ABSTRACT
During human pregnancy, cytotrophoblasts (CTBs) from the placenta differentiate into specialized subpopulations that play crucial roles in proper fetal growth and development. A subset of these CTBs differentiate along an invasive pathway, penetrating the decidua and anchoring the placenta to the uterus. A crucial hurdle in pregnancy is the ability of these cells to migrate, invade and remodel spiral arteries, ensuring adequate blood flow to nourish the developing fetus. Although advances continue in describing the molecular features regulating the differentiation of these cells, assessment of their global proteomic changes at mid-gestation remain undefined. Here, using sequential window acquisition of all theoretical fragment-ion spectra (SWATH), which is a data-independent acquisition strategy, we characterized the protein repertoire of second trimester human CTBs during their differentiation towards an invasive phenotype. This mass spectrometry-based approach allowed identification of 3026 proteins across four culture time points corresponding to sequential stages of differentiation, confirming the expression dynamics of established molecules and offering new information into other pathways involved. The availability of a SWATH CTB global spectral library serves as a beneficial resource for hypothesis generation and as a foundation for further understanding CTB differentiation dynamics.
KEY WORDS: Human, Placenta, Cytotrophoblast, Proteomics, SWATH-MS
Summary: The generation of a publicly available SWATH-MS global spectral library of human second-trimester primary villous cytotrophoblasts differentiating along an invasive pathway.
INTRODUCTION
The placenta is a transient organ that establishes and mediates maternal-fetal interactions during pregnancy. The creation of this interface depends on the proper development and function of the placenta's chorionic villi (Maltepe and Fisher, 2015). Throughout pregnancy, placental villi undergo extensive remodeling and branching, maturing into floating or anchoring villi. Most of the placental surface comprises floating villi that are in direct contact with, and suspended in, maternal blood. There, the exchange of nutrient, waste and gas is facilitated between the maternal and fetal compartments (Turco and Moffett, 2019). This is accomplished through the outermost epithelial layer of the villi, a multinuclear layer composed of syncitotrophoblasts (STBs) that is generated via fusion of the underlying cytotrophoblast (CTB) progenitors. In anchoring villi, these progenitors aggregate into cell columns that attach to the decidua. Invasive CTBs emanate from the cell columns and migrate deeply into the uterine wall. During this process, these extravillous trophoblasts remodel spiral arteries, which diverts blood flow to the placenta (Red-Horse et al., 2004). Proper regulation of CTB differentiation/invasion is crucial for successful pregnancy, and perturbations in this process are associated with several complications such as preeclampsia (PE) (Fisher, 2015), preterm labor (Romero et al., 2014) or placenta accreta spectrum (Jauniaux et al., 2018; McNally et al., 2020).
Given the unique structure and function of the human placenta (versus animal models), and the difficulty of in utero studies, primary human CTB models have been crucial for advancing our knowledge of how these cells function at a molecular level (Knöfler et al., 2019). Work from our group and others has revealed numerous components of the CTB repertoire that program differentiation/invasion, especially the vasculogenic component of this process. However, achieving a comprehensive global understanding of the dynamic changes that occur has yet to be realized. We previously characterized global gene expression changes in primary CTB cultures under conditions that promote acquisition of an invasive phenotype (Robinson et al., 2017). Numerous factors, however, can impact translational efficiency from transcript to protein (Tian et al., 2004). As a result, proteomic studies provide complementary biological insights as they carry out the majority of the cell's functions.
Mass spectrometry (MS)-based methods have developed rapidly. One such approach, Sequential Window Acquisition of All Theoretical spectra (SWATH), has emerged as a comprehensive, highly reproducible data acquisition workflow for proteomic studies (Collins et al., 2017). Additionally, this approach has the added advantage of enabling relative quantification without introducing a chemical labeling step. In SWATH-MS, spectra are recorded using an unbiased, data-independent acquisition (DIA) based on preselected mass range windows (‘swaths’). The results are then queried against a previously generated spectral library containing the spectrometric coordinates of proteins and peptides of interest (Ludwig et al., 2018). These ion libraries are a unique publicly available resource to which additional DIA data can be added over time, growing the repository and increasing its usefulness.
Here, we report the creation of a SWATH spectral library generated from a second-trimester human CTB culture model of differentiation/invasion that enabled the measurement of over 40,000 peptides from over 3000 proteins. We demonstrate the utility of this dataset by quantifying 3026 proteins across four culture time points. The dynamics of numerous molecules confirmed known expression patterns and revealed unique identification of molecules that could play roles in differentiation/invasion. Our study is the first reference SWATH library of second-trimester CTBs, as well as the first comprehensive description of the CTB proteome during differentiation towards an invasive phenotype.
RESULTS
Global proteomic analysis of human CTB differentiation
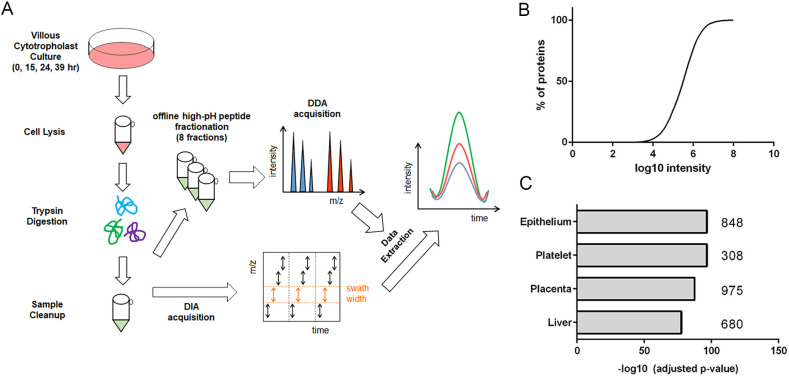
A simplified workflow is diagrammed in Fig. 1A. To profile CTB proteome dynamics during differentiation towards an invasive phenotype, primary human CTBs were isolated and purified from second trimester placentas (gestational ages 20 to 24 weeks; n=4) and grown in serum-free medium (Hunkapiller and Fisher, 2008) (Fig. S1). Cells were harvested at 0, 15, 24 and 39 h, a range of time points corresponding with the acquisition of an invasive phenotype in cultured second trimester CTBs (Damsky et al., 1994). For quantitative proteomic analyses using SWATH-MS, lysates were trypsin digested and a pooled sample was fractionated and analyzed by LC-MS/MS to create an ion library, which was annotated. Overall, 3026 proteins were quantified across all time points with relative protein abundances spanning over four orders of magnitude (Fig. 1B). A heatmap of Pearson scores among individual biological replicates illustrated a high degree of correlation (Fig. S2A). The coefficient of variation among samples for each time point was low, with a median of ∼0.3 (Fig. S2B). Tissue enrichment analysis using the Homo sapiens proteome as background showed that a large proportion of the CTB proteome included placental proteins (975) and, in keeping with their ectodermal origin, proteins (848) that are typically found in epithelia (Fig. 1C). Overlap between the CTB proteome and that of platelets and liver is consistent with the anti-coagulant and hepatic-like functions of these cells, respectively. Gene Ontology (GO) Enrichment analysis of the CTB proteome showed an abundance of proteins associated with localization (615) and transport (689) processes, with an overrepresentation of RNA binding (664) and cell-cell adhesion (228) (Fig. S2C). Additionally, with respect to cellular compartments, over one-third of the proteins had exosomal or vesicular annotations (1137).
Fig. 1.
Global proteomic profiling of cytotrophoblast differentiation/invasion. (A) Simplified workflow schematic of cytotrophoblast proteomic analysis using SWATH-MS. (B) Distribution of relative protein abundances, spanning four orders of magnitude. (C) Tissue enrichment analysis of annotated proteins from the dataset. n=4 biological replicates.
Protein dynamics during human CTB differentiation/invasion in vitro
To identify differentially expressed (DE) proteins during CTB differentiation (0-39 h), we applied an integrated linear model to our dataset (LIMMA) (Ritchie et al., 2015). In total, we identified 477 proteins that had significant changes in abundance relative to 0 h (P<0.05).
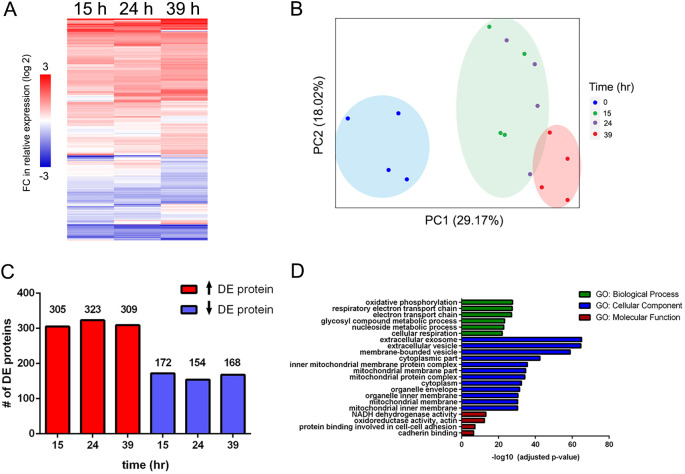
In general, a majority of the DE proteins showed upregulation (Fig. 2A). The magnitude of expression also increased with time, with ∼38% (181/477) of DE proteins at 39 h showing more than a twofold change (log2scale>|1|) (Fig. 3A-C). Principal components analysis (PCA) of the DE proteins showed their ability to distinctly cluster each sample according to culture time, with the lowest variance between 15 and 24 h, and the largest variance between 0 and 39 h (Fig. 2B). Fig. 2C shows the number of proteins whose abundances changed at each time point relative to CTBs prior to culture (0 h). With regard to functional analyses, DE proteins were most significantly enriched in metabolic and adhesive processes. Notably, nearly half of the DE proteins (48%) had exosomal or vesicular annotations, highlighting the importance of extracellular vesicles to CTB differentiation, including possible autocrine and/or paracrine roles (Fig. 2D).
Fig. 2.
Differentially expressed protein changes during cytotrophoblast differentiation/invasion. (A) Unsupervised hierarchical clustering of 477 differentially expressed (DE) proteins relative to 0 h. (B) Principal component analysis of DE proteins by biological replicate (n=4). (C) Distribution of DE protein expression changes at each time point relative to 0 h. (D) Gene ontology (GO) enrichment analysis of DE proteins. The most enriched categories from the three GO terms (biological process, cellular component and molecular function) were primarily matched to respiratory/metabolic processes, exosomal/vesicular components and metabolic/adhesive functions, respectively (Fisher's exact test; P<0.01, Benjamini-Hochberg corrected).
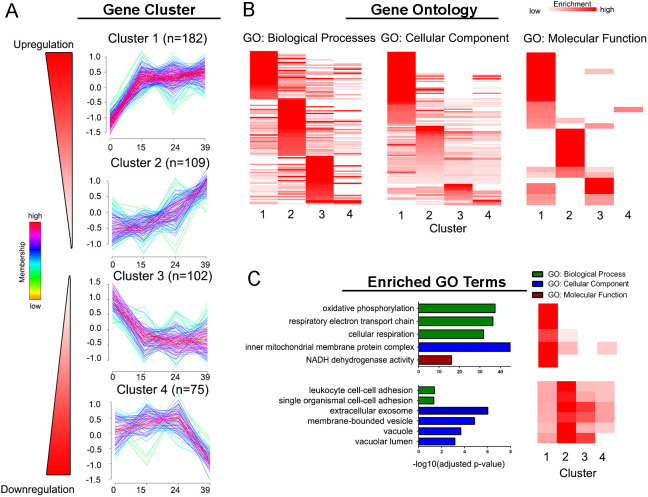
Fig. 3.
Dynamics of differentially expressed proteins during cytotrophoblast differentiation/invasion. (A) Unsupervised fuzzy clustering of 477 differentially expressed (DE) proteins identified four clusters with distinct expression profiles. n=number of proteins assigned to each cluster. (B) Heatmap of gene ontology (GO) enrichment analysis of each cluster. GO terms overrepresented in each cluster (Fisher's exact test; P<0.05, unadjusted) were visualized by fold enrichment as a qualitative comparison to functionalities in other clusters. (C) Representative GO terms enriched in clusters 1 and 2 (Fisher's exact test; P<0.05, Benjamini-Hochberg corrected) were visualized for biological processes, cellular component or molecular function (MF).
Of the proteins that were not significantly changing, enrichment analyses revealed a continued overrepresentation of proteasomal, ribosomal and transport processes, suggesting that the levels of proteins associated with many fundamental cellular processes remained relatively stable during CTB differentiation (Fig. 2D).
Patterns of temporal shifts in the CTB proteome
We performed an unsupervised fuzzy clustering of the DE proteins to stratify their temporal dynamics. Four distinct clusters were identified (Tibshirani et al., 2001): two upregulated (1 and 2) and two downregulated (3 and 4) (Fig. 3A). Many proteins in each cluster showed shared functionalities and cellular localization (Fig. 3B), with clusters 1 and 2 possessing significantly distinct biological processes at particular stages of differentiation (Fig. 3C).
Cluster 1 (1 and 2 proteins) had a pattern of rapid upregulation after 0 h followed by sustained expression to 39 h. This included proteins, e.g. HLA-G, that our group have shown are upregulated in the early stage of CTB differentiation (Mcmaster et al., 1995). Proteins in this cluster were predominantly mitochondrial and involved in oxidative phosphorylation. This is likely a reflection of the fact that oxygen tension plays a large role in regulating CTB invasion, and the increased abundance of proteins involved in oxidative phosphorylation is in line with the established importance of mitochondrial energetics in meeting the demands of trophoblast function. Conversely, abnormalities in mitochondrial morphology and/or function are associated with disease states such as pre-eclampsia, an example of a pregnancy complication that is associated with significant trophoblast dysfunction (Torbergsen et al., 1989).
Cluster 2 (109 proteins) consisted of proteins with a linear increase in expression from 0 h to 39 h. There was a significant enrichment in proteins with integral adhesive or migratory/invasive functions, such ESAM (Hirata et al., 2001) and CD44 (Goshen et al., 1996). Proteins in Cluster 2 were especially enriched exosomal and vacuole components. Molecules involved in autophagic vacuole formation or localization, such as LAMP-2, ATG16L1 and cathepsins S and D (Curtis et al., 2013). Autophagic activation is associated with the epithelial-to-mesenchymal transition (Gugnoni et al., 2016), a process that is one component of CTB differentiation to an invasive phenotype (DaSilva-Arnold et al., 2015; Saito and Nakashima, 2014).
Cluster 3 (102 proteins) contained proteins with a strong downregulation from 0 h to 15 h. A notable proportion of proteins (44/102) in this cluster were annotated as signal transduction molecules, including negative regulators of trophoblast migration and invasion such as endoglin (ENG) (Caniggia et al., 1997) and PAPPA2 (Winship et al., 2015).
Cluster 4 (75 proteins) contained proteins with a strong downregulation from 24 h to 39 h. Proteins in Cluster 4 showed overlap in enrichment with Cluster 3 functional processes, notably ER to Golgi transport. Although proteins in Cluster 4 did not show unique enrichment in origin, there was an enrichment in attribution to ‘acetylation’ (31/75) according to UniProtKB keywords. Interestingly, this contrasts with the upregulation of deacetylases (e.g. HDAC2) from Cluster 2. The relative activities of these enzymes remains to be determined.
Dynamics of functionally related proteins in cultured CTBs
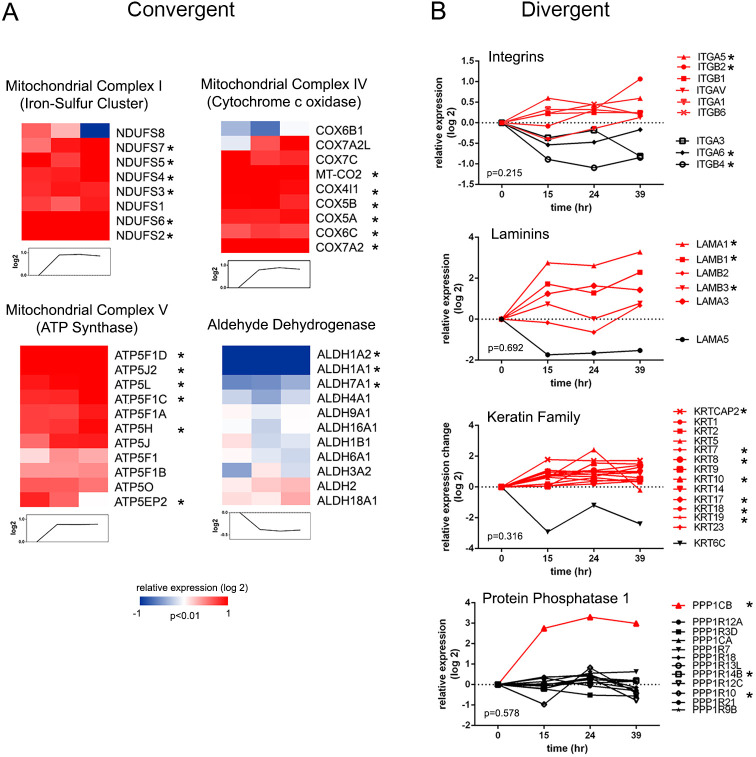
Next, we assessed the extent of temporal coordination in functionally related protein complexes and families in the proteins with significant changes in abundance over the 39 h culture period. Statistical assessment of protein clusters by annotation revealed broad coordination in expression dynamics (P<0.01) among protein and metabolic function subgroups. These included the iron-sulfur protein components of mitochondrial complex I, cytochrome c oxidase of mitochondrial complex IV and the ATP synthases of mitochondrial complex V (Fig. 4A), which showed significant coordinated upregulation during differentiation. Furthermore, the mitochondrial respiratory chain complex was largely captured in our analyses (Fig. S4A), broadly matching the expression profile of the energetic and metabolic processes of Cluster 1. The aldehyde dehydrogenase (ALDH) family showed coordinated downregulation matching the profile of Cluster 3, with ALDH1A1 and ALDH1A2 being the most highly differentially expressed. ALDH1A2 has previously been identified as a candidate tumor suppressor (Kim et al., 2005) and its downregulation accelerates an epithelial-to-mesenchymal transition (Seidensaal et al., 2015), an important component of CTB invasion. The annexins and cytochrome c oxidase proteins were also significantly upregulated as a group, respectively (Fig. 4B,C), underscoring previous observations of their roles in CTB function (Matsubara et al., 1997; Xu et al., 2019).
Fig. 4.
Protein expression profiles of families/complexes during cytotrophoblast differentiation/invasion. (A) Representative protein families/complexes with significant convergent expression profiles (P<0.01, distance measurement). (B) Representative protein families/complexes with significant divergent expression profiles (P<0.05, distance measurement). Asterisks indicate a protein that is significantly differentially expressed.
Conversely, the same statistical analysis revealed proteins that deviated from the expression profile of their corresponding complexes or families (P>0.05), potentially indicating a cell- or stage-specific function. The profile of the integrin subunits illustrated this phenomena. Integrin switching during CTB invasion/differentiation is a phenomenon that has been well-characterized by our group and others (Damsky et al., 1994). This was substantiated by our proteomic data, with significant upregulation of integrin α5/β1 and downregulation of α6/β4 (Fig. 4B) across culturing time. Laminins, which interact intimately with integrin ligands, are a key component of CTB extracellular matrix deposition (Blankenship et al., 1992; Yagel et al., 1988). Differential laminin distribution influences CTB function and differentiation; specifically, our data confirmed significant upregulation of the α1 and β1 chains (Korhonen and Virtanen, 1997) with time in culture. In contrast, the α5 chain tended toward downregulation, although the value did not reach significance (P=0.07).
A similar divergent pattern was observed with members of the serine protease inhibitor superfamily of serpins (Fig. S3D). The upregulation of SERPINE1 and SERPINB2, and the downregulation of SERPINA1 was observed in our dataset. SERPINE1 and SERPINB2, or plasminogen activator inhibitors (PAI) 1 and PAI2, are expressed by human trophoblasts in vitro and in vivo (Feinberg et al., 1989; Hofmann et al., 1994), and alterations in maternal circulation are associated with pregnancy complications with placental origins (Estellés et al., 1989; Wikström et al., 2009). SERPINA1, or α1 anti-trypsin, is an acute phase glycoprotein that may be locally synthesized and secreted by invading trophoblasts to dampen a maternal inflammatory response (Bergman et al., 1993). Although the secretome was not analyzed in this study to determine corresponding SERPINA1 release, its cellular downregulation suggests that expression may be extrinsically regulated during invasive differentiation.
Our dataset also provided insight into the expression of other protein families with divergent members. The keratins (KRT) have previously been used to profile distinct trophoblast subpopulations throughout pregnancy (Gauster et al., 2013; Mühlhauser et al., 1995; Pröll et al., 1997). Keratins were broadly represented in our dataset, with seven of the 14 detected showing significant, albeit modest, upregulation. These included KRT7, a widely used marker for villous trophoblast (Maldonado-Estrada et al., 2004). Others are associated with invasive cellular populations, e.g. KRT8, KRT17, KRT18 and KRT19. KRT6C displayed a distinct downregulated pattern; however, its associations to CTB differentiation is currently unexplored.
A unique expression pattern was observed with the serine/threonine phosphatase, protein phosphatase 1 (PPP1)’s, catalytic and regulatory subunits. Of the 12 PPP1 subunits detected, only catalytic subunit PPP1CB displayed a significant, robust upregulation. PPP1 is composed of three catalytic subunits, each with isoform-specific function and distribution (Bollen, 2001). The regulatory role of PPP1 regarding trophoblast differentiation is unclear at this time, but the expression pattern suggests that primary interactors of PPP1CB are important in this process.
Dynamics of proteins involved in CTB invasive and migration
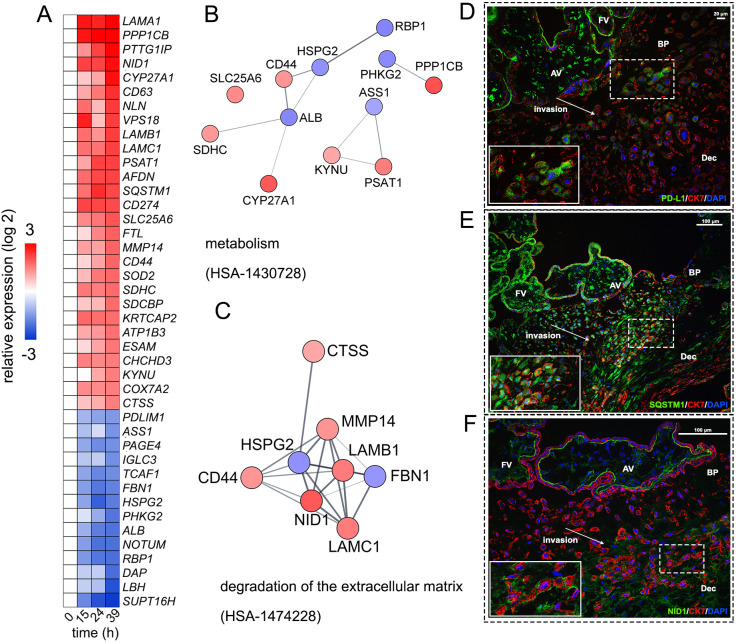
Given the number of DE proteins, we imposed additional cutoffs with the goal of visualizing the most highly robust proteins to play important roles in differentiation. Forty-two of the 477 DE proteins were stratified with more stringent criteria (P<0.01, absolute log 2 fold change ≥1.5 at 39 h) (Fig. 5A). STRING enrichment analysis of these proteins highlighted that robust changes occur in Reactome pathways associated with ‘metabolism’ (Fig. 5B) and ‘degradation of the extracellular matrix’ (Fig. 5C) (P<0.05, FDR corrected). Numerous molecules in these clusters have been associated with migratory or invasive roles, such as ASS1 (Huang et al., 2013), CD44 (Takahashi et al., 2014), CTSS (Yang et al., 2010) and MMP14 (Anacker et al., 2011). In addition, 50% of the proteins (21/42) had exosomal annotations, as determined by DAVID, highlighting the importance of their modulation during invasion.
Fig. 5.
Selected cytotrophoblast proteins with robust expression changes. Additional cutoffs (P<0.01, absolute log 2 fold change ≥|1.5|) were applied to the differentially expressed (DE) proteins. (A) Heatmap depicting expression changes of 42 robust DE proteins relative to 0 h. (B,C) Predicted protein-protein interaction networks of enriched pathways. Robust DE proteins were visualized in Cytoscape using the STRING database, and molecules in significantly enriched pathways were separated to individual clusters (P<0.05, FDR corrected). (D-F) Representative images from second trimester tissue sections of the basal plate (BP). Sections were stained with the anti-cytotrophoblast marker cytokeratin 7 (red), with DAPI (blue) and with (D) PD-L1, (E) SQSTM1 or (F) NID1 (green). n≥4 biological replicates. Scale bars: 20 µm in D; 100 µm in E,F. AV, anchoring villi; BP, basal plate; FV, floating villi; Dec, decidua.
To confirm the spatial distribution of robustly changing molecules of interest at the maternal-fetal interface, immunofluorescent staining experiments were performed on second-trimester biopsies. PD-L1, a negative regulator of T-cell signaling responsible for immune evasion (Keir et al., 2006), showed a pattern of upregulation in our proteomic data. Similar to previous observations in early and term placentas, staining patterns showed variable PD-L1 expression among CTBs, with the highest immunoreactivity occurring among the outer syncytiotrophoblast layer of villi, and more diffuse staining in subpopulations of invasive CTBs near the anchoring villi (Fig. 5D) (Veras et al., 2017).
Sequestosome 1 (SQSTM1/p62), a key regulator in the autophagic pathway (Katsuragi et al., 2015), was robustly upregulated during CTB differentiation/invasion. Autophagy-associated invasive trophoblast behavior has previously been reported to be mediated by ENG (Nakashima et al., 2013), a molecule closely associated with regulating CTB differentiation via inhibition of migratory and invasive function by TGFβ1 (Caniggia et al., 1997; Mano et al., 2011). Our proteomic data reinforce the importance of these molecules, with the concurrent upregulation of SQSTM1/p62 and the downregulation of ENG during our experimental window. DAP1, a negative regulator of autophagy, was also significantly downregulated (Koren et al., 2010). Matching previous observations in term biopsies (Nakashima et al., 2013), SQSTM1/p62 staining generally distributed in similar patterns to CK7, although CTBs deeper in the decidua exhibited weaker patterns of SQSTM1/p62 expression (Fig. 5E).
Nidogen 1 (NID1), a common component of basement membranes, also showed robust upregulation. Immunofluorescent staining of NID1 revealed a striking pattern, with nearly absent expression at the attachment site of the anchoring villi and tight spatial distribution around CTB populations deeper in the decidua (Fig. 5F). The contributions of Matrigel (Hughes et al., 2010), which are essential for the acquisition of an invasive phenotype in this model system, could not be fully discounted from our analysis. However, observations of NID1 secretion in trophoblast organoids (Turco et al., 2018) and the detection of laminin in CTB extracellular vesicles (Taylor et al., 2020), taken with the SWATH results in this study, suggest an intriguing possibility that, in select contexts, CTBs may modulate their microenvironment as part of their differentiation/invasion process. Further studies are necessary to explore this hypothesis.
Evaluating concordance between transcript-protein
We previously published a microarray study evaluating transcriptional dynamics of CTBs differentiating towards an invasive phenotype (Robinson et al., 2017). In total, 2805 genes overlapped with the 3026 proteins (92.6%) in our dataset. The global concordance of overlapping transcript-protein pairs between samples was modest, as calculated with Spearman's correlation (average rho=0.38, Fig. S5A). This low correlation is a widely recognized phenomenon (de Sousa Abreu et al., 2009; Liu et al., 2016; Payne, 2015). The directionality in mean log2 FC from 0 h was shared by 52% and 57% of transcript-protein pairs at 15 h and 39 h, respectively (Table S2). Shared directionality improved when considering only DE transcript-protein pairs derived from the proteomic dataset, increasing to 55% and 68.4% at 15 h and 39 h, respectively. Linear associations between transcript-protein FCs from 0 h were low, with Pearson correlation coefficients of 0.13 and 0.21 at 15 h and 39 h, respectively. Correlations modestly improved when considering only DE transcript-protein pairs (r=0.27, 15 h; r=0.4, 39 h).
To determine whether differences in transcript stability explained these differences, we assessed the mRNA half-lives of transcript-protein pairs using data generated from metabolic pulse labeling (Schwanhäusser et al., 2011). mRNA half-lives of DE transcript-protein pairs were not statistically different from the matched dataset (Wilcoxon rank-sum test, Fig. S5B). Overall, these results reinforce the notion that relative transcript and protein abundances are not predictive of one another with direct comparative measures. In addition to the regulatory repertoire (such as post-transcriptional and -translational mechanisms) that impacts these types of correlations in differentiated cells with a stable phenotype, we are studying differentiating cytotrophoblasts with a dynamically changing phenotype, which further complicates these comparisons. Another factor likely to degrade correlations is our focus on primary human cells, which doubtlessly exhibit sample-to-sample variations.
As an exploratory means of identifying potential co-regulatory modules that influence CTB differentiation between the datasets, we applied consensus network analysis to the overlapping transcript-protein pairs, defining each culture time point as a trait (Langfelder and Horvath, 2007; Zhang and Horvath, 2005). This analysis identified four unique modules, two each at 0 h and 39 h, that represent expression profiles unique to those time points (P<0.05) (Fig. S6A,B). Gene ontology analysis revealed enriched pathways unique to these modules, with processes such as ‘embryonic development’, ‘cell-cell adhesion’ and ‘mitochondrial transport’ being negatively correlated with 0 h, or an undifferentiated state, and processes such as ‘pattern specification’, ‘focal adhesion’ and ‘RNA binding’ positively correlated with 39 h, or a differentiated/invasive state (Fig. S6C). Genes with high significance measures within these modules includes known markers of differentiation/invasion, such as ITGA5, but also molecules of demonstrated importance in murine models, such as TLE3 (Gasperowicz et al., 2013) and S100A8 (Passey et al., 1999), suggesting underexplored roles in a human cell model (Horvath and Dong, 2008) (Table S3).
DISCUSSION
Primary cultures of villous CTBs recapitulate behaviors relevant to migration from the placental compartment, e.g. uterine invasion, and the changes in adhesive interactions that make this unusual differentiation program possible. Thus, this is a valuable model system for studying these processes in the context of pregnancy outcomes. In this study, we used SWATH-MS, a label-free data-independent acquisition method, to catalogue the global CTB proteome. This enabled a quantitative analysis of protein levels, which identified key molecules associated with CTB differentiation in culture. We discovered that 477 of the 3026 quantified proteins were differentially expressed across the time-course, with some of the most robustly changing proteins associated with ECM disassembly and migration, indicating remodeling of the CTB proteome towards an invasive phenotype. Our clustering analysis also revealed four distinct expression profiles, with notable modulation of proteins involved with oxidative phosphorylation and regulators of migratory/invasive functions. These data support the roles of numerous molecules previously identified by our group and others as regulators of CTB differentiation/invasion, and provide potential candidate molecules to be further explored in a similar context.
Three major observations emerged from our proteomic dataset. The first is the crucial role of oxidative phosphorylation in CTB differentiation/invasion. The multifaceted functions of the placenta necessitates substantial energy demands. According to GO Enrichment analysis, components of the electron transport chain (ETC) complexes were the most significantly enriched DE proteins in the global CTB proteome as a function of time in culture. Placental mitochondria have to be highly adaptable to the energy demands of their immediate environment, particularly in response to variable oxygen levels (Fisher et al., 2019). Prior to remodeling of the uterine vasculature, the placenta proper is maintained in a physiologically hypoxic environment, necessitating tight regulation of energy consumption relative to the developing fetus (Illsley et al., 2010). Although the relationship among oxygen tension, bioenergetics and CTB differentiation/invasion are important, the extent of the influence of each variable is not fully resolved. The role of oxygen in CTB differentiation is context driven (Treissman et al., 2020), with low and high levels promoting proliferation or differentiation/invasion, depending on factors such as gestational age (Caniggia et al., 2000; James et al., 2006; Wakeland et al., 2017), CTB maturation (Chang et al., 2018) and the culture system. However, the placenta is metabolically flexible, using glycolysis and oxidative phosphorylation to adapt to energy demands (Bax and Bloxam, 1997; Kolahi et al., 2017; Sferruzzi-Perri et al., 2019). Our data demonstrate that nearly all cataloged components of the ETC were upregulated in CTBs cultured under standard (20% O2) conditions, although the relevance of the downregulation of NDUFS8, a subunit of Complex I, and COX6B1, a subunit of Complex IV, is unclear. Given the role of metabolic programming in cellular differentiation, and the prevailing hypothesis that altered mitochondrial function is responsible for the pathogenesis of certain pregnancy complications, the proper modulation of ETC components during differentiation/invasion is likely to be crucial to normal pregnancy outcomes.
Second, the intriguing overlap between invasive human CTBs and cancer cells was reinforced. Invasive human CTBs use mechanisms that are implicated in the transition of benign tumor cells to a malignant phenotype (Ferretti et al., 2007). A significant difference is the tight regulation of CTB invasion, which is limited to the inner one-third of the myometrium. Thus, the dynamics of CTB expression of tumorigenic molecules often intersect those of cancer cells. For example, the divergent expression of RBP1 and SLC25A6, which is a cancer diagnostic, was observed in our dataset (Watkinson et al., 2008). Our results also demonstrate that ALDH1A2, a tumor suppressor gene that has a hypermethylated promoter in numerous cancers, is similarly downregulated in CTB differentiation/invasion. Meanwhile, GPRC5A, which can behave as an oncogene or tumor suppressor depending on context, was upregulated in CTBs with coincident downregulation of STAT3, suggesting that it may function as a tumor suppressor in this system (Chen et al., 2010). Another shared function between CTBs and tumor cells is the expression of immune inhibitory molecules, including HLA-G and PD-L1. Our lab has previously shown that HLA-G is restricted to CTBs differentiated along an invasive pathway (McMaster et al., 1995). Our proteomic data confirmed HLA-G expression while also observing sustained PD-L1 upregulation after culturing, affirming previous observations regarding expression of this molecule in second trimester samples (Holets et al., 2006).
Finally, our SWATH-MS experiments identified molecules whose functions are not fully known in the context of CTB differentiation, providing a valuable resource for future hypothesis generation. By assessing the global proteome of CTBs at various time points, the temporal coordination of pathways can provide insights into their functions. For example, SQSTM1/p62, a key regulator in the autophagic pathway (Katsuragi et al., 2015), was robustly upregulated during invasive CTB differentiation, along with associated components such as LC3 (MAP1LC3B) (Pankiv et al., 2007). The concurrent timing of the autophagic pathway with accompanying increases in oxidative phosphorylation and oxidative stress regulators (e.g. SOD2, GPX1) suggests further study regarding the influences of metabolic programming in CTB differentiation may be warranted (Shyh-Chang and Ng, 2017). In addition, our group has been interested in the role of histones and histone modifications in placental development. The progressive downregulation of SUPT16H, a major subunit of the FACT complex, may signal a maintenance or stabilization of chromatin structure as CTBs differentiate, with a decreased reliance on dedicated histone chaperones.
In conclusion, the compilation of the CTB global proteome at various stages of differentiation constitutes an important resource for investigators in the field that confirms previous results from our group and others, and highlights potentially fruitful areas for further mechanistic investigations of invasion. Our quantitative proteomics results give a comprehensive overview of protein abundances in time, thereby providing a unique database regarding protein expression patterns in cultured CTBs. Furthermore, given the nature of SWATH-MS, which allows continued building of a spectral library, our analyses constitute a foundation on which additional proteomic studies aimed at increasing our understanding of CTB differentiation dynamics can be constructed.
MATERIALS AND METHODS
Tissue collection
All methods were approved by the UCSF Institutional Review Board. Informed consent was obtained from all donors. Second trimester placentas were collected following elective terminations and placed in cytowash medium [DME/H-21 (Gibco), 12.5% fetal bovine serum (Hyclone), 1% glutamine plus (Atlanta Biologicals), 1% penicillin/streptomycin (Invitrogen) and 0.1% gentamicin (Gibco)]. Tissue samples were stored on ice prior to dissection.
Human primary villous cytotrophoblast culture
Cytotrophoblasts (CTBs) were isolated as previously described (Hunkapiller and Fisher, 2008). CTBs intended for proteomic analyses were cultured on precoated Matrigel (BD Biosciences) plates at a density of 104,167 cells/cm2 in medium containing DME/H-21, 2% Nutridoma (Roche), 1% sodium pyruvate (Sigma), 1% HEPES buffer (Invitrogen), 1% GlutaminePlus (Atlanta Biologicals) and 1% penicillin/streptomycin (Invitrogen). Cells were incubated at 37°C in 5% CO2/95% air. Cell culture purity was determined with cytokeratin (rat polyclonal; 1:100 (Damsky et al., 1992) and vimentin (rabbit monoclonal: 1:500) immunostaining after cytocentrifugation (800 g, 5 min); relative trophoblast purity above 85% was required for subsequent experimentation.
Protein extraction and digestion
Cultured CTBs were washed with 1×PBS and detached with Trypsin (∼5 min incubation) at 15, 24 and 39 h. Cell suspensions were centrifuged at 800 g for 5 min. The supernatant was removed and cells were washed twice with 1×PBS, centrifuging after each wash. After the final wash, supernatant was removed and cell pellets were snap-frozen and stored at −80°C. To prepare samples for mass spectrometry, samples were thawed and sonicated in lysis buffer (1% SDS in 50 mM ammonium bicarbonate) and protein concentrations were quantified using Micro BCA (Thermo Scientific). Equal amounts of protein (25 μg) were reduced in 5 mM TCEP (tris carboxyethylphospine) and incubated at 60°C for 45 min. Iodoacetic acid was added to the samples at a final concentration of 10 mM and incubated at room temperature for 15 min, covered. Trypsin (Promega) was added at a 1:20 ratio (w/w) and samples were digested overnight at 37°C. Detergent was removed from the samples with Pierce Detergent Removal Columns (Thermo Scientific) according to manufacturer's protocol.
Peptide fractionation and SWATH-MS
Peptides were separated offline into eight fractions using alkaline pH reversed-phase HPLC. Peptides from each fraction were separated using a nanoLC Ultra 2D Plus system (SCIEX) with the cHiPLC system interfaced with a 5600 Triple TOF mass spectrometer (SCIEX). The peptides were loaded onto a guard column (300 μm i.d.×5 mm, 5 μm particle size, 100 Å pore size; Acclaim PepMap300 C18; Thermo-Fisher) and were washed with aqueous phase composed of 2% solvent B [98% acetonitrile (ACN)/0.1% formic acid (FA)] in solvent A (2% ACN/0.1% FA), flow rate 12 μl/min for 3 min. The peptides were then separated on a C18 Acclaim PepMap100 column (75 μm i.d.×150 mm, 3 mm particle size, 100 Å pore size; Thermo Fisher Scientific) heated at 40°C. Peptides were eluted at a flow rate of 300 nl/min with a 90 min gradient of 2-35% solvent B. In positive-ion mode, MS scans from m/z 400-1200 were acquired followed by MS/MS scans of the 20 most abundant ions with an exclusion time of 15 s. SWATH-MS acquisition was performed with the instrumentation described above. A 50-variable-window setup for SWATH acquisitions was determined using the ‘variable window calculator’ algorithm of SCIEX. MS1 survey scans were acquired from 400-1200m/z for 250 ms and MS2 spectra were acquired in high-sensitivity mode from 100-1500m/z for 65 ms. The spectral library was constructed by processing the raw data-dependent fractionation files through ProteinPilot against a human reference proteome (UniProt). We extracted information that contains at least three transitions per peptide and three peptides per protein. Alignment of peaks was based on retention time of medium to high abundance endogenous peptides. Peak group detections were filtered at 1% FDR using the SWATH Replicates Analysis Template. Spectral libraries and SWATH-MS acquisitions were uploaded to the Illumina BaseSpace Cloud and analyses were performed in SCIEX Cloud OneOmics.
Data availability
Raw chromatogram files have been deposited in the MassIVE (https://massive.ucsd.edu) proteomic database under accession number MSV000086970. The relative abundances of quantified proteins, with annotations, can be found in Table S1.
Bioinformatic analysis
Normalized data were log2 transformed for downstream analysis. Gene ontology (GO) analysis was performed with DAVID (Dennis et al., 2003; Kuleshov et al., 2016) and Perseus (Tyanova and Cox, 2018) using the identified proteins in the study as the background list. Unsupervised hierarchical clustering was performed with MeV (MultiExperiment Viewer).
Transcriptomic and proteomic pairs were assessed with weighted gene co-expression analysis (WGCNA) (Zhang and Horvath, 2005). Consensus network modules were defined using a soft threshold power β=16, a minimum module size of 30 and a dynamic tree cut height of 0.65. Module-trait relationships were determined with the WGCNA package, and functional enrichment analysis of modules was determined using the WGCNA package and Enrichr (Chen et al., 2013). Modules can be found in Table S3.
Immunofluorescent staining
Briefly, tissue was fixed in 4% paraformaldehyde and dehydrated by passing through sequential increases in sucrose before embedding in OCT (Thermo Fisher Scientific). Sections were permeabilized with methanol and rinsed with PBS before incubation in blocking buffer (5% BSA and 0.05% Tween 20). CTBs were labeled with rat anti-cytokeratin 7 (1:100, Damsky et al., 1992) with: PD-L1 (1:100, Cell Signaling, #13684), p62/SQSTM1 (1:100, Novus Biologicals, #H00008878-M01) or NID1 (1:500, R&D Systems, AF2570) for 1 h at 37°C. After three additional PBS washes, primary antibodies were detected using species-specific secondary antibodies (1:1000, Jackson ImmunoResearch Labs, see Table S4). Sections were mounted using Vectashield with DAPI (Vector Laboratories). Images were acquired using a Leica DM5000 B inverted microscope. Antibodies and their sources are described in Table S4. Tissue sections from at least four placentas were evaluated for each condition.
Statistics
Graphs were created in R and Graphpad. Differential expression was determined using linear modeling [limma (Ritchie et al., 2015)]. Principal component analysis [FactoMineR (Lê et al., 2008)], gap statistic [NbClust (Charrad et al., 2014)], fuzzy clustering [Mfuzz (Kumar and Futschik, 2007)] and significance of protein profile similarities [proteinProfiles (Hansson et al., 2012)] were performed using their respective packages. For fuzzy clustering, relative abundance of quantified proteins was log10 transformed, Z-scored and clustered using a fuzzification value of two with a minimum membership value of 0.35. Network analysis was performed with Cytoscape using the STRING database (Shannon et al., 2003).
Supplementary Material
Acknowledgements
We thank the patient participants and recruiters. The authors also thank Drs Joanna Halkias, Adrian Erlebacher and Yan Zhou for their technical insight.
Footnotes
Competing interests
S.J.F. is a consultant for Novo Nordisk.
Author contributions
Conceptualization: H.C., K.E.W., J.F.R., S.J.F.; Methodology: H.C., K.E.W., M.K., J.F.R., S.J.F.; Software: J.F.R.; Validation: H.C., R.K.A.; Formal analysis: H.C., K.E.W., J.F.R.; Investigation: H.C., K.E.W., E.Y.K., M.K., K.A.P., R.K.A., J.F.R.; Resources: H.C., E.Y.K., M.K., K.A.P., J.F.R., S.J.F.; Data curation: H.C.; Writing - original draft: H.C., J.F.R., S.J.F.; Writing - review & editing: H.C., J.F.R., S.J.F.; Visualization: H.C., R.K.A.; Supervision: K.E.W., M.K., J.F.R., S.J.F.; Project administration: K.E.W., J.F.R., S.J.F.; Funding acquisition: S.J.F.
Funding
This work was supported by the generosity of the National Institutes of Health (K99ES030401-01A1 to H.C., R00ES023846 to J.F.R., and P30ES030284-01A1 to Tracey Woodruff and S.J.F.). Open access funding provided by the University of California, San Francisco. Deposited in PMC for immediate release.
Data availability
Raw chromatogram files have been deposited in the MassIVE (https://massive.ucsd.edu) proteomic database under accession number MSV000086970.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199561
References
- Anacker, J., Segerer, S. E., Hagemann, C., Feix, S., Kapp, M., Bausch, R. and Kämmerer, U. (2011). Human decidua and invasive trophoblasts are rich sources of nearly all human matrix metalloproteinases. Mol. Hum. Reprod. 17, 637-652. 10.1093/molehr/gar033 [DOI] [PubMed] [Google Scholar]
- Bax, B. E. and Bloxam, D. L. (1997). Energy metabolism and glycolysis in human placental trophoblast cells during differentiation. Biochim. Biophys. Acta 1319, 283-292. 10.1016/S0005-2728(96)00169-7 [DOI] [PubMed] [Google Scholar]
- Bergman, D., Kadner, S. S., Cruz, M. R., Esterman, A. L., Tahery, M. M., Young, B. K. and Finlay, T. H. (1993). Synthesis of α1-antichymotrypsin and α1-antitrypsin by human trophoblast. Pediatr. Res. 34, 312-317. 10.1203/00006450-199309000-00015 [DOI] [PubMed] [Google Scholar]
- Blankenship, T. N., Enders, A. C. and King, B. F. (1992). Distribution of Laminin, Type-Iv collagen, and fibronectin in the cell columns and trophoblastic shell of early macaque placentas. Cell Tissue Res. 270, 241-248. 10.1007/BF00328009 [DOI] [PubMed] [Google Scholar]
- Bollen, M. (2001). Combinatorial control of protein phosphatase-1. Trends Biochem. Sci. 26, 426-431. 10.1016/S0968-0004(01)01836-9 [DOI] [PubMed] [Google Scholar]
- Caniggia, I., Taylor, C. V., Ritchie, J. W. K., Lye, S. J. and Letarte, M. (1997). Endoglin regulates trophoblast differentiation along the invasive pathway in human placental villous explants. Endocrinology 138, 4977-4988. 10.1210/endo.138.11.5475 [DOI] [PubMed] [Google Scholar]
- Caniggia, I., Mostachfi, H., Winter, J., Gassmann, M., Lye, S. J., Kuliszewski, M. and Post, M. (2000). Hypoxia-inducible factor-1 mediates the biological effects of oxygen on human trophoblast differentiation through TGFbeta(3). J. Clin. Invest. 105, 577-587. 10.1172/JCI8316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.-W., Wakeland, A. K. and Parast, M. M. (2018). Trophoblast lineage specification, differentiation and their regulation by oxygen tension. J. Endocrinol. 236, R43-r56. 10.1530/JOE-17-0402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrad, M., Ghazzali, N., Boiteau, V. and Niknafs, A. (2014). NbClust: an R package for determining the relevant number of clusters in a data set. J. Stat. Softw. 61, 36. 10.18637/jss.v061.i06 [DOI] [Google Scholar]
- Chen, Y., Deng, J., Fujimoto, J., Kadara, H., Men, T., Lotan, D. and Lotan, R. (2010). Gprc5a deletion enhances the transformed phenotype in normal and malignant lung epithelial cells by eliciting persistent Stat3 signaling induced by autocrine leukemia inhibitory factor. Cancer Res. 70, 8917-8926. 10.1158/0008-5472.CAN-10-0518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, E. Y., Tan, C. M., Kou, Y., Duan, Q., Wang, Z., Meirelles, G., Clark, N. R. and Ma'ayan, A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, B. C., Hunter, C. L., Liu, Y., Schilling, B., Rosenberger, G., Bader, S. L., Chan, D. W., Gibson, B. W., Gingras, A.-C., Held, J. M.et al. (2017). Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 8, 291. 10.1038/s41467-017-00249-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, S., Jones, C. J. P., Garrod, A., Hulme, C. H. and Heazell, A. E. P. (2013). Identification of autophagic vacuoles and regulators of autophagy in villous trophoblast from normal term pregnancies and in fetal growth restriction. J. Maternal-Fetal Neonatal Med. 26, 339-346. 10.3109/14767058.2012.733764 [DOI] [PubMed] [Google Scholar]
- Damsky, C. H., Fitzgerald, M. L. and Fisher, S. J. (1992). Distribution patterns of extracellular matrix components and adhesion receptors are intricately modulated during first trimester cytotrophoblast differentiation along the invasive pathway, in vivo. J. Clin. Invest. 89, 210-222. 10.1172/JCI115565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky, C. H., Librach, C., Lim, K. H., Fitzgerald, M. L., Mcmaster, M. T., Janatpour, M., Zhou, Y., Logan, S. K. and Fisher, S. J. (1994). Integrin switching regulates normal trophoblast invasion. Development 120, 3657-3666. 10.1242/dev.120.12.3657 [DOI] [PubMed] [Google Scholar]
- Dasilva-Arnold, S., James, J. L., Al-Khan, A., Zamudio, S. and Illsley, N. P. (2015). Differentiation of first trimester cytotrophoblast to extravillous trophoblast involves an epithelial-mesenchymal transition. Placenta 36, 1412-1418. 10.1016/j.placenta.2015.10.013 [DOI] [PubMed] [Google Scholar]
- De Sousa Abreu, R., Penalva, L. O., Marcotte, E. M. and Vogel, C. (2009). Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512-1526. 10.1039/b908315d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, G., Jr, Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C. and Lempicki, R. A. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Estellés, A., Gilabert, J., Aznar, J., Loskutoff, D. J. and Schleef, R. R. (1989). Changes in the plasma levels of type 1 and type 2 plasminogen activator inhibitors in normal pregnancy and in patients with severe preeclampsia. Blood 74, 1332-1338. 10.1182/blood.V74.4.1332.1332 [DOI] [PubMed] [Google Scholar]
- Feinberg, R. F., Kao, L. C., Haimowitz, J. E., Queenan, J. T., Wun, T. C., Strauss, J. F. and , Kliman, H. J. (1989). Plasminogen-activator inhibitor type-1 and type-2 in human trophoblasts - Pai-1 is an immunocytochemical marker of invading trophoblasts. Lab. Invest. 61, 20-26. [PubMed] [Google Scholar]
- Ferretti, C., Bruni, L., Dangles-Marie, V., Pecking, A. P. and Bellet, D. (2007). Molecular circuits shared by placental and cancer cells, and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum. Reprod. Update 13, 121-141. 10.1093/humupd/dml048 [DOI] [PubMed] [Google Scholar]
- Fisher, S. J. (2015). Why is placentation abnormal in preeclampsia? Am. J. Obstet. Gynecol. 213, S115-S122. 10.1016/j.ajog.2015.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, J. J., Mckeating, D. R., Cuffe, J. S., Bianco-Miotto, T., Holland, O. J. and Perkins, A. V. (2019). Proteomic analysis of placental mitochondria following trophoblast differentiation. Front. Physiol. 10, 1536. 10.3389/Fphys.2019.01536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperowicz, M., Surmann-Schmitt, C., Hamada, Y., Otto, F. and Cross, J. C. (2013). The transcriptional co-repressor TLE3 regulates development of trophoblast giant cells lining maternal blood spaces in the mouse placenta. Dev. Biol. 382, 1-14. 10.1016/j.ydbio.2013.08.005 [DOI] [PubMed] [Google Scholar]
- Gauster, M., Blaschitz, A., Siwetz, M., Huppertz, B., Viodavsky, I., De Groot, N., Ben-Rafael, Z. and Stern, R. (2013). Keratins in the human trophoblast. Histol. Histopathol. 28, 817-825. 10.14670/HH-28.817 [DOI] [PubMed] [Google Scholar]
- Goshen, R., Ariel, I. I., Shuster, S., Hochberg, A., Viodavsky, I., de Groot, N., Ben-Rafael, Z., Stern, R. (1996). Hyaluronan, CD44 and its variant exons in human trophoblast invasion and placental angiogenesis. Mol. Hum. Reprod. 2, 685-691. 10.1093/molehr/2.9.685 [DOI] [PubMed] [Google Scholar]
- Gugnoni, M., Sancisi, V., Manzotti, G., Gandolfi, G. and Ciarrocchi, A. (2016). Autophagy and epithelial-mesenchymal transition: an intricate interplay in cancer. Cell Death Dis. 7, e2520. 10.1038/Cddis.2016.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson, J., Rafiee, M. R., Reiland, S., Polo, J. M., Gehring, J., Okawa, S., Huber, W., Hochedlinger, K. and Krijgsveld, J. (2012). Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2, 1579-1592. 10.1016/j.celrep.2012.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata, K.-I., Ishida, T., Penta, K., Rezaee, M., Yang, E., Wohlgemuth, J. and Quertermous, T. (2001). Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells. J. Biol. Chem. 276, 16223-16231. 10.1074/jbc.M100630200 [DOI] [PubMed] [Google Scholar]
- Hofmann, G. E., Glatstein, I., Schatz, F., Heller, D. and Deligdisch, L. (1994). Immunohistochemical localization of urokinase-type plasminogen activator and the plasminogen activator inhibitors 1 and 2 in early human implantation sites. Am. J. Obstet. Gynecol. 170, 671-676. 10.1016/S0002-9378(94)70246-2 [DOI] [PubMed] [Google Scholar]
- Holets, L. M., Hunt, J. S. and Petroff, M. G. (2006). Trophoblast CD274 (B7-H1) is differentially expressed across gestation: influence of oxygen concentration. Biol. Reprod. 74, 352-358. 10.1095/biolreprod.105.046581 [DOI] [PubMed] [Google Scholar]
- Horvath, S. and Dong, J. (2008). Geometric interpretation of gene coexpression network analysis. PLoS Comput. Biol. 4, e1000117. 10.1371/journal.pcbi.1000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, H.-Y., Wu, W.-R., Wang, Y.-H., Wang, J.-W., Fang, F.-M., Tsai, J.-W., Li, S.-H., Hung, H.-C., Yu, S.-C., Lan, J.et al. (2013). ASS1 as a novel tumor suppressor gene in myxofibrosarcomas: aberrant loss via epigenetic DNA methylation confers aggressive phenotypes, negative prognostic impact, and therapeutic relevance. Clin. Cancer Res. 19, 2861-2872. 10.1158/1078-0432.CCR-12-2641 [DOI] [PubMed] [Google Scholar]
- Hughes, C. S., Postovit, L. M. and Lajoie, G. A. (2010). Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics 10, 1886-1890. 10.1002/pmic.200900758 [DOI] [PubMed] [Google Scholar]
- Hunkapiller, N. M. and Fisher, S. J. (2008). Placental remodeling of the uterine vasculature. Method Enzymol. 445, 281. 10.1016/S0076-6879(08)03012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illsley, N. P., Caniggia, I. and Zamudio, S. (2010). Placental metabolic reprogramming: do changes in the mix of energy-generating substrates modulate fetal growth? Int. J. Dev. Biol. 54, 409-419. 10.1387/ijdb.082798ni [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, J. L., Stone, P. R. and Chamley, L. W. (2006). The effects of oxygen concentration and gestational age on extravillous trophoblast outgrowth in a human first trimester villous explant model. Hum. Reprod. 21, 2699-2705. 10.1093/humrep/del212 [DOI] [PubMed] [Google Scholar]
- Jauniaux, E., Collins, S. and Burton, G. J. (2018). Placenta accreta spectrum: pathophysiology and evidence-based anatomy for prenatal ultrasound imaging. Am. J. Obstet. Gynecol. 218, 75-87. 10.1016/j.ajog.2017.05.067 [DOI] [PubMed] [Google Scholar]
- Katsuragi, Y., Ichimura, Y. and Komatsu, M. (2015). p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 282, 4672-4678. 10.1111/febs.13540 [DOI] [PubMed] [Google Scholar]
- Keir, M. E., Liang, S. C., Guleria, I., Latchman, Y. E., Qipo, A., Albacker, L. A., Koulmanda, M., Freeman, G. J., Sayegh, M. H. and Sharpe, A. H. (2006). Tissue expression of PD-L1 mediates peripheral T cell tolerance. J. Exp. Med. 203, 883-895. 10.1084/jem.20051776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., Lapointe, J., Kaygusuz, G., Ong, D. E., Li, C., Van De Rijn, M., Brooks, J. D. and Pollack, J. R. (2005). The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 65, 8118-8124. 10.1158/0008-5472.CAN-04-4562 [DOI] [PubMed] [Google Scholar]
- Knöfler, M., Haider, S., Saleh, L., Pollheimer, J., Gamage, T. K. J. B. and James, J. (2019). Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell. Mol. Life Sci. 76, 3479-3496. 10.1007/s00018-019-03104-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolahi, K. S., Valent, A. M. and Thornburg, K. L. (2017). Cytotrophoblast, not syncytiotrophoblast, dominates glycolysis and oxidative phosphorylation in human term placenta. Sci. Rep. 7, 42941. 10.1038/srep42941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren, I., Reem, E. and Kimchi, A. (2010). DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr. Biol. 20, 1093-1098. 10.1016/j.cub.2010.04.041 [DOI] [PubMed] [Google Scholar]
- Korhonen, M. and Virtanen, I. (1997). The distribution of laminins and fibronectins is modulated during extravillous trophoblastic cell differentiation and decidual cell response to invasion in the human placenta. J. Histochem. Cytochem. 45, 569-581. 10.1177/002215549704500409 [DOI] [PubMed] [Google Scholar]
- Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., Koplev, S., Jenkins, S. L., Jagodnik, K. M., Lachmann, A.et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90-W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, L. and Futschik, M. E. (2007). Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5-7. 10.6026/97320630002005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder, P. and Horvath, S. (2007). Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 1, 54. 10.1186/1752-0509-1-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê, S., Josse, J. and Husson, F. (2008). FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25, 18. 10.18637/jss.v025.i01 [DOI] [Google Scholar]
- Liu, Y., Beyer, A. and Aebersold, R. (2016). On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535-550. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Ludwig, C., Gillet, L., Rosenberger, G., Amon, S., Collins, B. C. and Aebersold, R. (2018). Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol. Syst. Biol. 14, e8126. 10.15252/msb.20178126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Estrada, J., Menu, E., Roques, P., Barré-Sinoussi, F. and Chaouat, G. (2004). Evaluation of Cytokeratin 7 as an accurate intracellular marker with which to assess the purity of human placental villous trophoblast cells by flow cytometry. J. Immunol. Methods 286, 21-34. 10.1016/j.jim.2003.03.001 [DOI] [PubMed] [Google Scholar]
- Maltepe, E. and Fisher, S. J. (2015). Placenta: the forgotten organ. Annu. Rev. Cell Dev. Biol. 31, 523-552. 10.1146/annurev-cellbio-100814-125620 [DOI] [PubMed] [Google Scholar]
- Mano, Y., Kotani, T., Shibata, K., Matsumura, H., Tsuda, H., Sumigama, S., Yamamoto, E., Iwase, A., Senga, T. and Kikkawa, F. (2011). The loss of endoglin promotes the invasion of extravillous trophoblasts. Endocrinology 152, 4386-4394. 10.1210/en.2011-1088 [DOI] [PubMed] [Google Scholar]
- Matsubara, S., Minakami, H., Sato, I. and Saito, T. (1997). Decrease in cytochrome c oxidase activity detected cytochemically in the placental trophoblast of patients with pre-eclampsia. Placenta 18, 255-259. 10.1016/S0143-4004(97)80059-8 [DOI] [PubMed] [Google Scholar]
- Mcmaster, M. T., Librach, C. L., Zhou, Y., Lim, K. H., Janatpour, M. J., DeMars, R., Kovats, S., Damsky, C. and Fisher, S. J. (1995). Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J. Immunol. 154, 3771-3778. [PubMed] [Google Scholar]
- Mcnally, L., Zhou, Y., Robinson, J. F., Zhao, G., Chen, L., Chen, H., Kim, M. Y., Kapidzic, M., Gormley, M., Hannibal, R.et al. (2020). Up-regulated cytotrophoblast DOCK4 contributes to over-invasion in placenta accreta spectrum. Proc. Natl. Acad. Sci. USA 117, 15852-15861. 10.1073/pnas.1920776117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhauser, J., Crescimanno, C., Kasper, M., Zaccheo, D. and Castellucci, M. (1995). Differentiation of human trophoblast populations involves alterations in cytokeratin patterns. J. Histochem. Cytochem. 43, 579-589. 10.1177/43.6.7539466 [DOI] [PubMed] [Google Scholar]
- Nakashima, A., Yamanaka-Tatematsu, M., Fujita, N., Koizumi, K., Shima, T., Yoshida, T., Nikaido, T., Okamoto, A., Yoshimori, T. and Saito, S. (2013). Impaired autophagy by soluble endoglin, under physiological hypoxia in early pregnant period, is involved in poor placentation in preeclampsia. Autophagy 9, 303-316. 10.4161/auto.22927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv, S., Clausen, T. H., Lamark, T., Brech, A., Bruun, J.-A., Outzen, H., Øvervatn, A., Bjørkøy, G. and Johansen, T. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131-24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Passey, R. J., Williams, E., Lichanska, A. M., Wells, C., Hu, S., Geczy, C. L., Little, M. H. and Hume, D. A. (1999). A null mutation in the inflammation-associated S100 protein S100A8 causes early resorption of the mouse embryo. J. Immunol. 163, 2209-2216. [PubMed] [Google Scholar]
- Payne, S. H. (2015). The utility of protein and mRNA correlation. Trends Biochem. Sci. 40, 1-3. 10.1016/j.tibs.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröll, J., Blaschitz, A., Hartmann, M., Thalhamer, J. and Dohr, G. (1997). Cytokeratin 17 as an immunohistochemical marker for intramural cytotrophoblast in human first trimester uteroplacental arteries. Cell Tissue Res. 288, 335-343. 10.1007/s004410050819 [DOI] [PubMed] [Google Scholar]
- Red-Horse, K., Zhou, Y., Genbacev, O., Prakobphol, A., Foulk, R., Mcmaster, M. and Fisher, S. J. (2004). Trophoblast differentiation during embryo implantation and formation of the maternal-fetal interface. J. Clin. Invest. 114, 744-754. 10.1172/JCI200422991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W. and Smyth, G. K. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. F., Kapidzic, M., Gormley, M., Ona, K., Dent, T., Seifikar, H., Hamilton, E. G. and Fisher, S. J. (2017). Transcriptional dynamics of cultured human villous cytotrophoblasts. Endocrinology 158, 1581-1594. 10.1210/en.2016-1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, R., Dey, S. K. and Fisher, S. J. (2014). Preterm labor: one syndrome, many causes. Science 345, 760-765. 10.1126/science.1251816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, S. and Nakashima, A. (2014). A review of the mechanism for poor placentation in early-onset preeclampsia: the role of autophagy in trophoblast invasion and vascular remodeling. J. Reprod. Immunol. 101-102, 80-88. 10.1016/j.jri.2013.06.002 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., Chen, W. and Selbach, M. (2011). Global quantification of mammalian gene expression control. Nature 473, 337-342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Seidensaal, K., Nollert, A., Feige, A. H., Muller, M., Fleming, T., Gunkel, N., Zaoui, K., Grabe, N., Weichert, W., Weber, K.-J.et al. (2015). Impaired aldehyde dehydrogenase 1 subfamily member 2A-dependent retinoic acid signaling is related with a mesenchymal-like phenotype and an unfavorable prognosis of head and neck squamous cell carcinoma. Mol. Cancer 14, 204. 10.1186/S12943-015-0476-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sferruzzi-Perri, A. N., Higgins, J. S., Vaughan, O. R., Murray, A. J. and Fowden, A. L. (2019). Placental mitochondria adapt developmentally and in response to hypoxia to support fetal growth. Proc. Natl. Acad. Sci. USA 116, 1621-1626. 10.1073/pnas.1816056116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B. and Ideker, T.et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498-2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang, N. and Ng, H.-H. (2017). The metabolic programming of stem cells. Gene Dev. 31, 336-346. 10.1101/gad.293167.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, H., Takizawa, T., Matsubara, S., Ohkuchi, A., Kuwata, T., Usui, R., Matsumoto, H., Sato, Y., Fujiwara, H., Okamoto, A.et al. (2014). Extravillous trophoblast cell invasion is promoted by the CD44-hyaluronic acid interaction. Placenta 35, 163-170. 10.1016/j.placenta.2013.12.009 [DOI] [PubMed] [Google Scholar]
- Taylor, S. K., Houshdaran, S., Robinson, J. F., Gormley, M. J., Kwan, E. Y., Kapidzic, M., Schilling, B., Giudice, L. C. and Fisher, S. J. (2020). Cytotrophoblast extracellular vesicles enhance decidual cell secretion of immune modulators via TNF-α. Development 147, dev187013. 10.1242/dev.187013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Q., Stepaniants, S. B., Mao, M., Weng, L., Feetham, M. C., Doyle, M. J., Yi, E. C., Dai, H., Thorsson, V., Eng, J.et al. (2004). Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol. Cell. Proteomics 3, 960-969. 10.1074/mcp.M400055-MCP200 [DOI] [PubMed] [Google Scholar]
- Tibshirani, R., Walther, G. and Hastie, T. (2001). Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. B Stat. Methodol. 63, 411-423. 10.1111/1467-9868.00293 [DOI] [Google Scholar]
- Torbergsen, T., Øian, P., Mathiesen, E. and Borud, O. (1989). Pre-eclampsia--a mitochondrial disease? Acta Obstet. Gynecol. Scand. 68, 145-148. 10.3109/00016348909009902 [DOI] [PubMed] [Google Scholar]
- Treissman, J., Yuan, V., Baltayeva, J., Le, H. T., Castellana, B., Robinson, W. P. and Beristain, A. G. (2020). Low oxygen enhances trophoblast column growth by potentiating differentiation of the extravillous lineage and promoting LOX activity. Development 147, dev181263. 10.1242/dev.181263 [DOI] [PubMed] [Google Scholar]
- Turco, M. Y. and Moffett, A. (2019). Development of the human placenta. Development 146, dev163428. 10.1242/dev.163428 [DOI] [PubMed] [Google Scholar]
- Turco, M. Y., Gardner, L., Kay, R. G., Hamilton, R. S., Prater, M., Hollinshead, M. S., Mcwhinnie, A., Esposito, L., Fernando, R., Skelton, H.et al. (2018). Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 564, 263-267. 10.1038/s41586-018-0753-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova, S. and Cox, J. (2018). Perseus: a bioinformatics platform for integrative analysis of proteomics data in cancer research. In Cancer Systems Biology: Methods and Protocols (ed. von Stechow L.), pp. 133-148. New York, NY: Springer New York. [DOI] [PubMed] [Google Scholar]
- Veras, E., Kurman, R. J., Wang, T.-L. and Shih, I.-M. (2017). PD-L1 expression in human placentas and gestational trophoblastic diseases. Int. J. Gynecol. Pathol. 36, 146-153. 10.1097/PGP.0000000000000305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeland, A. K., Soncin, F., Moretto-Zita, M., Chang, C.-W., Horii, M., Pizzo, D., Nelson, K. K., Laurent, L. C. and Parast, M. M. (2017). Hypoxia directs human extravillous trophoblast differentiation in a hypoxia-inducible factor-dependent manner. Am. J. Pathol. 187, 767-780. 10.1016/j.ajpath.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson, J., Wang, X., Zheng, T. and Anastassiou, D. (2008). Identification of gene interactions associated with disease from gene expression data using synergy networks. BMC Syst. Biol. 2, 10. 10.1186/1752-0509-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikström, A.-K., Nash, P., Eriksson, U. J. and Olovsson, M. H. (2009). Evidence of increased oxidative stress and a change in the plasminogen activator inhibitor (PAI)-1 to PAI-2 ratio in early-onset but not late-onset preeclampsia. Am. J. Obstet. Gynecol. 201, 597.e1-8. 10.1016/j.ajog.2009.06.024 [DOI] [PubMed] [Google Scholar]
- Winship, A. L., Koga, K., Menkhorst, E., Van Sinderen, M., Rainczuk, K., Nagai, M., Cuman, C., Yap, J., Zhang, J.-G., Simmons, D.et al. (2015). Interleukin-11 alters placentation and causes preeclampsia features in mice. Proc. Natl. Acad. Sci. USA 112, 15928-15933. 10.1073/pnas.1515076112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Sui, L., Qiu, B., Yin, X., Liu, J. and Zhang, X. (2019). ANXA4 promotes trophoblast invasion via the PI3K/Akt/eNOS pathway in preeclampsia. Am. J. Physiol. Cell Physiol. 316, C481-C491. 10.1152/ajpcell.00404.2018 [DOI] [PubMed] [Google Scholar]
- Yagel, S., Parhar, R. S., Jeffrey, J. J. and Lala, P. K. (1988). Normal nonmetastatic human trophoblast cells share in vitro invasive properties of malignant cells. J. Cell. Physiol. 136, 455-462. 10.1002/jcp.1041360309 [DOI] [PubMed] [Google Scholar]
- Yang, Y., Lim, S.-K., Choong, L.-Y., Lee, H., Chen, Y., Chong, P.-K., Ashktorab, H., Wang, T. T., Salto-Tellez, M., Yeoh, K. Y.et al. (2010). Cathepsin S mediates gastric cancer cell migration and invasion via a putative network of metastasis-associated proteins. J. Proteome Res. 9, 4767-4778. 10.1021/pr101027g [DOI] [PubMed] [Google Scholar]
- Zhang, B. and Horvath, S. (2005). A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17. 10.2202/1544-6115.1128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw chromatogram files have been deposited in the MassIVE (https://massive.ucsd.edu) proteomic database under accession number MSV000086970. The relative abundances of quantified proteins, with annotations, can be found in Table S1.