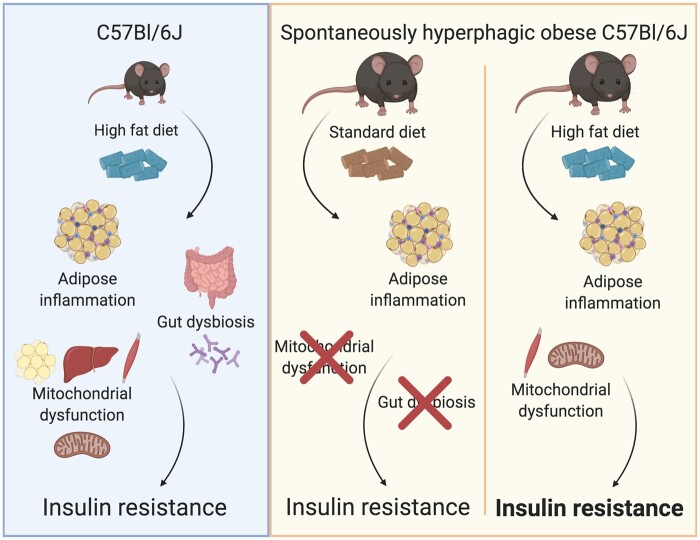
Abstract
Obesity is associated with adipose tissue hypertrophy, systemic inflammation, mitochondrial dysfunction, and intestinal dysbiosis. Rodent models of high-fat diet (HFD)-feeding or genetic deletion of multifunctional proteins involved in immunity and metabolism are often used to probe the etiology of obesity; however, these models make it difficult to divorce the effects of obesity, diet composition, or immunity on endocrine regulation of blood glucose. We, therefore, investigated the importance of adipose inflammation, mitochondrial dysfunction, and gut dysbiosis for obesity-induced insulin resistance using a spontaneously obese mouse model. We examined metabolic changes in skeletal muscle, adipose tissue, liver, the intestinal microbiome, and whole-body glucose control in spontaneously hyperphagic C57Bl/6J mice compared to lean littermates. A separate subset of lean and obese mice was subject to 8 weeks of obesogenic HFD feeding, or to pair feeding of a standard rodent diet. Hyperphagia, obesity, adipose inflammation, and insulin resistance were present in obese mice despite consuming a standard rodent diet, and these effects were blunted with caloric restriction. However, hyperphagic obese mice had normal mitochondrial respiratory function in all tissues tested and no discernable intestinal dysbiosis relative to lean littermates. In contrast, feeding mice an obesogenic HFD altered the composition of the gut microbiome, impaired skeletal muscle mitochondrial bioenergetics, and promoted poor glucose control. These data show that adipose inflammation and redox stress occurred in all models of obesity, but gut dysbiosis and mitochondrial respiratory dysfunction are not always required for obesity-induced insulin resistance. Rather, changes in the intestinal microbiome and mitochondrial bioenergetics may reflect physiological consequences of HFD feeding.
Keywords: inflammation, insulin resistance, microbiome, mitochondrial function, obesity, redox balance
Graphical Abstract
Graphical Abstract.
Introduction
Obesity and Type 2 diabetes are interlinked chronic diseases that affect a significant number of people; however, the etiology of connections between these diseases is poorly defined. A common working model suggests that overnutrition causes adipose tissue hypertrophy,1 which promotes hyperinsulinemia and insulin resistance through several cellular responses including endoplasmic reticulum (ER) stress, oxidative stress, and inflammation.2,3Obesity-related changes in adipose tissue function influence systemic metabolism through adipokines, dyslipidemia, ectopic lipid deposition, inflammation, and altered endocrine control of metabolism in other tissues such as the liver and muscle,4 ultimately influencing insulin and glucose homeostasis.
Many mechanistic insights into the etiology of obesity are derived from high-fat diet (HFD)-feeding rodent models, which rapidly result in whole-body glucose intolerance and insulin resistance, secondary to adipose tissue inflammation.5,6 The oversupply of fatty acids to peripheral tissues, particularly skeletal muscle, has been shown to directly impair insulin signaling and contribute to the development of glucose intolerance.7,8 In addition, in HFD-fed models, mitochondrial adenosine diphosphate (ADP) sensitivity and the ability of ADP to suppress mitochondrial reactive oxygen species (ROS) emission rates are impaired, indicating mitochondrial dysfunction.9 This response also appears linked to excess intramuscular lipids, as the presence of palmitoyl-CoA (P-CoA) dramatically decreases mitochondrial ADP sensitivity.10 Furthermore, HFD-induced alterations in the gut microbiome influence energy harvest from the diet, and can contribute to energy excess that promotes obesity.11,12 Altered composition of the intestinal microbiota during obesity coincides with the development of insulin resistance13; however, diet composition (e.g., high fat, lower fiber) is also highly influential in shaping the intestinal microbiome, independent of obesity.14 These findings highlight the difficulty in discerning the independent effects of obesity from lipid overload. It is, therefore, possible that changes in the gut microbiome, mitochondrial bioenergetics, and redox imbalance occurring with obesity may not all be necessary for the induction of insulin resistance, but instead reflect the physiological response to altered composition of the diet such as high fat intake.
In contrast to HFD approaches, genetic models targeting leptin or its receptor such as the ob/ob or db/db mouse have provided a hyperphagic model of obesity.15,16 These genetically obese mice are also characterized by adipose tissue hypertrophy, inflammation, and insulin resistance but do not require HFD feeding to induce this phenotype.17,18 However, leptin plays a role in immunity,19,20 which likely contributes to the altered intestinal microbiota composition in ob/ob mice independent of diet.21 While the microbiome is altered in db/db mice fed an ad libitum chow diet compared to lean controls, caloric restriction has been shown to prevent obesity in db/db mice without altering the gut microbiome composition.22 This suggests that leptin signaling has a direct influence on intestinal dysbiosis, and therefore it is difficult to determine the contribution of the gut microbiota to obesity-induced insulin resistance in these genetic models.
In this study, we characterize a spontaneous mutation in C57Bl/6J mice that leads to rapid development of an obese phenotype in the absence of changing dietary composition. Importantly, hyperphagia was not due decreased plasma leptin concentrations or mutations within the leptin receptor. We determined the hyperglycemic, hyperinsulinemic, and insulin-resistant phenotype of obese mice was associated with oxidative stress and inflammation within epididymal white adipose tissue (eWAT) but occurred without gut dysbiosis or skeletal muscle mitochondrial respiratory dysfunction. In contrast, HFD-feeding-induced insulin resistance in both spontaneously obese mice and lean littermates, while impairing skeletal muscle mitochondrial bioenergetics and altering the gut microbiome. Therefore, while adipose inflammation appears intrinsic to obesity, gut dysbiosis and mitochondrial respiratory dysfunction are consequences of HFD interventions and may not be directly required to induce obesity or insulin resistance.
Materials and Methods
Mice
C57Bl/6J mice were bred on site at the University of Guelph. Lean and obese littermates were cohoused with a 12:12 h light:dark cycle and fed a standard chow diet ad libitum. In a subset of experiments, obese mice were pair-fed to lean littermates. A group of both obese and lean animals was also subject to an 8-week HFD (60% fat, Research Diets D12492) or low-fat diet (LFD; 10% fat, Research Diets D12450J). In a separate subset of these mice (both HFD and LFD), tissues were removed and rapidly frozen in liquid nitrogen before (pre) and 15 min following (post) an intraperitoneal injection of 1 U/kg body weight of insulin (Novorapid). All procedures were approved by the Animal Care Committee at the University of Guelph and the University of Ottawa, and all mice were anesthetized at ∼16–24 weeks of age with an intraperitoneal injection of sodium pentobarbital (60 mg/kg). Thereafter key tissues and feces were removed, immediately snap-frozen in liquid nitrogen and stored at −80°C until analyses. Alternatively, mitochondrial bioenergetics was immediately analyzed on fresh tissue as described below.
For chow versus HFD comparison of mouse cecal microbiota, specific-pathogen-free C57Bl/6J littermate male mice were bred in house at McMaster University and aged to 8 weeks. All procedures were approved by McMaster University Animal Ethics Review Board. The HFD group was fed a 45% HFD (Research Diets D12451) for 16 weeks while the chow group continued on regular diet for this time (Teklad 22/5 diet, catalog #8640). After 16 weeks of chow or high-fat feeding, the mice were euthanized and cecums collected for DNA isolation and sequencing.
Whole Body Characterization
Fasting plasma glucose, insulin, leptin, and nonesterified fatty acids were determined as previously reported 5,23 using commercially available kits. On separate days, following an overnight fast, animals were injected intraperitoneally with either glucose (2 g/kg body weight) or insulin (1 U/kg body weight, Novorapid) to determine glucose and insulin tolerance, respectively, by monitoring tail blood glucose concentrations. Resting energy expenditure was determined by monitoring oxygen consumption (VO2) and carbon dioxide production (VCO2) over a 48 h period (Columbus Instruments, Columbus, OH), and subsequently calculating total carbohydrate (CHO) and fat oxidation as previously reported.24
Hyperinsulinemic–Euglycemic Clamp
Hyperinsulinemic–euglycemic clamps were performed as previously described.25 A catheter was inserted into the right jugular vein 5 days prior to the clamp, and on the day of the clamp, mice were infused with D-[3-3H]-glucose for 1 h. An insulin infusate (10 mU/kg/min) containing D-[3-3H]-glucose was then infused, and blood glucose levels were titrated with 50% dextrose to maintain euglycemia for a further 2 h. Basal and clamped rates of glucose disposal and hepatic glucose production (HGP) were calculated as previously described.25,26
Sequencing of the Mouse Genome
DNA was isolated from the gastrocnemius muscle from two obese and one lean animal using commercially available kits (Qiagen), frozen and shipped to The Centre for Applied Genomics (TCAG) at SickKids. Thereafter, the Next Generation sequencing facility utilized Illumina HiSeq X to sequence the genomes, compared to a known reference (mm10 C57Bl/6J), and filtered to determine common SNPs between the obese mice relative to the lean littermate. A full list of this returned annotated file is provided as an expanded view table.
Mitochondrial Bioenergetics
Standard methodology was utilized to determined mitochondrial respiration in isolated mitochondria (red gastrocnemius [RG] muscle, heart, and liver),27,28 permeabilized WAT,5 and permeabilized muscle fibres29 (Oroboros Oxygraph-2K: Oroboros Instruments, Innsbruck, Austria). In addition, mitochondrial H2O2 emission was determined in permeabilized muscle fibers as previously reported30 (Oroboros Oxygraph-2K: Oroboros Instruments, Innsbruck, Austria) and in permeabilized eWAT as previously reported.5
Western Blotting
Tissue was homogenized in lysis buffer, centrifuged for 15 min at 1500 g and 4°C, and diluted to 1 µg/µL protein content. Equal amounts of each sample were loaded for separation by SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. Membranes were incubated in commercially available primary antibodies to detect mitochondrial oxidative phosphorylation system (OXPHOS; 5µg protein; 1:1000; Mitosciences; ab110413), superoxide dismutase 2 (SOD2; 5µg protein; 1:5000; Abcam; ab13533), catalase (5µg protein; 1:2000; Abcam; ab16731), total c-Jun N-terminal kinase 1/2 (JNK1/2; 10µg protein; 1:1000; Cell Signaling; cs9252), phospho-JNK1/2 (15µg protein; 1:1000; Cell Signaling; cs4671), total extracellular-signal-regulated kinase 1/2 (ERK1/2; 5µg protein, 1:1000; Cell Signaling; cs9102), phospho-ERK1/2 (15µg protein, 1:1000; Cell Signaling; cs9101), total eukaryotic translation initiation factor 2A (EIF2α; 5µg protein; 1:1000; Cell Signaling; cs9772), phospho-EIF2α (15µg protein; 1:1000; Cell Signaling; cs9721), cytoplasmic FMR1-interacting protein 2 (CYFIP2; 5µg protein; 1:1000; Abcam; ab95969), phospho-Akt Ser473 (15µg protein; 1:1000; Cell Signaling; cs9271), phospho-Akt Thr308 (15µg protein; 1:1000; Cell Signaling; cs9275), total Akt (5µg protein; 1:1000; Cell Signaling; cs4691), and 4-hydroxynonenal (4HNE; 20µg protein; 1:1000; Alpha Diagnostics; HNE11-s). The α-tubulin (5µg protein, 1:1000; Abcam; ab7291) and Ponceau stains were used as a loading control. Western blots were quantified using FluorChem HD imaging chemiluminescence (Alpha Innotech, Santa Clara, US). All samples for each protein were loaded and detected on the same membrane.
Histology
Adipose tissue and liver were fixed, embedded, and stained as previously reported.23 Images were taken with an Olympus (Tokyo, Japan) microscope at ×40 magnification and analyzed with ImageJ 1.48 (National Institutes of Health) software obtained from http://rsb.info.nih.gov/ij/download.html.
Gene Expression
Total RNA was obtained from frozen WAT as previously described.31 Transcript expression was measured using TaqMan Assays with AmpliTaq Gold DNA polymerase (Thermo Fisher Scientific) in a Rotor-Gene Q real-time polymerase chain reaction (PCR) cycler (Qiagen). Target genes (Tnf, Ccl2, Cxcl1, Cxcl9, Cxcl10, Il1b, Il4, Il10, Emr1, Ccr7, Cd4, Cb8, and Cd11b) were compared to the Rplp0 housekeeping gene using the ΔΔCT method as previously described.31
Microbiome Bacterial Profiling
Genomic data were extracted from cecum segments and fecal samples as previously described.13 Illumina compatible PCR amplification of the variable 3 (V3) region of the 16S ribosomal RNA (rRNA) gene was completed on each sample and Illumina MiSeq was used to sequence DNA products of this amplification. Taxonomy was assigned to operational taxonomic units (OTUs) (based on 97% similarity). Ribosomal Database Project classifier in Quantitative Insights Into Microbial Ecology (QIIME)32 against the 2011 version of the Greengenes reference database.33 OTU assignments were converted to relative abundance before beta diversity calculations to account for depth of coverage and to normalize across samples. QIIME and R scripts were used to calculate beta diversity using the Bray–Curtis dissimilarity and principal coordinates analysis (PCoA), to generate plots of taxonomic data, and to perform statistical tests, as previously described.13
Statistics
Statistical analyses were completed using GraphPad Prism 8 software (GraphPad Software, Inc., LA Jolla, USA). Unpaired, two-tailed t-tests; one-way analysis of variance (ANOVA) with a lease significance differences (LSD) multiple comparison post hoc analysis; or two-way ANOVAs were used as appropriate (full details and sample sizes listed in figure legends). ADP titrations (drive on respiration and attenuation of H2O2) were analyzed with GraphPad Prism software as previously described.30,34 Statistically significance was determined as P < .05. Data expressed as mean ± SEM.
Analysis and data visualization of microbial 16S rRNA gene profiling was performed in R. The Wilcoxon rank-sum test was used for pairwise comparisons between two groups. Adjustment for the false discovery rate was calculated with the Benjamini–Hochberg method35 and statistical significance was accepted at P < .05. Custom R scripts used for data analysis are available from the corresponding author on reasonable request.
Results
Mouse Phenotype
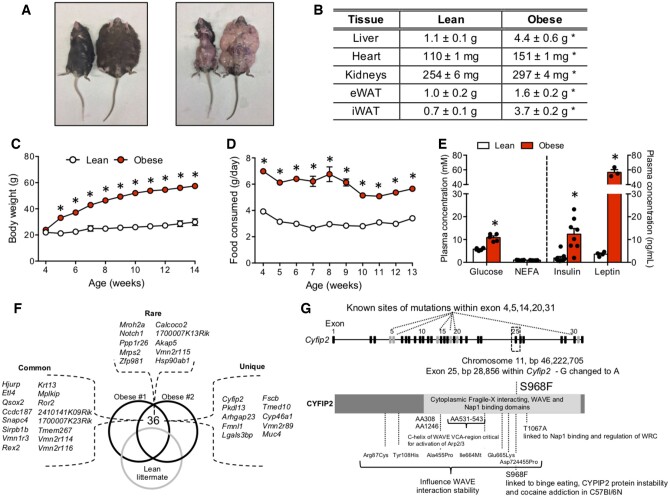
While breeding wildtype (WT) C57Bl/6J mice, a litter of pups spontaneously arose that resulted in lean and obese cagemates at maturity. Obese littermates at 20 weeks of age were up to 4 times heavier (∼80 g) compared to lean littermates (∼25 g; Figure 1A). The obese phenotype was associated with increased mass and size of WAT, liver, heart, and kidneys (Figure 1A and B; Figure S1A). We, therefore, tracked the time course of changes in body mass in subsequent litters of mice. While pups <4 weeks of age (i.e., during weaning) did not differ in body weight, some animals displayed a marked and rapid increase in body weight beginning at 5 weeks of age (∼10 g between weeks 5 and 6; Figure 1C). The obese phenotype was not sex specific, as a subset of both male and female mice displayed rapid weight gain. The increase in body weight was associated with hyperphagia when tracked over several weeks (Figure 1D), as well as hyperglycemia, hyperinsulinemia, and increased plasma leptin concentrations when assessed at ∼18 weeks of age (Figure 1E). Surprisingly, obesity occurred in the absence of higher plasma fatty acids (Figure 1E).
Figure 1.
Characterization of Spontaneous Obese Mice. (A, B) Some littermates displayed pronounced obesity and adiposity, which also manifested in increased mass of the liver, heart, kidney, and adipose tissue. (C, D) Obesity occurred between week 4 and 5 of age as a result of a marked increase in food intake. (E) Obesity was associated with hyperglycemia, hyperinsulinemia, increased leptin, but unaltered plasma NEFA concentrations. (F) In total, 36 nonsynonymous mutations were shared between two obese animals compared to one lean littermate, including 16 common mutations (occurring in >10 other species), 10 rare mutations (occurring in 2–5 other species), and 10 unique mutations (occurring in <2 other species). (G) The Cyfip2 gene appears to be a hotspot for mutations, including the S968F mutation and several others within the cytoplasmic fragile-X interacting, WAVE and Nap1 binding domains. Data expressed as mean ± SEM of results obtained from n = 3–10 for each genotype. *P < .05 versus lean (unpaired two-tailed Student’s t-test). Nap1, Nck-associated protein 1; NEFA, nonesterified fatty acids; WAVE, Wiskott–Aldrich syndrome protein–family verprolin-homologous protein.
Genetic Characterization
Given the hyperleptinemia in our spontaneously obese mice (Figure 1E), our model does not appear to replicate the ob/ob mouse. We next aimed to establish that this model was not simply recapitulating the db/db mouse (mutation in the leptin receptor in the presence of hyperleptinemia).36 To accomplish this, DNA was sequenced from two obese mice and one lean littermate mouse. No mutations were observed within the leptin receptor gene (Lepr), or within ∼2 million base pairs on chromosome 4 (Table S1). These data suggest that the current obesity model is independent of leptin deficiency or mutations within the leptin receptor. To examine other potential genetic targets, we next filtered the genomic data to probe for common mutations within two of our spontaneously obese mice. We also reasoned that our obesity-associated mutation would likely be uncommon in C57Bl/6J mice, and therefore we compared these animals to the mm10 reference genome (UCSD).37,38 This approach resulted in 16 common (previously identified in >5 species), 10 rare (previously identified in 2–5 species), and 10 unique (previously identified in <2 species) nonsynonymous mutations (Figure 1F;Table S1). Of the unique mutations found in our spontaneously obese mice, the CYFIP2 gene has been linked to binge eating and addiction behavior,39 and therefore could contribute to the development of hyperphagia and obesity in the present animals.
In support of a putative role of CYFIP2 in our spontaneously obese phenotype, the CYFIP2 gene appears to be a hotspot for mutations with downstream functional outcomes, as mutations in several exons (4, 5, 14, 20, and 31; Figure 1G) of the human CYFIP2 gene have been linked to the development of Prader–Willi syndrome.40 Intriguingly, the mutation we identified in our obese model (S968F) has previously been attributed to some of the genetic differences between C57Bl/6J and 6N mouse lines, as the Cyfip2 S968F mutation was introduced in the C57Bl/6N strain almost 70 years ago (Figure S1B).41 It was, therefore, possible that the mutation detected arose because our 6J mice were crossed with 6N mice and would imply the CYFIP2 mutation is unrelated to the observed obese phenotype in our strain of mice. However, other differences exist between the 6J and 6N mice, which we utilized to determine this possibility. Specifically, the 6J strain has a deletion in exon 7–11 of the nicotinamide nucleotide transhydrogenase (NNT) gene,42 coding a protein involved in mitochondrial antioxidant defense, and representing a mutation which is not present in the 6N strain (Figure S1B). We reasoned that if C57Bl/6 mouse strains were crossed, the full-length NNT gene would be introduced into the genome, and we would detect an insertion at the corresponding position of chromosome 13 (bp 119334317-119409011) within our obese genomic data. However, when we compared our mouse genomes to the mm10 genome as a reference (C57Bl/6J), no mutations were detected within the NNT gene (Figure S1C; Table S2). Therefore, it appears that the C57Bl/6J mice in this study spontaneously developed the Cyfip2 S968F mutation. Since litters are extremely small in number (n = 2–3), we cannot rely on Mendelian genetics to estimate the frequency of inheritance. However, as the obese animals sequenced were both homozygous and heterozygous for the S968F mutation, this suggests a dominant phenotype. In cell culture preparations, the Cyfip2 S968F mutation has been linked to decreased mRNA stability/half-life,41 and since the CYFIP2 protein is ubiquitously expressed within the brain, we examined CYFIP2 content in the cortex of 6N, lean 6J, and spontaneously obese 6J animals. Despite the S968F mutation, CYFIP2 protein content was not different between 6N and 6J genotypes, or in our obese animals (Figure S1D), suggesting total CYFIP2 protein content cannot explain the addictive behavior of 6N mice39 or the apparent hyperphagia in the obese mice used in our experiments. Regardless, since WT 6N mice do not develop obesity on a standard diet, it is evident that the present obese animals display a polygenic form of obesity not solely linked to CYFIP2.
As alternative targets involved in our polygenic obese model, nine other mutations appeared unique to this strain of mice (Figure 1F, Table S1). While polycystic kidney disease one like three (Pkdl13) represents one of these unique mutations, Pkdl13 has been linked to changes in kidney external appearance43; and as obese animals displayed smooth kidneys that appeared devoid of cysts (Figure S1A), it is therefore unlikely that this mutation contributes to the observed obese phenotype. In contrast, mutations in galectin-3-binding protein (Lgals3bp) have been associated with obesity in humans44 and a functioning transmembrane P24 trafficking protein 10 (Tmed10) appears required for normal ER function and preventing ER stress within the liver,45,46 suggesting mutations in these genes could be involved in an obese phenotype. Clearly, several genetic mutations could contribute to the observed spontaneous obesity occurring in our strain of C57Bl/6J mice. Regardless of solidifying which gene causes obesity, these mice represent a unique model to study the mechanisms associated with obesity-related glucose intolerance in the absence of HFD feeding or defects in leptin.
Insulin Sensitivity and Energy Expenditure
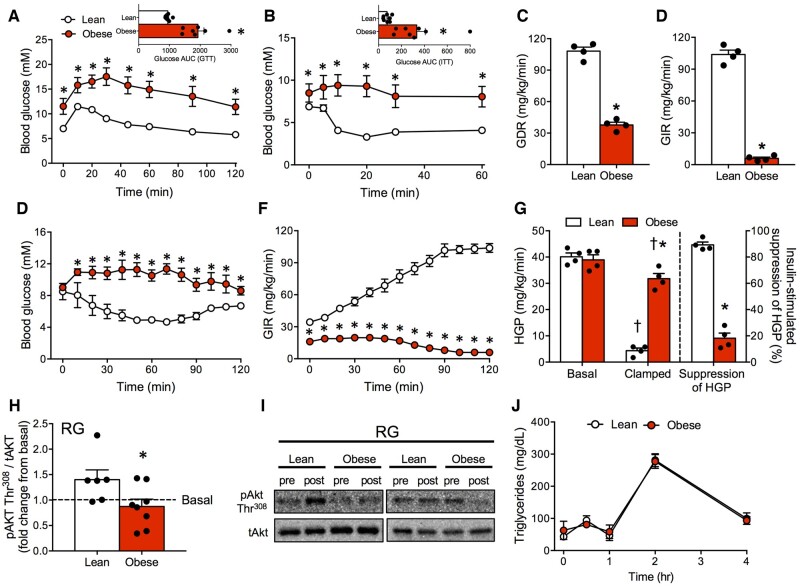
Insulin resistance and glucose intolerance are closely linked with obesity in genetic and diet-induced models; therefore, we examined several indices of glucose homeostasis in our line of obese mice. Compared to lean littermates, spontaneously obese mice had profound glucose intolerance and insulin resistance (Figure 2A and B). Weight gain and impaired glucose control were consistent in both males and females (Figure S2A–C). Furthermore, hyperinsulinemic–euglycemic clamps revealed a 3-fold lower glucose disposal rate, and 20-fold lower glucose infusion rate (Figure 2C–F) in obese mice, indicating robust peripheral and hepatic insulin resistance. While HGP was not different at rest, obese mice were resistant to the ability of insulin to suppress HGP (∼5-fold lower than lean mice, Figure 2G) which is not surprising given the marked liver hypertrophy (Figure 1B;Figure S1A). Furthermore, decreased insulin-stimulated phosphorylation of AktThr308 in the oxidative RG muscle of obese mice indicates skeletal muscle insulin resistance (Figure 2H and I). However, in contrast to the impairments in glucose control and insulin sensitivity, no alterations in plasma triglyceride levels before or after ingestion of lipids/oil were observed in obese mice (Figure 2J), indicating normolipidemia and the absence of changes in intestinal lipid absorption.
Figure 2.
Glucose Intolerance and Insulin Resistance in Spontaneous Obese Mice. (A, B) Spontaneously obese mice presented with profound glucose and insulin intolerance. (C–F) Hyperinsulinemic–euglycemic clamps revealed dramatically lower GDR and GIR in obese mice. (G) HGP was elevated in spontaneously obese mice, and the ability of insulin to suppress HGP was attenuated. (H, I) RG muscle of obese mice was insulin resistant, indicated by a reduction in AktThr308 phosphorylation following insulin stimulation. (J) Despite insulin resistance, lipid tolerance (triglyceride responses) following lipid ingestion was not altered in obese mice. Data expressed as mean ± SEM of results obtained from n = 4–8 for each genotype. *P < .05 versus lean, †P < .05 versus basal unstimulated (unpaired two-tailed Student’s t-test). AUC, area under curve; GDR, glucose disposal rate; GIR, glucose infusion rate; GTT, glucose tolerance test; ITT, insulin tolerance test.
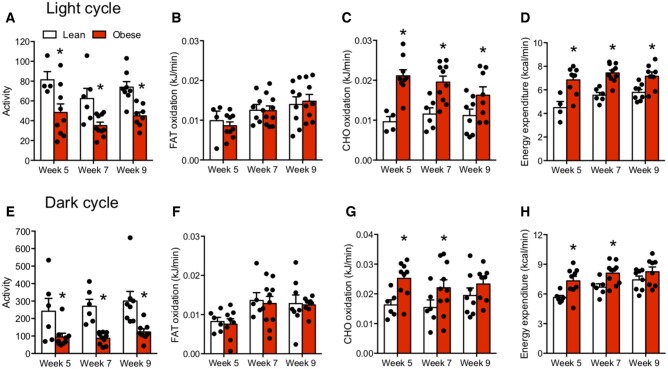
We next examined the possibility that a reduction in fat oxidation or total energy expenditure could contribute to the observed phenotype. However, despite reduced activity, absolute rates of fat oxidation were unaltered, while CHO oxidation and total energy expenditure were increased in both light and dark cycles in spontaneously obese animals (Figure 3A–H). Similar to body weight, these changes were not sex specific, as both male and female mice displayed similar increases in total energy expenditure, likely as a result of the increased body weight (Figure S2D–K). Altogether, the spontaneous mutation in C57Bl/6J mice resulted in obesity and severe insulin resistance due to hyperphagia in the absence of a reduction in energy expenditure or a direct link to a single genetic mutation.
Figure 3.
Decreased Fat Oxidation and Energy Expenditure Do Not Contribute to the Development of Obesity. (A–D) Total activity, fat oxidation, CHO oxidation, and energy expenditure in lean and obese mice when averaged over the middle of the second light cycle (ie, 10–13 h) during 48 h of indirect calorimetry. (E–H) Total activity, fat oxidation, CHO oxidation, and energy expenditure in lean and obese mice when averaged over the middle of the second dark cycle (ie, 22–1 h) during 48 h of indirect calorimetry. Data expressed as mean ± SEM of results obtained from n = 4–10 for each genotype. *P < .05 versus lean (unpaired two-tailed Student’s t-test).
Mitochondrial Bioenergetics
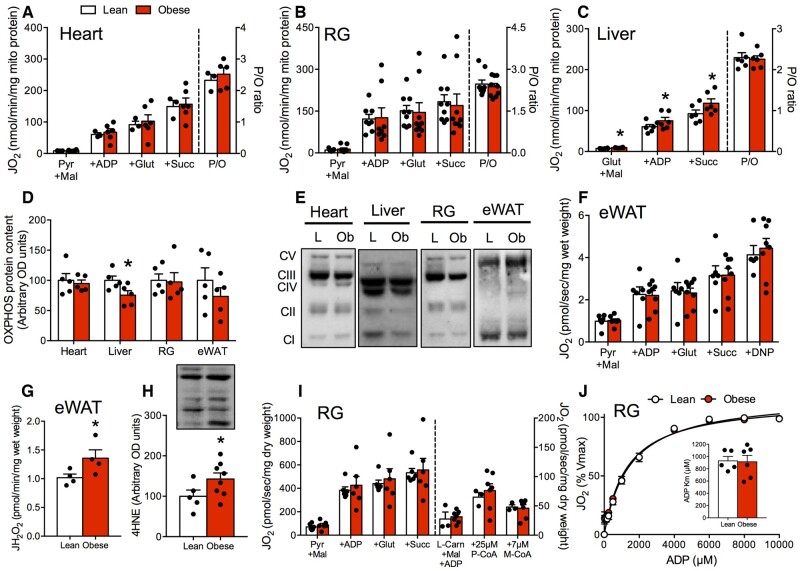
Given the link between mitochondrial bioenergetics and insulin resistance, we next examined if mitochondrial dysfunction contributed to poor glucose control and insulin resistance in obese mice. However, mitochondrial respiration, coupling rates, and leak respiration were not impaired in isolated mitochondria from the heart (Figure 4A) and skeletal muscle (Figure 4B). Intriguingly, respiration rates were subtly higher in the liver of obese mice (Figure 4C), however, this change was small in magnitude and the biological relevance remains unknown. Since these measurements are independent of mitochondrial content, we also assessed protein content of the OXPHOS within these tissues. However, obesity did not alter mitochondrial protein content with the exception of a reduction in OXPHOS in the liver (Figure 4D and E; Figure S3A–F). In addition, OXPHOS content was not reduced in eWAT of obese animals (Figure 4D and E) and mitochondrial respiration was not impaired (Figure 4F). Recently, increased mitochondrial ROS has been implicated as a cellular event more indicative of HFD-induced dysfunction within eWAT.47,48 Indeed, we determined that lipid-supported mitochondrial ROS emission rates were increased in eWAT of obese mice (Figure 4G), in association with increased 4HNE (marker of lipid peroxidation and redox stress; Figure 4H) compared to lean littermates.
Figure 4.
Mitochondrial Respiratory Dysfunction Is Not Required for the Development of Obesity. (A–C) Mitochondrial respiration was not reduced in mitochondria isolated from the heart, liver, or skeletal muscle of obese animals. (D, E) With the exception of a reduction in OXPHOS within the liver, mitochondrial content was not altered in obese mice. (F) Respiration in permeabilized eWAT was not influenced by obesity. (G, H) Mitochondrial lipid-supported ROS emission rates and 4HNE protein content were increased in eWAT of obese mice, indicating oxidative stress. (I, J) In permeabilized muscle fibers, maximal respiration and ADP sensitivity were not affected by obesity. Data expressed as mean ± SEM of results obtained from n = 5–10 per genotype. *P < .05 versus lean (unpaired two-tailed Student’s t-test). DNP, dinitrophenol; JH2O2, rate of hydrogen peroxide production; JO2, rate of oxygen utilization; Km, Michaelis–Menten constant; L, lean mice; L-Carn, L-carnitine; M, malate; M-CoA, malonyl-CoA; Ob, obese mice; P, pyruvate; P/O, the content of ADP utilized relative to oxygen; S, succinate; Vmax, maximal respiration rate.
In addition to altering OXPHOS capacity, HFD feeding has been associated with reductions in skeletal muscle mitochondrial ADP sensitivity9 and the capacity to utilize lipids.49 We, therefore, examined these parameters in permeabilized muscle fibers. Spontaneously obese animals displayed normal respiration and indices of mitochondrial ADP sensitivity, lipid-supported respiration, and malonyl-CoA sensitivity (Figure 4I and J). Furthermore, obese mice did not present with greater 4HNE or altered content of mitochondrial antioxidant proteins (SOD2, catalase) in skeletal muscle (Figure S3G and H). Combined, these data suggest the absence of skeletal muscle mitochondrial respiratory dysfunction in obese mice and show that impaired respiratory function or alterations in mitochondrial ADP sensitivity are not necessary in development of insulin resistance during obesity in mice.
Adipose Tissue and Liver Morphology, Inflammation and Intracellular Signaling
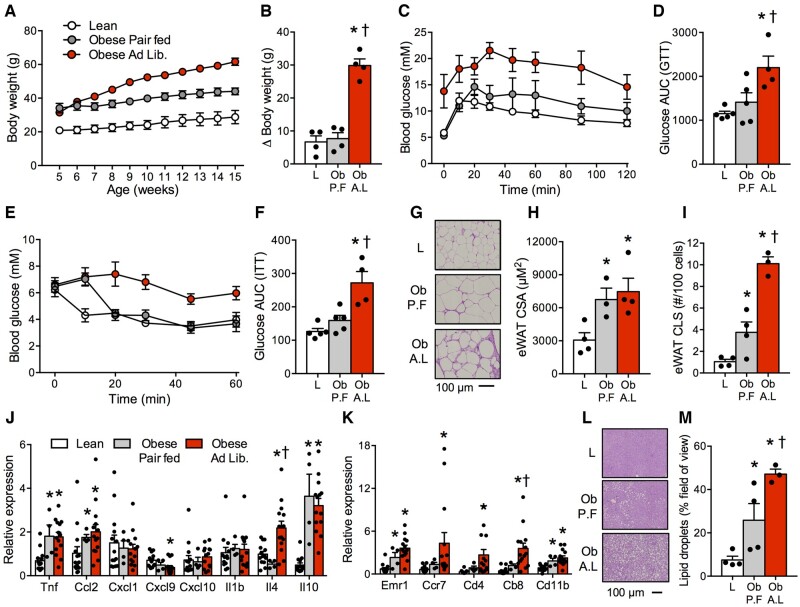
Adipose tissue expansion and inflammation is a hallmark of obesity and plays a fundamental role in the development of insulin resistance4; therefore, we next aimed to examine these processes in our spontaneous obese model. We randomized 5-week-old obese mice to consume food ad libitum or pair fed to lean littermate mice to examine the effect of restricting food intake. This approach resulted in similar weight gain in obese animals compared to lean mice once pair feeding commenced (ie, 5 weeks: Figure 5A and B). Moreover, food restriction mitigated glucose and insulin intolerance (Figure 5C–F). While pair feeding did not alter eWAT adipocyte cross-sectional area (CSA; Figure 5G and H), eWAT crown-like structures (CLS) were dramatically reduced following food restriction (Figure 5I). Within eWAT, pair feeding did not reduce macrophage/dendritic markers (Emr1, Cd11b; Figure 5J) or inflammatory protein markers (Figure S4A–C) associated with obesity. However, pair feeding partially reduced markers of cytotoxic, helper, and memory T-cells (Cd4, Cd8, Ccr7) in obese mice (Figure 5J and K). While liver steatosis was apparent in spontaneously obese mice, pair feeding dramatically reduced liver lipid droplets (Figure 5L and M), in association with reducing pEIF2α (Figure S4A, B, and D). Combined, these data strongly indicate that hyperphagia causes obesity, insulin resistance, and adipose inflammation, while food restriction mitigates several of these effects.
Figure 5.
Food Restriction Attenuates Many of the Phenotypic Characteristics of Obesity. (A, B) Pair feeding obese animals to match caloric intake of lean animals normalized weight gain. (C–F) Pair fed obese mice presented with normal glucose and insulin tolerance. (G–I) Histological images of eWAT revealed that pair feeding did not affect adipocyte CSA, but attenuated CLS. (J) Pair feeding did not affect the mRNA signature associated with inflammatory tone or macrophage/dendritic markers (Emr1, Cd11b) associated with obesity. (K) Food restriction partially improved markers of T-cells, including cytotoxic, helper, and memory T cells (Cd4, Cd8, Ccr7). (L, M) Histological images of the liver demonstrated that pair feeding reduced lipid droplet content. Data expressed as mean ± SEM of results obtained from n = 4–14 per genotype. *P < .05 versus lean, †P < .05 versus obese pair fed (one-way ANOVA with LSD post hoc). AUC, area under curve; A.L., ad libitum obese mice; GTT, glucose tolerance test; ITT, insulin tolerance test; L, lean mice; Ob, obese mice; P.F., pair fed obese mice.
HFD-Feeding Exacerbates the Obese Phenotype and Impairs Mitochondrial Bioenergetics
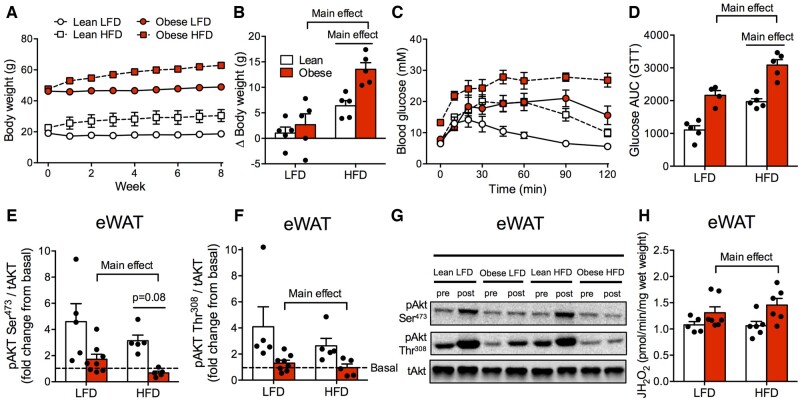
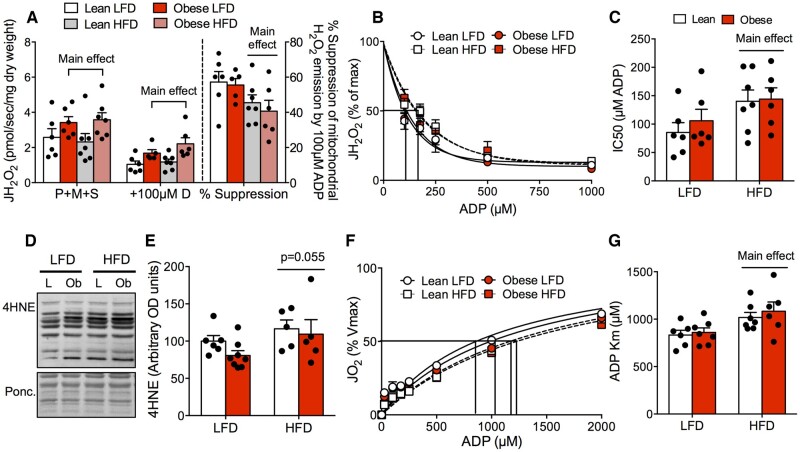
While adipose inflammation was directly linked to obesity, mitochondrial respiratory dysfunction was not present in any tissue studied in spontaneously obese mice. As a result, we subjected both lean littermate and spontaneously obese mice to an HFD or LFD to determine if HFD feeding would exacerbate insulin resistance while impairing mitochondrial bioenergetics. Regardless of genotype, HFD-feeding recapitulated the typical phenotype associated with this diet-induced model, including increased body weight (Figure 6A and B), impaired glucose homeostasis (Figure 6C and D), and reduced insulin-stimulated AktSer473 phosphorylation within eWAT (Figure 6E–G). In further support of an independent relationship between obesity and adipose tissue inflammation, regardless of diet, eWAT ROS emission rates (Figure 6H) and 4HNE protein content (Figure S4E) were higher in spontaneously obese mice, indicating cellular stress responses associated with insulin resistance in this tissue.5 Within skeletal muscle, regardless of diet, mitochondrial ROS emission rates were higher in obese mice (Figure 7A). While obese mice did not display overt oxidative stress on LFD (Figure S3G and H), the ability of ADP to suppress ROS was impaired following HFD feeding in both lean and obese mice (Figure 7B and C), which coincided with an increase in 4HNE content (Figure 7D and E). In addition, mitochondrial ADP sensitivity was impaired following HFD feeding (Figure 7F and G). This occurred in the absence of changes in mitochondrial ETC protein content in the RG (Figure S4F) or maximal Complexes I and II-supported mitochondrial respiratory capacity (Figure S4G). Combined, these data show that while obesity is associated with a conserved increase in mitochondrial H2O2, HFD feeding, but not obesity per se, impairs skeletal muscle mitochondrial respiration .
Figure 6.
HFD-Feeding Induces Insulin Resistance in Lean and Obese Mice. (A, B) Eight weeks of HFD-feeding increased body weight in lean and obese mice. (C, D) While obese mice fed an LFD were glucose intolerant compared to lean littermates, HFD-feeding impaired glycemic control in both genotypes. (E–G) eWAT of obese mice was insulin resistant (decreased Akt phosphorylation following insulin stimulation), and HFD further induced eWAT insulin resistance in both genotypes (AktSer473). (H) Lipid-supported (P-CoA + L-carnitine) mitochondrial ROS was increased in eWAT of obese mice compared to lean littermates, while HFD did not influence this parameter. Data expressed as mean ± SEM of results obtained from n = 5–8 per genotype/diet (two-way ANOVA with LSD post hoc). AUC, area under curve; GTT, glucose tolerance test; JH2O2, rate of hydrogen peroxide production; p, phosphorylated protein; t, total protein.
Figure 7.
HFD-Feeding Impairs Mitochondrial ADP Sensitivity and Attenuates the Ability of ADP to Suppress ROS Emission in Skeletal Muscle. (A–C) While maximal mitochondrial ROS emission rates were higher in obese mice compared to lean controls, HFD further impaired redox balance by attenuating the ability of ADP to suppress mitochondrial ROS (increased IC50) in both genotypes. (D, E) 4HNE protein content in RG was increased in both genotypes following HFD-feeding. (F, G) Mitochondrial ADP sensitivity was impaired following HFD in both genotypes. Data expressed as mean ± SEM of results obtained from n = 5–8 per genotype/diet (two-way ANOVA with LSD post hoc). IC50, half-maximal inhibitory concentration; JH2O2, rate of hydrogen peroxide production; JO2, oxygen consumption; L, lean mice; Ob, obese mice; P + M+S, pyruvate + malate + succinate.
Dissociation between the Microbiome and the Obese Phenotype
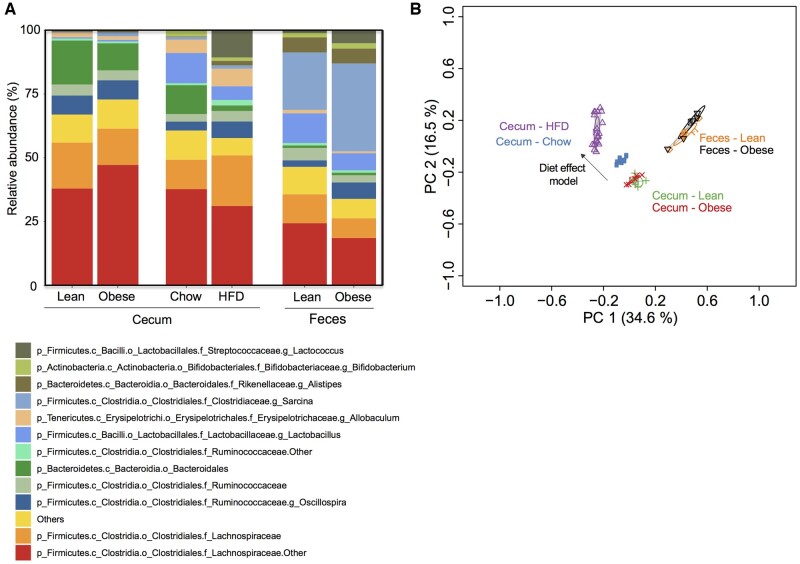
An alteration in the composition of the gut microbiome during obesity is commonly reported as a connection between inflammation, adipose tissue metabolism, and insulin resistance. Hence, we also examined the possibility that spontaneously obese mice had taxonomic alterations in the gut microbiome (cecum of males, feces of females) compared to lean cagemates. In the cecums, we detected 117 taxa, only 9 of which were different in lean versus spontaneously obese mice (5 increased, 4 decreased; Figure S5). In contrast, over 30 taxa were different in HFD-fed male mice compared to chow-fed littermates (Figure S6). These data suggest that profound obesity in spontaneously obese mice does not alter the beta diversity to the same extent as HFD feeding. This is further supported by similar relative abundances of the most abundant taxa and overlapping/superimposed PCoA blots in taxonomic results from both cecum and feces in spontaneously obese mice and lean littermate mice (Figure 8A and B). In contrast, we confirm an obesogenic HFD dramatically alters the microbial taxa in the cecum of C57Bl6/J mice, clearly resulting in divergent PCoA plots (Figure 8A and B; “Diet effect model”). Furthermore, we and others have shown that HFD feeding increases intestinal permeability50 detected by increased fluorescein isothiocyanate (FITC)-dextran in serum following a gavage, but this effect did not occur in spontaneously obese animals (3704 ± 1013 relative fluorescence; n = 3) compared to lean littermates (6040 ± 1319 relative fluorescence; n = 3; P = .2). These data suggest that spontaneously obese mice have minimal changes in the taxonomy of the gut and fecal microbiome and that alterations in the gut microbiota are not required for inflammation and insulin resistance in our non-HFD-fed model of obesity.
Figure 8.
The Gut Microbiome Is Altered with HFD-Feeding Compared to Lean Controls, but Intestinal Dysbiosis Is Not Evident in Spontaneously Obese Mice. (A) Stacked bar graph showing the relative abundance of the 13 most abundant bacterial taxa using Genus level assignments in the cecum (males only) and feces (females only) between lean and obese littermate mice. (B) PCoA of Bray–Curtis dissimilarity in the cecum (males only) and feces (females only) between lean and obese littermate mice. A comparator group of mice was fed a chow diet or HFD to demonstrate the effect of diet on Bray–Curtis dissimilarity in the PCoA plot. n = 5 for each lean/obese experiment. n = 7 for chow and n = 18 for HFD in comparator group. Individual differences in taxa are reported in Supplementary Figures.
Discussion
We characterized a new spontaneous mouse model of obesity in our C57Bl/6J colony and found that several mechanisms commonly purported to cause obesity-related insulin resistance in other mouse models are not necessary for insulin resistance. In spontaneously obese mice fed the same diet as lean littermates, adipose tissue displayed marked cellular stress and higher markers of inflammation; however, we found no discernable changes in the gut microbiome composition or mitochondrial respiratory function despite profound obesity, insulin resistance, adipose, and liver hypertrophy. These data suggest adipose tissue hypertrophy, inflammation, and redox imbalance can contribute to whole-body inflammation and insulin resistance in severely obese mice. In contrast, as impairments in skeletal muscle mitochondrial bioenergetics and gut dysbiosis only occurred following HFD feeding, these cellular processes may be driven by changes in diet rather than obesity per se, and therefore are not required for a certain level of insulin resistance.
Insulin resistance within adipose tissue impairs insulin-mediated suppression of lipolysis, causing lipid oversupply, reactive lipid accumulation, and redox stress in peripheral tissues germane to whole-body glucose homeostasis. As a result, adipose tissue has become a focus in the pursuit of a mechanistic understanding of the development of insulin resistance. Several processes have been emphasized in the induction of insulin resistance within WAT, as genetic models ablating mitochondrial ROS5 or JNK signaling51 prevent short-term HFD-induced WAT insulin resistance. In this study, despite consuming an LFD, spontaneously obese mice displayed marked obesity and insulin resistance in association with adipocyte hypertrophy (CSA), increased mitochondrial ROS, and inflammation/oxidative stress (CLS, macrophage mRNA expression, immune T-helper cell mRNA expression, 4HNE), phenocopying HFD models. These data suggest that certain aspects of adipose inflammation are intrinsic to obesity, particularly since food restriction mitigated some of these markers. Moreover, as key organelles involved in energy metabolism, impairments in mitochondrial content and/or function in several tissues have long been implicated in the development of obesity and insulin resistance.6,9,52 While markers of mitochondrial content in WAT are decreased in ob/ob mice,53 db/db mice,54 and HFD-fed mice,6 recent evidence indicates that impaired mitochondrial respiration in this tissue is not associated with HFD-induced WAT insulin resistance.47,48 This notion is supported by our current findings that mitochondrial respiration normalized to tissue weight was not reduced in eWAT of spontaneously obese mice. Furthermore, since CSA was increased in obese animals, a logical assumption would be that fewer adipose cells were added per gram of tissue.47,48 This would suggest that respiration per WAT cell would actually be higher in spontaneously obese mice, similar to previous findings in HFD-fed models.47,48 Combined, these data indicate obesity-induced increases in eWAT ROS and inflammation occur without a reduction in oxidative capacity and are independent of dietary composition.
Immune responses during obesity are also linked with changes in the gut microbiota. For example, changes in gut microbial composition during obesity have been associated with increased gut permeability, and as a result, increased circulating bacterial lipopolysaccharide and cell wall muropeptides capable of inducing inflammation, lipolysis, and insulin resistance.55–58 Furthermore, antibiotic treatment has been shown to attenuate the increase in mRNA expression of inflammatory markers (IL-1, tumor necrosis factor-α) and adipocyte hypertrophy in both HFD-fed WT mice and ob/ob mice, suggesting that changes in the composition of gut microbiota or microbial load are linked to tissue inflammation during obesity.58 However, in our study, we observed a robust increase in adipocyte hypertrophy (CSA) and inflammation in the absence of changes in microbiota composition in spontaneously obese mice. These results suggest that excessive caloric intake and energy imbalance are capable of inducing obesity and WAT inflammation independent of changes in the composition of the gut microbiome.
In addition to the role of microbiota in inflammation, considerable interest has recently been placed on the gut microbiome as a causative factor in the development of obesity-related insulin resistance. This stems from seminal findings that germ-free mice are leaner than conventionally colonized mice despite greater food intake, and colonizing germ-free mice with bacteria increased body fat and insulin resistance.11 Gut bacteria contribute to host energy storage by promoting digestion/extraction of ingested nutrients and uptake of CHOs and fatty acids.59 Furthermore, obesity has been shown to induce a shift in microbial phyla in obese compared to lean individuals, an effect which is mitigated by dietary restriction and weight loss.60 These microbial changes can occur rapidly in as little as 2 days following HFD consumption,14 appear to precede the development of chronic inflammation and insulin resistance,13 and are evident in genetic (ob/ob) models of obesity/insulin resistance.21 Combined, these data would suggest that the microbiome is a causative factor in the development of obesity and insulin resistance. However, it has been difficult to separate the effects of diet and/or host genetics in these models of obesity, as diet components or host immunity may be stand-alone factors that alter the gut microbiome. High fat intake itself, occurring even in the absence of obesity, can alter the composition of the gut microbiome,61 suggesting the shifts in microbial populations observed during HFD feeding may be a result of the high lipid/low fiber intake as opposed to obesity per se. In support of this notion, in this study, while HFD dramatically altered microbial communities in the cecums of WT mice compared to chow-fed lean animals, these changes were not evident in our spontaneously obese mice relative to lean littermates despite profound obesity and insulin resistance. Our results in spontaneously obese mice are in-line with others showing that pair feeding db/db mice normalized body weight without altering the gut microbiome from that of obese ad libitum fed db/db animals.22 Altogether, we provide evidence that obesity due to hyperphagia does not necessarily promote intestinal dysbiosis in male or female mice. As a result, a change in the microbial taxonomy may not be required for obesity and dysbiosis may in contrast be a better biomarker of diet than obesity.
Skeletal muscle is a key tissue contributing to whole-body glucose homeostasis and is highly sensitive to the detrimental effects of a high-fat environment.4,62 Indeed, reactive lipids have a strong association with the induction of skeletal muscle insulin resistance7,8 and mitochondria display a high sensitivity to lipid-induced ROS.63 Furthermore, genetic models decreasing the bioavailability of lipids (diacylglycerol acyltransferase (DGAT) overexpression)64 or promoting fatty acid oxidation within mitochondria (peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) overexpression)65 preserve insulin sensitivity. As result, mitochondria are a key organelle involved in energy metabolism/insulin sensitivity, and similar to WAT, impairments in skeletal muscle mitochondrial content/function have long been associated with the development of obesity and insulin resistance. In particular, impairments in submaximal mitochondrial bioenergetics are key events linked with HFD-induced insulin resistance, as mitochondrial respiration in the presence of physiological ADP concentrations, the sensitivity of mitochondria to ADP, and the ability of ADP to suppress mitochondrial ROS emissions are attenuated in HFD conditions and diabetes.9,66 However, in this study, the sensitivity of ADP to stimulate respiration and attenuate ROS was only affected by HFD and not the spontaneously obese phenotype; therefore, changes in submaximal mitochondrial respiration are not directly linked to obesity. In contrast (like eWAT), absolute mitochondrial ROS emission rates were increased in spontaneously obese mice regardless of diet, suggesting mitochondrial redox balance may be an important response inherent to obesity-induced insulin resistance. Thus, it seems that although changes in mitochondrial ADP kinetics can induce insulin resistance, these cellular responses are not required and may be dependent on P-CoA accumulation, a lipid moiety that has been shown to directly inhibit ANT-mediated mitochondrial ADP transport67 and impair mitochondrial ADP sensitivity.10 In support, previous findings have indicated that 7 days of bedrest68 and 14 days of limb immobilization69 impair insulin sensitivity in the absence of changes in skeletal muscle mitochondrial ADP sensitivity or the ability of ADP to suppress mitochondrial ROS. While speculative, the generation of 4HNE may be required for impairments in ADP sensitivity as ANT has been suggested to be susceptible to 4HNE-mediated redox changes.70,71 Moreover, HFD feeding9 and aging30 both increase 4HNE content while attenuating mitochondrial ADP responsiveness; cellular events which are absent following bed rest despite impaired insulin sensitivity.68 Together, these data suggest that different intracellular mechanisms may be involved in the induction of insulin resistance in the absence of HFD feeding and that HFD feeding may directly and independently influence mitochondrial ADP sensitivity.
Conclusion
We have characterized a spontaneous mutation in C57Bl/6J mice that resulted in profound obesity, whole-body insulin resistance, and adipose tissue hypertrophy, inflammation, and redox imbalance; effects which were blunted by pair feeding obese mice to that of lean controls. We provide evidence that skeletal muscle mitochondrial respiratory dysfunction and gut dysbiosis occur during HFD feeding but were not present in spontaneously obese mice. Our findings suggest gut dysbiosis and skeletal muscle mitochondrial respiratory dysfunction may be cellular events linked to HFD feeding and therefore not required for a certain level of obesity-induced insulin resistance, while in contrast, several aspects of adipose inflammation and redox imbalance appear intrinsic to the obese phenotype.
Supplementary Material
Acknowledgments
The graphical abstract was created using BioRender.com.
Supplementary Material
Supplementary material is available at the APS Function online.
Funding
This work was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) (G.P.H.; 400362) and a Canadian Institutes of Health Research (CIHR) Foundation grant (J.D.S.; FDN-154295), and Project grant (M.D.F.; PJT-148634) and New Investigator award (M.D.F.; MSH-141981). J.D.S. is a Canada Research Chair in Metabolism Inflammation. M.D.F. is also recipient of an Early Research Leadership Initiative grant from the Heart and Stroke Foundation of Canada and its partners and an Ontario Ministry of Research, Innovation and Science Early Researcher Award. H.L.P. is a recipient of an NSERC graduate scholarship.
Conflict of Interest Statement
All authors declare no conflict of interest.
Authors’ Contributions
H.L.P., K.P.F., J.D.S., and G.P.H. designed the study. H.L.P., K.P.F., S.Z., H.S.B., S.P., P.M.M., V.P.-B., C.O., D.J.P., M.D.F., J.D.S., and G.P.H. organized and performed experiments. All authors analyzed and interpreted the data. H.L.P., K.P.F., G.P.H., and J.D.S. drafted and edited the manuscript, and all authors approved the final version. G.P.H. and J.D.S. contributed equally as guarantors of the study.
H.L.P. and K.P. F. are primary authors who contributed equally.
J.D. S. and G.P.H. are senior authors who contributed equally.
References
- 1. Reilly SM, Saltiel AR. Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol 2017;13(11):633–643. [DOI] [PubMed] [Google Scholar]
- 2. Hotamisligil G. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 2010;140(6):900–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murdolo G, Piroddi M, Luchetti F, et al. Oxidative stress and lipid peroxidation by-products at the crossroad between adipose organ dysregulation and obesity-linked insulin resistance. Biochimie 2013;95(3):585–594. [DOI] [PubMed] [Google Scholar]
- 4. Cusi K. The role of adipose tissue and lipotoxicity in the pathogenesis of type 2 diabetes. Curr Diab Rep 2010;10(4):306–315. [DOI] [PubMed] [Google Scholar]
- 5. Paglialunga S, Ludzki A, Root-McCaig J, Holloway GP. In adipose tissue, increased mitochondrial emission of reactive oxygen species is important for short-term high-fat diet-induced insulin resistance in mice. Diabetologia 2015;58(5):1071–1080. [DOI] [PubMed] [Google Scholar]
- 6. Kusminski CM, Scherer PE. Mitochondrial dysfunction in white adipose tissue. Trends Endocrinol Metab 2012;23(9):435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Itani SI, Ruderman NB, Schmieder F, Boden G. Lipid-induced insulin resistance in human muscle is associated with changes in diacylglycerol, protein kinase C, and IkB-a. Diabetes 2002;51(7):2005–2011. [DOI] [PubMed] [Google Scholar]
- 8. Dubé JJ, Amati F, Stefanovic-Racic M, Toledo FGS, Sauers SE, Goodpaster BH. Exercise-induced alterations in intramyocellular lipids and insulin resistance: the athlete’s paradox revisited. Am J Physiol Endocrinol Metab 2008;294(5):E882–E888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miotto PM, LeBlanc PJ, Holloway GP. High fat diet causes mitochondrial dysfunction as a result of impaired ADP sensitivity. Diabetes 2018;67(11):2199–2205. [DOI] [PubMed] [Google Scholar]
- 10. Ludzki A, Paglialunga S, Smith BK, et al. Rapid repression of ADP transport by palmitoyl-CoA is attenuated by exercise training in humans: a potential mechanism to decrease oxidative stress and improve skeletal muscle insulin signaling. Diabetes 2015;64(8):2769–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004;101(44):15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008;3(4):213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Foley KP, Zlitni S, Denou E, et al. Long term but not short term exposure to obesity related microbiota promotes host insulin resistance. Nat Commun 2018;9(1):4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carmody RN, Gerber GK, Luevano JM, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015;17(1):72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ingalls AM, Dickie MM, Snell G. Obese, a new mutation in the house mouse. J Hered 1950;41(12):317–318. [DOI] [PubMed] [Google Scholar]
- 16. Hummel KP, Dickie MM, Coleman DL. Diabetes, a new mutation in the mouse. Science 1966;153(3740):1127–1128. [DOI] [PubMed] [Google Scholar]
- 17. Xu H, Barnes GT, Yang Q, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 2003;112(12):1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagpal R, Newman TM, Wang S, Jain S, Lovato JF, Yadav H. Obesity-linked gut microbiome dysbiosis associated with derangements in gut permeability and intestinal cellular homeostasis independent of diet. J Diabet Res 2018;2018:3462092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maurya R, Bhattacharya P, Dey R, Nakhasi HL. Leptin functions in infectious diseases. Front Immunol 2018;9:2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naylor C, Petri WAJ. Leptin regulation of immune responses. Trends Mol Med 2016;22(2):88–98. [DOI] [PubMed] [Google Scholar]
- 21. Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005;102(34):11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rajala MW, Patterson CM, Opp JS, Foltin SK, Young VB, Myers MG. Leptin acts independently of food intake to modulate gut microbial composition in male mice. Endocrinol 2014;155(3):748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beaudoin M, Snook LA, Arkell AM, Simpson JA, Holloway GP, Wright DC. Resveratrol supplementation improves white adipose tissue function in a depot-specific manner in Zucker diabetic fatty rats. Am J Physiol Regul Integr Comp Physiol 2013;305(5):R542–R551. [DOI] [PubMed] [Google Scholar]
- 24. Miotto PM, Holloway GP. In the absence of phosphate shuttling, exercise reveals the in vivo importance of creatine-independent mitochondrial ADP transport. Biochem J 2016;473(18):2831–2843. [DOI] [PubMed] [Google Scholar]
- 25. Fullerton MD, Galic S, Marcinko K, et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med 2013;19(12):1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steele R. Influences of glucose loading and of injected insulin on hepatic glucose output. Ann NY Acad Sci 1959;82(2):420–430. [DOI] [PubMed] [Google Scholar]
- 27. Smith BK, Jain SS, Rimbaud S, et al. FAT/CD36 is located on the outer mitochondrial membrane, upstream of long-chain acyl-CoA synthetase, and regulates palmitate oxidation. Biochem J 2011;437(1):125–134. [DOI] [PubMed] [Google Scholar]
- 28. Miotto PM, Steinberg GR, Holloway GP. Controlling skeletal muscle CPT-I malonyl-CoA sensitivity: the importance of AMPK-independent regulation of intermediate filaments during exercise. Biochem J 2017;474(4):557–569. [DOI] [PubMed] [Google Scholar]
- 29. Petrick HL, Holloway GP. High intensity exercise inhibits carnitine palmitoyltransferase-I sensitivity to L-carnitine. Biochem J 2019;476(3):547–558. [DOI] [PubMed] [Google Scholar]
- 30. Holloway GP, Holwerda AM, Miotto PM, Dirks ML, Verdijk LB, van Loon LJ. Age-associated impairments in mitochondrial ADP sensitivity contribute to redox stress in senescent human skeletal muscle. Cell Rep 2018;22(11):2837–2848. [DOI] [PubMed] [Google Scholar]
- 31. Cavallari JF, Anhê FF,, Foley KP, et al. Targeting macrophage scavenger receptor 1 promotes insulin resistance in obese male mice. Physiol Rep 2018;6(22):e13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Env Microbiol 2006;72(7):5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perry CGR, Kane DA, Lin C, et al. Inhibiting myosin-ATPase reveals a dynamic range of mitochondrial respiratory control in skeletal muscle. Biochem J 2011;437(2):215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Serc B Methdodol 1995;57(1):289–300. [Google Scholar]
- 36. Coleman D. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia 1978;14(3):141–148. [DOI] [PubMed] [Google Scholar]
- 37. Karolchik D, Hinrichs AS, Furey TS, et al. The UCSC table browser data retrieval tool. Nucl Acid Res 2004;32(9):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rosenbloom KR, Armstrong J, Barber GP, et al. The UCSC genome browser database: 2015 update. Nucl Acid Res 2015;43:670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kirkpatrick SL, Goldberg LR, Yazdani N, et al. Cytoplasmic FMR1-interacting protein 2 is a major genetic factor underlying binge eating. Biol Psychiatry 2018;81(9):757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Babbs RK, Ruan QT, Kelliher JC, et al. Cyfip1 haploinsufficiency increases compulsive-like behavior and modulates palatable food intake: Implications for Prader-Willi Syndrome. G3 2019;9(9):3009–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kumar V, Kim K, Joseph C, et al. C57BL/6N mutation in cytoplasmic FMR interacting protein 2 regulations cocaine response. Science 2013;342(6165):1508–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Freeman HC, Hugill A, Dear NT, Ashcroft FM, Cox RD. Deletion of nicotinamide nucleotide transhydrogenase: a new quantitive trait locus accounting for glucose interolance in C57BL/6J mice. Diabetes 2006;55(7):2153–2156. [DOI] [PubMed] [Google Scholar]
- 43. The European Polycystic Kidney Disease Consortium. The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell 1994;77(4):881–894. [DOI] [PubMed] [Google Scholar]
- 44. Dhana K, Braun KVE, Nano J, et al. An epigenome-wide association study of obesity-related traits. Am J Epidemiol 2018;187(8):1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hou W, Gupta S, Beauchamp M, Yuan L, Jerome-Majewska LA. Non-alcoholic fatty liver disease in mice with heterozygous mutation in TMED2. PLoS One 2017;12(8):e0182995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sasako T, Ohsugi M, Kubota N, et al. Hepatic Sdf2l1 controls feeding-induced ER stress and regulates metabolism. Nat Commun 2019;10(1):947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brunetta HS, Politis-Barber V, Petrick HL, et al. Nitrate attenuates HFD-induced glucose interolance in association with reduced epididymal adipose tissue inflammation and mitochondrial ROS emission. J Physiol 2020;598(16):3357–3371. [DOI] [PubMed] [Google Scholar]
- 48. Politis-Barber V, Brunetta HS, Paglialunga S, Petrick HL, Holloway GP. Long-term high-fat feeding exacerbates short-term increases in mitochondrial reactive oxygen species, without impairing respiratory function in adipose tissue. Am J Physiol Endocrinol Metab 2020;319(2):E376–E387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simoneau J-A, Veerkamp JH, Turcotte LP, Kelley DE. Markers of capacity to utilize fatty acids in human skeletal muscle: Relation to insulin resistance and obesity and effects of weight loss. FASEB J 1999;13(14):2051–2060. [DOI] [PubMed] [Google Scholar]
- 50. Johnson AMF, Costanzo A, Gareau MG, et al. High fat diet causes depletion of intestinal eosinophils associated with intestinal permeability. PLoS One 2015;10(4):e0122195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Becattini B, Zani F, Breasson L, et al. JNK1 ablation in mice confers long-term metabolic protection from diet-induced obesity at the cost of moderate skin oxidative damage. FASEB J 2016;30(9):3124–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim J, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res 2008;102(4):401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wilson-Fritch L, Nicoloro S, Chouinard M, et al. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. J Clin Invest 2004;114(9):1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Choo H, Kim J, Kwon O, et al. Mitochondria are impaired in the adipocytes of type 2 diabetic mice. Diabetologia 2006;49(2):784–791. [DOI] [PubMed] [Google Scholar]
- 55. Chi W, Dao D, Lau TC, et al. Bacterial peptidoglycan stimulates adipocyte lipolysis. PLoS One 2014;9(5):e97675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schertzer JD, Tamrakar AK, Magalhães JG, et al. NOD1 activators link innate immunity to insulin resistance. Diabetes 2011;60(9):2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chan KL, Tam TH, Boroumand P, et al. Circulating NOD1 activators and hematopoietic NOD1 contribute to metabolic inflammation and insulin resistance. Cell Rep 2017;18(10):2415–2426. [DOI] [PubMed] [Google Scholar]
- 58. Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet–induced obesity and diabetes in mice. Diabetes 2008;57(6):1470–1481. [DOI] [PubMed] [Google Scholar]
- 59. Musso G, Gambino R, Cassader M. Gut microbiota as a regulator of energy homeostasis and ectopic fat deposition: Mechanisms and implications for metabolic disorders. Curr Opin Lipidol 2010;21(1):76–83. [DOI] [PubMed] [Google Scholar]
- 60. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nat Commun 2006;444(7122):1022–1023. [DOI] [PubMed] [Google Scholar]
- 61. Hildebrandt MA, Hoffman C, Sherrill-Mix SA, et al. High fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 2009;137(5):1716–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Belfort R, Mandarino L, Kashyap S, et al. Dose-response effect of elevated plasma free fatty acid on insulin signaling. Diabetes 2005;54(6):1640–1648. [DOI] [PubMed] [Google Scholar]
- 63. Seifert EL, Estey C, Xuan JY, Harper ME. Electron transport chain-dependent and -independent mechanisms of mitochondrial H2O2 emission during long-chain fatty acid oxidation. J Biol Chem 2010;285(8):5748–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu L, Zhang Y, Chen N, Shi X, Tsang B, Yu Y. Upregulation of myocellular DGAT1 augments triglyceride synthesis in skeletal muscle and protects against fat-induced insulin resistance. J Clin Invest 2007;117(6):1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Benton CR, Nickerson JG, Lally J, et al. Modest PGC-1α overexpression in muscle in vivo is sufficient to increase insulin sensitivity and palmitate oxidation in subsarcolemmal, not intermyofibrillar, mitochondria. J Biol Chem 2008;283(7):4228–4240. [DOI] [PubMed] [Google Scholar]
- 66. Smith BK, Perry CGR, Herbst EAF, et al. Submaximal ADP-stimulated respiration is impaired in ZDF rats and recovered by resveratrol. J Physiol 2013;591(23):6089–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ciapaite J, Bakker SJL, Diamant M, et al. Metabolic control of mitochondrial properties by adenine nucleotide translocator determines palmitoyl-CoA effects: Implications for a mechanism linking obesity and type 2 diabetes. FEBS J 2006;273(23):5288–5302. [DOI] [PubMed] [Google Scholar]
- 68. Dirks ML, Miotto PM, Goossens GH, et al. Short-term bed rest-induced insulin resistance cannot be explained by increased mitochondrial H2O2 emission. J Physiol 2020;598(1):123–137. [DOI] [PubMed] [Google Scholar]
- 69. Miotto PM, McGlory C, Bahniwal R, Kamal M, Phillips SM, Holloway GP. Supplementation with dietary n-3 mitigates immobilization-induced reductions in skeletal muscle mitochondrial respiration in young women. FASEB J 2019;33(7):8232–8240. [DOI] [PubMed] [Google Scholar]
- 70. Azzu V, Parker N, Brand MD. High membrane potential promotes alkenal-induced mitochondrial uncoupling and influences adenine nucleotide translocase conformation. Biochem J 2008;413(2):323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yan LJ, Sohal RS. Mitochondrial adenine nucleotide translocase is modified oxidatively during aging. Proc Natl Acad Sci USA 1998;95(22):12896–12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.