ABSTRACT
Crop-associated microbiota are a key factor affecting host health and productivity. Most crops are grown within heterogeneous landscapes, and interactions between management practices and landscape context often affect plant and animal biodiversity in agroecosystems. However, whether these same factors typically affect crop-associated microbiota is less clear. Here, we assessed whether orchard management strategies and landscape context affected bacterial and fungal communities in pear (Pyrus communis) flowers. We found that bacteria and fungi responded differently to management schemes. Organically certified orchards had higher fungal diversity in flowers than conventional or bio-based integrated pest management (IPM) orchards, but organic orchards had the lowest bacterial diversity. Orchard management scheme also best predicted the distribution of several important bacterial and fungal genera that either cause or suppress disease; organic and bio-based IPM best explained the distributions of bacterial and fungal genera, respectively. Moreover, patterns of bacterial and fungal diversity were affected by interactions between management, landscape context, and climate. When examining the similarity of bacterial and fungal communities across sites, both abundance- and taxon-related turnovers were mediated primarily by orchard management scheme and landscape context and, specifically, the amount of land in cultivation. Our study reveals local- and landscape-level drivers of floral microbiome structure in a major fruit crop, providing insights that can inform microbiome management to promote host health and high-yielding quality fruit.
IMPORTANCE Proper crop management during bloom is essential for producing disease-free tree fruit. Tree fruits are often grown in heterogeneous landscapes; however, few studies have assessed whether landscape context and crop management affect the floral microbiome, which plays a critical role in shaping plant health and disease tolerance. Such work is key for identification of tactics and/or contexts where beneficial microbes proliferate and pathogenic microbes are limited. Here, we characterize the floral microbiome of pear crops in Washington State, where major production occurs in intermountain valleys and basins with variable elevation and microclimates. Our results show that both local-level (crop management) and landscape-level (habitat types and climate) factors affect floral microbiota but in disparate ways for each kingdom. More broadly, these findings can potentially inform microbiome management in orchards for promotion of host health and high-quality yields.
KEYWORDS: flower microbiome, integrated pest management, landscape heterogeneity, Pyrus communis
INTRODUCTION
Microbial communities affect plant health and productivity. For agricultural crops, microbes can affect nutrient mobilization and transport, often promoting plant growth and disease resistance (1–3). In turn, understanding and managing microbiome assembly could enhance agricultural sustainability by reducing reliance on external inputs, enhancing yields, and potentially contributing to the maintenance of both biodiversity and the functioning of agricultural landscapes (4–6). Yet, despite the growing recognition of the importance of the microbiome to crop productivity, processes governing the assembly of microbiomes for many crop species are still largely unclear (but see references 7 and 8).
Agricultural landscapes are often spatially heterogeneous. Accruing through shifts in land tenure over time, this heterogeneity reflects a landscape’s composition and configuration (9, 10). Specifically, crop production occurs on patches of land that exist within habitat mosaics containing patches of the same crop, alternative commodities, and seminatural vegetation. Such variation in land cover around a crop field may strongly affect local abiotic and biotic conditions. Most studies assessing the effects of spatial context, however, have focused primarily on plants (11) and animals (12), but effects of landscape-level drivers on plant-associated microbiomes have received less attention. This is a problematic knowledge gap, as microbes often disperse over long distances, and studies show that spillover of microbes from agricultural into natural habitats is affected by landscape context and dispersal ability of individual taxa (13). Many microbes are often affected strongly by environmental conditions, and abiotic variation across landscapes can sometimes predict outbreaks of pathogenic microbes (14).
At the orchard scale, management practices employed to control pests and disease can also shape microbiome assembly and structure. Agricultural producers often rely on agrochemicals to prevent establishment or directly suppress both pests and pathogens. As part of an integrated pest management (IPM) program, these practices can vary in intensity across orchards, including the frequency of application, the active ingredients of chemical controls, and how they are coupled with other biological or cultural control strategies (15). Indeed, the application of antibiotics, fungicides, or microbiological control agents can leave distinct signatures on the microbiome associated with tree fruits (16, 17). Though their application can often have direct, suppressive effects on the abundance of targeted, pathogenic taxa (16), nontarget effects on associated yeasts and bacteria have also been observed (17, 18).
Here, we assessed how local- and landscape-level processes affected the diversity and structure of microbe communities associated with pear (Pyrus communis) flowers in Washington State. We focused on microbes on flowers, as these ephemeral structures produce the fruit but are also the primary infection site for pathogens such as the bacterium Erwinia amylovora, the causal agent of fire blight (19). As a consequence, pear orchards are typically heavily managed during bloom to minimize disease risk while promoting pollination (16, 20). Such management tactics range from the use of managed honeybees to the application of diverse bactericides for control of fire blight. We predicted that floral microbiota would be impacted by orchard management practices and the abiotic and biotic landscape conditions. Such work provides important insights into microbial colonization and community structure pre- and postpollination, important windows for production.
RESULTS
Pear flower microbiome.
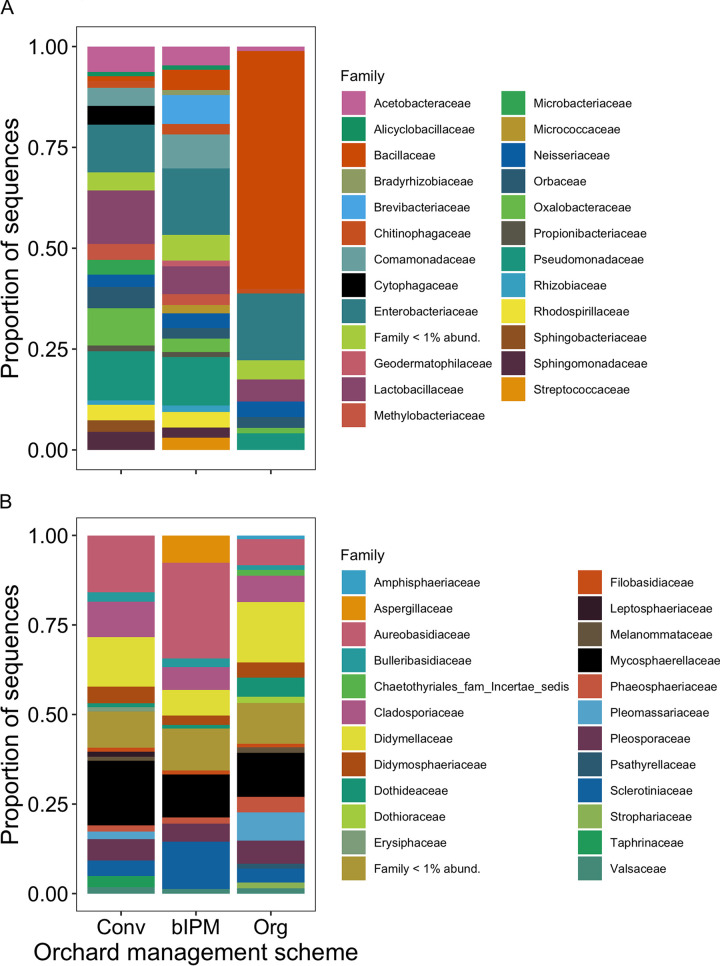
Our study sampled bacterial and fungal communities associated with pear flowers across 15 orchards with three management types (conventional, bio-based IPM, and organic; 5 sites of each). After bacterial (16S) and fungal (internal transcribed spacer [ITS]) gene sequencing, quality filtering, and processing, we detected 142 bacterial and 1,703 fungal amplicon sequence variants (ASVs) from the pear flowers. The bacterial community was dominated by members of Bacillaceae, Enterobacteriaceae, Lactobacillaceae, and Pseudomonadaceae (Fig. 1A), with each family comprising, on average, 22% (standard deviation [SD], ±0.32), 15% (SD, ±0.03), 9% (SD, ±0.4), and 9% (SD, ±0.05) of sequences, respectively. Beneficial bacteria associated with disease suppression in this system (i.e., Bacillus, Pantoea, and Pseudomonas) comprised 11% of taxa (ASVs) observed and 41% of the relative abundance. The fungal community was dominated by members of Aureobasidiaceae, Cladosporiaceae, Mycosphaerellaceae, and Sclerotiniaceae (Fig. 1B), with each family comprising, on average, 16% (SD, ±0.10), 8% (SD, ±0.02), 14% (SD, ±0.03), and 7% (SD, ±0.05) of sequences, respectively. Of the Aureobasidiaceae, four ASVs were identified to the species level as Aureobasidium pullulans, a beneficial fungus used for biological control of fire blight. Twenty-one additional ASVs were identified as belonging to genera Botrytis, Cladosporium, Monilinia, Mycosphaerella, or Penicillium, potentially important agents of pre- and postharvest disease.
FIG 1.
Relative abundance (proportion of sequences) of bacterial (A) and fungal (B) families associated with pear flowers. Flowers were collected from orchards that reflected three unique management schemes (conventional, bIPM, and organic).
Orchard management and landscape context affect bacterial and fungal alpha diversity.
Orchard pest management practices were significantly associated with pear flower bacterial and fungal diversity (Shannon index) (Table 1). Considered alone, conventional and biological-based integrated pest management (bIPM)-managed orchards were found to have ∼60% higher bacterial diversity than those managed organically (Fig. 2A), while organically managed orchards exhibited the highest fungal diversity (Fig. 2A). Yet the positive effects of organic management on fungal diversity were not significant in the multiple variate linear model when controlling for land cover and climate (Fig. 2B). In these linear regression models (Table 1), both organic and bIPM management styles reduced bacterial and fungal diversity, although the negative influence of organic management on fungal diversity was weak. Land cover was also associated with bacterial and fungal diversity: bacterial diversity declined with increasing proportion of habitat containing forest or pear, while fungal diversity increased with pear crop cover (Table 1). Microclimatic conditions were also associated with both bacterial and fungal diversity, though minimum temperature was the only variable of significant effect on fungal diversity and minimum vapor pressure deficit (VPD) for bacterial diversity in the top Akaike information criterion (AIC)-selected model (Table 1).
TABLE 1.
Multivariate linear regression models for bacterial and fungal diversity (Shannon index) associated with pear flowersb
| Variable | Estimate | SE | Pr(>t) | Model adjusted R2 | Pr(>F) |
|---|---|---|---|---|---|
| Bacteria | |||||
| Intercept | 0.869 | 0.274 | 0.005 | 0.522 | <0.001 |
| Organic management | −1.728 | 0.478 | 0.002 | ||
| bIPMa management | −0.964 | 0.369 | 0.016 | ||
| Proportion of landscape with forest | −0.597 | 0.186 | 0.004 | ||
| Proportion of landscape with pear | −0.506 | 0.187 | 0.013 | ||
| VPDa minimum (hPa) | −0.498 | 0.281 | 0.090 | ||
| VPD maximum (hPa) | −0.411 | 0.250 | 0.114 | ||
| Fungi | |||||
| Intercept | 0.460 | 0.294 | 0.131 | 0.426 | 0.002 |
| Organic management | −0.357 | 0.494 | 0.477 | ||
| bIPM management | −1.022 | 0.392 | 0.015 | ||
| Proportion of landscape with pear | 0.367 | 0.153 | 0.025 | ||
| VPD maximum (hPa) | −0.252 | 0.188 | 0.191 | ||
| Minimum temp (°C) | −0.411 | 0.184 | 0.035 |
bIPM, biological-based integrated pest management; VPD, vapor pressure deficit.
Top models were selected by AIC. Pr(>t), probability of the t statistic for each model coefficient; Pr(>F), probability of the F statistic for the overall model.
FIG 2.
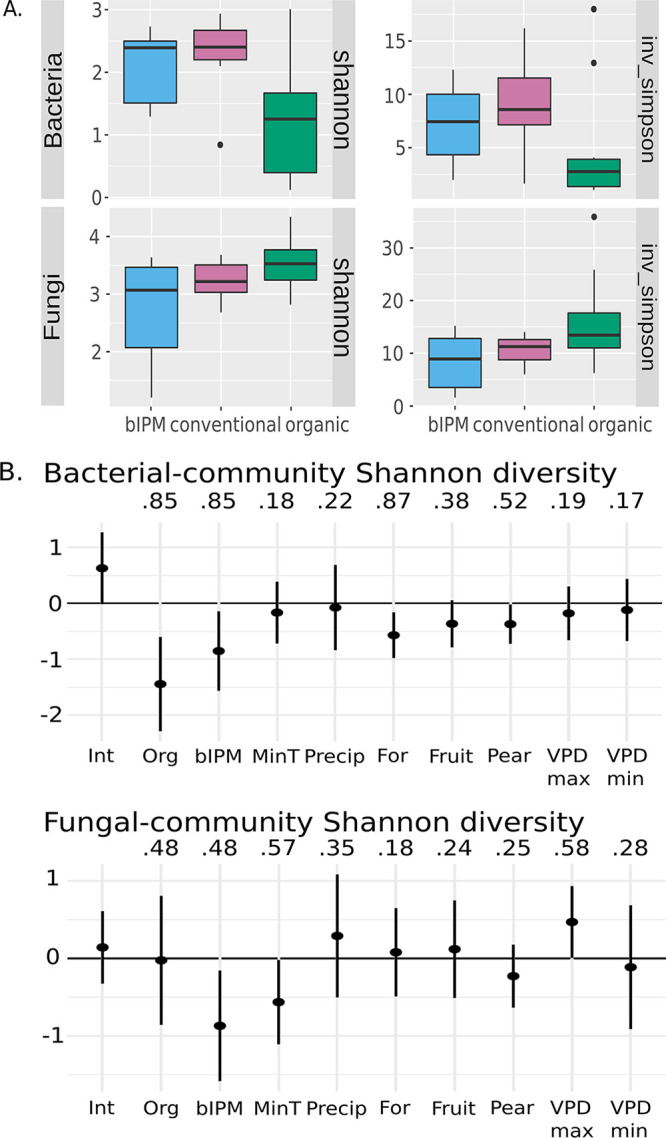
(A) Diversity (Shannon and inverse Simpson indices) of bacteria and fungi associated with pear flowers collected from orchards that vary in management scheme (conventional, bIPM, and organic). (B) Coefficients from the 90% confidence set of top multivariate models. Variable importance was evaluated as the number of models within the 90% confidence model set in which the factor was included.
Orchard management practices drive the distribution of pathogenic fungal species and the presence of bacterial genera associated with disease suppression.
Focal bacterial and fungal genera of concern were first investigated to assess the scale of spatial autocorrelation, as well as potential associations with aspects of landscape context. Positive spatial autocorrelation was exhibited for each of the nine taxa examined, but only at the shortest distances of less than 1 km. Using canonical correlation analysis to assess how landscape and management variables were associated with the microbial species composition, we found significant associations between predictors and bacterial (Fig. 3A; Table S1 in the supplemental material; Pillai’s trace P = 0.014) and fungal (Fig. 3B; Table S2; Pillai’s trace P = 0.005) communities. The three bacterial genera associated with disease suppression were distributed very differently in association with the factors of interest. More specifically, the relative abundance of Bacillus, bacteria commonly applied to suppress disease in pear, was most strongly associated with organic management (Fig. S2), followed closely by the amount of surrounding forest and then geographic distance. These top factors, aligned with axis 1, were negatively associated with Pseudomonas, while bIPM was the most important predictor of Pantoea (more aligned with axis 2). Similar to Pantoea, bIPM (+) and organic management (−) best predicted the presence of Aureobasidium, a beneficial fungus aligned with axis 1, and Monilinia to a lesser degree. Minimum temperature (+) and minimum VPD (+) best predicted Botrytis, Cladosporium, and Mycosphaerella, as well as Monilinia (−), all pathogenic fungi of concern for pears. Finally, the proportion of forest in the landscape and geographic distance were associated with the distribution of these fungal genera of interest (Table S2).
FIG 3.
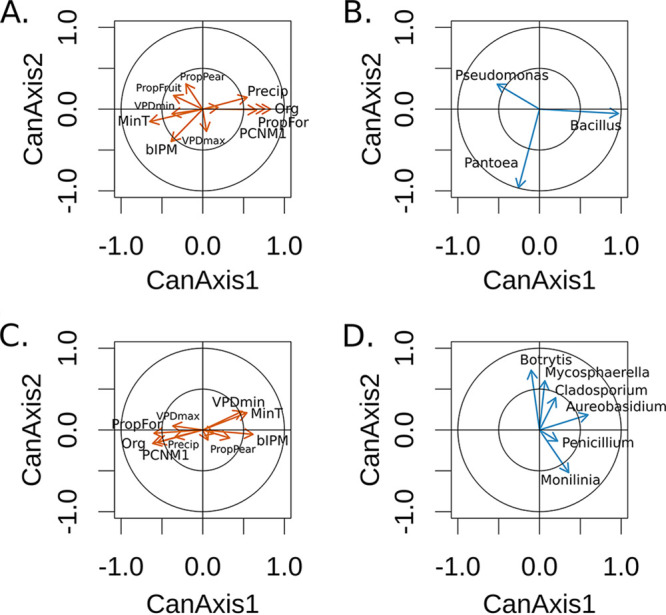
Canonical correlation analysis of three bacterial and five fungal taxa associated with both disease and disease prevention in pear flowers. The left panels depict the variance explained by the factors in the canonical axes for bacteria (A) and fungi (C), and the right panels depict the variance explained by the canonical axes in the bacterial (B) and fungal (D) taxa of interest.
Microbial beta diversity was affected by orchard management and landscape context.
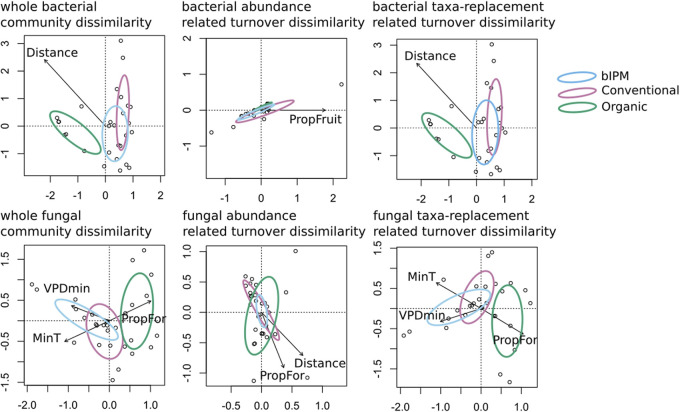
Orchard sites that were closer in proximity, or had the same management scheme, tended to be more similar in terms of bacterial community composition (Fig. 4; Table 2). In contrast, abundance-related turnover across sites was affected mainly by the proportion of landscape under fruit cultivation, namely, apple (Table 2). With respect to fungi (Fig. 4; Table 3), turnover of fungal communities across sites was associated with the amount of pear production in the landscape, temperature, and vapor pressure deficit (VPD). Temperature and VPD, along with surrounding forest, were important drivers of taxon-related turnover (Table 3). In contrast, abundance-related community turnover was associated with geographic distance and the proportion of landscape represented by forest around orchards (Table 3).
FIG 4.
Restricted distance-based analysis of pear flower bacterial and fungal community beta diversity, including explanatory variables in the top AIC-selected RDA models. Variance explained by each factor can be found in Tables 2 and 3.
TABLE 2.
Results from RDA of pear flower bacterial community beta diversitya
| Variable | Adjusted R2 | Pr(>F)b |
|---|---|---|
| Whole-community beta diversity | ||
| Orchard management scheme | 0.147 | 0.002 |
| Geographic distance (km) | 0.172 | 0.026 |
| Abundance-related community beta diversity | ||
| Proportion of landscape - fruit | 0.594 | 0.040 |
| Taxon-related community beta diversity | ||
| Orchard management scheme | 0.164 | 0.002 |
| Geographic distance (km) | 0.195 | 0.026 |
Top models were selected by AIC.
Pr(>F), probability of the F statistic.
TABLE 3.
Results from RDA of pear flower fungal community beta diversityb
| Variable | Adjusted R2 | Pr(>F)a |
|---|---|---|
| Whole-community beta diversity | ||
| Proportion of landscape with pear | 0.170 | 0.046 |
| Minimum temp (°C) | 0.092 | 0.002 |
| VPDa minimum (hPa) | 0.132 | 0.018 |
| Abundance-related community beta diversity | ||
| Geographic distance (km) | 0.266 | 0.008 |
| Proportion of landscape with forest | 0.579 | 0.002 |
| Taxon-related community beta diversity | ||
| Proportion of landscape with forest | 0.177 | 0.046 |
| Minimum temp (°C) | 0.089 | 0.002 |
| VPD minimum (hPa) | 0.134 | 0.026 |
VPD, vapor pressure deficit; Pr(>F), probability of the F statistic.
Top model selected by AIC.
DISCUSSION
The Pacific Northwest is responsible for ∼80% of pear production in the United States (60). Pre- and postharvest diseases that can take hold during bloom threaten production and the quality of yield, however (21). Here, we investigated how local orchard-level IPM practices interacted with landscape-level growing conditions to influence the structure and diversity of microbiota associated with pear flowers, potential sites for infection. Our analyses revealed that the orchard management scheme can significantly influence the structure and diversity of both bacterial and fungal communities. Beyond local orchard-level management, land cover and climate were also found to be significant predictors of microbe diversity, and bacterial and fungal communities were affected by different habitat types found in landscapes surrounding orchards. Finally, fungal alpha and beta diversity were more greatly affected by microclimatic conditions experienced in orchards than bacteria. In the sections that follow, we discuss these findings in light of understanding the key drivers of floral microbiome structure in this system.
Orchard management mediates microbial diversity.
Bacterial and fungal alpha diversity responded differently to orchard management schemes. Bacterial diversity was significantly higher in conventional and bIPM orchards than organic orchards; however, the opposite pattern was observed for fungi. Organic orchards had a high relative abundance of Bacillus, likely because of its application as a biological control agent. The strong effect of orchard management on bacterial diversity suggests that application of Bacillus reduced bacterial diversity, which may occur through resource competition, priority effects, or mass effects. Bacillus species have shown promise in limiting the establishment and development of the bacterial pathogen E. amylovora, the causal agent of fire blight (22, 23), and may also affect other floral microbes. Indeed, increased fungal diversity in organically managed orchards could be a consequence of Bacillus application, although we were unable to directly assess if fungal abundance was affected in our study. In contrast to bacteria applied for biological control, we observed that Aureobasidium had a higher relative abundance in conventional and bIPM orchards than organic ones (where it was applied in one orchard for biological control). Background levels of some microbial taxa may be high and more prevalent in the presence of particular landscape and climate conditions (e.g., higher precipitation and high proportion of forest; Tables S1 and S2 in the supplemental material). These patterns may represent preferential use of these biological treatments across orchards in our survey. Though unable to confirm whether ASVs recovered in our data set are these exact commercial strains, biologicals applied to pear flowers often have a high recovery rate in surveys (24, 25).
Land cover and microclimate shape microbial diversity.
Our results show that habitat patches with alternate tree fruit crops (apple, cherry) were negatively associated with both bacterial and fungal diversity on pear flowers and appeared to be primary drivers of microbial community structure (Tables 2 and 3). Pear orchards in the Wenatchee River Valley are primarily located in narrow, intermountain areas with highly variable elevation and land cover, including forest, additional pear orchards, and those dedicated to production of other deciduous fruits, namely, apple. Vegetation in and around orchards can be an important source of inocula via airborne dispersal (26, 27). Furthermore, previous work on apple and pear flowers has revealed considerable overlap in the identity of microbes associated with each host species (28–30). Such overlap, in addition to a reduction in diversity with increasing land cultivation, suggests a role for several key processes in shaping floral microbiomes in tree fruits. First, there is a high degree of shared usage of disease and pest management practices employed in pear and apple production systems, as both can suffer greatly from fire blight disease. Inputs applied in conventional and bIPM orchards, including antibiotics and fungicides (Table S3), can act as strong environmental filters on potential floral colonists (17, 18) or serve as a source for inocula when applied as biologicals, as observed in organic orchards. Second, both apple and pear systems rely considerably on honeybees (Apis mellifera) for pollination, which are known to leave a distinct imprint on floral microbiome diversity (31). Increased reliance on a single-pollinator species, combined with chemical and nonchemical inputs, are likely important contributors to patterns observed.
Bacterial and fungal community turnover and dispersal.
Orchard management scheme was a key determinant of bacterial community similarity across sites; however, other predictors often explained high levels of variance in community structure across sites. In particular, geographic distance explained a significant amount of variance in both whole-community and taxon-related beta diversity of bacteria. In contrast, for fungi, geographic distance was a significant predictor of only abundance-related turnover. Beyond geographic distance, climatic conditions also contributed significantly to explained variance in the beta diversity or community turnover of fungal communities. In particular, VPD and temperature were negatively associated with fungal diversity, suggesting both microclimate variables affect either species-specific patterns of growth and/or competition. Moisture availability is also an important determinant of microbial growth on the surface of plant tissues (32), with free water and humidity often being necessary for conidial germination, germ tube growth, and potential penetration of plant tissues, including floral organs. This has been frequently observed in other flowering systems of commercial value, including blueberries (33), raspberries (34), strawberries (35), and cut roses (36). Within these systems, infection of the gynoecium can be a primary route of disease development. Alternatively, infection of petals and other organs can facilitate secondary infections of fruits (37). Of the fungal genera examined in our study, Botrytis has been documented to successfully infect the mesocarp via stamen filaments (38). For the others of interest, it is unclear if there is a link between flower colonization and resulting development and pre- and postharvest diseases.
More broadly, our results provide insight into local- and landscape-level drivers of floral microbiome diversity in an important tree fruit commodity, pear. Given the critical link between flowers, yield, and disease, identifying such drivers across both spatial and temporal scales could improve the understanding of links between management, host microbiome structure, and potentially disease resistance or susceptibility. With growing appreciation for the role of host microbiota in affecting resistance against disease (3, 39), such information has potential to inform development of sustainable management practices in many different types of agroecosystems.
MATERIALS AND METHODS
Landscape survey.
We surveyed 15 orchards throughout the Wenatchee River Valley of central Washington (Fig. 5) in spring 2018. Within the United States, Washington State is the leading producer of deciduous tree fruit crops such as apples, pears, and cherries. These, as well as other commodities, are grown in variable intermountain river valleys and basins east of the Cascade Mountains. These production areas generally experience temperate, dry conditions, in addition to favorable access to irrigation water originating from streams and rivers fed by snowmelt (40). Given the diverse topography of this region, however, individual orchards range in elevation from 20 to 1,000 m above sea level (40). Key stages of fruit production, such as flower bloom, can thus experience considerable variation in microclimatic conditions among orchards, affecting bloom timing, fertilization, and fruit development (41, 42). As flowers are a habitat for diverse microbiota (43), including a number of pathogenic species that cause pre- and postharvest diseases of tree fruits (44), microclimatic conditions could affect habitat quality, as well as colonization dynamics and the resulting structure of the floral microbiome.
FIG 5.
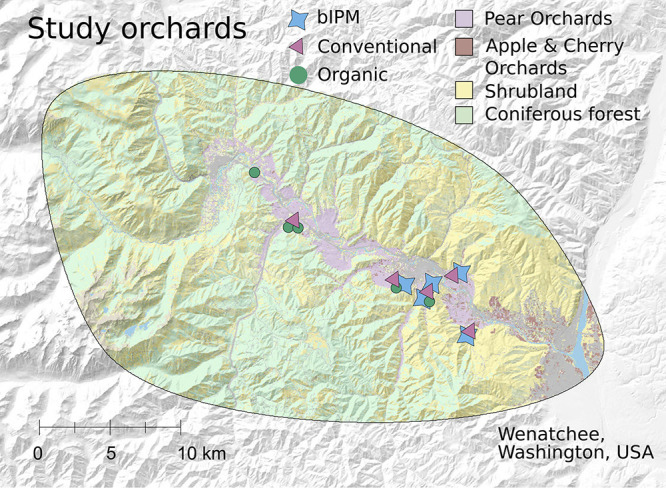
Geographic extent of survey, where 15 pear orchards in central Washington across variable landscape contexts were sampled during peak bloom. The study site map was created with ArcGIS (v10.8; ESRI Inc., Redlands, CA, USA) and Inkscape (v1.0.2; https://inkscape.org), with elevation and land cover shading based on the National Elevation Dataset (USDA NRCS) and Cropland Data Layer product (USDA NASS [https://www.nass.usda.gov/Research_and_Science/Cropland/SARS1a.php]), respectively.
Our survey assesses microbe communities in orchards that used one of three management schemes, with five replicates per scheme, which include organically certified, conventional, and biological-based integrated pest management (bIPM) (45). With each of these broad management types, growers were not restricted to a specific spray schedule, but each used a defined set of tools for pest and disease management (45). Conventional management followed a standard practice (e.g., application of synthetic pesticides), while organic orchards were all managed following USDA‐certified organic standards, which prohibits use of such synthetic chemicals. To control fire blight, organic producers often use Serenade Opti (Bayer Crop Science, St. Louis, MO, USA) at full bloom, a bio-based fungicide and bactericide that leverages Bacillus subtilis (strain QST 713) endospores and its metabolic by-products as active ingredients (22). Serenade is not the only bio-based product leveraged by producers for control of fire blight in pear, however, and other products such as Blossom Protect (Westbridge Agricultural Products, Vista, CA, USA) can be used across organic, bIPM, and conventional schemes. Blossom Protect is derived from air-dried spores of Aureobasidium pullulans (strains DSM 14940 and 14941) (25), an epiphytic or endophytic fungus associated with a wide range of plant species, including many tree fruits. For those orchards that employed the bIPM scheme, growers used a toolbox of cultural controls combined with pesticides with less documented negative impact on natural enemies and other beneficial organisms. Such products included lime sulfur, kaolin, spinosad, and biologicals applied at various stages of bloom (45).
Orchards were sampled once at peak bloom, either on 30 April or 1 May of 2018. At each orchard, 10 trees (Bradford variety) were sampled, 5 near the edge of the orchard and 5 in the interior. We chose this approach because previous studies suggest that seminatural habitat in the surrounding landscape can both support and increase rates of visitation by native pollinators such as bees and flies (46). Moreover, pollinators can be important dispersal agents for microbes (31, 47); thus, our aim was to detect potential contributions of pollinator visitation to flower microbiome assembly in orchards. For each site (i.e., edge or interior) and sampling event, 50 open flowers (n = 10 per tree) were collected using aseptic technique and pooled at the site level. Flowers with flat, fully reflexed petals that had been open for ∼3 days were collected. Once collected, flowers were placed in a cooler, transferred to the lab, and then stored at 4°C until processing.
Sample processing.
In a laboratory, whole flowers were washed with 20 ml of 1× 0.15% phosphate-buffered saline (PBS)-Tween solution, and samples were sonicated for 10 min to dislodge epiphytic microbes. After sonication, floral tissue debris was removed from sample tubes by pouring samples through autoclaved cheesecloth into a new, sterile Falcon tube. Falcon tubes containing debris-filtered samples were centrifuged at 3,000 rpm for 10 min at 4°C to pellet microbial cells. We poured off the supernatant, resuspended microbial cell pellets in 1 ml of autoclaved PBS solution, vortexed tubes, and then transferred the cell suspensions to new 1.7-ml microcentrifuge tubes.
DNA extraction and sequencing.
Genomic DNA was extracted from samples using a ZymoBIOMICS DNA microprep kit (Zymo Research, Irvine, CA, USA) following the manufacturer’s protocol. Extracted DNA was then used as the template for library preparation and amplicon sequencing following Comeau et al. (48), performed at the Centre for Comparative Genomics and Evolutionary Bioinformatics at Dalhousie University (Halifax, Nova Scotia, Canada). There, amplicon fragments were PCR- amplified from DNA in duplicate, using separate template dilutions (1:1 and 1:10) and high-fidelity Phusion polymerase (New England BioLabs Inc., Ipswich, MA, USA). A single round of PCR was performed using “fusion primers” (Illumina adaptors plus indices plus specific regions) targeting either the 16S V4-V5 (bacteria; primers 515FB and 926R [49, 50]) or ITS2 (fungi; primers ITS86 and ITS4 [51]) regions with multiplexing. PCR products were verified visually by running a high-throughput Invitrogen 96-well E-gel (Thermo Fisher Scientific Corp., Carlsbad, CA, USA). Any samples with failed PCRs (or spurious bands) were reamplified by optimizing PCR conditions to produce correct bands to complete a sample plate before continuing with sequencing. The PCRs from the same samples were pooled in one plate, cleaned, and then normalized using the high-throughput Invitrogen SequalPrep 96-well plate kit (Thermo Fisher Scientific Corp.). Samples were then pooled to make one library and then quantified fluorometrically before sequencing. Amplicon samples were then run on an Illumina MiSeq using 2× 300-bp paired-end V3 chemistry.
Demultiplexed sequences were trimmed of trailing low-quality bases using the DADA2 pipeline (v1.8.0) (52) in R (53). Paired-end reads were then quality filtered, error corrected, and assembled into ASVs. Once assembled, chimeras were detected and removed, and taxonomic information was then assigned to each ASV using the Ribosomal Database Project (RDP) naïve Bayesian classifier (54) trained to either the RDP training set (v14) or UNITE general FASTA release (v7.2) for bacteria or fungi, respectively. ASVs that failed to classify to kingdom or identified as chloroplast or mitochondrial sequences were discarded. Further, potential contaminant ASVs were identified through inclusion of negative controls during sample and sequence processing and then removed using the “prevalence” method with the decontam package in R (55). This filtering resulted in samples sequenced at a mean depth of 43,057 sequences per sample for bacteria and 25,890 for fungi. Samples were then rarefied (bacteria, 49; fungi, 14,920), with all but one bacterial sample (19orgedge) retained in the analyses that follow. Such a low cutoff for bacteria is a consequence of a large proportion of reads being identified as plastid DNA, which were removed from the data set. Despite this, we included bacterial data in our study because sampling curves indicate that we were able to identify the majority of bacterial taxa present in samples (Fig. S2 in the supplemental material). Moreover, previous characterization of microbial communities associated with flowers has frequently observed low species richness (43).
Landscape context.
Land cover within a 1-km buffer of each study orchard was classified into the following three habitat types: (i) pear orchard, (ii) other fruit orchard (apple and cherry), and (iii) forest. These classifications were performed using the Cropland Data Layer spatial product (https://www.nass.usda.gov/Research_and_Science/Cropland/SARS1a.php). Across our study region, pears were the dominant agricultural crop, although the habitat around individual study sites varied widely from 2% to 66% pear orchards. Other fruit crops had less variability, with 0% to 6% of surrounding land cover, while forest land was highly variable and ranged from 0% to 46%. Forest patches were primarily composed of evergreen trees.
To assess the role of abiotic factors, high-resolution climatic metrics for each site were obtained from publicly accessible PRISM data in April 2018. PRISM data are collected at a spatial resolution of 2.5 arcmin (∼4 km). An arcmin is an angular measurement equal to 1/60 of a degree. PRISM data used included elevation (m), minimum and maximum temperature (°C), minimum and maximum vapor pressure deficit (VPD [hPa]), and precipitation (mm). Vapor pressure deficit is the difference between the amount of moisture in the air and how much moisture the air can hold when saturated, where high VPD indicates drier conditions. As with land cover, the abiotic conditions where sites were located were variable, with elevation ranging from 1,152 to 1,526 m above sea level, April precipitation ranging from 4.2 to 5.3 cm, minimum temperatures ranging from 2.4 to 3.7°C, and maximum temperature ranging from 13.6 to 15.7°C.
Statistical analyses.
We used multivariate linear regression to assess effects of land cover, orchard management, and climate on the alpha diversity of pear flower microbiomes, using both the Shannon diversity and inverse Simpson index. We chose to include the latter metric to specifically isolate the evenness/dominance aspect of community structure from the taxonomic richness, which heavily contributes to the Shannon diversity metric. All analyses were conducted using R v3.6.1 (53). To reduce multicollinearity among predictors, we calculated variance inflation factors (VIFs) and used a threshold of 10 to eliminate variables with problematic covariance. This eliminated temperature, precipitation, and elevation from the alpha diversity models. We calculated multimodel average coefficients based on the 90% confidence interval of top models as well as the importance of each coefficient, which indicated the number of top models in which it appeared.
We also assessed effects of landscape, climate, and farm management on the dominance (relative abundance) of a few focal genera that are highly important for pre- and postharvest diseases of pear, including putative pathogens and beneficial taxa. These included fungal genera Aureobasidium, Botrytis, Cladosporium, Monilinia, Mycosphaerella, and Penicillium and beneficial bacteria, which included Bacillus, Pantoea, and Pseudomonas (24, 56, 57). One ASV (BactSeq29), identified as an Erwinia sp., was detected at a single orchard in our survey. Given such limited detection, we were unable to perform an analysis of links between variables of interest and Erwinia presence and abundance. However, to examine associations between microbial genera and predictors described earlier, we used canonical correlation analysis (CCA), an extension of linear regression that finds linear relationships between combinations of explanatory and response variables which maximize the correlation. Separate models were run on fungi and bacteria of interest.
Differences in species composition among sites could be affected by processes including substitution of taxa and variation in abundance of particular taxa, so we further evaluated the effects of farm management, land cover, and climate variables on abundance-related and taxon-related aspects of community turnover (microbial beta diversity) and the overall community dissimilarity (which incorporates both processes). Beta diversity was partitioned into abundance-related and taxa-related components of Bray-Curtis dissimilarity using the bray.part function in the betapart R package (58). The influence of explanatory variables on these two components of community turnover between sites, as well as their cumulative overall Bray-Curtis dissimilarity, was investigated using restricted distance-based analysis (RDA) and AIC model selection and executed using the capscale and ordiR2step functions in the vegan R package (59). The variance explained by factors included in the top AIC-selected models is included in the results.
Data availability.
Raw amplicon sequences are available on the NCBI Sequence Read Archive (SRA) under BioProject accession no. PRJNA659266.
ACKNOWLEDGMENTS
We are especially grateful to the growers who allowed us to sample on their properties and D.F.J. Sauza for assistance with data analysis.
This work was funded by USDA NIFA grants (2014-51106-22096 to D.W.C.; 2017-67012-26104 to R.N.S.), Western SARE (SW18-031 to D.W.C. and R.N.S.), the WSDA (number K1986 to S.T.D.), and the WSU BioAg program. This research was also supported by the Utah Agricultural Experiment Station, Utah State University, and approved as journal paper number 9450.
Footnotes
Supplemental material is available online only.
Contributor Information
Robert N. Schaeffer, Email: robert.schaeffer@usu.edu.
Irina S. Druzhinina, Nanjing Agricultural University
REFERENCES
- 1.Pii Y, Mimmo T, Tomasi N, Terzano R, Cesco S, Crecchio C. 2015. Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol Fertil Soils 51:403–415. 10.1007/s00374-015-0996-1. [DOI] [Google Scholar]
- 2.Vurukonda SSKP, Vardharajula S, Shrivastava M, SkZ A. 2016. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol Res 184:13–24. 10.1016/j.micres.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Berg M, Koskella B. 2018. Nutrient-and dose-dependent microbiome-mediated protection against a plant pathogen. Curr Biol 28:2487–2492.e3. 10.1016/j.cub.2018.05.085. [DOI] [PubMed] [Google Scholar]
- 4.Mueller UG, Sachs JL. 2015. Engineering microbiomes to improve plant and animal health. Trends Microbiol 23:606–617. 10.1016/j.tim.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, Morsy M, Eisen JA, Leach JE, Dangl JL. 2017. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15:e2001793. 10.1371/journal.pbio.2001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toju H, Peay KG, Yamamichi M, Narisawa K, Hiruma K, Naito K, Fukuda S, Ushio M, Nakaoka S, Onoda Y, Yoshida K, Schlaeppi K, Bai Y, Sugiura R, Ichihashi Y, Minamisawa K, Kiers ET. 2018. Core microbiomes for sustainable agroecosystems. Nat Plants 4:247–257. 10.1038/s41477-018-0139-4. [DOI] [PubMed] [Google Scholar]
- 7.Edwards J, Johnson C, Santos-Medellín C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V. 2015. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920. 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grady KL, Sorensen JW, Stopnisek N, Guittar J, Shade A. 2019. Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops. Nat Commun 10:1–10. 10.1038/s41467-019-11974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahrig L, Nuttle WK. 2005. Population ecology in spatially heterogeneous environments, p 95–118. In Lovett GM, Turner MG, Jones CG, Weathers KC (ed), Ecosystem function in heterogeneous landscapes. Springer, New York, NY. [Google Scholar]
- 10.Fahrig L, Baudry J, Brotons L, Burel FG, Crist TO, Fuller RJ, Sirami C, Siriwardena GM, Martin J-L. 2011. Functional landscape heterogeneity and animal biodiversity in agricultural landscapes. Ecol Lett 14:101–112. 10.1111/j.1461-0248.2010.01559.x. [DOI] [PubMed] [Google Scholar]
- 11.Smith OM, Cohen AL, Reganold JP, Jones MS, Orpet RJ, Taylor JM, Thurman JH, Cornell KA, Olsson RL, Ge Y, Kennedy CM, Crowder DW. 2020. Landscape context affects the sustainability of organic farming systems. Proc Natl Acad Sci U S A 117:2870–2878. 10.1073/pnas.1906909117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karp DS, Chaplin-Kramer R, Meehan TD, Martin EA, DeClerck F, Grab H, Gratton C, Hunt L, Larsen AE, Martínez-Salinas A, O'Rourke ME, Rusch A, Poveda K, Jonsson M, Rosenheim JA, Schellhorn NA, Tscharntke T, Wratten SD, Zhang W, Iverson AL, Adler LS, Albrecht M, Alignier A, Angelella GM, Zubair Anjum M, Avelino J, Batáry P, Baveco JM, Bianchi FJJA, Birkhofer K, Bohnenblust EW, Bommarco R, Brewer MJ, Caballero-López B, Carrière Y, Carvalheiro LG, Cayuela L, Centrella M, Ćetković A, Henri DC, Chabert A, Costamagna AC, De la Mora A, de Kraker J, Desneux N, Diehl E, Diekötter T, Dormann CF, Eckberg JO, Entling MH, et al. 2018. Crop pests and predators exhibit inconsistent responses to surrounding landscape composition. Proc Natl Acad Sci U S A 115:E7863–E7870. 10.1073/pnas.1800042115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell T, Tylianakis JM. 2016. Microbes in the Anthropocene: spillover of agriculturally selected bacteria and their impact on natural ecosystems. Proc R Soc B 283:20160896. 10.1098/rspb.2016.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith TJ, Pusey PL. 2010. Cougarblight 2010, a significant update of the Cougarblight fire blight infection risk model. Acta Hortic 896:331–336. 10.17660/ActaHortic.2011.896.45. [DOI] [Google Scholar]
- 15.Agrios GN. 2005. Plant pathology, 5th ed. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 16.Johnson KB, Stockwell VO. 1998. Management of fire blight: a case study in microbial ecology. Annu Rev Phytopathol 36:227–248. 10.1146/annurev.phyto.36.1.227. [DOI] [PubMed] [Google Scholar]
- 17.Schaeffer RN, Vannette RL, Brittain C, Williams NM, Fukami T. 2017. Non-target effects of fungicides on nectar-inhabiting fungi of almond flowers. Environ Microbiol Rep 9:79–84. 10.1111/1758-2229.12501. [DOI] [PubMed] [Google Scholar]
- 18.McGhee GC, Sundin GW. 2011. Evaluation of kasugamycin for fire blight management, effect on nontarget bacteria, and assessment of kasugamycin resistance potential in Erwinia amylovora. Phytopathology 101:192–204. 10.1094/PHYTO-04-10-0128. [DOI] [PubMed] [Google Scholar]
- 19.Vanneste JL. 2000. Fire blight: the disease and its causative agent, Erwinia amylovora. CABI Publishing, New York, NY. [Google Scholar]
- 20.McGregor SE. 1976. Insect pollination of cultivated crop plants. Agricultural Research Service, U.S. Department of Agriculture, Washington, DC. [Google Scholar]
- 21.Pscheidt JW, Ocamb CM. 2015. Pacific northwest plant disease management handbook. Oregon State University, Corvallis, OR. [Google Scholar]
- 22.Sundin GW, Werner NA, Yoder KS, Aldwinckle HS. 2009. Field evaluation of biological control of fire blight in the eastern United States. Plant Dis 93:386–394. 10.1094/PDIS-93-4-0386. [DOI] [PubMed] [Google Scholar]
- 23.Shemshura O, Alimzhanova M, Ismailova E, Molzhigitova A, Daugaliyeva S, Sadanov A. 2020. Antagonistic activity and mechanism of a novel Bacillus amyloliquefaciens MB40 strain against fire blight. J Plant Pathol 102:825–829. 10.1007/s42161-020-00515-4. [DOI] [Google Scholar]
- 24.Stockwell VO, Johnson KB, Sugar D, Loper JE. 2002. Antibiosis contributes to biological control of fire blight by Pantoea agglomerans strain Eh252 in orchards. Phytopathology 92:1202–1209. 10.1094/PHYTO.2002.92.11.1202. [DOI] [PubMed] [Google Scholar]
- 25.Johnson KB, Temple TN. 2013. Evaluation of strategies for fire blight control in organic pome fruit without antibiotics. Plant Dis 97:402–409. 10.1094/PDIS-07-12-0638-RE. [DOI] [PubMed] [Google Scholar]
- 26.Lindow SE, Andersen GL. 1996. Influence of immigration on epiphytic bacterial populations on navel orange leaves. Appl Environ Microbiol 62:2978–2987. 10.1128/AEM.62.8.2978-2987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lymperopoulou DS, Adams RI, Lindow SE. 2016. Contribution of vegetation to the microbial composition of nearby outdoor air. Appl Environ Microbiol 82:3822–3833. 10.1128/AEM.00610-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stockwell VO, McLaughlin RJ, Henkels MD, Loper JE, Sugar D, Roberts RG. 1999. Epiphytic colonization of pear stigmas and hypanthia by bacteria during primary bloom. Phytopathology 89:1162–1168. 10.1094/PHYTO.1999.89.12.1162. [DOI] [PubMed] [Google Scholar]
- 29.Pusey PL, Stockwell VO, Mazzola M. 2009. Epiphytic bacteria and yeasts on apple blossoms and their potential as antagonists of Erwinia amylovora. Phytopathology 99:571–581. 10.1094/PHYTO-99-5-0571. [DOI] [PubMed] [Google Scholar]
- 30.Smessaert J, Van Geel M, Verreth C, Crauwels S, Honnay O, Keulemans W, Lievens B. 2019. Temporal and spatial variation in bacterial communities of “Jonagold” apple (Malus x domestica Borkh.) and “Conference” pear (Pyrus communis L.) floral nectar. MicrobiologyOpen 8:e918. 10.1002/mbo3.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aizenberg-Gershtein Y, Izhaki I, Halpern M. 2013. Do honeybees shape the bacterial community composition in floral nectar? PLoS One 8:e67556. 10.1371/journal.pone.0067556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beattie GA. 2002. Leaf surface waxes and the process of leaf colonization by microorganisms, p 3–26. In Lindow SE, Elliott VJ, Hecht-Poinar EI (ed), Phyllosphere microbiology. American Phytopathological Society, St. Paul, MN. [Google Scholar]
- 33.Ngugi HK, Scherm H. 2004. Pollen mimicry during infection of blueberry flowers by conidia of Monilinia vaccinii-corymbosi. Physiol Mol Plant Pathol 64:113–123. 10.1016/j.pmpp.2004.08.004. [DOI] [Google Scholar]
- 34.McNicol RJ, Williamson B, Dolan A. 1985. Infection of red raspberry styles and carpels by Botrytis cinerea and its possible role in post‐harvest grey mould. Ann Appl Biol 106:49–53. 10.1111/j.1744-7348.1985.tb03093.x. [DOI] [Google Scholar]
- 35.Bulger MA, Ellis MA, Madden LV. 1987. Influence of temperature and wetness duration on infection of strawberry flowers by Botrytis cinerea and disease incidence of fruit originating from infected flowers. Phytopathology 77:1225–1230. 10.1094/Phyto-77-1225. [DOI] [Google Scholar]
- 36.Muñoz M, Faust JE, Schnabel G. 2019. Characterization of Botrytis cinerea from commercial cut flower roses. Plant Dis 103:1577–1583. 10.1094/PDIS-09-18-1623-RE. [DOI] [PubMed] [Google Scholar]
- 37.Petrasch S, Knapp SJ, Van Kan JA, Blanco‐Ulate B. 2019. Grey mould of strawberry, a devastating disease caused by the ubiquitous necrotrophic fungal pathogen Botrytis cinerea. Mol Plant Pathol 20:877–892. 10.1111/mpp.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kock SL, Holz G. 1992. Blossom-end rot of pears: systemic infection of flowers and immature fruit by Botrytis cinerea. J Phytopathol 135:317–327. 10.1111/j.1439-0434.1992.tb04317.x. [DOI] [Google Scholar]
- 39.Vannier N, Agler M, Hacquard S. 2019. Microbiota-mediated disease resistance in plants. PLoS Pathog 15:e1007740. 10.1371/journal.ppat.1007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith TJ. 2000. Overview of tree fruit production in the Pacific Northwest United States of America and southern British Columbia, Canada, p 25–30. In IV International Symposium on Mineral Nutrition of Deciduous Fruit Crops. ISHS Acta Horticulturae 564, Penticton, British Columbia, Canada. [Google Scholar]
- 41.Logan J, Mueller MA, Searcy MJ. 2000. Microclimates, peach bud phenology, and freeze risks in a topographically diverse orchard. Horttech 10:337–340. 10.21273/HORTTECH.10.2.337. [DOI] [Google Scholar]
- 42.Lopez G, DeJong TM. 2007. Spring temperatures have a major effect on early stages of peach fruit growth. J Hortic Sci Biotechnol 82:507–512. 10.1080/14620316.2007.11512266. [DOI] [Google Scholar]
- 43.Vannette RL. 2020. The floral microbiome: plant, pollinator, and microbial perspectives. Annu Rev Ecol Evol Syst 51:363–386. 10.1146/annurev-ecolsys-011720-013401. [DOI] [Google Scholar]
- 44.Ngugi HK, Scherm H. 2006. Biology of flower-infecting fungi. Annu Rev Phytopathol 44:261–282. 10.1146/annurev.phyto.44.070505.143405. [DOI] [PubMed] [Google Scholar]
- 45.DuPont ST, John Strohm C. 2020. Integrated pest management programmes increase natural enemies of pear psylla in Central Washington pear orchards. J Appl Entomol 144:109–122. 10.1111/jen.12694. [DOI] [Google Scholar]
- 46.Klein A-M, Brittain C, Hendrix SD, Thorp R, Williams N, Kremen C. 2012. Wild pollination services to California almond rely on semi-natural habitat: wild pollination services to California almond. J Appl Ecol 49:723–732. 10.1111/j.1365-2664.2012.02144.x. [DOI] [Google Scholar]
- 47.Vannette RL, Fukami T. 2017. Dispersal enhances beta diversity in nectar microbes. Ecol Lett 20:901–910. 10.1111/ele.12787. [DOI] [PubMed] [Google Scholar]
- 48.Comeau AM, Douglas GM, Langille MG. 2017. Microbiome helper: a custom and streamlined workflow for microbiome research. mSystems 2:e00127-16. 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 50.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Beeck MO, Lievens B, Busschaert P, Declerck S, Vangronsveld J, Colpaert JV. 2014. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One 9:e97629. 10.1371/journal.pone.0097629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 54.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. 2018. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson M, Lindow SE. 1993. Interactions between the biological control agent Pseudomonas fluorescens A506 and Erwinia amylovora in pear blossoms. Phytopathology 83:117–123. 10.1094/Phyto-83-117. [DOI] [Google Scholar]
- 57.Leibinger W, Breuker B, Hahn M, Mendgen K. 1997. Control of postharvest pathogens and colonization of the apple surface by antagonistic microorganisms in the field. Phytopathology 87:1103–1110. 10.1094/PHYTO.1997.87.11.1103. [DOI] [PubMed] [Google Scholar]
- 58.Baselga A, Orme CDL. 2012. betapart: an R package for the study of beta diversity. Methods Ecol Evol 3:808–812. 10.1111/j.2041-210X.2012.00224.x. [DOI] [Google Scholar]
- 59.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara R, Simpson GL, Solymos P, Henry M, Stevens H. 2014. Package ‘vegan.’ Community Ecology Package R Package Version 2. https://vegan.r-forge.r-project.org.
- 60.Northwest Horticultural Council. 2019. Industry facts—pear fact sheet. Northwest Horticultural Council, Yakima, WA. https://nwhort.org/industry-facts/pear-fact-sheet/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 and S2. Download AEM.00048-21-s0001.pdf, PDF file, 591 KB (590.3KB, pdf)
Table S1. Download AEM.00048-21-s0002.xlsx, XLSX file, 11 KB (10.6KB, xlsx)
Table S2. Download AEM.00048-21-s0003.xlsx, XLSX file, 11 KB (10.7KB, xlsx)
Table S3. Download AEM.00048-21-s0004.xlsx, PDF file, 15 KB (14KB, xlsx)
Data Availability Statement
Raw amplicon sequences are available on the NCBI Sequence Read Archive (SRA) under BioProject accession no. PRJNA659266.