Abstract
Membrane proteins play vital roles in living organisms, serving as targets for most currently prescribed drugs. Membrane protein structural biology aims to provide accurate structural information to understand their mechanisms of action. The advance of membrane protein structural biology has primarily relied on detergent-based methods over the past several decades. However, detergent-based approaches have significant drawbacks because detergents often damage the native protein–lipid interactions, which are often crucial for maintaining the natural structure and function of membrane proteins. Detergent-free methods recently have emerged as alternatives with a great promise, e.g. for high-resolution structure determinations of membrane proteins in their native cell membrane lipid environments. This minireview critically examines the current status of detergent-free methods by a comparative analysis of five groups of membrane protein structures determined using detergent-free and detergent-based methods. This analysis reveals that current detergent-free systems, such as the styrene-maleic acid lipid particles (SMALP), the diisobutyl maleic acid lipid particles (DIBMALP), and the cycloalkane-modified amphiphile polymer (CyclAPol) technologies are not better than detergent-based approaches in terms of maintenance of native cell membrane lipids on the transmembrane domain and high-resolution structure determination. However, another detergent-free technology, the native cell membrane nanoparticles (NCMN) system, demonstrated improved maintenance of native cell membrane lipids with the studied membrane proteins, and produced particles that were suitable for high-resolution structural analysis. The ongoing development of new membrane-active polymers and their optimization will facilitate the maturation of these new detergent-free systems.
Introduction
The structural complexity of cell membrane environments
Cell membranes are composed of two fundamental components — lipids and proteins. Cell membrane systems are crucial for all living organisms to survive and flourish. In the course of evolution, the eukaryotic cell membrane system has developed a more complicated architecture than that of prokaryotes. For example, many organelles such as mitochondria, chloroplast, lysosome, and endoplasmic rectum emerged in eukaryotic cells. The membrane surrounding these organelles is also uniquely featured by specific lipid components [1]. Furthermore, new types of lipids, such as cholesterol and sphingolipids, appeared in mammalian cell membrane environments [2]. Generally, biomembrane structure consists of two apposed leaflets formed mainly by phospholipids and a small portion of other lipids in the lipid bilayer; however, some archaeal membrane can be a monolayer composed of unique ether lipids [3]. The membrane proteins are embedded or adsorbed on the cell membrane system.
According to the fluid mosaic membrane model [4], the cell membrane is a laterally heterogeneous and anisotropic complex system. The complexity may be a result of coevolution between the cell membrane and membrane proteins [5]. The full picture of the cell membrane structure is more complicated when considering the diverse lipid components, various functional and structural membrane protein components, asymmetry of the lipid distribution, and how this asymmetry is employed to connect the cell membrane to the rest of the cell [6].
The structures and functions of membrane proteins depend on the native cell membrane environment
Native lipid environments are crucial for maintaining the natural structures and activities of many membrane proteins [7]. Therefore, how well we understand and can exploit the structure and function of those membrane proteins depends on how well we understand the interactions between membrane proteins and their associated native lipids. For example, receptors such as GPCRs often need cholesterol to maintain their natural structure and to elicit signaling [8–10]. Transporters, such as some members of the ABC family, need a cell membrane lipid environment to prevent uncoupled ATPase activity [11]. Channels, such as TRP and related proteins are regulated by specific lipids, such as phosphatidylinositol-4,5-bisphosphate (PIP2) [12]. Enzymes, such as respiratory complex I need lipid components to maintain their biological activity [13].
The need for dedicated detergent-free systems for membrane protein structural biology
As discussed above, the preservation of membrane protein stability and function requires a native lipid environment. Currently, for high-resolution structure determination, membrane proteins often need to be extracted first by using detergents. Indeed, for the past few decades, the advancement of membrane protein structural biology can primarily be attributed to detergents, and structure determination of membrane proteins using detergent-based approaches has led to four Nobel Prizes directly or indirectly [14–16]. However, detergent-based approaches have significant drawbacks because they often lead to over-delipidation of membrane proteins, subsequent destabilization, and loss of biological activity [15,17]. Furthermore, membrane protein stability can also be compromised by detergent interactions with soluble domains [18]. Structure-based drug discovery requires accurate structural information, and membrane proteins are today the main class of drug targets [19]. Potential structural problems of membrane proteins determined using NMR and X-ray crystallography have been recently reviewed [15,20,21]. Single-particle cryo-EM studies of membrane proteins have recently generated many new membrane protein structures, but as of now, potential structural denaturation problems have not been carefully examined. However, considering the predominant application of detergents, structural issues are inevitable. Many novel technologies have been developed to overcome the limitations of detergent-based approaches, including Nanodiscs [22], Amphipol [23], Saposin [24], and Peptidisc [25] methods. These technologies are still reliant on detergents to initially extract the membrane proteins; therefore, they are not truly detergent-free procedures. Although these novel methods can overcome some of the limitations rendered by the purely detergent methods, they still suffer from altering the native association states between membrane protein and cell membrane lipids. Considering the importance of the protein–lipid interactions for membrane protein structure and function, dedicated detergent-free systems that can extract membrane protein and associated lipids as functional units are clearly needed for membrane protein structural biology. Membrane-active polymers, such as styrene-maleic copolymers (SMAs) [26], diisobutylene maleic acid copolymers (DIBMA) [27], new Cycloalkane-Modified Amphiphilic Polymers (CyclAPol) [28], and related detergent-free technologies, such as the SMALP technology [17,26,29] and the NCMN system [30–32] have recently emerged as alternatives to detergent-based approaches that possess the great promise. Scheme 1 summarizes the current principal methods for membrane protein extraction and purification.
Scheme 1.
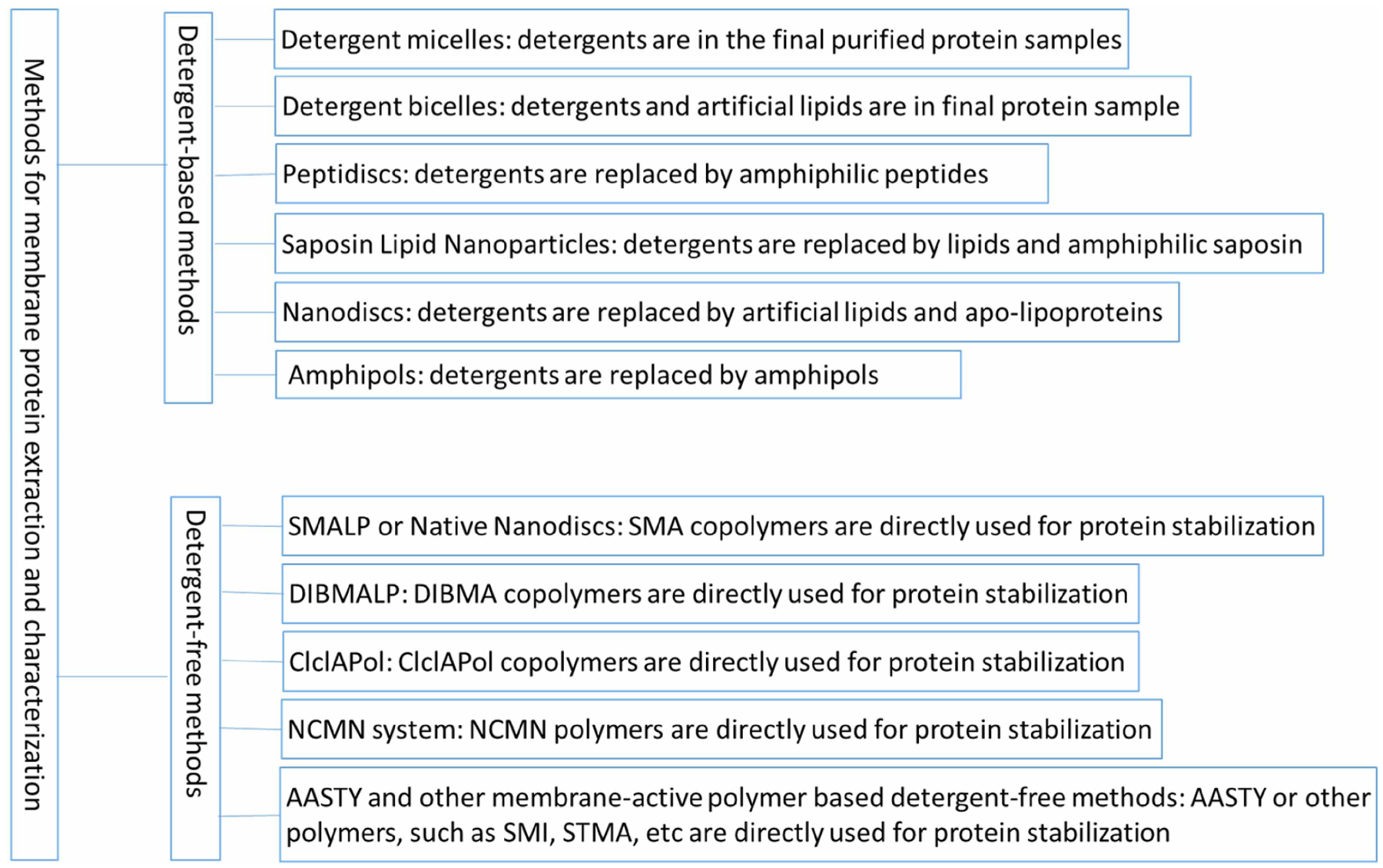
Main methods for membrane protein extraction and characterization.
Current status of the dedicated detergent-free systems
In contrast to detergents used for membrane protein structural biology, membrane-active polymers extract membrane protein but do not form micelles in aqueous solutions [33]. In 2009, the SMA was reported to extract membrane proteins [34], and thereafter considerable research was performed to solubilize and purify membrane proteins for enhanced stability and activity [35–40]. In 2017, the SMA was used to extract bacterial rhodopsin for crystallization in LCP [41]. One year later, styrene-maleic acid copolymers were demonstrated in successful high-resolution structure determinations of membrane proteins using single-particle cryo-EM [30,42]. Following this success, several high-resolution structures of different membrane proteins were reported, including the KimA potassium channel [43], acid-sensing ion channel 1 [44], and the Glycine receptor [45]. Simultaneously, novel membrane-active polymers such as DIBMA were also tested for membrane protein extraction and purification [46]. Recently, it has been reported that DIBMA could be used to determine the high-resolution cryo-EM structure of the mechanosensitive Channel YnaI from E. coli [47]. Moreover, the cycloalkane-modified amphiphile polymer (CyclAPol) has also been tested for the extraction of membrane proteins, and recent reports show that a novel CyclAPol, termed C8-C0-50, can be used for high-resolution structure determination of AcrB at 3.2 Å (Higgs A., et al., to be published). A high-resolution cryo-EM structure of human connexin 26 with associated cell membrane lipids using the NCMN system (Qiu, et al., to be published) was disclosed at the International SMALP Conferences 2020 (SMALP.net). Beyond these developments, other novel membrane-active polymers aiming to overcome the limitations of styrene-maleic acid copolymers have also been designed and demonstrated for membrane protein extraction [48,49].
To summarize, detergent-free technologies aim to overcome the limitations inherent in the detergent-based approaches, such as over-delipidation and destabilization of membrane proteins caused by detergents. In principle, a successful and dedicated detergent-free system should retain the natively associated lipids from the transmembrane domain of membrane proteins. It should also allow examining protein–lipid interactions to facilitate understanding of the structure–function relationships of membrane proteins. Correspondingly, the suitability of these new membrane-active polymers remains to be tested for high-resolution structure determination. The advance of detergent-free systems is quite promising, and clearly, all such methodologies generate some form of simulated biological construct. Thus, careful and critical examination of the structures solved with structural biological tools — single-particle cryo-EM, solid-state NMR, or X-crystallography — is required to assess the current status of detergent-free technologies.
This minireview will analyze five groups of membrane protein structures recently determined using four different detergent-free technologies. The analysis will focus on three themes: structural change, subunit association and protein–lipid interaction. The five groups of membrane proteins are AcrB, alternative complex III, KimA potassium channel, mechanosensitive channel YnaI and Glycine receptor. Relevant information on these five groups of proteins is summarized in Table 1. The four main detergent-free technologies include styrene-maleic lipid particles (SMALP), the native cell membrane nanoparticles (NCMN) system, diisobutylene maleic acid lipid particles (DIBMALP), and CyclAPols. All of these four detergent-free systems use membrane-active polymers to extract membrane proteins directly from cell membrane without using any detergent. The SMALP or alternatively Native Nanodiscs [50] is currently the most widely used detergent-free system. The NCMN system is distinct from SMALP technology: first, the NCMN system has a unique membrane-active polymer library. All the membrane-active polymers are purposely and individually developed for high-resolution structure determination of membrane proteins with diverse characteristics. Second, the NCMN system has a collection of protocols developed and optimized for each of the NCMN polymers for making high-quality NCMN particles. Third, the NCMN particles are only made from the native cell membrane, emphasizing the membrane proteins expressed by the cells from the same species. For example, human membrane proteins expressed in bacteria, yeast, or insect cell lines will not be appropriate for making NCMN particles and, therefore, will be avoided. Fourth, except for cryo-EM tomographic analysis, all downstream NCMN particle analyses, such as enzyme assays, single-particle cryo-EM, mass spectrometry, and proteoliposome reconstitution, require the use of homogenous samples that are confirmed with the electron microscope. Fifth, the NCMN system is not suitable for LCP crystallization because mixing NCMN particles with artificial lipids will disturb the native lipid–protein interaction. NCMN particles might be used for crystallization, just like soluble proteins. For comparison and evaluation of the detergent-free methods with detergent-based methods, each group of membrane proteins will be discussed in separate sections for the convenience of analysis.
Table 1.
Summary of the structures of five group of membrane proteins determined using detergent and detergent-free systems
| Group of proteins | Proteins | PDB/EMD | Function | Oligomeric states | Expression system | Species | Methods |
|---|---|---|---|---|---|---|---|
| AcrB and homologs | AcrB | 6BAJ/7074 | Transporter | Trimer | E.coli | E.coli | NCMN system |
| MtrDB | 6VKS/21228 | Transporter | Trimer | E.coli | N. onorrhoeae | Nanodisc | |
| AcrB | 6Z12/4460 | Transporter | Trimer | E.coli | S. enterica | SMALP | |
| AcrB | 6SGU/10185 | Transporter | Trimer | E.coli | E.coli | Saposin | |
| AcrB | 7B5P/12043 | Transporter | Trimer | E.coli | E.coli | CyclAPol | |
| AcrB | 1IWG | Transporter | Trimer | E.coli | E.coli | Detergent | |
| Alternative complexes | ACIII | 6BTM/7286 | Enzyme | Heterohexamer | F. johnsoniae | F. johnsoniae | SMALP |
| ACIII | 6LOD/0936 | Enzyme | Heterohexamer | R. castenholzii | R. astenholzii | Detergent | |
| ACIII | 6F0K/4165 | Enzyme | Heterohexmer | R. marinus | R. marinus | Detergent | |
| KimA | KimA | 6S3K/10092 | Transporter | Homodimer | E.coli | B. subtilis | SMALP |
| YnaI | YnaI | 6ZYD/11557 | Channel | Homoheptamer | E.coli | E.coli | DIBMA |
| YnaI | 6ZYE/11560 | Channel | Homoheptamer | E.coli | E.coli | DIBMA | |
| YnaI | 6URT/20862 | Channel | Homoheptamer | E.coli | E.coli | Detergent | |
| Glycine receptor | Glycine receptor | 6PLZ/20381 | Receptor/Channel | Homopentamer | S. frugiperda | D. rerio | SMALP |
| 6PXD/20518 | Receptor/Channel | Homopentamer | S. frugiperda | D. rerio | Detergent | ||
| 6PLS/20374 | Receptor/Channel | Homopentamer | S. frugiperda | D. rerio | Nonodisc |
Acrb and homologs
AcrB is a model membrane protein used for method development in several laboratories [24,30,51,52]. Structures of AcrB have been extensively investigated using detergent-based methods with both X-ray crystallography and cryo-EM [53–55]. However, even at a very high resolution of 1.8 Å [55], the lipid bilayer structure was lost in the detergent-prepared sample. The first single-particle cryo-EM structure of AcrB determined using SMALP technology only reached a poor resolution of 8.8 Å [56]. No lipid molecule density was observed in the reconstructed EM density map. In 2018, the first high-resolution single-particle cryo-EM structure of a lipid bilayer patch within AcrB was determined using the prototype NCMN system (Figure 1A) [30]. As the figure indicates, very robust EM density for native lipids can be observed in this structure. A novel amphipol, CyclAPol (C8-C0-50) was recently used to determine a high-resolution structure of AcrB at 3.2 Å (Higgs A., et al., to be published); yet, no lipid density was observed in the EM density. A single-particle cryo-EM structure of an E. coli AcrB homolog, gonococcal multidrug efflux pump, was solved at a high resolution of 2.7 Å using E. coli total lipids reconstituted nanodiscs (Figure 1B) [57]. In this structure, the hexagonal hydrocarbon pattern in the lipid plug’s inner leaflet position is observable; nevertheless, it is not complete, and the outer leaflet lipids were not well determined. A Salmonella AcrB cryo-EM structure at a resolution (4.6 Å) determined using SMALP technology has also been reported [58]. Again, no associated native lipid molecules were found in the EM density (Figure 1C). Similar to the lipid reconstitution using nanodisc technology, an E. coli AcrBZ complex reconstituted into saposin discs using E. coli lipids and saposin as scaffolding protein shows a similar hexagonal hydrocarbon tail pattern in the inner leaflet (Figure 1D,E) [59]. However, the outer leaflet is not well resolved, even at the same resolution of 3.2 Å. In both the lipid reconstitution cases using nanodisc and saposin technologies, although the lipids can be reconstituted to some extent, they were not the naturally associated lipids from the cell membrane. Therefore, artificial bias could be a potential concern when interpreting the protein–lipid interactions using the reconstituted system.
Figure 1. Structures of AcrB and homolog solved in detergent-based and detergent-free systems.
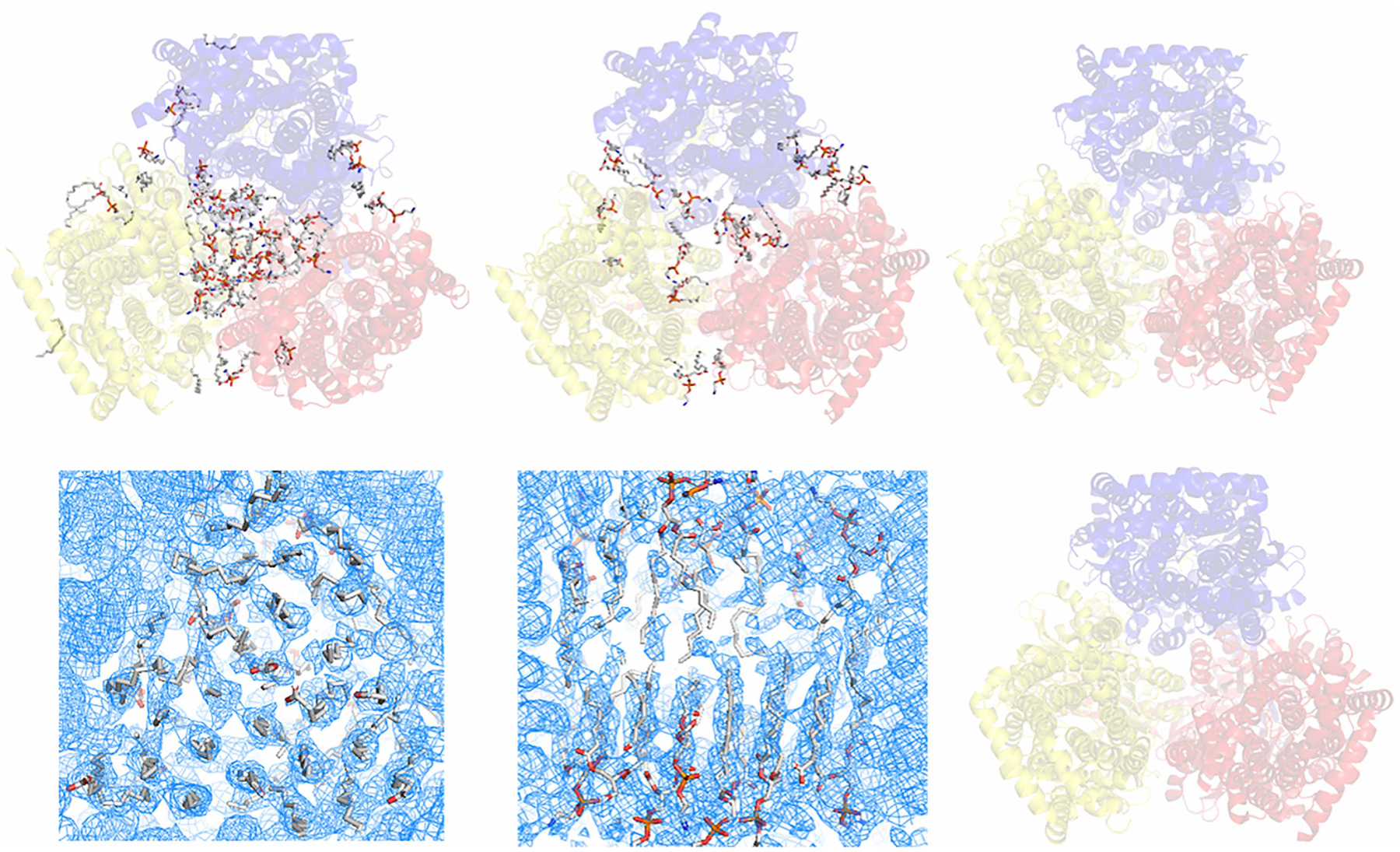
(A) Single-particle cryo-EM structure of E. coli AcrB determined using NCMN system (PDB: 6BAJ). The NCMN system retains native cell membrane lipids associated with AcrB, a native lipid bilayer patch composed of 24 lipid molecules can be unambiguously determined. (B) Single-particle cryo-EM structure of Neisseria gonorrhoeae MtrD (AcrB homolog) determined using Nanodisc technology (PDB: 6VKS). Nanodisc reconstitution using artificial lipids or total E. coli lipids could not reconstruct the native lipid bilayer patch within the lipid cavity of the transmembrane domain. (C) Single-particle cryo-EM structure of AcrB from Salmonella enterica subsp. enterica (PDB: 6Z12) determined using SMALP technology. (D,E) Single-particles cryo-EM density of E. coli AcrB determined using Saposin-lipid reconstitution (EMD:10185). Close-up view of the lipid plug (D) bottom view of the inner leaflet lipid-pattern with a similar orientation to the structure in panel A, but with a clockwise rotation around the Z-axis about 30° for a better view; (E) side view of the lipid bilayer plug, turn 90° clockwisely around the X-axis relative to the density in panel (D); top half: outer leaflet, bottom half: inner leaflet). Saposin-lipid reconstitution can partially reconstitute the lipid bilayer patch within the lipid cavity of the transmembrane domain of E. coli AcrB. However, it still could not recover the native state of the lipid bilayer structure. (F) Crystal structure of E. coli AcrB using detergent-based method. In this structure, almost all native lipids were washed away by detergents. In all the figures above, cartoons show the AcrB protein. Sticks show the lipid molecules.
Alternative complex III and homologs
Alternative complex III is an essential membrane protein component within the respiratory and/or photosynthetic electron transport chain in many bacteria [60]. Both detergent-based approaches and SMALP technology have been used to investigate the high-resolution structure of alternative complex III. The reported single-particle cryo-EM structure determined using SMALP technology was shown to retain some natively associated lipid from the cell membrane and to retain the supercomplex of alternative complex III and aa3-type cytochrome c oxidase (Figure 2A,D). Interestingly, the other high-resolution single-particle cryo-EM structure of alternative complex III prepared in detergents also shows essential structural native lipids in equivalent positions (Figure 2B). In the SMALP-derived structure, it was reported that alternative complex III forms Cyt-aa3 complex at a 1 : 1 ratio; however, this may not be the case. EM density displayed at contour level 5 (in sigma units) (Figure 2C) only shows a well-solved EM density of complex III but not Cyt-aa3. EM density displayed at contour level 3 (Figure 2D) does show EM density of Cyt-aa3 but is very noisy. There are two possibilities here. One is that Cyt-aa3 is structurally flexible; the other possibility is that in the SMALP prepared sample, only a small portion of particles contain Cyt-aa3, i.e. the alternative complex III/Cyt-aa3 ratio is much larger than 1. However, the natural complex III/Cyt-aa3 complex ratio on the cell membrane could be close to 1. Besides, because the Cyt-aa3 structure was solved at high-resolution separately [61], this structural flexibility can also be excluded; therefore, we can conclude that the poor density of Cyt-aa3 is because of the failure of SMALP technology to retain the alternative III/Cyt-aa3 supercomplex if these supercomplexes are predominant in the native cell membrane. Furthermore, in other research, an alternative complex III prepared in detergent also shows the same supercomplex [62].
Figure 2. Structures of alternative complex III/Cyt-aa3 supercomplex and homolog solved in detergent-based and detergent-free systems.
(A) Single-particle cryo-EM structure of alternative complex III SMALP technology (PDB: 6BTM). The tightly bound lipids within the complex were well solved. Blue colored EM density in mesh shows the location of lipids. (B) Single-particle cryo-EM structure of alternative complex III determined using detergent (PDB: 6LOD), in the structure, even with detergent, the tightly bind lipids within the complex were well resolved. Blue colored EM density in mesh shows the location of lipids. (C,D) Single-particle cryo-EM density of alternative complex III/Cyt-aa3supercomplex determined using SMALP technology. The density maps shown in (C,D) are in the same orientation, vertical to the plane of cell membrane. The density maps are displayed at contour levels 5 and 3 (in sigma units), respectively, using PyMol. At level 5, only alternative complex III is well resolved; at level 3, although EM density is observable, the density is too noisy to build the Cyt-aa3 structure.
Bacillus subtilis KimA potassium channel
Bacillus subtilis KimA potassium channel is a member of the KUP family. KUP transporters facilitate potassium uptake through the co-transport of protons [63]. Recently, a high-resolution single-particle cryo-EM structure of KimA has been solved using SMALP technology with the membrane-active polymer Xiran SL30010 P20 [43]. The solved single-particle cryo-EM structure does not have any associated native cell membrane lipids. However, molecular dynamic simulation analysis results suggest that the transmembrane domain of KimA dimer is associated with cell membrane lipids between the two subunits [43]. The KimA dimer structure might collapse once the lipid molecules were washed away by the membrane-active polymers, SZ30010. The KimA monomer was extracted with PyMol from the KimA homodimer coordinates to examine if the transmembrane region of the KimA dimer is consistent with the KimA monomer. Transmembrane region calculations of the dimeric and monomeric structure of KimA did show an apparent discrepancy: the tilting angle difference between the transmembrane helices in the KimA dimer and monomer is about 30° when calculated with the cell membrane as reference (Figure 3A,B). All the pieces of evidence indicate that SMALP technology failed to retain the lipids associated with the KimA potassium channel’s transmembrane domain and led to a non-natural structure of the KimA dimer.
Figure 3. Single-particles cryo-EM structures of KimA potassium channel determined using SMALP technology.
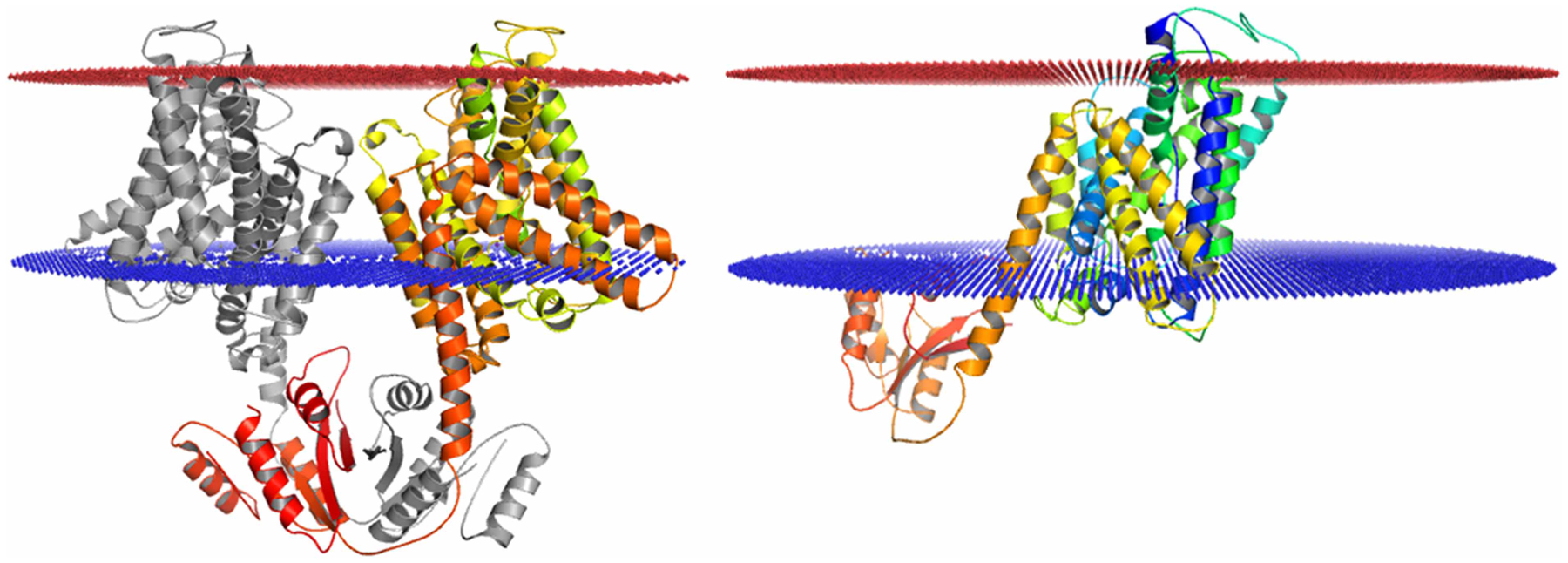
(A) Single-particle cryo-EM structure of KimA potassium channel dimer (PDB: 6S3K), in the structure, even without detergent, no lipids were resolved in the transmembrane domain. Transmembrane region analysis using PPM server showing the transmembrane domain of the dimer composed of two tilted subunits. (B) Transmembrane region analysis of a single subunit of KimA potassium channel dimer (PDB: 6S3K). The transmembrane domain region calculated from the monomer is drastically different from that of the KimA dimer.
Mechanosensitive channel YnaI
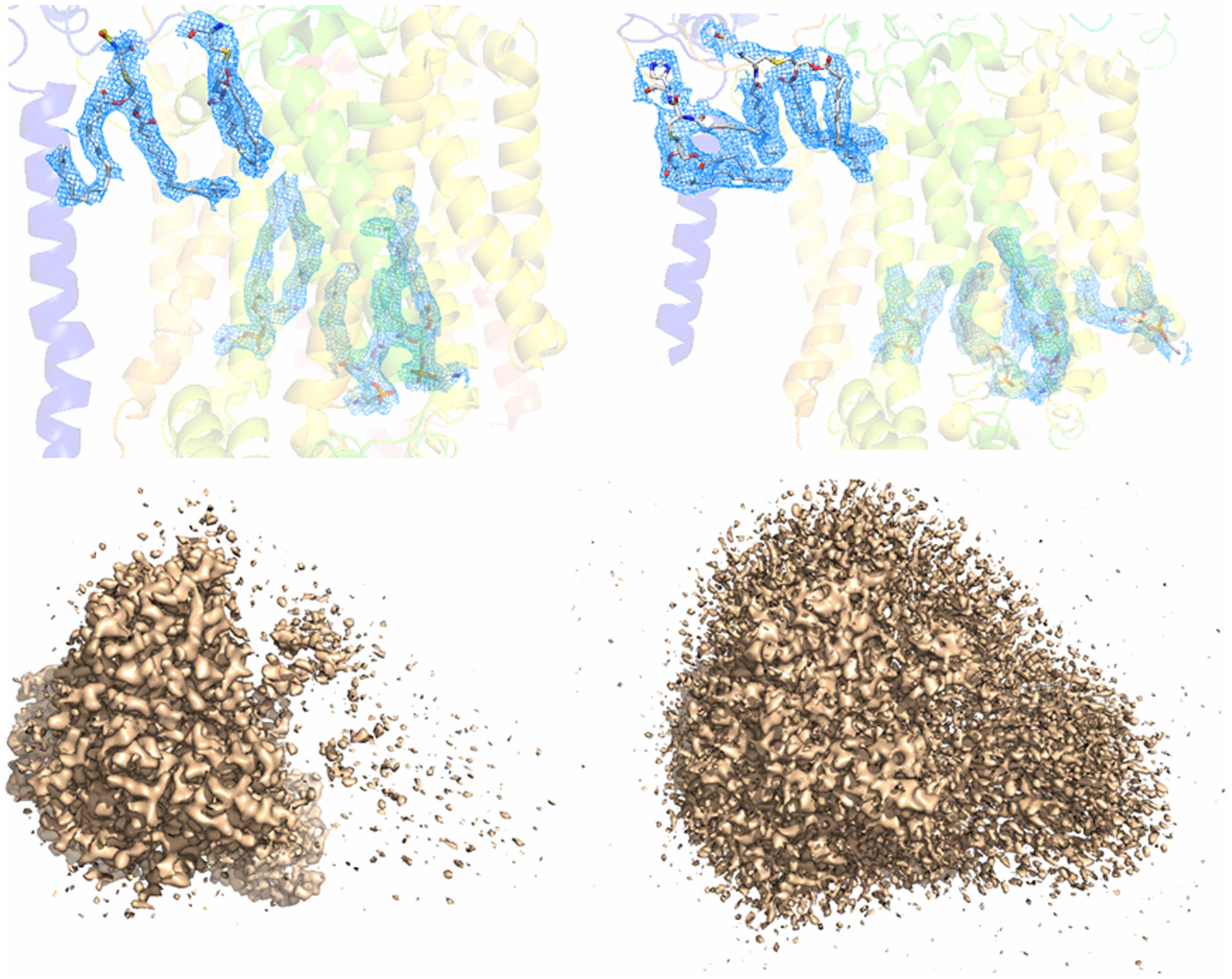
E. coli YnaI is a mechanosensitive channel [64] where protein–lipid interaction is crucial for gating [65,66]. Recently, DIBMA was used for high-resolution structure determinations for both the closed- and open states (Figure 4A,C) [47]. Examination of the EM density of YnaI solved with DIBMA revealed that in the claimed open state, the transmembrane domain has very poor EM density; no lipid molecule density can be assigned, and even the transmembrane helices could not be assigned unambiguously (Figure 4C). In the closed state, the (outer) transmembrane helices TM1 and TM2 EM density were not well resolved, and many of the native lipid molecules that are expected to be associated with the transmembrane domain were lost. Recently, the same YnaI channel single-particle cryo-EM structure determined using LMNG as detergent has also been reported (Figure 4B) (PDB: 6URT, EMD-20862, Hu, et al., to be published). In the detergent-based YnaI structure, all the transmembrane helices are well solved. Comparison between the cryo-EM structures of YnaI determined using DIBMA and LMNG shows drastic conformational differences in the transmembrane domain, especially for the transmembrane helices TM1 and TM2 (Figure 4D). Compared with the structure isolated via the detergent-based method, the DIBMA structure failed to show the advantage of retaining the natively associated lipid and maintain the native structure of membrane proteins. Here, it is also worth pointing out that current SMALP technology also failed initially in determining the cryo-EM structure of YnaI before the authors switched to DIBMALP. Related to this, it has also been reported that for human pannexin 1, samples prepared with SMALP technology produced a lower-resolution structure than samples prepared using detergent [67].
Figure 4. Single-particle cryo-EM analysis of E. coli mechanosensitive channel YnaI.
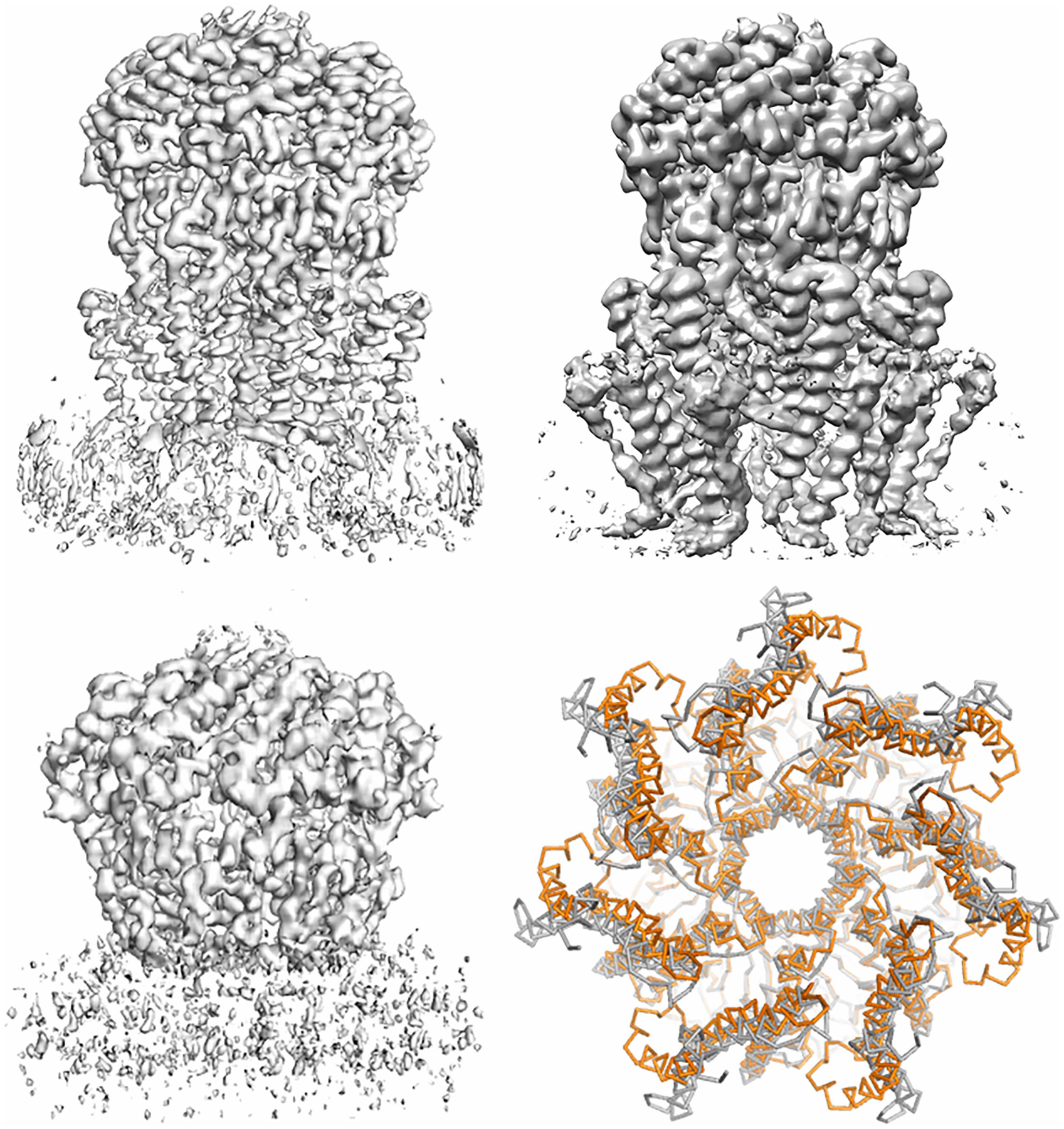
(A) Single-particle cryo-EM density of YnaI in a closed state. (EMD-11557) determined using DIBMA as a detergent-free method. The EM density in the transmembrane domain was not completely resolved; the EM density of TM1 and TM2 are not clear. (B) Single-particle cryo-EM structure of YnaI determined using detergent LMNG with addition of fluoridated Fos-choline 8 (EMD-20862). The transmembrane helices within the transmembrane domain are well resolved, however, no native lipid molecules were seen. (C) Single-particle cryo-EM density of YnaI in the claimed open state (EMD-11560). The EM density of transmembrane domain is not resolved at all, although the soluble domain is. (D) Structural comparison between YnaI determined using DIBMA (PDB: 6ZYD, orange colored) and LMNG (PDB: 6URT, gray colored). The two structures are significantly different for the TM1 and TM2 paddle, although the other portion is very similar; this reflects the potential bias of the structure determined using DIBMA resulting from the poor EM density in the transmembrane domain.
Glycine receptor
The glycine receptor belongs to ligand-gated ion channels, which mediate signal transduction at neural synapses in neurotransmitters’ responses [68]. Recently Eric Gouaux and colleagues reported 19 single-particle cryo-EM structures of the zebrafish glycine receptor in various states using a detergent-based method (1 structure), Nanodisc technology (5 structures), and SMALP technology (13 structures) [45]. Therefore, cryo-EM of glycine receptor structures serves as an excellent case for evaluation of the SMALP technology. Careful examination of the EM density maps corresponding to the reported 13 cryo-EM structures of glycine receptor reveals that none of them showed observable associated native cell membrane lipids density, although some structures were determined at high-resolution ~3.0 Å or better. Figure 5A shows a representative glycine receptor structure determined using the SMALP technology. Figure 5B shows the structure of glycine receptors determined using detergent. Both detergent-based and SMALP technology failed to retain native cell membrane lipids naturally associated with the glycine receptor. By contrast, all five cryo-EM structures of glycine receptors determined using Nanodisc technology show clear lipid density within the transmembrane domain. Although those lipids are not natively associated with cell membrane lipids, it indicates that glycine receptors are naturally associated with cell membrane lipid molecules. Figure 5C,D show cryo-EM structures determined using the Nanodisc technology in closed and desensitized states, respectively. The glycine receptor case study also tells us that current SMALP technology could not retain those associated native cell membrane lipids. In terms of high-structure determination, SMALP technology does not show the expected advantage of retaining native cell membrane lipids that are removed by detergents. However, the glycine receptor structures in the open and desensitized states solved using SMALP technology are consistent with the expected physiological conformations, despite the lack of resolved lipid density.
Figure 5. Single-particle cryo-EM analysis of zebra fish glycine receptor.
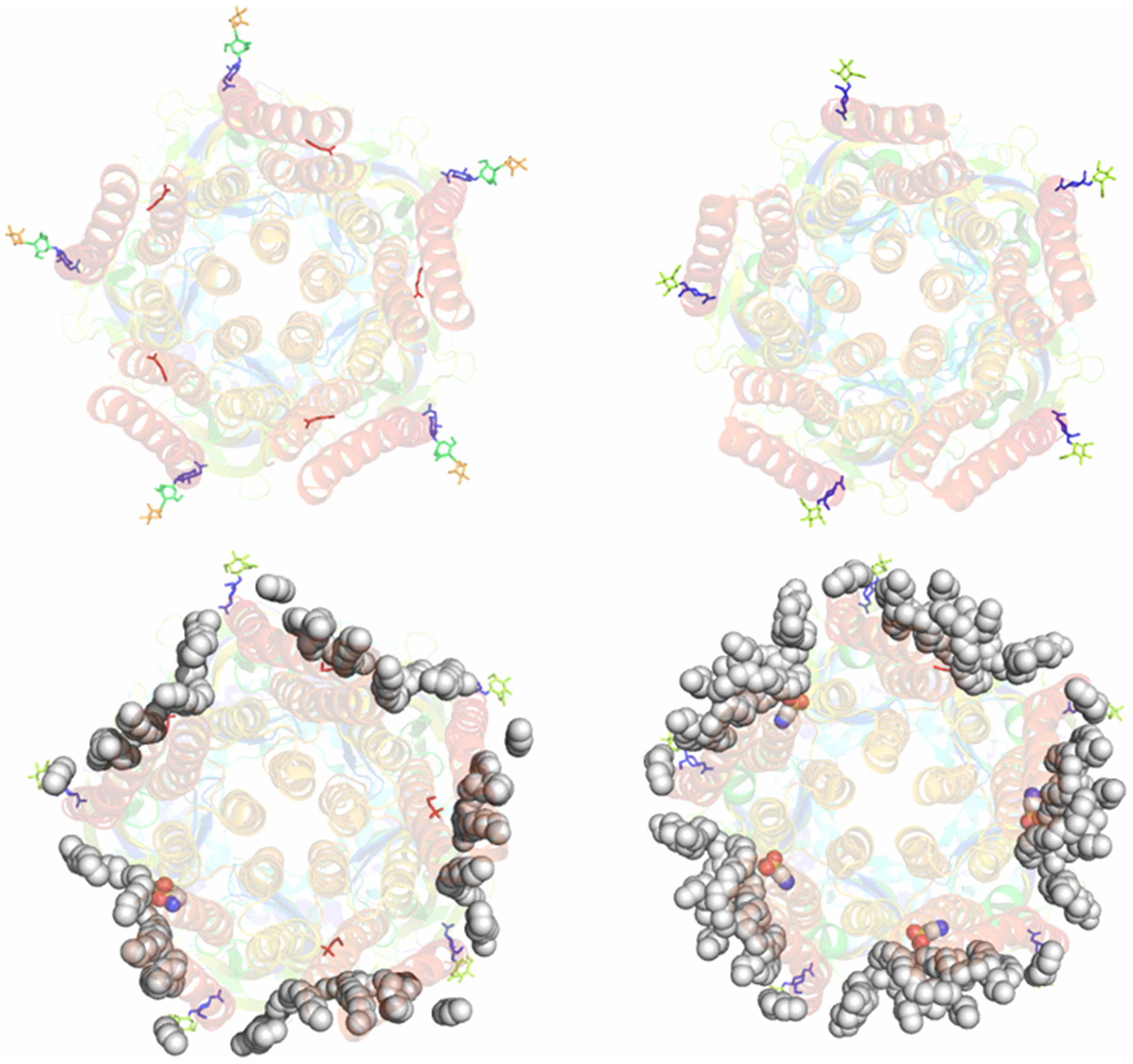
(A) Single-particle cryo-EM density of glycine receptor in a closed state (PDB: 6PLZ) determined using SMALP technology. No well-solved lipid molecule EM density observed in the transmembrane domain. (B) Single-particle cryo-EM structure of glycine receptor in apo state determined using detergent (PDB: 6PXD). No well-solved EM density was observed in the transmembrane domain. (C) Single-particle cryo-EM density of glycine receptor in a closed state determined using Nanodisc technology (PDB: 6PLT). Some well-resolved lipid density was observed in the transmembrane domain and lipid molecules can be built in the structural model. (D) Single-particle cryo-EM density of glycine receptor in a desensitized state determined using Nanodisc technology (PDB: 6PLS). Significant well-resolved lipid density was observed in the transmembrane domain and lipid molecules can be built in the structural model. The gray spheres show the lipid molecules. The colored spheres show taurine molecules. Sticks show the sugar moieties at the glycosylation sites. Cartoons show the glycine receptor protein.
Future directions
Significance of detergent-free technologies and current status
Protein–lipid interactions are crucial for the natural structure and the normal function of membrane proteins. Detergents have been predominantly used for membrane protein structural biology since 1985, when the first membrane protein crystal structure was reported [69]. Detergents have played a primary role in advancing membrane protein structural biology [14,16]. However, laborious detergent screening to find the best detergent for each study is performed by membrane protein structural biologists, and, unfortunately, this is often not successful. Furthermore, the detergent selected after such screening may still be deleterious to the study because of the over-delipidation of the membrane protein. Thus, detergent-free methods that can extract membrane proteins and the associated native cell membrane lipids are preferred. Considerable progress in detergent-free method development has been made during the past decade or so since the first report of using the styrene-maleic acid copolymer for membrane proteins extraction [34]. This was accelerated following the high-resolution structure determination of bacterial rhodopsin, AcrB, and alternative complex III using these copolymers. Some dedicated detergent-free systems such as SMALP, NCMN, DIBMALP technology, and CyclAPol have emerged as promising alternatives. In examining the recently reported single-particle cryo-EM structures of five groups of membrane proteins, it can be concluded that currently, detergent-free systems for membrane protein structural biology, especially for high-resolution structure determination, are still in their infancy. Except for the evolving studies using the NCMN system, the current detergent-free technologies failed to show many of the expected advantages over detergent-based approaches. Whether the NCMN system also works for other membrane proteins, especially for human homologs, is yet to be determined.
Below, we present some suggested guidelines for obtaining and studying structures with detergent-free methods.
The requirement for quality control of membrane-active polymers
All detergent-free systems depend on membrane-active polymers. Among these membrane-active polymers, SMA, DIBMA, and amphipols are most popular. Many derivatives based on these polymer backbones have also been developed [70–75]. Furthermore, new polymers with novel backbone structures have also been reported or are in development [48,49]. However, the suitability for high-resolution structure determination and the capability of retaining naturally associated and functionally important lipids remain to be examined for the many new membrane-active polymers. Systematic characterization and screening of these membrane-active polymers with model membrane proteins using single-particle cryo-EM analysis will be an excellent way to examine the quality of the new polymers. Only high-quality membrane-active polymers should be selected and used for membrane protein structural biology to guarantee the accuracy of the structural information of protein–lipid interactions.
Quality control of single-particles prepared with membrane-active polymers is also crucial in evaluating other biochemical and biophysical analysis results. Singe-particle cryo-EM or at least negative stain EM should also be used to check the prepared sample’s homogeneity. For example, lipid analysis of membrane protein samples using the detergent-free method could be misleading if the particles prepared are not of good quality and crucial native lipids were not successfully retained in the particles. In theory, membrane-active polymers should maintain the native cell membrane lipids and the membrane protein supercomplex; however, this may not be true in practice, as in the cases of AcrB, alternative complex III, KimA, YnaI, and the glycine receptor described in this minireview.
Systematic SAR (structure-activity relationships) studies of membrane-active polymers in conjunction with single-particle cryo-EM analysis will be the best way to accumulate experimental data for theoretical research and subsequent rational design of membrane-active polymers.
Protocols for the application of membrane-active polymers have to be optimized
Because of the structural diversity of membrane-active polymers, each polymer will have its unique set of properties. Therefore, an individual protocol should be developed for the application of each polymer for membrane proteins. Because of the diversity of cell membrane systems and local membrane protein–lipid interactions, it is probably impossible to have a single membrane-active polymer that can work well for all membrane proteins. However, some membrane-active polymers may be able to work broadly for many membrane proteins. Developing and screening such membrane-active polymers using model membrane proteins are urgent goals for developing the technology of detergent-free systems.
Detergent-free technology in conjunction with proteoliposome for functional studies
Proteoliposome is a powerful platform for functional studies of membrane proteins, especially for transporters and channels. Both SMALP technology and the NCMN system have been reported for successful proteoliposome reconstitution for the functional study of membrane proteins [32,50,76]. In theory, the detergent-free technology’s advantage is its capability to retain the native lipids with their membrane proteins and maintain the protein in its natural and functional state. However, it does not always work well, as in the case study of KcsA proteoliposome reconstitution where SMA2000 did not work well, and a novel NCMNP7-1 polymer was used for functional proteoliposome reconstitution [32], although SMA2000 was previously reported to be successful for reconstitution of KcsA onto a planar lipid bilayer [50]. This suggests the requirement that there be diversity of membrane-active polymers in practice.
Finally, I would acknowledge the remarkable achievements that have been made in understanding membrane protein structure and function using detergents. With caution, detergents will continue to make significant contributions, despite the drawbacks highlighted in this minireview.
Perspectives.
Importance of the field: Membrane proteins play crucial roles in many biological processes and serves as the primary drug targets of modern drugs. Membrane protein structure biology can provide accurate structural information of membrane proteins, but only when membrane proteins can be studied within the cell membrane lipid environment. Dedicated detergent-free technologies have been explicitly developed for the membrane protein structural biology within the cell membrane lipid environment.
Current thinking: The essentiality of detergent-free technology for membrane protein structural biology is well established as it is well known that detergent-based approaches have significant drawbacks. The detergent-free technologies such as SMALP, NCMN, DIBMALP, and CyclAPol are all promising platforms; however, they are all in their infancy.
Future directions: Many diverse membrane-active polymers will be required for the detergent-free technology to be universally applicable. High-level standards for quality control need to be established to select high-quality membrane-active polymers.
Acknowledgements
I am grateful to Glen Kellogg for his precious time and comments on this manuscript.
Funding
This research was funded by the National Institute of General Medical Sciences, grant number R01GM132329 (to Y.G.), and Virginia Commonwealth University (VCU) School of Pharmacy and Department of Medicinal Chemistry, through startup funds, and by the VCU Institute for Structural Biology, Drug Discovery and Development, through laboratory space and facilities. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
- Cryo-EM
cryogenic electron microscopy
- CyclAPol
cycloalkane-modified amphiphile polymer
- Cyt-aa3
cytochrome aa3
- DIBMALP
diisobutyl maleic acid lipid particles
- EMD
electron microscopy data resource
- GPCR
G protein-coupled receptor
- KUP
the potassium (K+) uptake permease
- LCP
lipidic cubic phase
- LMNG
lauryl maltose neopentyl glycol
- NCMN
native cell membrane nanoparticles
- NMR
nucleic magnetic resonance
- PDB
protein database bank
- PIP2
phosphatidylinositol-4,5-bisphosphate
- SAR
structure-activity relationships
- SMALP
styrene-maleic acid lipid particles
Footnotes
Competing Interests
Y.G. is the inventor of the NCMN system and a co-director of the SMALP network and has filed patents on NCMN polymers and corresponding protocols.
References
- 1.van Meer G, Voelker DR and Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol 9, 112–124 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown DA and London E (1998) Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol 14, 111–136 10.1146/annurev.cellbio.14.1.111 [DOI] [PubMed] [Google Scholar]
- 3.Koga Y and Morii H (2007) Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev 71, 97–120 10.1128/MMBR.00033-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singer SJ and Nicolson GL (1972) The fluid mosaic model of the structure of cell membranes. Science 175, 720–731 10.1126/science.175.4023.720 [DOI] [PubMed] [Google Scholar]
- 5.Mulkidjanian AY, Galperin MY and Koonin EV (2009) Co-evolution of primordial membranes and membrane proteins. Trends Biochem. Sci 34, 206–215 10.1016/j.tibs.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edidin M (2003) Timeline - lipids on the frontier: a century of cell-membrane bilayers. Nat. Rev. Mol. Cell Biol 4, 414–418 10.1038/nrm1102 [DOI] [PubMed] [Google Scholar]
- 7.Robertson JL (2018) The lipid bilayer membrane and its protein constituents. J. Gen. Physiol 150, 1472–1483 10.1085/jgp.201812153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prasanna X, Sengupta D and Chattopadhyay A (2016) Cholesterol-dependent conformational plasticity in GPCR dimers. Sci. Rep 6, 31858 10.1038/srep31858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiriakidi S, Kolocouris A, Liapakis G, Ikram S, Durdagi S and Mavromoustakos T (2019) Effects of cholesterol on GPCR function: insights from computational and experimental studies. Adv. Exp. Med. Biol 1135, 89–103 10.1007/978-3-030-14265-0_5 [DOI] [PubMed] [Google Scholar]
- 10.Luchetti G, Sircar R, Kong JH, Nachtergaele S, Sagner A, Byrne EF et al. (2016) Cholesterol activates the G-protein coupled receptor smoothened to promote hedgehog signaling. eLife 5, e20304 10.7554/eLife.20304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neumann J, Rose-Sperling D and Hellmich UA (2017) Diverse relations between ABC transporters and lipids: an overview. Biochim. Biophys. Acta Biomembr 1859, 605–618 10.1016/j.bbamem.2016.09.023 [DOI] [PubMed] [Google Scholar]
- 12.Duncan AL, Song WL and Sansom MSP (2020) Lipid-dependent regulation of ion channels and g protein-coupled receptors: insights from structures and simulations. Ann. Rev. Pharmacol. Toxicol 60, 31–50 10.1146/annurev-pharmtox-010919-023411 [DOI] [PubMed] [Google Scholar]
- 13.Jussupow A, Di Luca A and Kaila VRI (2019) How cardiolipin modulates the dynamics of respiratory complex I. Sci. Adv 5, eaav1850 10.1126/sciadv.aav1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrickson WA (2016) Atomic-level analysis of membrane-protein structure. Nat. Struct. Mol. Biol 23, 464–467 10.1038/nsmb.3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo YZ (2020) Be Cautious with crystal structures of membrane proteins or complexes prepared in detergents. Crystals 10, 86 10.3390/cryst10020086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garavito RM and Ferguson-Miller S (2001) Detergents as tools in membrane biochemistry. J. Biol. Chem 276, 32403–32406 10.1074/jbc.R100031200 [DOI] [PubMed] [Google Scholar]
- 17.Overduin M and Esmaili M (2019) Structures and interactions of transmembrane targets in native nanodiscs. SLAS Discov. 24, 943–952 10.1177/2472555219857691 [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Wang C, Zhou Q, An J, Hildebrandt E, Aleksandrov LA et al. (2014) Membrane protein stability can be compromised by detergent interactions with the extramembranous soluble domains. Protein Sci. 23, 769–789 10.1002/pro.2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin H and Flynn AD (2016) Drugging membrane protein interactions. Annu. Rev. Biomed. Eng 18, 51–76 10.1146/annurev-bioeng-092115-025322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chipot C, Dehez F, Schnell JR, Zitzmann N, Pebay-Peyroula E, Catoire LJ et al. (2018) Perturbations of native membrane protein structure in alkyl phosphocholine detergents: a critical assessment of NMR and biophysical studies. Chem. Rev 118, 3559–3607 10.1021/acs.chemrev.7b00570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cross TA, Sharma M, Yi M and Zhou HX (2011) Influence of solubilizing environments on membrane protein structures. Trends Biochem. Sci 36, 117–125 10.1016/j.tibs.2010.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denisov IG, Schuler MA and Sligar SG (2019) Nanodiscs as a new tool to examine lipid-protein interactions. Methods Mol. Biol 2003, 645–671 10.1007/978-1-4939-9512-7_25 [DOI] [PubMed] [Google Scholar]
- 23.Tribet C, Audebert R and Popot JL (1996) Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl Acad. Sci. U.S.A 93, 15047–15050 10.1073/pnas.93.26.15047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frauenfeld J, Löving R, Armache JP, Sonnen AF, Guettou F, Moberg P et al. (2016) A saposin-lipoprotein nanoparticle system for membrane proteins. Nat. Methods 13, 345–351 10.1038/nmeth.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlson ML, Young JW, Zhao Z, Fabre L, Jun D, Li J et al. (2018) The peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. eLife 7, e34085 10.7554/eLife.34085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SC, Knowles TJ, Postis VL, Jamshad M, Parslow RA, Lin YP et al. (2016) A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc 11, 1149–1162 10.1038/nprot.2016.070 [DOI] [PubMed] [Google Scholar]
- 27.Gulamhussein AA, Uddin R, Tighe BJ, Poyner DR and Rothnie AJ (2020) A comparison of SMA (styrene maleic acid) and DIBMA (di-isobutylene maleic acid) for membrane protein purification. Biochim. Biophys. Acta Biomembr 1862, 183281 10.1016/j.bbamem.2020.183281 [DOI] [PubMed] [Google Scholar]
- 28.Marconnet A, Michon B, Le Bon C, Giusti F, Tribet C and Zoonens M (2020) Solubilization and stabilization of membrane proteins by cycloalkane-modified amphiphilic polymers. Biomacromolecules 21, 3459–3467 10.1021/acs.biomac.0c00929 [DOI] [PubMed] [Google Scholar]
- 29.Simon KS, Pollock NL and Lee SC (2018) Membrane protein nanoparticles: the shape of things to come. Biochem. Soc. Trans 46, 1495–1504 10.1042/BST20180139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu W, Fu Z, Xu GG, Grassucci RA, Zhang Y, Frank J et al. (2018) Structure and activity of lipid bilayer within a membrane-protein transporter. Proc. Natl Acad. Sci. U.S.A 115, 12985–12990 10.1073/pnas.1812526115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kroeck KG, Qiu W, Catalano C, Trinh T and Guo Y (2020) Native cell membrane nanoparticles system for membrane protein-protein interaction analysis. J. Vis. Exp 161, 10.3791/61298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L, Yunyao Xu CC, Qiu W, Zhang D, McDermott A, Guo Y et al. (2021) A native cell membrane nanoparticles system allows for high-quality functional proteoliposome reconstitution. BBA Adv. 1, 100011 10.1016/j.bbadva.2021.100011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollock NL, Lee SC, Patel JH, Gulamhussein AA and Rothnie AJ (2018) Structure and function of membrane proteins encapsulated in a polymer-bound lipid bilayer. Biochim. Biophys. Acta Biomembr 1860, 809–817 10.1016/j.bbamem.2017.08.012 [DOI] [PubMed] [Google Scholar]
- 34.Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T and Overduin M (2009) Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc 131, 7484–7485 10.1021/ja810046q [DOI] [PubMed] [Google Scholar]
- 35.Jamshad M, Lin YP, Knowles TJ, Parslow RA, Harris C, Wheatley M et al. (2011) Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem. Soc. Trans 39, 813–818 10.1042/BST0390813 [DOI] [PubMed] [Google Scholar]
- 36.Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR and Watts A (2012) Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer lipodisq particles for functional and biophysical studies. Nano Lett. 12, 4687–4692 10.1021/nl3020395 [DOI] [PubMed] [Google Scholar]
- 37.Gulati S, Jamshad M, Knowles TJ, Morrison KA, Downing R, Cant N et al. (2014) Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem. J 461, 269–278 10.1042/BJ20131477 [DOI] [PubMed] [Google Scholar]
- 38.Swainsbury DJ, Scheidelaar S, van Grondelle R, Killian JA and Jones MR (2014) Bacterial reaction centers purified with styrene maleic acid copolymer retain native membrane functional properties and display enhanced stability. Angew. Chem. Int. Ed 53, 11803–11807 10.1002/anie.201406412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bell AJ, Frankel LK and Bricker TM (2015) High yield non-detergent isolation of photosystem I-lightharvesting chlorophyll II membranes from spinach thylakoids IMPLICATIONS FOR THE ORGANIZATION OF THE PS I ANTENNAE IN HIGHER PLANTS. J. Biol. Chem 290, 18429–18437 10.1074/jbc.M115.663872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smirnova IA, Sjöstrand D, Li F, Björck M, Schäfer J, Östbye H et al. (2016) Isolation of yeast complex IV in native lipid nanodiscs. Biochim. Biophys. Acta Biomembr 1858, 2984–2992 10.1016/j.bbamem.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broecker J, Eger BT and Ernst OP (2017) Crystallogenesis of membrane proteins mediated by polymer-bounded lipid nanodiscs. Structure 25, 384–392 10.1016/j.str.2016.12.004 [DOI] [PubMed] [Google Scholar]
- 42.Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J et al. (2018) Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 557, 123–126 10.1038/s41586-018-0061-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tascón I, Sousa JS, Corey RA, Mills DJ, Griwatz D, Aumüller N et al. (2020) Structural basis of proton-coupled potassium transport in the KUP family. Nat. Commun 11, 626 10.1038/s41467-020-14441-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoder N and Gouaux E (2020) The His-Gly motif of acid-sensing ion channels resides in a reentrant ‘loop’ implicated in gating and ion selectivity. eLife 9, e56527 10.7554/eLife.56527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J, Zhu H, Lape R, Greiner T, Du J, Lü W et al. (2021) Mechanism of gating and partial agonist action in the glycine receptor. Cell 184, 957–968.e21 10.1016/j.cell.2021.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adão R, Cruz PF, Vaz DC, Fonseca F, Pedersen JN, Ferreira-da-Silva F et al. (2020) DIBMA nanodiscs keep alpha-synuclein folded. Biochim. Biophys. Acta Biomembr 1862, 183314 10.1016/j.bbamem.2020.183314 [DOI] [PubMed] [Google Scholar]
- 47.Flegler VJ, Rasmussen A, Rao S, Wu N, Zenobi R, Sansom M et al. (2020) The mscS-like channel ynaI has a gating mechanism based on flexible pore helices. Proc. Natl Acad. Sci. U.S.A 117, 28754–28762 10.1073/pnas.2005641117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Esmaili M, Brown CJ, Shaykhutdinov R, Acevedo-Morantes C, Wang YL, Wille H et al. (2020) Homogeneous nanodiscs of native membranes formed by stilbene-maleic-acid copolymers. Nanoscale 12, 16705–16709 10.1039/D0NR03435E [DOI] [PubMed] [Google Scholar]
- 49.Smith AAA, Autzen HE, Faust B, Mann JL, Muir BW, Howard S et al. (2020) Lipid nanodiscs via ordered copolymers. Chem. 6, 2782–2795 10.1016/j.chempr.2020.08.004 [DOI] [Google Scholar]
- 50.Dörr JM, Koorengevel MC, Schäfer M, Prokofyev AV, Scheidelaar S, van der Cruijsen EA et al. (2014) Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc. Natl Acad. Sci. U.S.A 111, 18607–18612 10.1073/pnas.1416205112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Postis V, Rawson S, Mitchell JK, Lee SC, Parslow RA, Dafforn TR et al. (2015) The use of SMALPs as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim. Biophys. Acta Biomembr 1848, 496–501 10.1016/j.bbamem.2014.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao X, Fan X and Yan N (2020) Cryo-EM analysis of a membrane protein embedded in the liposome. Proc. Natl Acad. Sci. U.S.A 117, 18497–18503 10.1073/pnas.2009385117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murakami S, Nakashima R, Yamashita E and Yamaguchi A (2002) Crystal structure of bacterial multidrug efflux transporter acrB. Nature 419, 587–593 10.1038/nature01050 [DOI] [PubMed] [Google Scholar]
- 54.Yu EW, McDermott G, Zgurskaya HI, Nikaido H and Koshland DE Jr (2003) Structural basis of multiple drug-binding capacity of the acrB multidrug efflux pump. Science 300, 976–980 10.1126/science.1083137 [DOI] [PubMed] [Google Scholar]
- 55.Ababou A and Koronakis V (2016) Structures of gate loop variants of the acrB drug efflux pump bound by erythromycin substrate. PLoS ONE 11, e0159154 10.1371/journal.pone.0159154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parmar M, Rawson S, Scarff CA, Goldman A, Dafforn TR, Muench SP et al. (2018) Using a SMALP platform to determine a sub-nm single particle cryo-EM membrane protein structure. Biochim. Biophys. Acta. Biomembr 1860, 378–383 10.1016/j.bbamem.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyu M, Moseng MA, Reimche JL, Holley CL, Dhulipala V, Su CC et al. (2020) Cryo-EM structures of a gonococcal multidrug efflux pump illuminate a mechanism of drug recognition and resistance. Mbio 11, e00996–2 10.1128/mBio.00996-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson RM, Fais C, Parmar M, Cheruvara H, Marshall RL, Hesketh SJ et al. (2020) Cryo-EM structure and molecular dynamics analysis of the fluoroquinolone resistant mutant of the acrB transporter from salmonella. Microorganisms 8, 943 10.3390/microorganisms8060943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du D, Neuberger A, Orr MW, Newman CE, Hsu PC, Samsudin F et al. (2020) Interactions of a bacterial RND transporter with a transmembrane small protein in a lipid environment. Structure 28, 625–634.e6 10.1016/j.str.2020.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Refojo PN, Sousa FL, Teixeira M and Pereira MM (2010) The alternative complex III: a different architecture using known building modules. Biochim. Biophys. Acta Bioenerg 1797, 116 10.1016/j.bbabio.2010.04.347 [DOI] [PubMed] [Google Scholar]
- 61.Xu J, Ding Z, Liu B, Yi SM, Li J, Zhang Z et al. (2020) Structure of the cytochrome aa 3 −600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site. Proc. Natl Acad. Sci. U.S.A 117, 872–876 10.1073/pnas.1915013117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sousa JS, Calisto F, Langer JD, Mills DJ, Refojo PN, Teixeira M et al. (2018) Structural basis for energy transduction by respiratory alternative complex III. Nat. Commun 9, 1728 10.1038/s41467-018-04141-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bossemeyer D, Schlosser A and Bakker EP (1989) Specific cesium transport via the escherichia-Coli Kup (Trkd) K+ uptake system. J. Bacteriol 171, 2219–2221 10.1128/JB.171.4.2219-2221.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Booth IR, Miller S, Müller A and Lehtovirta-Morley L (2015) The evolution of bacterial mechanosensitive channels. Cell Calcium 57, 140–150 10.1016/j.ceca.2014.12.011 [DOI] [PubMed] [Google Scholar]
- 65.Rasmussen T, Rasmussen A, Yang L, Kaul C, Black S, Galbiati H et al. (2019) Interaction of the mechanosensitive channel, mscS, with the membrane bilayer through lipid intercalation into grooves and pockets. J. Mol. Biol 431, 3339–3352 10.1016/j.jmb.2019.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rasmussen T (2016) How do mechanosensitive channels sense membrane tension? Biochem. Soc. Trans 44, 1019–1025 10.1042/BST20160018 [DOI] [PubMed] [Google Scholar]
- 67.Ruan Z, Orozco IJ, Du J and Lü W (2020) Structures of human pannexin 1 reveal ion pathways and mechanism of gating. Nature 584, 646–651 10.1038/s41586-020-2357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lynch JW (2004) Molecular structure and function of the glycine receptor chloride channel. Physiol. Rev 84, 1051–1095 10.1152/physrev.00042.2003 [DOI] [PubMed] [Google Scholar]
- 69.Deisenhofer J, Epp O, Miki K, Huber R and Michel H (1985) Structure of the protein subunits in the photosynthetic reaction center of rhodopseudomonas-viridis at 3a resolution. Nature 318, 618–624 10.1038/318618a0 [DOI] [PubMed] [Google Scholar]
- 70.Di Mauro GM, La Rosa C, Condorelli M and Ramamoorthy A (2021) Benchmarks of SMA-copolymer derivatives and nanodisc integrity. Langmuir 37, 3113–3121 10.1021/acs.langmuir.0c03554 [DOI] [PubMed] [Google Scholar]
- 71.Esmaili M, Acevedo-Morantes C, Wille H and Overduin M (2020) The effect of hydrophobic alkyl sidechains on size and solution behaviors of nanodiscs formed by alternating styrene maleamic copolymer. Biochim. Biophys. Acta Biomembr 1862, 183360 10.1016/j.bbamem.2020.183360 [DOI] [PubMed] [Google Scholar]
- 72.Ravula T, Hardin NZ and Ramamoorthy A (2019) Polymer nanodiscs: advantages and limitations. Chem. Phys. Lipids 219, 45–49 10.1016/j.chemphyslip.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ravula T, Hardin NZ, Di Mauro GM and Ramamoorthy A (2018) Styrene maleic acid derivates to enhance the applications of bio-inspired polymer based lipid-nanodiscs. Eur. Polym. J 108, 597–602 10.1016/j.eurpolymj.2018.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ravula T, Hardin NZ, Bai J, Im SC, Waskell L and Ramamoorthy A (2018) Effect of polymer charge on functional reconstitution of membrane proteins in polymer nanodiscs. Chem. Commun. (Camb) 54, 9615–9618 10.1039/C8CC04184A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ravula T, Hardin NZ, Ramadugu SK, Cox SJ and Ramamoorthy A (2018) Formation of pH-resistant monodispersed polymer-lipid nanodiscs. Angew. Chem. Int. Ed. Engl 57, 1342–1345 10.1002/anie.201712017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 76.Smirnova IA, Adelroth P and Brzezinski P (2018) Extraction and liposome reconstitution of membrane proteins with their native lipids without the use of detergents. Sci. Rep 8, 14950 10.1038/s41598-018-33208-1 [DOI] [PMC free article] [PubMed] [Google Scholar]


