Abstract
With the rise in outbreaks of pathogenic bacteria in both food and water resulting in an increased instance of infection, there is a growing public health problem in both developed and developing countries. In this increasing threat the most effective method for control and prevention is rapid and cost-effective detection. Research has shifted in recent years towards the development of rapid and on-site assays for the detection of these kinds of bacteria. However, there are still some limitations in the implementation of these assays in the field. This article discusses the current on-site detection methods. Current scope of advancements and limitations in the development or use of these on-site technologies for food and waterborne bacterial detection is evaluated in this study. With the continued development of these technologies, on-site detection will continue to impact many areas of public health. As these methods continue to improve and diversify further, on-site detection could become more widely implemented in food and water analysis.
Keywords: Foodborne bacteria, point of care, assay development, biosensors, detection, food analysis
1.0. Introduction
Bacteria are commonly present in the environment and within the human body as naturally occurring flora that can be harmless. However, there are many species of bacteria that are pathogenic and can cause harm. Foodborne and waterborne illness are caused by consuming food or water that has been contaminated by these pathogens or their associated toxins. These can be present in many types of foods and in different environments (Bintsis, 2017). Some of the most common pathogens involved in these infections include Listeria monocytogenes, Salmonella enterica, Escherichia coli O157:H7, other Shiga toxin-producing E. coli (STEC), Staphylococcus aureus, Vibrio species, Bacillus cereus, Campylobacter jejuni, Clostridium, and others (Bintsis, 2017; Schirone, Visciano, Tofalo, & Suzzi, 2019). Contamination of these bacteria in food and water and their corresponding infections in humans have become a global problem with large outbreaks that occur even in circumstances where sanitization efforts were made. An overview of these different bacterial species can be seen in table 1. For example, within the United States alone, the CDC estimates that foodborne pathogens cause 9.4 million illnesses annually, with 55,961 of those leading to hospitalization and 1351 illnesses resulting in death (CDC, 2018). Furthermore, these kinds of statistics likely underestimate the totals, as evidence suggests that the reported incidences are a fraction of the real incidence rate (Scallan, et al., 2011). With more and more cases of bacterial illness presenting annually, these bacterial infections have created a high economic and health burden at a global level (Minor, et al., 2015). As a result, it is within global health interest to be able to identify and detect these pathogens at their respective sources before they can cause an outbreak that results in physical and economic suffering.
Table 1:
A comprehensive look at a variety of more commonly seen foodborne pathogens.
| Species | Sources | Infectious doses | Effects | References |
|---|---|---|---|---|
| Campylobacter | Chicken, unpasteurized milk, contact with animals, sausages, red meat, contaminated water | 800–106 cells | Local complications include cholecystitis, peritonitis, pancreatitis, and hemmorhage of the GI tract. Rarely manifests outside intestines. Can lead to Guillain - Barre’ syndrome post infection | (Acheson & Allos, 2001) |
| Listeria monocytogenes | Food processing (improper sanitization), deli meats, unpasteurized products | over 100 cells (often higher) | Listeriosis | (Buchanan, Gorris, Hayman, Jackson, & Whiting, 2017) |
| Escherichia coli O157 H:7 | Beef products, vegetables, water contamination. Can person to person transmit | 10–100 cells | Hemolytic uremic syndrome, hemorrhagic colitis | (Rahal, Kazzi, Nassar, & Matar, 2012) |
| Salmonella spp. | Poultry, eggs, dairy, fresh produce. | 10 cells | diarrhea, gastrointestinal illness, death | (Bell, et al., 2016) |
| Clostridium botulinum | Improperly canned foods, especially home-canned, cured, and fermented foods. | Detection is focused on the toxin as the bacteria itself is not a clear indicator of botulism. Needs to detect pM at least, if not lower. There is no permissible toxin level in food | Botulism, Muscle paralysis | (Thirunavukkarasu, et al., 2018) |
| Staphylococcus aureus | Improper food handling, unpasteurized product, ready to eat foods, processed foods | 0.5 ng/ mL of produced toxins can cause disease | Hypersalivation, nausea, vomiting, cramping | (Kadariya, Smith, & Thapaliya, 2014) |
| Bacillus cereus | Eggs, meat and dairy products. Rice, noodles, produce. | 105– 107 cells total | 2 types of illness. One is diarrheal. The other is vomiting primarily. Usually mild. | (Granum & Lund, 1997) |
| Vibrio cholerae | Contaminated Water | 108–1011 cells in healthy adults | Gastrointestinal distress - cramps, vomiting, diarrhea. Dehydration and metabolic acidosis. Circulatory collapse and death | (Nelson, Harris, Morris, Calderwood, & Camilli, 2009) |
| Vibrio parahaemolyticus/vulnificus | Seafood, especially molluscan shellfish | As low as 103 | Gastroenteritis, septicemia. Death | (Baker-Austin, et al., 2018) |
While for most pathogens the gold standard has been culturing followed by verification using other biological methods, this method is time consuming (Zhao, Lin, Wang, & Oh, 2014). As such, recent advances in detection technologies have shifted the emphasis toward more rapid screening methods. These technologies include a variety of biosensors, polymerase chain reaction (PCR) based technologies, and enzyme-linked immunoassays (ELISAs). While more definitive and faster than common culturing methods, these methods are still complex, expensive, and require skilled personnel for operation (Law, Ab Mutalib, Chan, & Lee, 2015). As such, assay development has sometimes traded high sensitivity for portability as assay design has shifted toward “on-site” detection platforms. Researchers have developed assays that are more rapid and capable of being transported to a potential source of contamination. Besides being less labor-intensive, these on-site detection methods hold promise for applications in a variety of areas such as food industry, clinical medicine, agriculture, water management, and more.
This review will summarize recent developments in on-site technologies that have been applied to the detection of waterborne and foodborne pathogens. By discussing the current state-of-the-art as well as future detection trends, this review seeks to provide readers with an informative guide to the rapidly changing landscape of on-site detection strategies.
2.0. Gold-standard method of pathogen detection
The gold standard and long-standing method for bacterial identification and detection has been culturing. These methods are the oldest bacterial detection methods to date and are still regarded as the gold standard. This is due to the relative inexpensiveness, sensitivity, and the ability to both qualify and quantitate level of bacteria in a sample. There have been new developments in culturing-based methods, particularly in the formulation of specific chromogenic agars. These agars are formulated to enhance the sensitivity and specificity of culture-based detection. These agars eliminate the need to add additional screening compounds to agar as done traditionally. An example of this was formulated for the detection of Vibrio parahaemolyticus – a seafood pathogen. ChromoVPagar was able to be more sensitive specific and accurate when compared to previous CHROMagar™ and thiosulfate- citrate- bile – salts (TCBS) agar (Lee, Azizah, & Kim, 2020). Additionally, some agars have been formulated to reduce the amount of turnaround time and improve the readout. CHROMagar Enterococcus (CHR) is a chromogenic medium to isolate Enterococcus from water samples. When compared to the commercially available mediums for the same purpose they were able to reduce the turnaround time to 18 hours. They also were able to increase the size of the colony morphology providing an easier read (Cho, Hiott, Woodley, Frye, & Jackson, 2020). While these culturing methods are accurate and reliable detection tools, they are also time-consuming and quite laborious, usually requiring a laboratory setting with personnel trained to perform the assays. Therefore, alternative methods such as immunoassays and nucleic acid-based methods have been accepted and widely used either as standalone techniques or supplementary techniques in conjunction with culturing for confirmation.
There have also been some instances where culturing has been combined with portable platforms for novel biosensors. In doing so there can be a reduction of turnaround time for results. For example, a paper based analytical device (PAD) was combined with a chromogenic medium for the detection of V. cholerae in water samples in Haiti. This system was able to reduce turnaround time to 18 – 24 hours compared to the standard Thiosulfate-citrate-bile salts-sucrose agar (TCBS) medium (Briquaire, et al., 2017). Another group integrated agar into a 3D microelectrode (Butler, Goel, Goodnight, Tadigadapa, & Ebrahimi, 2019). They were able to detect metabolism for 10,000 CFU/mL of K12 bacteria in only one hour. When ionic metabolites are released by actively metabolizing bacteria, the electrical characteristics of the culture media changes. The sensor can then measure the impedance changes caused by this. This serves as a general indication of bacterial contamination, rather than specifically detecting a species or strain. While the sensor was fabricated with uropathogenic E. coli in mind, this could be applied for foodborne pathogen detection, where rapid results are crucial. Additionally, further adaptation to enhance specificity would be even more beneficial in the context of food analysis.
Several unique challenges are seen with foodborne and waterborne bacterial detection. Given the perishable nature of many food sources – the turnaround time of culturing methods is too long to wait on results before allowing food to enter the market. This turnaround time can be further complicated when culturing methods are often accompanied by other methods. For example, it is common in the detection of Salmonella spp. to utilize culturing agars to confirm the species but then use nucleic acid based techniques to confirm particular serotyping (Bell, Jarvis, Ottesen, McFarland, & Brown, 2016). Foodborne and waterborne pathogens can often be present at lower concentrations and their infectious doses are often small. These infectious doses are highlighted in table 1 for several major pathogens. To further make matters more complicated, some bacteria are cumbersome to grow in culture such as L. monocytogenes. These bacteria grow very slowly and are often out-grown by other bacterial species in a sample (Gasanov, Hughes, & Hansbro, 2005). Detection limits need to be capable to detect small amounts of bacteria to ensure accurate detection results given that many bacteria cause infections in low doses. This can be a challenge, especially with large sample volumes. Furthermore, there is a huge diversity of food products which provide additional challenges in sample preparation. While lab-based techniques are sensitive enough, they are still too time-consuming. Previously developed point of care technologies have had challenges with achieving the adequate sensitivity. This is a key factor to consider and address when developing an on-site assay.
3.0. Developments in Sampling and Sample preparation Toward On-site Detection
Sample preparation is one of the most important steps in food analysis and is one that can play a direct role in how successful and sensitive an assay is. To make matters more challenging, sample conditions can vary between different bacterial species and different food types. The United States Department of Agriculture have published a Bacterial Analytical Manual that describes the best methods for food sampling. (FDA, 2020). While these methods were primarily considered for prepping samples for laboratory transport and laboratory analysis, these concepts have shaped how on-site technologies have tackled sampling. Proper and rigorous food sampling is crucial for analysis.
For sampling one of the biggest challenges remains in how much sampling needs to be done to guarantee the true absence of a pathogen. This is especially important when considering many of the technologies described utilize only microliters of sample to analyze. Currently the general standard is based off the concept of a representative sample in 25 g of food. Many on-site technologies that utilize small sample sizes can potentially bypass this sampling problem by using methods to reduce the volume of a sample such as centrifugation, concentration, or bacterial isolation. Additionally, by being sufficiently rapid and inexpensive assays can allow for several samples to be obtained. This could be an alternative option to allow the user to confidently say a larger lot is absent of bacteria through rigorous sampling. Once this sampling has been performed the sample must be prepared to be analyzed properly. One common method and an initial step for food preparation (especially for non-liquid food matrices) is the creation of a solution with some form of buffer and then homogenizing the sample. Homogenization is a key step to ensure that the collected sample is more representative. This can be achieved through means such as blending, grinding, pulsing, sonicating, and homogenizing by hand. A few articles have compared these and other sample preparation methods (Armstrong, et al., 2019; Rohde, Hammerl, Appel, Dieckmann, & Al Dahouk, 2015). Each preparation method varies in efficiency, and many of these methods result in some loss of bacteria in the sample (Kim, et al., 2012). Sample preparation is a major hurdle to address if assay sensitivity needs to be improved. One popular method is to remove bulk of the matrix by means of target extraction, especially in the context of nucleic acid isolation. One way this is done is using magnetic beads. These bind to the target bacteria or nucleic acids via the conjugation of specific probing moieties and allow for simple separation from the food matrices (Wang, et al., 2020). Magnetic nanoparticles can serve similar purposes (Xue, Zheng, Zhang, Jin, & Lin, 2018). In this case, the magnetic nanoparticles were modified with antibodies for immunogenic detection. This simple separation and recognition strategy improves sample extraction by simplifying the extraction process and allowing the user to isolate only the desired target.
Paper-based assays have utilized various means of filtration for sample preparation. These assays consist of either single or multiple layers of paper that can allow for all-in-one sample preparation, extraction, washing, and detection as the sample passes through the layers of paper. For example, many microfluidic paper analytical devices (μPAD) and other multilayered paper systems have incorporated layers that separate out contents of the sample (Eltzov & Marks, 2017). Additionally, some biosensing systems such as microfluidic based sensors also have built in means to separate out sample contents. These sample preparation methods either incorporated into the device or are done before sample analysis. This can include techniques such as the use of magnetism (Castillo-Torres, Arnold, & McLamore, 2019) or filtration (Zhao, et al., 2019). Microfluidics in the context of sample preparation has also been touched upon in a recent review (Kant, et al., 2018). Furthermore, some assays have even been able to detect bacteria in food samples without any need for extraction or sample preparation. For example, one group utilized an LED light and cell phone technology to measure the scatter signals from the surface of ground beef (Liang, Park, & Yoon, 2014). The addition of bacteria in comparison to the negative control exhibits a particular change in the light scatter angle – based on that the bacteria can be detected. This method was able to detect 10 CFU/mL of bacteria, was reagentless, and did not require any sample preparation. Additionally, Yousefi et al. created a sensing surface that generated a fluorescent signal in the presence of bacteria (Yousefi, Ali, Su, Filipe, & Didar, 2018). This material utilized microarrays at the picoliter size that have RNA-cleaving fluorogenic DNAzyme probes for detection. This material was able to detect as low as 103 CFU/mL in both meat and apple juice. This type of real-time monitoring is a great step towards simple and constant food monitoring that can be applied to various steps in the food production chain.
4.0. On-site methods for pathogen detection
4.1. REASSURED and ASSURED criteria for point-of-care assay development.
To be considered a point-of-care assay in the context of clinical analysis, a set of criteria known collectively by the acronym “ASSURED” has been set in place by the World Health Organization (WHO). ASSURED criteria specify that assays must be affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to an end user. Although, these do not apply fully or directly to food and waterborne pathogen detection, these criteria address assay concerns and form a standard that should be achieved for any pathogen detection. The ideal assay in this scenario should also be simple to perform with minimal training. Additionally, it should enable rapid results at first visit or sampling and should not require refrigerated or complicated storage (robust). This assay should use portable equipment as possible and then be able to be provided to areas that need to conduct analysis. More recently with the advancement these criteria have evolved to include two new points of consideration (Land, Boeras, Chen, Ramsay, & Peeling, 2019). This was then renamed REASSURED. The new criteria take into consideration real-time connectivity and ease of specimen collection. Real-time connectivity considers the use of a reader of smart device to power the reaction or read results and provide them to those who need them. Assays must also use non-invasive specimens. While not all assays can or need to fulfill the entire list of criteria, it is important to consider many of these ideals when designing an assay. Not all of these characteristics of point of care are necessarily crucial for foodborne detection. For example, there is no significant challenge in sample collection, but challenges exist in finding representative sample in the case of large bulky samples (such as a lot of a particular harvested produce). Analyzing food or water is noninvasive and easy to obtain. Instead, the bigger challenge is in sample preparation, which is not addressed specifically by these criteria but still important. Additionally, having real-time data, while not necessarily required, is a helpful consideration. Some papers have aimed to address real-time monitoring (Sun, Huang, Warden, & Ding, 2020; Yousefi, et al., 2018). In the context of food and waterborne infections, assay designs must not only be rapid and portable for on-site monitoring but also be highly sensitive and specific due to the lower concentrations and potential diversity of pathogens and other organisms present in the samples. Assays, therefore, need to be designed around the detection needs of a specific pathogen or related groups of pathogens. Meanwhile in the context of places such as developing countries where there may be a lack of refrigeration there is a higher need for robustness and affordability on top of the other criteria. These needs on top of the needs of the target user population will shape how the assays need to fulfill the different criteria. Furthermore, these criteria will drive and alter the development of these assays as efforts push towards the utilization of on-site assays.
4.2. Applications of cell-phone technologies
Cell phones have become of interest in the context of developing on site assays. They have been included with different pathogen detection technologies such as molecular methods and biosensors for monitoring the results. Cell phone technology can help improve portability of assays while maintaining decent sensitivity and allowing for inclusion of more technological methods such as electrochemical and fluorescent detection without the requirement for a bulky machine or an expensive portable model. Cellphone integration in assays takes advantage of either the imagers (camera) or the digital processors of the devices to conduct analyses such as image processing (Contreras-Naranjo, Wei, & Ozcan, 2016). Attachments can also include camera lens piece attachments and light boxes. For example, Adkins et al. used a cell phone in conjunction with a cardboard light box to visualize their paper-based assay and were able to detect down to 1 CFU/mL of pathogenic and non-pathogenic E. coli strains (Adkins, et al., 2017). A schematic of their design can be seen in figure 1. Another example utilizes a cassette with a microscopy set up. This assay targeted multi-drug resistant Staphylococcus aureus from milk samples. Fluorescent magnetic nanoparticles allowed for both sensitive visualization but also minimal sample processing. Magnetic capture allowed for easy processing. This system had a low limit of detection of 10 CFU/ml and obtained results in only 10 minutes (Shrivastava, Lee, & Lee, 2018). Combining on-site technologies such as paper-based systems (Wang, Gao, Wang, & Liu, 2020), nucleic acid amplification (Nguyen, Nguyen, Liu, & Seo, 2020), or biosensing systems (Sun, et al., 2020; Wang, et al., 2019) with smartphones can allow for more complex sensors to be developed that are still simple for an end user to operate and interpret accurately. For example, Jiang et al. fabricated a microfluidic impedance sensor utilizing a smartphone for the platform that combines sensing and pre-concentration to detect as low as 10 E. coli cells in a milliliter of water (Jiang, et al., 2014). The addition of cell-phone technologies also helps to potentially achieve lower limits of detection by allowing for more advanced and sensitive data acquisition and repeated analysis without adding expensive or bulky instrumentation, proving very useful for point-of-care test in the context of the ASSURED criteria. Taking it a step further, the addition of cell-phone technology has even allowed for sensitive and reagentless detection. Liang et al. used a near infrared LED that was irradiated and the gyro sensor and camera of the phone for measurements of the scattering signals from the surface of the sample (Liang, et al., 2014). No sample preparation was required, and they were able to detect E. coli at as low as 10 CFU/mL concentrations as described before. Cell-phone technology and its further utilization will dramatically alter and improve the landscape of on-site technologies.
Figure 1:
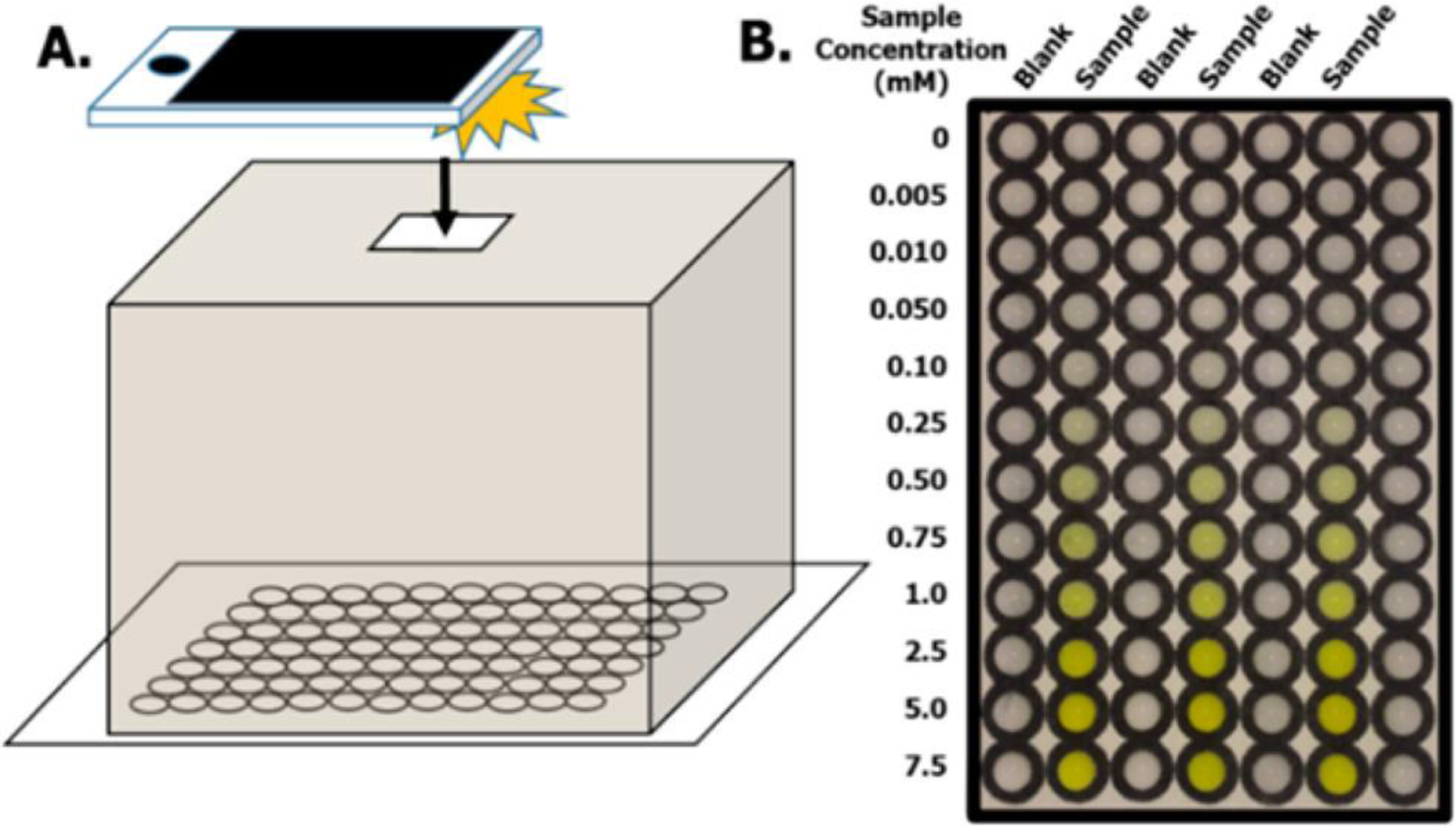
From Adkins et al. A) A mechanistic overview of their image capture process with a cell-phone and a light box serving to act similarly to a plate reading measurement. The light box has an opening to allow for the camera to see into the box. B) Calibration plate highlighting the blank versus a concentration of a target sample.
4.3. Paper-based Methods
Paper-based materials have been gaining popularity in the field of diagnostics. This strategy was applied to many other types of devices due to the versatility and low cost of paper materials. Paper is a broad definition and is intended for these purposes to include a porous surface such as nitrocellulose or cellulose derivatives that utilize the property of capillary action. Paper is an attractive material for applications in diagnostic assays because of its cost, abundance, and its mechanical properties. Most importantly, paper is biocompatible and biodegradable, attributes which serve well for disposable assays. These assays can be broken down into three general subtypes – which will be discussed in more detail in the following sections. A comprehensive overview of these assays can be seen in table 2.
Table 2:
Comprehensive list of discussed assays showcasing their limits of detections and assay run time.
| Detection method | Pathogen | Detection Limit | Time of assay | Reference |
|---|---|---|---|---|
| Traditional methods | ||||
| Plating (culturing) | 1 cell | 1–3 days | (de Boer & Beumer, 1999) | |
| Lateral Flow / paper strip | ||||
| Escherichia coli | ||||
| 10 CFU/mL | <3 hours | (Kim & Oh, 2019) | ||
| 12– 300 CFU/mL in foods | 40 mins | (Suaifan, et al., 2017) | ||
| 105 CFU/mL | 20 mins | (Shirshahi, et al., 2019) | ||
| 900 CFU/mL in milk | not directly stated but under 10 mins | (Han, et al., 2018) | ||
| Salmonella spp. | 5*105 CFU/mL | 5 mins | (Cam & Oktem, 2019) | |
| 80 CFU/mL | 11 mins | (Bu, et al., 2019) | ||
| 1 CFU/μL | 30 mins | (Wu, et al., 2020) | ||
| Multiple species | 100 cells | < 1 hour | (Peng & Chen, 2019) | |
| 1, 0.5, and 0.25 nM DNA | not fully discussed | (He, et al., 2019) | ||
| 1 CFU/mL with enrichment, 1.87 × 104 CFU and 1.47 × 104 CFU without | ~3 hours or 7 hours with enrichment | (Shin, et al., 2018) | ||
| 1 × 10−7 M to 1 × 10−9 M Quorum Sensing Molecules | (Wynn, et al., 2018) | |||
| Listeria monocytogenes | 53 cells | 5 mins | (Tasbasi, et al., 2019) | |
| Cronobacter sakazakii | 107 CFU/mL | 15 mins | (Scharinger, et al., 2017) | |
| Single Layer μPADs | ||||
| Vibrio cholerae | ~104 CFU/mL | 18–24 hours | (Briquaire, et al., 2017) | |
| Salmonella spp. | 100 CFU/mL | 90 mins | (Srisa-Art, et al., 2018) | |
| Escherichia coli and/ or Enterococcus | ~1 CFU/mL | 4–8 hours | (Adkins, et al., 2017) | |
| ~104 CFU/mL | <3 hours | (Pang, et al., 2018) | ||
| 100 CFU/mL with fluorescence, 44 CFU/mL with colorimetric detection | Not directly stated, <1 hour | (Wang, et al., 2020) | ||
| 3D μPADs | ||||
| Escherichia coli | 100 CFU/mL | <5 mins | (Eltzov & Marks, 2017) | |
| multiple species | 10 CFU/mL | <12 hours | (Kim, et al., 2019) | |
| 100 CFU/mL | not directly stated | (Ahn, et al., 2018) | ||
| 170 CFU/mL | 70 minutes | (Trinh & Lee, 2018) | ||
| Origami Paper Devices | ||||
| Escherichia coli | 103 CFU/mL | ~2 hours | (Trieu & Lee, 2019) | |
| 103 CFU/mL | 35 mins | (Sun, et al., 2019) | ||
| Salmonella spp. | 103 CFU/mL | ~2 hours | (Trieu & Lee, 2019) | |
| Biosensing systems | ||||
| Magnetic | Salmonella spp. | 1 CFU/mL | Not stated directly | (Zeinhom, et al., 2018) |
| Vibrio cholerae (toxin) | 0.2 ng/mL | ~2 hours | (Achtsnicht, et al., 2019) | |
| Escherichia coli | 100 CFU/100 mL | <45 mins | (Castillo-Torres, et al., 2019) | |
| Piezoelectric | Salmonella spp. | <1 CFU/mL | <4 hours | (Fulgione, et al., 2018) |
| Escherichia coli O157 H:7 | 1.46 * 103 CFU/mL | <1 hour | (Yu, et al., 2018) | |
| Optical | Escherichia coli | 2 CFU/mL | Not stated directly | (Duan, et al., 2020) |
| 40 CFU/mL | 1 hour | (Mou, et al., 2019) | ||
| 103 CFU/mL | Real time measurement | (Yousefi, et al., 2018) | ||
| 100 CFU/mL | Not stated directly | (Liang, et al., 2014) | ||
| Multiple spp. | ~105 CFU/mL | 10 mins | (Ledlod, et al., 2020) | |
| 10 CFU/mL | 1 hour | (Nguyen, et al., 2020) | ||
| 2.460 – 5.407 CFU/mL | <4 hours | (Wang, et al., 2020) | ||
| 1.0 × 109 CFU/mL (was proof of concept not detection limit) | Real time results | (Sun, et al., 2020) | ||
| Salmonella spp. | 58 CFU/mL | 2 hours | (Wang, et al., 2019) | |
| 11 CFU/mL | 2.5 hours | (Wang, et al., 2020) | ||
| 14 CFU/mL | 2 hours | (Xue, et al., 2018) | ||
| Pseudomonas aeruginosa | 30 μg/mL (proof of concept and not a detection limit) | 24 hours (incubation period) | (Gao, et al., 2021) | |
| Staphylococcus aureus | 10 CFU/mL | 10 mins | (Shrivastava, et al., 2018) | |
| Electrochemical | Escherichia coli | 2 CFU/mL | Not directly stated | (Shahrokhian & Ranjbar, 2018) |
| 15 CFU/mL | 30 mins | (Vu, et al., 2020) | ||
| 1400 cells in 25 μL | <1 hour | (Wang, et al., 2015) | ||
| 100 CFU/mL | <1 hour | (Li, et al., 2015) | ||
| 10 cells/mL | Real-time measurement | (Jiang, et al., 2014) | ||
| Vibrio parahaemolyticus | 5.74 CFU/mL | 30 mins | (Jiang, et al., 2021) | |
| 0.3 CFU / 25 g seafood | 45 mins | (Kampeera, et al., 2019) | ||
| Bacillus cereus | 100 CFU/mL | 5 mins | (Ait Lahcen, et al., 2018) |
4.3.1. Lateral Flow Assays (LFAs)
Lateral flow assays utilize the concepts of capillary action and selective analyte binding to distinguish the presence of a sample in a visual manner. These are typically made with nitrocellulose or cellulose membranes. These assays are recognized for their simplicity, low cost, portability, and rapid response time. Several recent reviews have discussed LFAs in detail (Mahmoudi, de la Guardia, & Baradaran, 2020; Nguyen, Song, Park, & Joo, 2020). Some groups have expanded and improved the utilization of nucleic acid amplification in conjunction to lateral flow assays, including isothermal techniques. As an example, one such technique is Recombinase Polymerase Amplification. Wu et al. utilized recombinase polymerase amplification with a lateral flow assay for the detection of Salmonella enterica typhimurium (Wu, et al., 2020). However, they were able to improve the use of this isothermal amplification technique by introducing base substitutions in the primer and probe sequences. By doing so they were able to eliminate primer-dependent artifacts. This system was able to detect 1 CFU/mL of unpurified culture in 30 minutes. This improvement can help prevent false positive signals that could result from primer-primer or primer-probe interactions. Interestingly, this assay did not use nor require DNA extraction, analyzing straight from a thermally inactivated sample. Additionally, a technology that has been rapidly advancing in recent years is the utilization of CRISPR with different cas protein systems (cas9, cas13, cas12a, etc.) This has been employed in the detection of several viral and bacterial targets (Wang, Shang, & Huang, 2020). Additionally, CRISPR/Cas9 has been employed in the Cas-EXPAR system which involves cas9 cleavage of the target followed by EXPAR strand extension directed by a polymerase and a nicking endonuclease inducing single strand nicking (Huang, Zhou, Wang, & Xing, 2018). Whole RNA was extracted utilizing a manufactured kit and reverse transcribed before using the CAS-EXPAR system. They were able to detect 2.5 and 1.5 μg of total L. monocytogenes mRNA using CAS-EXPAR.
Traditionally the most common reporter for LFAs is colloidal gold nanoparticles (AuNP) because it has an intense color and can be directly visualized without an additional reagent (Kim, et al., 2007). LFAs using colloidal gold have been applied to the detection of foodborne pathogens such as Salmonella spp. and E. coli O157 H:7 (Cam & Oktem, 2019; Kim & Oh, 2019). Unfortunately, LFAs using AuNPs and antibodies for bacterial detection often have high detection limits, partially determined by the binding affinity of recognition element pairs (Bishop, Hsieh, Gasperino, & Weigl, 2019). Unlabeled AuNPs have also been used to induce color change (Peng & Chen, 2019), and nucleic acid sequences have recently become commonplace as AuNP-conjugated DNA or RNA probes for detection. Nucleic acid based LFAs have been used to detect Salmonella spp. (He, et al., 2019), S. aureus (He, et al., 2019), and Vibrio spp. (Shin, et al., 2018).
Additionally, there has been considerable advances in nanoparticle mediated signal amplification. These improvements include enlarging nanoparticle aggregation, utilizing metals, and modifying with enzymes such as horseradish peroxidase (HRP) among others (Liu, Yang, & Liu, 2019). Aptamers have been also used in conjunction with nanoparticles for detection. For example, Tasbahi et al. used aptamer-gated silica nanoparticles to generate a detection limit of 53 L. monocytogenes cells per mL in 5 minutes (Tasbasi, et al., 2019). Aptamers are relatively short single stranded nucleic acid sequences that are capable of selective binding to targets such as proteins, small molecules, and even cells. The aptamer in this case allows for the targeting to the bacteria and its interaction in turn releases 3,3’,5,5’-Tetramethylbenzidine (TMB) which is a chromogenic substrate that creates a visible blue signal after its oxidation with peroxidases such as HRP. This combination allowed for a sensitive and rapid detection without the use of major equipment and without any amplification.
Outside of the typical colloidal gold for visualization, there are a variety of other visible reporters such as quantum dots (Gong, et al., 2017), biological dyes (Bu, et al., 2019) and magnetic beads (Suaifan, Alhogail, & Zourob, 2017) that can be used to achieve signal visualization in lateral flow assays. Often, these different visualizing technologies have been introduced as means to improve assay sensitivity. For example, Shirshahi et al. utilized functionalized reduced graphene oxide as a label for a lateral flow assay for E. coli O157 H:7 (Shirshahi, Tabatabaei, Hatamie, & Saber, 2019). They had a reported LOD around 105 CFU/mL. As means to improve LFA sensitivity, Han et al. used a “nanozyme” probe in the detection of E. coli O157 H:7 (Han, et al., 2018). These probes consisted of palladium-platinum nanoparticles labelled with antibody. Signal enhancement occurred by adding 3,3’,5,5’-tetramethylbenzidine (TMB) onto the test line where the nanozyme probe would accumulate. Their sensitivity was found to be 9.0 × 102 CFU/mL in milk samples, which is more sensitive than the traditional LFA based on colloidal gold. Another technique implemented in LFA is the combination of traditional microbiological staining with an LFA platform. Bacteria are targeted with both dyes and monoclonal antibodies for selective detection of S. enteritidis, producing a detection limit of 80 CFU/mL in 11 minutes (Bu, et al., 2019). This system was also applied to the detection of L. monocytogenes with a detection limit in this case of 104 CFU/mL.
Additionally, LFAs have been designed for multiplexing, which is advantageous for on-site assays by reducing the number of individual tests and samples to run. Several multiplexed LFA-based assays utilizing different reporter mechanisms such as gold nanoparticles, fluorescence, and bioluminescence have been developed for different foodborne pathogens such as Cronobacter sakazakii, E.coli O157 H:7, S.Typhimurium, S.aureus, B.cereus as well as overall food spoilage (Scharinger, Dietrich, Wittwer, Märtlbauer, & Schauer, 2017; Shin, et al., 2018; Wynn, et al., 2018). For example, one unique method for general foodborne contamination detection is the use of quorum sensing. Wynn et al. used a whole cell biosensing system on a paper-strip platform to detect two bacterial strains as model organisms for general food spoilage via the detection of quorum sensing molecules, which are acyl-homoserine lactones, AI-2s, or small peptides. These molecules are used by bacteria for cell-cell communication and are common to most Gram-negative bacteria. The assay used immobilized bacteria harboring plasmids that could allow for the sensing of acyl homoserine lactones since the AHLs can bind the regulatory protein in the plasmid and turns on the production of bioluminescent proteins and their substrate (encoded within the plasmids). The assay was able to detect as low as 10−9 M quorum sensing molecules in 3 hours for as little as $0.15 each.
4.3.2. Microfluidic Paper-Based Analytical Devices (μPADs).
μPADS are miniaturized lateral flow platforms typically composed of patterned, hydrophobic channels designed to utilize capillary action rather than external pumps to analyze small-volume biological samples. These devices enable a significant cost reduction over traditional microfluidic devices while adding the benefits of easy transport and disposability. Single layer μPADs have been used in the detection of several bacteria species such as V. cholerae (Briquaire, et al., 2017), Salmonella spp. (Srisa-Art, Boehle, Geiss, & Henry, 2018), E. coli spp.(Adkins, et al., 2017). For example, Srisa-Art et al. utilized immunomagnetic separation with beads conjugated to anti-Salmonella antibodies to capture the bacteria and then they utilized a sandwich assay with colorimetric detection similar to an ELISA but on paper. This had a limit of 100 CFU/mL with a limit of 1000 CFU/mL in milk samples.
By combining the technique of ELISA with paper microfluidics such as previously described, the complexity, cost, and runtime of the assay can be reduced while simultaneously maintaining a similar sensitivity. For example, Pang et al. developed a low-cost paper ELISA for the detection of E. coli O157 H:7 that was able to be completed in under 3 hours with only 5 μL of sample. Their limit of detection was found to be 1 × 104 CFU/mL (Pang, et al., 2018). This technology has seldom been used in bacterial detection; however, it does yield promise for potential future uses due to the enhanced portability and the maintenance of sensitivity. Further advancements for the increase of stability will strongly benefit these technologies for food analysis. A similar concept has been employed for viral detection with the use of microspots – similar to a well plate but on paper for detection (Zhang, et al., 2017). This system has a detection limit of 265 femtomoles and while used for the detection of Epstein-Barr virus from whole RNA extracts, could also be useful to develop foodborne pathogen assays.
There are also “3D” μPAD devices that utilize multiple, individually printed paper layers. These are advantageous because each layer can be individually functionalized with multiple reagents, allowing the integration of complex steps that would ordinarily require several single-layer devices (Kim, Kwon, Lee, & Noh, 2019). These sensors can also incorporate smaller sample volumes and can have shorter assay times than standard lateral flow assay strips. Further modification of μPAD devices enables the combination of ancillary reactions such as amplification and filtration without additional steps. For instance, amplification techniques such as Loop-Mediated Isothermal Amplification (LAMP) (Trinh & Lee, 2018) and Recombinase Polymerase Amplification (RPA) (Ahn, Batule, Seok, & Kim, 2018) are often incorporated into on-site assays because they can be run at lower, isothermal temperatures. Additionally, the multiple layers make it simple to create immunoassays with all-in-one approaches, creating a simpler assay. (Eltzov & Marks, 2017) 3D μPAD devices can enhance the end user experience through the integration and limiting of steps. Additionally, instead of colorimetric detection, there is also chronometric detection, which utilizes time as a marker of signal. Jangid et al. designed such an assay that utilizes a degradable biopolymer matrix (Jangid, et al., 2019). Enzymatic activation and amplification after sample injection allows for the biomatrix to be degraded and is concentration dependent – allowing for quantification. The assay is built in 3 layers/ zones and detection is contingent on the progression of the sample through the layers. The endpoint of this system is when the sample reaches a third layer/ zone (this is tracked via indicator dyes to see the progress of the sample). Concentration of the sample can be measured as the timing of the sample passing through the assay is able to be correlated to the concentration in the sample. This system could reach a limit of detection as low as 5 femtomoles. While this has not been applied to foodborne bacteria – this type of system could hold promising results in foodborne detection.
4.3.3. Origami Paper-Based Analytical Devices
Another popular form of paper-based assay design is the paper-origami which is a form of microfluidics where the paper devices take on a three-dimensional structure, similar to the concept of origami or paper folding. These assays are distinguishable from other three-dimensional (3D) assays because they are fabricated in a single layer and then assembled by folding. By being able to fabricate in a single layer there is a reduction in fabrication steps. This single layer is then folded in series to form a 3D structure. Additionally, by sequentially folding an assay different chemical reactions or processing steps can be performed by controlling the introduction of reagents to the sample (Govindarajan, Ramachandran, Vigil, Yager, & Böhringer, 2012). Different microfluidic origami devices have been implemented in the detection of foodborne pathogens such as E. coli (Sun, Chang, Zhang, & Liu, 2019; Trieu & Lee, 2019) and Salmonella spp (Trieu & Lee, 2019). Trieu et al. developed a 3D origami device to detect both E. coli O157 H:7 and Salmonella spp. The assay consists of a single paper layer embossed with microchannels and chambers to hold reagents for purification with chitosan (without any extraction method), amplification and colorimetric detection. These reactions are done separately through the sequential folding of the assay platform to introduce the sample to the lyophilized reaction reagents. This can be visualized in figure 2 B. Trieu et al. also compared their assay to tube-based detection for comparison as seen in figure 2A. Similarly, Sun et al. developed a origami device for the detection of E. coli K12 as a model utilizing isothermal amplification known as rolling circle amplification (RCA) (Sun, et al., 2019). Their device consisted of 4 panels, 3 of which are active. Panel B and A are folded together originally. The sample is then pipetted onto panel B on which lyophilized lysis reagents are stored to extract the RNA. This panel is then folded to together with panels C and D. Panel C is printed with a targeting moiety that cleaves the RNA. The sample travels through to panel D via capillary action where then an isothermal Rolling Circle Amplification (RCA) reaction is performed. An overview of this is seen in figure 2D. With this assay Sun et al. were able to detect as low as 103 CFU/mL of bacteria in 35 minutes. This data can be visualized in figure 2C. Their assay design allowed for the detection in complex sample matrices with the addition of the lysis reagents in the first panel and an original absorbent panel to filter and wash the sample before being passed through the other reagents. These origami devices are useful for being able to combine lysis, amplification, recognition, and detection in a single device and single step. These devices also have short run times of under an hour and perform with sensitivities similar to that of other portable assays.
Figure 2:
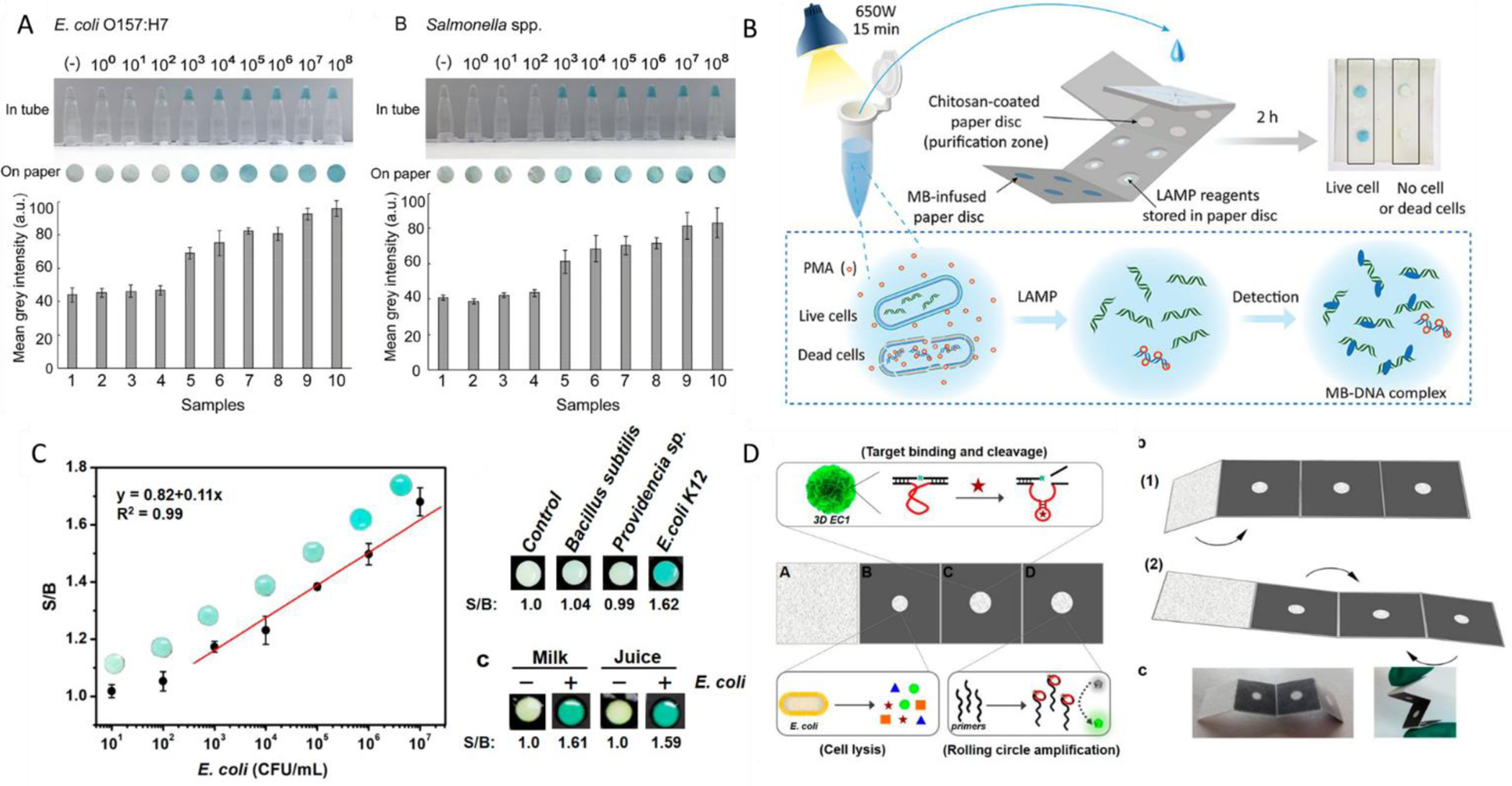
Taken from Trieu et al. A) Sensitivity of assay against both E. coli O157 H:7 and Salmonella spp. Authors investigated their detection limits in tube as well as on their paper platform in concentrations ranging from 108 – 100 CFU. B.) A comprehensive schematic overview of this paper-based assay from start to finish. Taken from Sun et al. C) Dose response curve, specificity, and spiked sampling data for their origami PAD assay utilizing several bacterial species and food matrices D) An overview schematic of the assay showing the individual reagent layers and the intended folding technique of the assay.
5.0. Developments in POC Biosensors/Biosensing Systems
Biosensors are analytical devices consisting minimally of a biological binder and a transducer element combined with a signal output reader. These binders can range from biological materials such as proteins, nucleic acids, whole cells to engineered biological moieties such as aptamers or biomimetics. Transducers can include optical, colorimetric, magnetic, electrochemical, and piezoelectric outputs (Turner, 2013). A signal output reader could be an analog or digital device. Biosensors are designed for selective target capture via a specific binder followed by the conversion of that binding event to a detectable output. Biosensor assays have the advantage of being simple and rapid, with portable designs that even include real-time sample data or multiplexed detection. Biosensors and their components in the context of pathogen detection have been reviewed before (Kaya, Cetin, Azimzadeh, & Topkaya, 2021; Kumar, Hu, Singh, & Mizaikoff, 2018) but for this review we aim to comprehensively highlight the different methods available and provide recent examples of advancements for on-site use biosensors.
5.1. Improvements in Biosensor Biorecognition Elements
Biorecognition is another critical step and limiting factor for bacterial detection. These biorecognition elements can include proteins, antibodies, and nucleic acid sequences and have been used in multiple biosensor designs. These standard recognition tools have been reviewed extensively in the literature (Morales & Halpern, 2018). This review will focus on the lesser covered recognition elements and cover the more recent innovations.
One of the biorecognition elements that is gaining traction in the detection of foodborne pathogens are lectins. Lectins have high specificity and can bind to multiple binding sites in many microorganisms. These elements are also relatively inexpensive and more stable than antibodies (Mi, et al., 2021). Additionally, these molecules are smaller in size than antibodies so in electrochemical biosensors they can more densely pack the electrode which in turn can provide a more sensitive detection. Alternatively, lectins can also serve as signal amplification tools. For example, Li et al. designed an electrochemical biosensor to rapidly detect E. coli O157 H:7. The sensor worked based on electrochemical impedance and novel screen-printed cross microelectrodes where the lectin wheat germ agglutinin was used as a signal amplification tool. This system was able to detect the bacteria with a limit of detection of 102 CFU/mL (Li, Fu, Fang, & Li, 2015).
Another useful recognition element is the molecularly imprinted polymer (MIP). These polymers bind to their target and are generated by polymerization in the presence of the target of interest. Upon the targets removal binding sites are conserved in the polymer. These have been used for the detection of bacteria and have been summarized in a recent review (Zhang, Wang, & Lu, 2021). This recognition element is often chosen for its high stability and sensitivity. One example of this technology was utilized by Lahcen et al. to detect the spores of B. cereus (Ait Lahcen, Arduini, Lista, & Amine, 2018). They utilized a conducting polymer that was fabricated in the presence of the bacterial spores for label-free detection. This electrochemical sensor was able to detect between 102 to 105 CFU/mL of bacterial spores.
Many sources have sought to improve these molecules to allow for increased binding, thereby improving the detection limit of the assay. Alternative efforts have also been made for stabilizing these recognition systems. This is crucial for the consideration of assays to be used on-site where conditions may not allow for proper long-term storage. There have been improvements for example in the stability of the microfluidic layers using bio-preservation to allow for long term storage of functionalized assays (Asghar, et al., 2016). Nanomaterials have also proven beneficial for stabilizing biorecognition elements. For example, while aptamers have proven to be valuable for their stability, engineered materials such as metal organic frameworks can help to further stabilize to increase the shelf life of assay components. An electrochemical biosensor for E. coli utilized amino-functionalized metal–organic frameworks (MOF) to achieve a limit of detection as low as 2 CFU/mL. An amine-modified aptamer against E. coli O157 H:7 containing adsorbed methylene blue is attached to the MOF. E. coli is introduced, releasing the methylene blue (which acts as a redox indicator), and the sensors measures the voltage of the MB peak charge before and after the E. coli introduction (Shahrokhian & Ranjbar, 2018).
Functionalizing these biorecognition elements has also proven to be beneficial in biosensor development. For example, Fulgione et.al developed a QCM method for the detection of S. typhimurium by functionalizing the QCM surface with anti-Salmonella antibodies that are UV-activated to improve sensitivity by limiting their conformations. This method allowed for the detection of S. typhimurium within 4 hours with a limit of detection of less than 1 CFU/mL in chicken samples. A pre-enrichment step is required which makes up almost half of the assay run time. While the enrichment step added time to their assay, it allowed for very sensitive limits of detection. Removing the enrichment step would be able to improve the assay run time but at a potential cost of sensitivity (Fulgione, et al., 2018). This assay and the corresponding data can be seen in figure 3.
Figure 3:
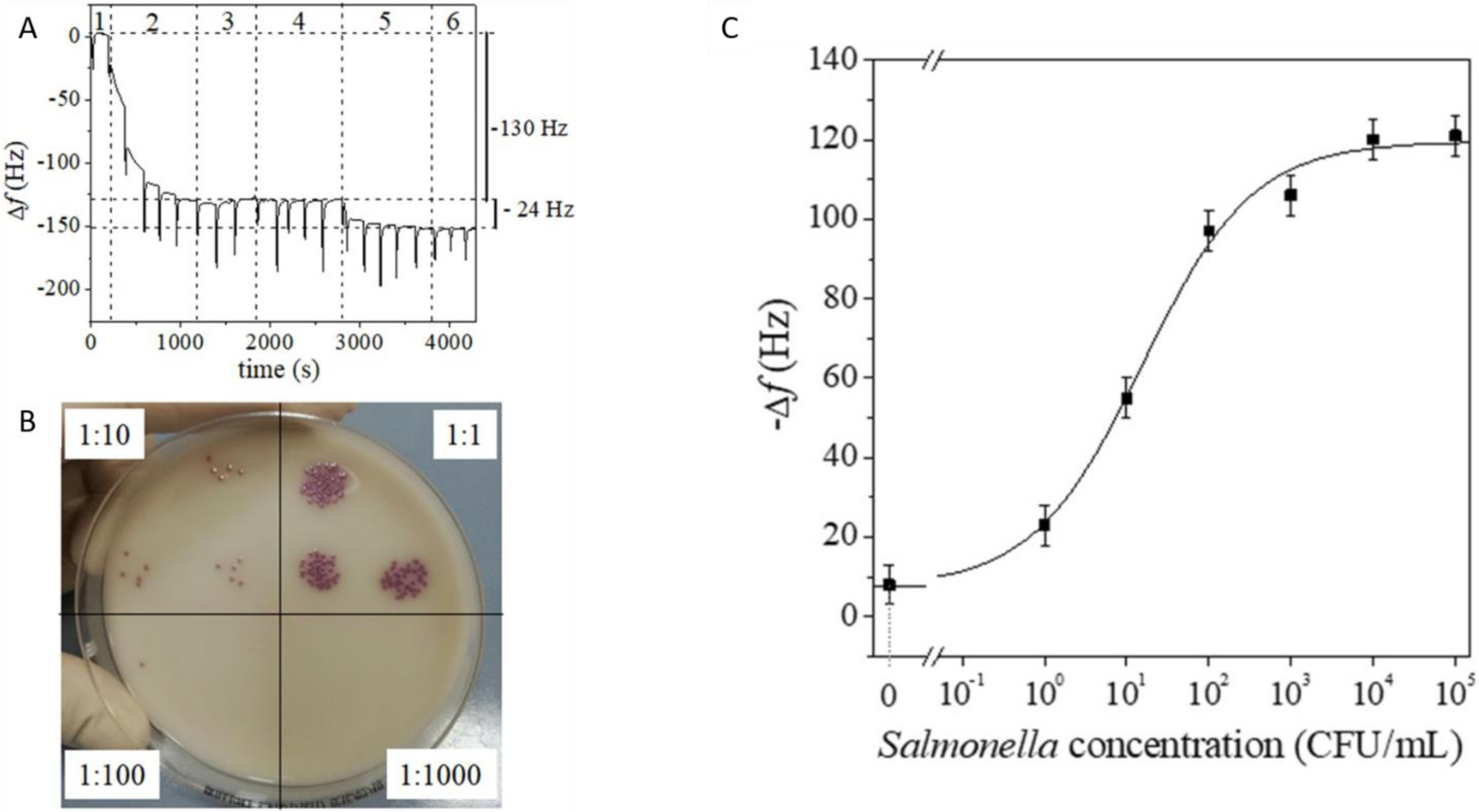
Taken from Fulgione et. al. A.) QCM Sensorgram after pre-enrichment. B.) Spot dilutions on Salmonella chromogenic agar base before pre-enrichment. Dilution factors are shown. C.) QCM Sensor dose-response curve referring to chicken meat contaminated with differing concentrations of S. typhimurium.
This functionalization can also be done with a variety of reporter moieties. These can include quantum dots, nanoparticles, and other excitable molecules. For example, a study shows the use of gold-coated magnetic disks as a method of isolation with fluorescent markers then being used for detection. This combination yields a detection limit of 102 CFU/100 mL and take about 45 minutes to perform without the need for an enrichment step (Castillo-Torres, et al., 2019). These gold-coated magnetic disks are functionalized with DNA aptamers specific to E. coli for target isolation. After capturing E. coli, the use of fluorescent labelling and viability staining with SYTO9 and propidium iodide (PI) allows for both detection and an assessment of bacterial viability. This gravity driven assay can be used for samples as large as 100 mL with the goal of rapid water sample analysis. While not currently developed for on-site, the use of cell-phone technology can drive this platform to be on-site. More importantly, the assay was able to include viability discrimination which is important when considering viable but not culturable bacteria being unrecognizable by traditional methods. Xue et al. combined magnetic separation with quantum dots for fluorescent detection also for E. coli O157 H:7. This method had a detection limit of 14 CFU/mL within two hours. The assay yielded a high recovery in spiked milk samples ranging from 95.92% to 108.15% (Xue, et al., 2018). Samples were able to be detected in 10 mL volumes and shows potential for larger sample volume. Additionally, the magnetic nanoparticles used for separation allowed for specific but efficient separation that made for a simplified sample preparation step. Quantum dots also have been used for multiplexed detection. Wang et al. used immunofluorescent quantum dot probes to detect E. coli O157 H:7, S. aureus, and V. parahaemolyticus with respective limits of detection in milk to be 6.66, 10.70, and 22.36 CFU/mL. This method could be performed in under 4 hours with high sensitivity and simplicity (Wang, et al., 2020). While not developed with on-site use in mind, this assay can be applicable for on-site technology with a portable fluorescent reader.
5.2. Improvements in Biosensor Transducers
Nanomaterial-based Sensors
With the increase in developments in nanoscience during recent years, attention has also shifted to nanomaterial-based sensors. Graphene has been used heavily in sensors due to its unique properties such as electroconductivity and quenching (Jiang, et al., 2020). Additionally, it is biocompatible. Functionalized nanomaterials have the ability to replace to increase signal, retain activity of biological molecules, and serve on their own in plasmonic and optical sensors (Yoon, Shin, Lee, & Choi, 2020). Some of the examples highlighted in this review utilize these nanomaterials to improve their sensors.
Gold nanoparticles in particular are commonly used as reporters in a variety of detection methods and are typically bound to aptamers or capture antibodies for recognition. For example, a study by Mou et al. demonstrated a click reaction using gold nanoparticles for the detection of E. coli. They found that pathogenic bacteria can reduce Cu2+ into Cu+. This Cu+ becomes a catalyst for a reaction between azide and alkyne functionalities appended to the surface of the gold nanoparticles. This reaction induced a visible color change from red to blue. The limit of detection was found to be 40 CFU/mL, and the assay could be visualized within 1 hour with assistance from a smartphone camera (Mou, et al., 2019). Another study was able to detect Salmonella spp., L.monocytogenes, and E.coli utilizing an aptamer Ap6 labelled to gold nanoparticles for colorimetric and UV absorbance detection in a single step with no preculturing or extraction methods. (Ledlod, Areekit, Santiwatanakul, & Chansiri, 2020). This aptamer binds to the surface of these bacteria. The authors reported high accuracy and specificities of 96% as well as an assay time of 10 minutes but with a sacrifice of sensitivity with a higher detection limit of 105 CFU/mL.
Alternatively, peroxidase-like activity of different kinds of nanostructures can be utilized for colorimetric detection. For example, Duan et al. used copper-metal organic framework nanoparticles as peroxidase mimics (Duan, Yang, Wu, Zou, & Wang, 2020).
These frameworks are porous crystalline materials that consist of metal ions and organic ligands. This nanoparticle consisting of copper-metal organic frameworks act as peroxidase mimetics and when modified with aptamer act as a signal probe. Aptamer coated on the microplate capture the bacteria that then interact with the aptamer on the nanoparticles, similar to a sandwich assay. These nanoparticles can cause the catalyzation of the oxidation of water and TMB resulting in a colorimetric detection visible with the naked eye. This is visualized in figure 4 A and the calibration curve with the bacteria can be seen in 4B. The combination of this colorimetric system with an aptasensor allowed for the detection limit of 2 CFU/mL and a limit of quantitation of 16 CFU/mL for E. coli. The assay can be integrated into a kit with all the needed materials and reagents when paired with a portable UV-reader.
Figure 4:
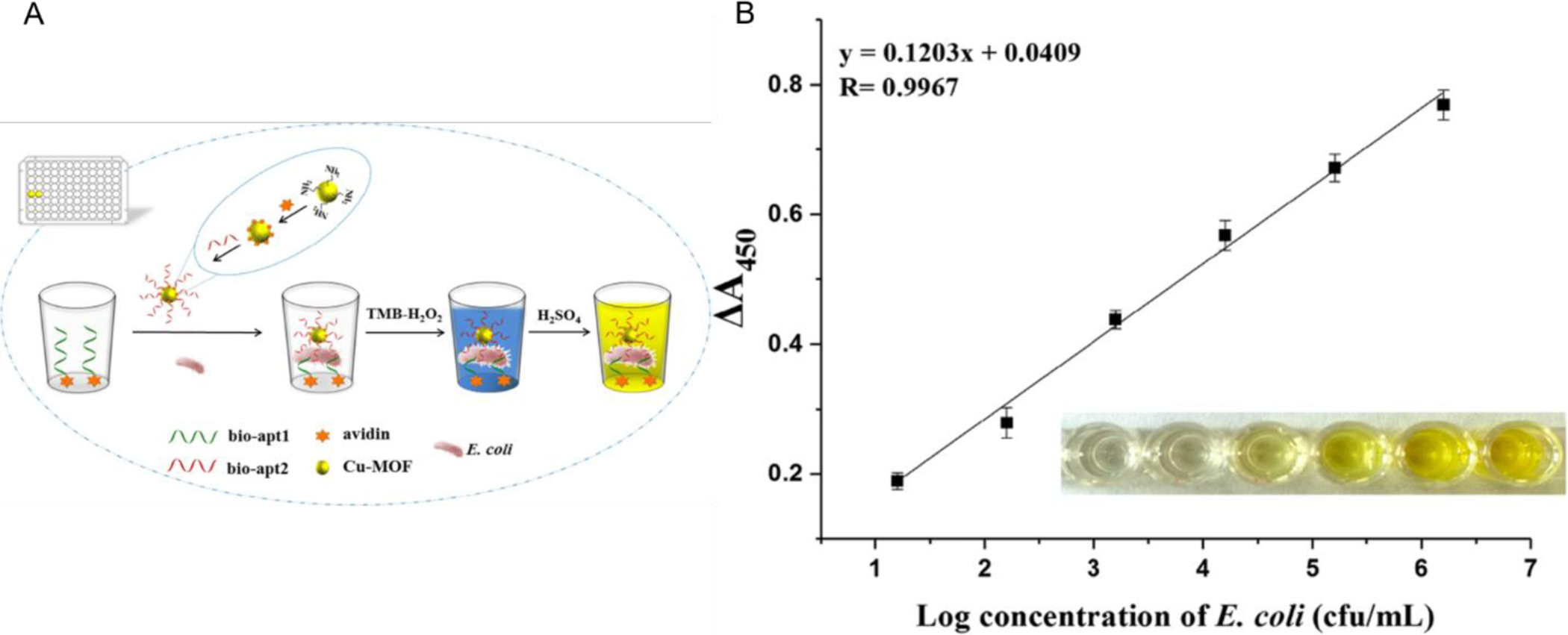
Taken from Duan et al. A.) Schematic of the designed colorimetric aptasensor utilizing copper metal organic frameworks in conjunction with aptamers and a colorimetric reaction. B. ) Linear calibration curve showing the absorbance response in relation to the concentration of E. coli O157 H:7. Inside the graph is a visual representation of the reaction that can be seen by the naked eye.
Magnetic Sensors
Magnetic sensors detect the interaction with the target by measuring the change in magnetic properties of the sensor. In recent years, rapid tests utilizing magnetic transducer elements have been able to detect samples in even shorter time intervals. (Achtsnicht, et al., 2019; Zeinhom, et al., 2018). Additionally, there is a method that instead of utilizing only beads or disks utilizes a magnetic grid separation column that can be used to detect Salmonella. This assay has a varying LOD of 11 CFU/mL - 104 CFU/mL and takes over 2.5 hours total to perform (Wang, et al., 2020). The separation column allows for continuous flow separation. As the bacteria pass through the column, the bacteria interact with and are bound by anti-Salmonella antibodies that are conjugated to magnetic particles that hold the antibodies to the magnetic column. Once the bacteria are bound, the PT@ZIF-8 nanocatalysts are injected and get conjugated to the bacterial cells. This serves as the reporting mechanism as these nanocatalysts mimic peroxidase activity to interact with TMB. The absorbance at 450 nm is then taken. This system with the separation column makes it simpler for larger volume (50 mL) analysis, which is often a major challenge. Furthermore, the capability of larger sample volume could be more beneficial for the generation of a proper representative sample.
Quartz Crystal Microbalance Sensors
One popular technique that utilizes piezoelectric transduction is referred to as quartz crystal microbalance (QCM), which measures the frequency change of quartz due to changes in mass cause by recognition molecules interacting with a target. QCM is useful in on-site diagnostics as it is rapid and able to be portable. Additionally, with the proper instructions QCM-based sensors do not need specialized personnel in order to obtain a measurement. Xiaofan Yu et. al developed a QCM method for the detection of E. coli. This study used the technique of whole-bacterium SELEX (systematic evolution of ligands by exponential enrichment) to select an aptasensor specific to E. coli. Briefly this technique uses in vitro selection to produce aptamers that bind to particular targets. The process begins with a random ssDNA library and is then incubated with the target bacteria that are immobilized and captured by magnetic beads. The unbound ssDNA and DNA bound to the magnetic capture beads are removed via negative selection and then ssDNA that binds to the bacteria is eluted. The sequences that bound the target are then amplified with PCR for following rounds of selection where this process is repeated but the stringency of elution conditions are increased to leave the highest binding aptamers left. Counter selection with non-target can also identify the aptamers that are not cross-reactive. This procedure is commonly used in the development of aptamer sequences. After 19 rounds of positive E. coli selection, other pathogenic bacteria were introduced for counter selection for 6 rounds. The QCM was then actualized by immobilization of streptavidin and biotinylated aptamer S1 to the surface. The sensor shows specificity and was able to provide results in under an hour with a LOD of 1.46 × 103 CFU/mL (Yu, Chen, Wang, & Li, 2018).
Electrochemical Sensors
Electrochemical biosensors utilize the interaction with the target to generate electrochemical signal in either resistance, impedance, current, or potential. Many materials can be utilized to fabricate electrochemical sensors. One cost-effective material in electrochemical electrodes, is carbon. Carbon electrodes were used in the detection of E. coli O157 H:7, where they were able to modify the electrode with gold nanoparticles (Vu, et al., 2020). Another popular option for screen printed electrodes are made of gold. These electrodes are cost-effective and are also easily manufactured. These were utilized by Wang et al. to detect E. coli O157 H:7 in conjunction with magnetic nanobeads for immunomagnetic separation. With this sensor they could detect as low as 1400 cells in 25 uL in under one hour (Wang, et al., 2015). Additionally, graphene screen-printed electrodes are cost-effective and sensitive choices for electrochemical detection. For example, Kampeera et al utilized graphene printed electrodes to detect as low as 0.3 CFU V. parahaemolyticus in seafood (Kampeera, et al., 2019). This sensor utilized LAMP for DNA amplification. This method is an isothermal amplification technic that is suitable for on-site detection due to its capability of being lyophilized. Using cost-effective materials in electrochemical sensors will potentially help increase the feasibility of on-site usage electrochemical sensors. Another interesting and novel electrode material is the use of thread. The electrodes are fabricated on nylon thread using conductive ink. These threads are pinned into position and then cotton thread was used to form microfluidic channels. This was used to detect V. parahaemolyticus in 30 minutes with a limit of detect as low as 5.74 CFU/mL (Jiang, Sun, Guo, & Weng, 2021). The usage of novel materials in fabrication can help drive down assay costs and create more sensitive assays.
Optical Methods
Optical methods exploit light interactions to produce a signal. A variety of optical biosensors are developed for pathogen detection and are discussed extensively in a recent review(Habimana, Ji, & Sun, 2018). Fluorescence and bioluminescence-based optical sensors have provided excellent sensitivity and are well developed. These sensors often are able to produce a signal that proportions to the concentration of the target. One example of an optical method utilized photonic hydrogels that become hydrolyzed in the presence of gelatinase from P. aeruginosa. This causes the gels to expand and causes a red-shift in the generated spectra. This method is advantageous as it is self-reporting and could be further investigated for use in on-site methods (Gao, et al., 2021).
6.0. Future Directions
Traditional methods for detecting foodborne and waterborne pathogens, despite their sensitivity are not particularly practical in the context of food and water contamination. These methods are too time consuming and bulky to be truly practical for frequent use in preventative and quality control testing. Newer methods for on-site use have begun to overcome this problem, with rapid and portable techniques being introduced for food and water analysis. While methods have significantly improved in recent years with respect to design and capability, some challenges and limitations still exist in regard to sensitivity, selectivity, and multiplexing capabilities. When considering on-site applications, care must be given to the criteria for point-of-care assays. While many assays have improved in certain aspects such as cost, time management, portability, or sensitivity, there is still need for more improvement. For example, while some assays have improved detection limits and sensitivities for particular pathogens, these detection limits are often still either above the infectious dose or the assay has become increasingly complex, which limits on-site use.
With paper diagnostics and other assays, there is potential to increase the sensitivity through non-colorimetric assay designs that include detection technologies such as fluorescence or electrochemistry. Oftentimes, these methods can lower detection limits as needed to be a viable option. However, despite their ability to achieve lower detection limits, these methods still are not particularly favorable due to the common requirements for expensive instrumentation and reagents, complex fabrication methods that raise the per-test cost, or the inclusion of complex steps to be carried out by the end user. Alternatively, the addition of amplification steps such as what is seen in nucleic acid amplification can greatly improve sensitivity without a major uptick in per-test cost or instrumentation. Some assays have been able to incorporate amplification steps in their assays without major additions to the assay user input. Further development of 3D PADs and biosensor chips have reduced the amount of user involvement. Furthermore, the availability and sophistication of cellphones and cellphone attachments have begun to improve the portability and costs of a variety of different assays and allow for more ways to analyze and measure assay data. Future development should focus on simplifying the user interface, decreasing the limits of detection, and further expansion of cell phone technologies for reduced assay costs without compromising sensitivity. For example, Trinh et al. developed a PAD based on LAMP amplification that integrated amplification and signal visualization via fluorescence from a probe infused into different layers of the paper device with only a single sample injection (Trinh & Lee, 2018). This technique can be visualized in figure 5. Samples could then be visualized with a UV lamp with a total run time of around a half hour with a detection limit of 1.7 × 102 CFU/mL for Salmonella spp. Furthermore, this assay was able to be multiplexed and identify target DNA of Salmonella spp., S. aureus, E. coli O157 H:7, and Cochlodinium polykrikoides. The combination of a sample to answer paper chip with fluorescence was able to generate a rapid and simple device that was also sensitive, passing most of the ASSURED criteria for point-of-care assays.
Figure 5:
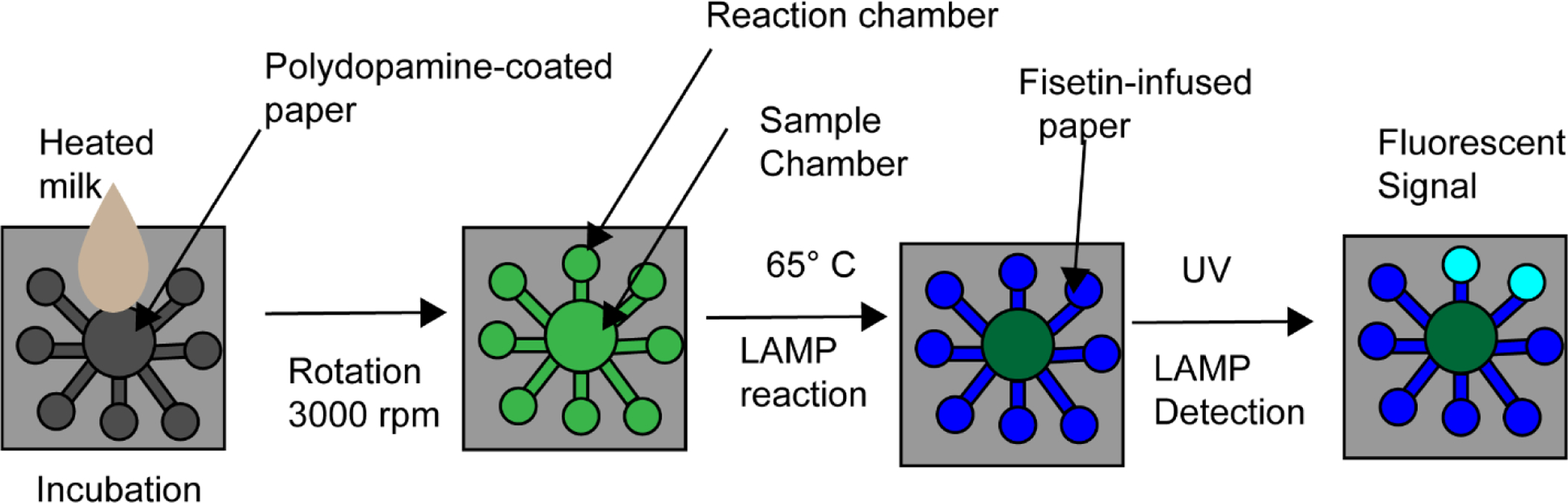
Adapted from Trinh et al. A schematic overview of their sample to answer PAD device. Briefly heated milk is injected into the center of the device and following rotation to spread the liquid across the device LAMP is performed within the device. From there the device is exposed to UV and fluorescent signal is determined visually.
Paper based assays are well suited for addressing cost issues while still maintaining sensitivity. Newer biosensor designs are also becoming increasingly sensitive without significant cost increases. Combining these technologies may provide enhanced sensitivity for paper-based assays without a significant cost penalty and might provide biosensors with an exceptionally user-friendly interface. With increasing cell-phone integration and design improvements in portable instrumentation, on-site assays will continue to improve in both field performance and implementation.
One of the bigger challenges that researchers face with the analysis of food matrices is sampling and sample preparation. All assays, both traditional and rapid, usually require some form of sample processing or sample enrichment. For foodborne contamination, sample preparation is often a limiting factor in the development for rapid methods. Food samples can vary widely in composition and, as such, make universal sample processing more challenging and inviting new research opportunities toward tackling matrix complexity. Oftentimes samples need to be large – however many technologies use small volumes to perform analysis. Sampling needs to be able to be performed enough times to be able to confidently state that the food is absent of any pathogens. Alternatively, improvements have been made in sample concentration and volume reduction. However there still is room for improvement, especially when on-site use is considered. Improvements have been made in sample preparation, even some advancements forgoing the need for major preparation or extraction. As the field of food analysis progresses, the advances in sample preparation will allow for more sensitive and enhanced detection of pathogens. Further work still needs to be done in this area, much like multiplexing. Sampling needs to be able to be performed enough times to be able to confidently state that the food is absent of any pathogens.
7.0. Conclusion
This article discusses the current state and recent innovations of available technologies for on-site detection of food and waterborne bacteria. To circumvent the laborious process of conventional pathogen detection assays, rapid methods have been developed that are able to be applied on-site. These methods are valuable as preventative measures to limit the spread of water- and food-borne disease outbreaks, especially in remote or low-resource areas. Further, these methods are valued for their efficiency, rapidness, and ease of use in comparison to conventional methods such as culturing. Despite current improvements and innovations in these assays, there is still untapped potential for further integration of technology – especially in relation to multiplexing and the cost per test. Cost per test is especially important if multiple tests are needed to survey a panel of common pathogens. Additionally, improvements in sensitivity/specificity as well as overall simplicity in design or methodology should also be considered when looking at the cost. Furthermore, to consider a sample to be truly absent of pathogen it is likely that many tests would need to be conducted to analyze several different samples. On-site technologies will continue to impact multiple areas of public health such as agriculture, water safety, and food processing. Having quick and simplistic methods to identify contamination will improve the response to bacterial outbreaks and could provide a means for reducing the overall number of outbreaks. This reduction not only reduces the healthcare burden and physical toll of an outbreak but also the economic impact by quickly identifying an outbreak before it can become widespread. On-site technologies carry the potential to mitigate a major global health threat while tangentially reducing the prevalence of antibiotic-resistant organisms. Through continuing improvements and novel combinations of these diverse technologies, on-site detection could easily become a normal implementation of food and water quality control in many industries.
Highlights.
Food and water contamination has increased in recent years, causing both health and economic effects globally.
On-site technologies for rapid detection are beneficial for rapid detection and mitigation of illness occurrence
Many technologies and innovations have been created or can be adapted for on-site application.
Review of the recent advances in on-site technologies for the detection of foodborne and waterborne pathogens
Acknowledgements
The authors would like to thank NIGMS (R01GM114321 and R01GM127706), the National Science Foundation (CBET-1841419), and the Sylvester Comprehensive Cancer Center for funding support. S.D. thanks the Miller School of Medicine of the University of Miami for the Lucille P. Markey Chair in Biochemistry and Molecular Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interests
The authors report no conflict of interests.
References
- Acheson D, & Allos BM (2001). Campylobacter jejuni Infections: Update on Emerging Issues and Trends. Clinical Infectious Diseases, 32, 1201–1206. [DOI] [PubMed] [Google Scholar]
- Achtsnicht S, Neuendorf C, Faßbender T, Nölke G, Offenhäusser A, Krause H-J, & Schröper F (2019). Sensitive and rapid detection of cholera toxin subunit B using magnetic frequency mixing detection. PLoS One, 14, e0219356–e0219356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins JA, Boehle K, Friend C, Chamberlain B, Bisha B, & Henry CS (2017). Colorimetric and Electrochemical Bacteria Detection Using Printed Paper- and Transparency-Based Analytic Devices. Analytical Chemistry, 89, 3613–3621. [DOI] [PubMed] [Google Scholar]
- Ahn H, Batule BS, Seok Y, & Kim MG (2018). Single-Step Recombinase Polymerase Amplification Assay Based on a Paper Chip for Simultaneous Detection of Multiple Foodborne Pathogens. Anal Chem, 90, 10211–10216. [DOI] [PubMed] [Google Scholar]
- Ait Lahcen A, Arduini F, Lista F, & Amine A (2018). Label-free electrochemical sensor based on spore-imprinted polymer for Bacillus cereus spore detection. Sensors and Actuators B: Chemical, 276, 114–120. [Google Scholar]
- Armstrong CM, Gehring AG, Paoli GC, Chen C-Y, He Y, & Capobianco JA (2019). Impacts of Clarification Techniques on Sample Constituents and Pathogen Retention. Foods (Basel, Switzerland), 8, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghar W, Yuksekkaya M, Shafiee H, Zhang M, Ozen MO, Inci F, Kocakulak M, & Demirci U (2016). Engineering long shelf life multi-layer biologically active surfaces on microfluidic devices for point of care applications. Scientific Reports, 6, 21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, & Martinez-Urtaza J (2018). Vibrio spp. infections. Nature Reviews Disease Primers, 4, 8. [DOI] [PubMed] [Google Scholar]
- Bell RL, Jarvis KG, Ottesen AR, McFarland MA, & Brown EW (2016). Recent and emerging innovations in Salmonella detection: a food and environmental perspective. Microbial Biotechnology, 9, 279–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintsis T (2017). Foodborne pathogens. AIMS Microbiol, 3, 529–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JD, Hsieh HV, Gasperino DJ, & Weigl BH (2019). Sensitivity enhancement in lateral flow assays: a systems perspective. Lab on a Chip, 19, 2486–2499. [DOI] [PubMed] [Google Scholar]
- Briquaire R, Colwell RR, Boncy J, Rossignol E, Dardy A, Pandini I, Villeval F, Machuron JL, Huq A, Rashed S, Vandevelde T, & Rozand C (2017). Application of a paper based device containing a new culture medium to detect Vibrio cholerae in water samples collected in Haiti. J Microbiol Methods, 133, 23–31. [DOI] [PubMed] [Google Scholar]
- Bu T, Huang Q, Yan L, Zhang W, Dou L, Huang L, Yang Q, Zhao B, Yang B, Li T, Wang J, & Zhang D (2019). Applicability of biological dye tracer in strip biosensor for ultrasensitive detection of pathogenic bacteria. Food Chem, 274, 816–821. [DOI] [PubMed] [Google Scholar]
- Buchanan RL, Gorris LGM, Hayman MM, Jackson TC, & Whiting RC (2017). A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control, 75, 1–13. [Google Scholar]
- Butler D, Goel N, Goodnight L, Tadigadapa S, & Ebrahimi A (2019). Detection of bacterial metabolism in lag-phase using impedance spectroscopy of agar-integrated 3D microelectrodes. Biosensors and Bioelectronics, 129, 269–276. [DOI] [PubMed] [Google Scholar]
- Cam D, & Oktem HA (2019). Development of rapid dipstick assay for food pathogens, Salmonella, by optimized parameters. J Food Sci Technol, 56, 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Torres KY, Arnold DP, & McLamore ES (2019). Rapid isolation of Escherichia coli from water samples using magnetic microdiscs. Sensors and Actuators B: Chemical, 291, 58–66. [Google Scholar]
- CDC. (2018). Burden of Foodborne Illness: Findings.
- Cho S, Hiott LM, Woodley TA, Frye JG, & Jackson CR (2020). Evaluation of a new chromogenic agar for the detection of environmental Enterococcus. Journal of Microbiological Methods, 178, 106082. [DOI] [PubMed] [Google Scholar]
- Contreras-Naranjo JC, Wei Q, & Ozcan A (2016). Mobile Phone-Based Microscopy, Sensing, and Diagnostics. IEEE Journal of Selected Topics in Quantum Electronics, 22, 1–14. [Google Scholar]
- de Boer E, & Beumer RR (1999). Methodology for detection and typing of foodborne microorganisms. International Journal of Food Microbiology, 50, 119–130. [DOI] [PubMed] [Google Scholar]
- Duan N, Yang W, Wu S, Zou Y, & Wang Z (2020). A Visual and Sensitive Detection of Escherichia coli Based on Aptamer and Peroxidase-like Mimics of Copper-Metal Organic Framework Nanoparticles. Food Analytical Methods, 13, 1433–1441. [Google Scholar]
- Eltzov E, & Marks RS (2017). Colorimetric stack pad immunoassay for bacterial identification. Biosensors and Bioelectronics, 87, 572–578. [DOI] [PubMed] [Google Scholar]
- FDA. (2020). Bacteriological Analytical Manual (BAM).
- Fulgione A, Cimafonte M, Della Ventura B, Iannaccone M, Ambrosino C, Capuano F, Proroga YTR, Velotta R, & Capparelli R (2018). QCM-based immunosensor for rapid detection of Salmonella Typhimurium in food. Sci Rep, 8, 16137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Chen Y, Li M, Jia L, Zhang L, & Zhu J (2021). Gelatin-based photonic hydrogels for visual detection of pathogenic Pseudomonas aeruginosa. Sensors and Actuators B: Chemical, 329, 129137. [Google Scholar]
- Gasanov U, Hughes D, & Hansbro PM (2005). Methods for the isolation and identification of Listeria spp. and Listeria monocytogenes: a review. FEMS Microbiology Reviews, 29, 851–875. [DOI] [PubMed] [Google Scholar]
- Gong X, Cai J, Zhang B, Zhao Q, Piao J, Peng W, Gao W, Zhou D, Zhao M, & Chang J (2017). A review of fluorescent signal-based lateral flow immunochromatographic strips. J Mater Chem B, 5, 5079–5091. [DOI] [PubMed] [Google Scholar]
- Govindarajan AV, Ramachandran S, Vigil GD, Yager P, & Böhringer KF (2012). A low cost point-of-care viscous sample preparation device for molecular diagnosis in the developing world; an example of microfluidic origami. Lab on a Chip, 12, 174–181. [DOI] [PubMed] [Google Scholar]
- Granum PE, & Lund T (1997). Bacillus cereus and its food poisoning toxins. FEMS Microbiology Letters, 157, 223–228. [DOI] [PubMed] [Google Scholar]
- Habimana J. d. D., Ji J, & Sun X (2018). Minireview: Trends in Optical-Based Biosensors for Point-Of-Care Bacterial Pathogen Detection for Food Safety and Clinical Diagnostics. Analytical Letters, 51, 2933–2966. [Google Scholar]
- Han J, Zhang L, Hu L, Xing K, Lu X, Huang Y, Zhang J, Lai W, & Chen T (2018). Nanozyme-based lateral flow assay for the sensitive detection of Escherichia coli O157:H7 in milk. J Dairy Sci, 101, 5770–5779. [DOI] [PubMed] [Google Scholar]
- He X, Liu Z, Yang Y, Li L, Wang L, Li A, Qu Z, & Xu F (2019). Sensitivity Enhancement of Nucleic Acid Lateral Flow Assays through a Physical–Chemical Coupling Method: Dissoluble Saline Barriers. ACS Sensors, 4, 1691–1700. [DOI] [PubMed] [Google Scholar]
- Huang M, Zhou X, Wang H, & Xing D (2018). Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Analytical Chemistry, 90, 2193–2200. [DOI] [PubMed] [Google Scholar]
- Jangid AR, Strong EB, Escamilla E, Lore BA, Tod NJ, Thiel R, Martinez AW, & Martinez NW (2019). Chronometric Quantitation of Analytes in Paper-Based Microfluidic Devices (MicroPADs) via Enzymatic Degradation of a Metastable Biomatrix. Inventions, 4. [Google Scholar]
- Jiang H, Sun Z, Guo Q, & Weng X (2021). Microfluidic thread-based electrochemical aptasensor for rapid detection of Vibrio parahaemolyticus. Biosensors and Bioelectronics, 182, 113191. [DOI] [PubMed] [Google Scholar]
- Jiang J, Wang X, Chao R, Ren Y, Hu C, Xu Z, & Liu GL (2014). Smartphone based portable bacteria pre-concentrating microfluidic sensor and impedance sensing system. Sensors and Actuators B: Chemical, 193, 653–659. [Google Scholar]
- Jiang Z, Feng B, Xu J, Qing T, Zhang P, & Qing Z (2020). Graphene biosensors for bacterial and viral pathogens. Biosensors and Bioelectronics, 166, 112471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadariya J, Smith TC, & Thapaliya D (2014). Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. BioMed research international, 2014, 827965–827965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampeera J, Pasakon P, Karuwan C, Arunrut N, Sappat A, Sirithammajak S, Dechokiattawan N, Sumranwanich T, Chaivisuthangkura P, Ounjai P, Chankhamhaengdecha S, Wisitsoraat A, Tuantranont A, & Kiatpathomchai W (2019). Point-of-care rapid detection of Vibrio parahaemolyticus in seafood using loop-mediated isothermal amplification and graphene-based screen-printed electrochemical sensor. Biosensors and Bioelectronics, 132, 271–278. [DOI] [PubMed] [Google Scholar]
- Kant K, Shahbazi MA, Dave VP, Ngo TA, Chidambara VA, Than LQ, Bang DD, & Wolff A (2018). Microfluidic devices for sample preparation and rapid detection of foodborne pathogens. Biotechnol Adv, 36, 1003–1024. [DOI] [PubMed] [Google Scholar]
- Kaya HO, Cetin AE, Azimzadeh M, & Topkaya SN (2021). Pathogen detection with electrochemical biosensors: Advantages, challenges and future perspectives. Journal of Electroanalytical Chemistry, 882, 114989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kwon C, Lee BS, & Noh H (2019). One-step sensing of foodborne pathogenic bacteria using a 3D paper-based device. Analyst, 144, 2248–2255. [DOI] [PubMed] [Google Scholar]
- Kim J-H, & Oh S-W (2019). Development of a filtration-based LAMP–LFA method as sensitive and rapid detection of E. coli O157:H7. Journal of Food Science and Technology, 56, 2576–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Lee GG, Park JS, Jung YH, Kwak HS, Kim SB, Nam YS, & Kwon ST (2007). A novel multiplex PCR assay for rapid and simultaneous detection of five pathogenic bacteria: Escherichia coli O157:H7, Salmonella, Staphylococcus aureus, Listeria monocytogenes, and Vibrio parahaemolyticus. J Food Prot, 70, 1656–1662. [DOI] [PubMed] [Google Scholar]
- Kim SR, Yoon Y, Kim WI, Park KH, Yun HJ, Chung DH, Yun JC, & Ryu KY (2012). Comparison of sample preparation methods for the recovery of foodborne pathogens from fresh produce. J Food Prot, 75, 1213–1218. [DOI] [PubMed] [Google Scholar]
- Kumar N, Hu Y, Singh S, & Mizaikoff B (2018). Emerging biosensor platforms for the assessment of water-borne pathogens. Analyst, 143, 359–373. [DOI] [PubMed] [Google Scholar]
- Land KJ, Boeras DI, Chen X-S, Ramsay AR, & Peeling RW (2019). REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nature microbiology, 4, 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JW-F, Ab Mutalib N-S, Chan K-G, & Lee L-H (2015). Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Frontiers in Microbiology, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledlod S, Areekit S, Santiwatanakul S, & Chansiri K (2020). Colorimetric aptasensor for detecting Salmonella spp., Listeria monocytogenes, and Escherichia coli in meat samples. Food Science and Technology International, 1082013219899593. [DOI] [PubMed] [Google Scholar]
- Lee J-M, Azizah RN, & Kim K-S (2020). Comparative evaluation of three agar media-based methods for presumptive identification of seafood-originated Vibrio parahaemolyticus strains. Food Control, 116, 107308. [Google Scholar]
- Li Z, Fu Y, Fang W, & Li Y (2015). Electrochemical Impedance Immunosensor Based on Self-Assembled Monolayers for Rapid Detection of Escherichia coli O157:H7 with Signal Amplification Using Lectin. Sensors, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P-S, Park TS, & Yoon J-Y (2014). Rapid and reagentless detection of microbial contamination within meat utilizing a smartphone-based biosensor. Scientific Reports, 4, 5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yang D, & Liu G (2019). Signal amplification strategies for paper-based analytical devices. Biosensors and Bioelectronics, 136, 60–75. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T, de la Guardia M, & Baradaran B (2020). Lateral flow assays towards point-of-care cancer detection: A review of current progress and future trends. TrAC Trends in Analytical Chemistry, 125, 115842. [Google Scholar]
- Mi F, Guan M, Hu C, Peng F, Sun S, & Wang X (2021). Application of lectin-based biosensor technology in the detection of foodborne pathogenic bacteria: a review. Analyst, 146, 429–443. [DOI] [PubMed] [Google Scholar]
- Minor T, Lasher A, Klontz K, Brown B, Nardinelli C, & Zorn D (2015). The Per Case and Total Annual Costs of Foodborne Illness in the United States. Risk Anal, 35, 1125–1139. [DOI] [PubMed] [Google Scholar]
- Morales MA, & Halpern JM (2018). Guide to Selecting a Biorecognition Element for Biosensors. Bioconjugate Chemistry, 29, 3231–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou X-Z, Chen X-Y, Wang J, Zhang Z, Yang Y, Shou Z-X, Tu Y-X, Du X, Wu C, Zhao Y, Qiu L, Jiang P, Chen C, Huang D-S, & Li Y-Q (2019). Bacteria-Instructed Click Chemistry between Functionalized Gold Nanoparticles for Point-of-Care Microbial Detection. ACS Applied Materials & Interfaces, 11, 23093–23101. [DOI] [PubMed] [Google Scholar]
- Nelson EJ, Harris JB, Morris JG Jr., Calderwood SB, & Camilli A (2009). Cholera transmission: the host, pathogen and bacteriophage dynamic. Nature reviews. Microbiology, 7, 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HV, Nguyen VD, Liu F, & Seo TS (2020). An Integrated Smartphone-Based Genetic Analyzer for Qualitative and Quantitative Pathogen Detection. ACS Omega, 5, 22208–22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V-T, Song S, Park S, & Joo C (2020). Recent advances in high-sensitivity detection methods for paper-based lateral-flow assay. Biosensors and Bioelectronics, 152, 112015. [DOI] [PubMed] [Google Scholar]
- Pang B, Zhao C, Li L, Song X, Xu K, Wang J, Liu Y, Fu K, Bao H, Song D, Meng X, Qu X, Zhang Z, & Li J (2018). Development of a low-cost paper-based ELISA method for rapid Escherichia coli O157:H7 detection. Anal Biochem, 542, 58–62. [DOI] [PubMed] [Google Scholar]
- Peng H, & Chen IA (2019). Rapid Colorimetric Detection of Bacterial Species through the Capture of Gold Nanoparticles by Chimeric Phages. ACS Nano, 13, 1244–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahal E, Kazzi N, Nassar F, & Matar G (2012). Escherichia coli O157:H7—Clinical aspects and novel treatment approaches. Frontiers in Cellular and Infection Microbiology, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Hammerl JA, Appel B, Dieckmann R, & Al Dahouk S (2015). Sampling and Homogenization Strategies Significantly Influence the Detection of Foodborne Pathogens in Meat. BioMed research international, 2015, 145437–145437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, & Griffin PM (2011). Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis, 17, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharinger EJ, Dietrich R, Wittwer T, Märtlbauer E, & Schauer K (2017). Multiplexed Lateral Flow Test for Detection and Differentiation of Cronobacter sakazakii Serotypes O1 and O2. Frontiers in Microbiology, 8, 1826–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirone M, Visciano P, Tofalo R, & Suzzi G (2019). Editorial: Foodborne Pathogens: Hygiene and Safety. Front Microbiol, 10, 1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrokhian S, & Ranjbar S (2018). Aptamer immobilization on amino-functionalized metal–organic frameworks: an ultrasensitive platform for the electrochemical diagnostic of Escherichia coli O157:H7. Analyst, 143, 3191–3201. [DOI] [PubMed] [Google Scholar]
- Shin JH, Hong J, Go H, Park J, Kong M, Ryu S, Kim K-P, Roh E, & Park J-K (2018). Multiplexed Detection of Foodborne Pathogens from Contaminated Lettuces Using a Handheld Multistep Lateral Flow Assay Device. Journal of Agricultural and Food Chemistry, 66, 290–297. [DOI] [PubMed] [Google Scholar]
- Shirshahi V, Tabatabaei SN, Hatamie S, & Saber R (2019). Functionalized reduced graphene oxide as a lateral flow immuneassay label for one-step detection of Escherichia coli O157:H7. J Pharm Biomed Anal, 164, 104–111. [DOI] [PubMed] [Google Scholar]
- Shrivastava S, Lee W-I, & Lee N-E (2018). Culture-free, highly sensitive, quantitative detection of bacteria from minimally processed samples using fluorescence imaging by smartphone. Biosensors and Bioelectronics, 109, 90–97. [DOI] [PubMed] [Google Scholar]
- Srisa-Art M, Boehle KE, Geiss BJ, & Henry CS (2018). Highly Sensitive Detection of Salmonella typhimurium Using a Colorimetric Paper-Based Analytical Device Coupled with Immunomagnetic Separation. Anal Chem, 90, 1035–1043. [DOI] [PubMed] [Google Scholar]
- Suaifan G, Alhogail S, & Zourob M (2017). Paper-based magnetic nanoparticle-peptide probe for rapid and quantitative colorimetric detection of Escherichia coli O157:H7. Biosens Bioelectron, 92, 702–708. [DOI] [PubMed] [Google Scholar]
- Sun J, Huang J, Warden AR, & Ding X (2020). Real-time detection of foodborne bacterial viability using a colorimetric bienzyme system in food and drinking water. Food Chemistry, 320, 126581. [DOI] [PubMed] [Google Scholar]
- Sun Y, Chang Y, Zhang Q, & Liu M (2019). An Origami Paper-Based Device Printed with DNAzyme-Containing DNA Superstructures for Escherichia coli Detection. Micromachines (Basel), 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasbasi BB, Guner BC, Sudagidan M, Ucak S, Kavruk M, & Ozalp VC (2019). Label-free lateral flow assay for Listeria monocytogenes by aptamer-gated release of signal molecules. Anal Biochem, 587, 113449. [DOI] [PubMed] [Google Scholar]
- Thirunavukkarasu N, Johnson E, Pillai S, Hodge D, Stanker L, Wentz T, Singh B, Venkateswaran K, McNutt P, Adler M, Brown E, Hammack T, Burr D, & Sharma S (2018). Botulinum Neurotoxin Detection Methods for Public Health Response and Surveillance. Frontiers in bioengineering and biotechnology, 6, 80–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu PT, & Lee NY (2019). Paper-Based All-in-One Origami Microdevice for Nucleic Acid Amplification Testing for Rapid Colorimetric Identification of Live Cells for Point-of-Care Testing. Anal Chem, 91, 11013–11022. [DOI] [PubMed] [Google Scholar]
- Trinh TND, & Lee NY (2018). A rapid and eco-friendly isothermal amplification microdevice for multiplex detection of foodborne pathogens. Lab Chip, 18, 2369–2377. [DOI] [PubMed] [Google Scholar]
- Turner AP (2013). Biosensors: sense and sensibility. Chem Soc Rev, 42, 3184–3196. [DOI] [PubMed] [Google Scholar]
- Vu QK, Tran QH, Vu NP, Anh T-L, Dang TTL, Matteo T, & Nguyen THH (2020). A label-free electrochemical biosensor based on screen-printed electrodes modified with gold nanoparticles for quick detection of bacterial pathogens. Materials Today Communications, 101726. [Google Scholar]
- Wang C, Gao X, Wang S, & Liu Y (2020). A smartphone-integrated paper sensing system for fluorescent and colorimetric dual-channel detection of foodborne pathogenic bacteria. Analytical and Bioanalytical Chemistry, 412, 611–620. [DOI] [PubMed] [Google Scholar]
- Wang D, Lian F, Yao S, Liu Y, Wang J, Song X, Ge L, Wang Y, Zhao Y, Zhang J, Zhao C, & Xu K (2020). Simultaneous Detection of Three Foodborne Pathogens Based on Immunomagnetic Nanoparticles and Fluorescent Quantum Dots. ACS Omega, 5, 23070–23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Huo X, Zheng L, Cai G, Wang Y, Liu N, Wang M, & Lin J (2020). An ultrasensitive biosensor for colorimetric detection of Salmonella in large-volume sample using magnetic grid separation and platinum loaded zeolitic imidazolate Framework-8 nanocatalysts. Biosensors and Bioelectronics, 150, 111862. [DOI] [PubMed] [Google Scholar]
- Wang R, Lum J, Callaway Z, Lin J, Bottje W, & Li Y (2015). A Label-Free Impedance Immunosensor Using Screen-Printed Interdigitated Electrodes and Magnetic Nanobeads for the Detection of E. coli O157:H7. Biosensors, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zheng L, Cai G, Liu N, Liao M, Li Y, Zhang X, & Lin J (2019). A microfluidic biosensor for online and sensitive detection of Salmonella typhimurium using fluorescence labeling and smartphone video processing. Biosensors and Bioelectronics, 140, 111333. [DOI] [PubMed] [Google Scholar]
- Wang X, Shang X, & Huang X (2020). Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods. Emerging microbes & infections, 9, 1682–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhao P, Yang X, Li J, Zhang J, Zhang X, Zeng Z, Dong J, Gao S, & Lu C (2020). A Recombinase Polymerase Amplification and Lateral Flow Strip Combined Method That Detects Salmonella enterica Serotype Typhimurium With No Worry of Primer-Dependent Artifacts. Front Microbiol, 11, 1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn D, Raut N, Joel S, Pasini P, Deo SK, & Daunert S (2018). Detection of bacterial contamination in food matrices by integration of quorum sensing in a paper-strip test. Analyst, 143, 4774–4782. [DOI] [PubMed] [Google Scholar]
- Xue L, Zheng L, Zhang H, Jin X, & Lin J (2018). An ultrasensitive fluorescent biosensor using high gradient magnetic separation and quantum dots for fast detection of foodborne pathogenic bacteria. Sensors and Actuators B: Chemical, 265, 318–325. [Google Scholar]
- Yoon J, Shin M, Lee T, & Choi J-W (2020). Highly Sensitive Biosensors Based on Biomolecules and Functional Nanomaterials Depending on the Types of Nanomaterials: A Perspective Review. Materials, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi H, Ali MM, Su H-M, Filipe CDM, & Didar TF (2018). Sentinel Wraps: Real-Time Monitoring of Food Contamination by Printing DNAzyme Probes on Food Packaging. ACS Nano, 12, 3287–3294. [DOI] [PubMed] [Google Scholar]
- Yu X, Chen F, Wang R, & Li Y (2018). Whole-bacterium SELEX of DNA aptamers for rapid detection of E.coli O157:H7 using a QCM sensor. Journal of Biotechnology, 266, 39–49. [DOI] [PubMed] [Google Scholar]
- Zeinhom MMA, Wang Y, Sheng L, Du D, Li L, Zhu M-J, & Lin Y (2018). Smart phone based immunosensor coupled with nanoflower signal amplification for rapid detection of Salmonella Enteritidis in milk, cheese and water. Sensors and Actuators B: Chemical, 261, 75–82. [Google Scholar]
- Zhang D, Broyles D, Hunt EA, Dikici E, Daunert S, & Deo SK (2017). A paper-based platform for detection of viral RNA. Analyst, 142, 815–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang Y, & Lu X (2021). Molecular imprinting technology for sensing foodborne pathogenic bacteria. Analytical and Bioanalytical Chemistry. [DOI] [PubMed] [Google Scholar]
- Zhao F, Lee EY, Noh GS, Shin J, Liu H, Qiao Z, & Shin Y (2019). A robust, hand-powered, instrument-free sample preparation system for point-of-care pathogen detection. Scientific Reports, 9, 16374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Lin CW, Wang J, & Oh DH (2014). Advances in rapid detection methods for foodborne pathogens. J Microbiol Biotechnol, 24, 297–312. [DOI] [PubMed] [Google Scholar]


