Abstract
Accurate and efficient quantification of cardiac motion offers promising biomarkers for non-invasive diagnosis and prognosis of structural heart diseases. Cine cardiac magnetic resonance imaging remains one of the most advanced imaging tools to provide image acquisitions needed to assess and quantify in-vivo heart kinematics. The majority of cardiac motion studies are focused on human data, and there remains a need to develop and implement an image-registration pipeline to quantify full three-dimensional (3D) cardiac motion in mice where ideal image acquisition is challenged by the subject size and heart rate and the possibility of traditional tagged imaging is hampered. In this study, we used diffeomorphic image registration to estimate strains in the left ventricular wall in two wild-type mice and one diabetic mouse. Our pipeline resulted in a continuous and fully 3D strain map over one cardiac cycle. The estimation of 3D regional and transmural variations of strains is a critical step towards identifying mechanistic biomarkers for improved diagnosis and phenotyping of structural left heart diseases including heart failure with reduced or preserved ejection fraction.
Keywords: Cardiac magnetic resonance imaging, left ventricle, wall strain, small animals
1. Introduction
The social and economic burden of cardiovascular diseases (CVDs), specifically acute cardiac events, has continued to increase over the past decade (3). Accurate assessment of the cardiac contractile function is crucial to improving the morbidity and mortality associated with adverse cardiac events. While ejection fraction (EF) has been used to quantify the myocardial contractility and to classify the heart failures (HF), similar to most lumped parameters, it cannot provide etiological clues or information on the underlying mechanisms and ignores the regional variations in left ventricular diastolic and contractile behavior (2; 9). Considering the spatial heterogeneity of the pathologies which impair the left ventricle (LV) (1), the need to account for the local variations in the left heart motion in the diagnosis, studying, and treatment of these conditions still remains.
Myocardial strains, measured through non-invasive means, can be used to quantify cardiac diastolic and systolic functions. Over the past two decades, advances in cardiac magnetic resonance (cMR) tissue tagging, cine magnetic resonance imaging (cine-MRI), and speckle tracking echocardiography (STE) have paved the way for the non-invasive estimation of cardiac strains. While cMR tagging is accepted as the validated reference method for strain imaging (22; 10; 7; 17), the need for specialized software, relatively complex protocols, and low temporal resolution have kept cMR tagging mostly a research tool for human and large animal data and not suitable for studying small animal models with high heart rate. STE, on the other hand, is widely accessible and easy to use. In 2D-STE, the cardiac strains are estimated by tracking the image texture and contours (14; 8). While this approach offers higher spatial and temporal resolutions, compared to cMR, reproducing the acquisition planes is difficult and it cannot capture out-of-plane motions. Although 3D echocardiography methods, such as 3D-STE, enable tracing speckles in all direction and therefore can address the latter issue (6), similar to tagged cMR, low spatial and temporal resolutions limit the accuracy of their predictions (21; 13). These issues become even more pronounced when it comes to studying LV dysfunction in small animal models. This gap has motivated interests in alternative techniques which are not limited by the complex imaging protocols, can capture out-of-plane motions, and offer reasonable spatial and temporal resolutions (15).
Cine-MRI, that provides detailed 3D anatomical images of the heart, offers great potential for the estimation of cardiac strains. Broadly speaking, one can divide the strain estimation methods which utilize cine-MRI into two categories: (a) mechanics-based nonlinear image registration algorithms, such as hyperelastic warping, which use finite element methods to simulate the myocardium and rely on assumptions about the passive and/or active mechanical properties of the tissue (19; 20; 23) and (b) deformable image registration methods that do not use finite element simulations (4; 11; 12). In this study, our goal was to develop a MATLAB pipeline that enables the estimation of 3D cardiac strain using cardiac cine-MRI. We used the diffeomorphic elastic image registration algorithm, integrated with interpolation schemes, to estimate the displacement fields from short-axis (SA) cine-MRI scans and then calculated complete Green-Lagrange strain tensors at each point in the myocardial wall over one cardiac cycle. Importantly, our pipeline is not restricted to in-plane strain calculations which is a typical limitation with available commercial software using ”feature tracking” technique. We were able to track out-of-plane movement of pixels within the stack of SA planes enabling the calculation of 3D strains at each point. We applied our pipeline to calculate the LV strains in two wild-type (WT) mice and one diabetic mouse exhibiting mild LV diastolic dysfunction (LVDD) and compared our predictions to those of two commercial software.
2. Materials and Methods
2.1. Image acquisition
Short-axis cine-MRI scans of healthy WT female (n=2, 21 weeks old, WT-1 and WT-2) and pathological male B6.BKS(D)-Lepr db/J (n=1, 14 weeks old, db-1) mice were acquired under anesthesia using a vertical wide-bore 9.4T Bruker Avance III HD scanner with a 36 mm proton volume coil (Bruker BioSpin MRI GmbH) for the WT and a 7T Biospec system (Bruker, Billerica, MA) ultizing a 72 mm ID linear volume transmit coil for db-1 mice. The imaging protocol was approved by the University of Cincinnati’s Animal Care and Use Committee (UC IACUC Protocol 19–10-03–01 and 2018–0054). We used ECG signal to trigger image acquisition (R-R over 3 periods) and acquired a set of 10 short-axis scan with the following parameters: Fast Low Angle Shot, TE = 1.8 ms for the 9.4T and 1.6 ms for the 7T, TR = 9 ms, bit depth = 16, flip angle = 30 for the 9.4T and 20 for the 7T, bandwidth = 65789 Hz, resolution = 200 μm, slice thickness = 1 mm, matrix = 160×160, averages=4. In the db-1 mouse, the plasma levels of insulin increases at 10 to 14 days and they become obese at 3 to 4 weeks of age (5). They are expected to exhibit LVDD at 4 months of age and older.
2.2. Image registration
The 3D image registration was performed using a custom MATLAB (MathWorks, MA, USA) library. The major steps in our pipeline included: (i) semi-automatically producing the region of interest (ROI) in each cine-MRI slice; the ROI comprises two contours defining the LV endocardial and epicardial borders, (ii) performing pre-processing that consists of cropping the stack of 2D images and linear resampling of the images, (iii) the 3D image registration over one complete cardiac cycle to calculate displacement field at each timepoint, and (iv) calculating strains from displacement field. The ROI borders were traced using a segmentation software.
We used the diffeomorphic demons (DD) registration algorithm to, successively, match the stream of images in 3D. In this algorithm, the object boundaries are considered as semi-permeable membranes and deformable grids in the fixed and the moving images, respectively (16; 18). The DD algorithm assumes that the pixel intensity of the moving image remains constant over time and identifies the transformation which aligns the moving image, , with the fixed image, , through minimizing the global energy function W, expressed as
| (1) |
where σi and σx represent the image intensity noise and the spatial uncertainty on the correspondences respectively, σT controls the amount of the regularization, u is the spatial transformation, and c is the non-parametric spatial transformation (18). Also, here, we used a diffusion-like regularization, and a histogram matching filter was used prior to the image registration step. The endocardial and epicardial contours were used to: (i) isolate the ROI after the estimation of the displacement fields and (ii) calculate the endocardial volume at each timepoint within the cardiac cycle.
2.3. Strain calculations
We calculated the Green-Lagrange strain tensor, E, from the deformation gradient tensor, F, defined as
| (2) |
and
| (3) |
where x is the reference frame, X is the target frame, and I is the identity tensor. The strain values were transferred from the Cartesian to the polar cylindrical coordinates (Ep = Q·E·QT, where Q is a rotation matrix) at each slice and the strain estimations were compared to values obtained from the strain calculation module commercially available in the Segment and CVi42 software packages.
3. Results
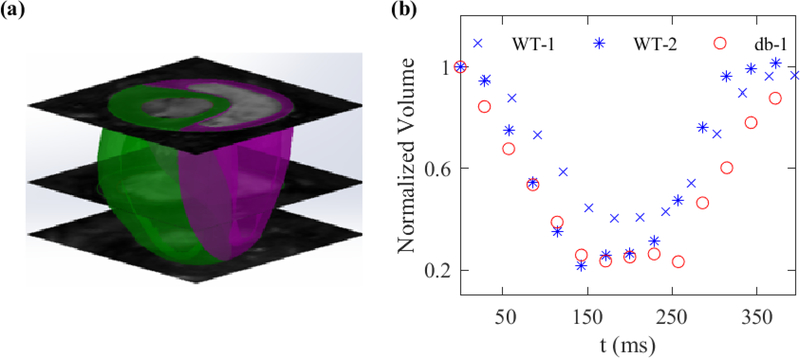
Assessments of LV chamber volume throughout a representative cardiac cycle, calculated by summing over the volume of all the intra-chamber pixels, is shown in Fig. 1. The volume was reduced by more than 50% at end-systole (ES) timepoint in all three mice.
Fig. 1:
(a) Biventricular reconstruction of a representative mouse heart using SA acquisitions. (b) Volume of the LV chamber drops by more than 50% at ES in WT-1, WT-2, and db-1 mice.
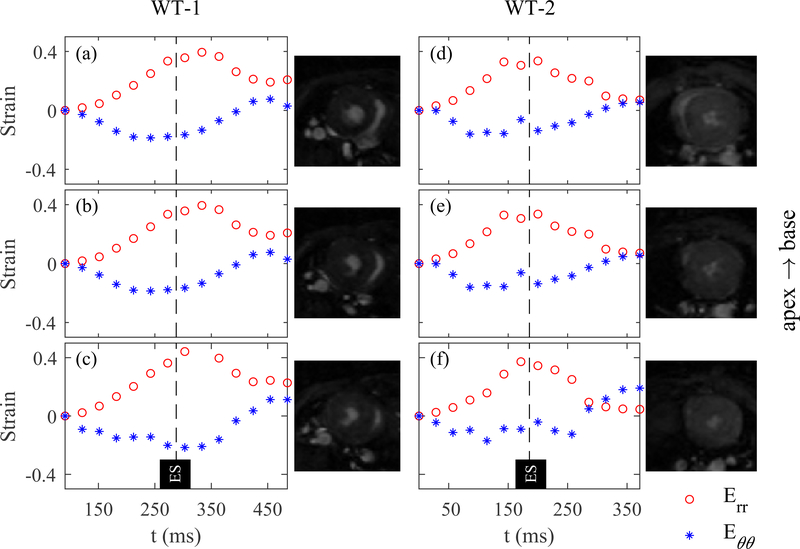
Average sectional strains for three representative short-axis sections over one cardiac cycle is shown for WT-1 and WT-2 in Fig. 2. The sections were chosen from basal, mid, and apical regions and normalized average of radial and circumferential strains were calculated in each section. The maximums for the radial and circumstantially strains in both mice appeared to occur close to the global ES timepoint and nearly maintained their values across multiple SA planes from base to apex. Also, both radial and circumferential strains exhibited consistent patterns across the SA planes except for the circumferential strain in the apical plane in WT-2 where the maximum strain occurred twice, before and after the global ES timepoint.
Fig. 2:
Variation of radial and circumferential strains over one cardiac cycle in WT-1 and WT-2. The average radial (Err) and circumferential (Eθθ) strains were calculated at basal (a and d), mid (b and e), and apical (c and f) left ventricular cross-sections for two WT mice.
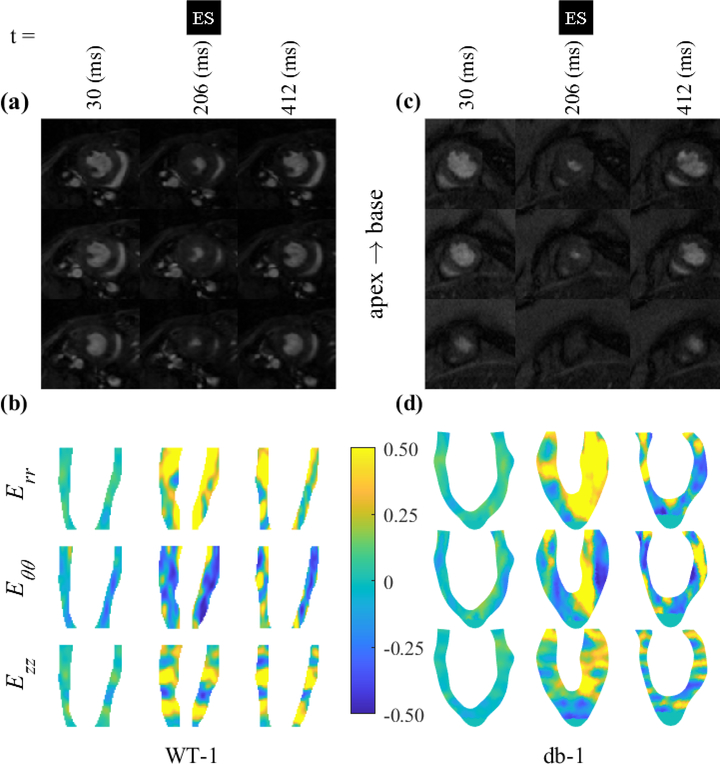
Regional and transmural strain variations across an intra-myocardial long-axis cross-section are shown for WT-1 and db-1 in Fig. 3. The section is fixed in the space and the measurements for radial, circumferential, and longitudinal strains are given at three representative timepoints including ES. The interpolation scheme along the LV long axis used in our pipeline enabled the delivery of a continuous strain map across seven short-axis measurements originally used in the calculations. As depicted by representative LV short-axis slices, the chosen long-axis sections contain the basal and mid-wall regions and part of the apical region. As expected, spatial maps of strains qualitatively suggested that radial and circumferential strains exhibit notable tensile and compressive strains at ES timepoint, respectively, with radial positive strains being predominately present in the endocardial region. In contrast, the longitudinal strains exhibited stronger regional variations and tensile/compressive strain pattern in the given section with the basal regions remaining consistently in positive strains.
Fig. 3:
Contour plots of the left ventricular radial, circumferential, and longitudinal strains in WT-1 (a,b) and db-1 (c,d) mice.
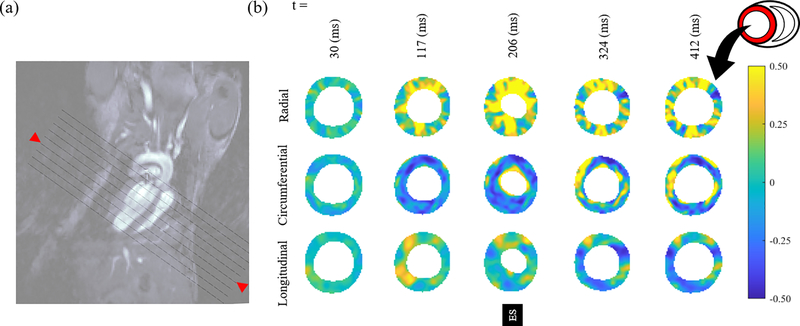
Regional variations in SA views were continuously quantified for all the strains (Fig. 4 shows the results for a chosen mid-section). The radial strain in the short-axis view showed the most pronounced variation followed by circumferential, and longitudinal strains. The regional variations at ES timepoint were not significant, leading to similar levels of strain for anterior, posterior, lateral, and septal regions on average. In contrast, all strains showed stronger regional variations prior and subsequent to ES timepoint. A significant wall thickening was observed as a synergistic result of large tensile and compressive strains in the radial and circumferential directions, respectively.
Fig. 4:
(a) Short-axis slices shown in the long-axis view with oblique lines. The red arrows identify the location of strain contours shown in panel b. (b) Distribution of radial, circumferential, and longitudinal strains in the LV in WT-1.
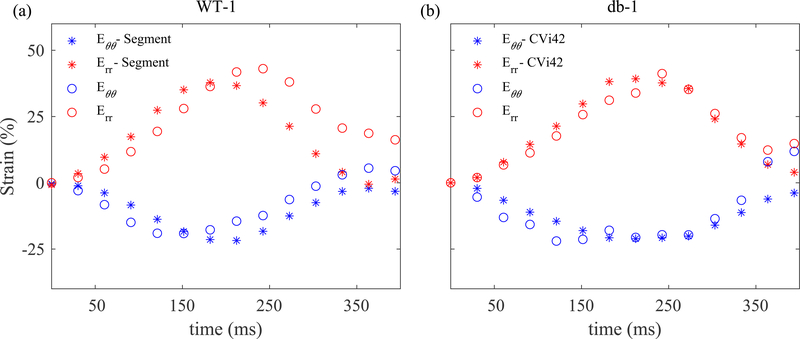
Two commercially available software that can calculate 2D strains from cine cardiac MRI are CVi42 and Segment. These software use feature tracking and can quantify strains from both clinical and pre-clinical images. We assessed our strain estimations against those calculated by both software (Fig. 5). We found an excellent agreement between our results for average radial and circumferential strains on a mid-wall short-axis section and corresponding results from Segment and CVi42 for WT-1 and db-1, respectively. An apparent discrepancy between our results and Segment estimation occurred in the radial strain following ES timepoint. Part of this discrepancy is likely due to the fact that the images representing one complete cardiac cycle are indeed averaged from different cycles, but at appropriate normalized timepoints, during data acquisition. Given that ventricular pressure waves slightly differ from one cycle to another, it is likely that the calculated strains from deformable image registration may not go back to zero at the end of the cycle. However, additional algorithms in commercial software, such as Segment and CVi42, are used to enforce equal strains at the beginning and end of each cycle.
Fig. 5:
Strain values obtained from the MATLAB pipeline match reasonably well with corresponding estimates from Segment and CVi42. Radial and circumferential strains in WT-1 (a) and db-1 (b) mice are compared with values obtained from two commercial software.
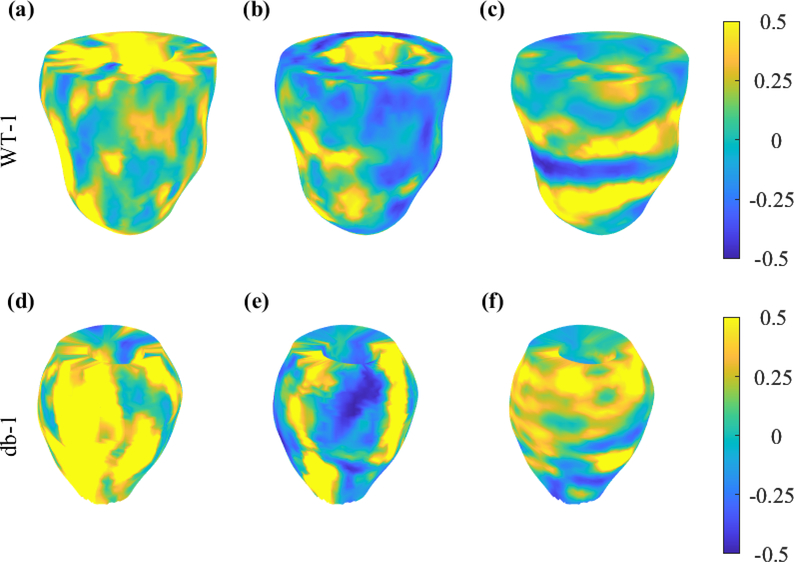
Finally, using all the SA acquisitions, 3D strain maps were created for radial, circumferential, and longitudinal strains (Fig. 6 shows the results for WT-1 and db-1).
Fig. 6:
3D representation of radial (a and d), circumferential (b and e), and longitudinal (c and f) strains at ES in WT-1 and db-1 mice.
4. Discussion
Efficient non-invasive quantification of full 3D LV kinematics offers a comprehensive set of biomarkers that go beyond traditional measures of LV function and anatomy and can serve as insightful diagnostic and treatment-planning tools in the clinic. Surprisingly, despite significant advances in cine cardiac imaging, to the best of authors knowledge, an open-source, non-commercial package to quantify cardiac strains from un-tagged cine MRI remains elusive. The need to develop such a technology remains significant, especially for small animals where the acquisition of high quality imaging data is challenged by the subject size and heart rate. To summarize, in this work we proposed and implemented an image-registration pipeline to estimate full 3D cardiac strains from cine MRI in mice. Unlike some of existing advanced warping methods, the pipeline calculates the strains by processing raw image acquisitions without the need of finite-element simulations.
Using the deformable image registration framework, we provided 3D and continuous map of strain field in the LV myocardial wall. The generated deformation map allowed us to obtain strain at any given cross section and also with respect to an arbitrary coordinates. The results were presented in the clinically-relevant radial, circumferential, and longitudinal directions. Regional and transmural variations were evident and well-captured in the results. As expected, the radial and circumferential strains tended to be predominantly positive and negative, respectively, that consistently led to a pronounced wall thickening. Although not presented here, the shear strains including longitudinal-radial strains as well as principal strains can be easily obtained from the deformation map.
4.1. Overall findings
4.2. Application to HFpEF mice model
Remodeling of the heart left ventricle is a key and common consequence in the setting of several structural heart diseases including myocardial infarction (MI) and heart failure with preserved ejection fraction (HFpEF). The remodeling events take place at multiple scales in the LV potentially altering tissue architecture and biomechanics collectively leading to functional adaptation of the LV. Therefore, traditional measures such as LV dilation and thickness used to characterize the LV remodeling provide limited information on cardiac performance. New biomechanics-driven biomarkers that reveal and quantify regional variations in LV remodeling are needed for early diagnosis and accurate prognosis of structural LV diseases.
In particular, we used our pipeline for a diabetic mouse with mild LVDD (db-1). Our results suggested a noticeable LV hypertrophy with slightly reduced circumferential strains in db-1. We plane to improve our understanding of how the LV adapts in HFpEF and quantify the regional variations in biomechanical measures of LVDD which is a key component in the progression of HFpEF. The pipeline in this work will allow us to study the tissue- and organ-level development of LVDD in murine models of HFpEF in a serial manner. Such explorations are needed to identify key LV kinematic metrics that can be used as clinical biomarkers for early diagnosis and phenotyping of HFpEF that tend to be an impeding factor in the development of efficient treatments for this disease.
5. Summary and implications
Progressive myocardial remodeling events in several LV diseases lead to sizeable changes in the kinematics of the LV that, in turn, mount to LV dysfunction. The ability of non-invasive, high-fidelity, and 3D quantification of cardiac motion in small animals will severe as a significant tool to conduct longitudinal study of LV remodeling in mice. In this work, we developed and implemented a pipeline that uses diffeomorphic image registration technique to estimate 3D cardiac strains in the LV wall in WT mice. We presented and analyzed the regional and transmural distributions of radial, circumferential, and longitudinal strains in both long-axis and short-axis views. In future works, we will use our pipeline to identify LV kinematics-related biomarkers that can be used for early and image-based diagnosis of LV diastolic dysfunction.
7. Acknowledgements
This work was supported by the National Institutes of Health R00HL138288 to R.A. Dr. Sadayappan has received support from National Institutes of Health grants R01 HL130356, R01 HL105826, R01 AR078001, and R01 HL143490; American Heart Association 2019 Institutional Undergraduate Student (19UFEL34380251) and transformation (19TPA34830084) awards.
Footnotes
Declaration of competing interest
Dr. Sadayappan provided consulting and collaborative research studies to the Leducq Foundation, Red Saree Inc., Greater Cincinnati Tamil Sangam, AstraZeneca, MyoKardia, Merck and Amgen, but such work is unrelated to the content of this manuscript. No other disclosures are reported.
References
- [1].Abhayaratna WP, Marwick TH, Smith WT, Becker NG: Characteristics of left ventricular diastolic dysfunction in the community: an echocardiographic survey. Heart 92(9), 1259–1264 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Amzulescu MS, De Craene M, Langet H, Pasquet A, Vancraeynest D, Pouleur AC, Vanoverschelde JL, Gerber B: Myocardial strain imaging: review of general principles, validation, and sources of discrepancies. European Heart Journal-Cardiovascular Imaging 20(6), 605–619 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Benjamin EJ, Muntner P, Alonso A, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Das SR, et al. : Heart disease and stroke statistics—2019 update: a report from the american heart association. Circulation 139(10), e56–e528 (2019) [DOI] [PubMed] [Google Scholar]
- [4].Bistoquet A, Oshinski J, Škrinjar O: Myocardial deformation recovery from cine mri using a nearly incompressible biventricular model. Medical image analysis 12(1), 69–85 (2008) [DOI] [PubMed] [Google Scholar]
- [5].Coleman DL: Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia 14(3), 141–148 (1978) [DOI] [PubMed] [Google Scholar]
- [6].De Craene M, Marchesseau S, Heyde B, Gao H, Alessandrini M, Bernard O, Piella G, Porras A, Tautz L, Hennemuth A, et al. : 3d strain assessment in ultrasound (straus): A synthetic comparison of five tracking methodologies. IEEE transactions on medical imaging 32(9), 1632–1646 (2013) [DOI] [PubMed] [Google Scholar]
- [7].Garot J, Bluemke DA, Osman NF, Rochitte CE, McVeigh ER, Zerhouni EA, Prince JL, Lima JA: Fast determination of regional myocardial strain fields from tagged cardiac images using harmonic phase mri. Circulation 101(9), 981–988 (2000) [DOI] [PubMed] [Google Scholar]
- [8].Geyer H, Caracciolo G, Abe H, Wilansky S, Carerj S, Gentile F, Nesser HJ, Khandheria B, Narula J, Sengupta PP: Assessment of myocardial mechanics using speckle tracking echocardiography: fundamentals and clinical applications. Journal of the American Society of Echocardiography 23(4), 351–369 (2010) [DOI] [PubMed] [Google Scholar]
- [9].Konstam MA, Abboud FM: Ejection fraction: misunderstood and overrated (changing the paradigm in categorizing heart failure). Circulation 135(8), 717–719 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lima JA, Jeremy R, Guier W, Bouton S, Zerhouni EA, McVeigh E, Buchalter MB, Weisfeldt ML, Shapiro EP, Weiss JL: Accurate systolic wall thickening by nuclear magnetic resonance imaging with tissue tagging: correlation with sonomicrometers in normal and ischemic myocardium. Journal of the American College of Cardiology 21(7), 1741–1751 (1993) [DOI] [PubMed] [Google Scholar]
- [11].Mansi T, Pennec X, Sermesant M, Delingette H, Ayache N: ilogdemons: A demons-based registration algorithm for tracking incompressible elastic biological tissues. International journal of computer vision 92(1), 92–111 (2011) [Google Scholar]
- [12].Mansi T, Peyrat JM, Sermesant M, Delingette H, Blanc J, Boudjemline Y, Ayache N: Physically-constrained diffeomorphic demons for the estimation of 3d myocardium strain from cine-mri. In: International Conference on Functional Imaging and Modeling of the Heart. pp. 201–210. Springer; (2009) [Google Scholar]
- [13].Mor-Avi V, Lang RM, Badano LP, Belohlavek M, Cardim NM, Derumeaux G, Galderisi M, Marwick T, Nagueh SF, Sengupta PP, et al. : Current and evolving echocardiographic techniques for the quantitative evaluation of cardiac mechanics: Ase/eae consensus statement on methodology and indications endorsed by the japanese society of echocardiography. European Journal of Echocardiography 12(3), 167–205 (2011) [DOI] [PubMed] [Google Scholar]
- [14].Perk G, Tunick PA, Kronzon I: Non-doppler two-dimensional strain imaging by echocardiography–from technical considerations to clinical applications. Journal of the American Society of Echocardiography 20(3), 234–243 (2007) [DOI] [PubMed] [Google Scholar]
- [15].Schuster A, Hor KN, Kowallick JT, Beerbaum P, Kutty S: Cardiovascular magnetic resonance myocardial feature tracking: concepts and clinical applications. Circulation: Cardiovascular Imaging 9(4), e004077 (2016) [DOI] [PubMed] [Google Scholar]
- [16].Thirion JP: Image matching as a diffusion process: an analogy with maxwell’s demons. Medical image analysis 2(3), 243–260 (1998) [DOI] [PubMed] [Google Scholar]
- [17].Thomas D, Ferrari V, Janik M, Kim D, Pickup S, Glickson J, Zhou R: Quantitative assessment of regional myocardial function in a rat model of myocardial infarction using tagged mri. Magnetic Resonance Materials in Physics, Biology and Medicine 17(3–6), 179–187 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Vercauteren T, Pennec X, Perchant A, Ayache N: Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage 45(1), S61–S72 (2009) [DOI] [PubMed] [Google Scholar]
- [19].Veress AI, Phatak N, Weiss JA: Deformable image registration with hyperelastic warping. In: Handbook of biomedical image analysis, pp. 487–533. Springer; (2005) [Google Scholar]
- [20].Veress AI, Weiss JA, Huesman RH, Reutter BW, Taylor SE, Sitek A, Feng B, Yang Y, Gullberg GT: Measuring regional changes in the diastolic deformation of the left ventricle of shr rats using micropet technology and hyperelastic warping. Annals of biomedical engineering 36(7), 1104–1117 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Voigt JU, Pedrizzetti G, Lysyansky P, Marwick TH, Houle H, Baumann R, Pedri S, Ito Y, Abe Y, Metz S, et al. : Definitions for a common standard for 2d speckle tracking echocardiography: consensus document of the eacvi/ase/industry task force to standardize deformation imaging. European Heart Journal-Cardiovascular Imaging 16(1), 1–11 (2015) [DOI] [PubMed] [Google Scholar]
- [22].Zerhouni EA, Parish DM, Rogers WJ, Yang A, Shapiro EP: Human heart: tagging with mr imaging–a method for noninvasive assessment of myocardial motion. Radiology 169(1), 59–63 (1988) [DOI] [PubMed] [Google Scholar]
- [23].Zou H, Leng S, Xi C, Zhao X, Koh AS, Gao F, Le Tan J, Tan RS, Allen JC, Lee LC, et al. : Three-dimensional biventricular strains in pulmonary arterial hypertension patients using hyperelastic warping. Computer methods and programs in biomedicine 189, 105345 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]