Abstract
The generation of organismal form (i.e., morphogenesis) arises from forces produced at the cellular level. In animal cells, much of this force is produced by the actin cytoskeleton. Here, we review how mechanisms of actin-based force generation are deployed during animal morphogenesis to sculpt organs and organisms. Furthermore, we discuss how cytoskeletal forces are coupled through cell adhesions to propagate across tissues, and cases where cytoskeletal force or adhesion is patterned across a tissue to direct shape changes. Together, our review highlights a conceptual framework to reflect our current understanding of animal morphogenesis and provides perspectives on future opportunities of study.
Introduction:
A fundamental goal of developmental biology is to determine how the cells of the embryo generate the exquisite structures of the adult body. This process, morphogenesis, or “the creation of ordered form”, has intrigued scientists for centuries because it represents the ‘nuts and bolts’ mechanism of how our bodies, and the bodies of other animals, are constructed – how cells move to generate new structures, how embryonic tissues morph into organs. The more we learn about morphogenesis, the closer we come to knowing how our bodies are built, which is of vital importance for human health1. Congenital malformations resulting from defects in morphogenesis are the leading cause of infant mortality in the United States, and pose a significant risk for children of all ages2-5.
From its outset, the field of experimental embryology has been a quest to link the movements and shape changes of cells within the embryo to the generation of adult body form. Wilhelm His observed neural tube closure in the chick embryo and theorized that a mechanical process driven by the mitotic divisions and motility of the cells could be responsible for the folding of this tissue6. Wilhelm Roux expanded on this idea with his concept of ‘developmental mechanics’, in which he merged cellular descriptions of developmental processes with experimental manipulations to infer causal relationships and identify ‘active’ components of embryonic tissues7. The works of His and Roux, and the later thinking of D’arcy Thompson, marked a transition from the view that an external ‘vital force’ sculpted embryonic tissues, to the recognition that morphogenetic processes rely on quantifiable physical forces generated by embryonic cells8. While the underlying molecular mechanism was unknowable in the 19th century, we now know that the actin cytoskeleton, a meshwork of filamentous actin (F-actin) and various accessory proteins, including the molecular motor non-muscle myosin type II (Myo-II), is a key force generating machine that powers cell movements9. Furthermore, we know that these mechanisms are evolutionarily conserved and shared by all animals (and likely beyond)9-11.
In this review, we examine mechanisms by which animal cells change their shape using the actin cytoskeleton, and how cell shape changes are coordinated to restructure tissues. Because tissue morphogenesis results from forces generated from the molecular to tissue scales, it can be broken down into several components, including: (1) the molecular mechanisms of force generation and how they act locally within the cell to produce shape changes, and (2) the physical mechanisms that connect or adhere the cells of a tissue. The nature and patterns of intercellular linkages in a tissue must also be considered, including (a) the mechanical integration of force generating machines and adhesion, and (b) the spatial and temporal organization of the force generating machines and how they connect across a tissue.
Lastly, other cytoskeletal components, such as microtubules and intermediate filaments, also contribute significantly to morphogenetic processes. The interaction between these cytoskeletal systems and the actomyosin cytoskeleton is an exciting and active area of research. However, our main focus here is on morphogenetic mechanisms which utilize the actin cytoskeleton, because this seems to be the predominant mode of force generation in animal morphogenesis9. For a detailed overview of microtubule-dependent mechanisms of morphogenesis and intermediate filaments, we recommend other excellent reviews12-14,16.
Cytoskeletal mechanisms of cell shape change:
Cells utilize a variety of mechanisms to generate the force necessary to change shape, but these transformations can be placed in a simple framework in which cells push or pull in order to expand or contract, respectively. These transformations can be organized in one or two dimensions, such as modifying the shape of a single cell edge/interface or of a cellular surface, or in three dimensions, such as changes to multiple cellular surfaces or a change in volume. Here, we will provide a brief overview of actin cytoskeletal force-generating mechanisms associated with pushing and pulling forces, and then discuss how these mechanisms are deployed in a few ‘case studies’ of tissue remodeling during development.
Molecular mechanisms of pushing and pulling:
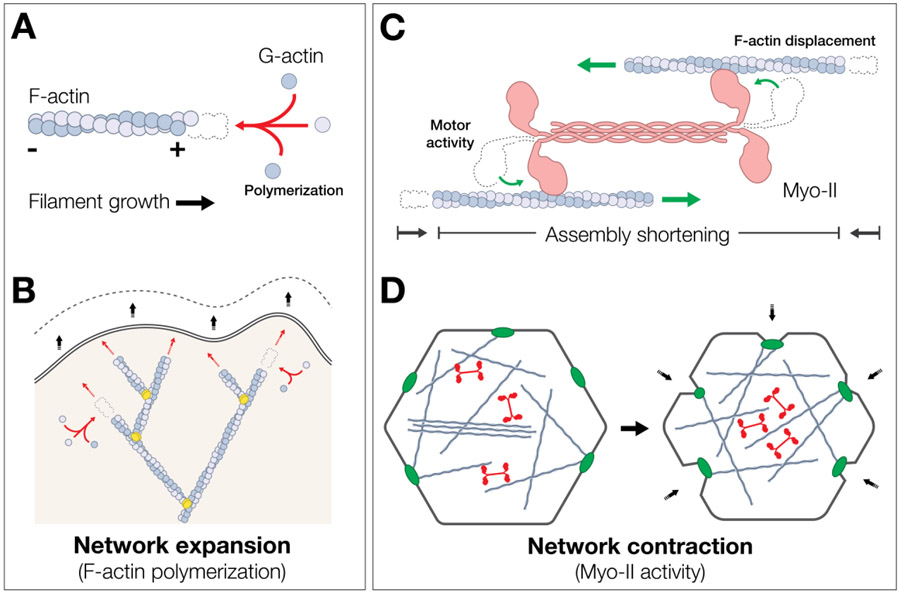
Pushing forces are primarily generated by F-actin network polymerization. F-actin is a semi-flexible polymer that forms from the controlled polymerization of monomeric actin subunits. Actin has an intrinsic polarity, such that monomers preferentially add to the growing (plus or barbed) end (Fig. 1A) (For a detailed review on mechanisms of actin polymerization and turnover; refer to15,17). When filament growth is oriented towards the plasma membrane, such as in the lamellipodium of a migrating cell, actin monomer addition can push the cell edge forward (Fig. 1B)15,18,19. This generates a modest pushing force at the molecular level, on the order of ~1 pN per polymerizing filament when measured in vitro (Table 1).
Figure 1: Molecular mechanisms of pushing and pulling:
(A) polymerization of F-actin by the preferential addition of actin monomers to the plus end creates a small pushing force (~1.3 pN). (B) In a branched actin network at the leading edge of the cell, the force of actin polymerization is sufficient to push the plasma membrane forward. (C) pulling forces are produced by the action of Myo-II minifilaments (pink) pulling against opposing F-actin filaments to generate tension (~4 pN). (D) polarized contraction of Myo-II (red) within an actomyosin network can pull cell junctions (green), or other points of connection to the membrane, inwards and contract a surface of a cell.
Table 1: magnitude of forces generated at molecular, cellular, and tissue scales.
values are approximations based on the literature, and provided to allow for ballpark comparison as opposed to a definitive reference (for a more detailed accounting of forces at the cellular level, please see Ananthakrishnan and Erlicher, 2007 197).
| Molecule | Stress type | Force (pN) | Citation(s): | |
|---|---|---|---|---|
| molecules | Myo-II | tension (pulling) | 3 – 5 | 25,26 |
| F- actin (polymerization) | compression (pushing) | 1 – 2 | 191,192 | |
| Within the cell | Force (pN μm−2) | |||
| Lamellipodium (keratinocytes and fibroblasts in culture) | compression | 1000 – 2500 | 34,35 | |
| Actomyosin cortex (embryos) | tension | 100 - 500 | 36,37 | |
| Actomyosin cortex (cell culture) | 600 – 800 | 38 | ||
| Traction at focal adhesion (Fibroblast) | 1000 – 2000 | 193 | ||
| Tug across a single cell-cell contact (adherens junction) | 500 - 1000 | 39,194 | ||
| Between cells in tissue culture | Cell Type | |||
| Epithelial | tension | 3000 – 4000 | 41 | |
| Epithelial (during collective migration) | 300 – 1000 | 195 | ||
| Mesenchymal | 1000 – 1500 | 41 | ||
| Between cells in embryos | Species | |||
| Drosophila | tension | 100 | 40 | |
| Zebrafish | 20 - 60 | 103 | ||
| Xenopus | 5 | 196 | ||
| Mouse | 1600 | 41 |
The core mechanism for producing pulling forces is mediated by F-actin and Myo-II. In its active, phosphorylated state, Myo-II oligomerizes into a bipolar mini-filament that can bind to opposing actin filaments and “walk” along them in an ATP-dependent manner (Fig. 1C)20-22. This myosin motor activity produces actomyosin contractility: movement of the myosin head domain slides F-actin filaments in opposing directions, shortening the total length of the actomyosin assembly (for details of actomyosin mechanics, see22-24). At the molecular level, a single myosin molecule generates a force on the order of several piconewtons (~3-5pN)25,26. In the cell, actomyosin networks can generate a pulling force that tugs on points of connection between the cytoskeleton and integral membrane proteins, such as those at cell junctions. Actomyosin assemblies can be organized in one dimension, such as in a bundle or fiber (e.g., stress fibers) that can shorten or bear stress27,28. It can also be organized in two dimensions (2D), as in the lamella of a migrating cell or the surface of an epithelial cell (i.e., apical, basal, lateral). On such 2D surfaces, actomyosin is organized as a meshwork that can draw the cell edges towards the cell center through contraction (Fig. 1D)19,29-32.
At the scale of cells and tissues, single-molecule forces can compound significantly to produce larger forces, but the extent to which this happens depends on tissue context. For instance, the force of actin polymerization is compounded within a branched F-actin network composed of many growing filaments, such that forces can be more than three orders of magnitude greater than a single filament across an entire protruding cell surface (~1 nN μm−2)33-35. Similarly, contractility of a larger actomyosin network containing many Myo-II mini-filaments (dozens to hundreds) generates tension in the cell cortex that is approximately two orders of magnitude greater than the force generated by a single Myo-II mini-filament (~100 - 800 pN μm−2)36-40. This tension is largely dependent on actomyosin contractility, because inhibition of myosin activity decreases cortical tension, and increasing contractility or inhibiting Arp2/3-mediated F-actin polymerization increases tension38. Tissue-level tension can be an order of magnitude greater than cellular tension (thousands of pN per square micron)39-41, but levels of tension can vary greatly between tissues composed of different cell types, and between model organisms: tension measurements range over three orders of magnitude when comparing between species (~5 – 4000 pN μm−2; Table 1). Thus, it is clear that many hundreds or even thousands of motors work together to generate force in a given cell. However, because these force generating mechanisms (actin polymerization and actomyosin) are oriented and exist within a network with complex architecture, the relationship between force and motor number is not simple.
F-actin polymerization, depolymerization, and contraction happen continuously in cells, even when cells are not actively changing shape. Homeostatic levels of actomyosin contractility and actin polymerization and depolymerization (actin turnover) are part of the normal state of the actomyosin cortex24, and occur regularly in the cortical F-actin cytoskeleton of multiple cell states (epithelial and mesenchymal), independent of developmental signaling or mechanical cues from cell-cell or cell-ECM adhesion42,43. There is inherent antagonism between contractility and turnover – maintaining cytoskeletal network cohesion requires a certain amount of actin turnover to prevent Myo-II contractility from fragmenting the actomyosin network, however, actin turnover can dissipate stress generated through Myo-II contractility44-48. Shape change occurs when these processes are up- or down-regulated locally outside of the range of this typical ‘resting state’ of normal cytoskeletal dynamics, such that a force imbalance results.
Actin-dependent force generation is evolutionarily ancient and broadly required across all animals and is also present in our unicellular eukaryotic relatives. Actins and myosins are not unique to animals; they are ancient, pan-eukaryotic protein families that are found in other multicellular eukaryotes, including plants49, and are particularly diverse in the Holozoa (animals plus our closes unicellular relatives)50. Actomyosin-based contractility mechanisms likely arose before the advent of animal multicellularity – they appear to underlie a collective contractile mechanism for colony morphogenesis in choanoflagellates, the unicellular protists most closely related to animals, suggesting they were present in our common ancestor11. The mechanisms for cell shape changes based on F-actin polymerization are also quite ancient. The Excavate protist, Naegleria, which shared a common ancestor with humans more than 1 billion years ago, exists predominantly in an amoeboid state that completely lacks a microtubule cytoskeleton, and relies on a branched actin network regulated by many of the same actin regulators found in our own cells for protrusive motility and phagocytosis51. If, as this evidence suggests, actin-based mechanisms of cell shape change are part of the common inheritance of animals, a key outstanding question is how the fantastic variation of forms seen between species are created from a conserved set of cytoskeletal effectors. It is perhaps the evolution of complexity in localizing these processes to different areas of the cell and their coordination between cells that has generated the vast complexity in tissue form from a common set of cytoskeletal components. In the next section, we will explain how localized contraction and expansion mediate morphogenesis. We will not explain the particulars of each individual system but focus on the broader geometrical framework for understanding how tissues change shape.
Localized contraction and expansion in epithelial morphogenesis:
Epithelial tissues are widespread in animals and have polarity52. Epithelial cells have multiple surfaces that can contract or expand, such as apical (lumen facing), basal (ECM facing), and lateral (between neighboring cells). Furthermore, epithelial cells can exhibit a vectorial polarity with respect to the epithelial plane, called planar cell polarity53. Because of the intrinsic polarity in the plasma membrane components and underlying actin cortex, proteins can be differentially localized and/or activated to induce distinct apical, basal, or lateral domain behaviors.
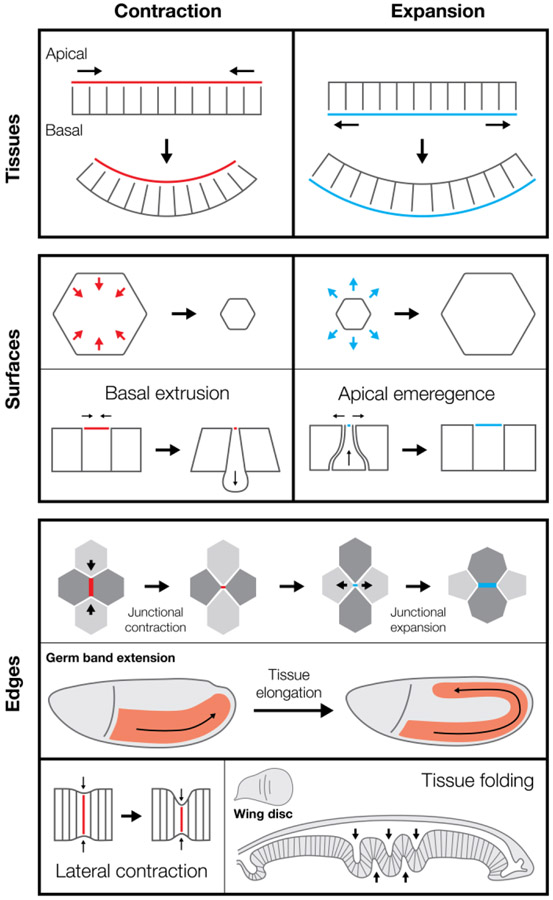
Collective contraction or expansion of apical or basal surfaces induces epithelial curvature. Apical and basal constriction converts epithelial cell shape from columnar to wedged, which changes local tissue curvature when happening in a population of cells (Figure 2, tissues). Apical constriction is implicated in mammalian intestinal crypt invagination54, vertebrate lens placode invagination55, Drosophila mesoderm and endoderm invagination56, and Drosophila salivary gland invagination57. Basal constriction induces the opposite curvature with respect to the apical-basal axis of an epithelium, generating the midbrain-hindbrain boundary folds in zebrafish58 and optic cup in zebrafish59. Expansion can also induce curvature. In the Drosophila wing disc, local basal cortex relaxation results in basal expansion that induces inward epithelial bending independently of apical constriction60. Thus, like differential expansion of one metal on a bimetallic strip changing strip curvature61, apical or basal constriction/expansion changes epithelial curvature by changing the length of one surface relative to the other. For this mechanism to be effective, the apical and basal surfaces must be mechanically coupled through lateral edges. In addition to apical constriction, contractility along lateral edges is necessary for gastrulation in Drosophila, and in ascidians62,64,87.
Figure 2: Shape change by contraction and expansion:
(tissues, top) polarized contraction or expansion on apical or basal surfaces can generate tissue folding. (surfaces, center) Shape change of individual cellular surfaces can also alter tissue geometry, as in basal extrusion or apical emergence. (edges, bottom) Coordinated shape change of individual edges or cellular interfaces can drive tissue movements, such as the convergent extension movements that elongates the Drosophila germ band (red). Contraction of lateral interfaces can also shorten edges between neighboring cells along the apical-basal axis, promoting tissue folding, as is seen in the Drosophila wing disc epithelium (right).
Constriction or expansion of individual epithelial cell surfaces also changes tissue architecture (Figure 2, surfaces). For example, unbalanced apical contractility (i.e., a cell is more contractile than its neighbors) can induce basal epithelial cell extrusion, such as during C. elegans gastrulation29,63, Drosophila neuroblast ingression65,66, and Drosophila dorsal closure67. In cell culture, epithelial cell extrusion results when apical tension relaxes relative to neighboring cells, suggesting that the force imbalance is critical for a cell to leave the epithelium68. Consistent with the force balance argument, cell-autonomous actin-based protrusion causes apical emergence (i.e. the opposite of extrusion), as is seen as multiciliated cells are added to an epithelium in Xenopus 69,70. Thus, apical/basal contraction or expansion have distinct effects depending on whether they occur at the single cell or population level.
Lateral surfaces between two cells (i.e., bicellular junctions) can contract or expand leading to cell rearrangements. When these rearrangements are planar cell polarized they lead to convergence and extension movements that elongate tissues, such as in the vertebrate neural plate71,72 and the Drosophila germband73 (Figure 2, edges). Planar polarized Myo-II activation can contract junctions71,74-77,80 – elevating tension along distinct junctional interfaces in the Drosophila germband78,79. Mediolaterally polarized basolateral protrusive activity in the cells at junctional vertices cooperates with junctional contractility to shrink these edges81. When a bicellular junction contracts to create a 4-cell interface, expansion must occur to create a new bicellular junction orthogonal to the ‘old’ junction. In cell culture, Rac-mediated protrusions expand junctional interfaces82 and actin polymerization-mediated pushing has been observed to counteract contractility and expand epithelial junctions in vivo83. In addition to pushing forces, contractile forces can promote junction growth by pulling at the poles of the new junction during Drosophila germ band extension84,85. Thus, convergence and extension movements illustrate how contractility and protrusive forces in a local cell neighborhood cooperate to elicit a complicated morphogenetic movement. Lateral edges can also contract along the apical-basal axis, which shortens cells and promotes tissue folding, as is in the Drosophila wing and leg discs60,86 (Figure 2, edges).
Indeed, morphogenetic processes are often quite complex, with multiple changes happening simultaneously within the cell or in different regions of the embryo. Contraction and expansion can coincide in space or time, sometimes with synergistic effects. For example, constriction on one side of the epithelium is often associated with expansion of the other side. In Drosophila mesoderm invagination, basal expansion closely follows apical constriction and is associated with the invagination87. Additionally, inhibition of contractility in neighboring ectoderm cells is important for gastrulation, as it allows these cells to soften, stretch, and move in order to accommodate the cell shape changes associated with mesoderm invagination88,89. During C. elegans gastrulation, cell ingression is assisted by neighboring cells pushing with Arp2/3-mediated actin protrusions90. Mitotic cell entry has also been shown to be associated with apical relaxation, which can promote apical constriction and invagination of neighboring contractile cells37,91. Thus, understanding morphogenetic events requires investigating both local properties of the tissue undergoing shape change, and the properties of the surrounding environment because both can contribute to movement.
Cell Adhesion:
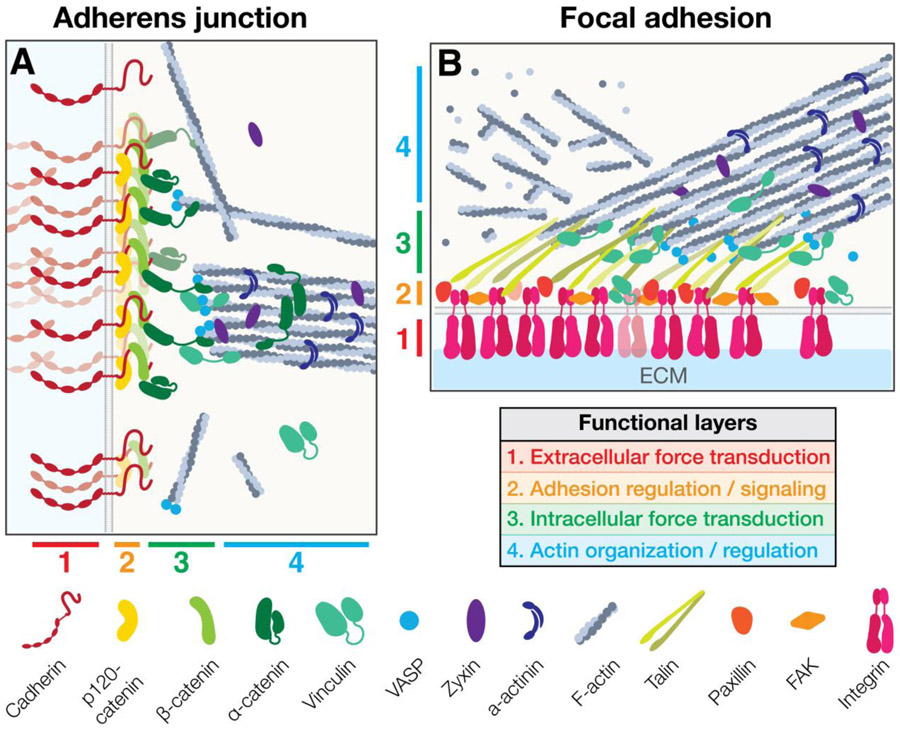
The formation of robust mechanical contacts to other cells and to the extracellular environment enables cytoskeletal force transmission to power tissue shape changes. The two main adhesive structures involved in actomyosin force propagation are adherens junctions, which mediate cell-cell adhesion, and focal adhesions, which mediate cell-ECM adhesion (Figure 3). The core adhesion receptors in the adherens junction are cadherins, which are composed of extracellular calcium-dependent adhesion domain repeats that homotypically interact with cadherins on adjacent cells92. In mammalian embryos, loss-of-function mutations in cadherins disrupt cohesion and compaction of the early embryo, and result in failures in later morphogenetic events, including gastrulation, neurulation, and organogenesis93-95. The principal adhesion receptors in focal adhesions are integrins, which are heterodimers containing an α- and β-subunit, that interact with a variety of ECM components via their large extracellular domains96,97. In addition to serving as critical signaling centers that are important for cell survival and cell polarity, cell-ECM adhesions are also crucial for morphogenesis, such as during zebrafish optic cup morphogenesis and mouse neural tube closure98-100.
Figure 3: Structure of junctions that link adhesion to the actomyosin cytoskeleton:
Adherens junction (A), and focal adhesions (B) share common organizational principles that facilitate dynamic attachment to the F-actin cytoskeleton, including stratification into a similar set of functionally distinct compartments that enable: (1) extracellular force transduction, (2) adhesion regulation, (3) intracellular force transduction, and (4) F-actin regulation.
There are physical constraints to the load-bearing capacity of adhesion molecules. Estimates of the maximum rupture forces of individual adhesion molecules are variable across the literature but are generally on the order of hundreds of piconewtons, compared to the ~5 pN generated by a single Myo-II mini-filament (Table 2). Adhesive strength varies between individual adhesion receptors, and therefore also varies between cell types expressing different complements of receptors. For example, the interaction between two E-cadherin molecules is, on average, an order of magnitude stronger than the N-cadherin:N-cadherin interaction (~200 vs. 40 pN)101. Similar differences exist between different combinations of α- and β-integrin subunits and ECM components (Table 2). Corresponding differences in adhesive capacity are seen between germ layers in the zebrafish embryo and could reflect either differences in adhesion proteins or different organization/concentration of the same adhesion protein (Table 3)102. These differences in adhesion strength are important, as they can alter the mechanical properties of a tissue103,104, and drive morphogenetic movements, such as cell sorting102,105. They also suggest that different tissues have inherently different tension-bearing capacities, which is useful to consider when comparing different morphogenetic processes.
Table 2: Examples of unbinding forces required to break or rupture bonds within mechanical connections between the actomyosin cytoskeleton and adhesion complexes.
(this represents only a partial list; for more extensive information, please see Weisel et al. 2003, and Rocha-Cusachs et al. 2012) 132,210.
| Molecular interaction | Stress type | Rupture force (pN) |
Citation: | |
|---|---|---|---|---|
| Adhesions | α5β1 integrin : fibronectin | Tension | 60 – 100 | 198,199 |
| α2β1 : collagen | 100 – 160 | 200,201 | ||
| E-cadherin : E-Cadherin | 70 - 200 | 101,202 | ||
| N-Cadherin : N-Cadherin | 30 – 40 | 101 | ||
| Adaptors | Talin : Actin | 2 | 203 | |
| α-catenin : Actin | 5-10 | 146 | ||
| Vinculin : Actin | 4-8 | 148 | ||
| Cytoskeleton | Actin : Myosin | 5 - 15 | 204 | |
| Actin : Actin (within F-actin) | 400 – 600 | 205,206 | ||
| Torsion | 100 – 300 | 205 | ||
| Compression | 0.16 – 4 | 207-209 |
Table 3: Rupture forces required to break adhesions in cells from different germ layers of a vertebrate embryo.
| Organism | Germ layer | Rupture force (pN μm−2) | Citation |
|---|---|---|---|
| Zebrafish | Endoderm | 400 – 600 | 102 |
| Mesoderm | 700 – 1500 | ||
| Ectoderm | 1500 – 3000 |
Adhesions can mature or strengthen due to structural changes in adhesion receptors in response to force or due to prolonged physical contact. Many protein binding interactions exhibit a ‘slip bond’ behavior, in which bond lifetimes decrease under tension. In contrast, many adhesion protein interactions are strengthened (i.e., become longer lived) under tension, a phenomenon called ‘catch bond’ behavior. The extracellular domains of E-cadherin can bind in two distinct conformations, X-dimers or strand-swapped dimers. These conformations differ in their response to tensile force: X-dimers behave like catch bonds, whereas strand-swap dimers behave like slip bonds106. The catch bond effect of the X-dimer is activated at forces greater than ~20 pN, which would require the coordinated activity of multiple Myo-II motors connected to a single cadherin. This suggests that cadherin heterodimer conformation responds to local tensile forces generated by actomyosin within a tissue, switching from X-dimer catch bonds that grip strongly under load, progressing to form more robust strand-swap dimers that have a high affinity in the absence of force. Integrins also exhibit catch bond behavior that is dependent on Myo-II contractility, as well as the stiffness of the underlying ECM. When tension is applied across the α5β1 integrin heterodimer, the extracellular integrin headpiece shifts to an activated conformation that can bind fibronectin for longer durations107,108. In addition, integrin adhesion force increases with substrate stiffness, as increased stiffness allows for interaction of the integrin headpiece with an additional synergy site adjacent to the primary binding site in fibronectin107.
Organization of adhesive junctions:
The strength and stability of adhesion also depends on the organization of cell junctions. Studies at the nanometer scale have shown that both cell-cell109-111 and cell-matrix112 adhesions are composed of smaller clusters of adhesion proteins, on the order of 50 – 100 nm, that represent a modular unit of organization that may be a general feature of cell adhesion receptors (for in-depth reviews on cadherin and integrin clustering, see113,114). Receptor clustering stabilizes junctions and can increase the force bearing capacity of adhesions115. While clustering is not necessary to establish adhesions, it locally increases adhesion receptor density, and, thereby, promotes adhesion formation116. Both cadherin and integrin heterodimer cluster formation are driven by multiple factors, including ligand binding, extracellular domain cis-interactions, and cytoplasmic interactions with cytoskeletal linking proteins like α-Catenin and Talin115,117-120. Integrin clusters have up to a 6-fold increase in tension threshold compared to individual integrin heterodimers117, and clustered E-cadherin also has a more robust mechanical connection to the actin cytoskeleton that can resist higher tensile forces115. Interestingly, not all adhesion molecules within an adhesion cluster are loaded equally – in a focal adhesion cluster, there is heterogeneity of force loading on integrin heterodimers, with a majority experiencing only weak forces (~1-10pN), and a subpopulation experiencing more substantial loads121. Similarly, within a cadherin cluster, only a subset of cadherins (~50%) have adhesive trans interactions and can propagate force between cells122. This implies that most adhesion receptors are experiencing approximately single-molecule levels of force, but more additive forces are channeled through a subset of adhesion molecules within a cluster.
Super-resolution microscopy has also demonstrated that in addition to the two-dimensional organization of adhesion complexes into clusters, there is stratification relative to the plasma membrane. This stratification has been thoroughly demonstrated in integrin adhesions123-125, and more recently for cadherin adhesions126. Together, these data suggest a general model for the organization of actin-linked adhesion complexes (Figure 3): (1) an extracellular adhesion layer, which has cis interactions governing cluster formation, and a mechanosensitive element that promotes stronger adhesions under force; (2) a membrane-proximal signaling layer, that also contributes to clustering, activation of adhesion, and regulation of turnover; (3) a force transduction and cytoskeletal adaptor layer, that contains a second mechanosensitive element to meter interactions with F-actin; and (4) an F-actin regulatory layer that contains a variety of actin-binding proteins that modulate the state of the cytoskeleton (bundling, polymerization, nucleation, etc.). Importantly, while layers 1 and 2 contain components that are distinct between cell-cell and cell-ECM adhesions, the layers 3 and 4 are partially shared by both, with many proteins (Vinculin in layer 3, and Zyxin, VASP, α-Actinin, etc. in layer 4) localizing to adherens junctions and focal adhesions123,126,129.
Coupling of force generation to adhesions:
Robust mechanical connections between adhesions and the cytoskeleton are essential for morphogenesis127,128,130. When adhesion is compromised, the point of failure is often not breakage of extracellular adhesions, but instead rupture of the connection between adhesion complexes and the actin cortex102. This link is comprised of a ‘core’ set of adaptor proteins capable of bridging the cytoplasmic tails of adhesion receptors and F-actin. For cadherin, these adaptor proteins are α- and β-Catenin131, and for integrin, a major adaptor protein is Talin132. Each of these linking proteins are broadly conserved133-135, and inhibition or mutation of them produce severe defects in adhesion96,131. In both adherens junctions and focal adhesions, there is a larger repertoire of interacting ‘adhesome’ proteins that play redundant, modulatory, or supporting roles in facilitating linkage, on the order of 125 adhesome proteins for cadherins, and 200 for integrins136-138. Characterizing the composition, organization, and force dependence of the network of proteins mediating cytoskeletal linkage to adhesions will continue to be an exciting area of research with implications for morphogenesis.
The linkage between the cytoskeleton and the adhesions is dynamic and can be actively regulated during development. A general model that has emerged is the concept of a molecular clutch, in which junctional adaptor proteins operate in an analogous manner to the clutch of a mechanical engine – when the clutch is ‘engaged’, pushing or pulling forces generated by the actin cytoskeleton are physically coupled to cell adhesions, resulting in force propagation to neighboring cells or the ECM139,140. This mechanical ‘clutch’ model for the regulated coupling between an adhesive complex and the actin cytoskeleton originated from detailed study of focal adhesions and actin filament movement, where it was shown that variable traction stress relates to the lamellipodial actin retrograde flow141,142. However, a regulated attachment between the actin cortex and adherens junctions was also shown for apically constricting cells in C. elegans gastrulation29.
Part of the underlying mechanism of the molecular clutch appears to be force-sensitive elements within the structure of adaptor proteins. For example, when force is applied to Talin, it induces a shift in structural conformation that exposes a binding site for the recruitment of Vinculin, which reinforces its interaction with F-actin118,124,143. A similar force-dependent conformational switch exists in α-Catenin, which controls vinculin recruitment at adherens junctions144,145. The α-Catenin : F-actin interaction operates as a catch bond, increasing in binding affinity under force, suggesting multiple mechanisms for mechanosensation and linkage adaptation may be operating simultaneously146. Indeed, many adaptor proteins have recently been shown to exhibit catch bonds, including Talin147 and Vinculin148, which suggests that mechanosensation may be a general property of adaptor proteins involved in cytoskeletal-junctional linkage. Some of these proteins, such as Vinculin and Afadin may enable strengthening of this connection in response to force149-151. In addition, actin turnover can strengthen this connection. In Drosophila gastrulation, F-actin turnover promotes stable connection of the contractile machinery to junctions during apical constriction152. In the Drosophila pupal wing epithelium, F-actin turnover also recruits additional factors, such as Canoe/Afadin, to strengthen the connection between actomyosin and the junction153.
The properties and mechanisms of cytoskeleton-junctional connectivity can vary between different regions of the cell, between tissues, and between organisms. Adhesome composition can vary with cell type and between adhesion molecule subtypes and can depend on tension. For example, the N-cadherin adhesome in cardiomyocytes is distinct from the E-Cadherin adhesome of epithelial cells154. Proteomics approaches have also revealed that as many as 400 proteins bind to integrin adhesions in a force dependent manner and may be involved in mechanotransduction155. Characterizing the composition of junctional adhesomes under different mechanical states will be critical to our understanding of force propagation in morphogenesis. The recruitment of additional adaptor proteins, such as Vinculin or p120-catenin, has been shown to modulate the total force transmitted between adhesion complexes and actomyosin156, as well as the stability of junctional complexes by regulating rates of adhesion receptor endocytosis157.
Macroscopic patterning of force generation in morphogenesis:
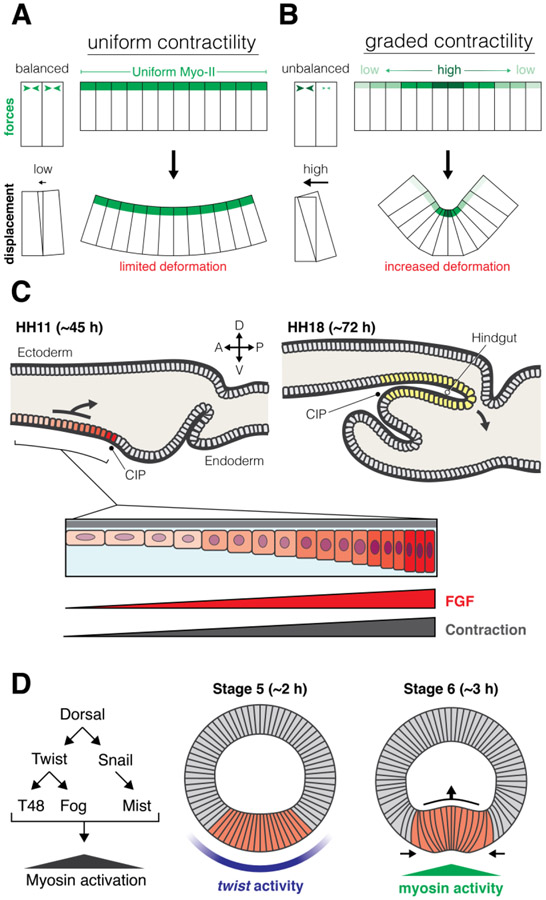
Because cytoskeletal systems are connected between cells, the macroscopic organization of cytoskeletal components, and how forces are patterned across an entire tissue is critical to morphogenesis. Similar to how morphogens set up a gradient of signaling activity to specify unique cell fate, signaling gradients can also specify unique force generating properties in cells across a tissue, which can promote morphogenesis. As opposed to cases of uniform contractility (Fig. 4A), spatial gradients of Myo-II activity (Fig. 4B) can create an imbalance of forces within a tissue that enhance tissue deformation. In addition, the actomyosin cytoskeleton can form structures that are interconnected across tissues (i.e., supracellular), sometimes including hundreds or even thousands of cells.
Figure 4: Spatial patterning of contractility:
(A) When contractility is uniform forces between cells are balanced, and deformation is limited. In contrast, when contractility is organized in a gradient (B), force imbalance can allow for increased deformation and tissue folding. (C) In chick hindgut development, an FGF signaling gradient patterns graded contractility and cell movement to produce tissue folding: endoderm anterior to the caudal intestinal portal (CIP) receives graded levels of FGF, which induces a corresponding gradient in contractility levels. (D) In Drosophila gastrulation, the dorsal-ventral patterning cascade (left) produces a gradient of Myo-II activity. A graded pattern of the transcription factor, twist, on the ventral side of the embryo (center) proceeds a gradient of Myo-II activity that powers invagination of the mesoderm (right, red).
Recent evidence suggests that force imbalances may be genetically patterned by the same signaling mechanisms involved in cell fate specification. Fibroblast growth factor (FGF) signaling stimulates cell motility and is required for axis elongation of the chick embryo158. FGF signaling also stimulates actomyosin contractility and apical constriction in the zebrafish lateral line and the chick otic vesicle159-161. In the developing chick hindgut, a gradient of FGF signaling establishes a contractility gradient that is required for the polarized collective cell migration that forms the hindgut (Fig. 4C)162. In this case, hindgut cells move from low tension to high tension, towards the source of highest FGF signal, and this movement, in conjunction with cell contraction, generates a fold in the endoderm that gives rise to the hindgut. Contractility gradients have also been observed in other systems, such as Drosophila gastrulation (Fig. 4D). During Drosophila gastrulation, the transcription factor twist controls mesoderm differentiation and its target genes that induce apical actomyosin activity are expressed in a gradient around the ventral midline163-165. This gene expression gradient results in a gradient of apical constriction and myosin activity that is highest at the midline of the invagination and decreases with distance from the midline56,163,164,166. Drosophila endoderm invagination also exhibits a wave of apical constriction, but in this case the spatial patterning results from mechanotransduction. In the endoderm, there is an integrin-mediated anchorage of cells at the primordium edge and a mechanical relay in which cells tug and stretch their neighbors inducing them to apically constrict167,168.
Actomyosin cytoskeletal structures can also be organized at larger spatial scales within the embryo. In many morphogenetically active tissues, junctional actomyosin is oriented and linked into supracellular ‘cables’ or meshworks stretching over many cell diameters that serve important functions in tissue-scale processes such as wound healing and morphogenesis169. F-actin cables or rings form in a variety of morphogenetic processes in diverse organisms, such as during mouse neural tube closure and eyelid closure170,171, chick amniogenesis and lens placode invagination 172,173, zebrafish epiboly and rhombomere boundary formation174-176, and Drosophila dorsal closure, compartment boundary formation, and germband extension79,177,178, which suggests that supracellular actomyosin cables may be an evolutionarily conserved mechanism of transmitting forces at tissue scales. However, there is some debate as to the function of supracellular actomyosin structures – do they contract or constrict structures, just at larger scales? Or do they have alternative functions, such as modulating tissue mechanical properties?
An intuitive model for the function of supracellular actomyosin networks is to generate tissue-scale contractility in order to constrict or shorten surfaces or edges. For example, in the ‘purse string’ model of dorsal closure in the Drosophila embryo, a supracellular actomyosin cable was thought to constrict in order to ‘zip’ the epidermis closed (Fig. 5A)177,179. However, other evidence has argued against this model, instead suggesting that the actin cable creates tension to straighten tissues edges, to allow for uniform tissue closure180-182. Actomyosin cables have been shown to play a similar role in straightening edges during the formation of compartment boundaries in multiple tissues within the Drosophila embryo – Myo-II contractility at these boundaries creates a local increase in tension that biases junctional rearrangements to suppress transient mixing between compartments (Fig. 5B)178,183,184. A similar phenomenon also occurs at compartment boundaries in the developing zebrafish brain, where actomyosin cables refine boundaries between rhombomeres176. In the early Drosophila embryo, planar polarized Myo-II is organized into a supracellular “ribbon” that functions as a ‘denoising’ mechanism to ensure morphogenetic precision in creating the cephalic fold that separates future head structures from the rest of the embryo (Fig. 5C)185. Similarly, in Drosophila gastrulation, actomyosin organized into a supracellular meshwork, as opposed to a cable, promotes directional tissue stiffening and robust folding during mesoderm invagination (Fig. 5D)186. In the Drosophila wing disc, multicellular actomyosin cables form in a tension-dependent manner in order to stiffen the tissue and prevent mechanical stress from degrading tissue integrity187. Taken together, these findings suggest that supracellular actomyosin structures may operate macroscopically to generate tissue stiffness and/or confer robustness, and do not always function to contract tissue surfaces.
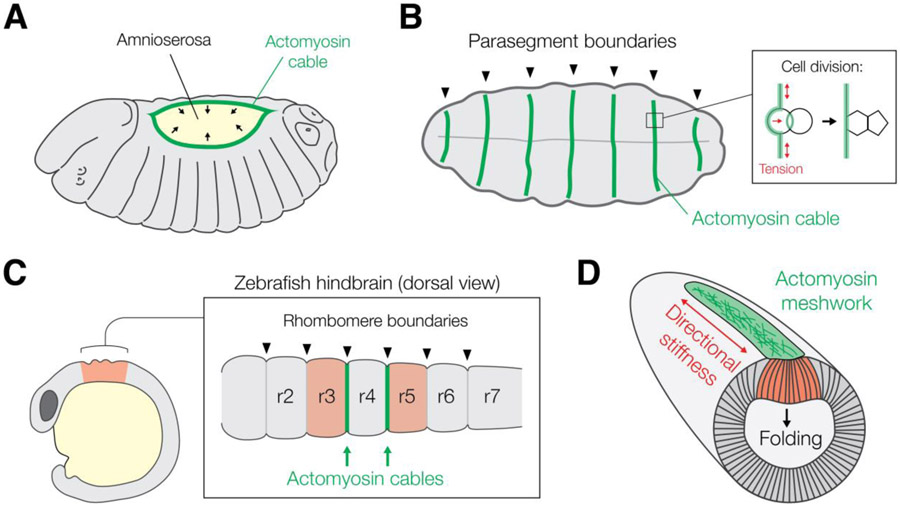
Figure 5: Supracellular actomyosin structures in embryos:
In Drosophila embryos, supracellular actomyosin cables (green) are seen during dorsal closure (A) and cases of compartment boundary formation, such as between parasegments in the early embryo (B). One function of actomyosin cables is to generate tension to straighten tissue edges and prevent mixing across boundaries (B, inset). (C) A similar phenomenon is seen in the Zebrafish hindbrain, where actomyosin cables promote boundary formation between the developing rhombomere brain segments and inhibit cell mixing. Supracellular actomyosin structures can also confer proof-reading or robustness to morphogenetic processes, such as the actomyosin meshwork present during Drosophila gastrulation (D). Here, the meshwork (green) creates directional tissue stiffness along the anterior-posterior axis, which resists bending along the long axis of the embryo, and supports folding and internalization of the mesoderm (red).
How do such extensive cytoskeletal structures form? In the Drosophila pupal dorsal thorax and wing disc, evidence suggests that supracellular actomyosin structures are, in part, organized by the adhesion apparatus, but it is unclear whether this is a general organizing principal across tissues27,104. During Drosophila germ band extension, contractile actomyosin flows are biased towards sites of increased adhesion, which amplifies asymmetries in junctional contractility and generates anisotropic cell deformation188. Alternatively, the actomyosin cytoskeleton itself could respond to extrinsic forces or constraints that are present in the tissue. Indeed it has been shown in numerous developmental contexts that actomyosin fibers are assembled or stabilized in response to stretch or tension 79,187. More work is necessary to determine how these structures form, and interpret their function and importance for tissue morphogenesis.
Conclusions and future directions:
Overall, it is clear that organisms liberally utilize cytoskeletal contraction and protrusion and a multitude of mechanisms to polarize force-generating machines to generate the wondrous diversity of organismal form. In addition to thinking about morphogenesis as a sum of cellular building blocks, there is clear importance to understanding how cells interconnect and transmit force across a tissue and how this is patterned at a macroscopic level. Understanding how cytoskeletal structures link between cells and how forces are transmitted from cell to cell in a tissue will be a fruitful area of further investigation. In addition to force generation, supracellular cytoskeletal structures are also likely to affect the mechanical state of tissues in a developing embryo, such as whether it behaves like a solid or liquid. Indeed, recent evidence suggests that tissue fluidity can be modulated by multicellular actomyosin patterns, and that this is important for gap closure in the context of wound healing and normal developmental processes189,190. Further studies linking supracellular actomyosin structures to tissue material properties in diverse systems will be necessary to unravel this connection.
It will be important to couple this macroscopic view of morphogenesis to higher resolution visualization of the machines that carry out force generation. Some outstanding questions are: 1) how is cytoskeletal filament alignment, which determines force polarity, controlled in a developing tissue? and 2) what is the importance of dynamic cytoskeletal behaviors, such as waves, pulses, and flows? We find the coupling of the macro-scale tissue analysis to the nano- and micro-scale organization within cells to be one of the most exciting future opportunities that will advance our understanding of morphogenesis.
Acknowledgements:
We would like to thank members of the Martin lab for helpful conversations and critical feedback during the writing of this manuscript. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health, grant number R01-GM105984 (to A.C.M.), and fellowship F32-GM134577 (to D.N.C.).
References:
- 1.Barresi MJF, and Gilbert SF (2020). Developmental biology. [Google Scholar]
- 2.Wallingford JB (2019). We Are All Developmental Biologists. Dev. Cell 50, 132–137. [DOI] [PubMed] [Google Scholar]
- 3.Khokha MK, Mitchell LE, and Wallingford JB (2017). White paper on the study of birth defects. Birth Defects Res. 109, 180–185. [DOI] [PubMed] [Google Scholar]
- 4.Murphy SL, Xu J, Kochanek KD, and Arias E (2017). Mortality in the United States, 2017 Key findings Data from the National Vital Statistics System. [Google Scholar]
- 5.Khokha MK, Liu KJ, and Wallingford JB (2020). Challenges and opportunities at the interface of birth defects, human genetics and developmental biology. Development 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.His W (1874). Unsere Körperform und das physiologische Problem ihrer Entstehung: Briefe an einen befreundeten Naturforscher (FCW Vogel). [Google Scholar]
- 7.Roux W (1888). Beiträge zur Entwickelungsmechanik des Embryo. Arch. für Pathol. Anat. und Physiol. und für Klin. Med 114, 246–291. [Google Scholar]
- 8.Thompson DW (1917). On growth and form (Cambridge university press; ). [Google Scholar]
- 9.Quintin S, Gally C, and Labouesse M (2008). Epithelial morphogenesis in embryos: asymmetries, motors and brakes. Trends Genet. 24, 221–230. [DOI] [PubMed] [Google Scholar]
- 10.Pukhlyakova E, Aman AJ, Elsayad K, and Technau U (2018). β-Catenin-dependent mechanotransduction dates back to the common ancestor of Cnidaria and Bilateria. Proc. Natl. Acad. Sci. U. S. A 115, 6231–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunet T, Larson BT, Linden TA, Vermeij MJA, McDonald K, and King N (2019). Light-regulated collective contractility in a multicellular choanoflagellate. Science 366, 326–334. [DOI] [PubMed] [Google Scholar]
- 12.Röper K (2020). Microtubules enter centre stage for morphogenesis. Philos. Trans. R. Soc. B Biol. Sci 375, 20190557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matis M (2020). The Mechanical Role of Microtubules in Tissue Remodeling. BioEssays 42, 1900244. [DOI] [PubMed] [Google Scholar]
- 14.Pan X, Hobbs RP, and Coulombe PA (2013). The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr. Opin. Cell Biol 25, 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollard TD, and Borisy GG (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- 16.Sanghvi-Shah R, and Weber GF (2017). Intermediate filaments at the junction of mechanotransduction, migration, and development. Front. Cell Dev. Biol 5, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siton-Mendelson O, and Bernheim-Groswasser A (2017). Functional Actin Networks under Construction: The Cooperative Action of Actin Nucleation and Elongation Factors. Trends Biochem. Sci 42, 414–430. [DOI] [PubMed] [Google Scholar]
- 18.Koestler SA, Auinger S, Vinzenz M, Rottner K, and Small JV (2008). Differentially oriented populations of actin filaments generated in lamellipodia collaborate in pushing and pausing at the cell front. Nat. Cell Biol 10, 306–313. [DOI] [PubMed] [Google Scholar]
- 19.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, and Danuser G (2004). Two distinct actin networks drive the protrusion of migrating cells. Science (80-. ) 305, 1782–1786. [DOI] [PubMed] [Google Scholar]
- 20.Vasquez CG, and Martin AC (2016). Force transmission in epithelial tissues. Dev. Dyn 245, 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vicente-Manzanares M, Ma X, Adelstein RS, and Horwitz AR (2009). Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol 10, 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murrell M, Oakes PW, Lenz M, and Gardel ML (2015). Forcing cells into shape: The mechanics of actomyosin contractility. Nat. Rev. Mol. Cell Biol 16, 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stricker J, Falzone T, and Gardel ML (2010). Mechanics of the F-actin cytoskeleton. J. Biomech 43, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salbreux G, Charras G, and Paluch E (2012). Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol. 22, 536–545. [DOI] [PubMed] [Google Scholar]
- 25.Finer JT, Simmons RM, and Spudich JA (1994). Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature 368, 113–119. [DOI] [PubMed] [Google Scholar]
- 26.Molloy JE, Burns JE, Kendrick-Jones B, Tregear RT, and White DCS (1995). Movement and force produced by a single myosin head. Nature 378, 209–212. [DOI] [PubMed] [Google Scholar]
- 27.López-Gay JM, Nunley H, Spencer M, di Pietro F, Guirao B, Bosveld F, Markova O, Gaugue I, Pelletier S, Lubensky DK, et al. (2020). Apical stress fibers enable a scaling between cell mechanical response and area in epithelial tissue. Science 370. [DOI] [PubMed] [Google Scholar]
- 28.Lee S, Kassianidou E, and Kumar S (2018). Actomyosin stress fiber subtypes have unique viscoelastic properties and roles in tension generation. Mol. Biol. Cell 29, 1992–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roh-Johnson M, Shemer G, Higgins CD, McClellan JH, Werts AD, Tulu US, Gao L, Betzig E, Kiehart DP, and Goldstein B (2012). Triggering a cell shape change by exploiting preexisting actomyosin contractions. Science (80-. ) 335, 1232–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burnette DT, Manley S, Sengupta P, Sougrat R, Davidson MW, Kachar B, and Lippincott-Schwartz J (2011). A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Biol 13, 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason FM, Tworoger M, and Martin AC (2013). Apical domain polarization localizes actin-myosin activity to drive ratchet-like apical constriction. Nat. Cell Biol 15, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanchard GB, Murugesu S, Adams RJ, Martinez-Arias A, and Gorfinkiel N (2010). Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development 137, 2743–2752. [DOI] [PubMed] [Google Scholar]
- 33.Dmitrieff S, and Nédélec F (2016). Amplification of actin polymerization forces. J. Cell Biol 212, 763–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prass M, Jacobson K, Mogilner A, and Radmacher M (2006). Direct measurement of the lamellipodial protrusive force in a migrating cell. J. Cell Biol 174, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abraham VC, Krishnamurthi V, Lansing Taylor D, and Lanni F (1999). The actin-based nanomachine at the leading edge of migrating cells. Biophys. J 77, 1721–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maître JL, Niwayama R, Turlier H, Nedelec F, and Hiiragi T (2015). Pulsatile cell-autonomous contractility drives compaction in the mouse embryo. Nat. Cell Biol 17, 849–855. [DOI] [PubMed] [Google Scholar]
- 37.Godard BG, Dumollard R, Munro E, Chenevert J, Hebras C, McDougall A, and Heisenberg C-P (2020). Apical Relaxation during Mitotic Rounding Promotes Tension-Oriented Cell Division. Dev. Cell [DOI] [PubMed] [Google Scholar]
- 38.Cartagena-Rivera AX, Logue JS, Waterman CM, and Chadwick RS (2016). Actomyosin Cortical Mechanical Properties in Nonadherent Cells Determined by Atomic Force Microscopy. Biophys. J 110, 2528–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, and Chen CS (2010). Mechanical tugging force regulates the size of cell-cell junctions. Proc. Natl. Acad. Sci. U. S. A 107, 9944–9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bambardekar K, Clément R, Blanc O, Chardès C, and Lenne PF (2015). Direct laser manipulation reveals the mechanics of cell contacts in vivo. Proc. Natl. Acad. Sci. U. S. A 112, 1416–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campàs O, Mammoto T, Hasso S, Sperling RA, O’connell D, Bischof AG, Maas R, Weitz DA, Mahadevan L, and Ingber DE (2014). Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods 11, 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baird MA, Billington N, Wang A, Adelstein RS, Sellers JR, Fischer RS, and Waterman CM (2017). Local pulsatile contractions are an intrinsic property of the myosin 2A motor in the cortical cytoskeleton of adherent cells. Mol. Biol. Cell 28, 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelkar M, Bohec P, and Charras G (2020). Mechanics of the cellular actin cortex: From signalling to shape change. Curr. Opin. Cell Biol 66, 69–78. [DOI] [PubMed] [Google Scholar]
- 44.Mak M, Zaman MH, Kamm RD, and Kim T (2016). Interplay of active processes modulates tension and drives phase transition in self-renewing, motor-driven cytoskeletal networks. Nat. Commun 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McFadden WM, McCall PM, Gardel ML, and Munro EM (2017). Filament turnover tunes both force generation and dissipation to control long-range flows in a model actomyosin cortex. PLoS Comput. Biol 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clément R, Dehapiot B, Collinet C, Lecuit T, and Lenne PF (2017). Viscoelastic Dissipation Stabilizes Cell Shape Changes during Tissue Morphogenesis. Curr. Biol 27, 3132–3142.e4. [DOI] [PubMed] [Google Scholar]
- 47.Chen Q, and Pollard TD (2011). Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J. Cell Biol 195, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verkhovsky AB, Svitkina TM, and Borisy GG (1997). Polarity sorting of actin filaments in cytochalasin-treated fibroblasts. J. Cell Sci 110 ( Pt 1, 1693–1704. [DOI] [PubMed] [Google Scholar]
- 49.Madison SL, and Nebenführ A (2013). Understanding myosin functions in plants: Are we there yet? Curr. Opin. Plant Biol 16, 710–717. [DOI] [PubMed] [Google Scholar]
- 50.Sebé-Pedrós A, Grau-Bové X, Richards TA, and Ruiz-Trillo I (2014). Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol. Evol 6, 290–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Velle KB, and Fritz-Laylin LK (2020). Conserved actin machinery drives microtubule-independent motility and phagocytosis in Naegleria. J. Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Boulan E, and Macara IG (2014). Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol 15, 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butler MT, and Wallingford JB (2017). Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol 18, 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sumigray KD, Terwilliger M, and Lechler T (2018). Morphogenesis and Compartmentalization of the Intestinal Crypt. Dev. Cell 45, 183–197.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plageman TF, Chauhan BK, Yang C, Jaudon F, Shang X, Zheng Y, Lou M, Debant A, Hildebrand JD, and Lang RA (2011). A trio-rhoA-shroom3 pathway is required for apical constriction and epithelial invagination. Development 138, 5177–5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sweeton D, Parks S, Costa M, and Wieschaus E (1991). Gastrulation in Drosophila: the formation of the ventral furrow and posterior midgut invaginations. Development 112. [DOI] [PubMed] [Google Scholar]
- 57.Myat MM, and Andrew DJ (2000). Fork head prevents apoptosis and promotes cell shape change during formation of the Drosophila salivary glands. Development 127. [DOI] [PubMed] [Google Scholar]
- 58.Gutzman JH, Graeden E, Brachmann I, Yamazoe S, Chen JK, and Sive H (2018). Basal constriction during midbrain–hindbrain boundary morphogenesis is mediated by Wnt5b and focal adhesion kinase. Biol. Open 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sidhaye J, and Norden C (2017). Concerted action of neuroepithelial basal shrinkage and active epithelial migration ensures efficient optic cup morphogenesis. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sui L, Alt S, Weigert M, Dye N, Eaton S, Jug F, Myers EW, Jülicher F, Salbreux G, and Dahmann C (2018). Differential lateral and basal tension drive folding of Drosophila wing discs through two distinct mechanisms. Nat. Commun 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Timoshenko S (1925). Analysis of Bi-Metal Thermostats. J. Opt. Soc. Am 11, 233. [Google Scholar]
- 62.Sherrard K, Robin F, Lemaire P, and Munro E (2010). Sequential activation of apical and basolateral contractility drives ascidian endoderm invagination. Curr. Biol 20, 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JY, Marston DJ, Walston T, Hardin J, Halberstadt A, and Goldstein B (2006). Wnt/Frizzled Signaling Controls C. elegans Gastrulation by Activating Actomyosin Contractility. Curr. Biol 16, 1986–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gracia M, Theis S, Proag A, Gay G, Benassayag C, and Suzanne M (2019). Mechanical impact of epithelial–mesenchymal transition on epithelial morphogenesis in Drosophila. Nat. Commun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.An Y, Xue G, Shaobo Y, Mingxi D, Zhou X, Yu W, Ishibashi T, Zhang L, and Yan Y (2017). Apical constriction is driven by a pulsatile apical myosin network in delaminating Drosophila neuroblasts. Dev. 144, 2153–2164. [DOI] [PubMed] [Google Scholar]
- 66.Simões S, Oh Y, Wang MFZ, Fernandez-Gonzalez R, and Tepass U (2017). Myosin II promotes the anisotropic loss of the apical domain during Drosophila neuroblast ingression. J. Cell Biol 216, 1387–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toyama Y, Peralta XG, Wells AR, Kiehart DP, and Edwards GS (2008). Apoptotic force and tissue dynamics during Drosophila embryogenesis. Science (80-. ) 321, 1683–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teo JL, Tomatis VM, Coburn L, Lagendijk AK, Schouwenaar I-M, Budnar S, Hall TE, Verma S, McLachlan RW, Hogan BM, et al. (2020). Src kinases relax adherens junctions between the neighbors of apoptotic cells to permit apical extrusion. Mol. Biol. Cell, mbc.E20-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sedzinski J, Hannezo E, Tu F, Biro M, and Wallingford JB (2017). RhoA regulates actin network dynamics during apical surface emergence in multiciliated epithelial cells. J. Cell Sci 130, 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sedzinski J, Hannezo E, Tu F, Biro M, and Wallingford JB (2016). Emergence of an Apical Epithelial Cell Surface In Vivo. Dev. Cell 36, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams M, Yen W, Lu X, and Sutherland A (2014). Distinct Apical and Basolateral Mechanisms Drive Planar Cell Polarity-Dependent Convergent Extension of the Mouse Neural Plate. Dev. Cell 29, 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ybot-Gonzalez P, Savery D, Gerrelli D, Signore M, Mitchell CE, Faux CH, Greene NDE, and Copp AJ (2007). Convergent extension, planar-cell-polarity signaling and initiation of mouse neural tube closure. Development 134, 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Irvine KD, and Wieschaus E (1994). Cell intercalation during Drosophila germband extension and its regulation by pair-rule segmentation genes. Development 120, 827–841. [DOI] [PubMed] [Google Scholar]
- 74.Shindo A, and Wallingford JB (2014). PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science (80-. ) 343, 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Butler MT, and Wallingford JB (2018). Spatial and temporal analysis of PCP protein dynamics during neural tube closure. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zallen JA, and Wieschaus E (2004). Patterned gene expression directs bipolar planar polarity in Drosophila. Dev. Cell 6, 343–355. [DOI] [PubMed] [Google Scholar]
- 77.Bertet C, Sulak L, and Lecuit T (2004). Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature 429, 667–671. [DOI] [PubMed] [Google Scholar]
- 78.Rauzi M, Verant P, Lecuit T, and Lenne PF (2008). Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nat. Cell Biol 10, 1401–1410. [DOI] [PubMed] [Google Scholar]
- 79.Fernandez-Gonzalez R, de M. Simoes S, Röper JC, Eaton S, and Zallen JA (2009). Myosin II Dynamics Are Regulated by Tension in Intercalating Cells. Dev. Cell 17, 736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blankenship JT, Backovic ST, Sanny JSSP, Weitz O, and Zallen JA (2006). Multicellular Rosette Formation Links Planar Cell Polarity to Tissue Morphogenesis. Dev. Cell 11, 459–470. [DOI] [PubMed] [Google Scholar]
- 81.Huebner RJ, and Wallingford JB (2018). Coming to Consensus: A Unifying Model Emerges for Convergent Extension. Dev. Cell 46, 389–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamada S, and Nelson WJ (2007). Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J. Cell Biol 178, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Del Signore SJ, Cilla R, and Hatini V (2018). The WAVE Regulatory Complex and Branched F-Actin Counterbalance Contractile Force to Control Cell Shape and Packing in the Drosophila Eye. Dev. Cell 44, 471–483.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Collinet C, Rauzi M, Lenne PF, and Lecuit T (2015). Local and tissue-scale forces drive oriented junction growth during tissue extension. Nat. Cell Biol 17, 1247–1258. [DOI] [PubMed] [Google Scholar]
- 85.Yu JC, and Fernandez-Gonzalez R (2016). Local mechanical forces promote polarized junctional assembly and axis elongation in Drosophila. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Monier B, Gettings M, Gay G, Mangeat T, Schott S, Guarner A, and Suzanne M (2015). Apico-basal forces exerted by apoptotic cells drive epithelium folding. Nature 518, 245–248. [DOI] [PubMed] [Google Scholar]
- 87.Polyakov O, He B, Swan M, Shaevitz JW, Kaschube M, and Wieschaus E (2014). Passive mechanical forces control cell-shape change during drosophila ventral furrow formation. Biophys. J 107, 998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perez-Mockus G, Mazouni K, Roca V, Corradi G, Conte V, and Schweisguth F (2017). Spatial regulation of contractility by Neuralized and Bearded during furrow invagination in Drosophila. Nat. Commun 8, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rauzi M, Krzic U, Saunders TE, Krajnc M, Ziherl P, Hufnagel L, and Leptin M (2015). Embryo-scale tissue mechanics during Drosophila gastrulation movements. Nat. Commun 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roh-Johnson M, and Goldstein B (2009). In vivo roles for Arp2/3 in cortical actin organization during C. elegans gastrulation. J. Cell Sci 122, 3983–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ko CS, Kalakuntla P, and Martin AC (2020). Apical Constriction Reversal upon Mitotic Entry Underlies Different Morphogenetic Outcomes of Cell Division. Mol. Biol. Cell 31, 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shapiro L, and Weis WI (2009). Structure and biochemistry of cadherins and catenins. Cold Spring Harb. Perspect. Biol 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Riethmacher D, Brinkmanni V, and Birchmeier C (1995). A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc. Natl. Acad. Sci. U. S. A 92, 855–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bondow BJ, Faber ML, Wojta KJ, Walker EM, and Battle MA (2012). E-cadherin is required for intestinal morphogenesis in the mouse. Dev. Biol 371, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, and Hynes RO (1997). Developmental Defects in Mouse Embryos Lacking N-Cadherin. [DOI] [PubMed] [Google Scholar]
- 96.Hynes RO (2002). Integrins: Bidirectional, allosteric signaling machines. Cell 110, 673–687. [DOI] [PubMed] [Google Scholar]
- 97.Humphries JD, Byron A, and Humphries MJ (2006). Integrin ligands at a glance. J. Cell Sci 119, 3901–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bryan CD, Casey MA, Pfeiffer RL, Jones BW, and Kwan KM (2020). Optic cup morphogenesis requires neural crest-mediated basement membrane assembly. Dev. 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Molè MA, Galea GL, Rolo A, Weberling A, Nychyk O, De Castro SC, Savery D, Fässler R, Ybot-González P, Greene NDE, et al. (2020). Integrin-Mediated Focal Anchorage Drives Epithelial Zippering during Mouse Neural Tube Closure. Dev. Cell 52, 321–334.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Berrier AL, and Yamada KM (2007). Cell-matrix adhesion. J. Cell. Physiol 213, 565–573. [DOI] [PubMed] [Google Scholar]
- 101.Panorchan P, Thompson MS, Davis KJ, Tseng Y, Konstantopoulos K, and Wirtz D (2006). Single-molecule analysis of cadherin-mediated cell-cell adhesion. J. Cell Sci 119, 66–74. [DOI] [PubMed] [Google Scholar]
- 102.Maître JL, Berthoumieux H, Krens SFG, Salbreux G, Jülicher F, Paluch E, and Heisenberg CP (2012). Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science (80-. ) 338, 253–256. [DOI] [PubMed] [Google Scholar]
- 103.Mongera A, Rowghanian P, Gustafson HJ, Shelton E, Kealhofer DA, Carn EK, Serwane F, Lucio AA, Giammona J, and Campàs O (2018). A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 561, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bosveld F, Bonnet I, Guirao B, Tlili S, Wang Z, Petitalot A, Marchand R, Bardet PL, Marcq P, Graner F, et al. (2012). Mechanical control of morphogenesis by fat/dachsous/four-jointed planar cell polarity pathway. Science (80-. ) 336, 724–727. [DOI] [PubMed] [Google Scholar]
- 105.Y-C Tsai T, Sikora M, Xia P, Colak-Champollion T, Knaut H, Heisenberg C-P, and Megason SG An adhesion code ensures robust pattern formation during tissue morphogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rakshit S, Zhang Y, Manibog K, Shafraz O, and Sivasankar S (2012). Ideal, catch, and slip bonds in cadherin adhesion. Proc. Natl. Acad. Sci. U. S. A 109, 18815–18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Friedland JC, Lee MH, and Boettiger D (2009). Mechanically activated integrin switch controls α5β 1 function. Science (80-. ) 323, 642–644. [DOI] [PubMed] [Google Scholar]
- 108.Kong F, García AJ, Mould AP, Humphries MJ, and Zhu C (2009). Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol 185, 1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu Y, Kanchanawong P, and Zaidel-Bar R (2015). Actin-Delimited Adhesion-Independent Clustering of E-Cadherin Forms the Nanoscale Building Blocks of Adherens Junctions. Dev. Cell 32, 139–154. [DOI] [PubMed] [Google Scholar]
- 110.Indra I, Choi J, Chen CS, Troyanovsky RB, Shapiro L, Honig B, and Troyanovsky SM (2018). Spatial and temporal organization of cadherin in punctate adherens junctions. Proc. Natl. Acad. Sci. U. S. A 115, E4406–E4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Truong Quang BA, Mani M, Markova O, Lecuit T, and Lenne PF (2013). Principles of E-cadherin supramolecular organization in vivo. Curr. Biol 23, 2197–2207. [DOI] [PubMed] [Google Scholar]
- 112.Changede R, Xu X, Margadant F, and Sheetz MP (2015). Nascent Integrin Adhesions Form on All Matrix Rigidities after Integrin Activation. Dev. Cell 35, 614–621. [DOI] [PubMed] [Google Scholar]
- 113.Yap AS, Gomez GA, and Parton RG (2015). Adherens Junctions Revisualized: Organizing Cadherins as Nanoassemblies. Dev. Cell 35, 12–20. [DOI] [PubMed] [Google Scholar]
- 114.Changede R, and Sheetz M (2017). Integrin and cadherin clusters: A robust way to organize adhesions for cell mechanics. BioEssays 39, e201600123. [DOI] [PubMed] [Google Scholar]
- 115.Strale PO, Duchesne L, Peyret G, Montel L, Nguyen T, Png E, Tampé R, Troyanovsky S, Hénon S, Ladoux B, et al. (2015). The formation of ordered nanoclusters controls cadherin anchoring to actin and cell-cell contact fluidity. J. Cell Biol 210, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang Y, Sivasankar S, Nelson WJ, and Chu S (2009). Resolving cadherin interactions and binding cooperativity at the single-molecule level. Proc. Natl. Acad. Sci. U. S. A 106, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Roca-Cusachs P, Gauthier NC, Del Rio A, and Sheetz MP (2009). Clustering of α5β1 integrins determines adhesion strength whereas αvβ3 and talin enable mechanotransduction. Proc. Natl. Acad. Sci. U. S. A 106, 16245–16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Elosegui-Artola A, Oria R, Chen Y, Kosmalska A, Pérez-González C, Castro N, Zhu C, Trepat X, and Roca-Cusachs P (2016). Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol 18, 540–548. [DOI] [PubMed] [Google Scholar]
- 119.Cluzel C, Saltel F, Lussi J, Paulhe F, Imhof BA, and Wehrle-Haller B (2005). The mechanisms and dynamics of αvβ3 integrin clustering in living cells. J. Cell Biol 171, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Thompson CJ, Su Z, Vu VH, Wu Y, Leckband DE, and Schwartz DK (2020). Cadherin clusters stabilized by a combination of specific and nonspecific cis-interactions. Elife 9, 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chang AC, Mekhdjian AH, Morimatsu M, Denisin AK, Pruitt BL, and Dunn AR (2016). Single Molecule Force Measurements in Living Cells Reveal a Minimally Tensioned Integrin State. ACS Nano 10, 10745–10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Erami Z, Timpson P, Yao W, Zaidel-Bar R, and Anderson KI (2015). There are four dynamically and functionally distinct populations of E-cadherin in cell junctions. Biol. Open 4, 1481–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, and Waterman CM (2010). Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Case LB, and Waterman CM (2015). Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol 17, 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Case LB, Baird MA, Shtengel G, Campbell SL, Hess HF, Davidson MW, and Waterman CM (2015). Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol 17, 880–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bertocchi C, Wang Y, Ravasio A, Hara Y, Wu Y, Sailov T, Baird MA, Davidson MW, Zaidel-Bar R, Toyama Y, et al. (2017). Nanoscale architecture of cadherin-based cell adhesions. Nat. Cell Biol 19, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sawyer JK, Harris NJ, Slep KC, Gaul U, and Peifer M (2009). The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol 186, 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, and Wieschaus EF (2005). Folded gastrulation, cell shape change and the control of myosin localization. Development 132, 4165–4178. [DOI] [PubMed] [Google Scholar]
- 129.Tang VW, and Brieher WM (2012). α-Actinin-4/FSGS1 is required for Arp2/3-dependent actin assembly at the adherens junction. J. Cell Biol 196, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gorfinkiel N, and Arias AM (2007). Requirements for adherens junction components in the interaction between epithelial tissues during dorsal closure in Drosophila. J. Cell Sci 120, 3289–3298. [DOI] [PubMed] [Google Scholar]
- 131.Halbleib JM, and Nelson WJ (2006). Cadherins in development: Cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 20, 3199–3214. [DOI] [PubMed] [Google Scholar]
- 132.Roca-Cusachs P, Iskratsch T, and Sheetz MP (2012). Finding the weakest link-exploring integrin-mediated mechanical molecular pathways. J. Cell Sci 125, 3025–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Miller PW, Clarke DN, Weis WI, Lowe CJ, and Nelson WJ (2013). The evolutionary origin of epithelial cell-cell adhesion mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nathaniel Clarke D, Lowe CJ, and James Nelson W (2019). The cadherin-catenin complex is necessary for cell adhesion and embryogenesis in Nematostella vectensis. Dev. Biol 447, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hynes RO, and Zhao Q (2000). The evolution of cell adhesion. J. Cell Biol 150, 89–95. [DOI] [PubMed] [Google Scholar]
- 136.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, and Geiger B (2007). Functional atlas of the integrin adhesome. Nat. Cell Biol 9, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zaidel-Bar R (2013). Cadherin adhesome at a glance. J. Cell Sci 126, 373–378. [DOI] [PubMed] [Google Scholar]
- 138.Toret CP, D’Ambrosio MV, Vale RD, Simon MA, and Nelson WJ (2014). A genome-wide screen identifies conserved protein hubs required for cadherin-mediated cell-cell adhesion. J. Cell Biol 204, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mitchison T, and Kirschner M (1988). Cytoskeletal dynamics and nerve growth. Neuron 1, 761–772. [DOI] [PubMed] [Google Scholar]
- 140.Elosegui-Artola A, Trepat X, and Roca-Cusachs P (2018). Control of Mechanotransduction by Molecular Clutch Dynamics. Trends Cell Biol. 28, 356–367. [DOI] [PubMed] [Google Scholar]
- 141.Gardel ML, Sabass B, Ji L, Danuser G, Schwarz US, and Waterman CM (2008). Traction stress in focal adhesions correlates biphasically with actin retrograde fl ow speed. J. Cell Biol 183, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lin CH, and Forscher P (1995). Growth cone advance is inversely proportional to retrograde F-actin flow. Neuron 14, 763–771. [DOI] [PubMed] [Google Scholar]
- 143.Del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, and Sheetz MP (2009). Stretching single talin rod molecules activates vinculin binding. Science (80-. ) 323, 638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yao M, Qiu W, Liu R, Efremov AK, Cong P, Seddiki R, Payre M, Lim CT, Ladoux B, Mège RM, et al. (2014). Force-dependent conformational switch of α -catenin controls vinculin binding. Nat. Commun 5. [DOI] [PubMed] [Google Scholar]
- 145.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, and Shibata M (2010). α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol 12, 533–542. [DOI] [PubMed] [Google Scholar]
- 146.Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, and Dunn AR (2014). The minimal cadherin-catenin complex binds to actin filaments under force. Science (80-. ) 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Owen LM, Bax NA, Weis WI, and Dunn AR (2020). The C-terminal actin binding domain of talin forms an asymmetric catch bond with F-actin. bioRxiv, 2020.09.01.276568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Huang DL, Bax NA, Buckley CD, Weis WI, and Dunn AR (2017). Vinculin forms a directionally asymmetric catch bond with F-actin. Science (80-. ) 357, 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yu HH, and Zallen JA (2020). Abl and Canoe/Afadin mediate mechanotransduction at tricellular junctions. Science (80-. ) 370, eaba5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, and Waterman CM (2010). Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol 188, 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, and De Rooij J (2010). Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J. Cell Biol 189, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jodoin JN, Coravos JS, Chanet S, Vasquez CG, Tworoger M, Kingston ER, Perkins LA, Perrimon N, and Martin AC (2015). Stable Force Balance between Epithelial Cells Arises from F-Actin Turnover. Dev. Cell 35, 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ikawa K, and Sugimura K (2018). AIP1 and cofilin ensure a resistance to tissue tension and promote directional cell rearrangement. Nat. Commun 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li Y, Merkel CD, Zeng X, Heier JA, Cantrell PS, Sun M, Stolz DB, Watkins SC, Yates NA, and Kwiatkowski AV (2019). The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics. J. Cell Sci 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kuo JC, Han X, Hsiao C. Te, Yates JR, and Waterman CM (2011). Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol 13, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tan SJ, Chang AC, Anderson SM, Miller CM, Prahl LS, Odde DJ, and Dunn AR (2020). Regulation and dynamics of force transmission at individual cell-matrix adhesion bonds. Sci. Adv 6, eaax0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Iyer KV, Piscitello-Gómez R, Paijmans J, Jülicher F, and Eaton S (2019). Epithelial Viscoelasticity Is Regulated by Mechanosensitive E-cadherin Turnover. Curr. Biol 29, 578–591.e5. [DOI] [PubMed] [Google Scholar]
- 158.Bénazéraf B, Francois P, Baker RE, Denans N, Little CD, and Pourquié O (2010). A random cell motility gradient downstream of FGF controls elongation of an amniote embryo. Nature 466, 248–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sai XR, and Ladher RK (2008). FGF Signaling Regulates Cytoskeletal Remodeling during Epithelial Morphogenesis. Curr. Biol 18, 976–981. [DOI] [PubMed] [Google Scholar]
- 160.Lecaudey V, Cakan-Akdogan G, Norton WHJ, and Gilmour D (2008). Dynamic Fgf signaling couples morphogenesis and migration in the zebrafish lateral line primordium. Development 135, 2695–2705. [DOI] [PubMed] [Google Scholar]
- 161.Harding MJ, and Nechiporuk AV (2012). Fgfr-Ras-MAPK signaling is required for apical constriction via apical positioning of Rho-associated kinase during mechanosensory organ formation. Dev. 139, 3130–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Nerurkar NL, Lee CH, Mahadevan L, and Tabin CJ (2019). Molecular control of macroscopic forces drives formation of the vertebrate hindgut. Nature 565, 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Heer NC, Miller PW, Chanet S, Stoop N, Dunkel J, and Martin AC (2017). Actomyosin-based tissue folding requires a multicellular myosin gradient. J. Cell Sci 130, 1876–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lim B, Levine M, and Yamakazi Y (2017). Transcriptional Pre-patterning of Drosophila Gastrulation. Curr. Biol 27, 286–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rahimi N, Averbukh I, Carmon S, Schejter ED, Barkai N, and Shilo BZ (2019). Dynamics of Spaetzle morphogen shuttling in the Drosophila embryo shapes gastrulation patterning. Development 146. [DOI] [PubMed] [Google Scholar]
- 166.Oda H, Tsukita S, and Takeichi M (1998). Dynamic behavior of the cadherin-based cell-cell adhesion system during Drosophila gastrulation. Dev. Biol 203, 435–450. [DOI] [PubMed] [Google Scholar]
- 167.Münster S, Jain A, Mietke A, Pavlopoulos A, Grill SW, and Tomancak P (2019). Attachment of the blastoderm to the vitelline envelope affects gastrulation of insects. Nature 568, 395–399. [DOI] [PubMed] [Google Scholar]
- 168.Bailles A, Collinet C, Philippe JM, Lenne PF, Munro E, and Lecuit T (2019). Genetic induction and mechanochemical propagation of a morphogenetic wave. Nature 572, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Röper K (2013). Supracellular actomyosin assemblies during development. Bioarchitecture 3, 45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Galea GL, Cho YJ, Galea G, Molè MA, Rolo A, Savery D, Moulding D, Culshaw LH, Nikolopoulou E, Greene NDE, et al. (2017). Biomechanical coupling facilitates spinal neural tube closure in mouse embryos. Proc. Natl. Acad. Sci. U. S. A 114, E5177–E5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Heller E, Kumar KV, Grill SW, and Fuchs E (2014). Forces generated by cell intercalation tow epidermal sheets in mammalian tissue morphogenesis. Dev. Cell 28, 617–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tipping N, and Wilson D (2011). Chick amniogenesis is mediated by an actin cable. Anat. Rec 294, 1143–1149. [DOI] [PubMed] [Google Scholar]
- 173.Houssin NS, Martin JB, Coppola V, Yoon SO, and Plageman TF (2020). Formation and contraction of multicellular actomyosin cables facilitate lens placode invagination. Dev. Biol 462, 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chai J, Hamilton AL, Krieg M, Buckley CD, Riedel-Kruse IH, and Dunn AR (2015). A Force Balance Can Explain Local and Global Cell Movements during Early Zebrafish Development. Biophys. J 109, 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Behrndt M, Salbreux G, Campinho P, Hauschild R, Oswald F, Roensch J, Grill SW, and Heisenberg CP (2012). Forces driving epithelial spreading in zebrafish gastrulation. Science (80-. ) 338, 257–260. [DOI] [PubMed] [Google Scholar]
- 176.Calzolari S, Terriente J, and Pujades C (2014). Cell segregation in the vertebrate hindbrain relies on actomyosin cables located at the interhombomeric boundaries. EMBO J. 33, 686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Young PE, Richman AM, Ketchum AS, and Kiehart DP (1993). Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 7, 29–41. [DOI] [PubMed] [Google Scholar]
- 178.Landsberg KP, Farhadifar R, Ranft J, Umetsu D, Widmann TJ, Bittig T, Said A, Jülicher F, and Dahmann C (2009). Increased Cell Bond Tension Governs Cell Sorting at the Drosophila Anteroposterior Compartment Boundary. Curr. Biol 19, 1950–1955. [DOI] [PubMed] [Google Scholar]
- 179.Martin P, and Lewis J (1992). Actin cables and epidermal movement in embryonic wound healing. Nature 360, 179–183. [DOI] [PubMed] [Google Scholar]
- 180.Ducuing A, and Vincent S (2016). The actin cable is dispensable in directing dorsal closure dynamics but neutralizes mechanical stress to prevent scarring in the Drosophila embryo. Nat. Cell Biol 18, 1149–1160. [DOI] [PubMed] [Google Scholar]
- 181.Pasakarnis L, Frei E, Caussinus E, Affolter M, and Brunner D (2016). Amnioserosa cell constriction but not epidermal actin cable tension autonomously drives dorsal closure. Nat. Cell Biol 18, 1161–1172. [DOI] [PubMed] [Google Scholar]
- 182.Jacinto A, Wood W, Woolner S, Hiley C, Turner L, Wilson C, Martinez-Arias A, and Martin P (2002). Dynamic analysis of actin cable function during Drosophila dorsal closure. Curr. Biol 12, 1245–1250. [DOI] [PubMed] [Google Scholar]
- 183.Umetsu D, Aigouy B, Aliee M, Sui L, Eaton S, Jülicher F, and Dahmann C (2014). Local increases in mechanical tension shape compartment boundaries by biasing cell intercalations. Curr. Biol 24, 1798–1805. [DOI] [PubMed] [Google Scholar]
- 184.Monier B, Pélissier-Monier A, Brand AH, and Sanson B (2010). An actomyosin-based barrier inhibits cell mixing at compartmental boundaries in Drosophila embryos. Nat. Cell Biol 12, 60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Eritano AS, Bromley CL, Bolea Albero A, Schütz L, Wen F-L, Takeda M, Fukaya T, Sami MM, Shibata T, Lemke S, et al. (2020). Tissue-Scale Mechanical Coupling Reduces Morphogenetic Noise to Ensure Precision during Epithelial Folding. Dev. Cell [DOI] [PubMed] [Google Scholar]
- 186.Yevick HG, Miller PW, Rn Dunkel J, and Martin AC (2019). Structural Redundancy in Supracellular Actomyosin Networks Enables Robust Tissue Folding Article Structural Redundancy in Supracellular Actomyosin Networks Enables Robust Tissue Folding. Dev. Cell 50, 586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Duda M, Kirkland NJ, Khalilgharibi N, Tozluoglu M, Yuen AC, Carpi N, Bove A, Piel M, Charras G, Baum B, et al. (2019). Polarization of Myosin II Refines Tissue Material Properties to Buffer Mechanical Stress. Dev. Cell 48, 245–260.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Levayer R, and Lecuit T (2013). Oscillation and Polarity of E-Cadherin Asymmetries Control Actomyosin Flow Patterns during Morphogenesis. Dev. Cell 26, 162–175. [DOI] [PubMed] [Google Scholar]
- 189.Jain A, Ulman V, Mukherjee A, Prakash M, Cuenca MB, Pimpale LG, Münster S, Haase R, Panfilio KA, Jug F, et al. (2020). Regionalized tissue fluidization is required for epithelial gap closure during insect gastrulation. Nat. Commun 11, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tetley RJ, Staddon MF, Heller D, Hoppe A, Banerjee S, and Mao Y (2019). Tissue fluidity promotes epithelial wound healing. Nat. Phys 15, 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Footer MJ, Kerssemakers JWJ, Theriot JA, and Dogterom M (2007). Direct measurement of force generation by actin filament polymerization using an optical trap. Proc. Natl. Acad. Sci. U. S. A 104, 2181–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kovar DR, and Pollard TD (2004). Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. U. S. A 101, 14725–14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, and Chen CS (2010). Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods 7, 969–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Maruthamuthu V, Sabass B, Schwarz US, and Gardel ML (2011). Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc. Natl. Acad. Sci. U. S. A 108, 4708–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Trepat X, Wasserman M, Angelini T, physics, E.M.-N., and 2009, undefined Physical forces during collective cell migration. nature.com. [Google Scholar]
- 196.Zhou J, Pal S, Maiti S, and Davidson LA (2015). Force production and mechanical accommodation during convergent extension. Dev. 142, 692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ananthakrishnan R, and Ehrlicher A (2007). The forces behind cell movement. Int. J. Biol. Sci 3, 303–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li F, Redick SD, Erickson HP, and Moy VT (2003). Force measurements of the α5β1 integrin-fibronectin interaction. Biophys. J 84, 1252–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Galior K, Liu Y, Yehl K, Vivek S, and Salaita K (2016). Titin-Based Nanoparticle Tension Sensors Map High-Magnitude Integrin Forces within Focal Adhesions. Nano Lett. 16, 341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Taubenberger A, Cisneros DA, Friedrichs J, Puech PH, Muller DJ, and Franz CM (2007). Revealing early steps of α2β1 integrin-mediated adhesion to collagen type I by using single-cell force spectroscopy. Mol. Biol. Cell 18, 1634–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Niland S, Westerhausen C, Schneider SW, Eckes B, Schneider MF, and Eble JA (2011). Biofunctionalization of a generic collagenous triple helix with the α2β1 integrin binding site allows molecular force measurements. Int. J. Biochem. Cell Biol 43, 721–731. [DOI] [PubMed] [Google Scholar]
- 202.Chu YS, Thomas WA, Eder O, Pincet F, Perez E, Thiery JP, and Dufour S (2004). Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J. Cell Biol 167, 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jiang G, Giannone G, Critchley DR, Fukumoto E, and Sheet MP (2003). Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424, 334–337. [DOI] [PubMed] [Google Scholar]
- 204.Nishizaka T, Miyata H, Yoshikawa H, Ishiwata S, and Kinosita K (1995). Unbinding force of a single motor molecule of muscle measured using optical tweezers. Nature 377, 251–254. [DOI] [PubMed] [Google Scholar]
- 205.Tsuda Y, Yasutake H, Ishuima A, and Yanaogida T (1996). Torsional rigidity of single actin filaments and actin-actin bond breaking force under torsion measured directly by in vitro micromanipulation. Proc. Natl. Acad. Sci. U. S. A 93, 12937–12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kishino A, and Yanagida T (1988). Force measurements by micromanipulation of a single actin filament by glass needles. Nature 334, 74–76. [DOI] [PubMed] [Google Scholar]
- 207.Howard J (2001). Mechanics of motor proteins and the cytoskeleton. [Google Scholar]
- 208.Murrell MP, and Gardel ML (2012). F-actin buckling coordinates contractility and severing in a biomimetic actomyosin cortex. Proc. Natl. Acad. Sci. U. S. A 109, 20820–20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lenz M, Gardel ML, and Dinner AR (2012). Requirements for contractility in disordered cytoskeletal bundles. New J. Phys 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Weisel JW, Shuman H, and Litvinov RI (2003). Protein-protein unbinding induced by force: Single-molecule studies. Curr. Opin. Struct. Biol 13, 227–235. [DOI] [PubMed] [Google Scholar]