Abstract
SNP arrays are powerful tools for high-resolution studies of the genetic basis of complex traits, facilitating both selective breeding and population genomic research. The European seabass (Dicentrarchus labrax) and the gilthead seabream (Sparus aurata) are the two most important fish species for Mediterranean aquaculture. While selective breeding programmes increasingly underpin stock supply for this industry, genomic selection is not yet widespread. Genomic selection has major potential to expedite genetic gain, particularly for traits practically impossible to measure on selection candidates, such as disease resistance and fillet characteristics. The aim of our study was to design a combined-species 60 K SNP array for European seabass and gilthead seabream, and to test its performance on farmed and wild populations from numerous locations throughout the species range. To achieve this, high coverage Illumina whole-genome sequencing of pooled samples was performed for 24 populations of European seabass and 27 populations of gilthead seabream. This resulted in a database of ~20 million SNPs per species, which were then filtered to identify high-quality variants and create the final set for the development of the ‘MedFish’ SNP array. The array was then tested by genotyping a subset of the discovery populations, highlighting a high conversion rate to functioning polymorphic assays on the array (92% in seabass; 89% in seabream) and repeatability (99.4–99.7%). The platform interrogates ~30 K markers in each species, includes features such as SNPs previously shown to be associated with performance traits, and is enriched for SNPs predicted to have high functional effects on proteins. The array was demonstrated to be effective at detecting population structure across a wide range of fish populations from diverse geographical origins, and to examine the extent of haplotype sharing among Mediterranean farmed fish populations. In conclusion, the new MedFish array enables efficient and accurate high-throughput genotyping for genome-wide distributed SNPs for each fish species, and will facilitate stock management, population genomics approaches, and acceleration of selective breeding through genomic selection.
Keywords: European seabass, gilthead seabream, SNP array, Aquaculture
Highlights
-
•
Α 60 K SNP array (MedFish) was designed for European seabass and gilthead seabream from wild and domesticated populations.
-
•
The array exhibited a high conversion rate (92% in seabass; 89% in seabream) and repeatability (99.4 and 99.7%).
-
•
The MedFish array is expected to facilitate stock management and acceleration of selective breeding via genomic selection.
1. Introduction
Modern aquaculture selective breeding programmes are embracing the availability of genomic technologies to sustainably increase genetic gain. Genomic tools can also facilitate improvements to methods for forming base populations for breeding programmes by computing well-characterized genetic variability and relationships, which is important for many aquaculture species still in the process of domestication [1,2]. To achieve these goals in target species typically requires the generation of genome-wide genetic marker data (usually SNP markers) across large numbers of individuals. When paired with trait recording on the genotyped individuals, such datasets can be applied to examine the genetic architecture of production traits of interest, including detection of quantitative trait loci (QTL) using genome-wide association studies (GWAS). If the detected QTL are of sufficiently large effect, flanking markers can be utilized to select candidates with favourable alleles at the QTL, also known as Marker-Assisted Selection (MAS). While MAS has been successfully applied for a small number of traits, such as resistance to infectious pancreatic necrosis in Atlantic salmon [3,4], most traits of interest for aquaculture are underpinned by a polygenic architecture [1,5,6]. For such traits, genome-wide SNP markers combined with phenotype data on a reference population can be used to estimate genomic breeding values for selection candidates [7]. Genomic selection is predicted to result in a notably higher selection accuracy and therefore genetic gain in aquaculture breeding programmes, as has also been demonstrated in early studies in several aquaculture species [1,8], including European seabass (Dicentrarchus labrax) [9] and gilthead seabream (Sparus aurata) [10,11].
The European seabass and the gilthead seabream are the two most important fish species in Mediterranean aquaculture. At the European level, they rank third and fourth, respectively, in terms of value after Atlantic salmon and rainbow trout [12]. Substantial genomic tools have been developed for both species, including the assembly and characterization of high-quality reference genomes [13, 14]. Medium or high-density SNP arrays have been developed for several other important finfish aquaculture species such as rainbow trout (Oncorhynchus mykiss) [15], Atlantic salmon (Salmo salar) [16,17], catfish (Ictalurus furcatus and I. punctatus) [18,19], common carp (Cyprinus carpio) [20], Arctic charr (Salvelinus alpinus) [21], and Nile tilapia (Oreochromis niloticus) [[22], [23], [24]], which have been used for studies into population structure, genetic diversity, signatures of domestication, the genetic architecture of traits of interest, and testing of genomic selection. A 57 K SNP array was also recently developed for European seabass [25] and has been applied to assess the genetic basis of resistance to viral nervous necrosis. However, this array is only available on request from the GeneSea consortium. Therefore, from both an aquaculture and population genetics perspective, there is a need for a publicly available high-throughput genotyping platform for European seabass and gilthead seabream.
Herein, an extensive and comprehensive SNP database was generated for European seabass and gilthead seabream across Europe by extensive sampling and pooled sequencing of ~25 populations per species from wild and aquaculture sites. From this SNP database, a subset of ~60 K SNPs was chosen based on several filtering criteria to give thorough coverage of each species' genome. The SNP array was created and tested on several of the discovery populations, including highlighting its potential utility for detecting population structure and excess haplotype sharing between farmed populations. This open-access tool will provide new opportunities to the scientific community and industry for genome-scale research and application to improve selective breeding in these two focal European aquaculture species.
2. Materials and methods
2.1. Samples for SNP discovery
A diverse range of farmed and wild populations of European seabass (n = 24) and gilthead seabream (n = 27) were collected for SNP discovery. A farmed population was defined as that composed of fish originating from the same commercial hatchery or established farm. A total of 538 European seabass individuals were sampled from 14 farmed and 10 wild populations distributed across the Mediterranean Sea, and a total of 642 gilthead seabream individuals were sampled from 12 farmed and 15 wild populations from the Mediterranean and the Atlantic (Table 1). Fin clips were collected from 11 to 30 individuals per population and stored in absolute ethanol until transportation to either the University of Edinburgh (UK), the Hellenic Centre for Marine Research (Greece) or the University of Padova (Italy) for DNA extraction.
Table 1.
Summary of the European seabass and gilthead seabream populations sampled for sequencing and SNP discovery.
| Species | Origin | Region | Country | Pool ID | № individuals per pool | N° pools prepared |
|---|---|---|---|---|---|---|
| European seabass | Farmed | Mediterranean | France | Sba_farm_1 | 12 | 1 |
| Spain | Sba_farm_2 | 25 | 2 | |||
| Spain | Sba_farm_3 | 25 | 2 | |||
| Italy | Sba_farm_4 | 25 | 2 | |||
| Croatia | Sba_farm_5 | 25 | 2 | |||
| Croatia | Sba_farm_6 | 25 | 2 | |||
| Greece | Sba_farm_7 | 25 | 2 | |||
| Greece | Sba_farm_8 | 25 | 2 | |||
| Greece | Sba_farm_9 | 25 | 2 | |||
| Greece | Sba_farm_10 | 25 | 2 | |||
| Greece | Sba_farm_11 | 25 | 2 | |||
| Greece | Sba_farm_12 | 23 | 1 | |||
| Cyprus | Sba_farm_13 | 25 | 2 | |||
| Egypt |
Sba_farm_14 |
15 |
1 |
|||
| Wild | Mediterranean | France | Sba_wild_1 | 25 | 2 | |
| Spain | Sba_wild_2 | 11 | 1 | |||
| Morocco | Sba_wild_3 | 25 | 2 | |||
| Italy | Sba_wild_4 | 25 | 2 | |||
| Croatia | Sba_wild_5 | 12 | 1 | |||
| Greece | Sba_wild_6 | 25 | 2 | |||
| Greece | Sba_wild_7 | 25 | 2 | |||
| Cyprus | Sba_wild_8 | 15 | 1 | |||
| Turkey | Sba_wild_9 | 25 | 2 | |||
| Turkey | Sba_wild_10 | 25 | 2 | |||
|
Total N° pools |
42 |
|||||
| Gilthead seabream | Farmed | Mediterranean | France | Sbr_farm_1 | 25 | 2 |
| Spain | Sbr_farm_2 | 25 | 2 | |||
| Spain | Sbr_farm_3 | 25 | 2 | |||
| Italy | Sbr_farm_4 | 25 | 2 | |||
| Croatia | Sbr_farm_5 | 25 | 2 | |||
| Greece | Sbr_farm_6 | 14 | 1 | |||
| Greece | Sbr_farm_7 | 13 | 1 | |||
| Greece | Sbr_farm_8 | 25 | 2 | |||
| Greece | Sbr_farm_9 | 25 | 2 | |||
| Greece | Sbr_farm_10 | 25 | 2 | |||
| Israel | Sbr_farm_11 | 25 | 2 | |||
| Egypt |
Sbr_farm_12 |
15 |
1 |
|||
| Wild | Atlantic | France | Sbr_wild_1 | 25 | 2 | |
| Spain | Sbr_wild_2 | 25 | 2 | |||
| Spain |
Sbr_wild_3 |
25 |
2 |
|||
| Mediterranean | Spain | Sbr_wild_4 | 25 | 2 | ||
| Spain | Sbr_wild_5 | 25 | 2 | |||
| Tunisia | Sbr_wild_6 | 25 | 2 | |||
| Italy | Sbr_wild_7 | 25 | 2 | |||
| Italy | Sbr_wild_8 | 25 | 2 | |||
| Greece | Sbr_wild_9 | 25 | 2 | |||
| Greece | Sbr_wild_10 | 25 | 2 | |||
| Greece | Sbr_wild_11 | 25 | 2 | |||
| Greece | Sbr_wild_12 | 25 | 2 | |||
| Greece | Sbr_wild_13 | 25 | 2 | |||
| Turkey | Sbr_wild_14 | 25 | 2 | |||
| Turkey | Sbr_wild_15 | 25 | 2 | |||
| Total N° pools | 51 |
2.2. DNA extraction and pooling for sequencing
High quality genomic DNA was isolated from each fin-clip using a salt-based extraction method [26]. The integrity of the DNA extractions was assessed by performing an agarose gel electrophoresis. DNA purity was evaluated by using a NanoDrop ND-1000 (Thermo Fisher Scientific) spectrophotometer. The extracted DNA was quantified in duplicate using the fluorescent-based Qubit® quantitation assay (Thermo Fischer Scientific, cat #Q32850). DNA stocks were diluted to 10–30 ng/ul and then combined at equimolar concentrations into pools of 11–25 individuals per population. The majority of populations had a sample size of 25, and for these populations DNA pools were prepared twice (technical replicates). For the remaining few populations with less individuals (6 and 3 populations in the European seabass and gilthead seabream, respectively), a single population pool was prepared (Table 1).
2.3. Library construction and sequencing
Two sequencing facilities provided the library preparation and sequencing services – the Norwegian Sequencing Centre (NSC) (Oslo, Norway) and Edinburgh Genomics (University of Edinburgh, UK). Both facilities followed the TruSeq® PCR-free library preparation protocol to generate sequencing libraries from the pooled genomic DNA samples. Almost all European seabass population pools were sequenced on a HiSeq 4000 instrument (2 × 150 bp) at NSC, whereas all gilthead seabream pools were sequenced on a HiSeq X Ten platform (2 × 150 bp) at Edinburgh Genomics.
2.4. Bioinformatics analysis for SNP discovery
The sequencing reads of the population pools – 42 and 51 pools for the European seabass and gilthead seabream, respectively (see Table 1) – were processed separately for each species using identical software and parameter values. These reads were filtered using the fastp software v 0.20.0 [27]. Reads with a minimum length of 80 bp for which <20% of their bases showed a BQ ≤ 20 were retained. Cleaned paired-end reads from each population pool were then aligned to either the European seabass [13] or the gilthead seabream [14] genome assemblies using BWA v 0.7.8 [28]. Only primary alignments to the relevant reference genome were retained for further analysis. PCR duplicates were removed from the alignment files using SAMtools v 1.6 [29]. Variants were called separately for each species across all population pools using Freebayes v 1.20 [30] with GNU Parallel [31]. Freebayes was set to call a variant if either (i) a minimum of 3 reads supporting the non-reference allele was observed, or (ii) the allele frequency in the pool was above 0.05, after excluding alignments with a MQ < 20. This SNP calling pipeline led to the discovery of ~17 and 34 million putative polymorphisms in the European seabass and gilthead seabream genomes, respectively.
This initial list of variants was then filtered using vcflib (https://github.com/vcflib/vcflib) to keep bi-allelic SNPs that (i) showed supporting reads on both strands, (ii) a sequence coverage ranging from 17× to 90× for the European seabass and from 25× to 100× for the gilthead seabream, (iii) at least two reads ‘balanced’ to each side of the variant site, (iv) >90% of the observed alternate and reference alleles supported by properly paired reads, and (v) the ratio of mapping qualities between reference and alternate allele was between 0.9 and 1.1. SNPs were retained only if they had no interfering polymorphic sites within <35 bp upstream and downstream of the variant. The purpose of this filter was to identify markers compatible with array design and eliminate SNPs that could fail the assay due to flanking polymorphisms interfering with probe annealing. The minor allele frequency (MAF) was estimated for all SNPs that were successfully genotyped in more than 18 population pools per species, after averaging the estimated MAF for population pools with technical replicates. To avoid spurious SNPs resulting from sequence differences between paralogues, only SNPs with a MAF between 0.05 and 0.45 were retained for further SNP selection. From this list of candidate markers (~1 million high-quality markers for each fish species), 35 bp probes were extracted downstream and upstream from each SNP. The 71-mer nucleotide sequences were then submitted to Thermo Fisher Scientific for further quality check and in silico probe scoring.
2.5. SNP selection
As a first filtering step, and as recommended by Thermo Fisher Scientific, the remaining SNPs were filtered to avoid A/T and C/G polymorphisms because they require twice the number of probes for genotyping compared to other types of SNP polymorphisms. The remaining SNPs were divided into selection tiers and were sequentially included in the MedFish platform based on the following hierarchy of importance.
First, SNPs were included as high priority markers based on evidence of their association with relevant production traits. For the European seabass, markers associated with mandibular prognathism [32], resistance to viral nervous necrosis [9], and sex [33] were included. For the gilthead seabream, the set of markers of this type comprised SNPs associated with production traits of high economic importance – i.e., fat content, weight, tag weight and length to width ratio [34] – and resistance to photobacteriosis [11]. Importantly, if the aforementioned SNPs were not identified through our pool-sequencing experiment, they were not included directly on the platform. Instead, the economically relevant marker was substituted by a proxy SNP that was chosen by screening the surrounding region for the closest high quality variant present in our dataset.
A second group of SNPs included in the MedFish SNP array is shared with other platforms that were developed in parallel by the GeneSea consortium [25]. The purpose of including a subset of markers from the existing platforms was to facilitate backward compatibility and cross-study comparison, especially via the use of genotype imputation.
A third criterion for inclusion of SNPs on the MedFish platform was based on their predicted effect on protein-coding genes. SNPs on genes may affect protein function, for example, by causing truncated proteins. To potentially target variants with a potential functional effect, which may have a direct impact on relevant phenotypes, the list of high confidence variants identified in the European seabass and the gilthead seabream genomes were annotated with SNPEff v 4.3 [35]. For both species, SNPs that were predicted to have a HIGH functional effect on proteins were considered important and included as high priority markers in the array.
Fourthly, from the total number of ~1 million SNPs per fish species that were submitted as 71-mers to Thermo Fisher Scientific for in silico probe evaluation, only those that were categorized as either ‘recommended’ or ‘neutral’ became the pool from which array SNPs were selected. From the substantial SNP database generated in this study, markers were selected to achieve good coverage of the reference genomes of the European seabass and gilthead seabream following [33]. In brief, markers were selected along each fish chromosome at a variable density depending on the estimated local nucleotide diversity (π), as in European seabass [13] and other fish species [36] a positive correlation between nucleotide diversity and recombination rate has been observed. For SNPs that were mapped to the “UN” chromosome of the European seabass, the synthetic chromosome was split into contigs that had been previously concatenated by 100 consecutive Ns. The contigs were isolated and the SNPs located within them were remapped with the contigs starting position set to 1. The genomes of both fish species were divided into 70 Kb (for European seabass) or 85 Kb (for gilthead seabream) non-overlapping windows and local nucleotide diversity was estimated with VCFtools v 0.1.15 [37]. Genomic windows were categorized into one of the following classes depending on their estimated π value: π ≤0.001 (Class 1), 0.001 < π ≤0.002 (Class 2), 0.002 < π ≤0.003 (Class 3), 0.003 < π ≤0.004 (Class 4) and π >0.004 (Class 5). SNPs were chosen to cover all chromosomes of both fish species with a variable SNP density – ranging from 1 to 5 SNPs – depending on the diversity class assigned to each region. For the SNP selection process carried out within each type of diversity class window, two factors were considered as the main inclusion criteria: (i) the MAF for the SNPs in the window and (ii) the physical distance between markers. All discovered markers were divided into three different MAF categories (>0.3, 0.3–0.2 and 0.2–0.1). SNP markers within the MAF >0.3 category were prioritized across all five window classes such that at least 50% of the markers selected for each type of diversity class window came from the most informative SNP category (Table 2). Within each window, SNPs were selected successively from each MAF category by requiring a minimum inter-marker distance of 10,000 bp with any other previously chosen set of markers. To fill the remaining target of ~30 K SNPs per species, the physical distance between pairs of pre-selected SNPs was calculated, and intervals then sorted by length in decreasing order. SNP markers were then included sequentially (one SNP per interval) irrespective of its MAF.
Table 2.
Summary of SNP selection approach. A variable number of SNPs was selected along chromosomes according to the local nucleotide diversity (π) estimates for non-overlapping genomic windows.
| Genomic window diversity class | Range | № of SNPs sampled per window | № SNPs sampled per MAF category per window |
||
|---|---|---|---|---|---|
| >0.3 | 0.2–0.3 | 0.1–0.2 | |||
| Class 1 | π ≤0.001 | 1 | 1 | 0 | 0 |
| Class 2 | 0.001 < π ≤0.002 | 2 | 2 | 0 | 0 |
| Class 3 | 0.002 < π ≤0.003 | 3 | 2 | 1 | 0 |
| Class 4 | 0.003 < π ≤0.004 | 4 | 2 | 2 | 0 |
| Class 5 | π >0.004 | 5 | 2 | 2 | 1 |
A final list of ~70 K SNPs was sent to Thermo Fisher Scientific for the creation of the 60 K SNP array. This 384-format genotyping array was called the MedFish array, reflecting the two European Union funded consortium projects MedAID and PerformFish (see the ‘Acknowledgements’ section for details).
2.6. Testing of the MedFish array through population genomic analyses
2.6.1. Genotyping
A subset of 502 European seabass and 478 gilthead seabream fin clips from the same populations used for SNP discovery was sent to IdentiGEN (Ireland) for DNA extraction and genotyping with the MedFish SNP array (Table 3). To assess the repeatability and quantify the putative error rate of the platform, a single replicate sample (one per species) was genotyped twelve times across three different arrays. Only SNPs with genotype calls across all replicate samples were considered for evaluation (26,569 SNPs for European seabass and 25,547 SNPs for gilthead seabream). The proportion of SNPs at which the (replicate) individuals shared identical-by-state (IBS) alleles was calculated.
Table 3.
Fish samples genotyped using the combined species MedFish SNP array.
| Species | Origin | Population ID | Country | N° individuals typed | N° individuals passing QC |
|---|---|---|---|---|---|
| European seabass | Farmed | Sba_farm_1 | France | 12 | 8 |
| Sba_farm_2 | Spain | 25 | 18 | ||
| Sba_farm_3 | Spain | 25 | 16 | ||
| Sba_farm_4 | Italy | 24 | 24 | ||
| Sba_farm_6 | Croatia | 25 | 16 | ||
| Sba_farm_7 | Greece | 25 | 18 | ||
| Sba_farm_8 | Greece | 25 | 16 | ||
| Sba_farm_9 | Greece | 24 | 16 | ||
| Sba_farm_10 | Greece | 25 | 17 | ||
| Sba_farm_11 | Greece | 24 | 11 | ||
| Sba_farm_12 | Greece | 23 | 6 | ||
| Sba_farm_13 | Cyprus | 23 | 14 | ||
| Sba_farm_14 |
Egypt |
14 |
12 |
||
| Wild |
Sba_wild_Mediterranean |
208 |
184 |
||
| gilthead seabream | Farmed | Sbr_farm_1 | France | 24 | 19 |
| Sbr_farm_2 | Spain | 18 | 17 | ||
| Sbr_farm_3 | Spain | 25 | 19 | ||
| Sbr_farm_5 | Croatia | 19 | 19 | ||
| Sbr_farm_6 | Greece | 13 | 12 | ||
| Sbr_farm_7 | Greece | 13 | 13 | ||
| Sbr_farm_8 | Greece | 21 | 19 | ||
| Sbr_farm_9 | Greece | 24 | 22 | ||
| Sbr_farm_10 | Greece | 20 | 17 | ||
| Sbr_farm_11 | Israel | 13 | 9 | ||
| Sbr_farm_12 |
Egypt |
15 |
14 |
||
| Wild | Sbr_wild_Atlantic | 28 | 27 | ||
| Sbr_wild_Mediterranean | 245 | 221 |
SNP quality control and genotype calling from the intensity files was performed using the Axiom Analysis Suite software v 2.0.035 at default parameter values for diploid species (call rate (CR) > 97; dish QC (DQC) >0.82). Because a significant fraction of the European seabass samples had a CR below the default value of 97 (201 individuals), the threshold was reduced to 93, allowing to recover genotypes for 460 individual samples.
2.6.2. Evaluation of SNP ascertainment bias
SNP markers genotyped with SNP arrays may suffer from a type of bias that is introduced during the array design process at the SNP selection stage. Markers to be included on a platform are typically selected (ascertained) from a larger pool of polymorphisms – discovered in a variable number of individuals from a variable number of populations – based on specific criteria (e.g. MAF threshold, equidistant spacing along the genome, etc.). Particularly when the size and number of SNP discovery populations are limited, this approach can lead to a final panel of markers in which rare SNPs, population-specific SNPs, and more recent SNPs are underrepresented [38]. In turn, this can lead to bias in several downstream population genetic analyses [39], particularly where these analyses are dependent on allele frequencies. To assess whether the SNP selection strategy followed for the development of the MedFish array affects population genetic inferences in European seabass and gilthead seabream, a commonly used summary statistic (FST) was estimated for pairs of populations (pairwise FST) based on the pooled whole-genome sequence data before (‘non-ascertained’ panel) and after SNP selection (‘ascertained’ panel). The ‘non-ascertained’ dataset (i.e. initial SNP discovery panel) reflects an unbiased representation of the ‘true’ genomic variation and allele frequency spectra present in the sampled populations of both fish species, as measured in this study. This dataset comprised ~1.1 million high quality SNPs called across 24 European seabass population pools and ~9 million SNPs genotyped across 27 gilthead seabream population pools. For populations for which two replicate pools were available (see Table 1), only one was selected for evaluation. The SNP QC filters applied to the variants called from the alignment files were the same as described in the ‘Bioinformatics analysis for SNP discovery’ section, except for the removal of markers with interfering SNPs in the flanking region. To generate the ‘ascertained’ datasets (one for each fish species), the SNP positions of the array markers were sub-sampled from the former (‘non-ascertained’) datasets using VCFtools v0.1.15 [37]. The four vcf files – ‘non-ascertained’ and ‘ascertained’ SNP panels for both European seabass and gilthead seabream – were imported to the R package poolfstat v1.2.0 [40] for pairwise FST estimation using the ANOVA method. For each species, the correlation between the pairwise FST matrices generated from the two datasets – ‘non-ascertained’ and ‘ascertained’ SNP panels – was assessed with a Mantel test. For visualization, a principal coordinate analysis (PCoA) was performed based on the Euclidean distances of the pairwise FST matrices using the R package LabDSV [41].
2.6.3. Population structure
The combined species ~60 K MedFish array was tested by performing a metric multidimensional scaling (MDS) analysis of a wide range of Mediterranean (and a few Atlantic) European seabass and gilthead seabream farmed and wild populations typed with the platform. These individuals were part of the same set of samples used for the SNP discovery process from Pool-seq data (Table 3). A QC-filtered SNP dataset was created by applying the following filters in PLINK v2.0 [42]. Bi-allelic SNPs were retained for analysis if they had (i) a call rate > 0.95, (ii) MAF > 0.01, (iii) HWE test p-value ≥1e-4 (estimated separately for each population) and (iv) no pairs with a squared LD correlation (r2) > 0.2 occurred within a 100 Kb window. For duplicated or related individuals with a kinship coefficient (KING-rob) > 0.177 (first-degree relatives or closer), only one member of a pair was retained for further analysis. All individuals to be evaluated required to have <10% missing genotypes. The relationship among individuals and populations was visualized using a MDS analysis based on the genome-wide IBS pairwise distances as implemented in PLINK.
2.6.4. Analysis of haplotype sharing among farms
To assess the ability of the SNP array to identify historical connections between farmed populations, a haplotype sharing analysis was performed on the farmed population samples (13 European seabass farms; 11 gilthead seabream farms). A SNP dataset in which all individual and SNP QC filters had been applied (see ‘Population structure’ section), except the removal of markers based on linkage disequilibrium (measured as r2) was used for the analysis. Markers that were not located on the assembled chromosomes of the reference genome assemblies (i.e. those located on unplaced scaffolds) were removed from the dataset. Haplotypes were inferred for each individual using the software fastPHASE v1.4.8 [43]. All individuals were phased together in a single analysis, taking into consideration their population labels during the model fitting procedure. For both fish species, the number of random starts of the EM algorithm (T) was set to 20, the number of iterations (C) was set to 35, and the number of haplotype clusters (K) to 8.
The reference genomes of both species were divided into 1 Mb non-overlapping windows using BEDTools v2.25 [44]. SNP-based haplotype variants were defined for each window. The last window of each chromosome was excluded from the analysis. Since the number of haplotypes can be influenced by sample size, the same number of individuals were randomly chosen from each farmed population (6 individuals for the European seabass and 9 for the gilthead seabream). For each individual within a farm, the two haplotypes at any given locus were used to screen the whole dataset for an exact match. All matches with other individuals from a different farm were recorded. The totals were then summed across all individuals that belonged to the same farm, and the proportion of shared haplotypes across farms calculated.
To assess whether a pair of farms had excess haplotype sharing, 1000 permutations were performed. For each permutation, all individuals from a fish species were randomly assigned to an arbitrary farm.
2.7. Ethics statement
The fish fin clip collected in this study were obtained from commercial samples or specific sampling efforts managed and sampled in accordance with the European directive 2010/63/UE on the protection of animals used for scientific purposes.
3. Results
3.1. SNP array development
The pooled DNA sequencing of 24 European seabass and 27 gilthead seabream populations and their replicate pools (see Table 1) produced 8205 and 23,784 million paired-end reads, respectively. The alignment of the post-quality filtered reads of the population pools against each species reference genome resulted in the discovery of ~17 million polymorphisms in the European seabass and ~34 million putative polymorphisms in the gilthead seabream genomes (including both SNPs and indels). The generated sequence led to an average coverage at SNP variant sites of 36× in the European seabass and 63× in the gilthead seabream (in both cases, averaged across all pools). After applying the QC filters on the variant call set (see ‘Bioinformatics analysis for SNP discovery’ section), a pool of 1,056,218 and 1,015,264 high confidence SNPs in the European seabass and the gilthead seabream, respectively, remained for SNP selection. The QC filter that removed the largest amount of data was the restriction to retaining SNPs without other polymorphisms in close proximity (within 35 bp on either side). This filter alone removed 88% and 96% of the SNPs discovered in the European seabass and gilthead seabream, respectively.
Following the submission and evaluation of these filtered SNPs by Thermo Fisher Scientific, high-value markers – i.e. those associated with production traits, with an effect on proteins or shared with another array – were tiled on the MedFish array with two sets of probes. The remaining set of SNP from the platform (~24 K in European seabass and ~ 26 K in gilthead seabream) were tiled with a single probe and were obtained from a sampling along chromosomes based on the strategy of selecting SNPs in proportion to the putative local recombination rate (measured as π) of the genomic region. Notably, and in comparison to the European seabass, a particularly high number of polymorphisms were initially discovered along the chromosomes of the gilthead seabream, particularly towards the terminal ends of the chromosome-level scaffolds. The most likely cause was that in this species the higher average sequencing depth of 63× (compared to 36× in the European seabass population pools) enabled the discovery of variants segregating at a lower frequency. Consequently, when the QC filter that removed SNPs with interfering markers in close proximity was applied to the gilthead seabream dataset, a substantial number of markers were filtered out from regions of the genome exhibiting higher levels of genetic polymorphisms. This led to fewer SNPs left to choose from for assay design in regions of the gilthead seabream genome that showed putative higher recombination (e.g. chromosome ends) and for which a higher number of SNPs had to be sampled based on our SNP selection strategy. Therefore, the SNP selection strategy led to a more even sampling of SNPs along the gilthead seabream genome. While in European seabass, the array SNPs followed the expected pattern of the SNP selection strategy, with more markers being assayed towards the terminal ends of the chromosome-level scaffolds (Fig. 1).
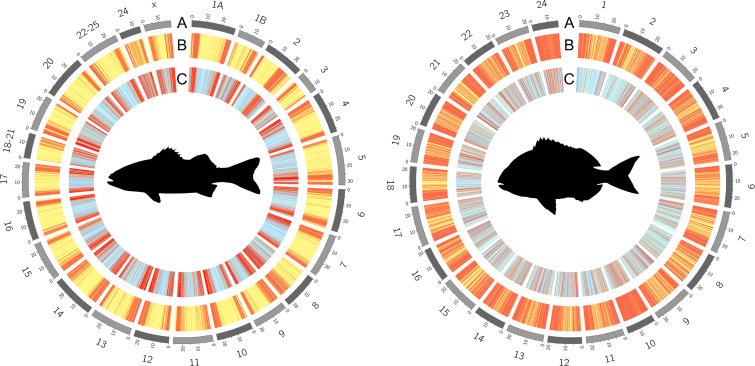
Fig. 1.
Schematic representation of the distribution of array markers in the European seabass (left) and gilthead seabream (right) genomes after following a SNP selection strategy based on local nucleotide diversity. (A) Chromosome number. (B) Levels of diversity (π) estimated over 70 Kb and 85 Kb windows in the European seabass and gilthead seabream, respectively. Red bars represent regions with high nucleotide diversity. (C) Genome-wide distribution of markers on the combined-species SNP chip. Light blue bars represent windows for which 1–3 SNPs were selected. Red bars represent windows for which more than four SNPs were selected.
The final MedFish SNP array was designed to interrogate 29,888 SNPs in the European seabass genome and 29,807 SNPs in the gilthead seabream genome. Among these markers, 4560 SNPs (15%) in the European seabass and 3208 SNPs (11%) in the gilthead seabream are shared with other SNP arrays that were being developed at the time of this study [25]. A significant fraction of the SNPs on the platform are located in genes (46% seabass; 32% seabream), among which 107 and 179 SNPs, respectively, were predicted in silico to have high functional effects on proteins. For the SNPs included on the array, the physical distance between consecutive markers was similar for both species and averaged 20 Kb in the European seabass and 18 Kb in the gilthead seabream (density plot of inter-marker distances shown in Fig. S1). The largest gaps between markers (200–300 Kb) represented a small fraction of the platform and comprised five regions on chromosomes 1, 4, 9, 13 and 16 of the gilthead seabream, which summed up to ~1.4% of the genome [14]. Detailed examination revealed that these regions lacked suitable markers matching our SNP selection criteria. No large regions in the European seabass genome were devoid of assays, with the highest inter-marker distance being ~120 Kb, which comprised two pairs of markers on chromosomes 5 and 20, and represented <0.05% of the European seabass genome [13].
Two metrics were used to assess the performance of the assays on the array: (i) conversion rate and (ii) platform error rate. Here the conversion rate is defined to be the fraction of probes that yielded strong signals with high-quality clusters discerning different genotypes. The conversion rate of the European seabass fraction of the array was 91.9%, whereas the gilthead seabream assays on the array had a conversion rate of 88.8% (Table 4). In terms of the informativeness of the markers on the platform, for 99.8% of the validated loci in the European seabass the MAF was >5%. In the gilthead seabream, 98.7% of the markers had a MAF > 5%. The process of calculating the platform error rate involved genotyping two samples (one per species) twelve times each. For European seabass, one replicate sample failed to generate a CEL file; consequently, eleven samples remained for evaluation. The repeatability of the assays, after excluding loci with at least one missing value across replicates, was 99.4% for the European seabass and 99.8% for the gilthead seabream. Taken together, these metrics support the high quality and reliability of the genotype data generated by the MedFish SNP array.
Table 4.
Number of SNPs for each species of each Axiom quality class.
| Conversion typea | № European seabass (%) | № gilthead seabream (%) |
|---|---|---|
| Polymorphic high resolution | 26,466 (88.55%) | 26,369 (88.47%) |
| No minor homozygote | 993 (3.32%) | 75 (0.25%) |
| Total high quality polymorphic | 27,459 (91.87%) | 26,444 (88.72%) |
| Monomorphic high resolution | 26 (0.09%) | 36 (0.12%) |
| Off-target-variant (OTV) | 50 (0.17%) | 78 (0.26%) |
| Call rate below threshold (97%) | 889 (2.97%) | 1292 (4.33%) |
| Other | 1464 (4.90%) | 1957 (6.57%) |
| Total SNPs on the array | 29,888 (100%) | 29,807 (100%) |
The categories are based on cluster properties and QC metrics.
aThe Conversion type follows Thermo Fisher's terminology:
PolyHighResolution = Class with the highest quality probes. SNP is polymorphic and the presence of both the major and minor homozygous clusters is observed.
NoMinorHom = similar to a PolyHighResolution, but no evidence of individuals with minor homozygous genotypes, presumably due to a low genotype frequency.
MonoHighResolution = SNP can reliably be scored as monomorphic.
Off-target variant (OTV) = SNPs where additional (i.e. more than three) clusters are observed, making genotype calling ambiguous.
CallRateBelowThreshold = SNP with the expected number of clusters (usually 3, one for each possible genotype), but where the proportion of samples scored at the SNP falls below a user-defined threshold.
Other = SNPs that do not fall in any of the above categories.
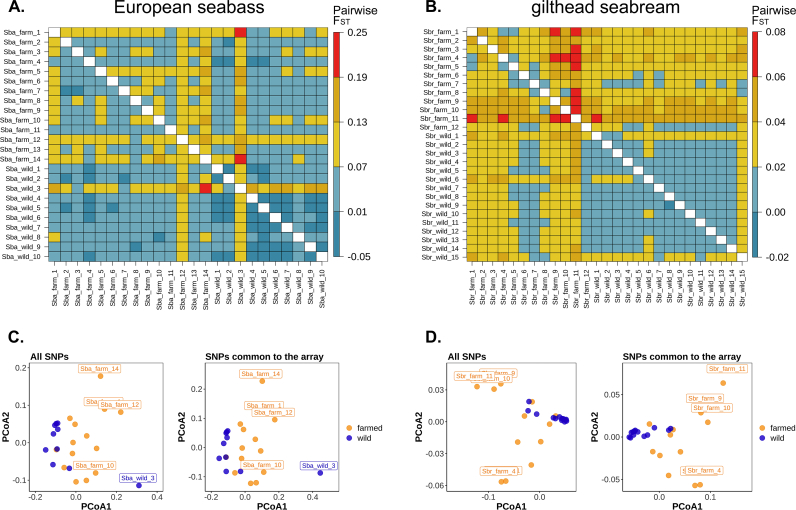
To evaluate whether the SNP selection strategy followed for the design of the MedFish array could bias FST estimates, SNPs common to the array (~30 K for each species) were selected from the whole-genome sequence data of the population pools (consisting of 11–25 individuals each) to mimic array-derived genotype data. Pairwise FST were calculated from this ‘ascertained’ dataset and then compared to those estimates obtained for the same population pools but by including all sites discovered through the re-sequencing experiment in the analysis (~1.1 million SNPs in the European seabass; ~9 million SNPs in the gilthead seabream) (‘non-ascertained’ dataset). Overall, a high correlation between the pairwise FST matrices calculated from the ‘non-ascertained’ and ‘ascertained’ SNP panels was observed, with an r = 0.97 for both European seabass and gilthead seabream (Fig. 2A and B). Although for both fish species most of the pairwise FST values were slightly higher when the estimations were based on the ‘ascertained’ dataset (Fig. S2), both ‘ascertained’ and ‘non-ascertained’ datasets yielded similar population clustering patterns, as observed across the first and second dimension of a PCoA (Fig. 2C and D).
Fig. 2.
Evaluation of SNP ascertainment on pairwise FST estimates in European seabass and gilthead seabream pooled population samples. Pairwise FST values obtained for (A) European seabass and (B) gilthead seabream populations based on a ‘non-ascertained’ marker panel (below the diagonal) and an ‘ascertained’ panel containing the array SNPs (above the diagonal). Principle coordinate analysis of pairwise FST among (C) European seabass and (D) gilthead seabream populations, and comparison between results obtained from a ‘non-ascertained’ (left panel) vs ‘ascertained’ (right panel) set of SNPs.
3.2. Population structure
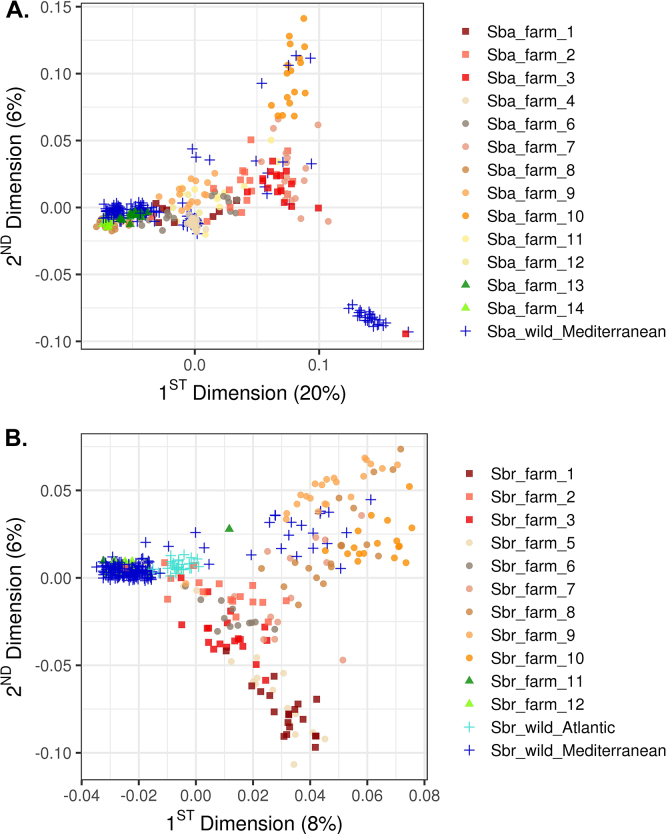
To test the SNP array and to gain a general overview on the population structure within each species, a MDS analysis was performed. The two first dimensions explained 26% and 14% of the total variance for European seabass and gilthead seabream, respectively (Fig. 3).
Fig. 3.
MDS analysis performed on individuals from farmed and wild (A) European seabass and (B) gilthead seabream populations. The different point symbols separate samples by origin in (i) farms from the West of the Mediterranean (■), (ii) farms from the centre of the Mediterranean (●), and (iii) farms from the East of the Mediterranean (▲), from (iv) wild populations (+).
In European seabass, most of the sampled farmed populations form a loose cluster along D1, which explains 20% of the variance. No geographical cline is observed as farms from the West (France, Spain), centre (Italy, Croatia, Greece) and East (Cyprus, Egypt) of the Mediterranean cluster at least partially in this dimension. On the other hand, three distinctive clusters are recognized for the wild European seabass populations, with a few exceptions corresponding to individuals clustering near farmed populations instead. D2 explains 6% of the total variation and mainly separates (i) a single well-defined wild population cluster, (ii) a large group containing most of the farmed and wild seabass populations, and (iii) a group of individuals that belong to a farm sampled from the centre of the Mediterranean (farm № 10 sampled from a Greek hatchery) (Fig. 3A).
For the gilthead seabream analysis, the sampled farmed populations appear to form a continuum along D1 rather than discrete units. Although the majority of gilthead seabream wild populations were sampled from the Mediterranean Sea, a few populations from the Atlantic coast of France and Spain were included in the analysis. Individuals sampled from a wide range of wild populations tend to group by origin into either a Mediterranean or Atlantic cluster on one extreme of the D1 axis. D2 accounts for 6% of the variance and distinguishes two groups of overlapping farmed populations that partially coincide with their macro-region of origin. The first group is composed only of farms located in the centre of the Mediterranean (i.e. Greece). A few wild gilthead seabream individuals co-localize with this group of farmed samples. The second group is comprised of a mixture of all three farms sampled from the West of the Mediterranean (i.e. from either France or Spain) and a few populations sampled from farms located in the centre of the Mediterranean, namely farms № 5 (from Croatia) and № 6 (from Greece) (Fig. 3B).
3.3. Haplotype sharing analysis among farms
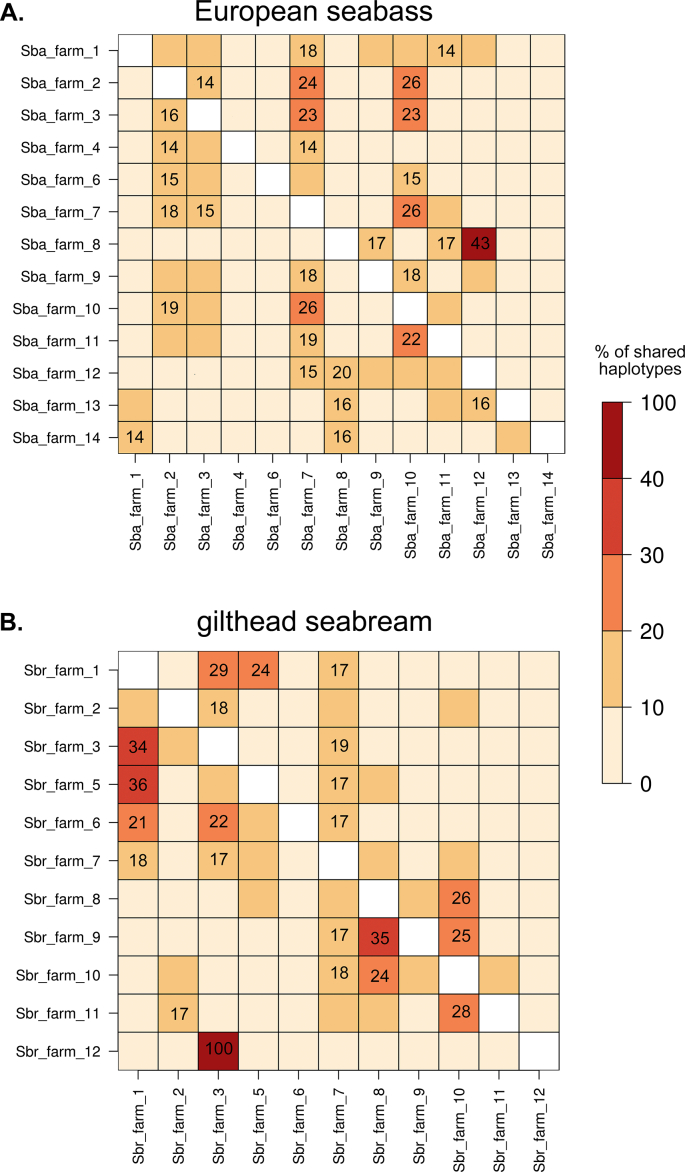
After applying QC filters, a total of 21,822 SNPs in European seabass and 24,765 SNPs in the gilthead seabream remained for the assessment of haplotype sharing between pairs of Mediterranean fish farms.
The pairwise comparison among European seabass farmed population samples revealed that all populations showed an excess of haplotype sharing with at least one other Mediterranean farm (Fig. 4A). The highest percentage of haplotype sharing (43%) was found between two Greek seabass farms (farm № 8 vs farm № 12). The reverse relationship between these two farms (i.e. farm № 12 vs farm № 8) is also significant but is ranked 9th (20%) in terms of haplotype-sharing between all pairs of farmed populations. This difference in reciprocal comparisons is explained by differences in the total numbers of shared haplotypes identified within each farm (File S1). Haplotypes from individuals of a European seabass farm located in Greece (farm № 7) were present at significant frequencies in all farms sampled from the West of the Mediterranean (i.e. farms of French or Spanish origin), and most of the seabass farms sampled from the centre of the Mediterranean (i.e. either from Italy, Croatia or Greece).
Fig. 4.
Heatmaps indicating the percentage of shared haplotypes between pairs of (A) European seabass and (B) gilthead seabream farmed populations. The colorbar on the right indicates the percentage of shared haplotypes. Entries are shown for pairs of farms with a statistically significant excess of shared haplotypes (p-value <0.05).
In common with the results observed for European seabass, all gilthead seabream farms evaluated show an excessive sharing of haplotypes with at least one other Mediterranean farm (Fig. 4B). In gilthead seabream, a clear geographical break (with farm № 7 forming the boundary) separates the farms from the West and centre of the Mediterranean in two groups of farms between which reduced haplotype sharing is observed. One group includes five farms – farms № 1, 2, 3, 5, and 6 – from diverse geographical origins (i.e. France, Spain, Croatia and Greece). The second group comprises commercial farms exclusively based in Greece – farms № 8, 9 and 10. The farm in the boundary of both groups – farm № 7 – had haplotypes that were present at significant levels in farms belonging to both aforementioned groups. A seabream farm sampled from the East of the Mediterranean (farm № 12) had the lowest total number of shared haplotypes among all commercial farms evaluated from both fish species. Most haplotypes identified in farm № 12 were unique and specific to the farm, which showed complete absence of shared variants with all but one Mediterranean farm (i.e. farm № 3), with which it shared only two haplotypes in total (File S1).
4. Discussion
4.1. Properties of the combined species MedFish SNP array
A publicly available, combined species SNP chip that assays ~30 K SNPs throughout the genome of two prominent Mediterranean fish species - the European seabass and the gilthead seabream – was developed. To evaluate the performance of the MedFish SNP array two metrics were analyzed: conversion rate and platform error rate. The conversion rate is a measure of the number of SNPs successfully assayed by a technology and reflects the quality of both the chosen SNPs and the technology used to score them [45]. Conversion rates were high for the SNP array regardless of the fish species. The assay conversion rate of the European seabass part of the array was 91.9%, while for the gilthead seabream part it was 88.8%. These values are slightly lower than assays designed for terrestrial livestock species (e.g. 92.6% in cattle and 97.5% in pigs), however, they are comparatively higher than similar assays developed for aquatic organisms (e.g. 72.5% in oysters and 86.1% in catfish) [18,[46], [47], [48]], and in the upper range of high performing arrays for fish species [15,25]. As a second metric to assess performance, the platform error rate was calculated based on the genotype concordance of repeated assays on the same individual. By this metric, the MedFish platform shows high genotype accuracy, with a repeatability ranging from 99.4% to 99.7%. These levels of accuracy are comparable to those achieved with Illumina GoldenGate assays in humans (99.7%) or Affymetrix SNP chips in trout (99.4%) and pig (100%) [15,46,49]. Compared to other SNP arrays developed for aquaculture species, the MedFish platform has among the best performance in terms of genotype accuracy and repeatability. To assess potential ascertainment bias arising in chip-derived genotype data, pairwise FST was estimated for the same populations using a ‘non-ascertained’ and an 'ascretained' SNP panel for comparison. The patterns of pairwise FST among populations were highly concordant when results based on the markers on the MedFish platform were compared to the full SNP discovery panel (Fig. 2), which supports the utility of the platform for population genetic studies. However, a slight upward bias of FST values was observed in the dataset that mimics the array-derived markers and is likely caused by prioritizing common high-frequency markers, at the expense of SNPs with rarer alleles, in the SNP selection process. While the differences between FST estimates between both datasets (‘ascertained’ versus ‘non-ascertained’) were minor (Fig. S2), additional fish populations that were not part of the ascertainment panel should be evaluated to accurately assess the impact of chip-based genotyping data on population genetics estimators for these species [50]. Until recently, high-throughput genotyping analysis was typically only achievable in these fish species by means of reduced-representation sequencing approaches [9,11,34]. Although these genotyping by sequencing techniques can be particularly cost-effective for small numbers of samples and should also display minimal ascertainment bias compared to SNP arrays [51] (however, see [52]), they can suffer from inconsistent marker recovery across experiments and a comparatively lower robustness to low quality input DNA [53]. Hence, the development of this combined species SNP array represents a powerful alternative for high-throughput genotyping in European seabass and gilthead seabream, which may facilitate molecular breeding applications, genetic stock identification and population and evolutionary studies in these emblematic fish species. Moreover, the fact that the two species are represented on the same platform increases the overall volume of arrays that can be purchased, which should reduce the cost of the array per sample due to economy of scale. While it should be noted that the number of arrays required to reach this lower per-sample cost is high (multiples of 384 in the case of the MedFish platform), this improved cost-effectiveness is likely to be important for the uptake of the platform by aquaculture breeding and production companies for the routine application of genomic selection.
As part of the SNP array design, ~25 farmed and wild populations (>500 individuals) per species were sequenced and screened for informative markers. By following a DNA pooling approach, reliable genome-wide allele frequency information was obtained for several fish populations at a fraction of the effort of individual sequencing. Given that the majority of the samples genotyped with the SNP array were also part of the SNP discovery process, metrics such as number and mean MAF of polymorphic markers reflected the performance of the SNP selection strategy. Despite the relatively small DNA pools (11–25 individuals), we were able to reliably identify and select informative markers for inclusion in our SNP array. The number of informative markers (MAF > 0) was high for both fish species. For the European seabass, 23,900 SNPs (99.8%) of the validated markers were polymorphic, whereas for the gilthead seabream, this type of markers comprised 26,017 SNPs (99.3%) of the data. The number of polymorphic markers was remarkably similar in wild and farmed populations of both species (90–99% across populations), demonstrating the efficacy of the SNP selection strategy for recovering highly informative markers in Pool-seq data sequenced at a high to moderate coverage across a wide range of different populations. When evaluating the MAF across European seabass and gilthead seabream populations, the allele frequency profiles were similar within species and did not vary significantly by origin (either wild or farmed) (Fig. S3). The mean MAF across the European seabass (0.33) and gilthead seabream (0.31) populations was higher than that reported when validating SNP arrays in Nile tilapia (0.29) and rainbow trout (0.25) [15,22]. However, the high average MAF observed in this study is most likely influenced by the fact that most of the discovery populations were also used for the validation of the SNP chip. Nonetheless, it should be noted that the discovery population samples cover a large portion of the distribution range in the wild and include the majority of commercial hatcheries for the two species.
A significant obstacle to the uptake of high-throughput genotyping technologies by the industry is the risk that a low fraction of a pre-built platform yields useful information. Indeed ascertainment bias is a common issue for genotyping arrays and can be caused when designing platforms based on a reduced number of individuals [50]. Due to the fact that the MedFish 60 K array was developed based on the screening of genetic data derived from extensive sampling of dozens of Mediterranean fish populations and hundreds of fish of each species, it is tailored to maximize the retrieval of genetic information and provide an increased resolution for the analysis of farmed or wild stocks from this region.
4.2. Population structure and haplotype sharing analysis
To test the MedFish SNP array, the genotyping data obtained from typing a diverse range of wild and farmed European seabass and gilthead seabream fish were used to perform a MDS and haplotype sharing analysis.
Regarding the European seabass populations, the two first dimensions explained 26% of the genotypic variation. Interestingly, the wild Mediterranean populations span a continuum across the range of D1, but has a rather smaller dispersal across the D2 range. However, this continuum in D1 has gaps, and the wild populations seem to be divided into two clearly differentiated clusters located at each end of this major axis. These two clusters are represented by (i) distinct groups of wild individuals showing low levels of genetic differentiation, consistent with a sub-division (albeit weak) of the European seabass Mediterranean lineage [54]; and (ii) a wild population sampled from Morocco, which based on previous findings [54] is expected to cluster with Atlantic rather than Mediterranean populations. All farmed European seabass populations have a more limited distribution across the range of the two dimensions compared to the wild populations, forming clear clusters although less dense than the wild populations, probably due to their smaller sample sizes (Table 3). Most farm populations fall within the range of the wild populations, with overlap among each other. A few of the wild individuals fall clearly within the range of farm populations, which could indicate the presence of escapees from fish farms in the set of wild samples, reflecting a well-known phenomenon occurring in the Mediterranean [[55], [56], [57]]. Only a single farmed fish population seems to cluster separately from the other farm samples (Fig. 3A; farm № 10), suggesting either founder effects, stronger artificial selection, higher number of generations of selection, or any combination thereof. European seabass farms of different geographical origin tend to cluster together in the plot. For instance, farms № 2 and 3 (from Spain) group with farm № 7 (from Greece). This observation is consistent with the haplotype sharing analysis, as a significant number of 1 Mb SNP-based haplotype variants were jointly present in these farms. A high frequency of shared haplotypes between pairs of populations provides information about their historical relationship, reflecting either a common ancestry and/or gene flow between populations. In the context of aquaculture farming, a high frequency of shared haplotypes between farms might indicate (i) animal transfer between farms or (ii) the recent establishment of these farmed populations from the same wild source (i.e. recent population divergence). Since the MDS analysis revealed that wild populations of European seabass form tight and distinctive clusters, it is likely that pairs of European seabass farms sharing a high frequency of haplotype variants are derived from human-mediated translocations of eggs or juveniles.
For the gilthead seabream populations, MDS results explained much less of the observed genetic variance (only 14% covered by both D1 and D2 summed up), showing a less clear structure than European seabass for most of the populations sampled in this study. In this case, wild populations were sampled from two regions, the Mediterranean and Atlantic. Wild individuals segregate into two closely bound Mediterranean and Atlantic clusters, which is consistent with previous findings indicating a low genetic differentiation between regions [58,59]. Similar to European seabass, a few wild individuals are found scattered throughout farmed populations, likely representing escapees from local fish farms. Farmed gilthead seabream populations seem to be much more differentiated compared to their wild counterparts, with two broader clusters forming a gradient of overlapping farmed populations. The first group is composed only of farms located in Greece. The second cluster groups a mixture of all three farms sampled from the West of the Mediterranean (either France or Spain) and a few farms from Croatia and Greece (Fig. 3B). This pattern may reflect artificial selection and/or different degrees of admixture between farms. The haplotype sharing analysis mirrors this finding and reinforces the idea that most seabream farms from the Mediterranean separate in two clusters, between which a reduced recent contact is observed. However, while the results for both the European seabass and gilthead seabream highlight the potential utility of the SNP array for detecting and studying population structure, more extensive studies are required to further assess these phenomena in the two species.
5. Conclusions
A medium density SNP array suitable for genotyping both the European seabass and the gilthead seabream was developed. The MedFish SNP array is a robust resource with a high assay performance, as demonstrated by its high conversion rate (92% in the European seabass; 89% in the gilthead seabream) and repeatability (99.4% in the European seabass; 99.8% in the gilthead seabream). The platform interrogates ~30 K markers in each fish species and includes features such as SNPs previously shown to be associated with performance traits and enrichment for SNPs predicted to have high functional effects on proteins. The SNP array was highly informative when tested on the majority of the discovery population samples and was further validated by performing a population structure and haplotype sharing analysis across a wide range of fish populations from diverse geographical backgrounds. This recently developed platform will allow the efficient and accurate high-throughput genotyping of ~30 K SNPs across the genomes of each fish species, facilitating population genomic research and the application of genomic selection for the acceleration of genetic improvement in European seabass and gilthead seabream breeding programs.
Data availability
Raw sequence reads from the European seabass and gilthead seabream population pools analyzed for SNP discovery have been deposited in NCBI's Sequence Read Archive (SRA) under accession number PRJEB40423. Details of the allele frequencies of the SNPs on the MedFish array can be found in the Mendeley Data Repository (http://dx.doi.org/10.17632/7w4cb4mdd4.1).
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
This study was made possible by the EU projects MedAID (H2020 grant agreement No 727315) and PerformFish (H2020 grant agreement No 727610). We are grateful to both projects' partners that contributed with samples for SNP discovery as well as the projects' coordinators (A. López-Francos, B. Basurco, D. Furones and K. Moutou) and the H2020 project officer for enhancing a fruitful interaction between research consortia. The authors would also like to thank Christos Palaiokostas and Sara Faggion for providing additional information for trait-associated variants included in the SNP array. CP and RH are supported by funding from the BBSRC Institute Strategic Programme Grants BB/P013759/1 and BB/P013740/1. We also give thanks to the sequencing facilities Edinburgh Genomics (University of Edinburgh, UK) and the Norwegian Sequencing Centre (University of Oslo, Norway). TM would like to thank George Nomikos for valuable support on data analysis. This research was supported through computational resources provided by IMBBC (Institute of Marine Biology, Biotechnology, and Aquaculture) of the HCMR (Hellenic Centre for Marine Research). Funding for establishing the IMBBC HPC has been received by the MARBIGEN (EU Regpot) project, LifeWatchGreece RI, and the CMBR (Centre for the study and sustainable exploitation of Marine Biological Resources) RI. We also thank the GeneSea consortium (n° R FEA 4700 16 FA 100 0005) for providing some variants allowing interoperability with existing arrays.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ygeno.2021.04.038.
Contributor Information
R.D. Houston, Email: ross.houston@roslin.ed.ac.uk.
C.S. Tsigenopoulos, Email: tsigeno@hcmr.gr.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Houston R.D., Bean T.P., Macqueen D.J., Gundappa M.K., Jin Y.H., Jenkins T.L., Selly S.L.C., Martin S.A.M., Stevens J.R., Santos E.M., Davie A., Robledo D. Harnessing genomics to fast-track genetic improvement in aquaculture. Nat. Rev. Genet. 2020;21:389–409. doi: 10.1038/s41576-020-0227-y. [DOI] [PubMed] [Google Scholar]
- 2.Fernández J., Toro M.Á., Sonesson A.K., Villanueva B. Optimizing the creation of base populations for aquaculture breeding programs using phenotypic and genomic data and its consequences on genetic progress. Front. Genet. 2014;5 doi: 10.3389/fgene.2014.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moen T., Baranski M., Sonesson A.K., Kjøglum S. Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics. 2009;10:368. doi: 10.1186/1471-2164-10-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houston R.D., Haley C.S., Hamilton A., Guy D.R., Tinch A.E., Taggart J.B., McAndrew B.J., Bishop S.C. Major quantitative trait loci affect resistance to infectious pancreatic necrosis in Atlantic Salmon (Salmo salar) Genetics. 2008;178:1109–1115. doi: 10.1534/genetics.107.082974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraslin C., Quillet E., Rochat T., Dechamp N., Bernardet J.-F., Collet B., Lallias D., Boudinot P. Combining multiple approaches and models to dissect the genetic architecture of resistance to infections in fish. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horn S.S., Ruyter B., Meuwissen T.H.E., Moghadam H., Hillestad B., Sonesson A.K. GWAS identifies genetic variants associated with omega-3 fatty acid composition of Atlantic salmon fillets. Aquaculture. 2020;514:734494. [Google Scholar]
- 7.Meuwissen T.H., Hayes B.J., Goddard M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zenger K.R., Khatkar M.S., Jones D.B., Khalilisamani N., Jerry D.R., Raadsma H.W. Genomic selection in aquaculture: application, limitations and opportunities with special reference to marine shrimp and pearl oysters. Front. Genet. 2019;9 doi: 10.3389/fgene.2018.00693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palaiokostas C., Cariou S., Bestin A., Bruant J.S., Haffray P., Morin T., Cabon J., Allal F., Vandeputte M., Houston R.D. Genome-wide association and genomic prediction of resistance to viral nervous necrosis in European sea bass (Dicentrarchus labrax) using RAD sequencing. Genet. Sel. Evol. 2018;50:30. doi: 10.1186/s12711-018-0401-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palaiokostas C., Ferraresso S., Franch R., Houston R.D., Bargelloni L. Genomic prediction of resistance to pasteurellosis in gilthead sea bream (Sparus aurata) using 2b-RAD sequencing. G3: Genes|Genomes|Genetics. 2016;6:3693–3700. doi: 10.1534/g3.116.035220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aslam M.L., Carraro R., Bestin A., Cariou S., Sonesson A.K., Bruant J.-S., Haffray P., Bargelloni L., Meuwissen T.H.E. Genetics of resistance to photobacteriosis in gilthead sea bream (Sparus aurata) using 2b-RAD sequencing. BMC Genet. 2018;19:43. doi: 10.1186/s12863-018-0631-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eurostat . Eurostat Statistical Books. 2019. Agriculture, forestry and fishery statistics. [Google Scholar]
- 13.Tine M., Kuhl H., Gagnaire P.-A., Louro B., Desmarais E., Martins R.S.T., Hecht J., Knaust F., Belkhir K., Klages S., Dieterich R., Stueber K., Piferrer F., Guinand B., Bierne N., Volckaert F.A.M., Bargelloni L., Power D.M., Bonhomme F., Canario A.V.M., Reinhardt R. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat. Commun. 2014;5:5770. doi: 10.1038/ncomms6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pauletto M., Manousaki T., Ferraresso S., Babbucci M., Tsakogiannis A., Louro B., Vitulo N., Quoc V.H., Carraro R., Bertotto D., Franch R., Maroso F., Aslam M.L., Sonesson A.K., Simionati B., Malacrida G., Cestaro A., Caberlotto S., Sarropoulou E., Mylonas C.C., Power D.M., Patarnello T., Canario A.V.M., Tsigenopoulos C., Bargelloni L. Genomic analysis of Sparus aurata reveals the evolutionary dynamics of sex-biased genes in a sequential hermaphrodite fish. Commun. Biol. 2018;1:119. doi: 10.1038/s42003-018-0122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palti Y., Gao G., Liu S., Kent M.P., Lien S., Miller M.R., Rexroad C.E., 3rd, Moen T. The development and characterization of a 57K single nucleotide polymorphism array for rainbow trout. Mol. Ecol. Resour. 2015;15:662–672. doi: 10.1111/1755-0998.12337. [DOI] [PubMed] [Google Scholar]
- 16.Yáñez J.M., Naswa S., Lopez M.E., Bassini L., Correa K., Gilbey J., Bernatchez L., Norris A., Neira R., Lhorente J.P., Schnable P.S., Newman S., Mileham A., Deeb N., Di Genova A., Maass A. Genomewide single nucleotide polymorphism discovery in Atlantic salmon (Salmo salar): validation in wild and farmed American and European populations. Mol. Ecol. Resour. 2016;16:1002–1011. doi: 10.1111/1755-0998.12503. [DOI] [PubMed] [Google Scholar]
- 17.Houston R.D., Taggart J.B., Cézard T., Bekaert M., Lowe N.R., Downing A., Talbot R., Bishop S.C., Archibald A.L., Bron J.E., Penman D.J., Davassi A., Brew F., Tinch A.E., Gharbi K., Hamilton A. Development and validation of a high density SNP genotyping array for Atlantic salmon (Salmo salar) BMC Genomics. 2014;15:90. doi: 10.1186/1471-2164-15-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Q., Fu Q., Li Y., Waldbieser G., Bosworth B., Liu S., Yang Y., Bao L., Yuan Z., Li N., Liu Z. Development of a 690 K SNP array in catfish and its application for genetic mapping and validation of the reference genome sequence. Sci. Rep. 2017;7:40347. doi: 10.1038/srep40347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S., Sun L., Li Y., Sun F., Jiang Y., Zhang Y., Zhang J., Feng J., Kaltenboeck L., Kucuktas H., Liu Z. Development of the catfish 250K SNP array for genome-wide association studies. BMC Res. Notes. 2014;7:135. doi: 10.1186/1756-0500-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J., Zhao Z., Zhang X., Zheng X., Li J., Jiang Y., Kuang Y., Zhang Y., Feng J., Li C., Yu J., Li Q., Zhu Y., Liu Y., Xu P., Sun X. Development and evaluation of the first high-throughput SNP array for common carp (Cyprinus carpio) BMC Genomics. 2014;15:307. doi: 10.1186/1471-2164-15-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nugent C.M., Leong J.S., Christensen K.A., Rondeau E.B., Brachmann M.K., Easton A.A., Ouellet-Fagg C.L., Crown M.T.T., Davidson W.S., Koop B.F., Danzmann R.G., Ferguson M.M. Design and characterization of an 87k SNP genotyping array for Arctic charr (Salvelinus alpinus) PLoS One. 2019;14 doi: 10.1371/journal.pone.0215008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peñaloza C., Robledo D., Barría A., Trịnh T.Q., Mahmuddin M., Wiener P., Benzie J.A.H., Houston R.D. Development and validation of an open access SNP array for Nile tilapia (Oreochromis niloticus) G3: Genes|Genomes|Genetics. 2020;10:2777–2785. doi: 10.1534/g3.120.401343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joshi R., Árnyasi M., Lien S., Gjøen H.M., Alvarez A.T., Kent M. Development and validation of 58K SNP-array and high-density linkage map in Nile Tilapia (O. niloticus) Front. Genet. 2018;9 doi: 10.3389/fgene.2018.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yáñez J.M., Yoshida G., Barria A., Palma-Véjares R., Travisany D., Díaz D., Cáceres G., Cádiz M.I., López M.E., Lhorente J.P., Jedlicki A., Soto J., Salas D., Maass A. High-throughput single nucleotide polymorphism (SNP) discovery and validation through whole-genome resequencing in Nile tilapia (Oreochromis niloticus) Mar. Biotechnol. 2020;22:109–117. doi: 10.1007/s10126-019-09935-5. [DOI] [PubMed] [Google Scholar]
- 25.Griot R., Allal F., Phocas F., Brard-Fudulea S., Morvezen R., Bestin A., Haffray P., François Y., Morin T., Poncet C., Vergnet A., Cariou S., Brunier J., Bruant J.-S., Peyrou B., Gagnaire P.-A., Vandeputte M. Genome-wide association studies for resistance to viral nervous necrosis in three populations of European sea bass (Dicentrarchus labrax) using a novel 57k SNP array DlabChip. Aquaculture. 2021;530:735930. [Google Scholar]
- 26.Aljanabi S.M., Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S., Zhou Y., Chen Y., Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. S. Genome project data processing, the sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrison E., Marth G. arXiv. 2012. Haplotype-based variant detection from short-read sequencing. [Google Scholar]
- 31.Tange O. Login: The USENIX Magazine, Frederiksberg, Denmark. 2011. GNU parallel - the command-line power tool; p. 5. [Google Scholar]
- 32.Babbucci M., Ferraresso S., Pauletto M., Franch R., Papetti C., Patarnello T., Carnier P., Bargelloni L. An integrated genomic approach for the study of mandibular prognathism in the European seabass (Dicentrarchus labrax) Sci. Rep. 2016;6:38673. doi: 10.1038/srep38673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faggion S., Vandeputte M., Chatain B., Gagnaire P.-A., Allal F. Population-specific variations of the genetic architecture of sex determination in wild European sea bass Dicentrarchus labrax L. Heredity (Edinb) 2019;122:612–621. doi: 10.1038/s41437-018-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyriakis D., Kanterakis A., Manousaki T., Tsakogiannis A., Tsagris M., Tsamardinos I., Papaharisis L., Chatziplis D., Potamias G., Tsigenopoulos C.S. Scanning of genetic variants and genetic mapping of phenotypic traits in gilthead sea bream through ddRAD sequencing. Front. Genet. 2019;10 doi: 10.3389/fgene.2019.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leitwein M., Guinand B., Pouzadoux J., Desmarais E., Berrebi P., Gagnaire P.-A. G3 (Bethesda, Md.) 2017. A dense brown trout (Salmo trutta) linkage map reveals recent chromosomal rearrangements in the salmo genus and the impact of selection on linked neutral diversity; pp. 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R., G. Genomes Project Analysis The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lachance J., Tishkoff S.A. SNP ascertainment bias in population genetic analyses: why it is important, and how to correct it. BioEssays: News Rev. Mol. Cell. Dev. Biol. 2013;35:780–786. doi: 10.1002/bies.201300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malomane D.K., Reimer C., Weigend S., Weigend A., Sharifi A.R., Simianer H. Efficiency of different strategies to mitigate ascertainment bias when using SNP panels in diversity studies. BMC Genomics. 2018;19:22. doi: 10.1186/s12864-017-4416-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hivert V., Leblois R., Petit E.J., Gautier M., Vitalis R. Measuring genetic differentiation from pool-seq data. Genetics. 2018;210:315–330. doi: 10.1534/genetics.118.300900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts D.W. 2012. Package ‘LabDSV’. [Google Scholar]
- 42.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheet P., Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardenbol P., Yu F., Belmont J., MacKenzie J., Bruckner C., Brundage T., Boudreau A., Chow S., Eberle J., Erbilgin A., Falkowski M., Fitzgerald R., Ghose S., Iartchouk O., Jain M., Karlin-Neumann G., Lu X., Miao X., Moore B., Moorhead M., Namsaraev E., Pasternak S., Prakash E., Tran K., Wang Z., Jones H.B., Davis R.W., Willis T.D., Gibbs R.A. Highly multiplexed molecular inversion probe genotyping: over 10,000 targeted SNPs genotyped in a single tube assay. Genome Res. 2005;15:269–275. doi: 10.1101/gr.3185605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramos A.M., Crooijmans R.P.M.A., Affara N.A., Amaral A.J., Archibald A.L., Beever J.E., Bendixen C., Churcher C., Clark R., Dehais P., Hansen M.S., Hedegaard J., Hu Z.-L., Kerstens H.H., Law A.S., Megens H.-J., Milan D., Nonneman D.J., Rohrer G.A., Rothschild M.F., Smith T.P.L., Schnabel R.D., Van Tassell C.P., Taylor J.F., Wiedmann R.T., Schook L.B., Groenen M.A.M. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS ONE. 2009;4:e6524. doi: 10.1371/journal.pone.0006524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi H., Song K., Li C., Wang W., Li B., Li L., Zhang G. Construction and evaluation of a high-density SNP array for the Pacific oyster (Crassostrea gigas) PLoS One. 2017;12 doi: 10.1371/journal.pone.0174007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matukumalli L.K., Lawley C.T., Schnabel R.D., Taylor J.F., Allan M.F., Heaton M.P., O’Connell J., Moore S.S., Smith T.P.L., Sonstegard T.S., Van Tassell C.P. Development and characterization of a high density SNP genotyping assay for cattle. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan J.B., Oliphant A., Shen R., Kermani B.G., Garcia F., Gunderson K.L., Hansen M., Steemers F., Butler S.L., Deloukas P., Galver L., Hunt S., McBride C., Bibikova M., Rubano T., Chen J., Wickham E., Doucet D., Chang W., Campbell D., Zhang B., Kruglyak S., Bentley D., Haas J., Rigault P., Zhou L., Stuelpnagel J., Chee M.S. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 2003;68:69–78. doi: 10.1101/sqb.2003.68.69. [DOI] [PubMed] [Google Scholar]
- 50.Albrechtsen A., Nielsen F.C., Nielsen R. Ascertainment biases in SNP chips affect measures of population divergence. Mol. Biol. Evol. 2010;27:2534–2547. doi: 10.1093/molbev/msq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu J., Zhao Y., Beier S., Schulthess A.W., Stein N., Philipp N., Röder M.S., Reif J.C. Suitability of single-nucleotide polymorphism arrays versus genotyping-by-sequencing for genebank genomics in wheat. Front. Plant Sci. 2020;11 doi: 10.3389/fpls.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnold B., Corbett-Detig R.B., Hartl D., Bomblies K. RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol. Ecol. 2013;22:3179–3190. doi: 10.1111/mec.12276. [DOI] [PubMed] [Google Scholar]
- 53.Robledo D., Palaiokostas C., Bargelloni L., Martínez P., Houston R. Applications of genotyping by sequencing in aquaculture breeding and genetics. Rev. Aquac. 2018;10:670–682. doi: 10.1111/raq.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Souche E.L., Hellemans B., Babbucci M., MacAoidh E., Guinand B., Bargelloni L., Chistiakov D.A., Patarnello T., Bonhomme F., Martinsohn J.T., Volckaert F.A.M. Range-wide population structure of European sea bass Dicentrarchus labrax. Biol. J. Linn. Soc. 2015;116:86–105. [Google Scholar]
- 55.Brown C., Miltiadou D., Tsigenopoulos C. Prevalence and survival of escaped European seabass Dicentrarchus labrax in Cyprus identified using genetic markers. Aquac. Environ. Interact. 2015;7 [Google Scholar]
- 56.Polovina E.-S., Kourkouni E., Tsigenopoulos C.S., Sanchez-Jerez P., Ladoukakis E.D. Genetic structuring in farmed and wild gilthead seabream and European seabass in the Mediterranean Sea: implementations for detection of escapees. Aquat. Living Resour. 2020;33:7. [Google Scholar]
- 57.Šegvić-Bubić T., Grubišić L., Trumbić Ž., Stanić R., Ljubković J., Maršić-Lučić J., Katavić I. Genetic characterization of wild and farmed European seabass in the Adriatic Sea: assessment of farmed escapees using a Bayesian approach. ICES J. Mar. Sci. 2016;74:369–378. [Google Scholar]
- 58.Maroso F., Gkagkavouzis K., De Innocentiis S., Hillen J., do Prado F., Karaiskou N., Taggart J.B., Carr A., Nielsen E., Triantafyllidis A., Bargelloni L., C. the Aquatrace Genome-wide analysis clarifies the population genetic structure of wild gilthead sea bream (Sparus aurata) PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0236230. e0236230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alarcón J.A., Magoulas A., Georgakopoulos T., Zouros E., Alvarez M.C. Genetic comparison of wild and cultivated European populations of the gilthead sea bream (Sparus aurata) Aquaculture. 2004;230:65–80. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2
Data Availability Statement
Raw sequence reads from the European seabass and gilthead seabream population pools analyzed for SNP discovery have been deposited in NCBI's Sequence Read Archive (SRA) under accession number PRJEB40423. Details of the allele frequencies of the SNPs on the MedFish array can be found in the Mendeley Data Repository (http://dx.doi.org/10.17632/7w4cb4mdd4.1).