Abstract
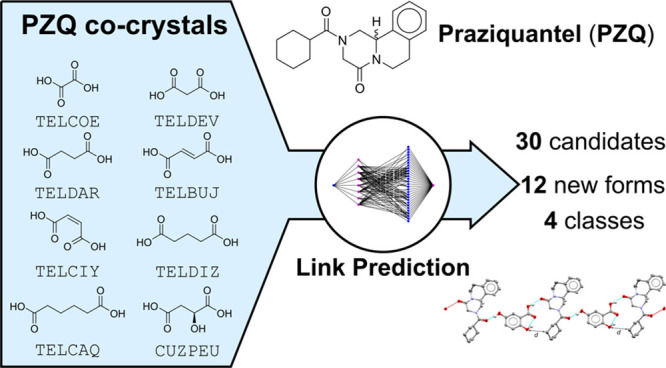
Cocrystallization has been promoted as an attractive early development tool as it can change the physicochemical properties of a target compound and possibly enable the purification of single enantiomers from racemic compounds. In general, the identification of adequate cocrystallization candidates (or coformers) is troublesome and hampers the exploration of the solid-state landscape. For this reason, several computational tools have been introduced over the last two decades. In this study, cocrystals of Praziquantel (PZQ), an anthelmintic drug used to treat schistosomiasis, are predicted with network-based link prediction and experimentally explored. Single crystals of 12 experimental cocrystal indications were grown and subjected to a structural analysis with single-crystal X-ray diffraction. This case study illustrates the power of the link-prediction approach and its ability to suggest a diverse set of new coformer candidates for a target compound when starting from only a limited number of known cocrystals.
Short abstract
In this study, cocrystals for Praziquantel (PZQ), an anthelmintic drug used to treat schistosomiasis, are predicted with network-based link prediction. Single crystals of 12 cocrystal indications were grown and subjected to a structural analysis with single-crystal X-ray diffraction.
1. Introduction
Upon their discovery, active pharmaceutical ingredients (APIs) have physicochemical properties that are often different from those desired in the final drug product. As a variety of these properties depend on the crystal structure of the drug candidate, several methods have been proposed to address this issue by adjusting the underlying solid-state characteristics. These methods include screening for polymorphic forms,1,2 multicomponent crystals,3−5 and (co)amorphous phases.6,7 Among these options, cocrystallization has been identified as a flexible and reliable way to explore different solid forms of an API due to the abundance of possible cocrystal formers (called coformers) and the general thermodynamic stability of cocrystalline phases.8
For APIs containing one or more stereogenic centers, the administration of the drug in racemic formulations (i.e., containing both enantiomers in a stoichiometric ratio) can introduce unwanted side effects originating from one of the API’s enantiomers (the so-called distomer). Because the current industrial climate has shifted toward the acceptance of only chirally pure drugs,9,10 a severe threat is posed to drugs that are only obtainable as racemic compounds (covering 90–95% of the cases)11 and are inherently hard to separate.12 However, the addition of an achiral coformer can prompt the formation of a conglomerate of enantiopure cocrystals13−16 (i.e., a physical mixture of crystals containing a single enantiomer and the coformer). This opens a window of possibilities for crystallization-based separation techniques, such as preferential crystallization17 and deracemization methods,18−22 as was recently demonstrated in two separate case studies for cocrystals of racemic compound-forming targets.23,24 Moreover, introducing a chiral coformer can lead either to the formation of a pair of diastereomeric cocrystals or the formation of an enantiospecific cocrystal (where only one of the target’s enantiomers interacts with the chiral coformer). The latter approach has previously been shown to be a viable strategy for achieving enantiomerically pure substances.25−27 Therefore, the exploration of new cocrystals of racemic compounds with both achiral and chiral coformers can yield new forms relevant for chiral separation.
Praziquantel28 (PZQ; Figure 1) is an anthelmintic drug used in the preventive treatment of schistosomiasis, a disease caused by parasitic flatworms, and suffers from a number of the above-mentioned issues. Due to its poor solubility in aqueous media, the drug is classified as a BCS class II druga, and efforts to improve its solubility and dissolution profile by screening for polymorphs,29−34 hydrates,35 and cocrystals29,36,37 have been previously reported. Additionally, while PZQ is currently administered as a racemate, the (R)-enantiomer possesses the desired therapeutic activity38−41 and the (S)-enantiomer, the distomer, is held responsible for the drug’s bitter taste and other unwanted side effects.42 Earlier studies with the aim of resolving PZQ’s enantiomers have been reported using a diastereomeric salt of its amine precursor43 and more recently by the deracemization of a derivative of the amine precursor.44 Finding a suitable conglomerate-forming multicomponent crystal of PZQ itself could be a worthwhile addition to the set of solutions applicable for chiral separation, either using preferential crystallization or deracemization methods. As PZQ does not contain readily ionizable functional groups, it is precluded from salt formation; therefore, cocrystallization is the method of choice for the further exploration of new multicomponent solid forms. Cocrystals of PZQ with various dicarboxylic acids29,36 and other pharmaceutically acceptable compounds37 have previously been reported, and the structures of several combinations were resolved with single-crystal X-ray diffraction (SC-XRD) and deposited in the Cambridge Structural Database (CSD).45 The discovery of a new set of PZQ cocrystals could potentially form a breakthrough in the formulation of the drug product and the cost-effective purification of the desired enantiomer.
Figure 1.
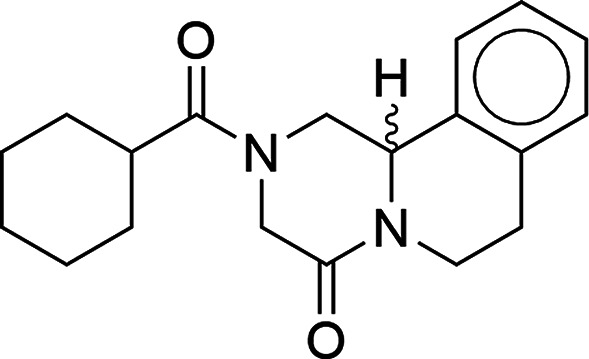
Praziquantel.
In the present research, we evaluate the formation of cocrystals of PZQ with 30 coformers that were predicted using network-based link prediction.46 By representing the information on cocrystals in the CSD as a network consisting of coformers with their cocrystals acting as links,47 new cocrystals can be predicted in the network using mathematical link-prediction algorithms. A requirement for such algorithms to function is the availability of some experimental cocrystallization data for the target compound. As for PZQ cocrystal data is indeed available in the CSD, it is an excellent “real-life” candidate to apply the link-prediction approach. This article describes the application of the network-based link-prediction method to predict 30 new coformer candidates for PZQ and the structural characterization and classification of 12 new cocrystalline forms of PZQ that were discovered with this approach.
2. Methods
To predict new cocrystals for PZQ with network-based link prediction,
a network of coformers must be constructed. By searching with ConQuest48 (CSD ver. 5.39, November 2017, two updates)
for CSD entries containing two chemical residues that were organic,
not ionic or polymeric, error-free, and had determined three-dimensional
coordinates (including disorder), a list of 34 555 two-component
(binary) cocrystals, solvates, and crystals containing a gas molecule
were found. The components in these entries were found after the conversion
of the corresponding structure data files (SD) to canonical SMILES
strings with OpenBabel,49 and 9141 cocrystals
were found by comparing these components to lists of common gases
and solvents.46 To construct a network
of coformers, a set of 7141 unique coformers N was
gathered from the above-mentioned list of cocrystals, and an index
was assigned to each coformer. Next, an adjacency matrix A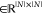 , which is the
mathematical basis of the
network, was built, and all binary cocrystals with coformer indices i and j were added by setting Ai,j and Aj,i to a value of 1.47
, which is the
mathematical basis of the
network, was built, and all binary cocrystals with coformer indices i and j were added by setting Ai,j and Aj,i to a value of 1.47
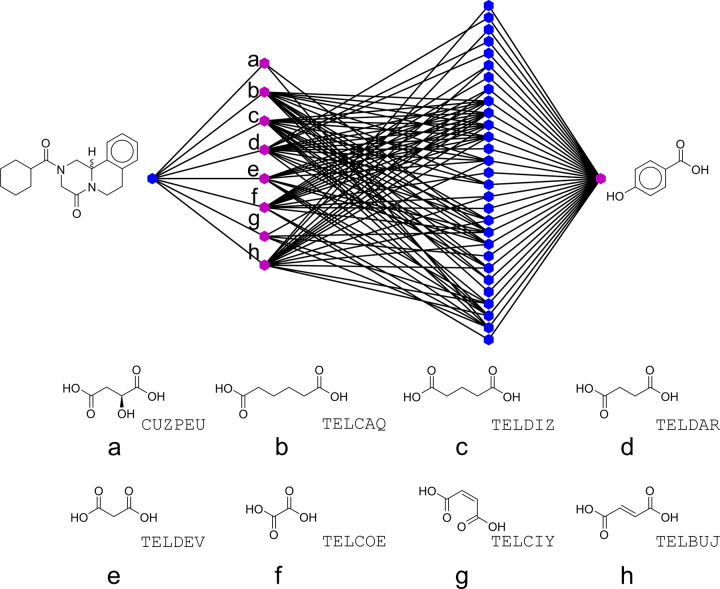
As the CSD contains only the collection of cocrystals that have been successfully determined up to this point in time, it is likely that an abundance of cocrystals are yet to be discovered. In other words, the cocrystal network is probably, to a large extent, incomplete. In fact, it is mathematically possible to identify the missing links (i.e., plausible combinations between coformers) with link-prediction algorithms based on the structure of the network (via the adjacency matrix A).46 Such algorithms look for coformer candidates that exhibit a similar tendency to form cocrystals as those known to interact with the target. This is visualized in Figure 2 for PZQ (the target, blue node) and 4-hydroxybenzoic acid (the coformer candidate, pink node). Here, the known coformers of the target compound (PZQ), l-malic acid (compound a(36)) and other dicarboxylic acids (compounds b–h29) (middle pink nodes), have multiple cocrystallization partners (middle blue nodes) in common with the coformer candidate 4-hydroxybenzoic acid.
Figure 2.
A network containing, from left to right, the target PZQ (blue), coformers known to form cocrystals with PZQ (pink, a–h), and the coformers (blue) interconnecting PZQ’s known coformers to the candidate 4-hydroxybenzoic acid (pink). The molecular structures of PZQ’s neighbors are also shown with the six-character codes that correspond to their cocrystal entries in the CSD.
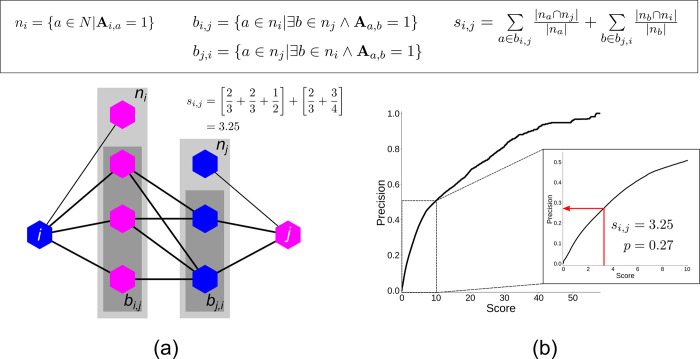
To quantify with link prediction
how likely two coformers are to
form a cocrystal, several score indices were proposed and tested.46 Finally, the bipartite resource allocation score
index (see Figure 3a) was selected, as it performed best on the validation data. The
calculation of this score index is based on properties of the intermediate
network between the two coformers and can loosely be interpreted as
a summation of similarities between nodes of equal color in Figure 3a. The score values
obtained with link prediction can be compared to those of known cocrystals
during a validation experiment (cross validation, for a detailed explanation
see ref (46)) and can
be related to a precision 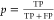 , where TP is the number of true positives
and FP is the number of false positives for the validation set at
that score value (see Figure 3b). The procedures for the construction of the coformer network
and network-based link prediction were all performed using the Python
programming language (ver. 3.7.9).
, where TP is the number of true positives
and FP is the number of false positives for the validation set at
that score value (see Figure 3b). The procedures for the construction of the coformer network
and network-based link prediction were all performed using the Python
programming language (ver. 3.7.9).
Figure 3.
(a) Visualization of a local intermediate network formed between two coformers i and j, properties of the local network (n and b), and the bipartite resource allocation score index si,j. (b) Relation between the score and precision values obtained during the cross validation of the method (see ref (46)).
For the present research, the network-based link-prediction approach was used to compute the scores for PZQ and the 7140 remaining available coformers in the network. Based on their scores, which ranged from 0 to 9.31, and corresponding precisions, which ranged from 0 to 49%, the 30 highest-scoring coformers were selected as candidates for cocrystal screening with racemic PZQ, taking the estimated number of new cocrystal forms to be discovered into account. This resulted in 17 new indications for cocrystal formation, and crystals suitable for single-crystal X-ray diffraction (SC-XRD) could be grown for a subset of these indications (experimental details are available in the ESI). An experimental screening strategy, covering liquid-assisted grinding, solvent evaporation, and the saturation temperatures of specific component mixtures, was used, which proved to be essential for a robust screening and discovery of the newly found cocrystals and will form the topic of a future publication.
3. Results
3.1. Coformer Candidates for PZQ Proposed with Network-Based Link Prediction
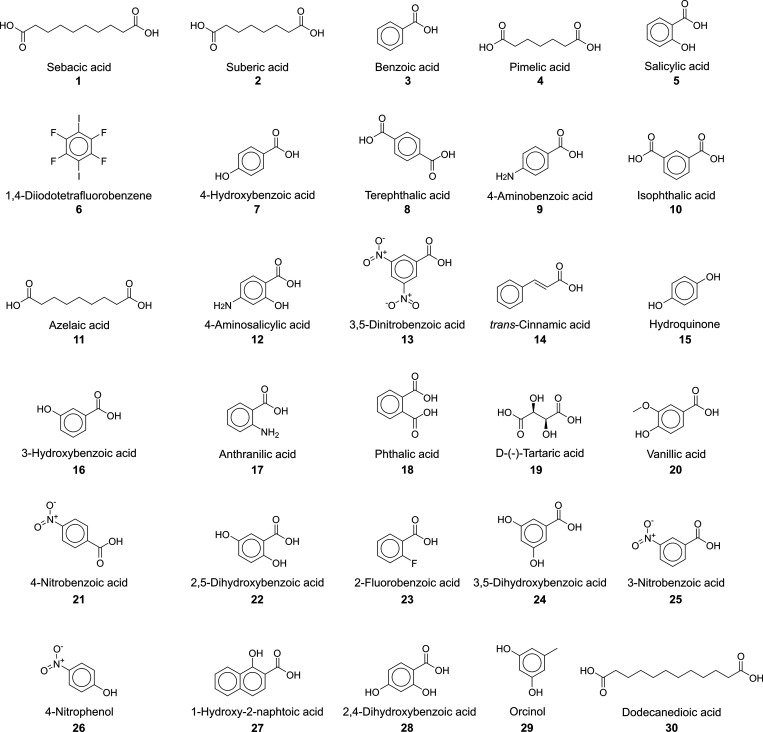
Using the knowledge gained from eightb cocrystal structures of Praziquantel found in the CSD (shown in Figure 2), the link-prediction algorithm was used to predict 30 new coformer candidates (Figure 4 and Table 1). Scores are calculated for each coformer (ranked in decreasing order in Table 1) using the local networks between these coformer candidates and PZQ, which each correspond to an expected precision value after model validation.
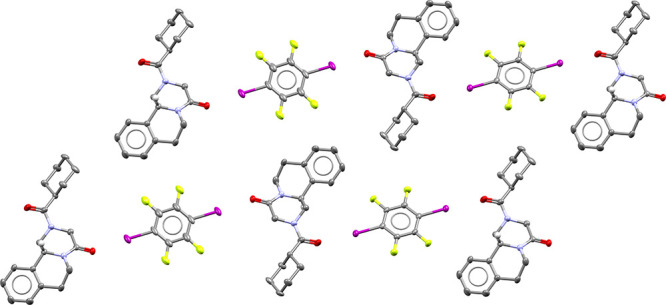
Figure 4.
The 30 coformers predicted with link prediction for PZQ.
Table 1. Overview of the Coformers Predicted and Screened for Racemic Praziquantela.
| rank | coformer | score | precision | screening (PXRD) | SC-XRD |
|---|---|---|---|---|---|
| 1 | sebacic acid | 9.31 | 0.49 | x | |
| 2 | suberic acid | 9.23 | 0.49 | x | |
| 3 | benzoic acid | 8.00 | 0.47 | x | |
| 4 | pimelic acid | 7.54 | 0.46 | √ | |
| 5 | salicylic acid | 6.91 | 0.44 | √ | √(+H2O) |
| 6 | 1,4-diiodotetrafluorobenzene | 5.82 | 0.40 | √ | √ |
| 7 | 4-hydroxybenzoic acid | 5.26 | 0.38 | √ | √ |
| 8 | terephthalic acid | 5.22 | 0.38 | x | |
| 9 | 4-aminobenzoic acid | 4.79 | 0.36 | x | |
| 10 | isophthalic acid | 3.82 | 0.31 | x | |
| 11 | azelaic acid | 3.75 | 0.30 | x | |
| 12 | 4-aminosalicylic acid | 3.45 | 0.28 | √ | √(+MeCN) |
| 13 | 3,5-dinitrobenzoic acid | 3.34 | 0.28 | √ | √ |
| 14 | trans-cinnamic acid | 3.22 | 0.27 | x | |
| 15 | hydroquinone | 3.09 | 0.26 | √, √ | √ |
| 16 | 3-hydroxybenzoic acid | 3.02 | 0.26 | x | |
| 17 | anthranilic acid | 2.85 | 0.25 | x | |
| 18 | phthalic acid | 2.68 | 0.24 | x | |
| 19 | d-(−)-tartaric acid | 2.67 | 0.24 | x | |
| 20 | vanillic acid | 2.55 | 0.23 | √, √ | √ |
| 21 | 4-nitrobenzoic acid | 2.33 | 0.22 | x | |
| 22 | 2,5-dihydroxybenzoic acid | 2.21 | 0.21 | √, √ | √, √(+MeCN) |
| 23 | 2-fluorobenzoic acid | 2.12 | 0.20 | x | |
| 24 | 3,5-dihydroxybenzoic acid | 2.10 | 0.20 | √ | √(+MeCN) |
| 25 | 3-nitrobenzoic acid | 2.02 | 0.19 | x | |
| 26 | 4-nitrophenol | 1.98 | 0.19 | x | |
| 27 | 1-hydroxy-2-naphtoic acid | 1.94 | 0.19 | x | |
| 28 | 2,4-hydroxybenzoic acid | 1.86 | 0.18 | √, √, √ | √ |
| 29 | orcinol | 1.72 | 0.17 | √ | √ |
| 30 | dodecanedioic acid | 1.69 | 0.17 | x |
√, new phase observed with powder X-ray diffraction (PXRD) or single-crystal X-ray diffraction (SC-XRD); x, physical mixture or oil of the constituents observed with powder X-ray diffraction (PXRD).
Besides the five aliphatic dicarboxylic acids (four of which were in fact already considered by Espinosa-Lara et al.29), the list of 30 predicted coformers includes rather different compounds, such as benzoic acid derivatives and aromatic compounds with hydroxyl, amine, and nitro-groups, all of which present the possibility to form strong intermolecular interactions with Praziquantel. Salicylic acid and an enantiomer of tartaric acid are also present in this list, and indications for cocrystal formation with these compounds were reported earlier by Cugovčan et al.;37 however, their crystal structures were not revealed. Remarkably, 1,4-diiodotetrafluorobenzene was also predicted as a coformer for PZQ. This compound is very dissimilar to any of the other predicted coformer candidates or coformers already reported for PZQ in the CSD.
It is noteworthy that the predicted score values of the 30 coformers
correspond to relatively low precisions, the highest being 49% and
lowest being 17%. One would therefore expect around nine new cocrystals
to emerge from this list (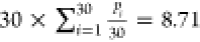 , with pi being the precision associated with coformer i in Table 1). Yet,
as already explained in ref (46), this precision is underestimated and is in fact the lower
limit for its actual value. A larger number of new cocrystals is therefore
expected to emerge from the subsequent experimental screening. Therefore,
a unique advantage of this approach is that it not only predicts a
set of suitable coformers but also estimates how many cocrystals are
expected to emerge from an experimental study.
, with pi being the precision associated with coformer i in Table 1). Yet,
as already explained in ref (46), this precision is underestimated and is in fact the lower
limit for its actual value. A larger number of new cocrystals is therefore
expected to emerge from the subsequent experimental screening. Therefore,
a unique advantage of this approach is that it not only predicts a
set of suitable coformers but also estimates how many cocrystals are
expected to emerge from an experimental study.
From the top 30 coformer candidates, 12 yielded one or several new phases (see Table 1), resulting in 17 indications for cocrystal formation. The experimental proof of these new forms, presented as powder diffractograms, can be found in Figures S1–S14, and the experimental conditions used for their synthesis can be found in Table S2 (see the ESI). This article focuses on the application of network-based link prediction, briefly introduces the newly identified phases, and investigates their crystal structures. A thorough description of the various screening methods, their thermodynamic nature, and the corresponding results will be discussed in a future article.
3.2. Structural Analysis of Newly Discovered Cocrystals for PZQ
Single crystals of 12 new cocrystals of racemic PZQ were successfully grown and analyzed (Table 1). PZQ molecules are always found in their anti-conformation where the carbonyl groups face opposite directions, in some cases with a 90° rotation of the cyclohexyl ring. The crystal structures can be classified into four classes, based on the established intermolecular interaction patterns and the packing of the PZQ enantiomers.
The most frequently occurring class is populated by cocrystals characterized by one-dimensional enantiopure chains (Figures 5 and 6) where both carbonyl groups of PZQ molecules always interact with the coformer through hydrogen bonds. The crystal structures are centrosymmetric and therefore also contain chains of the opposite chirality. The different chains are held together by interactions weaker than the hydrogen bonds within the chains. Figure 5 shows four cocrystals characterized by the formation of these similar chains, where the coformers can be regarded as interchangeable agents for cocrystal formation. The cocrystal solvates containing 2,5-dihydroxybenzoic acid (Figure 5b) and 4-aminosalicylic acid (Figure 5d) are isostructural, and their enantiopure chains lie in the same crystallographic direction ([111], the overlay is shown in Figure S20). The chains formed in the orcinol cocrystal (Figure 6a), on the other hand, do not resemble those shown in Figure 5. While hydrogen bonds are still formed to both of the carbonyl groups of PZQ via its hydroxyl groups, a zigzag chain is visible (Figure 6b), and chains with an equal chirality stack on top of each other.
Figure 5.
Structures of four PZQ cocrystals exhibiting similar enantiopure chains. The (S)-enantiomer of PZQ is shown in each structure, and all hydrogen atoms except those involved in hydrogen bonding interactions and those explanatory for the absolute configuration are omitted. For clarity, only the major conformation is shown in case of disorder (ORTEP plots with disorder are in the ESI). (a) Cocrystal with 2,5-dihydroxybenzoic acid (22). (b) Cocrystal solvate with 2,5-dihydroxybenzoic acid (22) and MeCN. (c) Cocrystal with 4-hydroxybenzoic acid (7). (d) Cocrystal solvate with 4-aminosalicylic acid (12) and MeCN.
Figure 6.
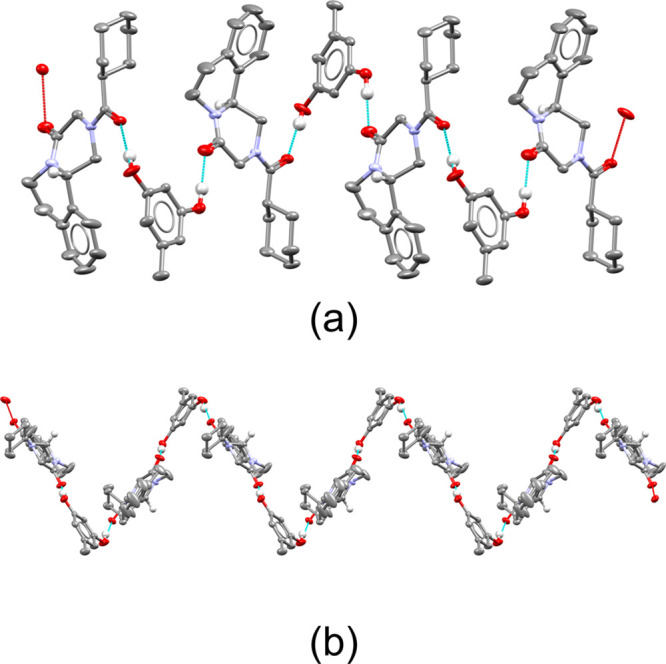
(a) Structure of the cocrystal containing (S)-PZQ and orcinol (29). (b) Enantiopure zigzag chain of (S)-PZQ and orcinol running along the [010] direction. Chains with an identical chirality stack on top of each other. Only PZQ’s major conformation and the hydrogen atoms relevant for hydrogen bonding and chirality are shown (see the ESI for more details).
For the cocrystals (and cocrystal solvate) with hydroquinone (Figure 7a), 3,5-dihydroxybenzoic acid (Figure 7b) and 2,4-dihydroxybenzoic acid (Figure 7c), hydrogen bonding patterns induce the formation of chains containing both enantiomers of PZQ (in a 1:1 molar ratio) and the coformer. Similar to the above-mentioned enantiopure chains, both carbonyl groups of the (alternating) PZQ enantiomers take part in hydrogen bonding interactions with the coformer.
Figure 7.
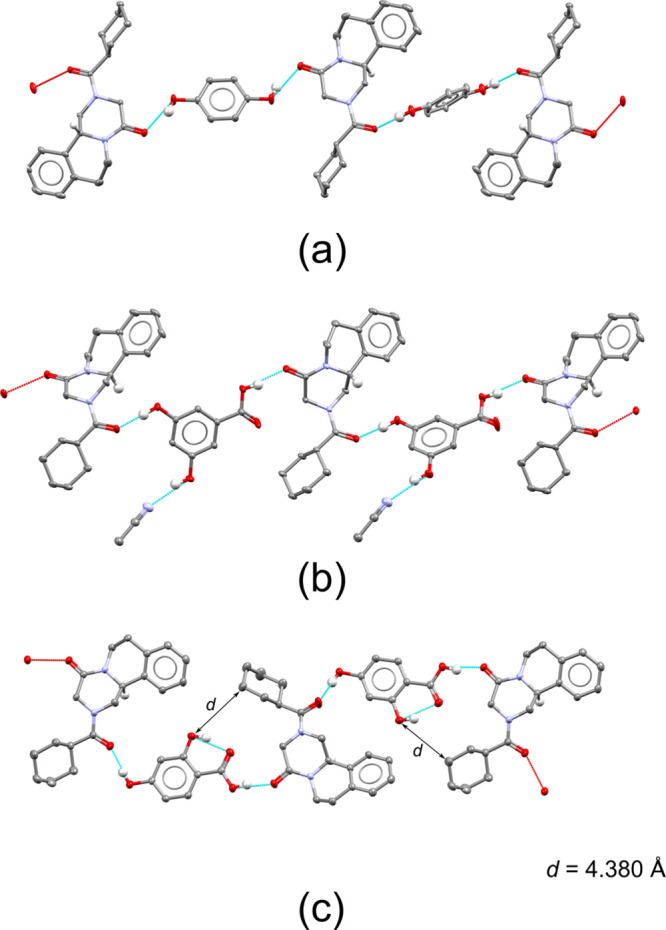
Cocrystals of PZQ connected by racemic chains. Only hydrogen atoms involved in hydrogen bonding interactions and those explanatory for the absolute configuration are displayed. For clarity, only the major conformation is shown in case of disorder (ORTEP plots with disorder and details are available in the ESI). (a) Cocrystal with hydroquinone (15). (b) Cocrystal solvate with 3,5-dihydroxybenzoic acid (24) and MeCN. (c) Cocrystal with 2,4-hydroxybenzoic acid (28).
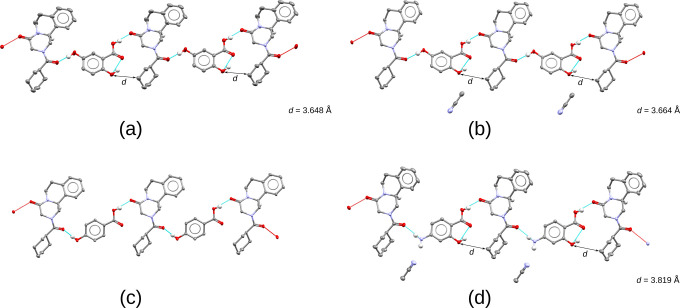
Contrary to the above-mentioned enantiopure and racemic chains, a class of so-called racemic pair cocrystals was identified where the (R)- and (S)-enantiomers of PZQ interact via hydrogen bonding interactions similar to those found in the racemic compound polymorph of pure PZQ with refcode TELCEU0132 (Figure 8a). The formation of this pair was observed for the cocrystals with vanillic acid (Figure 8c) and, albeit shifted, 3,5-dinitrobenzoic acid (Figure 8d). For the cocrystal with salicylic acid, a hydrate is formed where the enantiomers are bridged in a similar fashion as that for TELCEU01. However, they are now bridged via two water molecules (Figure 8b).
Figure 8.
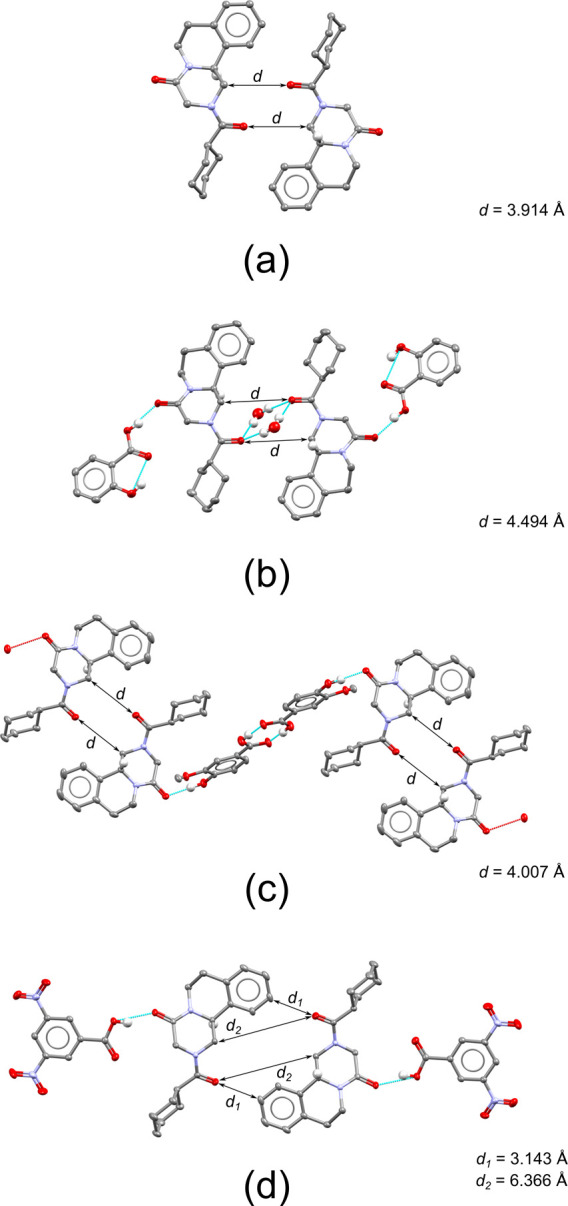
Structures of a racemic compound polymorph of PZQ (TELCEU01) and three cocrystals resembling its racemic pair formation. (a) Racemic polymorph of Praziquantel (TELCEU01).32 (b) Cocrystal hydrate with salicylic acid (5). (c) Cocrystal with vanillic acid (20). (d) Cocrystal with 3,5-dinitrobenzoic acid (13). Only PZQ’s major conformation and the hydrogen atoms relevant for hydrogen bonding and chirality are shown (detail in the ESI).
In contrast to the other coformers suggested by the network-based link prediction, 1,4-diiodotetrafluorobenzene (6) is the only molecule for which hydrogen bonding is precluded, leaving halogen bonding and π–π interactions as plausible alternatives for cocrystal formation. Unlike the apparent zero- or one-dimensional nature of the interactions observed in the former three classes, the structure of the cocrystal of 1,4-diiodotetrafluorobenzene reveals a two-dimensional network (Figure 9). Besides the alternation of PZQ enantiomers halogen-bonded to the coformer via carbonyl-iodide interactions on either side, the fluorine atoms of the coformer interact with the hydrogens of the aromatic and cyclohexyl rings of PZQ.
Figure 9.
Racemic halogen-bonded network structure obtained for the cocrystal of PZQ and 1,4-diiodotetrafluorobenzene (6). Only the hydrogen atoms relevant for the determination of the absolute configuration are shown. The cyclohexyl ring of PZQ is disordered, and its major conformation is shown (details in the ESI).
4. Discussion
The availability of cocrystallization data facilitates the application of the network-based link-prediction method. Using eight known PZQ cocrystals as present in the CSD, the method is able to propose a variety of chemically diverse new coformers for PZQ. Sometimes related, yet in some cases rather different (e.g., 1,4-diiodotetrafluorobenzene) coformers are suggested. The algorithm performs a targeted search in the cocrystal network for coformer candidates that exhibit a similar tendency to form cocrystals as the coformers found in the list of known cocrystals for PZQ and ranks them according to their estimated scores or precision values. The latter can be interpreted as the success rate of finding a new cocrystal form. Given a sufficiently large test set, the expected number of new cocrystals can be calculated by a weighted summation of the precision values. The number of hits, i.e., the new cocrystals for PZQ, agrees with this expected number of nine. Eight new binary cocrystals and an additional four cocrystal solvates were discovered, all of which were characterized by SC-XRD. With this new extended information on cocrystals for PZQ, the link-prediction method can be applied again, and an updated and more precise list of coformer candidates can be generated. To illustrate this concept, the coformer update was performed once for PZQ, and the resulting top 50 predictions are presented in Table S7. Alternatively, the obtained information on both positive and negative indications for cocrystal formation can be fed into a cocrystal prediction framework based on artificial neural networks,50 further guiding the search for adequate coformers. The latter classifies coformers into plausible and unlikely candidates and finds its primary use in the earliest stages of cocrystal screening when no information about a molecule’s ability to form cocrystals is known. Once experimental data become available, however, a combined approach using the ranking and precisions obtained from link prediction and the discrimination from neural networks may be employed to guide the screening process.
In the set of 12 new cocrystal structures for Praziquantel, four structural classes can be identified based on the packing of PZQ molecules and the hydrogen bonding patterns present. Similar to the PZQ cocrystals with adipic, glutaric, succinic, oxalic, maleic, and fumaric acid reported by Espinosa-Lara et al.29 (coformers b–d and f–h, respectively, in Figure 2), coformers containing two hydrogen bond donors usually fulfill an effective bridging function as part of enantiopure chains of PZQ molecules. Some of the coformers in these chains appear to be interchangeable, leading to remarkable similarities in the obtained unit cell parameters and intermolecular distances. Apparently, this feature, which is not exclusive to cocrystal forms of PZQ, is embedded in the coformer network and can successfully be extracted via link prediction. Additionally, these forms highlight a reliable pathway for the cocrystal formation of PZQ with coformers characterized by a similar hydrogen bond donor site separation and geometry. Differences in the substitution pattern of hydroxybenzoic acid derivatives can, however, result in a change in the structural class (e.g., containing racemic instead of enantiopure chains), demonstrating the flexibility of PZQ to adopt various packings and adapt to a specific coformer. Surprisingly, this adaptability of PZQ for different types of coformers is also stored in the network.
It can be envisaged that, besides solvate and hydrate formation, cocrystal polymorphism is possible for PZQ given the various structural classes observed. Whereas single-donor molecules such as salicylic (5) and 3,5-dinitrobenzoic acid (13) must rely on the aggregation of PZQ as in Figure 8a, cocrystal formation with vanillic acid (20) can in principle lead to different crystal packings. The different phases found for vanillic acid during cocrystal screening are therefore probably the result of cocrystal polymorphism. The same could be assumed for 2,4-dihydroxybenzoic acid (28).
The structural exploration of the cocrystals of PZQ that were predicted by link prediction revealed the dynamics and flexibility of PZQ when it forms cocrystals. It also demonstrates that information on cocrystal formation, such as that present in the coformer network derived from the CSD, can not always easily be replaced by chemical intuition. This underlines the added value of our data-driven cocrystal prediction method based on link prediction applied to CSD data.
The cocrystal structures with achiral coformers studied here are all centrosymmetric (containing both the (R)- and (S)-enantiomer of PZQ). For the purpose of enantioseparation using preferential crystallization or deracemization methods, enantiomorphic structural classes different from those discussed here should be discovered. Also, whereas not all of the 30 considered coformers can be found on the so-called GRAS listc, this study has unveiled several examples that contain acceptable or drug-like molecules (e.g., salicylic acid, 4-hydroxybenzoic acid, and 2,4- and 2,5-dihydroxybenzoic acid) for which the pharmaceutical applicability may be further investigated.
5. Conclusion
Using the information on eight known PZQ cocrystals present in the CSD, network-based link prediction was successfully used to propose several new coformers for PZQ. Seventeen experimental indications for cocrystals were obtained, and single crystals for 12 of the indications were resolved by SC-XRD. Of these new cocrystalline materials, eight were found to be two-component cocrystals, and four of them were found to be cocrystal solvates, which is in line with the estimated nine new combinations statistically expected from the predicted precision values. A classification of the PZQ cocrystals was performed based on the packing of the enantiomers and intermolecular interactions between the coformers and PZQ, demonstrating the concept of interchangeable, similar coformers.
This case study for PZQ showed how knowledge of a limited set of known coformers for a target compound can be used to generate meaningful network-based predictions for new coformers given the cocrystal information in the CSD. Experimental cocrystallization data gained from subsequent cocrystal screening studies can easily be introduced to the method as additional information, generating an even larger and more precise list of predictions. We therefore envisage the approach to be useful to researchers aiming to expand the cocrystal landscape of target compounds on the basis of a limited availability of in-house cocrystallization data.
Acknowledgments
The authors thank Sander van Lith for his contribution to the experimental work and Dr. David Maillard (Merck) for his involvement and fruitful discussions. This research received funding as part of the CORE ITN Project by the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie Grant Agreement no. 722456 CORE ITN.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.cgd.1c00211.
Materials, experimental techniques and conditions, powder diffractograms for new conformer hits, crystallographic data, and discussion of the new crystal structures (PDF)
Accession Codes
CCDC 2054486–2054497 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif, or by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44 1223 336033.
The authors declare no competing financial interest.
Footnotes
The Biopharmaceutics Classification System classifies drugs into four categories based on their solubility and permeability. Class II corresponds to low solubility and high permeability.
In principle, nine PZQ cocrystals can be found in the CSD at the time of writing this article, seven of which are with dicarboxylic acids, one of which is with l-malic acid, and one of which is with d-malic acid. However, the network approach represents cocrystals with chiral molecules by using a single enantiomer as one of its nodes (see ref (46)), reducing the total number to eight.
Generally recognized as safe. This is a list published by the FDA specifying which substances are approved to be added to drugs or food.
Supplementary Material
References
- Karpinski P. H. Polymorphism of Active Pharmaceutical Ingredients. Chem. Eng. Technol. 2006, 29, 233–237. 10.1002/ceat.200500397. [DOI] [Google Scholar]
- Newman A. Specialized Solid Form Screening Techniques. Org. Process Res. Dev. 2013, 17, 457–471. 10.1021/op300241f. [DOI] [Google Scholar]
- Newman A. W.; Childs S. L.; Cowans B. A.. Salt and Cocrystal Form Selection. In Preclinical Development Handbook; Gad S. C., Ph.D., D.A.B.T., Ed.; John Wiley & Sons, Ltd: Hoboken, NJ, 2008; pp 455–481. [Google Scholar]
- Aitipamula S.; et al. Polymorphs, Salts, and Cocrystals: What’s in a Name?. Cryst. Growth Des. 2012, 12, 2147–2152. 10.1021/cg3002948. [DOI] [Google Scholar]
- Grothe E.; Meekes H.; Vlieg E.; ter Horst J. H.; de Gelder R. Solvates, Salts, and Cocrystals: A Proposal for a Feasible Classification System. Cryst. Growth Des. 2016, 16, 3237–3243. 10.1021/acs.cgd.6b00200. [DOI] [Google Scholar]
- Disordered Pharmaceutical Materials; Descamps M, Ed.; Wiley-VCH: Weinheim, Germany, 2016. [Google Scholar]
- Dengale S. J.; Grohganz H.; Rades T.; Löbmann K. Recent advances in co-amorphous drug formulations. Adv. Drug Delivery Rev. 2016, 100, 116–125. 10.1016/j.addr.2015.12.009. [DOI] [PubMed] [Google Scholar]
- Taylor C. R.; Day G. M. Evaluating the Energetic Driving Force for Cocrystal Formation. Cryst. Growth Des. 2018, 18, 892–904. 10.1021/acs.cgd.7b01375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H.From Racemates to Single Enantiomers - Chiral Synthetic Drugs over the Last 20 Years. In Novel Optical Resolution Technologies; Sakai K., Hirayama N., Tamura R., Eds.; Springer Berlin Heidelberg: Berlin, Germany, 2007; pp 273–299. [DOI] [PubMed] [Google Scholar]
- Calcaterra A.; D'Acquarica I. The market of chiral drugs: Chiral switches versus de novo enantiomerically pure compounds. J. Pharm. Biomed. Anal. 2018, 147, 323–340. 10.1016/j.jpba.2017.07.008. [DOI] [PubMed] [Google Scholar]
- Jacques J.; Collet A.; Wilen S. H.. Enantiomers, Racemates, And Resolutions; Krieger Publishing Company: Malabar, FL, 1994. [Google Scholar]
- Lorenz H.; Seidel-Morgenstern A. Processes To Separate Enantiomers. Angew. Chem., Int. Ed. 2014, 53, 1218–1250. 10.1002/anie.201302823. [DOI] [PubMed] [Google Scholar]
- Elacqua E.Supramolecular chemistry of molecular concepts: Tautomers, chirality, protecting groups, trisubstituted olefins, cyclophanes, and their impact on the organic solid state. Ph.D. Dissertation, The University of Iowa, Iowa City, IA, 2012. 10.17077/etd.a63mwy7p. [DOI] [Google Scholar]
- Neurohr C.; Marchivie M.; Lecomte S.; Cartigny Y.; Couvrat N.; Sanselme M.; Subra-Paternault P. Naproxen-Nicotinamide Cocrystals: Racemic and Conglomerate Structures Generated by CO2 Antisolvent Crystallization. Cryst. Growth Des. 2015, 15, 4616–4626. 10.1021/acs.cgd.5b00876. [DOI] [Google Scholar]
- Villamil O. F.Preferential crystallization of a racemic compound via its conglomerate co-crystals. M.S. Thesis, Technische Universiteit Delft, Delft, The Netherlands, 2016. http://resolver.tudelft.nl/uuid:14154bd2-d5a5-46bb-8d37-172725d6953b. [Google Scholar]
- Harfouche L. C.; Couvrat N.; Sanselme M.; Brandel C.; Cartigny Y.; Petit S.; Coquerel G. Discovery of New Proxyphylline-Based Chiral Cocrystals: Solid State Landscape and Dehydration Mechanism. Cryst. Growth Des. 2020, 20, 3842–3850. 10.1021/acs.cgd.0c00149. [DOI] [Google Scholar]
- Coquerel G.Preferential Crystallization. In Novel Optical Resolution Technologies; Sakai K., Hirayama N., Tamura R., Eds.; Springer Berlin Heidelberg: Berlin, Germany, 2007; pp 1–51. [Google Scholar]
- Noorduin W.; Vlieg E.; Kellogg R.; Kaptein B. From Ostwald Ripening to Single Chirality. Angew. Chem., Int. Ed. 2009, 48, 9600–9606. 10.1002/anie.200905215. [DOI] [PubMed] [Google Scholar]
- Suwannasang K.; Flood A. E.; Rougeot C.; Coquerel G. Using Programmed HeatingCooling Cycles with Racemization in Solution for Complete Symmetry Breaking of a Conglomerate Forming System. Cryst. Growth Des. 2013, 13, 3498–3504. 10.1021/cg400436r. [DOI] [Google Scholar]
- Sögütoglu L.-C.; Steendam R. R. E.; Meekes H.; Vlieg E.; Rutjes F. P. J. T. Viedma ripening: a reliable crystallisation method to reach single chirality. Chem. Soc. Rev. 2015, 44, 6723–6732. 10.1039/C5CS00196J. [DOI] [PubMed] [Google Scholar]
- Li W. W.; Spix L.; de Reus S. C. A.; Meekes H.; Kramer H. J. M.; Vlieg E.; ter Horst J. H. Deracemization of a Racemic Compound via Its Conglomerate-Forming Salt Using Temperature Cycling. Cryst. Growth Des. 2016, 16, 5563–5570. 10.1021/acs.cgd.6b01034. [DOI] [Google Scholar]
- Belletti G.; Tortora C.; Mellema I. D.; Tinnemans P.; Meekes H.; Rutjes F. P. J. T.; Tsogoeva S. B.; Vlieg E. Photoracemization-Based Viedma Ripening of a BINOL Derivative. Chem. - Eur. J. 2020, 26, 839–844. 10.1002/chem.201904382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfouche L. C.; Brandel C.; Cartigny Y.; Petit S.; Coquerel G. Resolution by Preferential Crystallization of Proxyphylline by Using Its Salicylic Acid Monohydrate Co-Crystal. Chem. Eng. Technol. 2020, 43, 1093–1098. 10.1002/ceat.202000040. [DOI] [Google Scholar]
- Buol X.; Caro Garrido C.; Robeyns K.; Tumanov N.; Collard L.; Wouters J.; Leyssens T. Chiral Resolution of Mandelic Acid through Preferential Cocrystallization with Nefiracetam. Cryst. Growth Des. 2020, 20, 7979–7988. 10.1021/acs.cgd.0c01236. [DOI] [Google Scholar]
- Harmsen B.; Leyssens T. Enabling Enantiopurity: Combining Racemization and Dual-Drug Co-crystal Resolution. Cryst. Growth Des. 2018, 18, 3654–3660. 10.1021/acs.cgd.8b00438. [DOI] [Google Scholar]
- Harmsen B.; Leyssens T. Dual-Drug Chiral Resolution: Enantiospecific Cocrystallization of (S)-Ibuprofen Using Levetiracetam. Cryst. Growth Des. 2018, 18, 441–448. 10.1021/acs.cgd.7b01431. [DOI] [Google Scholar]
- Guillot M.; Meester J. d.; Huynen S.; Collard L.; Robeyns K.; Riant O.; Leyssens T. Cocrystallization-Induced Spontaneous Deracemization: A General Thermodynamic Approach to Deracemization. Angew. Chem., Int. Ed. 2020, 59, 11303–11306. 10.1002/anie.202002464. [DOI] [PubMed] [Google Scholar]
- Andrews P. Praziquantel: mechanisms of anti-schistosomal activity. Pharmacol. Ther. 1985, 29, 129–156. 10.1016/0163-7258(85)90020-8. [DOI] [PubMed] [Google Scholar]
- Espinosa-Lara J. C.; Guzman-Villanueva D.; Arenas-García J. I.; Herrera-Ruiz D.; Rivera-Islas J.; Román-Bravo P.; Morales-Rojas H.; Höpfl H. Cocrystals of Active Pharmaceutical Ingredients - Praziquantel in Combination with Oxalic, Malonic, Succinic, Maleic, Fumaric, Glutaric, Adipic, And Pimelic Acids. Cryst. Growth Des. 2013, 13, 169–185. 10.1021/cg301314w. [DOI] [Google Scholar]
- Borrego-Sánchez A.; Viseras C.; Aguzzi C.; Sainz-Díaz C. I. Molecular and crystal structure of praziquantel. Spectroscopic properties and crystal polymorphism. Eur. J. Pharm. Sci. 2016, 92, 266–275. 10.1016/j.ejps.2016.04.023. [DOI] [PubMed] [Google Scholar]
- Borrego-Sánchez A.; Hernández-Laguna A.; Sainz-Díaz C. I. Molecular modeling and infrared and Raman spectroscopy of the crystal structure of the chiral antiparasitic drug Praziquantel. J. Mol. Model. 2017, 23, 106. 10.1007/s00894-017-3266-3. [DOI] [PubMed] [Google Scholar]
- Zanolla D.; Perissutti B.; Passerini N.; Chierotti M. R.; Hasa D.; Voinovich D.; Gigli L.; Demitri N.; Geremia S.; Keiser J.; Cerreia Vioglio P.; Albertini B. A new soluble and bioactive polymorph of praziquantel. Eur. J. Pharm. Biopharm. 2018, 127, 19–28. 10.1016/j.ejpb.2018.01.018. [DOI] [PubMed] [Google Scholar]
- Zanolla D.; Perissutti B.; Passerini N.; Invernizzi S.; Voinovich D.; Bertoni S.; Melegari C.; Millotti G.; Albertini B. Milling and comilling Praziquantel at cryogenic and room temperatures: Assessment of the process-induced effects on drug properties. J. Pharm. Biomed. Anal. 2018, 153, 82–89. 10.1016/j.jpba.2018.02.018. [DOI] [PubMed] [Google Scholar]
- Zanolla D.; Perissutti B.; Vioglio P. C.; Chierotti M. R.; Gigli L.; Demitri N.; Passerini N.; Albertini B.; Franceschinis E.; Keiser J.; Voinovich D. Exploring mechanochemical parameters using a DoE approach: Crystal structure solution from synchrotron XRPD and characterization of a new praziquantel polymorph. Eur. J. Pharm. Sci. 2019, 140, 105084. 10.1016/j.ejps.2019.105084. [DOI] [PubMed] [Google Scholar]
- Salazar-Rojas D.; Maggio R. M.; Kaufman T. S. Preparation and characterization of a new solid form of praziquantel, an essential anthelmintic drug. Praziquantel racemic monohydrate. Eur. J. Pharm. Sci. 2020, 146, 105267. 10.1016/j.ejps.2020.105267. [DOI] [PubMed] [Google Scholar]
- Sánchez-Guadarrama O.; Mendoza-Navarro F.; Cedillo-Cruz A.; Jung-Cook H.; Arenas-García J. I.; Delgado-Díaz A.; Herrera-Ruiz D.; Morales-Rojas H.; Höpfl H. Chiral Resolution of RS-Praziquantel via Diastereomeric Co-Crystal Pair Formation with l-Malic Acid. Cryst. Growth Des. 2016, 16, 307–314. 10.1021/acs.cgd.5b01254. [DOI] [Google Scholar]
- Cugovčan M.; Jablan J.; Lovrić J.; Cinčić D.; Galic N.; Jug M. Biopharmaceutical characterization of praziquantel cocrystals and cyclodextrin complexes prepared by grinding. J. Pharm. Biomed. Anal. 2017, 137, 42–53. 10.1016/j.jpba.2017.01.025. [DOI] [PubMed] [Google Scholar]
- Liu Y. H.; Qian M. X.; Wang X. G.; Jia J.; Wang Q. N.; Jiang Y. F.; Wang R. Q.; Yan S. H.; Chen B. Y.; Li J. S.; et al. Comparative efficacy of praziquantel and its optic isomers in experimental therapy of schistosomiasis japonica in rabbits. Chin. Med. J. 1986, 99, 935–940. [PubMed] [Google Scholar]
- Tanaka M.; Ohmae H.; Utsunomiya H.; Nara T.; Irie Y.; Yasuraoka K. A comparison of the antischistosomal effect of levo- and dextro-praziquantel on Schistosoma japonicum and S. mansoni in mice. Am. J. Trop. Med. Hyg. 1989, 41, 198–203. 10.4269/ajtmh.1989.41.198. [DOI] [PubMed] [Google Scholar]
- Shu-Hua X.; Catto B. A. Comparative in vitro and in vivo activity of racemic praziquantel and its levorotated isomer on Schistosoma mansoni. J. Infect. Dis. 1989, 159, 589–592. 10.1093/infdis/159.3.589. [DOI] [PubMed] [Google Scholar]
- Xu Z.-Y.; Hu L.-S.; Wu M.-H.; Zhang S.-J.; Chen M.; Yuan H.-C.; Wei R.-M.; Wang C.-Z.; Wu Z.-S.; Lian W.-N.; Wei C.-C.; Liu Z.-D.; Jiang Q.-W.; Yuan S.-J.; Yang Q.-J. Comparison of the therapeutic efficacy and side effects of a single dose of levo-praziquantel with mixed isomer praziquantel in 278 cases of schistosomiasis japonica. Am. J. Trop. Med. Hyg. 1991, 45, 345–349. 10.4269/ajtmh.1991.45.345. [DOI] [PubMed] [Google Scholar]
- Meyer T.; Sekljic H.; Fuchs S.; Bothe H.; Schollmeyer D.; Miculka C. Taste, A New Incentive to Switch to (R)-Praziquantel in Schistosomiasis Treatment. PLoS Neglected Trop. Dis. 2009, 3, e357. 10.1371/journal.pntd.0000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle M.; Seerden J.-P.; de Gooijer J.; Pouwer K.; Olliaro P.; Todd M. H. Resolution of Praziquantel. PLoS Neglected Trop. Dis. 2011, 5, e1260. 10.1371/journal.pntd.0001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti G.; Tinnemans P.; Baglai I.; Noorduin W. L.; Kaptein B.; Leeman M.; ter Horst J. H.; Kellogg R. M. Combining incompatible processes for deracemization of a Praziquantel derivative under flow conditions. Angew. Chem., Int. Ed. 2021, 60, 5279. 10.1002/anie.202013502. [DOI] [PubMed] [Google Scholar]
- Groom C. R.; Bruno I. J.; Lightfoot M. P.; Ward S. C. The Cambridge Structural Database. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2016, B72, 171–179. 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devogelaer J. J.; Brugman S. J. T.; Meekes H.; Tinnemans P.; Vlieg E.; de Gelder R. Cocrystal design by network-based link prediction. CrystEngComm 2019, 21, 6875–6885. 10.1039/C9CE01110B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devogelaer J. J.; Meekes H.; Vlieg E.; de Gelder R. Cocrystals in the Cambridge Structural Database: a network approach. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2019, B75, 371–383. 10.1107/S2052520619004694. [DOI] [PubMed] [Google Scholar]
- Bruno I. J.; Cole J. C.; Edgington P. R.; Kessler M.; Macrae C. F.; McCabe P.; Pearson J.; Taylor R. New software for searching the Cambridge Structural Database and visualizing crystal structures. Acta Crystallogr., Sect. B: Struct. Sci. 2002, B58, 389–397. 10.1107/S0108768102003324. [DOI] [PubMed] [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 33. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devogelaer J. J.; Meekes H.; Tinnemans P.; Vlieg E.; de Gelder R. Co-crystal Prediction by Artificial Neural Networks. Angew. Chem., Int. Ed. 2020, 59, 21711–21718. 10.1002/anie.202009467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.