Abstract
We describe cross-reactive human antibodies recognizing influenza B viruses spanning nearly 80 years of antigenic drift. Structures show that they engage the receptor-binding site (RBS) of the viral hemagglutinin with strong similarities to their influenza A counterparts, despite structural differences between the RBS of influenza A and B. Our data show that these antibodies readily cross-react with both influenza B Victoria and Yamagata lineages. We also note that all antibodies are encoded by IGHV3-9/IGK1-33. Future research will provide insight into the prevalence of these antibodies in the human population.
Keywords: influenza vaccine, hemagglutinin, B-cell memory
Graphical Abstract
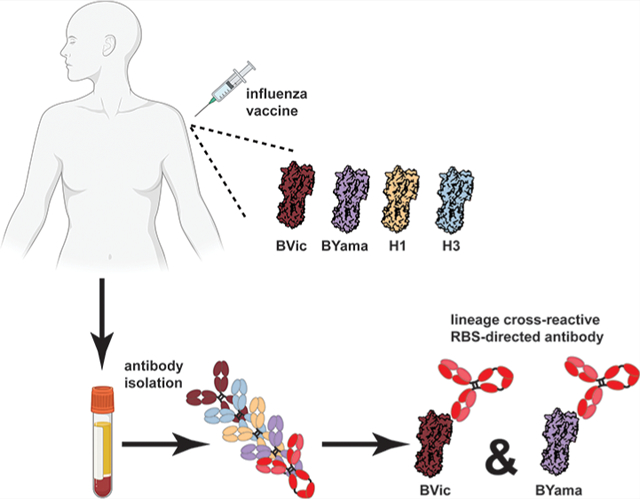
The substantial morbidity and mortality from influenza viral infections have prompted intensive efforts to design more broadly effective vaccines.1 Two influenza A subtypes, H1N1 and H3N2, and two influenza B lineages, Victoria and Yamagata, currently cocirculate in the human population.2 Influenza B viruses derive from a common ancestral strain that evolved into two antigenically distinct lineages in the 1980s.3 Influenza B infections have recently increased and now surpass those by H1N1 influenza A viruses, especially in infants.4 Traditional vaccine approaches have historically centered on the circulating H1 and H3 influenza A strains, but influenza B viruses now elicit almost equal attention.
Influenza hemagglutinin (HA) is both the attachment protein recognizing sialic acid on host cells and the viral fusogen;5,6 it is the more abundant of the two glycoproteins on the virion surface.7 The characterization of B-cell responses to HAs of influenza A has identified conserved epitopes on the viral glycoprotein—the receptor-binding site (RBS), the head interface, and the membrane-proximal stem—and has yielded antibodies, the so-called broadly protective antibodies (bpAbs), that recognize a wide range of strains.8–15 We, and others, have identified bpAbs that target the receptor-binding site (RBS)12,13,15 or the head interface epitope on influenza A HA.8,9,14 For the former class, we showed that these Abs mimic the HA receptor, sialic acid, by providing a critical dipeptide on the tip of their heavy-chain complementarity determining region 3 (HCDR3). For the latter class, we have found diverse ways to recognize a core epitope in the 220-loop of HA. Comparably detailed structural analyses of RBS-directed antibodies against influenza B virus HA have not yet been reported.
We examined paired heavy- and light-chain antibody sequences from plasmablasts of human donors administered trivalent, inactivated seasonal vaccines from 2007 to 2008 (H1 Solomon Islands/03/2006, H3 Wisconsin/67/2005, and B/Malaysia/06/2004) or 2008 to 2009 (H1 Brisbane/59/2007, H3 Uruguay/716/2007, and B/Florida/04/2006). We previously reported influenza A-reactive antibodies from donors in this cohort.16,17 From these donors, we identified antibodies that bind HAs from both the Yamagata and Victoria influenza B lineages (Figures 1A and S1). Using vaccine HA components B/Malaysia/06/2004 and B/Florida/04/2006, we identified a three-membered antibody lineage “1261” comprising antibodies H1207, H1209, and H1235 (Figure 1C) as well as two “orphan” Abs H1272 and H2365. We selected one 1261 lineage member and the two orphan Abs for further biochemical characterization. We expressed and purified Fabs (to avoid any avidity effects) and measured affinities to monomeric HA1 “heads” using biolayer interferometry (BLI). All three Fabs cross-reacted with B/Yamagata and B/Victoria lineages and bound all HAs tested with variable affinities ranging from low nM to μM (Figures 1B and S2).
Figure 1.
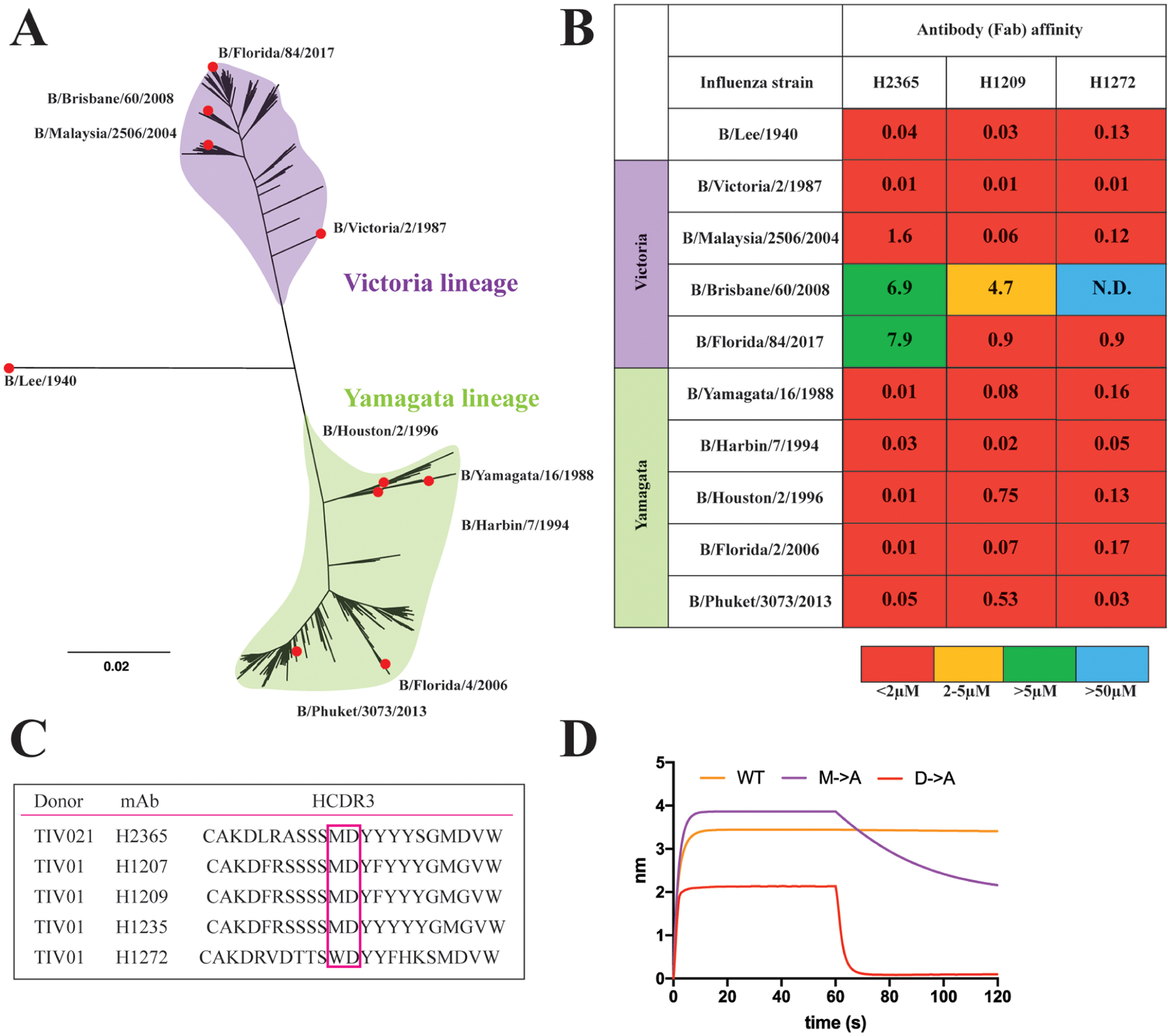
Influenza B hemagglutinin phylogeny and cross-lineage binding antibodies. (A) Phylogenetic tree of influenza B viruses rooted on the ancestral B/Lee/1940 sequence. The divergent, cocirculating lineages Victoria and Yamagata are highlighted in purple and green, respectively. At the tips of the branches, highlighted with red circles, are the influenza B seasonal strains whose recombinant HA proteins were tested for binding with the antibodies. (B) Affinity measurements of the Fab to monomeric HA heads. The “heatmap” color scheme is an arbitrary visualization aid. Warm colors are high affinity and cool colors, low affinity. The calculated KD values are reported in μM. (C) Sequence alignment of the antibody heavy complementarity determining region 3 (HCDR3) loops of the 5 antibodies isolated from 2 donors. The critical dipeptide motif is highlighted. (D) Biolayer interferometry binding isotherms for the H2365 wild-type (WT) and its mutants Met102Ala (M → A) and Asp103Ala (D → A) binding to the B/Phuket/3073/2013 HA head.
The isolated antibodies provide exceptional breadth by recognizing historical HAs from the 1940s to today. Both the lineage and orphan Abs have relatively long, 20-residue HCDR3s, with a central dipeptide motif of a hydrophobic and an acidic residue at its tip (Figure 1C). As in the case of influenza A RBS-directed Abs, this dipeptide would likely make RBS contacts that mimic those made by sialic acid. We therefore tested the impact on affinity to HA by replacing the hydrophobic (Met) or acidic (Asp) residues in H2365 with alanine (Figure 1D). Both substitutions lowered the affinity (i.e., increased overall KD) by accelerating the dissociation rate, indicating that both the hydrophobic and the acidic moieties in the HCDR3 are necessary for strong binding to the HA RBS. The Asp substitution had a more pronounced effect than the hydrophobic change.
We determined the crystal structures of all three antibodies in complex with HA (Figure 2C,E,G, Table S1). The Abs engage the viral RBS, converging on a mode of molecular recognition similar to the modes of their influenza A counterparts (compare Figure 2C,E,G with Figure 2A). The central recognition element is a crucial dipeptide at the tip of HCDR3 that mimics many of the sialic acid contacts (Figures 2D,F,H and S4). The antigen-combining site includes additional contacts between HCDR2 and the HA 190-helix and between LCDR3 and the HA 150-loop (Figure S3). In the critical dipeptide, Asp supplies hydrogen bonds to the conserved Ser140 and to Gln239, thus mimicking the carboxylic acid moiety of sialic acid (Figure 2B), while the hydrophobic residue Met or Trp of the antibody contacts the conserved Trp158 at the base of the RBS, mimicking the –CH3 of the sialic acid acetamido group (Figure 2D,F,H). A noted difference in the influenza B HA RBS is a Phe at position 95 (Figure 2D,F,H) instead of the Tyr conserved in influenza A. B HAs thus contain one less hydrogen bond donor in their RBS. This evolutionarily conserved substitution reduces the affinity for the sialic acid receptor.18 Our study suggests this has no effect on eliciting the RBS-directed antibodies.
Figure 2.
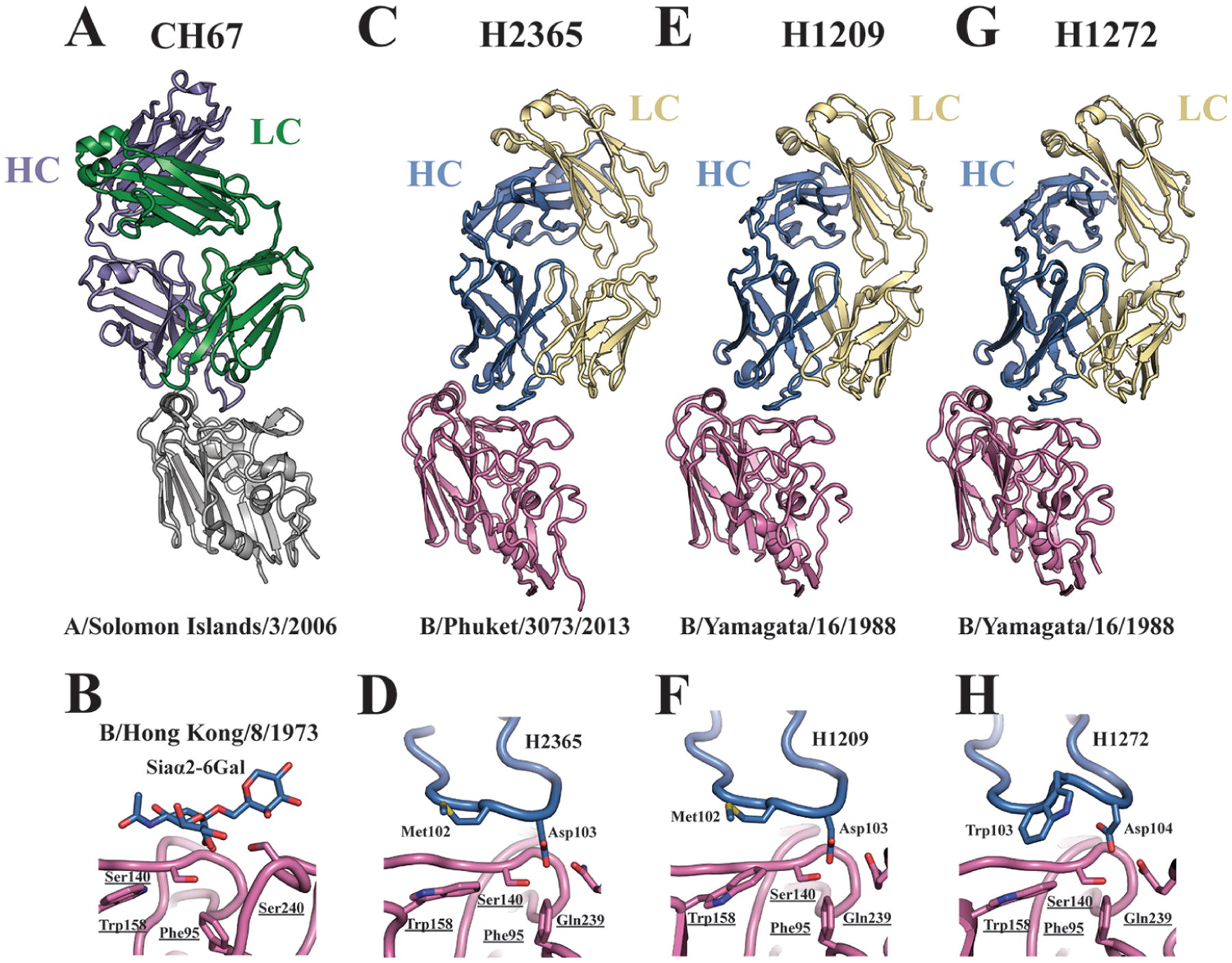
Structural analyses of the antibody–hemagglutinin complexes. CH67 Fabs in complex with an influenza A HA head (PDB ID 4HKX) is shown for reference in (A). Antibody receptor mimicry through the critical dipeptide motif is made in comparison with a sialic acid bound HA structure of B/Hong Kong/8/1973 (PDB ID: 2RFU) shown in (B). Overall structures of the H2365 (C), H1209 (E), and H1272 (G) Fabs in complex with influenza B HA heads. Essential HCDR3 amino acid contacts are detailed in (D), (F), and (H). The orientation of the structures in (D), (F), and (H) is as looking from the left, i.e., from the 190-helix in (C), (E), and (G), respectively. Essential HA residues are underlined.
It is worth noting that the contemporary B Victoria lineage viruses evolved during the 2016/2017 season to incorporate two (K162/N163) or three (K162/N163/D164) amino acid deletions. Recent studies showed that these viruses are antigenically distinct from each other and from the progenitor virus that lacks the deletions.19 One representative virus of the K162/N163 cluster is the B/Florida/84/2017 isolate (Figure S5A). All antibodies tested in this study preserve binding to that isolate (Figure 1B) indicating that individuals with such antibodies would most likely still be protected against the current B Victoria strains. Another feature of this strain is a putative N-linked glycosylation site at position 197 (Figure S5A) that could potentially interfere with antibody binding (Figure S5B). The historical HAs used in this study do not have a glycosite at that position, but the structures presented here indicate that, even if that particular site were glycosylated, it would most likely not impede the binding of our antibodies. For reference, we found that a previously characterized Crucell antibody CR8033 failed to bind the B/Florida/84/2017 HA (Figure S5C).
All 5 antibodies identified in this study derive from the VH3-9 gene recombined with JH6. We also note that, in all cases, the antibody heavy-chains pair with the same light kappa-chain IGK1-33. We speculate that the potential germline bias stems from two principal requirements: (1) engagement of the 190-helix through VH3-9 encoded HCDR2 and (2) the length of the HCDR3 provided by the JH6 gene segment. Further studies on much larger donor cohorts are necessary to corroborate these speculations.
Our study identifies the molecular signatures of human antibodies that engage influenza B HA by receptor mimicry and suggests a therapeutic potential for such RBS-directed antibodies. We also raise the possibility of a “universal” vaccine that elicits RBS-directed antibodies against both A and B influenza viruses.
METHODS
Expression and Purification of HA.
Influenza B HA1 “head” constructs were cloned into a pFastBac vector for insect cell expression (Hi5 cells) as previously described.13,20 Of note, these insect cell produced HAs do not contain sialic acids on their N-linked glycans and are therefore better mimics of HAs on flu viruses than would be the case had they been expressed in mammalian cells. All constructs were confirmed by DNA sequencing at the DNA Sequencing Core Facility at the Dana Farber Cancer Institute. For biolayer interferometry (BLI) and crystallography, the HA1 head constructs contained an HRV 3C-cleavable C-terminal 6xHis tag. All constructs were purified from the supernatants by passage over Co2+-NTA TALON resin (Takara) followed by gel filtration chromatography on a Superdex 200 Increase (GE Healthcare) in 10 mM Tris-HCl and 150 mM NaCl at pH 7.5. For BLI and crystallography, the tags were removed using HRV 3C protease (ThermoScientific) and the protein was repurified using the Co2+-NTA TALON resin to remove the protease, tag, and noncleaved protein.
Fab Expression and Purification.
For Fab production, the genes for the heavy- and light-chain variable domains were synthesized and codon optimized by Integrated DNA Technologies and subcloned into pVRC protein expression vectors containing human heavy- and light-chain constant domains, as previously described.13,20 Heavy-chain constructs for Fab production contained an HRV 3C-cleavable 6xHis tag. Constructs were confirmed by sequencing at the DNA Sequencing Core Facility at the Dana Farber Cancer Institute. Fabs were produced by transient transfection in a suspension of 293F cells using polyethylenamine (PEI, Polysciences). Supernatants were harvested 4–5 days later and clarified by centrifugation. Fabs were purified using Co2+-NTA TALON resin (Takara) followed by gel filtration chromatography on a Superdex 200 Increase (GE Healthcare) in 10 mM Tris-HCl and 150 mM NaCl at pH 7.5. For BLI and crystallography, the tags were removed using HRV 3C protease (ThermoScientific) and the protein was repurified using Co2+-NTA TALON resin to remove the protease, tag, and noncleaved protein.
Interferometry Binding Experiments.
Interferometry experiments were performed using a BLItz instrument (fortéBIO Pall Corporation). Histidine-tagged HA heads or full-length HAs were immobilized on a Ni2+-NTA biosensor, and cleaved Fabs were then applied to obtain binding affinities. Single-hit concentrations were tested at 20 μM for binding. All measurements were repeated in independent experiments. KD was obtained through local fit of the curves by applying a 1:1 binding isotherm model using vendor-supplied software. All experiments were performed in 10 mM Tris-HCl and 150 mM NaCl at pH 7.5 and at room temperature.
Crystallization and Data Collection.
Influenza B HA1 heads and Fabs were incubated at a 1:1.5 molar ratio, respectively. The complex was isolated by size exclusion chromatography using a 24 mL Superdex Increase equilibrated in 10 mM Tris-HCl and 150 mM NaCl. Crystallization was achieved at 15–18 mg/mL of the complex by hanging drop vapor diffusion at 18 °C. Crystals were grown as follows: the H1209 complex in 200 mM sodium citrate, pH 7.0, with 20% (wt/vol) PEG 3350; the H2365 complex in 100 mM HEPES, pH 7.5, with 25% (wt/vol) PEG 3350; the H2365 complex in 100 mM bis-Tris, pH 6.5, with 20% PEG MME 5000. Crystals were cryoprotected in mother liquor supplemented with 25% (v/v) glycerol and flash-frozen in liquid nitrogen. Data were collected at 0.999 Å with a rotation of 1° per image on the 8.2.2 beamline, Advanced Light Source, at Berkley National Laboratory or on the beamline ID-24-C at the Advanced Photon Source (Argonne National Laboratory).
Structure Determination and Analysis.
X-ray diffraction data were processed using XDS.21 While CC1/2 (at 0.1% significance level) was used to select the resolution cutoff for all data sets, unusually high R factors were noted for the H1209 antibody complex. Indeed, the data processing and structure refinement statistics become poor at resolutions better than 5 Å. The sequence and the structure similarity with the H2365 complex, for which the diffraction data were of better quality, however, make us confident about the structural analyses and the conclusions we draw from such analyses. We calculated the composite annealed omit maps for the H2365 and the H1209 complexes (Figure S2) to illustrate the confidence of tracing the Fab HCDR3 loops in the electron density. The structures were determined by molecular replacement using PHASER22,23 with the B/Florida/4/2006 HA1 head (PDB ID: 4FQJ) and Fab CR8033 (PDB ID: 4FQL) as search models.24 Refinement was performed using PHENIX.25 Model building was done in COOT26 and assessed with MolProbity.27 N-Linked glycan stereochemistry was assessed with Privateer.28 Figures were generated using the PyMOL Molecular Graphics System (v2.4.0; Schrödinger LLC).
Supplementary Material
ACKNOWLEDGMENTS
We thank Kevin McCarthy and Aaron Schmidt for helpful discussions and Tori Duback and Max Maron for help with the binding measurements. BLI affinity measurements were carried out in the Center for Macromolecular Interactions at Harvard Medical School, directed by Kelly Arnett. We thank the beamline staff at the Advanced Light Source (Lawrence Berkeley Laboratory) and at the Advanced Photon Source (Argonne National Laboratory) for assistance in recording of the X-ray diffraction data. Sequencing reactions were carried out with an ABI3730xl DNA analyzer at the DNA Resource Core of the Dana-Farber/Harvard Cancer Center (funded in part by NCI Cancer Center support grant 2P30CA006516-48). This research was supported by NIH grant P01 AI089618 (to S.C.H.).
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00726.
Antibody sequence alignments; details of the affinity measurements and molecular interactions; composite annealed omit maps of the Fab HCDR3; antigenic evolution of contemporary B Victoria HAs; data collection and refinement statistics (PDF)
Accession Codes
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 7KQG, 7KQH, and 7KQI for the H1272, H2365, and H1209 complexes, respectively.
Complete contact information is available at: https://pubs.acs.org/10.1021/acsinfecdis.0c00726
Contributor Information
Goran Bajic, Laboratory of Molecular Medicine, Boston Children’s Hospital and Department of Pediatrics, Harvard Medical School, Boston, Massachusetts 02115, United States;.
Stephen C. Harrison, Laboratory of Molecular Medicine, Boston Children’s Hospital, Harvard Medical School, Boston, Massachusetts 02115, United States; Howard Hughes Medical Institute, Boston, Massachusetts 02115, United States
REFERENCES
- (1).Krammer F, and Palese P (2015) Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discovery 14 (3), 167–182. [DOI] [PubMed] [Google Scholar]
- (2).Taubenberger JK, and Morens DM (2010) Influenza: the once and future pandemic. Public Health Rep. 125 (Suppl 3), 15–26. [PMC free article] [PubMed] [Google Scholar]
- (3).Yamashita M, Krystal M, Fitch WM, and Palese P (1988) Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology 163 (1), 112–22. [DOI] [PubMed] [Google Scholar]
- (4).Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, and Fukuda K (2004) Influenza-associated hospitalizations in the United States. JAMA 292 (11), 1333–40. [DOI] [PubMed] [Google Scholar]
- (5).Skehel JJ, and Wiley DC (2000) Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem 69, 531–69. [DOI] [PubMed] [Google Scholar]
- (6).Matrosovich MN, Gambaryan AS, and Chumakov MP (1992) Influenza viruses differ in recognition of 4-O-acetyl substitution of sialic acid receptor determinant. Virology 188 (2), 854–8. [DOI] [PubMed] [Google Scholar]
- (7).Wasilewski S, Calder LJ, Grant T, and Rosenthal PB (2012) Distribution of surface glycoproteins on influenza A virus determined by electron cryotomography. Vaccine 30 (51), 7368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, Sempowski GD, Takahashi Y, Kelsoe G, Kuraoka M, and Schmidt AG (2019) Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe 25 (6), 827–835.E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, Gilchuk I, Turner HL, García-Sastre A, Li S, Ward AB, Wilson IA, and Crowe JE Jr (2019) A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell 177 (5), 1136–1152.E18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, and Lanzavecchia A (2011) A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333 (6044), 850–6. [DOI] [PubMed] [Google Scholar]
- (11).Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, and Wilson IA (2009) Antibody recognition of a highly conserved influenza virus epitope. Science 324 (5924), 246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, Kepler TB, Schmidt AG, Kelsoe G, and Harrison SC (2018) Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity 48 (1), 174–184.E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, and Harrison SC (2015) Viral receptor-binding site antibodies with diverse germline origins. Cell 161 (5), 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, McGee CE, Sempowski GD, Feng F, Urick P, Kepler TB, Takahashi Y, Harrison SC, and Kelsoe G (2019) Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell 177 (5), 1124–1135.E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, and Harrison SC (2011) Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A 108 (34), 14216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, Chen X, Munshaw S, Marshall DJ, Whitesides JF, Drinker MS, Amos JD, Gurley TC, Eudailey JA, Foulger A, DeRosa KR, Parks R, Meyerhoff RR, Yu JS, Kozink DM, Barefoot BE, Ramsburg EA, Khurana S, Golding H, Vandergrift NA, Alam SM, Tomaras GD, Kepler TB, Kelsoe G, Liao HX, and Haynes BF (2011) H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One 6 (10), No. e25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Schmidt AG, Do KT, McCarthy KR, Kepler TB, Liao HX, Moody MA, Haynes BF, and Harrison SC (2015) Immunogenic Stimulus for Germline Precursors of Antibodies that Engage the Influenza Hemagglutinin Receptor-Binding Site. Cell Rep. 13 (12), 2842–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Matrosovich MN, Gambaryan AS, Tuzikov AB, Byramova NE, Mochalova LV, Golbraikh AA, Shenderovich MD, Finne J, and Bovin NV (1993) Probing of the receptor-binding sites of the H1 and H3 influenza A and influenza B virus hemagglutinins by synthetic and natural sialosides. Virology 196 (1), 111–21. [DOI] [PubMed] [Google Scholar]
- (19).Shu B, Kirby MK, Warnes C, Sessions WM, Davis WG, Liu J, Wilson MM, Lindstrom S, Wentworth DE, and Barnes JR (2020) Detection and discrimination of influenza B Victoria lineage deletion variant viruses by real-time RT-PCR. Euro Surveill 25 (41), 1900652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, Schmidt AG, and Harrison SC (2018) Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. U. S. A 115 (1), 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kabsch W (2010) Acta Crystallogr., Sect. D: Biol. Crystallogr 66 (Pt 2), 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).McCoy AJ (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr., Sect. D: Biol. Crystallogr 63 (Pt 1), 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007) Phaser crystallographic software. J. Appl. Crystallogr 40 (Pt 4), 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin Ö, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, and Friesen RH (2012) Highly conserved protective epitopes on influenza B viruses. Science 337 (6100), 1343–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, and Adams PD (2019) Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct Biol 75 (Pt 10), 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr 60 (Pt 12), 2126–2132. [DOI] [PubMed] [Google Scholar]
- (27).Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr 66 (Pt 1), 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Agirre J, Iglesias-Fernández J, Rovira C, Davies GJ, Wilson KS, and Cowtan KD (2015) Privateer: software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol 22 (11), 833–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


