Lett 2011, 21 (17), 4989–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goon S; Schilling B; Tullius MV; Gibson BW; Bertozzi CR, Metabolic incorporation of unnatural sialic acids into Haemophilus ducreyi lipooligosaccharides. Proc Natl Acad Sci U S A 2003, 100 (6), 3089–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark EL; Emmadi M; Krupp KL; Podilapu AR; Helble JD; Kulkarni SS; Dube DH, Development of rare bacterial monosaccharide analogs for metabolic glycan labeling in pathogenic bacteria. ACS Chem Biol 2016, 11 (12), 3365–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams DA; Pradhan K; Paul A; Olin IR; Tuck OT; Moulton KD; Kulkarni SS; Dube DH, Metabolic inhibitors of bacterial glycan biosynthesis. Chemical Science 2020, 11 (7), 1761–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andolina G; Wei R; Liu H; Zhang Q; Yang X; Cao H; Chen S; Yan A; Li XD; Li X, Metabolic labeling of pseudaminic acid-containing glycans on bacterial surfaces. ACS Chem Biol 2018, 13 (10), 3030–3037. [DOI] [PubMed] [Google Scholar]
- 55.Salah Ud-Din AIM; Roujeinikova A, Flagellin glycosylation with pseudaminic acid in Campylobacter and Helicobacter: prospects for development of novel therapeutics. Cell Mol Life Sci 2018, 75 (7), 1163–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moulton KD; Adewale AP; Carol HA; Mikami SA; Dube DH, Metabolic glycan labeling-based screen to identify bacterial glycosylation genes. ACS Infect Dis 2020, 6 (12), 3247–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fura JM; Kearns D; Pires MM, D-amino acid probes for penicillin binding protein-based bacterial surface labeling. J Biol Chem 2015, 290 (51), 30540–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsu YP; Hall E; Booher G; Murphy B; Radkov AD; Yablonowski J; Mulcahey C; Alvarez L; Cava F; Brun YV; Kuru E; VanNieuwenhze MS, Fluorogenic D-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays. Nat Chem 2019, 11 (4), 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hsu YP; Rittichier J; Kuru E; Yablonowski J; Pasciak E; Tekkam S; Hall E; Murphy B; Lee TK; Garner EC; Huang KC; Brun YV; VanNieuwenhze MS, Full color palette of fluorescent D-amino acids for in situ labeling of bacterial cell walls. Chem Sci 2017, 8 (9), 6313–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuru E; Hughes HV; Brown PJ; Hall E; Tekkam S; Cava F; de Pedro MA; Brun YV; VanNieuwenhze MS, In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl 2012, 51 (50), 12519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radkov AD; Hsu YP; Booher G; VanNieuwenhze MS, Imaging bacterial cell wall biosynthesis. Annu Rev Biochem 2018, 87, 991–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qiao Y; Lebar MD; Schirner K; Schaefer K; Tsukamoto H; Kahne D; Walker S, Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J Am Chem Soc 2014, 136 (42), 14678–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegrist MS; Whiteside S; Jewett JC; Aditham A; Cava F; Bertozzi CR, (D)-Amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chem Biol 2013, 8 (3), 500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baranowski C; Welsh MA; Sham LT; Eskandarian HA; Lim HC; Kieser KJ; Wagner JC; McKinney JD; Fantner GE; Ioerger TR; Walker S; Bernhardt TG; Rubin EJ; Rego EH, Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cameron TA; Anderson-Furgeson J; Zupan JR; Zik JJ; Zambryski PC, Peptidoglycan synthesis machinery in Agrobacterium tumefaciens during unipolar growth and cell division. mBio 2014, 5 (3), e01219–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liechti G; Kuru E; Packiam M; Hsu YP; Tekkam S; Hall E; Rittichier JT; VanNieuwenhze M; Brun YV; Maurelli AT, Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog 2016, 12 (5), e1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liechti GW; Kuru E; Hall E; Kalinda A; Brun YV; VanNieuwenhze M; Maurelli AT, A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 2014, 506 (7489), 507–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Montón Silva A; Otten C; Biboy J; Breukink E; VanNieuwenhze M; Vollmer W; den Blaauwen T, The fluorescent D-amino acid NADA as a tool to study the conditional activity of transpeptidases in Escherichia coli. Front Microbiol 2018, 9, 2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kuru E; Radkov A; Meng X; Egan A; Alvarez L; Dowson A; Booher G; Breukink E; Roper DI; Cava F; Vollmer W; Brun Y; VanNieuwenhze MS, Mechanisms of incorporation for D-amino acid probes that target peptidoglycan biosynthesis. ACS Chem Biol 2019, 14 (12), 2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.García-Heredia A; Pohane AA; Melzer ES; Carr CR; Fiolek TJ; Rundell SR; Lim HC; Wagner JC; Morita YS; Swarts BM; Siegrist MS, Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jankute M; Cox JA; Harrison J; Besra GS, Assembly of the mycobacterial cell wall. Annu Rev Microbiol 2015, 69, 405–23. [DOI] [PubMed] [Google Scholar]
- 72.Abrahams KA; Besra GS, Mycobacterial cell wall biosynthesis: a multifaceted antibiotic target. Parasitology 2018, 145 (2), 116–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kavunja HW; Biegas KJ; Banahene N; Stewart JA; Piligian BF; Groenevelt JM; Sein CE; Morita YS; Niederweis M; Siegrist MS; Swarts BM, Photoactivatable glycolipid probes for identifying mycolate-protein interactions in live mycobacteria. J Am Chem Soc 2020, 142 (17), 7725–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rhee HW; Zou P; Udeshi ND; Martell JD; Mootha VK; Carr SA; Ting AY, Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339 (6125), 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roux KJ; Kim DI; Raida M; Burke B, A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 2012, 196 (6), 801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han S; Li J; Ting AY, Proximity labeling: spatially resolved proteomic mapping for neurobiology. Curr Opin Neurobiol 2018, 50, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beck DB; Bonasio R, Photoactivated in vivo proximity labeling. Curr Protoc Chem Biol 2017, 9 (2), 128–146. [DOI] [PubMed] [Google Scholar]
- 78.Calabretta PJ; Hodges HL; Kraft MB; Marando VM; Kiessling LL, Bacterial cell wall modification with a glycolipid substrate. J Am Chem Soc 2019, 141 (23), 9262–9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kalograiaki I; Euba B; Fernández-Alonso MDC; Proverbio D; St Geme JW 3rd; Aastrup T; Garmendia J; Cañada FJ; Solís D, Differential recognition of Haemophilus influenzae whole bacterial cells and isolated lipooligosaccharides by galactose-specific lectins. Sci Rep 2018, 8 (1), 16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Breitenbach Barroso Coelho LC; Marcelino Dos Santos Silva P; Felix de Oliveira W; de Moura MC; Viana Pontual E; Soares Gomes F; Guedes Paiva PM; Napoleão TH; Dos Santos Correia MT, Lectins as antimicrobial agents. J Appl Microbiol 2018, 125 (5), 1238–1252. [DOI] [PubMed] [Google Scholar]
- 81.Prado Acosta M; Lepenies B, Bacterial glycans and their interactions with lectins in the innate immune system. Biochem Soc Trans 2019, 47 (6), 1569–1579. [DOI] [PubMed] [Google Scholar]
- 82.Dam TK; Brewer CF, Lectins as pattern recognition molecules: the effects of epitope density in innate immunity. Glycobiology 2010, 20 (3), 270–9. [DOI] [PubMed] [Google Scholar]
- 83.Chen L; Li J; Yang G, A comparative review of intelectins. Scand J Immunol 2020, 92 (1), e12882. [DOI] [PubMed] [Google Scholar]
- 84.Chen L; Yan J; Shi J; Sun W; Chen Z; Yu J; Qi J; Du Y; Zhang H; Feng L, Zebrafish intelectin 1 (zITLN1) plays a role in the innate immune response. Fish Shellfish Immunol 2018, 83, 96–103. [DOI] [PubMed] [Google Scholar]
- 85.Lee JK; Schnee J; Pang M; Wolfert M; Baum LG; Moremen KW; Pierce M, Human homologs of the Xenopus oocyte cortical granule lectin XL35. Glycobiology 2001, 11 (1), 65–73. [DOI] [PubMed] [Google Scholar]
- 86.Wesener DA; Dugan A; Kiessling LL, Recognition of microbial glycans by soluble human lectins. Curr Opin Struct Biol 2017, 44, 168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wangkanont K; Wesener DA; Vidani JA; Kiessling LL; Forest KT, Structures of Xenopus embryonic epidermal lectin reveal a conserved mechanism of microbial glycan recognition. J Biol Chem 2016, 291 (11), 5596–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen L; Yan J; Sun W; Zhang Y; Sui C; Qi J; Du Y; Feng L, A zebrafish intelectin ortholog agglutinates both Gram-negative and Gram-positive bacteria with binding capacity to bacterial polysaccharide. Fish Shellfish Immunol 2016, 55, 729–36. [DOI] [PubMed] [Google Scholar]
- 89.Sýkorová P; Novotná J; Demo G; Pompidor G; Dubská E; Komárek J; Fujdiarová E; Houser J; Hároníková L; Varrot A; Shilova N; Imberty A; Bovin N; Pokorná M; Wimmerová M, Characterization of novel lectins from Burkholderia pseudomallei and Chromobacterium violaceum with seven-bladed β-propeller fold. Int J Biol Macromol 2020, 152, 1113–1124. [DOI] [PubMed] [Google Scholar]
- 90.Thai Le S; Malinovska L; Vašková M; Mező E; Kelemen V; Borbás A; Hodek P; Wimmerová M; Csávás M, Investigation of the binding affinity of a broad array of L-fucosides with six fucose-specific lectins of bacterial and fungal origin. Molecules 2019, 24 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li PC, Overview of microarray technology. Methods Mol Biol 2016, 1368, 3–4. [DOI] [PubMed] [Google Scholar]
- 92.Briard JG; Jiang H; Moremen KW; Macauley MS; Wu P, Cell-based glycan arrays for probing glycan-glycan binding protein interactions. Nat Commun 2018, 9 (1), 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jégouzo SAF; Nelson C; Hardwick T; Wong STA; Lau NKK; Neoh GKE; Castellanos-Rueda R; Huang Z; Mignot B; Hirdaramani A; Howitt A; Frewin K; Shen Z; Fox RJ; Wong R; Ando M; Emony L; Zhu H; Holder A; Werling D; Krishnan N; Robertson BD; Clements A; Taylor ME; Drickamer K, Mammalian lectin arrays for screening host-microbe interactions. J Biol Chem 2020, 295 (14), 4541–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dupin L; Noël M; Bonnet S; Meyer A; Géhin T; Bastide L; Randriantsoa M; Souteyrand E; Cottin C; Vergoten G; Vasseur JJ; Morvan F; Chevolot Y; Darblade B, Screening of a library of oligosaccharides targeting lectin LecB of Pseudomonas Aeruginosa and synthesis of high affinity oligoglycoclusters. Molecules 2018, 23 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang N; Hirata A; Nokihara K; Fukase K; Fujimoto Y, Peptidoglycan microarray as a novel tool to explore protein-ligand recognition. Biopolymers 2016, 106 (4), 422–9. [DOI] [PubMed] [Google Scholar]
- 96.Götze S; Reinhardt A; Geissner A; Azzouz N; Tsai YH; Kurucz R; Varón Silva D; Seeberger PH, Investigation of the protective properties of glycosylphosphatidylinositol-based vaccine candidates in a Toxoplasma gondii mouse challenge model. Glycobiology 2015, 25 (9), 984–91. [DOI] [PubMed] [Google Scholar]
- 97.Reinhardt A; Yang Y; Claus H; Pereira CL; Cox AD; Vogel U; Anish C; Seeberger PH, Antigenic potential of a highly conserved Neisseria meningitidis lipopolysaccharide inner core structure defined by chemical synthesis. Chem Biol 2015, 22 (1), 38–49. [DOI] [PubMed] [Google Scholar]
- 98.Broecker F; Hanske J; Martin CE; Baek JY; Wahlbrink A; Wojcik F; Hartmann L; Rademacher C; Anish C; Seeberger PH, Multivalent display of minimal Clostridium difficile glycan epitopes mimics antigenic properties of larger glycans. Nat Commun 2016, 7, 11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schumann B; Hahm HS; Parameswarappa SG; Reppe K; Wahlbrink A; Govindan S; Kaplonek P; Pirofski LA; Witzenrath M; Anish C; Pereira CL; Seeberger PH, A semisynthetic Streptococcus pneumoniae serotype 8 glycoconjugate vaccine. Sci Transl Med 2017, 9 (380). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Seeberger PH, The logic of automated glycan assembly. Acc Chem Res 2015, 48 (5), 1450–63. [DOI] [PubMed] [Google Scholar]
- 101.Eller S; Collot M; Yin J; Hahm HS; Seeberger PH, Automated solid-phase synthesis of chondroitin sulfate glycosaminoglycans. Angew Chem Int Ed Engl 2013, 52 (22), 5858–61. [DOI] [PubMed] [Google Scholar]
- 102.Campanero-Rhodes MA L. E; Bengoechea JA; Solis D , Bacteria microarrays as sensitive tools for exploring pathogen surface epitopes and recognition by host receptors. RSC Adv 2015, 5, 7173–7181. [Google Scholar]
- 103.Kapil S; Petit C; Drago VN; Ronning DR; Sucheck SJ, Synthesis and in vitro characterization of trehalose-based inhibitors of mycobacterial trehalose 6-phosphate phosphatases. Chembiochem 2019, 20 (2), 260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wolber JM; Urbanek BL; Meints LM; Piligian BF; Lopez-Casillas IC; Zochowski KM; Woodruff PJ; Swarts BM, The trehalose-specific transporter LpqY-SugABC is required for antimicrobial and anti-biofilm activity of trehalose analogues in Mycobacterium smegmatis. Carbohydr Res 2017, 450, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kalograiaki I; Euba B; Proverbio D; Campanero-Rhodes MA; Aastrup T; Garmendia J; Solís D, Combined bacteria microarray and quartz crystal microbalance approach for exploring glycosignatures of nontypeable Haemophilus influenzae and recognition by host lectins. Anal Chem 2016, 88 (11), 5950–7. [DOI] [PubMed] [Google Scholar]
- 106.Alemka A; Nothaft H; Zheng J; Szymanski CM, N-glycosylation of Campylobacter jejuni surface proteins promotes bacterial fitness. Infect Immun 2013, 81 (5), 1674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chatterjee AN; Young FE, Regulation of the bacterial cell wall: isolation and characterization of peptidoglycan mutants of Staphylococcus aureus. J Bacteriol 1972, 111 (1), 220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Howard SL; Jagannathan A; Soo EC; Hui JP; Aubry AJ; Ahmed I; Karlyshev A; Kelly JF; Jones MA; Stevens MP; Logan SM; Wren BW, Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect Immun 2009, 77 (6), 2544–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smedley JG 3rd; Jewell E; Roguskie J; Horzempa J; Syboldt A; Stolz DB; Castric P, Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. Infect Immun 2005, 73 (12), 7922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weidenmaier C; Peschel A; Xiong YQ; Kristian SA; Dietz K; Yeaman MR; Bayer AS, Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J Infect Dis 2005, 191 (10), 1771–7. [DOI] [PubMed] [Google Scholar]
- 111.Woodruff PJ; Carlson BL; Siridechadilok B; Pratt MR; Senaratne RH; Mougous JD; Riley LW; Williams SJ; Bertozzi CR, Trehalose is required for growth of Mycobacterium smegmatis. J Biol Chem 2004, 279 (28), 28835–43. [DOI] [PubMed] [Google Scholar]
- 112.Naclerio GA; Karanja CW; Opoku-Temeng C; Sintim HO, Antibacterial small molecules that potently inhibit Staphylococcus aureus lipoteichoic acid biosynthesis. ChemMedChem 2019, 14 (10), 1000–1004. [DOI] [PubMed] [Google Scholar]
- 113.De Schutter JW; Morrison JP; Morrison MJ; Ciulli A; Imperiali B, Targeting bacillosamine biosynthesis in bacterial pathogens: development of inhibitors to a bacterial amino-sugar acetyltransferase from Campylobacter jejuni. J Med Chem 2017, 60 (5), 2099–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Walvoort MT; Lukose V; Imperiali B, A modular approach to phosphoglycosyltransferase inhibitors inspired by nucleoside antibiotics. Chemistry 2016, 22 (11), 3856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Madec AGE; Schocker NS; Sanchini S; Myratgeldiyev G; Das D; Imperiali B, Facile solid-phase synthesis and assessment of nucleoside analogs as inhibitors of bacterial UDP-sugar processing enzymes. ACS Chem Biol 2018, 13 (9), 2542–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Solanki V; Tiwari M; Tiwari V, Host-bacteria interaction and adhesin study for development of therapeutics. Int J Biol Macromol 2018, 112, 54–64. [DOI] [PubMed] [Google Scholar]
- 117.Machida T; Novoa A; Gillon É; Zheng S; Claudinon J; Eierhoff T; Imberty A; Römer W; Winssinger N, Dynamic cooperative glycan assembly blocks the binding of bacterial lectins to epithelial cells. Angew Chem Int Ed Engl 2017, 56 (24), 6762–6766. [DOI] [PubMed] [Google Scholar]
- 118.Fu TK; Ng SK; Chen YE; Lee YC; Demeter F; Herczeg M; Borbás A; Chiu CH; Lan CY; Chen CL; Chang MD, Rhamnose binding protein as an anti-bacterial agent-targeting biofilm of Pseudomonas aeruginosa. Mar Drugs 2019, 17 (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gottesmann M; Paraskevopoulou V; Mohammed A; Falcone FH; Hensel A, BabA and LPS inhibitors against Helicobacter pylori: pectins and pectin-like rhamnogalacturonans as adhesion blockers. Appl Microbiol Biotechnol 2020, 104 (1), 351–363. [DOI] [PubMed] [Google Scholar]
- 120.Phillips ZN; Husna AU; Jennings MP; Seib KL; Atack JM, Phasevarions of bacterial pathogens - phase-variable epigenetic regulators evolving from restriction-modification systems. Microbiology 2019, 165 (9), 917–928. [DOI] [PubMed] [Google Scholar]
- 121.Howlett R; Read N; Varghese A; Kershaw C; Hancock Y; Smith MCM, Streptomyces coelicolor strains lacking polyprenol phosphate mannose synthase and protein O-mannosyl transferase are hyper-susceptible to multiple antibiotics. Microbiology 2018, 164 (3), 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Becker K; Haldimann K; Selchow P; Reinau LM; Dal Molin M; Sander P, Lipoprotein glycosylation by protein-O-mannosyltransferase (MAB_1122c) contributes to low cell envelope permeability and antibiotic resistance of Mycobacterium abscessus. Front Microbiol 2017, 8, 2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mello SS; Van Tyne D; Lebreton F; Silva SQ; Nogueira MCL; Gilmore MS; Camargo I, A mutation in the glycosyltransferase gene lafB causes daptomycin hypersusceptibility in Enterococcus faecium. J Antimicrob Chemother 2020, 75 (1), 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chiosis G; Boneca IG, Selective cleavage of D-Ala-D-Lac by small molecules: re-sensitizing resistant bacteria to vancomycin. Science 2001, 293 (5534), 1484–7. [DOI] [PubMed] [Google Scholar]
- 125.Yarlagadda V; Konai MM; Manjunath GB; Ghosh C; Haldar J, Tackling vancomycin-resistant bacteria with ‘lipophilicvancomycin-carbohydrate conjugates’. J Antibiot 2015, 68 (5), 302–12. [DOI] [PubMed] [Google Scholar]
- 126.Roy R; Tiwari M; Donelli G; Tiwari V, Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9 (1), 522–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Høiby N; Bjarnsholt T; Givskov M; Molin S; Ciofu O, Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 2010, 35 (4), 322–32. [DOI] [PubMed] [Google Scholar]
- 128.Yoshii Y; Okuda KI; Yamada S; Nagakura M; Sugimoto S; Nagano T; Okabe T; Kojima H; Iwamoto T; Kuwano K; Mizunoe Y, Norgestimate inhibits staphylococcal biofilm formation and resensitizes methicillin-resistant Staphylococcus aureus to β-lactam antibiotics. NPJ Biofilms Microbiomes 2017, 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Su Z; Peng L; Worthington RJ; Melander C, Evaluation of 4,5-disubstituted-2-aminoimidazole-triazole conjugates for antibiofilm/antibiotic resensitization activity against MRSA and Acinetobacter baumannii. ChemMedChem 2011, 6 (12), 2243–51. [DOI] [PubMed] [Google Scholar]
- 130.Si Z; Lim HW; Tay MYF; Du Y; Ruan L; Qiu H; Zamudio-Vazquez R; Reghu S; Chen Y; Tiong WS; Marimuthu K; De PP; Ng OT; Zhu Y; Gan YH; Chi YR; Duan H; Bazan GC; Greenberg EP; Chan-Park MB; Pethe K, A glycosylated cationic block poly(β-peptide) reverses intrinsic antibiotic resistance in all ESKAPE Gram-negative bacteria. Angew Chem Int Ed Engl 2020, 59 (17), 6819–6826. [DOI] [PubMed] [Google Scholar]
- 131.Lyu Y; Yang X; Goswami S; Gorityala BK; Idowu T; Domalaon R; Zhanel GG; Shan A; Schweizer F, Amphiphilic tobramycin-lysine conjugates sensitize multidrug resistant Gram-negative bacteria to rifampicin and minocycline. J Med Chem 2017, 60 (9), 3684–3702. [DOI] [PubMed] [Google Scholar]
- 132.Reddick LE; Alto NM, Bacteria fighting back: how pathogens target and subvert the host innate immune system. Mol Cell 2014, 54 (2), 321–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fura JM; Sabulski MJ; Pires MM, D-amino acid mediated recruitment of endogenous antibodies to bacterial surfaces. ACS Chem Biol 2014, 9 (7), 1480–9. [DOI] [PubMed] [Google Scholar]
- 134.Feigman MS; Kim S; Pidgeon SE; Yu Y; Ongwae GM; Patel DS; Regen S; Im W; Pires MM, Synthetic immunotherapeutics against Gram-negative pathogens. Cell Chem Biol 2018, 25 (10), 1185–1194.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.DeMeester KE; Liang H; Jensen MR; Jones ZS; D’Ambrosio EA; Scinto SL; Zhou J; Grimes CL, Synthesis of functionalized N-acetyl muramic acids to probe bacterial cell wall recycling and biosynthesis. J Am Chem Soc 2018, 140 (30), 9458–9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Taylor JA; Bratton BP; Sichel SR; Blair KM; Jacobs HM; DeMeester KE; Kuru E; Gray J; Biboy J; VanNieuwenhze MS; Vollmer W; Grimes CL; Shaevitz JW; Salama NR, Distinct cytoskeletal proteins define zones of enhanced cell wall synthesis in Helicobacter pylori. Elife 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Swarts BM; Holsclaw CM; Jewett JC; Alber M; Fox DM; Siegrist MS; Leary JA; Kalscheuer R; Bertozzi CR, Probing the mycobacterial trehalome with bioorthogonal chemistry. J Am Chem Soc 2012, 134 (39), 16123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Foley HN; Stewart JA; Kavunja HW; Rundell SR; Swarts BM, Bioorthogonal chemical reporters for selective in situ probing of mycomembrane components in mycobacteria. Angew Chem Int Ed Engl 2016, 55 (6), 2053–7. [DOI] [PubMed] [Google Scholar]
- 139.Wang W; Zhu Y; Chen X, Selective imaging of Gram-negative and Gram-positive microbiotas in the mouse gut. Biochemistry 2017, 56 (30), 3889–3893. [DOI] [PubMed] [Google Scholar]
- 140.Hudak JE; Alvarez D; Skelly A; von Andrian UH; Kasper DL, Illuminating vital surface molecules of symbionts in health and disease. Nat Microbiol 2017, 2, 17099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zlitni A; Gowrishankar G; Steinberg I; Haywood T; Sam Gambhir S, Maltotriose-based probes for fluorescence and photoacoustic imaging of bacterial infections. Nat Commun 2020, 11 (1), 1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rodriguez-Rivera FP; Zhou X; Theriot JA; Bertozzi CR, Visualization of mycobacterial membrane dynamics in live cells. J Am Chem Soc 2017, 139 (9), 3488–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Groenevelt JM; Meints LM; Stothard AI; Poston AW; Fiolek TJ; Finocchietti DH; Mulholand VM; Woodruff PJ; Swarts BM, Chemoenzymatic synthesis of trehalosamine, an aminoglycoside antibiotic and precursor to mycobacterial imaging probes. J Org Chem 2018, 83 (15), 8662–8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Peña-Zalbidea S; Huang AY; Kavunja HW; Salinas B; Desco M; Drake C; Woodruff PJ; Vaquero JJ; Swarts BM, Chemoenzymatic radiosynthesis of 2-deoxy-2-[(18)F]fluorod-trehalose ([(18)F]-2-FDTre): A PET radioprobe for in vivo tracing of trehalose metabolism. Carbohydr Res 2019, 472, 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Samuel B; Dirk R; Frederic D; Michael K, Fluorine-18 chemistry for PET: a concise introduction. Current Radiopharmaceuticals 2010, 3 (2), 68–80. [Google Scholar]
- 146.Barrios-Lopez B; Bergstrom K, Radiolabeled sugars used for PET and SPECT imaging. Curr Radiopharm 2016, 9 (3), 180–186. [DOI] [PubMed] [Google Scholar]
- 147.Hodges HL; Brown RA; Crooks JA; Weibel DB; Kiessling LL, Imaging mycobacterial growth and division with a fluorogenic probe. Proc Natl Acad Sci U S A 2018, 115 (20), 5271–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Holmes NJ; Kavunja HW; Yang Y; Vannest BD; Ramsey CN; Gepford DM; Banahene N; Poston AW; Piligian BF; Ronning DR; Ojha AK; Swarts BM, A FRET-based fluorogenic trehalose dimycolate analogue for probing mycomembrane-remodeling enzymes of mycobacteria. ACS Omega 2019, 4 (2), 4348–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hsu YP; Booher G; Egan A; Vollmer W; VanNieuwenhze MS, D-amino acid derivatives as in situ probes for visualizing bacterial peptidoglycan biosynthesis. Acc Chem Res 2019, 52 (9), 2713–2722. [DOI] [PubMed] [Google Scholar]
- 150.Boersma MJ; Kuru E; Rittichier JT; VanNieuwenhze MS; Brun YV; Winkler ME, Minimal peptidoglycan (PG) turnover in wild-type and PG hydrolase and cell division mutants of Streptococcus pneumoniae D39 growing planktonically and in host-relevant biofilms. J Bacteriol 2015, 197 (21), 3472–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Benito-Peña E; Moreno-Bondi MC; Orellana G; Maquieira A; van Amerongen A, Development of a novel and automated fluorescent immunoassay for the analysis of beta-lactam antibiotics. J Agric Food Chem 2005, 53 (17), 6635–42. [DOI] [PubMed] [Google Scholar]
- 152.Gee KR; Kang HC; Meier TI; Zhao G; Blaszcak LC, Fluorescent bocillins: synthesis and application in the detection of penicillin-binding proteins. Electrophoresis 2001, 22 (5), 960–5. [DOI] [PubMed] [Google Scholar]
- 153.June CM; Vaughan RM; Ulberg LS; Bonomo RA; Witucki LA; Leonard DA, A fluorescent carbapenem for structure function studies of penicillin-binding proteins, β-lactamases, and β-lactam sensors. Anal Biochem 2014, 463, 70–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lakaye B; Damblon C; Jamin M; Galleni M; Lepage S; Joris B; Marchand-Brynaert J; Frydrych C; Frere JM, Synthesis, purification and kinetic properties of fluorescein-labelled penicillins. Biochem J 1994, 300 (Pt 1) (Pt 1), 141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Morikawa Y; Kitazato M; Mitsuyama J; Mizunaga S; Minami S; Watanabe Y, In vitro activities of piperacillin against beta-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob Agents Chemother 2004, 48 (4), 1229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stone MRL; Butler MS; Phetsang W; Cooper MA; Blaskovich MAT, Fluorescent antibiotics: new research tools to fight antibiotic resistance. Trends Biotechnol 2018, 36 (5), 523–536. [DOI] [PubMed] [Google Scholar]
- 157.McInerney MP; Roberts KD; Thompson PE; Li J; Nation RL; Velkov T; Nicolazzo JA, Quantitation of polymyxin-lipopolysaccharide interactions using an image-based fluorescent probe. J Pharm Sci 2016, 105 (2), 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Akram AR; Chankeshwara SV; Scholefield E; Aslam T; McDonald N; Megia-Fernandez A; Marshall A; Mills B; Avlonitis N; Craven TH; Smyth AM; Collie DS; Gray C; Hirani N; Hill AT; Govan JR; Walsh T; Haslett C; Bradley M; Dhaliwal K, In situ identification of Gram-negative bacteria in human lungs using a topical fluorescent peptide targeting lipid A. Sci Transl Med 2018, 10 (464). [DOI] [PubMed] [Google Scholar]
- 159.Mao W; Qian X; Zhang J; Xia L; Xie H, Specific Detection of extended-spectrum β-lactamase activities with a ratiometric fluorescent probe. Chembiochem 2017, 18 (20), 1990–1994. [DOI] [PubMed] [Google Scholar]
- 160.Sharma S; Erickson KM; Troutman JM, Complete tetrasaccharide repeat unit biosynthesis of the immunomodulatory Bacteroides fragilis capsular polysaccharide A. ACS Chem Biol 2017, 12 (1), 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Scott PM; Erickson KM; Troutman JM, Identification of the functional roles of six key proteins in the biosynthesis of Enterobacteriaceae colanic acid. Biochemistry 2019, 58 (13), 1818–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kocaoglu O; Carlson EE, Progress and prospects for small-molecule probes of bacterial imaging. Nat Chem Biol 2016, 12 (7), 472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parker MFL; Flavell RR; Luu JM; Rosenberg OS; Ohliger MA; Wilson DM, Small molecule sensors targeting the bacterial cell wall. ACS Infect Dis 2020, 6 (7), 1587–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Woo CM; Iavarone AT; Spiciarich DR; Palaniappan KK; Bertozzi CR, Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat Methods 2015, 12 (6), 561–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hubbard SC; Boyce M; McVaugh CT; Peehl DM; Bertozzi CR, Cell surface glycoproteomic analysis of prostate cancer-derived PC-3 cells. Bioorg Med Chem Lett 2011, 21 (17), 4945–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nilsson J; Rüetschi U; Halim A; Hesse C; Carlsohn E; Brinkmalm G; Larson G, Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Methods 2009, 6 (11), 809–11. [DOI] [PubMed] [Google Scholar]
- 167.Zaro BW; Yang YY; Hang HC; Pratt MR, Chemical reporters for fluorescent detection and identification of O GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4–1. Proc Natl Acad Sci U S A 2011, 108 (20), 8146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang H; Li XJ; Martin DB; Aebersold R, Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat Biotechnol 2003, 21 (6), 660–6. [DOI] [PubMed] [Google Scholar]
- 169.Zaia J, On-line separations combined with MS for analysis of glycosaminoglycans. Mass Spectrom Rev 2009, 28 (2), 254–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Woo CM; Felix A; Byrd WE; Zuegel DK; Ishihara M; Azadi P; Iavarone AT; Pitteri SJ; Bertozzi CR, Development of IsoTaG, a chemical glycoproteomics technique for profiling intact N- and O-glycopeptides from whole cell proteomes. J Proteome Res 2017, 16 (4), 1706–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Woo CM; Bertozzi CR, Isotope targeted glycoproteomics (IsoTaG) to characterize intact, metabolically labeled glycopeptides from complex proteomes. Curr Protoc Chem Biol 2016, 8 (1), 59–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Woo CM; Felix A; Zhang L; Elias JE; Bertozzi CR, Isotope-targeted glycoproteomics (IsoTaG) analysis of sialylated N- and O-glycopeptides on an orbitrap fusion tribrid using azido and alkynyl sugars. Anal Bioanal Chem 2017, 409 (2), 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Han Z; Thuy-Boun PS; Pfeiffer W; Vartabedian VF; Torkamani A; Teijaro JR; Wolan DW, Identification of an N-acetylneuraminic acid-presenting bacteria isolated from a human microbiome. Sci Rep 2021, 11 (1), 4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sanapala SR; Kulkarni SS, Expedient route to access rare deoxy amino L-sugar building blocks for the assembly of bacterial glycoconjugates. J Am Chem Soc 2016, 138 (14), 4938–47. [DOI] [PubMed] [Google Scholar]
- 175.Behera A; Kulkarni SS, Chemical synthesis of rare, deoxy-amino sugars containing bacterial glycoconjugates as potential vaccine candidates. Molecules 2018, 23 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Johnson KF, Synthesis of oligosaccharides by bacterial enzymes. Glycoconj J 1999, 16 (2), 141–6. [DOI] [PubMed] [Google Scholar]
- 177.Hanson S; Best M; Bryan MC; Wong CH, Chemoenzymatic synthesis of oligosaccharides and glycoproteins. Trends Biochem Sci 2004, 29 (12), 656–63. [DOI] [PubMed] [Google Scholar]


