ABSTRACT
Cell shape regulation is important, but the mechanisms that govern shape are not fully understood, in part due to limited experimental models in which cell shape changes and underlying molecular processes can be rapidly and non-invasively monitored in real time. Here, we used an optogenetic tool to activate RhoA in the middle of mononucleated macrophages to induce contraction, resulting in a side with the nucleus that retained its shape and a non-nucleated side that was unable to maintain its shape and collapsed. In cells overexpressing focal adhesion kinase (FAK; also known as PTK2), the non-nucleated side exhibited a wide flat morphology and was similar in adhesion area to the nucleated side. In cells overexpressing fascin, an actin-bundling protein, the non-nucleated side assumed a spherical shape and was similar in height to the nucleated side. This effect of fascin was also observed in fibroblasts even without inducing furrow formation. Based on these results, we conclude that FAK and fascin work together to maintain cell shape by regulating adhesion area and height, respectively, in different cell types.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Cell shape, Fascin, Focal adhesion kinase, Microscopy, Optogenetics
Summary: Fascin and focal adhesion kinase work in conjunction to regulate cell morphology during shape-altering processes such as furrow formation by controlling height and adhesion area, respectively.
INTRODUCTION
Cells have different shapes and sizes that often influence their functions. A cell's shape is defined by the geometrical space that it occupies, and encompasses parameters such as aspect ratio, size and membrane curvature (Paluch et al., 2006; Chen and Gumbiner, 2006). Regulation of cell shape is necessary to sustain a number of vital processes. Studying the mechanisms behind shape regulation is, therefore, of great importance.
In processes like cytokinesis (Théry and Bornens, 2006; Wildwater et al., 2011), migration (Bodor et al., 2020; Driscoll et al., 2012; Bretscher, 2008; Pasqualato et al., 2013) and apical contraction (Sawyer et al., 2010), biochemical signals induce cell contraction through cytoskeletal networks. In cytokinesis, the cell needs to contract at the midpoint to form the cleavage furrow, while during cell migration the cell produces membrane extensions such as lamellipodia and filopodia through actin polymerization (Bodor et al., 2020; Théry and Bornens, 2006). Cytoskeletal contraction is, therefore, an important part for cell shape changes (Paluch and Heisenberg, 2009; Cadart et al., 2014). Cell shape also depends on the cell's interaction with the surrounding environment, and it has been proposed that a cell mechano-senses external signals through adhesions to the extracellular matrix and responds to these signals through cell shape changes (Ohashi et al., 2017; Taneja et al., 2016; Ron et al., 2017). It follows that shape regulation involves a combination of biochemical and mechano-transduction signals (Martino et al., 2018; Wolfenson et al., 2019; Dahl et al., 2008; Stephens et al., 2019), but the exact role of these signals and how they work together is unclear.
Identifying the signaling pathways involved in cell shape maintenance has been difficult, because much of the research in this area has relied on theoretical and mathematical models (Marshall et al., 2012; Mohan et al., 2015; Srivastava et al., 2016; Marée et al., 2012). The existing experimental methods for studying cell shape in processes like cell division often involve growing cells on surfaces of different shapes and topographies (Ron et al., 2017) or confining cells to specific areas (Chen et al., 2003). Additionally, biochemical methods, such as disruption of the cytoskeleton with inhibitors or knockdown of small GTPase activity (Mammoto et al., 2007; Vega et al., 2011; Sero and Bakal, 2017) have been used in cell shape studies. These methods may allow researchers to see the recruitment and localization of certain proteins and observe cell shape changes after periods of time, but they do not allow for precise spatiotemporal manipulation of cell shape to study the dynamic cellular responses in real time.
Optogenetic approaches provide a powerful alternative, as they have previously been used to induce and study dynamic processes such as polarization and migration (Karunarathne et al., 2015; Meshik et al., 2018; O'Neill et al., 2018; Berlew et al., 2020), cytokinesis (Castillo-Badillo et al., 2020; Wagner and Glotzer, 2016) and transport (Adrian et al., 2017; Duan et al., 2015). Previously, we found that we can induce furrow and intercellular bridge formation in multinucleated macrophage cells by optogenetically activating RhoA in the middle of the cell to generate actomyosin contraction (Castillo-Badillo et al., 2020). Here, we used the same optogenetic approach in mononucleated cells to induce the cell shape changes associated with furrow formation. This results in one side of the cell containing a nucleus and another side lacking a nucleus. Because the nucleus is absent during native late anaphase, when the cleavage furrow creates nascent daughter cells and cannot provide mechanical support to maintain cell shape, this method allowed us to use the non-nucleated side to identify hypothetical mechanisms that are involved in cell shape maintenance, as nascent daughter cells form during furrow formation.
Using this optogenetic regime, we found that, in mononucleated macrophages, focal adhesion kinase (FAK; also known as PTK2) is involved in regulating the adhesion area during optogenetically induced furrow formation, while the actin-bundling protein fascin is involved in regulating the height. This allows the cell to generate and maintain the cell shape. Both proteins work downstream of RhoA (Del Re et al., 2008; Villari et al., 2015; Torsoni et al., 2005).
We also found that fascin regulates cell height in flat and highly adherent fibroblasts when actomyosin contraction is induced globally, demonstrating that the role of fascin in cell shape regulation is not restricted to nascent daughter cells that result from furrow formation.
RESULTS
Optogenetic tool for inducing cell shape changes
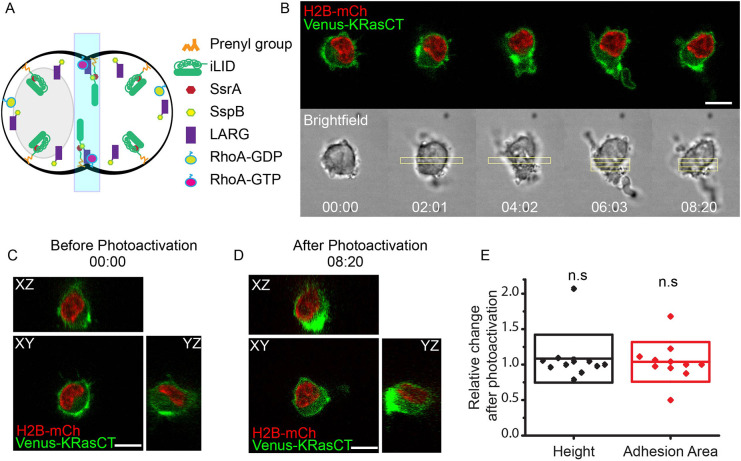
Previously, we found that we could optogenetically induce cleavage furrow formation in multinucleated macrophage cells by optically activating RhoA in the middle to generate actomyosin contraction (Castillo-Badillo et al., 2020). This optogenetic tool consists of a light-inducible dimerization (iLID) component (Guntas et al., 2015) containing the peptide SsrA and attached to a CaaX motif for plasma membrane localization, and a second component composed of a guanine-nucleotide-exchange factor (GEF) for RhoA, LARG (also known as ARHGEF12), fused to the peptide SspB. After stimulation with blue light, iLID undergoes a conformational change, which exposes SsrA, which then dimerizes with SspB (Fig. 1A). In this manner LARG is recruited to the plasma membrane, where it activates RhoA (Castillo-Badillo et al., 2020; O'Neill et al., 2018). Because this optogenetic approach was able to effectively induce the complex shape changes associated with cleavage furrow and intercellular bridge formation in multinucleated cells, we utilized it here to examine the mechanisms regulating cell shape.
Fig. 1.
Non-nucleated side collapses after RhoA-induced contraction in the middle of mononucleated control cells. (A) Diagram of optical control of cell shape changes during cleavage furrow formation by RhoA activation using the light-inducible dimerization (iLID) system. (B) Confocal images of cell shape changes via optogenetic activation of RhoA. Mononucleated cell is expressing LARG-mTurq-SspB, iLID-CaaX, the nuclear marker H2B-mCh (red) and the plasma membrane marker Venus-KRasCT (green). Image is representative of 45 single confocal images of different cells. (C) Orthogonal views of the cell in B prior to RhoA photoactivation. (D) Orthogonal views of the cell in B after RhoA photoactivation. (E) Relative change in cell shape parameters in response to photoactivation. ‘Relative change after photoactivation’ refers to the value of height or adhesion area at t=10 min divided by the value at t=0. The box represents s.d. n=11. n.s., not significant; P=0.42566 (height), P=0.65533 (adhesion area), one-sample Student's t-test, mean=1. For these and all subsequent data shown, experiments were repeated on at least two separate days. Rectangles represent the area of photoactivation. Scale bars: 10 μm. Time is in min:s.
We transfected RAW 264.7 mononucleated macrophage cells with iLID-CaaX, LARG-mTurq-SspB, membrane marker Venus-KRasCT and nuclear marker H2B-mCherry (mCh). We subsequently refer to these cells as control cells because they express native levels of structural proteins and cannot form a furrow. RhoA was optically activated in the middle of mononucleated cells to induce the cell shape changes associated with furrow formation, and images were acquired every 5 s. As in our previous work (Castillo-Badillo et al., 2020), we added a second photoactivation region after the furrow formed in order to promote the movement of the two sides in opposite directions and maintain the furrow shape. A 51-plane z-stack of the cell was acquired at the beginning and end of the experiment.
Upon RhoA activation, the cells rapidly contract in the middle, and the contraction causes the nucleus to be pushed to one side of the photoactivated region. After several minutes, the non-nucleated side collapses completely, forming a structure resembling a uropod (Fig. 1B; Movie 1). The cell then migrates in the direction away from the light, as in the case of optogenetically induced amoeboid migration (O'Neill et al., 2018). The non-nucleated side collapses, but the nucleated side maintains a morphology similar to that of the cell before photoactivation, with comparable height and adhesion area to the pre-photoactivated cell (Fig. 1B–E). This provided a model system to investigate the role of adhesion and cytoskeletal elements in maintaining the shape of the cell in the absence of potential mechanical support from the nucleus. By expressing molecules involved in adhesion and cytoskeletal rigidity, we investigated the role of these elements in conferring cell shape stability.
FAK regulates cell adhesion area through adhesion turnover
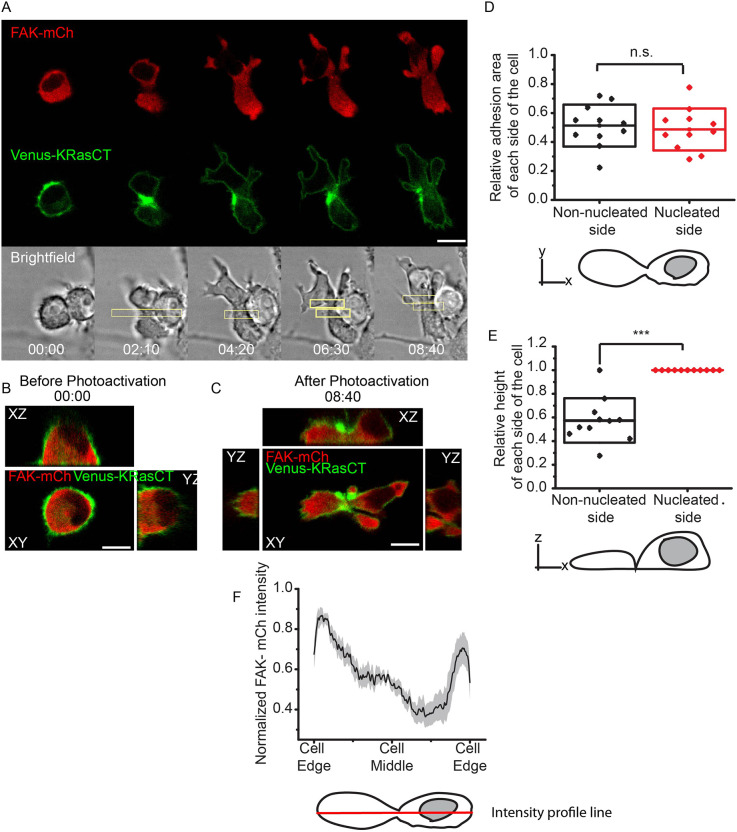
Focal adhesions are complexes composed of regulatory and structural proteins that control the cell's attachment to the extracellular matrix (Mitra et al., 2005). Overexpression of proteins involved in focal adhesions allows us to test whether the degree of adhesions is involved in cell shape maintenance. FAK is an attractive candidate, because it plays a key role in focal adhesion turnover (Hamadi et al., 2005; Hu et al., 2014). We expressed FAK in macrophage cells and again optically induced contraction in the middle. Prior to RhoA photoactivation, FAK-overexpressing cells exhibit a similar morphology to that of control cells (Figs 1C and 2B; Fig. S2). However, after RhoA photoactivation, the non-nucleated side in FAK-overexpressing cells does not collapse after furrow formation. Both the nucleated and non-nucleated sides begin with relatively rounded shapes, turning significantly flatter and more laterally elongated with significant membrane protrusions upon continued photoactivation. The FAK-mCh fluorescence appears higher in regions distal to the furrow, indicating a greater presence of FAK in these regions (Fig. 2A,F; Movie 2). In contrast to the cells with wild-type levels of FAK (Fig. 1B; Movie 1), the two sides appear fairly symmetrical after 8 min of photoactivation (Fig. 2A–C). To determine the extent of this symmetry, we measured each side's height and adhesion area, or the area of the plane of the cell in contact with the glass, using the 3D image acquired after photoactivation (Fig. 2C).
Fig. 2.
FAK overexpression maintains adhesion area during cell shape changes associated with furrow formation. (A) Confocal images of cell shape changes via optogenetic activation of RhoA. Mononucleated cell is expressing LARG-mTurq-SspB, iLID-CaaX, the nuclear marker FAK-mCh (red) and Venus-KRasCT (green). Image is representative of 42 of 46 single confocal images of different cells. (B) Orthogonal views of the cell in A prior to RhoA photoactivation. Images were reconstructed from a 51-plane z-stack. (C) Orthogonal views of the cell in A after RhoA photoactivation. Images were reconstructed from a 51-plane z-stack. (D) Relative adhesion area of non-nucleated and nucleated side after photoactivation. ‘Relative adhesion area’ refers to the adhesion area of the side in question divided by the total adhesion area of the cell. n=11. The box represents s.d. n.s., not significant; P=0.766, paired Student's t-test. (E) Relative height of non-nucleated and nucleated side after photoactivation. ‘Relative height’ is defined as the height of the side in question divided by the maximum height of the cell. The box represents s.d. n=11, 51-plane z-stack images. ***P=0.0000206, paired Student's t-test. (F) Normalized FAK-mCh intensity profile along the length of the cell. n=18. Shaded region represents s.e.m. The scheme shows the intensity profile line in red, which is used to quantify the FAK distribution across the cell. Rectangles represent the area of photoactivation. Scale bars: 10 μm. Time is in min:s.
The adhesion areas of the nucleated and non-nucleated sides show no statistically significant difference (Fig. 2E), indicating that the two sides are symmetrical when it comes to adhesion area. The simplest explanation is that RhoA activates FAK through ROCK protein, and this activates focal adhesion turnover (Mierke et al., 2017; Mitra et al., 2005; Seo et al., 2011). If FAK is the limiting step in this process, then overexpressing FAK increases the adhesion turnover after RhoA photoactivation, as the cell is undergoing shape changes. The cell is able to regulate adhesion turnover in regions distal to the furrow, as evidenced by the greater presence of FAK there (Fig. 2F). This increased adhesion turnover gives stability to the cell, allowing it to maintain the furrow and prevent the non-nucleated side from collapsing.
Unlike adhesion area, the height of the two sides is significantly different, with the non-nucleated side's height being around half of the nucleated side's height (Fig. 2D). It appears that the increased FAK-mediated adhesion turnover and cell elongation promote a flatter morphology in the absence of a nucleus. This suggests that, while FAK is maintaining the non-nucleated side's adhesion area to promote symmetry during shape changes, it does not maintain symmetry when it comes to height. We hypothesized that the height maintenance of the cell could instead be mediated by cytoskeletal proteins and their regulation.
Fascin regulates the height of the cell by conferring rigidity to the cytoskeleton
Actin-bundling proteins are known to control cytoskeletal rigidity, and their expression has been shown to increase during cell shape perturbation (Ron et al., 2017). We reasoned that actin-bundling proteins could be involved in maintaining cell shape changes during processes like furrow formation, similar to FAK. Fascin is an actin-bundling protein that has been shown to regulate membrane protrusions (Jayo et al., 2016; Yamashiro et al., 1998; Fletcher and Mullins, 2010), and its inhibition results in a more loose or less rigid cytoskeleton (Elkhatib et al., 2014; Huang et al., 2015). We therefore hypothesized that fascin could be involved in regulating cell shape through the cytoskeleton.
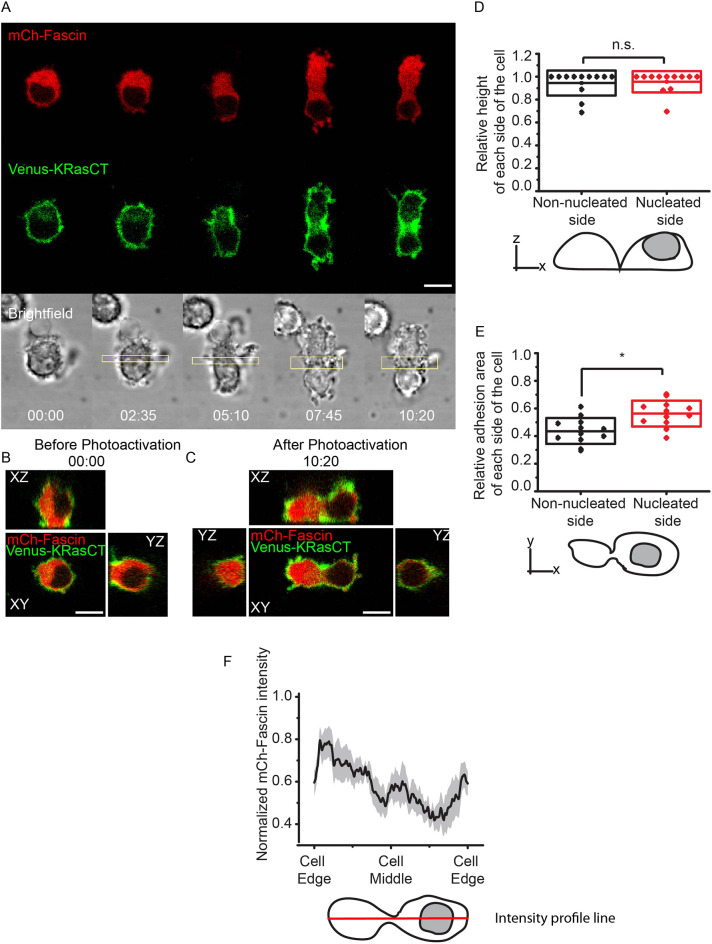
We found that, like FAK-overexpressing cells, fascin-overexpressing cells are morphologically similar to control cells (Figs 1C and 3B; Fig. S2) prior to RhoA photoactivation. However, when RhoA is photoactivated in the middle of mononucleated cells overexpressing fascin-mCh, the cells are able to maintain the furrow and, in some cases, a small intercellular bridge. The fascin-mCh fluorescence is higher in the non-nucleated side of the cell (Fig. 3A,F; Movie 3), suggesting that fascin is preferentially recruited to that side. Like the FAK-overexpressing cells, the nucleated and non-nucleated sides appear fairly symmetrical, but exhibit a more spherical morphology compared to cells overexpressing FAK (Fig. 3A–C). Quantitative analysis showed that the two sides are not significantly different in height (Fig. 3D), but the nucleated side has a larger adhesion area (Fig. 3E), suggesting that fascin-mediated cytoskeletal regulation helps to maintain cellular height.
Fig. 3.
Fascin overexpression maintains cell height during cell shape changes associated with furrow formation. (A) Confocal images of cell shape changes via optogenetic activation of RhoA. Mononucleated cell is expressing LARG-mTurq-SspB, iLID-CaaX, the nuclear marker fascin-mCh (red) and Venus-KRasCT (green). Image is representative of 47 out of 51 single confocal images of different cells. (B) Orthogonal views of the cell in A prior to photoactivation. Images were reconstructed from a 51-plane z-stack. (C) Orthogonal views of the cell in A after photoactivation. Images were reconstructed from a 51-plane z-stack. (D) Relative height of non-nucleated and nucleated side after photoactivation. ‘Relative height’ is defined as the height of the side in question divided by the maximum height of the cell. The box represents s.d. n=12, 51-plane z-stack images. n.s., not significant; P=0.819, paired Student's t-test. (E) Relative adhesion area of non-nucleated and nucleated side after photoactivation. ‘Relative adhesion area’ is defined as the adhesion area of the side in question divided by the total adhesion area of the cell. The box represents s.d. n=12. *P=0.0411, paired Student's t-test. (F) Normalized fascin-mCh intensity profile along the length of the cell. n=14. Shaded region represents s.e.m. The scheme shows the intensity profile line in red, to quantify the fascin distribution across the cell. Rectangles represent the area of photoactivation. Scale bars: 10 μm. Time is in min:s.
The results showing that fascin overexpression results in a more spherical shape with a greater height are logical, given that macrophages in their basal state display a spherical morphology (Fig. 1C,D) when compared to highly adherent flat cells like fibroblasts. Because of fascin's known role in conferring cytoskeletal rigidity (Elkhatib et al., 2014; Jayo et al., 2016; Huang et al., 2015), cells expressing higher levels of fascin can more easily maintain their native height while undergoing shape changes. RhoA activation induces actomyosin contraction in the middle of the cell, and a greater presence of fascin allows for more actin bundling and, therefore, increased stabilization of the furrow shape. The finding that fascin seems to be preferentially polarized to the non-nucleated side, as opposed to the side on which the nucleus is conferring the height and spherical shape, suggests that the cell could be maintaining symmetry by recruiting structural proteins such as fascin to regions where they are most needed.
It is known that fascin localizes to focal adhesions and restricts their growth (Elkhatib et al., 2014), which explains why the cell does not extend and flatten like with FAK overexpression. These results provide insight into how FAK and fascin regulate different features of cell shape. Importantly, our optogenetic model allows us to use the non-nucleated part of the cell, which resembles the cell in native anaphase, to uncover the dynamics of the cell shape regulation during furrow formation and potentially other processes.
FAK overexpression increases adhesions during cell shape changes associated with furrow formation
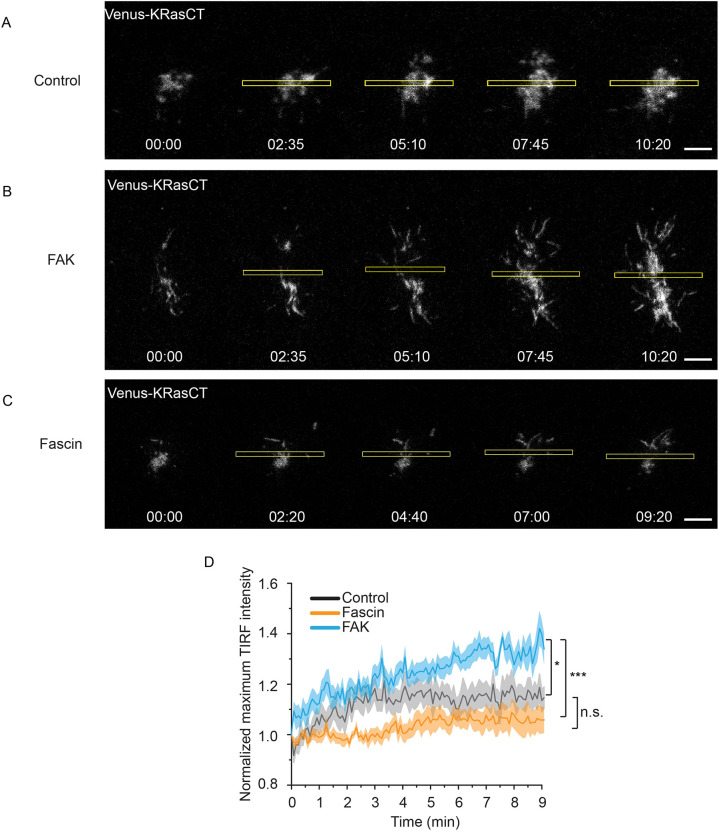
To further study whether FAK is indeed maintaining cell shape by providing stability through adhesions, we used total internal reflection fluorescence (TIRF) imaging to visualize adhesion dynamics in real time while optogenetically inducing shape changes. TIRF microscopy is ideal for visualizing events such as adhesions, which occur at the plasma within ∼100 nm above the substrate (Fish, 2009; Parhamifar and Moghimi, 2012; Poulter et al., 2015). We used the fluorescence of membrane marker Venus-KRasCT to visualize and quantify adhesions in TIRF, because the association of plasma membrane with the substrate is indicative of adhesions.
Control cells, which express wild-type levels of FAK and fascin and are unable to maintain the furrow (Fig. 1), show a brief increase in adhesions upon initiation of photoactivation, followed by a plateau (Fig. 4A,D; Movie 4). Likewise, fascin-overexpressing cells display a minimal change in adhesions (Fig. 4C,D; Movie 4). In contrast, FAK-overexpressing cells exhibit an immediate increase in membrane at the substrate upon optical induction of cell shape changes by RhoA photoactivation, indicative of increased adhesions. The membrane continues to flatten and extend as the cell becomes more adherent (Fig. 4B,D; Movie 4), consistent with our above results showing that FAK overexpression results in flatter cells. These results support the model that FAK is involved in cell shape regulation by stabilization of cell adhesions, whereas fascin works through independent means.
Fig. 4.
Total internal reflection fluorescence (TIRF) visualization of adhesion dynamics during cell shape changes associated to furrow formation. (A) Adhesion dynamics during optogenetic activation of RhoA in control cells. Cell is expressing LARG-mTurq-SspB, iLID-CaaX, H2B-mCh and Venus-KRasCT (gray). Data are representative of 10 out of 10 cells. (B) Adhesion dynamics during optogenetic activation of RhoA in FAK-overexpressing cells. Cell is expressing LARG-mTurq-SspB, iLID-CaaX, FAK-mCh and Venus-KRasCT (gray). Data are representative of 26 out of 30 cells. (C) Adhesion dynamics during optogenetic activation of RhoA in fascin-overexpressing cells. Cell is expressing LARG-mTurq-SspB, iLID-CaaX, fascin-mCh and Venus-KRasCT (gray). Data are representative of 11 out of 13 cells. (D) Normalized maximum intensity of Venus-KRasCT in the TIRF plane. Shaded regions represent s.e.m. Control cells, n=11; FAK-overexpressing cells, n=11; fascin-overexpressing cells, n=13. *P=0.00136, ***P=0.000291; n.s., not significant; P=0.28339; unpaired two-sample Student's t-test. Rectangles represent the area of photoactivation. Scale bars: 10 μm. Time is in min:s.
Inhibition of FAK prevents cell shape changes associated with furrow formation
The above results with FAK and fascin overexpression show that these proteins contribute to different aspects of cell shape, but the extent of their individual requirement is less clear. To determine the extent to which FAK activity is required for cell shape regulation, we used FAKi14, a small molecule inhibitor of FAK autophosphorylation that prevents adhesion formation (Golubovskaya et al., 2008).
To verify that FAKi14 inhibits FAK activity in the cells used here, we observed FAK phosphorylation at the Tyr397 site via immunoblotting. RAW 264.7 cells transfected with LARG-mTurq-SspB and iLID-CaaX were incubated for 30 min with 10 μM FAKi14 and globally photoactivated to activate FAK downstream of RhoA. Cells with no photoactivation showed basal levels of phospho-FAK, while cells after RhoA photoactivation showed an increase in FAK phosphorylation (Fig. S1). Cells treated with FAKi14 after RhoA photoactivation showed a decrease in phospho-FAK (Fig. S1). This result confirms the effectiveness of FAK inhibition by FAKi14 and provides additional evidence that FAK is downstream of RhoA.
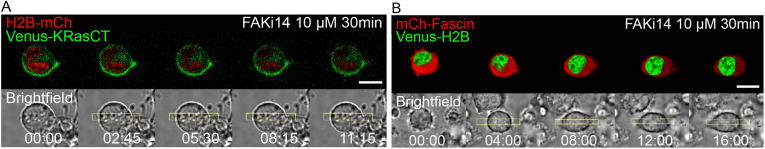
After incubating cells for 30 min with 10 μM FAKi14, we photoactivated RhoA in the middle of mononucleated cells. We found that, in cells with wild-type levels of FAK and fascin, FAK inhibition prevents the shape changes associated with furrow formation in 10 out of 10 cells (Fig. 5A; Movie 5). This is consistent with previous studies showing that FAK works downstream of RhoA (Torsoni et al., 2005; Hirakawa et al., 2004), and FAK-mediated adhesion turnover is necessary to induce cell shape changes.
Fig. 5.
Inhibition of FAK autophosphorylation prevents furrow formation and the associated cell shape changes. (A) Optogenetic activation of RhoA after 30 min incubation with 10 µM FAK inhibitor FAKi14. Cell is expressing LARG-mTurq-SspB, iLID-CaaX, H2B-mCh (red) and Venus-KRasCT (green). Data are representative of 10 out of 10 cells. (B) Optogenetic activation of RhoA in fascin-overexpressing cells after 30 min incubation with 10 µM FAKi14. Cell is expressing LARG-mTurq-SspB, iLID-CaaX, fascin-mCh (red) and Venus-H2B (green). Data are representative of 23 out of 30 cells. Rectangles represent the area of photoactivation. Scale bars: 10 μm. Time is in min:s.
Interestingly, inhibition of FAK autophosphorylation also prevents furrow formation in 23 out of 30 cells overexpressing fascin (Fig. 5B; Movie 5). This shows that fascin alone is not sufficient to induce and maintain shape changes in the cell, and FAK-mediated adhesion turnover is necessary. This formation of new adhesions is required to confer the stability needed to regulate cell shape.
Inhibition of fascin prevents cell shape changes associated with furrow formation
To determine the extent to which fascin activity is required for cell shape regulation, we used G2, a small molecule that inhibits the actin-bundling activity of fascin. It has been shown to block processes that rely on cytoskeletal dynamics, such as filopodial formation, tumor cell migration and invasion (Huang et al., 2015, 2018).
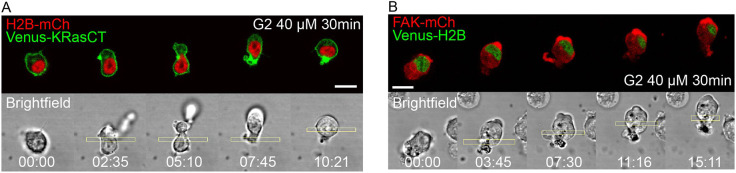
We incubated cells with 40 µM G2 for 30 min. In cells with wild-type levels of FAK and fascin, RhoA activation in the middle initially induces a slight contraction. This furrow cannot be maintained, however, causing the non-nucleated side to collapse and the cell to begin migrating (Fig. 6A; Movie 6). This was the case in 10 out of 10 cells assayed. The migration is amoeboid in nature, as evidenced by plasma membrane accumulation at the back and lack of protrusions. This shows that, like FAK activity, fascin activity is required to generate and maintain cell shape changes. The ability of the cells to migrate in the presence of G2, despite previous studies showing that G2 inhibits filipodial migration, is likely due to RhoA-driven migration being amoeboid in nature, which relies on membrane flow and not on actin-mediated protrusions at the leading edge (O'Neill et al., 2018).
Fig. 6.
Inhibition of fascin-mediated actin bundling prevents furrow formation and the associated cell shape changes. (A) Optogenetic activation of RhoA after 30 min incubation with 40 µM fascin inhibitor G2. Cell is expressing LARG-mTurq-SspB, iLID-CaaX, H2B-mCh (red) and Venus-KRasCT (green). Data are representative of 10 out of 10 cells. (B) Optogenetic activation of RhoA in FAK-overexpressing cells after 30 min incubation with 40 µM G2. Cell is expressing LARG-mTurq-SspB, iLID-CaaX, FAK-mCh and Venus-H2B (green). Data are representative of 28 out of 35 cells. Rectangles represent the area of photoactivation. Scale bars: 10 μm. Time is in min:s.
Likewise, cells overexpressing FAK are also unable to maintain the furrow in the presence of G2 and migrate instead (Fig. 6B; Movie 6). This was the case in 28 out of 35 cells. FAK is recruited to the front of the moving cells, consistent with our earlier results showing FAK at the extending edges distal to the region of photoactivation (Fig. 2A). These results show that FAK activity is independent of fascin, at least partially. Overall, the results of fascin inhibition show that the cell is unable to generate and maintain cell shape changes in the absence of fascin-mediated actin bundling and cytoskeletal dynamics. The fascin and FAK inhibition results together show that these two proteins are independently required to induce and regulate cellular shape changes.
RhoA activation induces fascin-mediated height increase in naturally flat cells
The optogenetic approach of activating RhoA in the middle of mononucleated macrophages to induce furrow formation is a convenient way to rapidly and efficiently generate morphological changes in the cell, while observing the contribution of signaling proteins such as FAK and fascin in real time. Specifically, the non-nucleated side provides insight into cell shape maintenance during native late anaphase. Our finding that cells expressing higher levels of FAK are able to sustain shape changes by maintaining a greater adhesion area is consistent with previous findings on the role of FAK in the formation of strong adhesions (Michael et al., 2009).
The finding that fascin maintains cell height, however, is less intuitive, and we therefore aimed to obtain further evidence of fascin's role in cell shape regulation. Specifically, we tested whether fascin's role in height regulation applies to the general process of cell shape maintenance and not exclusively to furrow formation. This was achieved by inducing RhoA-mediated contraction globally over a cell rather than at the midpoint.
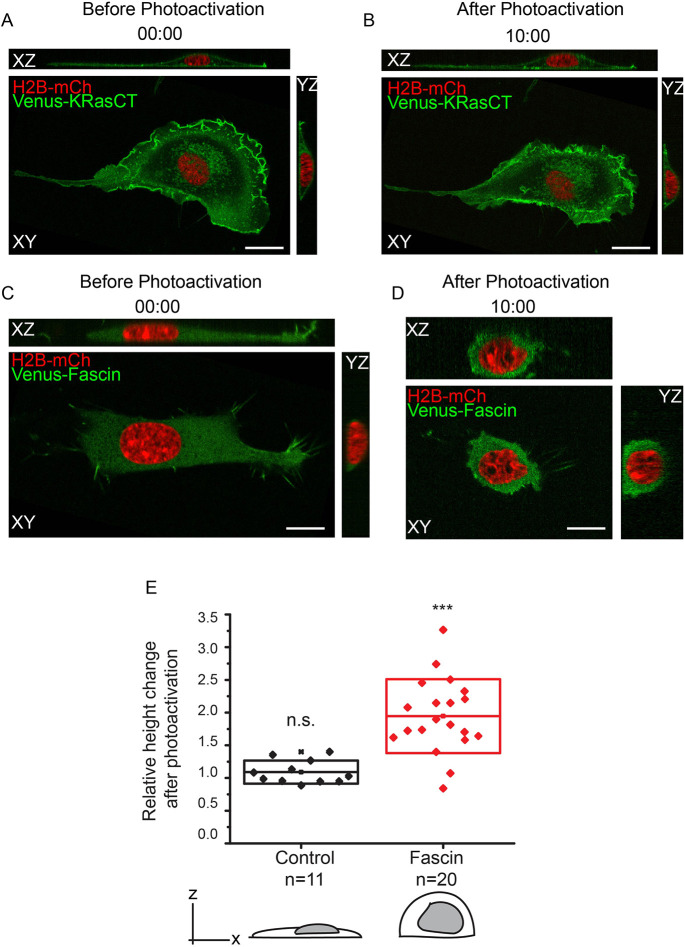
NIH3T3 fibroblast cells exhibit a significantly flatter and more elongated morphology compared to macrophages, making them ideal for testing the effect of fascin on height. We transfected NIH3T3 cells with the optogenetic RhoA-activation tool and globally photoactivated the entire cell for 5 min, acquiring a 51-plane z-stack before and after photoactivation. Cells with wild-type levels of fascin do not exhibit a statistically significant height change in response to global photoactivation (Fig. 7A,B,E; Movie 7). This shows that RhoA activation is not sufficient to produce shape changes in highly adherent and flat cells like fibroblasts expressing wild-type levels of fascin.
Fig. 7.
Fascin-overexpressing fibroblasts increase in height upon global RhoA activation. (A) Orthogonal views of an NIH3T3 cell expressing LARG-mTurq-SspB, iLID-CaaX, H2B-mCh (red) and Venus-KRasCT (green) prior to photoactivation. Data are representative of 12 out of 12 cells. (B) Orthogonal views of the cell in A after 5 min of global RhoA photoactivation. (C) Orthogonal views of an NIH3T3 cell, expressing LARG-mTurq-SspB, iLID-CaaX, H2B-mCh (red) and Venus-fascin (green) prior to photoactivation. Data are representative of 21 out of 21 cells. (D) Orthogonal views of the cell in B after 5 min of global RhoA photoactivation. (E) Relative height change in control and fascin-overexpressing fibroblasts after global RhoA activation. ‘Relative height change’ is defined as the height after 5 min of photoactivation divided by the initial height. The box represents s.d. ***P=0.000000438; n.s., not significant, P=0.12193; one-sample Student's t-test, mean=1. Scale bars: 10 μm. Time is in min:s.
On the other hand, fibroblasts overexpressing fascin exhibit almost a twofold increase in height in response to global RhoA activation. The cells lose their flat elongated morphology and acquire a more spherical shape (Fig. 7C–E; Movie 7). We reasoned that, in these flat cells, like in macrophages, RhoA-mediated actomyosin contractility recruits fascin. When fascin is overexpressed, it is more readily available, making the contraction more efficient. This confers more rigidity to the cytoskeleton, allowing the cell to acquire a more spherical shape and increase its height.
These results again show that RhoA-mediated fascin activity provides the cytoskeletal stability necessary for cells to generate and maintain shape changes, specifically height. Importantly, these data demonstrate that our model can be applied to processes that induce a variety of shape perturbations, both global and localized. They also show that the mechanisms of cell shape regulation are conserved across multiple cell types.
DISCUSSION
The regulation of cell shape is a dynamic response to an interaction between expansive and contractile forces and the structural integrity of the cytoskeleton (Marshall, 2016; Haupt and Minc, 2018). We found that expressing FAK or fascin results in different effects on cell shape by influencing adhesion or cytoskeletal geometry.
FAK is activated by RhoA through ROCK protein to regulate assembly and disassembly of adhesions (Seo et al., 2011; Huveneers and Danen, 2009; Khalil et al., 2014). Activated FAK forms a complex with adhesion proteins like integrins, as well as with paxillin and talin, which link the adhesion complex to the actin cytoskeleton. Inactivation of FAK promotes the disassembly of adhesion complexes, resulting in detachment of the cell from the extracellular matrix (Huveneers and Danen, 2009; Mitra et al., 2005), and inhibition of FAK is known to increase the strength of adhesions by preventing their disassembly (Taneja et al., 2016).
Although the role of FAK in adhesion turnover is quite well established, its role as a cell shape regulator has not been previously studied. The results here suggest that FAK-mediated adhesion turnover regulates cell geometry during shape-altering processes such as furrow formation. A static cell in its basal state requires less adhesion turnover than a cell undergoing dynamic shape changes. Upon furrow formation, the disassembly of adhesions in the middle of the cell allows it to effectively contract and ingress. The dynamic disassembly of existing adhesions and formation of new adhesions in the two sides of the cell, particularly in areas distal to the furrow, allows the cell to form new attachments to the substrate. This provides the cell with more stability in the bottom plane, giving the cell a flatter and more elongated morphology. This is consistent with our findings using TIRF, which showed that FAK-overexpressing cells display higher levels of dynamic adhesions upon RhoA-induced cell shape alteration. Thus, FAK likely allows the cell to undergo complex shape changes by regulating its adhesion area (Fig. 8).
Fig. 8.
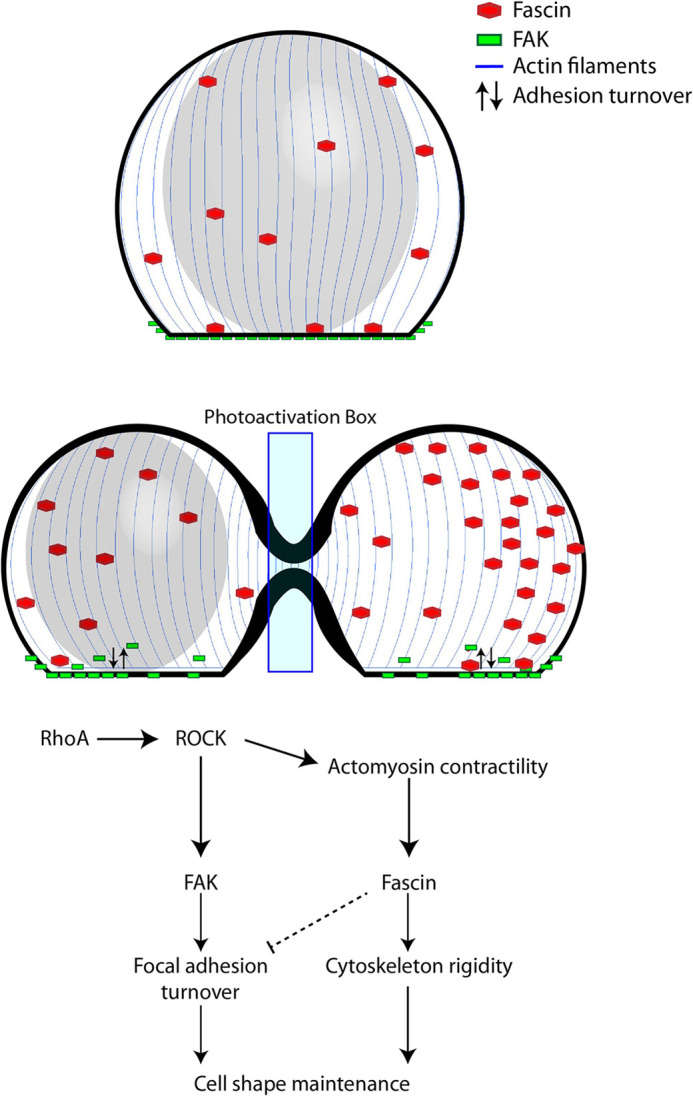
Coordinated regulation of cell shape by FAK and fascin. RhoA activates ROCK protein, which activates both FAK and actomyosin contraction, resulting in increased fascin activity at the cytoskeleton. FAK regulates adhesion area by promoting focal adhesion turnover, while fascin regulates height by increasing cytoskeletal rigidity. Fascin can also interact with FAK to inhibit focal adhesion turnover (Villari et al., 2015).
Fascin plays a different, yet equally important role in cell shape regulation. It is part of the actin-bundling family of proteins, which associate with parallel actin filaments to form rigid arrays (Stevenson et al., 2012). In previous studies, expression of actin-bundling proteins was shown to be higher in cells grown on shape-altering substrates, and these actin-bundling proteins were localized to the cell peripheries, where the shape alteration was most extreme (Ron et al., 2017).
The specific role of fascin, however, when it comes to cell shape regulation, has not been studied in detail. There is evidence of fascin's involvement in nuclear shape regulation (Jayo et al., 2016), and studies have shown that actin filaments become more flexible upon dissociation from fascin (Tanaka et al., 2019), suggesting that fascin can potentially regulate cell shape. Other actin-crosslinking and -bundling proteins have been shown to be involved in cortical contraction, which is an essential part of processes such as migration and cell division (Logue et al., 2015; Ennomani et al., 2016), Here, for the first time to our knowledge, we show that fascin regulates shape by maintaining the height of the cell. According to our model, RhoA- and ROCK protein-mediated actomyosin contraction in the middle of the cell induces increased recruitment of fascin to the cytoskeleton, where it creates rigid arrays of parallel actin filaments. This results in a cytoskeleton with greater height and spherical shape (Fig. 8). In our experiments with optogenetic furrow formation by RhoA photoactivation in mononucleated cells, fascin is present in greater amounts in the non-nucleated side, as opposed to the side on which the nucleus confers the height. This suggests that the cell can direct structural proteins to regions where they are most needed for shape maintenance. Studies have shown that fascin can also interact with FAK to restrict turnover of focal adhesions (Villari et al., 2015; Elkhatib et al., 2014; Jansen et al., 2011; Van Audenhove et al., 2015), which could explain why, in our case, fascin-overexpressing cells display a smaller adhesion area after furrow formation by RhoA photoactivation. Overall, our findings about fascin's role in cell shape regulation introduce the possibility that other actin-bundling and -crosslinking proteins play a key role in this process.
Our model demonstrates, for the first time, that FAK and fascin work in conjunction and in parallel downstream of RhoA to regulate different features of cell shape. This coordination between adhesions and the cytoskeleton has been observed in other processes. For instance, in epithelial cells, adhesions and cortical tension have opposing effects on the interfacial tension at cell–cell boundaries, but a positive feedback loop between signals helps to stabilize the length of the boundaries (Paluch and Heisenberg, 2009; Chanet and Martin, 2014). Our results with fibroblasts also indicate that the pathway proposed here is not unique to macrophages or to furrow formation. Thus, our model can be used to explain how, in different cell types, cell shape is regulated by common mechanisms.
MATERIALS AND METHODS
DNA constructs
The following constructs were purchased from Addgene: FAK-mCh (#55044, deposited by Michael Davidson), H2B-mCh (#20972, deposited by Robert Benezra) and Venus-H2B (#20971, deposited by Robert Benezra). To make Venus-fascin, pcDNA3.1 was cut with NheI and BamH1. A three-way ligation was performed with pcDNA3.1 cut with NheI and BamH1, a PCR fragment of Venus cut with NheI and BsrG1, and a fascin fragment cut out from Addgene #57177 (deposited by Michael Davidson) with BsrG1 and BamH1 (Table S1). There is a spacer generated between Venus and fascin in this construct. To make fascin-mCh, pcDNA 3.1 cut with NheI and BamH1 was ligated to a PCR fragment of mCh with NheI and XmaI, and a fascin fragment cut out of Addgene #51777 using BspE1 and BamH1. XmaI and BspE1 are compatible, and there is also a spacer between the mCherrry and fascin in this construct (Table S1).
LARG-mTurq-SspB (Addgene #90460, deposited by Narasimhan Gautam) and Venus-KRasCT (Addgene #99016, deposited by Narasimhan Gautam) were synthesized as previously described (O'Neill et al., 2018). iLID-CaaX (Addgene #85680, deposited by Narasimhan Gautam) was synthesized as previously described (O'Neill et al., 2016).
Cell culture and transfections
RAW 264.7 and NIH3T3 cells (American Type Culture Collection, Manassas, VA) were grown at 37°C and 5% CO2 in high-glucose Dulbecco's modified Eagle medium (Sigma-Aldrich, St Louis, MO) with 10% dialyzed fetal bovine serum (Atlanta Biologicals) and 1% penicillin–streptomycin. Cells were passaged between five and 15 times before transfection.
Cells were transiently transfected with Lonza Nucleofector 2b electroporator using previously described protocols (Meshik et al., 2018). Around 3 million cells and 0.2–4 μg of each plasmid DNA were used per transfection (4 μg LARG-mTurq-SspB, 2 μg iLID-CaaX, 2 μg FAK-mCh, 2 μg fascin-mCh, 1 μg Venus-KRasCT). Electroporated cells were plated on glass-bottom dishes and imaged 4–10 h after transfection, because cells do not appear healthy after 10 h.
Reagents
The FAK inhibitor FAKi14 (Millipore Sigma, St Louis, MO) was resuspended and stored in deionized H2O and diluted to its working concentration in Hanks’ balanced salt solution (HBSS) containing 1 g/l glucose. Cells were incubated with 10 μM FAKi14 for 30 min prior to imaging.
The fascin inhibitor FASCIN-G2 (Xcess Biosciences, San Diego, CA) was resuspended and stored in dimethyl sulfoxide and diluted to its working concentration in HBSS containing 1 g/l glucose. Cells were incubated with 40 μM FASCIN-G2 for 30 min prior to imaging.
Imaging and photoactivation
A portion of the imaging experiments was performed on an Andor Revolution spinning disk confocal system with a Leica DMI6000B microscope, Yokogawa CSU-X1 spinning disk unit, Andor iXon camera, Andor FRAPPA unit for localized photoactivation and a Leica 63× 1.4 NA oil immersion objective. The system was controlled through Andor iQ2 software. For optical activation of iLID system, a 445 nm laser at 145 nW power was scanned across the selected region at 0.9 ms/μm2. For imaging of fluorescent proteins, 515 nm and 594 nm solid state lasers were used together with Venus 528/20 nm and mCh 628/20 nm emission filters.
The remaining imaging experiments were performed on an Andor Dragonfly spinning disk confocal system with a Nikon Eclipse Ti2 inverted microscope, Andor Zyla and iXon Lite EMCCD cameras, Andor Mosaic 3 DMD array unit for localized photoactivation and Nikon APO TIRF 60× 1.49 NA oil immersion objective. Photoactivation was performed with a CoolLED pE-4000 LED illumination system using a 460 nm wavelength LED at 4.16 mW/mm2. Imaging was performed using 514 nm and 561 nm solid state lasers. For TIRF experiments, penetration was set to 100 nm.
For time series experiments, photoactivation and image acquisition were performed every 5 s. For experiments involving z-stacks, 51 planes were captured for each cell.
All imaging was performed at 37°C and 5% CO2. The culture medium was replaced with HBSS containing 1 g/l prior to imaging.
Quantitative analysis
All image analysis was performed with ImageJ. To quantify cell height, 3D z-stack images were thresholded and the top and bottom planes of the cell were identified using the 3D ROI Manager plugin (Ollion et al., 2013). The number of planes comprising the cell was multiplied by the distance between planes to obtain the height. To quantify adhesion area, the 3D ROI Manager plugin was used to measure the area of the thresholded image corresponding to the bottom plane of the cell. For quantification of TIRF intensity, the maximum intensity value was obtained at each time point using the Measure Stack feature in ImageJ. Intensity profiles along the length of the cell were obtained by drawing a line connecting the distal edges and measuring the fluorescence intensity along the line using the Plot Profile feature in ImageJ. Plots were normalized to account for differences in cell size.
Statistical analysis was performed using Origin Pro. Because the data followed a normal distribution, Student's t-test was used to determine statistical significance.
Western blotting
Three pools of ∼6 million cells each were transiently transfected as described above. We used 0.2–4 μg of each plasmid DNA per transfection (4 μg LARG-mTurq-SspB, 2 μg iLID-CaaX). Electroporated cells were plated on T25 flasks. Cells in the first condition were untreated and kept in the dark to avoid photoactivation. Cells in the second condition were globally photoactivated for 15 min with the CoolLED pE-4000 LED illumination system using a 460 nm wavelength LED at 4.16 mW/mm2. Cells in the third condition were incubated for 30 min with 10 μM FAKi14 and then globally photoactivated for 15 min. Cells were washed with ice-cold HBSS buffer and lysed for 5 min with phosphate-buffered saline containing 25 mM NaF, 5 μM suberoylanilide hydroxamic acid, 5 μM nicotinamide, phosphatase inhibitor cocktail 2 and 3, 1% Triton X-100 and 2% SDS on ice. Lysates were sonicated with five pulses of 30 s and centrifuged at 12,700 g for 15 min. Proteins in supernatants were separated by electrophoresis on 10% SDS-PAGE. Proteins were electrotransferred to nitrocellulose membranes and immunoblotting was performed using the same membranes. Incubation with primary selective antibody anti-FAK(Tyr397) (Cell Signaling Technology, #3283S), 1:500 dilution, in 5% BSA TBS buffer was conducted for 12 h at 4°C and with the secondary antibody for 30 min at room temperature. Super signal-enhanced chemiluminescence kit was employed, exposing the membranes in a Bio-Rad ChemiDoc™ MP System.
Supplementary Material
Acknowledgements
We thank Vani Kalyanaraman for DNA constructs and Xenia Meshik for helpful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: J.A.C.-B.; Formal analysis: J.A.C.-B.; Investigation: J.A.C.-B.; Writing - original draft: J.A.C.-B.; Project administration: N.G.
Funding
This work was supported by the National Institute of General Medical Sciences (R35 GM122577). Deposited in PMC for release after 12 months.
Peer review history
The peer review history is available online at https://journals.biologists.com/jcs/article-lookup/doi/10.1242/jcs.258321
References
- Adrian, M., Nijenhuis, W., Hoogstraaten, R. I., Willems, J. and Kapitein, L. C. (2017). A phytochrome-derived photoswitch for intracellular transport. ACS Synth. Biol. 6, 1248-1256. 10.1021/acssynbio.6b00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlew, E. E., Kuznetsov, I. A., Yamada, K., Bugaj, L. J. and Chow, B. Y. (2020). Optogenetic Rac1 engineered from membrane lipid-binding RGS-LOV for inducible lamellipodia formation. Photochem. Photobiol. Sci. 19, 353-361. 10.1039/C9PP00434C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodor, D. L., Pönisch, W., Endres, R. G. and Paluch, E. K. (2020). Of cell shapes and motion: the physical basis of animal cell migration. Dev. Cell 52, 550-562. 10.1016/j.devcel.2020.02.013 [DOI] [PubMed] [Google Scholar]
- Bretscher, M. S. (2008). On the shape of migrating cells--a ‘front-to-back’ model. J. Cell Sci. 121, 2625-2628. 10.1242/jcs.031120 [DOI] [PubMed] [Google Scholar]
- Cadart, C., Zlotek-Zlotkiewicz, E., Le Berre, M., Piel, M. and Matthews, H. K. (2014). Exploring the function of cell shape and size during mitosis. Dev. Cell 29, 159-169. 10.1016/j.devcel.2014.04.009 [DOI] [PubMed] [Google Scholar]
- Castillo-Badillo, J. A., Bandi, A. C., Harlalka, S. and Gautam, N. (2020). SRRF-stream imaging of optogenetically controlled furrow formation shows localized and coordinated endocytosis and exocytosis mediating membrane remodeling. ACS Synth. Biol. 9, 902-919. 10.1021/acssynbio.9b00521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanet, S. and Martin, A. C. (2014). Mechanical force sensing in tissues. Prog. Mol. Biol. Transl. Sci. 126, 317-352. 10.1016/B978-0-12-394624-9.00013-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. and Gumbiner, B. M. (2006). Paraxial protocadherin mediates cell sorting and tissue morphogenesis by regulating C-cadherin adhesion activity. J. Cell Biol. 174, 301-313. 10.1083/jcb.200602062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. S., Alonso, J. L., Ostuni, E., Whitesides, G. M. and Ingber, D. E. (2003). Cell shape provides global control of focal adhesion assembly. Biochem. Biophys. Res. Commun. 307, 355-361. 10.1016/S0006-291X(03)01165-3 [DOI] [PubMed] [Google Scholar]
- Dahl, K. N., Ribeiro, A. J. S. and Lammerding, J. (2008). Nuclear shape, mechanics, and mechanotransduction. Circ. Res. 102, 1307-1318. 10.1161/CIRCRESAHA.108.173989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Re, D. P., Miyamoto, S. and Brown, J. H. (2008). Focal adhesion kinase as a RhoA-activable signaling scaffold mediating Akt activation and cardiomyocyte protection. J. Biol. Chem. 283, 35622-35629. 10.1074/jbc.M804036200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll, M. K., Mccann, C., Kopace, R., Homan, T., Fourkas, J. T., Parent, C. and Losert, W. (2012). Cell shape dynamics: from waves to migration. PLoS Comput. Biol. 8, e1002392. 10.1371/journal.pcbi.1002392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, L., Che, D., Zhang, K., Ong, Q., Guo, S. and Cui, B. (2015). Optogenetic control of molecular motors and organelle distributions in cells. Chem. Biol. 22, 671-682. 10.1016/j.chembiol.2015.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhatib, N., Neu, M. B., Zensen, C., Schmoller, K. M., Louvard, D., Bausch, A. R., Betz, T. and Vignjevic, D. M. (2014). Fascin plays a role in stress fiber organization and focal adhesion disassembly. Curr. Biol. 24, 1492-1499. 10.1016/j.cub.2014.05.023 [DOI] [PubMed] [Google Scholar]
- Ennomani, H., Letort, G., Guérin, C., Martiel, J.-L., Cao, W., Nédélec, F., de La Cruz, E. M., Théry, M. and Blanchoin, L. (2016). Architecture and connectivity govern actin network contractility. Curr. Biol. 26, 616-626. 10.1016/j.cub.2015.12.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish, K. N. (2009). Total internal reflection fluorescence (TIRF) microscopy. Curr. Protoc. Cytom. 12, 12.18. 10.1002/0471142956.cy1218s50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, D. A. and Mullins, R. D. (2010). Cell mechanics and the cytoskeleton. Nature 463, 485-492. 10.1038/nature08908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubovskaya, V. M., Nyberg, C., Zheng, M., Kweh, F., Magis, A., Ostrov, D. and Cance, W. G. (2008). A small molecule inhibitor, 1,2,4,5-benzenetetraamine tetrahydrochloride, targeting the y397 site of focal adhesion kinase decreases tumor growth. J. Med. Chem. 51, 7405-7416. 10.1021/jm800483v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas, G., Hallett, R. A., Zimmerman, S. P., Williams, T., Yumerefendi, H., Bear, J. E. and Kuhlman, B. (2015). Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. USA 112, 112-117. 10.1073/pnas.1417910112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamadi, A., Bouali, M., Dontenwill, M., Stoeckel, H., Takeda, K. and Rondé, P. (2005). Regulation of focal adhesion dynamics and disassembly by phosphorylation of FAK at tyrosine 397. J. Cell Sci. 118, 4415-4425. 10.1242/jcs.02565 [DOI] [PubMed] [Google Scholar]
- Haupt, A. and Minc, N. (2018). How cells sense their own shape - mechanisms to probe cell geometry and their implications in cellular organization and function. J. Cell Sci. 131, jcs214015. 10.1242/jcs.214015 [DOI] [PubMed] [Google Scholar]
- Hirakawa, M., Oike, M., Karashima, Y. and Ito, Y. (2004). Sequential activation of RhoA and FAK/paxillin leads to ATP release and actin reorganization in human endothelium. J. Physiol. 558, 479-488. 10.1113/jphysiol.2004.065334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y.-L., Lu, S., Szeto, K. W., Sun, J., Wang, Y., Lasheras, J. C. and Chien, S. (2014). FAK and paxillin dynamics at focal adhesions in the protrusions of migrating cells. Sci. Rep. 4, 6024. 10.1038/srep06024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, F.-K., Han, S., Xing, B., Huang, J., Liu, B., Bordeleau, F., Reinhart-King, C. A., Zhang, J. J. and Huang, X.-Y. (2015). Targeted inhibition of fascin function blocks tumour invasion and metastatic colonization. Nat. Commun. 6, 7465. 10.1038/ncomms8465 [DOI] [PubMed] [Google Scholar]
- Huang, J., Dey, R., Wang, Y., Jakoncic, J., Kurinov, I. and Huang, X.-Y. (2018). Structural Insights into the Induced-fit Inhibition of Fascin by a Small-Molecule Inhibitor. J. Mol. Biol. 430, 1324-1335. 10.1016/j.jmb.2018.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huveneers, S. and Danen, E. H. J. (2009). Adhesion signaling - crosstalk between integrins, Src and Rho. J. Cell Sci. 122, 1059-1069. 10.1242/jcs.039446 [DOI] [PubMed] [Google Scholar]
- Jansen, S., Collins, A., Yang, C., Rebowski, G., Svitkina, T. and Dominguez, R. (2011). Mechanism of actin filament bundling by fascin. J. Biol. Chem. 286, 30087-30096. 10.1074/jbc.M111.251439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayo, A., Malboubi, M., Antoku, S., Chang, W., Ortiz-Zapater, E., Groen, C., Pfisterer, K., Tootle, T., Charras, G., Gundersen, G. G.et al. (2016). Fascin regulates nuclear movement and deformation in migrating cells. Dev. Cell 38, 371-383. 10.1016/j.devcel.2016.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathne, W. K. A., O'Neill, P. R. and Gautam, N. (2015). Subcellular optogenetics - controlling signaling and single-cell behavior. J. Cell Sci. 128, 15-25. 10.1242/jcs.154435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, B. D., Hanna, S., Saykali, B. A., el-Sitt, S., Nasrallah, A., Marston, D., el-Sabban, M., Hahn, K. M., Symons, M. and el-Sibai, M. (2014). The regulation of RhoA at focal adhesions by StarD13 is important for astrocytoma cell motility. Exp. Cell Res. 321, 109-122. 10.1016/j.yexcr.2013.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue, J. S., Cartagena-Rivera, A. X., Baird, M. A., Davidson, M. W., Chadwick, R. S. and Waterman, C. M. (2015). Erk regulation of actin capping and bundling by Eps8 promotes cortex tension and leader bleb-based migration. eLife 4, e08314. 10.7554/eLife.08314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto, A., Huang, S. and Ingber, D. E. (2007). Filamin links cell shape and cytoskeletal structure to Rho regulation by controlling accumulation of p190RhoGAP in lipid rafts. J. Cell Sci. 120, 456-467. 10.1242/jcs.03353 [DOI] [PubMed] [Google Scholar]
- Marée, A. F. M., Grieneisen, V. A. and Edelstein-Keshet, L. (2012). How cells integrate complex stimuli: the effect of feedback from phosphoinositides and cell shape on cell polarization and motility. PLoS Comput. Biol. 8, e1002402. 10.1371/journal.pcbi.1002402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W. F. (2016). Cell geometry: how cells count and measure size. Annu. Rev. Biophys. 45, 49-64. 10.1146/annurev-biophys-062215-010905 [DOI] [PubMed] [Google Scholar]
- Marshall, W. F., Young, K. D., Swaffer, M., Wood, E., Nurse, P., Kimura, A., Frankel, J., Wallingford, J., Walbot, V., Qu, X.et al. (2012). What determines cell size? BMC Biol. 10, 101. 10.1186/1741-7007-10-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino, F., Perestrelo, A. R., Vinarský, V., Pagliari, S. and Forte, G. (2018). Cellular mechanotransduction: from tension to function. Front. Physiol. 9, 824. 10.3389/fphys.2018.00824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshik, X., O'Neill, P. R. and Gautam, N. (2018). Optogenetic control of cell migration. Methods Mol. Biol. 1749, 313-324. 10.1007/978-1-4939-7701-7_22 [DOI] [PubMed] [Google Scholar]
- Michael, K. E., Dumbauld, D. W., Burns, K. L., Hanks, S. K. and García, A. J. (2009). Focal adhesion kinase modulates cell adhesion strengthening via integrin activation. Mol. Biol. Cell 20, 2508-2519. 10.1091/mbc.e08-01-0076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierke, C. T., Fischer, T., Puder, S., Kunschmann, T., Soetje, B. and Ziegler, W. H. (2017). Focal adhesion kinase activity is required for actomyosin contractility-based invasion of cells into dense 3D matrices. Sci. Rep. 7, 42780. 10.1038/srep42780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, S. K., Hanson, D. A. and Schlaepfer, D. D. (2005). Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56-68. 10.1038/nrm1549 [DOI] [PubMed] [Google Scholar]
- Mohan, K., Luo, T., Robinson, D. N. and Iglesias, P. A. (2015). Cell shape regulation through mechanosensory feedback control. J. R Soc. Interface 12, 20150512. 10.1098/rsif.2015.0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi, K., Fujiwara, S. and Mizuno, K. (2017). Roles of the cytoskeleton, cell adhesion and rho signalling in mechanosensing and mechanotransduction. J. Biochem. 161, 245-254. 10.1093/jb/mvw082 [DOI] [PubMed] [Google Scholar]
- Ollion, J., Cochennec, J., Loll, F., Escudé, C. and Boudier, T. (2013). TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840-1841. 10.1093/bioinformatics/btt276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, P. R., Kalyanaraman, V. and Gautam, N. (2016). Subcellular optogenetic activation of Cdc42 controls local and distal signaling to drive immune cell migration. Mol. Biol. Cell 27, 1442-1450. 10.1091/mbc.E15-12-0832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, P. R., Castillo-Badillo, J. A., Meshik, X., Kalyanaraman, V., Melgarejo, K. and Gautam, N. (2018). Membrane flow drives an adhesion-independent amoeboid cell migration mode. Dev. Cell 46, 9-22.e4. 10.1016/j.devcel.2018.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch, E. and Heisenberg, C.-P. (2009). Biology and physics of cell shape changes in development. Curr. Biol. 19, R790-R799. 10.1016/j.cub.2009.07.029 [DOI] [PubMed] [Google Scholar]
- Paluch, E., Sykes, C., Prost, J. and Bornens, M. (2006). Dynamic modes of the cortical actomyosin gel during cell locomotion and division. Trends Cell Biol. 16, 5-10. 10.1016/j.tcb.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Parhamifar, L. and Moghimi, S. M. (2012). Total internal reflection fluorescence (TIRF) microscopy for real-time imaging of nanoparticle-cell plasma membrane interaction. Methods Mol. Biol. 906, 473-482. 10.1007/978-1-61779-953-2_38 [DOI] [PubMed] [Google Scholar]
- Pasqualato, A., Lei, V., Cucina, A., Dinicola, S., D'anselmi, F., Proietti, S., Masiello, M. G., Palombo, A. and Bizzarri, M. (2013). Shape in migration: quantitative image analysis of migrating chemoresistant HCT-8 colon cancer cells. Cell Adh. Migr. 7, 450-459. 10.4161/cam.26765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter, N. S., Pitkeathly, W. T. E., Smith, P. J. and Rappoport, J. Z. (2015). The physical basis of total internal reflection fluorescence (TIRF) microscopy and its cellular applications. Methods Mol. Biol. 1251, 1-23. 10.1007/978-1-4939-2080-8_1 [DOI] [PubMed] [Google Scholar]
- Ron, A., Azeloglu, E. U., Calizo, R. C., Hu, M., Bhattacharya, S., Chen, Y., Jayaraman, G., Lee, S., Neves-Zaph, S. R., Li, H.et al. (2017). Cell shape information is transduced through tension-independent mechanisms. Nat. Commun. 8, 2145. 10.1038/s41467-017-02218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, J. M., Harrell, J. R., Shemer, G., Sullivan-Brown, J., Roh-Johnson, M. and Goldstein, B. (2010). Apical constriction: a cell shape change that can drive morphogenesis. Dev. Biol. 341, 5-19. 10.1016/j.ydbio.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, C. H., Furukawa, K., Montagne, K., Jeong, H. and Ushida, T. (2011). The effect of substrate microtopography on focal adhesion maturation and actin organization via the RhoA/ROCK pathway. Biomaterials 32, 9568-9575. 10.1016/j.biomaterials.2011.08.077 [DOI] [PubMed] [Google Scholar]
- Sero, J. E. and Bakal, C. (2017). Multiparametric analysis of cell shape demonstrates that β-PIX directly couples YAP activation to extracellular matrix adhesion. Cell Syst. 4, 84-96.e6. 10.1016/j.cels.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, V., Iglesias, P. A. and Robinson, D. N. (2016). Cytokinesis: Robust cell shape regulation. Semin. Cell Dev. Biol. 53, 39-44. 10.1016/j.semcdb.2015.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, A. D., Liu, P. Z., Kandula, V., Chen, H., Almassalha, L. M., Herman, C., Backman, V., O'halloran, T., Adam, S. A., Goldman, R. D.et al. (2019). Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 30. mbcE19050286T. 10.1091/mbc.E19-05-0286 [DOI] [PubMed] [Google Scholar]
- Stevenson, R. P., Veltman, D. and Machesky, L. M. (2012). Actin-bundling proteins in cancer progression at a glance. J. Cell Sci. 125, 1073-1079. 10.1242/jcs.093799 [DOI] [PubMed] [Google Scholar]
- Tanaka, M., Fujii, Y., Hirano, K., Higaki, T., Nagasaki, A., Ishikawa, R., Okajima, T. and Katoh, K. (2019). Fascin in lamellipodia contributes to cell elasticity by controlling the orientation of filamentous actin. Genes Cells 24, 202-213. 10.1111/gtc.12671 [DOI] [PubMed] [Google Scholar]
- Taneja, N., Fenix, A. M., Rathbun, L., Millis, B. A., Tyska, M. J., Hehnly, H. and Burnette, D. T. (2016). Focal adhesions control cleavage furrow shape and spindle tilt during mitosis. Sci. Rep. 6, 29846. 10.1038/srep29846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, M. and Bornens, M. (2006). Cell shape and cell division. Curr. Opin. Cell Biol. 18, 648-657. 10.1016/j.ceb.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Torsoni, A. S., Marin, T. M., Velloso, L. A. and Franchini, K. G. (2005). RhoA/ROCK signaling is critical to FAK activation by cyclic stretch in cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 289, H1488-H1496. 10.1152/ajpheart.00692.2004 [DOI] [PubMed] [Google Scholar]
- van Audenhove, I., Debeuf, N., Boucherie, C. and Gettemans, J. (2015). Fascin actin bundling controls podosome turnover and disassembly while cortactin is involved in podosome assembly by its SH3 domain in THP-1 macrophages and dendritic cells. Biochim. Biophys. Acta Mol. Cell Res. 1853, 940-952. 10.1016/j.bbamcr.2015.01.003 [DOI] [PubMed] [Google Scholar]
- Vega, F. M., Fruhwirth, G., Ng, T. and Ridley, A. J. (2011). RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J. Cell Biol. 193, 655-665. 10.1083/jcb.201011038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villari, G., Jayo, A., Zanet, J., Fitch, B., Serrels, B., Frame, M., Stramer, B. M., Goult, B. T. and Parsons, M. (2015). A direct interaction between fascin and microtubules contributes to adhesion dynamics and cell migration. J. Cell Sci. 128, 4601-4614. 10.1242/jcs.175760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E. and Glotzer, M. (2016). Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. J. Cell Biol. 213, 641-649. 10.1083/jcb.201603025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater, M., Sander, N., de Vreede, G. and van den Heuvel, S. (2011). Cell shape and Wnt signaling redundantly control the division axis of C. elegans epithelial stem cells. Development 138, 4375-4385. 10.1242/dev.066431 [DOI] [PubMed] [Google Scholar]
- Wolfenson, H., Yang, B. and Sheetz, M. P. (2019). Steps in mechanotransduction pathways that control cell morphology. Annu. Rev. Physiol. 81, 585-605. 10.1146/annurev-physiol-021317-121245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro, S., Yamakita, Y., Ono, S. and Matsumura, F. (1998). Fascin, an actin-bundling protein, induces membrane protrusions and increases cell motility of epithelial cells. Mol. Biol. Cell 9, 993-1006. 10.1091/mbc.9.5.993 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.