ABSTRACT
The kinesin-4 member KIF7 plays critical roles in Hedgehog signaling in vertebrate cells. KIF7 is an atypical kinesin as it binds to microtubules but is immotile. We demonstrate that, like conventional kinesins, KIF7 is regulated by auto-inhibition, as the full-length protein is inactive for microtubule binding in cells. We identify a segment, the inhibitory coiled coil (inhCC), that is required for auto-inhibition of KIF7, whereas the adjacent regulatory coiled coil (rCC) that contributes to auto-inhibition of the motile kinesin-4s KIF21A and KIF21B is not sufficient for KIF7 auto-inhibition. Disease-associated mutations in the inhCC relieve auto-inhibition and result in strong microtubule binding. Surprisingly, uninhibited KIF7 proteins did not bind preferentially to or track the plus ends of growing microtubules in cells, as suggested by previous in vitro work, but rather bound along cytosolic and axonemal microtubules. Localization to the tip of the primary cilium also required the inhCC, and could be increased by disease-associated mutations regardless of the auto-inhibition state of the protein. These findings suggest that loss of KIF7 auto-inhibition and/or altered cilium tip localization can contribute to the pathogenesis of human disease.
KEY WORDS: Microtubule, Kinesin, KIF7, Acrocallosal syndrome, Joubert syndrome
Summary: KIF7 is regulated by auto-inhibition, and human disease-associated mutations cause uniform binding along cytosolic microtubules and/or increased primary cilium localization.
INTRODUCTION
Microtubule-based motors of the kinesin superfamily play essential roles in cell division, cell motility, intracellular trafficking, control of microtubule dynamics and ciliary function (Hirokawa et al., 2009). Kinesin proteins are defined by the presence of a kinesin motor domain that contains signature sequences for nucleotide and microtubule binding. Sequence differences within this core motor domain provide family-specific microtubule-based properties. The ‘conventional’ kinesin property of ATP-dependent processive motility along the microtubule surface is a characteristic feature of members of the kinesin-1, kinesin-2 and kinesin-3 families, with sequence changes providing family-specific outputs in terms of speed, processivity and force generation (Cochran, 2015; Hunter and Allingham, 2020). Unconventional kinesin properties include the sliding of microtubules by kinesin-5 motors and the regulation of microtubule dynamics by members of the kinesin-4, kinesin-8 and kinesin-13 families (Friel and Welburn, 2018; Mann and Wadsworth, 2019; Singh et al., 2018).
KIF7 is a member of the kinesin-4 family and plays critical roles in Hedgehog signaling in vertebrates (Bangs and Anderson, 2017). In mice, knockout of Kif7 is perinatal lethal with early embryonic defects, including preaxial polydactyly, exencephaly, agenesis of the corpus callosum, and microphthalmia (Endoh-Yamagami et al., 2009; Cheung et al., 2009; Putoux et al., 2019). In fish, knockdown of KIF7 function results in altered Hedgehog pathway gene expression, disruption of left-right asymmetry and scoliosis (Wilson et al., 2009; Maurya et al., 2013; Terhune et al., 2020; Liang et al., 2020). In humans, mutations in KIF7 have been shown to cause severe ciliopathies, including Joubert, hydrolethalus, acrocallosal, Meckel-Gruber and Bardet-Biedl syndromes (Ali et al., 2012; Barakeh et al., 2015; Bachmann-Gagescu et al., 2015; Karaer et al., 2015; Putoux et al., 2011, 2012; Tunovic et al., 2015; Walsh et al., 2013; Ibisler et al., 2015; Dafinger et al., 2011; Asadollahi et al., 2018; Subramanian et al., 2019; Liang et al., 2020; Terhune et al., 2020; Dahl et al., 2020; Niceta et al., 2020). Recent work has suggested that KIF7 also plays a role in cell proliferation during development and disease (Coles et al., 2015; Wong et al., 2017; Lau et al., 2017; Hu et al., 2020; Li et al., 2012).
In the vertebrate Hedgehog signaling pathway, KIF7 plays both positive and negative roles in Hedgehog signaling by regulating the abundance of Gli2 and Gli3 transcription factors, as well as the balance between their activator and repressor forms (Endoh-Yamagami et al., 2009; Cheung et al., 2009; Liem et al., 2009). In mice, KIF7 activity depends on the presence of the primary cilium, and in response to Hedgehog pathway activation, KIF7 localization to the tip of the primary cilium increases, and this is thought to facilitate the localization of Hedgehog effectors to the same location (Endoh-Yamagami et al., 2009; Cheung et al., 2009; Liem et al., 2009). KIF7 has also been suggested to regulate the length of the primary cilium and organization of the cilium tip (He et al., 2014). In some cases, humans and mice with Kif7 mutations have longer cilia (He et al., 2014; Emechebe et al., 2016; Putoux et al., 2011, 2016). These ciliary functions of KIF7 are thought to be due to the ability of the motor to bind selectively to the plus ends of microtubules and regulate microtubule dynamics (He et al., 2014; Jiang et al., 2019). However, this model is based on in vitro characterization of the microtubule-based activities of KIF7, and whether it binds selectively to the plus ends of microtubules in cells has not been demonstrated.
As a kinesin protein, KIF7 has several unique properties. Most striking is the fact that KIF7 is not capable of processive motility but rather interacts statically with the microtubule (He et al., 2014; Yue et al., 2018). The mechanistic basis of this immotile behavior was found to be an inability to respond to microtubule binding with an alteration in nucleotide binding or hydrolysis (Yue et al., 2018; Jiang et al., 2019). Still unclear is how the microtubule binding of KIF7 is regulated and how the immotile behavior of KIF7 relates to its functions in Hedgehog signaling.
Kinesin activity must be tightly regulated in cells to prevent non-productive ATP hydrolysis and misregulation of microtubule-based activities. A general model in the field is that kinesin proteins are regulated by an auto-inhibition mechanism, in which non-motor regions of the protein act in cis to block ATP and/or microtubule binding (Verhey and Hammond, 2009). Here we demonstrate that full-length KIF7 is regulated by auto-inhibition and identify a coiled-coil (CC) segment, the inhibitory CC (inhCC), as critical for auto-inhibition of the N-terminal motor domain. Several mutations associated with human disease are found within the inhCC, and we show that mutation of Q1136P in mouse KIF7 results in a loss of auto-inhibition, whereas mutation of H1115P has no effect on auto-inhibition. These results indicate that loss of auto-inhibition may be causative for disease. Finally, we find that active versions of KIF7 that have lost auto-inhibition do not bind specifically to the plus ends of microtubules in a cellular context, suggesting that microtubule binding at the ciliary tip is not required for the cellular functions of KIF7.
RESULTS
Full-length KIF7 is auto-inhibited
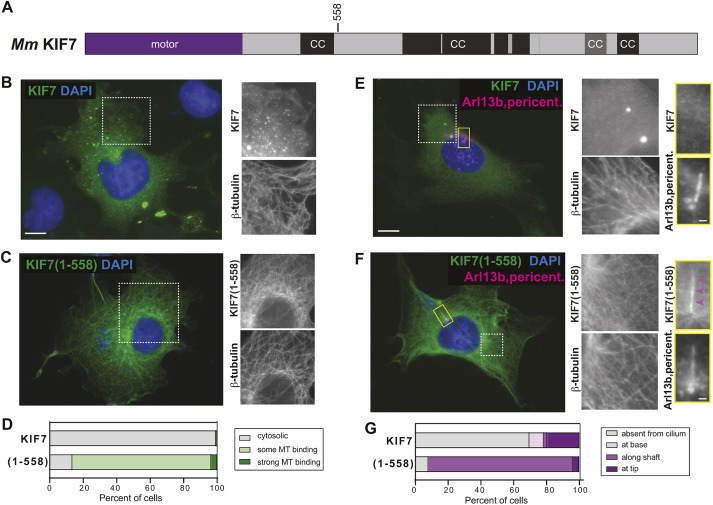
To test whether KIF7 is regulated by auto-inhibition, we compared the localization of full-length mouse KIF7 to that of a previously studied truncated version, KIF7(1-558), which binds to microtubules but is immotile in in vitro single-molecule motility assays (He et al., 2014; Yue et al., 2018). Both proteins were tagged at the C-terminus with the fluorescent protein monomeric Citrine (mCit). The truncated KIF7(1-558)-mCit protein decorated microtubules in COS-7 cells (Fig. 1C,D; Fig. S1), consistent with its ability to bind to microtubules in in vitro assays (He et al., 2014; Yue et al., 2018). In contrast, the full-length KIF7 protein localized in a diffuse manner throughout the cytoplasm (Fig. 1B,D). The full-length protein also accumulated in puncta of various sizes (Fig. 1B,D), as previously observed for GFP-tagged proteins expressed in C3H10T1/2, MEF and NIH-3T3 cells (Coles et al., 2015; Endoh-Yamagami et al., 2009; He et al., 2014; Liem et al., 2009; Liu et al., 2014), and for the endogenous protein in fish and mouse cells (Coles et al., 2015; He et al., 2014; Maurya et al., 2013); the puncta do not colocalize with organelle markers and thus may be aggregates, as has been noted previously (Maurya et al., 2013). The inability of full-length KIF7 to bind to microtubules suggests that the motor is regulated by auto-inhibition.
Fig. 1.
KIF7 is regulated by auto-inhibition. (A) Domain organization of full-length KIF7. The position of truncation at amino acid 558 is indicated on top. CC, coiled coil; black box, CC prediction >90%; gray box, CC prediction >80%, according to COILS software. (B-D) COS-7 cells expressing mCit-tagged full-length (B) or truncated (1-558) (C) versions of KIF7 were fixed and stained with an antibody against β-tubulin to mark microtubules and with DAPI to mark the nucleus. The white boxed region is shown on the right with separate channels for KIF7-mCit and microtubules. (D) Quantification of microtubule (MT) binding. n=136 (KIF7) and 88 [KIF7(1-558)] cells across at least three independent experiments. (E-G) NIH-3T3 cells expressing mCit-tagged full-length (E) or truncated (1-558) (F) versions of KIF7 were fixed and stained with antibodies against β-tubulin to mark microtubules, Arl13b to mark the primary cilium, pericentrin to mark the basal body and DAPI to mark the nucleus. The white boxed region is shown to the right with separate channels for KIF7-mCit and microtubules. The primary cilium in the yellow boxed region is shown on the far right with separate images of KIF7 and the cilium markers. Purple arrowheads indicate localization along the cilium shaft. (G) Quantification of cilium localization. n=46 (KIF7) and 25 [KIF7(1-558)] cells across at least three independent experiments. Scale bars: 10 µm (B,C,E,F); 1 µm (E,F, far right panels).
To extend these results, we expressed the full-length and truncated KIF7(1-558) proteins in two other cell types. First, we expressed full-length and truncated (1-558) KIF7 in CAD neuronal cells where the microtubule minus and plus ends are spatially segregated to the cell body and neurite tips, respectively, and the accumulation of kinesin motor proteins in the neurite tips has been used as a readout of plus end-directed motility (Hammond et al., 2009; Huang and Banker, 2012). When expressed in neuronal CAD cells, truncated KIF7(1-558) bound to microtubules throughout the cell body and proximal region of the neurite (Fig. S2B), whereas full-length KIF7 localized diffusely throughout the cell and to puncta in the cell body (Fig. S2A). Neither protein accumulated in the neurite tips, suggesting that neither the full-length nor the truncated motor is capable of using processive motility to reach the distal neurite.
Second, we expressed full-length and truncated KIF7(1-558) in NIH-3T3 cells, which generate primary cilia during the G1 phase of the cell cycle and are used as a model system for Hedgehog signaling. Similar to the results in COS-7 cells, the truncated KIF7(1-558) motor decorated microtubules in NIH-3T3 cells, suggesting that it has lost auto-inhibition of microtubule binding (Fig. 1F). In general, the expressed KIF7(1-558) protein maintained more of a cytosolic pool in NIH-3T3 cells compared to the near-total microtubule binding observed in COS-7 cells, suggesting that differences in cellular environment can shift motor behavior. The truncated KIF7(1-558) protein also localized to microtubules in the primary cilium, with localization along the cilium shaft in 88% of cells and accumulation at the tip of the cilium in 4% of cells (Fig. 1F,G; Fig. S1C). In contrast, full-length KIF7 displayed a diffuse localization and accumulation in bright puncta in the cytoplasm of NIH-3T3 cells (Fig. 1E). The full-length motor also accumulated at the tip of the primary cilium in 20% of cells (Fig. 1G; Fig. S1C), consistent with previous work (Endoh-Yamagami et al., 2009; He et al., 2014; Liem et al., 2009). Together, these results indicate that full-length KIF7 is auto-inhibited, whereas the truncated KIF7(1-558) protein has lost this regulation and can interact with all microtubules in cells, including axonemal microtubules in the shaft of the primary cilium.
Auto-inhibition requires the last coiled-coil segments of KIF7
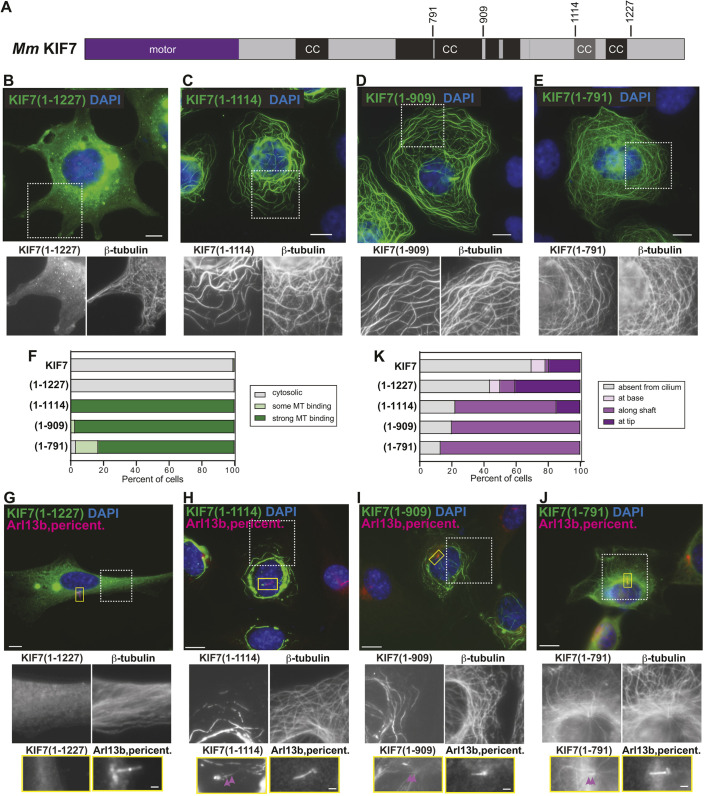
Domain analysis of the KIF7 sequence revealed an N-terminal motor domain, several regions of predicted coiled coil (CC), and a C-terminal tail domain (Fig. 1A; Fig. S3). To determine the regions of KIF7 involved in auto-inhibition, we created truncated versions that removed the tail domain [KIF7(1-1227)], the C-terminal CC segments [KIF7(1-1114)], half of the long central CC domain [KIF7(1-909)] or nearly all of the central CC domain [KIF7(1-791)] (Fig. 2A), and expressed the truncated proteins in COS-7 cells. Similar to the full-length protein, the truncated motor KIF7(1-1227) displayed a diffuse cytosolic localization in 100% of the cells (Fig. 2B,F), with some accumulation in puncta of various sizes (Fig. 2B). In contrast, the truncated motor KIF7(1-1114) displayed strong microtubule localization in 100% of the cells (Fig. 2C,F), suggesting that the C-terminal CC segments are critical for the auto-inhibition of KIF7. We thus refer to this region of KIF7 (amino acids 1115-1227) as the inhibitory CC (inhCC). Further truncations to remove portions of the central CC also resulted in truncated proteins that showed strong microtubule binding in 97% of cells for KIF7(1-909) (Fig. 2D,F) and 83% of cells for KIF7(1-791) (Fig. 2E,F). As these truncated proteins showed stronger microtubule binding than KIF7(1-558) (Fig. 1C), these results suggest that there may be sequences in the central coiled coil of KIF7 that bind directly to microtubules or facilitate microtubule binding of the motor domain.
Fig. 2.
Truncations that remove the inhCC are active for microtubule binding. (A) Domain organization of full-length KIF7. The positions of the truncations at amino acids 791, 909, 1114 and 1227 are indicated on top. CC, coiled coil; black box, CC prediction >90%; gray box, CC prediction >80%, according to COILS program. (B-F) COS-7 cells expressing the indicated mCit-tagged truncated versions of KIF7 were fixed and stained with an antibody against β-tubulin to mark microtubules and with DAPI to mark the nucleus. The white boxed region is shown below with separate channels for KIF7-mCit and microtubules. (F) Quantification of microtubule (MT) binding. n=136 (KIF7), 132 [KIF7(1-1227)], 123 [KIF7(1-1114)], 149 [KIF7(1-909)] and 125 [KIF7(1-791)] cells across at least three independent experiments. (G-K) NIH-3T3 cells expressing the indicated mCit-tagged truncated versions of KIF7 were fixed and stained with antibodies against β-tubulin to mark microtubules, Arl13b to mark the primary cilium, pericentrin to mark the basal body and DAPI to mark the nucleus. The white boxed region is shown below with separate channels for KIF7-mCit and microtubules. The primary cilium in the yellow boxed region is shown below with separated images of KIF7 and the cilium markers. Purple arrowheads indicate localization along the cilium shaft. (K) Quantification of cilium localization. n=46 (KIF7), 32 [KIF7(1-1227)], 27 [KIF7(1-1114)], 25 [KIF7(1-909)] and 23 [KIF7(1-791)] cells across at least three independent experiments. Scale bars: 10 µm (B-E,G-J); 1 µm (G-J, bottom panels).
To examine the effects of the truncations on the ability of the motor to localize to primary cilia, we expressed the truncated proteins in NIH-3T3 cells. Compared to the full-length motor, removal of the tail domain resulted in an increased percentage of cells with KIF7 at the tip of the cilium [41% of cells for KIF7(1-1227) versus 20% of cells for full-length KIF7] (Fig. 2G,K). This result suggests that the tail domain is not required for ciliary targeting of KIF7 but may be involved in KIF7 removal from the cilium via an unknown pathway. Further truncations resulted in localization of the proteins to cytosolic microtubules and to the shaft of the cilium rather than the tip of the cilium, with shaft localization observed in 63% of cells for KIF7(1-1114), 80% of cells for KIF7(1-909) and 87% of cells for KIF7(1-791) (Fig. 2H-K). These results confirm that the inhCC is involved in the regulation of the microtubule binding of KIF7, as removal of this region results in a KIF7 protein that binds tightly to microtubules in the cytosolic and ciliary compartments. These results also indicate that loss of auto-inhibition correlates with loss of cilium tip localization. Thus, an important function of auto-inhibition may be to enable KIF7 accumulation at the tip of rather than along the shaft of primary cilia. An alternative possibility is that the inhCC region (amino acids 1115-1227) contains sequences necessary for KIF7 localization at the tip of the primary cilium.
A disease-associated mutation in the inhCC segment relieves auto-inhibition
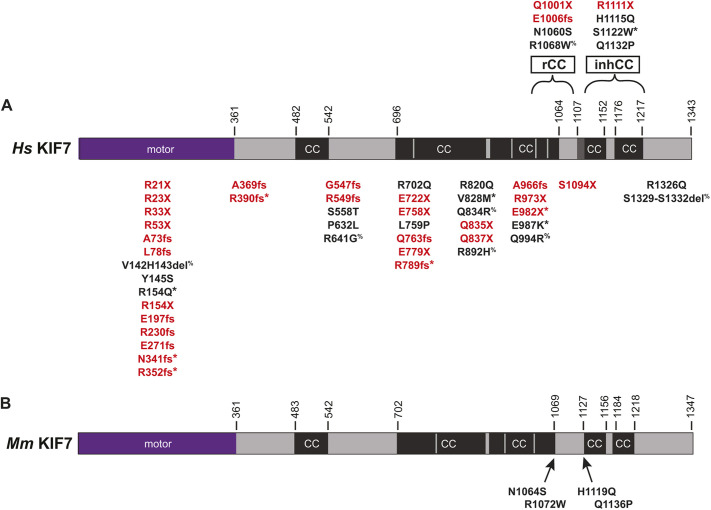
To distinguish between these possibilities, we sought to create point mutations in the inhCC that would relieve auto-inhibition within the context of the full-length protein. We thus searched the literature for disease-associated point mutations in human KIF7. We also searched for mutations reported to be pathogenic or likely pathogenic in ClinVar. Interestingly, 58.8% (30/51) of the mutations in KIF7 are nonsense or frameshift mutations that result in a truncated protein lacking the inhCC (Fig. 3A, red text). Based on our finding that the truncated mouse protein KIF7(1-1114) results in a loss of auto-inhibition (Fig. 2), we infer that these disease-associated human proteins have also lost their auto-inhibition mechanism, suggesting that truncation of the protein and release of auto-inhibition have implications for human disease. The remaining KIF7 mutations (Fig. 3A, black text) involve deletions or missense mutations that occur in a homozygous or heterozygous manner (5/51) as a compound heterozygote of two different KIF7 mutations (4/51), as a compound heterozygote with another ciliopathy gene (7/51), or unreported zygosity (5/51).
Fig. 3.
Mutations in KIF7 associated with human disease. (A) Location of disease-associated KIF7 mutations mapped on the domain structure of human KIF7. Red text, nonsense or frameshift mutations; black text, missense or deletion mutations. *Present as compound heterozygote of two different KIF7 alleles; %Present as compound heterozygote with another ciliopathy gene. inhCC, inhibitory coiled coil; rCC, regulatory coiled coil. (B) Location of human disease-associated missense mutations introduced into mouse KIF7.
To test whether point mutations within the inhCC can relieve auto-inhibition of KIF7, we focused on two missense mutations in this region. First, H1115Q, which was identified as a heterozygous variant in patients with acrocallosal syndrome, hydrolethalus syndrome 2 and idiopathic scoliosis (Putoux et al., 2011; Terhune et al., 2020), and second, Q1132P, which lacks a clinical description but has been described as likely pathogenic based on the absence of this variant in large population cohorts (ClinVar VCV000451950.2). We thus introduced the corresponding mutations H1119Q and Q1136P into the sequence of our mouse KIF7 protein (Figs 3B, 4A), which is 88% identical to the human KIF7 protein (Fig. S4).
Fig. 4.
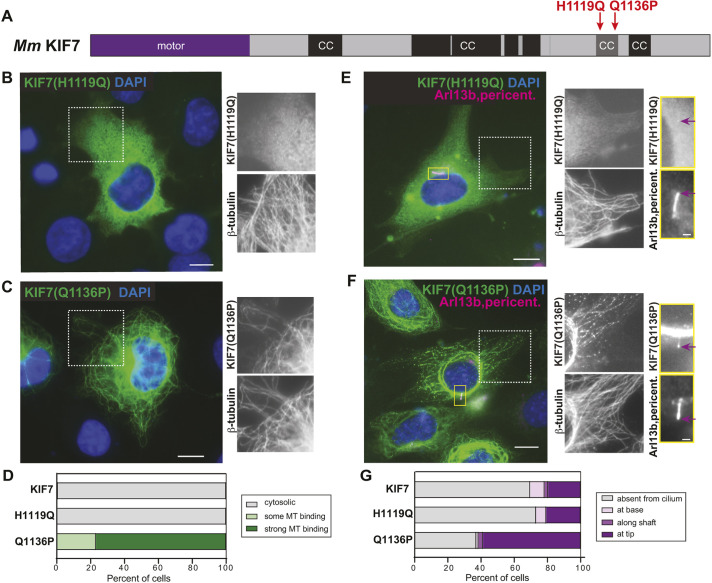
The disease-associated mutation Q1136P in the inhCC relieves auto-inhibition. (A) Domain organization of full-length KIF7. The positions of the disease-associated mutations H1119Q and Q1136P are indicated on top. CC, coiled coil; black box, CC prediction >90%; gray box, CC prediction >80%, according to COILS software. (B-D) COS-7 cells expressing mCit-tagged H1119Q (B) or Q1136P (C) mutant versions of KIF7 were fixed and stained with an antibody against β-tubulin to mark microtubules (MT) and with DAPI to mark the nucleus. The white boxed region is shown to the right with separated channels for KIF7-mCit and microtubules. (D) Quantification of microtubule binding. n=136 (KIF7), 223 [KIF7(H1119Q)] and 177 [KIF7(Q1136P)] cells across at least three independent experiments. (E-G) NIH-3T3 cells expressing mCit-tagged H1119Q (E) or Q1136P (F) mutant versions of KIF7 were fixed and stained with antibodies against β-tubulin to mark microtubules, Arl13b to mark the primary cilium, pericentrin to mark the basal body and DAPI to mark the nucleus. The white boxed region is shown to the right with separate images of KIF7-mCit and microtubules. The primary cilium in the yellow boxed region is shown to the far right with separate images of KIF7 and the cilium markers. Purple arrows indicate localization at the cilium tip. (G) Quantification of cilium localization. n=46 (KIF7), 82 [KIF7(H1119Q)] and 78 [KIF7(Q1136P)] cells across at least three independent experiments. Scale bars: 10 µm (B,C,E,F); 1 µm (E,F, far right panels).
When expressed in COS-7 cells, the KIF7(H1119Q) mutant protein behaved similarly to the wild-type protein, with a diffuse cytosolic localization in 100% of cells (Fig. 4B,D). In contrast, the KIF7(Q1136P) mutant protein displayed strong microtubule localization in 77% of cells (Fig. 4C,D), similar to the active truncation mutant KIF7(1-1114) (Fig. 2K), suggesting that the single point mutation at Q1136 results in a release of auto-inhibition.
When expressed in NIH-3T3 cells, the KIF7(H1119Q) mutant protein displayed a diffuse localization in the cytoplasm (Fig. 4E), consistent with an auto-inhibited state, and localized to the tips of primary cilia in 21% of cells, similar to the wild-type protein (Fig. 4G). The KIF7(Q1136P) mutant protein showed strong microtubule binding in the cytoplasm (Fig. 4F), consistent with loss of auto-inhibition. However, the uninhibited Q1136P protein did not localize to microtubules in the ciliary shaft like the active KIF7(1-1114) protein (Fig. 2H,K), but rather localized to the tip of the cilium in 59% of cells (Fig. 4E,G). Thus, although truncations that remove the inhCC result in loss of auto-inhibition and loss of ciliary tip localization, the Q1136P point mutation results only in loss of auto-inhibition. These results suggest that separate sequences within the inhCC are involved in the auto-inhibition of microtubule binding and ciliary tip localization.
Disease-associated mutations in the regulatory coiled coil do not relieve auto-inhibition
The KIF7 stalk segment identified here as critical for auto-inhibition, the inhCC, differs from previous work that predicted that amino acids 998-1077 in human KIF7 were critical for auto-inhibition (Bianchi et al., 2016). This predication was based on the findings that auto-inhibition of KIF21A and KIF21B requires a regulatory coiled-coil (rCC) segment in the stalk domain of these kinesin-4 family members (Bianchi et al., 2016; Cheng et al., 2014; van der Vaart et al., 2013; van Riel et al., 2017). Sequence alignment suggested that rCC-like sequences exist in the stalk regions of KIF7 and KIF27, and may therefore also regulate the auto-inhibition of these kinesin-4 members (Bianchi et al., 2016). Whether the rCC region forms a coiled-coil segment in the human and mouse KIF7 proteins depends on the prediction program (Fig. S3).
To test the hypothesis that the rCC-like sequence plays a role in the auto-inhibition of KIF7, we took advantage of disease-associated mutations in this region. N1060S was identified as a homozygous variant in a patient with macrocephaly, multiple epiphyseal dysplasia and distinctive facies (Ali et al., 2012), and notably, an asparagine residue is conserved at this position across the kinesin-4 family members KIF21A, KIF21B, KIF7, and KIF27 (Fig. S5). We thus generated the equivalent mutation N1064S in our full-length mouse KIF7 protein (Fig. 5A). R1068W was identified in a patient with Bardet–Biedl syndrome as a compound heterozygote with mutations in BBS9 (Putoux et al., 2011). R1068 occupies the same position in the predicted rCC as a KIF21A mutation identified in a patient associated with congenital fibrosis of the extraocular muscles type 1 (CFEOM1) (Bianchi et al., 2016; Cheng et al., 2014; van der Vaart et al., 2013), and a KIF21B mutation identified in a patient with global developmental delay and mild to moderate intellectual disability (Asselin et al., 2020), suggesting that R1068 resides at a position that is sensitive to perturbation (Fig. S5). We thus introduced the corresponding mutation, R1072W, into the sequence of our mouse KIF7 protein (Fig. 5A).
Fig. 5.
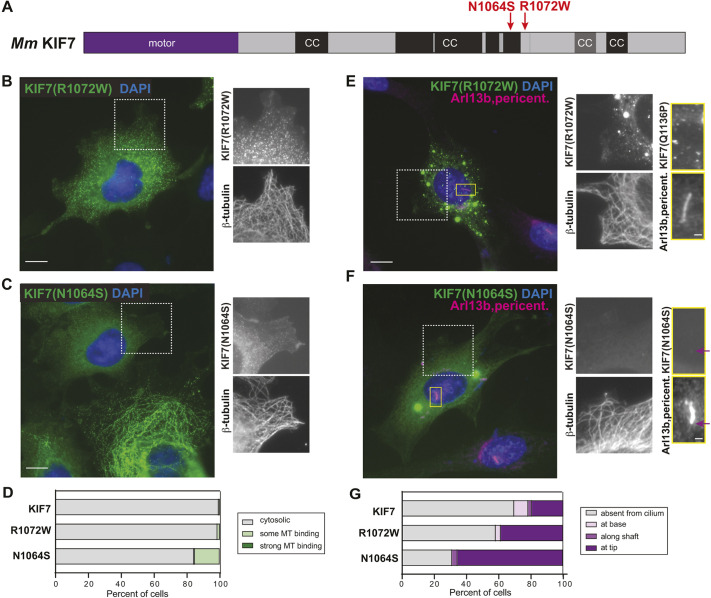
Disease-associated mutations in rCC do not relieve auto-inhibition. (A) Domain organization of full-length KIF7. The positions of the mutations N1064S and R1072W are indicated on top. CC, coiled coil; black box, CC prediction >90%; gray box, CC prediction >80%, according to COILS software. (B-D) COS-7 cells expressing mCit-tagged R1072W (B) or N1064S (C) mutant versions of KIF7 were fixed and stained with an antibody against β-tubulin to mark microtubules and with DAPI to mark the nucleus. The white boxed region is shown to the right with separate images of KIF7-mCit and microtubules. (D) Quantification of microtubule (MT) binding. n=136 (KIF7), 140 [KIF7(R1072W)] and 476 [KIF7(N1064S)] cells across at least three independent experiments. (E-G) NIH-3T3 cells expressing mCit-tagged R1072W (E) or N1064S (F) mutant versions of KIF7 were fixed and stained with antibodies against β-tubulin to mark microtubules, Arl13b to mark the primary cilium, pericentrin to mark the basal body and DAPI to mark the nucleus. The white boxed region is shown to the right with separate images of KIF7-mCit and microtubules. The primary cilium in the yellow boxed region is shown to the far right with separate images of KIF7 and the cilium markers. Purple arrows indicate localization at the cilium tip. (G) Quantification of the cilium localization. n=46 (KIF7), 31 [KIF7(R1072W)] and 29 [KIF7(N1064S)] cells across at least three independent experiments. Scale bars: 10 µm (B,C,E,F); 1 µm (E,F, far right panels).
When expressed in COS-7 cells, the KIF7(R1072W) mutant protein behaved similarly to the wild-type protein, with a diffuse cytosolic localization in 99% of the cells (Fig. 5B,D). This result suggests that KIF7(R1072W) retains the capacity for auto-inhibition and, thus, its pathogenic role in patients is unlikely to be due to a loss of auto-inhibition. The KIF7(N1064S) mutant protein also displayed a largely diffuse localization when expressed in COS-7 cells (84% of cells, Fig. 5C,D). Weak microtubule binding was detected in 14% of cells expressing KIF7(N1064S) (Fig. 5D), suggesting that this mutation may alter protein folding and/or contribute to intramolecular auto-inhibitory interactions.
To examine the impact of these disease-associated mutations on the ability of KIF7 to localize to the primary cilium, the R1072W and N1064S mutant proteins were expressed in NIH-3T3 cells. The KIF7(R1072W)-mCit mutant protein localized to the tips of primary cilia in 39% of cells (Fig. 5E,G), and the KIF7(N1064S)-mCit localized to the cilium tip in 66% of cells (Fig. 5F,G), as compared to the wild-type protein, which localized to the cilium tip in 20% of cells (Fig. 5G). Thus, although the R1072W and N1064S mutations do not relieve auto-inhibition, they appear to facilitate KIF7 localization to the tip or hinder its removal from the primary cilium.
Uninhibited KIF7 does not track the plus ends of microtubules in cells
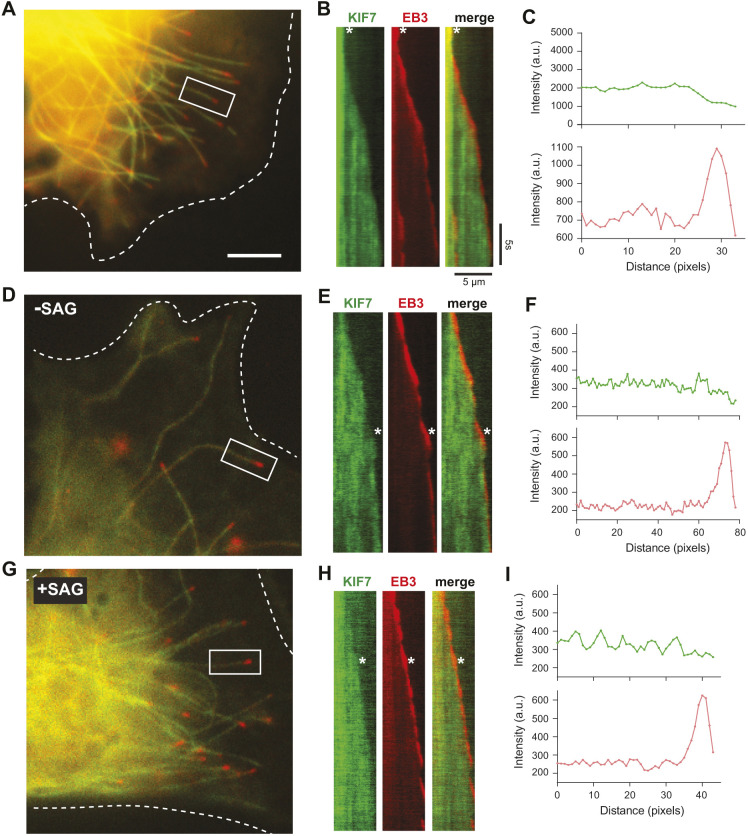
Previous work using purified truncated versions of mouse KIF7(1-560) or human KIF7(1-543) in in vitro assays suggested that KIF7 binds preferentially to GTP-tubulin-containing microtubules and tracks with their growing plus ends (He et al., 2014; Jiang et al., 2019). The ability to localize to the plus ends of microtubules was then postulated to provide a role for KIF7 in regulating ciliary length and organizing the ciliary tip compartment (He et al., 2014). However, when examined in fixed COS-7, NIH-3T3 or CAD cells, mouse KIF7(1-558) bound to all microtubules in the cytoplasm and the cilium, and showed no preference for any subset of microtubules or region, such as the plus ends (Fig. 1; Fig. S2). Similar results were obtained for other uninhibited versions of KIF7, such as the truncated protein KIF7(1-1114) (Fig. 2) and the full-length mutant protein KIF7(Q1136P) (Fig. 4). As fixation could result in the loss of plus-end labeling, we used live-cell imaging to examine whether KIF7(1-558) could track the growing ends of microtubules in cells. KIF7(1-558) was tagged with monomeric NeonGreen (mNG) and co-expressed in COS-7 cells with EB3-mCherry, which marks the growing plus ends of microtubules. In live cells, KIF7(1-558)-mNG was observed along the shaft of all microtubules (Fig. 6A) and did not localize preferentially to or track with the growing plus ends marked by EB3-mCherry (Fig. 6B,C). Although we cannot rule out the possibility that individual KIF7(1-558)-mNG motors bound to the plus ends of microtubules, we did not observe a significant enrichment or preferential interaction of KIF7(1-558)-mNG with microtubule plus ends.
Fig. 6.
KIF7 does not track the plus ends of microtubules in cells. (A-C) Microtubule dynamics in COS-7 cells expressing KIF7(1-558)-mNG together with EB3-mCherry. (A) Representative image. The periphery of the transfected cell is indicated by white dotted lines. Scale bar: 10 µm. (B) Representative kymographs of live-cell imaging of KIF7(1-558)-mNG and EB3-mCherry on the microtubule in the white boxed region of A. Time is on the y-axis (scale bar: 5 s) and distance is on the x-axis (scale bar: 5 µm). The white asterisks indicate the time point used for line-scan analysis of KIF7 (green) and EB3 (red) fluorescence intensity (C). (D-I) Microtubule dynamics in NIH-3T3 cells expressing KIF7(1-558)-mNG together with EB3-mCherry and in the absence (D-F) or presence of SAG treatment for 2-4 h (G-I). (D,G) Representative images. The periphery of the transfected cells is indicated by white dotted lines. (E,H) Representative kymographs of live-cell imaging of KIF7(1-558)-mNG and EB3-mCherry on the microtubule in the white boxed region. The white asterisks indicate the time point used for line-scan analysis of KIF7(1-558) (green) and EB3-mCherry (red) fluorescence intensity along the growing microtubule (F,I). a.u., arbitrary units.
There are several reasons why truncated KIF7 proteins could bind preferentially to and track the growing microtubule plus ends in in vitro assays (He et al., 2014; Jiang et al., 2019) but not in COS-7 cells (Fig. 6A-C). One possibility is that the preference of KIF7 for GTP-tubulin and/or growing plus ends may be actively inhibited (e.g. via a post-translational modification) in the cytoplasm of mammalian cells and relieved under specific conditions (e.g. following hedgehog stimulation). To test this possibility, we co-expressed KIF7(1-558)-mNG with EB3-mCherry in NIH-3T3 cells and carried out live-cell imaging under unstimulated conditions or after Hedgehog pathway activation by the Sonic Hedgehog agonist SAG. Similar to the results in COS-7 cells, KIF7(1-558)-mNG bound along the shaft of all microtubules (Fig. 6D) and was not observed to track the growing microtubule plus ends under unstimulated conditions (Fig. 6E,F) or under SAG-stimulated conditions (Fig. 6G-I). We verified that SAG stimulation resulted in Hedgehog pathway activation by confirming an increase in the number of cells with KIF7 at the tip of the primary cilium (Fig. S6E,F). We also tested whether the full-length protein could track the plus ends of microtubules under basal or SAG-stimulated states but did not observe any microtubule association of KIF7 under these conditions (Fig. S6A-D).
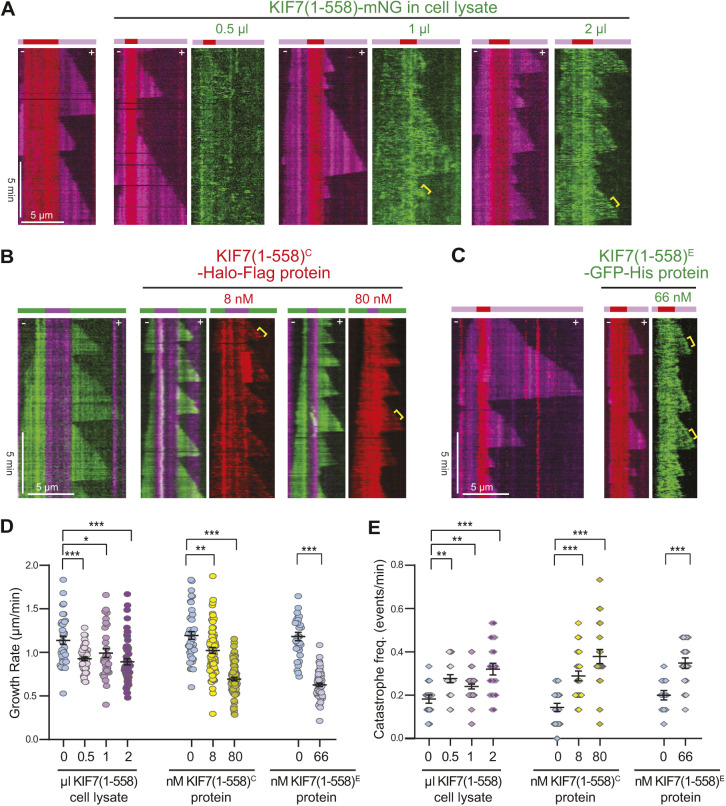
A second possibility is that there are factors (e.g. +TIP proteins) that physically block KIF7 from tracking plus ends in cells but were absent in the previous work with in vitro microtubule dynamics assays (He et al., 2014; Jiang et al., 2019). To test this possibility, we carried out in vitro microtubule dynamics assays with KIF7(1-558) tagged with monomeric NeonGreen (mNG) and present in diluted COS-7 cell extracts. Addition of increasing amounts of KIF7(1-558)-mNG in cell lysates resulted in a decrease in the microtubule growth rate and an increase in the microtubule catastrophe rate (Fig. 7A,D,E), consistent with previous work (He et al., 2014; Jiang et al., 2019; Yue et al., 2018). However, we did not observe preferential binding to the GMPCPP-tubulin seed or the GTP-tubulin plus end (Fig. 7A).
Fig. 7.
KIF7 alters microtubule dynamics in vitro without tracking the microtubule plus ends. (A) Representative kymographs of microtubule dynamics in the absence or with increasing concentrations of cell lysate containing KIF7(1-558)-mNG. Red, GMPCPP-containing microtubule seeds; magenta, growing microtubules; green, KIF7(1-558)-mNG. (B) Representative kymographs of microtubule dynamics in the absence or with increasing concentrations of KIF7(1-558)-Halo-Flag protein purified from COS-7 cells [KIF7(1-558)-Halo-FlagC]. Magenta, GMPCPP-containing microtubule seeds; green, growing microtubules; red, KIF7(1-558)-Halo-FlagC (JF552 ligand). (C) Representative kymographs of microtubule dynamics in the absence or with increasing concentrations of KIF7(1-558)-eGFP-His protein purified from E. coli [KIF7(1-558)-eGFP-HisE]. Red, GMPCPP-containing microtubule seeds; magenta, growing microtubules; green, KIF7(1-558)-eGFP-HisE. For all kymographs, time is on the y-axis (scale bar: 5 min) and distance is on the x-axis (scale bar: 5 µm). Yellow brackets indicate events with a slight enrichment of KIF7(1-558) at the microtubule plus end. (D,E) Quantification of microtubule growth rates (D) and catastrophe frequency (E) in the absence or presence of the indicated KIF7(1-558) protein. KIF7(1-558)-mNG: n=30-60 events in D and n=15-40 microtubules in E from two independent experiments; KIF7(1-558)-Halo-FlagC: n=43-84 events in D and n=19-24 microtubules in E from two independent experiments; KIF7(1-558)-eGFP-HisE: n=33-63 events in D and n=12-18 microtubules in E from two independent experiments. Data are reported as mean±s.e.m. *P<0.05; **P<0.01; ***P<0.001 compared to microtubule dynamics in the absence of KIF7 (two-tailed unpaired Student's t-test).
It is also possible that the ability of KIF7 to recognize the growing microtubule plus ends is actively inhibited in mammalian cells by a mechanism (e.g. a KIF7 binding partner) that is lacking in bacterial and insect cell expression systems. To examine these possibilities, we purified KIF7(1-558) from COS-7 cells [KIF7(1-558)-Halo-FlagC] or from Escherichia coli cells [KIF7(1-558)-EGFPE], and examined the localization of the proteins in microtubule dynamics assays. Both purified proteins suppressed microtubule growth rates and increased microtubule catastrophes (Fig. 7B-E) but did not bind preferentially to the GMPCPP-tubulin seed or the GTP-tubulin plus end (Fig. 7B,C). Although we did sometimes observe a slight enrichment of KIF7(1-558) at the growing plus end (Fig. 7, brackets), the KIF7(1-558) proteins bound all along the microtubules.
We noted that our assay conditions differed slightly from the previous work and thus repeated the experiments with purified KIF7(1-558)-Halo-FlagC (Fig. S7A) under different assay conditions. Using our conditions of BRB80 buffer and 1 mg/ml casein as a blocking agent, we did not observe purified KIF7(1-558)-Halo-FlagC protein binding preferentially to or tracking the plus ends of growing microtubules at high (Fig. S7D) or low (Fig. S7G) concentrations. We repeated these experiments to mimic the buffer conditions used in the previous studies (He et al., 2014; Jiang et al., 2019), and did observe preferential binding of KIF7(1-558) to the GMPCPP-containing microtubule seed (Fig. S7B,C,E). The critical component in these assays appears to be the amount of casein present as KIF7(1-558) bound equally to the seed and growing microtubule with 1.0 mg/ml casein as the blocking agent (Fig. S7F). We thus conclude that binding of truncated KIF7 to the plus ends of microtubules is not an inherent property of KIF7 but rather can be observed under specific in vitro conditions.
DISCUSSION
The kinesin-4 member KIF7 is an atypical kinesin as it is immotile. How the microtubule binding of KIF7 is regulated and what role microtubule binding plays in KIF7 function are not known. Here, we demonstrate that KIF7 is regulated by an auto-inhibition mechanism and suggest that loss of KIF7 auto-inhibition can be pathogenic for humans. We demonstrate that the inhCC segment is critical for auto-inhibition and that mutant versions that have lost auto-inhibition localize uniformly along cytosolic microtubules. Although uninhibited forms of KIF7 can impact dynamics at the microtubule plus ends in in vitro assays, regulation of microtubule dynamics is not likely to be a primary function of KIF7 in mammalian cells. Localization of KIF7 to the tip of the primary cilium also requires the inhCC segment but is independent of microtubule binding. These results suggest that mutations that alter the cilium tip localization, and thus likely the function of KIF7, can also be pathogenic for humans.
The inhCC plays a critical role in KIF7 auto-inhibition
A general model for kinesin regulation involves auto-inhibition of the motor domain by non-motor regions of the protein (Verhey and Hammond, 2009). We demonstrate that this model applies to KIF7, as the full-length protein is unable to bind to microtubules, whereas variants that lack or carry mutations in the inhCC are competent for microtubule binding. For kinesin-1, early work demonstrated that the globular C-terminal tail domain is critical for the auto-inhibition of the N-terminal motor domain of kinesin-1 (Coy et al., 1999; Friedman and Vale, 1999; Hackney and Stock, 2000; Verhey et al., 1998). Recent work has shown that for a growing number of kinesins in the kinesin-2, kinesin-3 and kinesin-4 families, the auto-inhibition mechanism uses a centrally located coiled-coil segment to interact with and inhibit the N-terminal motor domain (Brunnbauer et al., 2010; Hammond et al., 2010; Imanishi et al., 2006; Farkhondeh et al., 2015; Hammond et al., 2009; Huo et al., 2012; Lee et al., 2004; Niwa et al., 2016; Ren et al., 2018; Siddiqui et al., 2019; Yamada et al., 2007; Bianchi et al., 2016; Cheng et al., 2014; van der Vaart et al., 2013; van Riel et al., 2017). Our results demonstrate that KIF7 uses this latter mechanism of stalk-mediated auto-inhibition.
Previous work suggested that the rCC could be critical for the auto-inhibition of KIF7 (Bianchi et al., 2016) as the rCC is critical for the auto-inhibition mechanism of the kinesin-4 family members KIF21A and KIF21B. Although full-length KIF21A displays little to no microtubule-based activity, suggesting a relatively strong auto-inhibition, full-length KIF21B can be observed to bind to and move along microtubules, suggesting that the protein is not completely inhibited (Asselin et al., 2020; Ghiretti et al., 2016; van Riel et al., 2017). Nevertheless, for both KIF21A and KIF21B, truncations that remove the rCC result in a dramatic increase in the number of motility events, as well as an increase in the velocity and run length of those events (Cheng et al., 2014; Ghiretti et al., 2016; van der Vaart et al., 2013; van Riel et al., 2017). In the case of KIF7, we suggest that the inhCC rather than the rCC is critical for KIF7 auto-inhibition based on two lines of evidence. First, truncations that remove the inhCC are sufficient to relieve auto-inhibition of microtubule binding, and further truncations that remove the rCC do not alter this phenotype. Second, a single point mutation in the inhCC of mouse KIF7, Q1136P, results in a release of auto-inhibition based on the strong microtubule binding phenotype observed for the mutant full-length protein. These results support the conclusion that the inhCC is required for the auto-inhibition of KIF7, whereas the rCC is not sufficient.
Uninhibited versions of KIF7 do not bind preferentially to or track growing microtubule plus ends
Loss of auto-inhibition due to truncation or mutation of KIF7 resulted in proteins that bound to all microtubules in cells, including cytosolic and axonemal microtubules (Figs 1, 2 and 4). High expression of uninhibited KIF7 proteins also resulted in the bundling of microtubules and collapse of the cytosolic microtubule network around the nucleus, perhaps due to stabilization of KIF7-bound microtubules. This is consistent with the previous results of He et al. (2014) who reported that KIF7(1-560)-EGFP bound to cytosolic microtubules and caused their bundling in cells. The ability of uninhibited KIF7 to cause microtubule reorganization and bundling in cells is reminiscent of previous observations of altered microtubule morphology upon overexpression of microtubule-associated proteins (e.g. Barlow et al., 1994; Gruss et al., 2002; Lewis et al., 1989; Masson and Kreis, 1993).
In contrast to previous in vitro studies (He et al., 2014; Jiang et al., 2019), we found that uninhibited versions of KIF7 did not bind preferentially to the plus ends of microtubules in cells, including COS-7 fibroblast-like, NIH-3T3 fibroblast or CAD neuronal cells. In live cells, we confirmed that truncated KIF7 proteins do not track the plus ends of dynamic microtubules marked by EB3 (also known as Mapre3), regardless of cell type or Hedgehog pathway stimulation (Fig. 6). These results highlight the importance of verifying in vitro findings using cellular assays.
We also did not observe KIF7(1-558) binding selectively to the plus ends of microtubules in in vitro microtubule dynamics assays regardless of tag, purification state or source of protein (E. coli or mammalian cells) (Fig. 7). For some events, we did observe a slight enrichment of KIF7(1-558) at the GMPCPP-tubulin microtubule seed and/or the GTP-containing microtubule cap; however, even in these cases, KIF7(1-558) was also observed along the shaft of the microtubule. The most likely explanation for the differences between the results presented here and previous work relates to assay conditions. At lower concentrations of casein as a blocking agent, we found that purified KIF7(1-558) protein bound preferentially to the GMPCPP-containing microtubule seeds, consistent with previous work (He et al., 2014; Jiang et al., 2019). However, at higher concentrations of casein, KIF7(1-558) uniformly coated the microtubule lattice. We note that the ability of KIF7 to bind preferentially to GTP-like microtubules has also been shown to be sensitive to assay conditions in previous work. For example, in microtubule dynamics assays with AMPPNP added to the assay buffer, purified mouse KIF7(1-560) and human KIF7(1-543) decorate the GDP-lattice rather than the GMPCPP-seed (He et al., 2014; Jiang et al., 2019). Furthermore, a monomeric version of human KIF7 (amino acids 1-386) bound preferentially to the GTP-like seed and growing ends of dynamic microtubules at 1× BRB80 but to the GDP-containing tubulin lattice at 0.5× BRB80 (Jiang et al., 2019). Taken together, these results suggest that localization to the growing plus ends of microtubules is not an inherent property of KIF7 as proteins that have lost their auto-inhibition due to truncation or mutation do not localize to microtubule plus ends in cells or under all in vitro conditions.
The discrepancy concerning the ability of KIF7 to bind preferentially to microtubule plus ends is reminiscent of recent work studying the microtubule-associated protein TPX2, which localized to the spindle poles in metaphase and to the spindle midzone in late anaphase (Gruss et al., 2002). In microtubule dynamics assays, TPX2 reduces the catastrophe frequency and promotes nucleation of microtubules (Reid et al., 2016; Roostalu et al., 2015; Wieczorek et al., 2015). However, GFP-TPX2 was shown to bind preferentially to GMPCPP-containing seeds and GTP-containing plus ends in one study (Roostalu et al., 2015), whereas mCherry-TPX2 bound along the microtubule with only a slight enrichment at the growing plus end in another study (Reid et al., 2016), a discrepancy that was attributed to different experimental conditions. Similar to our observations with KIF7, GFP-TPX2 localizes along microtubules and does not preferentially associate with the microtubule plus ends when expressed in cultured mammalian cells or added to spindle formation assays in Xenopus extracts (Brunet et al., 2004; Gruss et al., 2002). These results highlight the importance of testing different in vitro conditions and verifying in vitro findings using cellular assays.
Localization of KIF7 to the tip of the primary cilium also requires the inhCC but is independent of microtubule binding
Removal of the C-terminal tail domain (e.g. 1-1227) did not alter the auto-inhibition or ciliary tip localization of KIF7, whereas further truncations that remove the inhCC (e.g. 1-1114) resulted in loss of both auto-inhibition and ciliary tip localization. This result suggests that sequences in the inhCC contribute to both auto-inhibition and ciliary tip localization. It appears that separate sequences in the inhCC contribute to auto-inhibition and ciliary tip localization, as mutation of Q1136P resulted in a loss of auto-inhibition but increased ciliary tip localization. Interestingly, mutations of N1064S and R1072W in the rCC did not release auto-inhibition but resulted in an increase in ciliary tip localization. Thus, ciliary tip localization of KIF7 is not related to its ability to bind to microtubules. Further work is needed to determine whether the increased ciliary tip localization of these point mutants is due to their increased entry into the cilium or their increased retention at the cilium tip.
The identification of point mutations that result in increased KIF7 tip localization will be beneficial for investigating the role of KIF7 at the ciliary tip. KIF7 has been proposed to play a role in regulating the dynamics of axonemal microtubules and organizing the ciliary tip compartment (He et al., 2014). Consistent with this hypothesis, in mice, loss of KIF7 expression, or an L130P point mutation in Kif7, resulted in increased cilium length and mislocalization of IFT components (He et al., 2014; Emechebe et al., 2016; Putoux et al., 2011, 2016). In zebrafish carrying an insertion plus a missense mutation in the motor domain of Kif7, the cilia in the central canal were longer than those in the central canal of wild-type animals (Terhune et al., 2020). However, in fibroblast lines from patients with acrocallosal syndrome, the cilia were longer in cells from one patient but were of normal length and ultrastructural organization from a second patient (Putoux et al., 2011). Thus, the influence of KIF7 on cilium length may depend on the context of the mutation. As the control of cilium length is complex (Mirvis et al., 2018; Sánchez and Dynlacht, 2016) and uninhibited versions of KIF7 do not localize to the plus ends of microtubules, the role of KIF7 in regulating ciliary length is still unclear.
Release of auto-inhibition may contribute to human disease
Our data suggest that loss of KIF7 auto-inhibition can be pathogenic for human disease. Interestingly, a majority of variants in human patients reported in the literature, or described as pathogenic or likely pathogenic in ClinVar, involve nonsense or frameshift mutations that would result in a KIF7 protein lacking the inhCC segment. Furthermore, mutation of Q1136P in the inhCC results in an uninhibited protein that binds strongly to microtubules in the cytosol of mammalian cells. Mutation of the corresponding residue in the human protein, Q1132P, has been reported as an allele of germline origin, with no clinical condition provided, and designated as a likely pathogenic variant as it is not observed in large population cohorts (ClinVar VCV000451950.2). Our data suggest that release of auto-inhibition could provide an explanation for the pathogenicity of the Q1132P variant. In contrast, we find that another point mutation in the inhCC, H1119Q, had no effect on KIF7 auto-inhibition. The corresponding human mutation, H1115Q, was identified in patients with acrocallosal syndrome, hydrolethalus syndrome 2 and idiopathic scoliosis (Putoux et al., 2011; Terhune et al., 2020), but was predicted to be benign based on allele frequency data from public databases (ClinVar VCV000129415.4), and our data are consistent with this designation. Thus, our study extends the work in the field, demonstrating that loss of auto-inhibition can provide an explanation for the pathogenic variants of kinesin motor proteins (Asselin et al., 2020; Cheng et al., 2014; Chiba et al., 2019; Kelliher et al., 2018; Moua et al., 2011; Niwa et al., 2016; van der Vaart et al., 2013; Williams et al., 2014).
MATERIALS AND METHODS
Plasmids and antibodies
Plasmids for expression of full-length and truncated (amino acids 1-558) versions of mouse KIF7 in mammalian cells have been described previously (Yue et al., 2018). Additional truncated, mutant or chimeric versions were generated by PCR or gene synthesis, and subcloned using appropriate restriction enzymes. KIF7(1-558) was subcloned for E. coli expression with EGFP at its C-terminus for fluorescence microscopy and His-tag for purification. All plasmids were verified by DNA sequencing. The following antibodies were purchased for immunofluorescence staining: Arl13b (1:1000, Protein Tech Group, 17711-1-AP), Pericentrin (1:500, AbCam, ab4448), β-tubulin (1:5000, clone E7, Developmental Studies Hybridoma Bank), acetylated α-tubulin (1:10,000, Sigma-Aldrich, T6793).
Cell culture, transfection and lysate preparation
COS-7 cells [monkey kidney fibroblast, American Type Culture Collection (ATCC), RRID:CVCL_0224] and NIH-3T3 cells (ATCC; RRID:CVCL_0594) were cultured in Dulbecco's modified Eagle medium (DMEM, Gibco) with 10% (v/v) Fetal Clone III (HyClone) and GlutaMAX (Gibco) at 37°C with 5% CO2. Mouse central nervous system catecholaminergic CAD cells (RRID:CVCL_0199) were grown in a 1:1 mixture of F12:DMEM (Gibco) plus 10% fetal bovine serum (HyClone) and GlutaMAX (Gibco) at 37°C with 5% CO2. Cells were tested annually for mycoplasma contamination, and COS-7 and NH-3T3 cells were authenticated through mass spectrometry (the protein sequences exactly match those in the African green monkey and mouse genomes, respectively). For NIH-3T3 cells, cells were switched to low serum medium at the time of transfection to arrest cells in G1. Forty-eight hours post-transfection, the cells were treated with or without 500 nM SAG for 3 h and then fixed and stained. For CAD cells, 12 h after transfection, cells were switched to differentiation medium (F12:DMEM without serum) to induce neurite outgrowth, and 48 h after transfection, cells were fixed and stained. Transfections were carried out using Trans-IT LT1 (Mirus) in Opti-MEM (Gibco) according to the manufacturer's instructions.
To prepare extracts for microtubule dynamics assays, COS-7 cells 16 h post-transfection were harvested and centrifuged at low speed at 4°C. The cell pellet was washed with PBS and resuspended in ice-cold BRB80 buffer [80 mM PIPES/KOH (pH 6.8), 1 mM MgCl2 and 1 mM EGTA] freshly supplemented with 1 mM ATP, 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors cocktail. The cells were lysed by sonication (Fisher Scientific, Sonic Dismembrator Model 500) using 10% power, 4×10 s on ice. After centrifugation for 10 min at 20,000 g at 4°C, aliquots of the supernatant were snap-frozen in liquid nitrogen and stored at −80°C until further use.
Immunofluorescence and image analysis
COS-7 cells were fixed with 3.7% formaldehyde in PBS, treated with 50 mM NH4Cl in PBS to quench unreacted formaldehyde, permeabilized with 0.2% Triton X-100 in PBS, and then blocked in blocking solution (0.2% fish skin gelatin in PBS). NIH-3T3 cells were fixed with ice-cold methanol for 10 min, rinsed with PBS and blocked in blocking solution (0.2% fish skin gelatin in PBS). Primary and secondary antibodies were placed in blocking solution at room temperature for 1 h each. Nuclei were stained with 10.9 μM DAPI and cover glasses were mounted in ProlongGold (Life Technologies). Images were collected on an inverted epifluorescence microscope (Nikon TE2000E) equipped with a 60×1.40 numerical aperture (NA) oil-immersion objective and a 1.5× tube lens on a Photometrics CoolSnapHQ camera driven by NIS-Elements (Nikon) software.
Only cells expressing low to medium levels of exogenous motor were selected for image capture and analysis based on the criteria in Fig. S1A. Quantification of exogenous protein expression was performed using Fiji/ImageJ2 (www.imagej.net/Fiji). Using the polygon tool, expressing cells were outlined and the average fluorescence signal was measured for each cell. The average background fluorescence was measured in an area adjacent to the expressing cell and was subtracted from the average fluorescence value. Values were normalized to compare data collected across different experimental repeats with varying optical (illumination) configurations.
The microtubule and cilium localization were scored visually based on the criteria in Fig. S1B,C. In COS-7 cells, if the exogenous protein exhibited a cytoplasmic/diffuse localization, the phenotype was defined as ‘no MT binding’. If the exogenous protein could be seen both in the cytoplasm and on microtubules within the cell, this was defined as ‘weak microtubule binding’. Constructs for which the exogenous protein could only be seen on microtubules were defined as exhibiting ‘strong microtubule binding’. In NIH-3T3 cells, if the exogenous protein could not be detected in the cilium, this was classified as ‘absent from cilium’. If the exogenous protein was elevated at the base of the cilium (defined by the presence of pericentrin staining), this was classified as ‘at base’. If the exogenous protein was elevated at the tip of the cilium (defined by the absence of pericentrin staining), the localization was classified as ‘at tip’. If the exogenous protein was distributed uniformly along the cilium, the localization was classified as ‘along shaft’.
Protein expression and purification from COS-7 cells
COS-7 cells were transfected with plasmids for the expression of KIF7(1-558)-Halo-Flag using Trans-IT LT1 (Mirus), and the protein was fluorescently labeled by the inclusion of 50 nM JF552 Halo ligand (Tocris Bioscience) in the medium. Cells from two 10 cm dishes were harvested 24 h after transfection and lysed in 1 ml lysis buffer [25 mM HEPES, 115 mM KOAc, 5 mM NaOAc, 5 mM MgCl2, 0.5 mM EGTA, 1% Triton X-100 (pH 7.4, with KOH)] supplemented with protease inhibitor cocktail, 1 mM PMSF, 1 mM ATP and 1 mM DTT. After centrifugation at 21,000 g for 10 min at 4°C, the supernatant was incubated with 50 μl anti-Flag M2 agarose beads (Sigma-Aldrich) for 1.5 h at 4°C with rotation. Bound protein was washed with wash buffer [150 mM KCl, 20 mM Imidazole (pH 7.5), 5 mM MgCl2, 1 mM EDTA and 1 mM EGTA] supplemented with protease inhibitor cocktail, 1 mM PMSF, 1 mM DTT and 3 mM ATP. The protein was eluted by incubation with 80 μl BRB80 buffer [80 mM PIPES/KOH (pH 6.8), 1 mM MgCl2 and 1 mM EGTA] supplemented with protease inhibitor cocktail, 1 mM PMSF, 0.5 mM DTT, 0.1 mM ATP and 0.15 mg/ml 3xFlag peptide (Sigma-Aldrich) for 2 h on ice. The protein was collected by centrifugation at 1000 g for 5 min at 4°C. Aliquots were snap-frozen and stored at −80°C.
Protein expression and purification from E. coli cells
Plasmids were transformed into BL21-CodonPlus(DE3)-RIPL competent cells (Agilent Technologies, 230280). A single colony was inoculated in 3 ml terrific broth with 100 μg/ml carbenicillin and shaken at 37°C overnight. The culture was then inoculated into 400 ml of terrific broth with 100 μg/ml carbenicillin and incubated at 37°C with shaking for 4-5 h. After cooling on ice for 1 h, isopropyl β-d-1-thiogalactopyranoside was added to the culture to a final 0.1 mM concentration to induce expression, and the culture was incubated at 18°C with shaking overnight. The cells were harvested by centrifugation at 3000 g for 10 min at 4°C. The supernatant was discarded, and the cells were resuspended in 5 ml of B-PER complete bacterial protein extraction reagent (Thermo Fisher Scientific, 89821) with 2 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1 mM ATP, 2 mM PMSF and 10% glycerol. The resuspended cells were snap-frozen and stored at −80°C.
For protein purification, the frozen cells were quickly thawed at 37°C and nutated at room temperature for 20 min. The thawed solution was then dounced for 10 stokes on ice to lyse the cells. The cell lysate was centrifuged at 278,288 g for 10 min at 4°C using a Beckman Coulter tabletop centrifuge unit. The supernatant was incubated with 200 μl of Ni-nitrilotriacetic acid resin (Roche complete His-Tag purification resin, MilliporeSigma, 5893682001) at 4°C for 1 h. Afterwards, the resin was washed with wash buffer [50 mM HEPES, 300 mM KCl, 2 mM MgCl2, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 0.1 mM ATP, 0.1% Triton X-100 and 10% glycerol (pH 7.2)] and the protein was then eluted with elution buffer (wash buffer with 250 mM imidazole). The eluate was snap-frozen and stored at −80°C.
Live-cell imaging of microtubule dynamics
COS-7 cells seeded onto glass-bottomed dishes (MatTek Corporation) were co-transfected with plasmids for the expression of EB3-mCherry and KIF7 (1-558)-mNG. After ∼5 h, the cells were washed once and then incubated in Leibovitz's L-15 medium (Gibco). NIH-3T3 cells seeded onto glass-bottomed dishes (MatTek Corporation) were serum starved for 24 h and then co-transfected with plasmids for co-expression of EB3-mCherry with KIF7 (1-558)-mNG or KIF7(FL)-mCitrine. After 3 h, the cells were treated with 500 nM SAG (Enzo Life Sciences) for 2-4 h to activate Hedgehog signaling. Five to seven hours post-transfection, the cells were washed once and then incubated in Leibovitz's L-15 medium (Gibco). The cells were imaged at 37°C in a temperature-controlled and humidified live-imaging chamber (Tokai Hit) mounted on an inverted total internal reflection fluorescence (TIRF) microscope Ti-E/B (Nikon) equipped with a 100×1.49 N.A. oil immersion TIRF objective (Nikon), three 20 mW diode lasers (488 nm, 561 nm and 640 nm) and electron multiplying charged-couple device detector (iXon X3DU897, Andor). The angle of illumination was adjusted for maximum penetration of the evanescent field into the cell. Image acquisition was controlled with Nikon Element software. Images were acquired in both 488- and 561-nm channels every 200 ms for 60 s. Kymographs (width=3 pixels) of KIF7 and EB3 along microtubules were generated using Fiji/ImageJ2. The fluorescence intensity of KIF7(1-558)-mNG and EB3-mCherry along individual microtubules was analyzed using line-scan analysis in Fiji/ImageJ2.
In vitro microtubule dynamics assays
A flow cell (∼10 μl volume) was assembled by attaching a clean #1.5 coverslip (Fisher Scientific) to a glass slide (Fisher Scientific) with two strips of double-sided tape. Microtubule seeds containing 10% biotin-labeled tubulin and either 10% X-rhodamine-labeled or 20% Hilyte647-labeled tubulins (Cytoskeleton Inc.) were generated by polymerization in the presence of the non-hydrolysable GTP analog GMPCPP (NU-405S, Jena Bioscience), and then immobilized on the coverslip by incubating the flow chamber sequentially with the following solutions: (1) 1 mg/ml bovine serum albumin (BSA)-biotin (A8549, Sigma-Aldrich); (2) blocking buffer (1 mg/ml BSA in BRB80); (3) 0.5 mg/ml NeutrAvidin (31000, Thermo Fisher Scientific); (4) blocking buffer; (5) GMPCPP-stabilized microtubule seeds; and (6) blocking buffer.
Microtubule growth was initiated by flowing in (condition #1) 20 µM tubulin containing 7% Hilyte647-labeled tubulin (Cytoskeleton Inc.) together with cell extract in reaction buffer #1 [BRB80 supplemented with 2.5 mM GTP, 2.5 mM ATP, 2 mM MgCl2, 2 mM DTT, 0.2 mg/ml BSA, 2 mg/ml casein, 0.2% methylcellulose (Sigma-Aldrich) and oxygen-scavenging system (20 mM glucose, 0.16 mg/ml catalase and 0.4 mg/ml glucose oxidase)]. Alternatively, microtubule growth was initiated by flowing in (condition #2) 18.8 µM tubulin containing 10% Hilyte488-labeled tubulin (Cytoskeleton Inc.) together with purified KIF7 protein in reaction buffer #2 [BRB80 supplemented with 1 mM GTP, 1 mM ATP, 3 mM MgCl2, 0.1% methylcellulose and oxygen-scavenging system (16 mM glucose, 0.7 mg/ml catalase and 0.3 mg/ml glucose oxidase)]. To examine whether the reaction buffer affects the affinity of KIF7 protein for microtubule seeds and plus ends, reaction buffer #2 was supplemented with 3% sucrose, 20 mM KCl, 0.5% β-mercaptoethanol or 2.5 mM DTT and 0.2 or 1.0 mg/ml casein.
The flow cells were sealed with molten paraffin wax and imaged by TIRF microscopy. The temperature was set at 35°C (for cell lysate) or 30°C (for KIF7 protein) in a temperature-controlled chamber (Tokai Hit). Time-lapse images were acquired in 488 nm, 561 nm and 640 nm channels at a rate of every 5 s for 15 min. To determine the growth rate and catastrophe frequency of microtubule plus ends, maximum intensity projections were generated and kymographs (width=3 pixels) were prepared using Fiji/ImageJ2, and displayed with time on the y-axis and distance on the x-axis. Only growth events with a slope over a three-pixel length were analyzed. Catastrophe frequency was obtained by dividing the total number of catastrophes observed by the imaging time (15 min).
Protein sequence analysis
Coiled coil predictions were carried out using MarCoil (Delorenzi and Speed, 2002), PCOIL (Gruber et al., 2006), and COILS (Lupas, 1996). Sequence alignments were carried out using T-COFFEE (version 11.00) (Di Tommaso et al., 2011) and edited in JalView (2.11.1.4) (Waterhouse et al., 2009) and Adobe Illustrator.
Statistical analysis
For microtubule dynamics assays, comparisons for each KIF7 condition were made to the same assay in the absence of KIF7 using a two-tailed t-test. Statistical analysis and graph construction were performed using Prism software (GraphPad 8.0.0).
Supplementary Material
Acknowledgements
We thank members of the Verhey and Gennerich labs for helpful discussions. We also thank Takashi Hotta for help with preparing sonicated cell exacts.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: T.L.B., Y.Y., K.J.V.; Formal analysis: T.L.B., Y.Y.; Investigation: T.L.B., Y.Y., R.P.; Resources: X.L., A.G., K.J.V.; Data curation: T.L.B., Y.Y.; Writing - original draft: K.J.V.; Writing - review & editing: T.L.B., Y.Y., X.L., A.G., K.J.V.; Visualization: T.L.B., Y.Y., R.P.; Supervision: A.G., K.J.V.; Project administration: K.J.V.; Funding acquisition: A.G., K.J.V.
Funding
The work was funded by the National Institutes of Health (R01GM070862, R35GM131744 and R01GM118751 to K.J.V., and R01GM098469 and R01NS114636 to A.G.). Deposited in PMC for release after 12 months.
References
- Ali, B. R., Silhavy, J. L., Akawi, N. A., Gleeson, J. G. and Al-Gazali, L. (2012). A mutation in KIF7 is responsible for the autosomal recessive syndrome of macrocephaly, multiple epiphyseal dysplasia and distinctive facial appearance. Orphanet J. Rare Dis. 7, 27. 10.1186/1750-1172-7-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadollahi, R., Strauss, J. E., Zenker, M., Beuing, O., Edvardson, S., Elpeleg, O., Strom, T. M., Joset, P., Niedrist, D., Otte, C.et al. (2018). Clinical and experimental evidence suggest a link between KIF7 and C5orf42-related ciliopathies through Sonic Hedgehog signaling. Eur. J. Hum. Genet. 26, 197-209. 10.1038/s41431-017-0019-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin, L., Rivera Alvarez, J., Heide, S., Bonnet, C. S., Tilly, P., Vitet, H., Weber, C., Bacino, C. A., Baranaño, K., Chassevent, A.et al. (2020). Mutations in the KIF21B kinesin gene cause neurodevelopmental disorders through imbalanced canonical motor activity. Nat. Commun. 11, 2441. 10.1038/s41467-020-16294-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann-Gagescu, R., Dempsey, J. C., Phelps, I. G., O'roak, B. J., Knutzen, D. M., Rue, T. C., Ishak, G. E., Isabella, C. R., Gorden, N., Adkins, J.et al. (2015). Joubert syndrome: a model for untangling recessive disorders with extreme genetic heterogeneity. J. Med. Genet. 52, 514-522. 10.1136/jmedgenet-2015-103087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangs, F. and Anderson, K. V. (2017). Primary cilia and mammalian Hedgehog signaling. Cold Spring Harb. Perspect. Biol. 9, a028175. 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakeh, D., Faqeih, E., Anazi, S., S Al-Dosari, M., Softah, A., Albadr, F., Hassan, H., Alazami, A. M. and Alkuraya, F. S. (2015). The many faces of KIF7. Hum. Genome Var. 2, 15006. 10.1038/hgv.2015.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow, S., Gonzalez-Garay, M. L., West, R. R., Olmsted, J. B. and Cabral, F. (1994). Stable expression of heterologous microtubule-associated proteins (MAPs) in Chinese hamster ovary cells: evidence for differing roles of MAPs in microtubule organization. J. Cell Biol. 126, 1017-1029. 10.1083/jcb.126.4.1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, S., Van Riel, W. E., Kraatz, S. H. W., Olieric, N., Frey, D., Katrukha, E. A., Jaussi, R., Missimer, J., Grigoriev, I., Olieric, V.et al. (2016). Structural basis for misregulation of kinesin KIF21A autoinhibition by CFEOM1 disease mutations. Sci. Rep. 6, 30668. 10.1038/srep30668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet, S., Sardon, T., Zimmerman, T., Wittmann, T., Pepperkok, R., Karsenti, E. and Vernos, I. (2004). Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol. Biol. Cell 15, 5318-5328. 10.1091/mbc.e04-05-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunnbauer, M., Mueller-Planitz, F., Kosem, S., Ho, T. H., Dombi, R., Gebhardt, J. C. M., Rief, M. and Okten, Z. (2010). Regulation of a heterodimeric kinesin-2 through an unprocessive motor domain that is turned processive by its partner. Proc. Natl. Acad. Sci. USA 107, 10460-10465. 10.1073/pnas.1005177107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L., Desai, J., Miranda, C. J., Duncan, J. S., Qiu, W., Nugent, A. A., Kolpak, A. L., Wu, C. C., Drokhlyansky, E., Delisle, M. M.et al. (2014). Human CFEOM1 mutations attenuate KIF21A autoinhibition and cause oculomotor axon stalling. Neuron 82, 334-349. 10.1016/j.neuron.2014.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, H. O.-L., Zhang, X., Ribeiro, A., Mo, R., Makino, S., Puviindran, V., Law, K. K. L., Briscoe, J. and Hui, C.-C. (2009). The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci. Signal. 2, ra29. 10.1126/scisignal.2000405 [DOI] [PubMed] [Google Scholar]
- Chiba, K., Takahashi, H., Chen, M., Obinata, H., Arai, S., Hashimoto, K., Oda, T., Mckenney, R. J. and Niwa, S. (2019). Disease-associated mutations hyperactivate KIF1A motility and anterograde axonal transport of synaptic vesicle precursors. Proc. Natl. Acad. Sci. USA 116, 18429-18434. 10.1073/pnas.1905690116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran, J. C. (2015). Kinesin motor enzymology: chemistry, structure, and physics of nanoscale molecular machines. Biophys. Rev. 7, 269-299. 10.1007/s12551-014-0150-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles, G. L., Baglia, L. A. and Ackerman, K. G. (2015). KIF7 controls the proliferation of cells of the respiratory airway through distinct microtubule dependent mechanisms. PLoS Genet. 11, e1005525. 10.1371/journal.pgen.1005525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy, D. L., Hancock, W. O., Wagenbach, M. and Howard, J. (1999). Kinesin's tail domain is an inhibitory regulator of the motor domain. Nat. Cell Biol. 1, 288-292. 10.1038/13001 [DOI] [PubMed] [Google Scholar]
- Dafinger, C., Liebau, M. C., Elsayed, S. M., Hellenbroich, Y., Boltshauser, E., Korenke, G. C., Fabretti, F., Janecke, A. R., Ebermann, I., Nürnberg, G.et al. (2011). Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J. Clin. Invest. 121, 2662-2667. 10.1172/JCI43639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, S., Pettersson, M., Eisfeldt, J., Schröder, A. K., Wickström, R., Teär Fahnehjelm, K., Anderlid, B.-M. and Lindstrand, A. (2020). Whole genome sequencing unveils genetic heterogeneity in optic nerve hypoplasia. PLoS ONE 15, e0228622. 10.1371/journal.pone.0228622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorenzi, M. and Speed, T. (2002). An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617-625. 10.1093/bioinformatics/18.4.617 [DOI] [PubMed] [Google Scholar]
- Di Tommaso, P., Moretti, S., Xenarios, I., Orobitg, M., Montanyola, A., Chang, J.-M., Taly, J.-F. and Notredame, C. (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13-W17. 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emechebe, U., Kumar, P. P., Rozenberg, J. M., Moore, B., Firment, A., Mirshahi, T. and Moon, A. M. (2016). T-box3 is a ciliary protein and regulates stability of the Gli3 transcription factor to control digit number. eLife 5, e07897. 10.7554/eLife.07897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh-Yamagami, S., Evangelista, M., Wilson, D., Wen, X., Theunissen, J.-W., Phamluong, K., Davis, M., Scales, S. J., Solloway, M. J., De Sauvage, F. J.et al. (2009). The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol. 19, 1320-1326. 10.1016/j.cub.2009.06.046 [DOI] [PubMed] [Google Scholar]
- Farkhondeh, A., Niwa, S., Takei, Y. and Hirokawa, N. (2015). Characterizing KIF16B in neurons reveals a novel intramolecular “stalk inhibition” mechanism that regulates its capacity to potentiate the selective somatodendritic localization of early endosomes. J. Neurosci. 35, 5067-5086. 10.1523/JNEUROSCI.4240-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, D. S. and Vale, R. D. (1999). Single-molecule analysis of kinesin motility reveals regulation by the cargo-binding tail domain. Nat. Cell Biol. 1, 293-297. 10.1038/13008 [DOI] [PubMed] [Google Scholar]
- Friel, C. T. and Welburn, J. P. (2018). Parts list for a microtubule depolymerising kinesin. Biochem. Soc. Trans. 46, 1665-1672. 10.1042/BST20180350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiretti, A. E., Thies, E., Tokito, M. K., Lin, T., Ostap, E. M., Kneussel, M. and Holzbaur, E. L. F. (2016). Activity-dependent regulation of distinct transport and cytoskeletal remodeling functions of the dendritic kinesin KIF21B. Neuron 92, 857-872. 10.1016/j.neuron.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, M., Söding, J. and Lupas, A. N. (2006). Comparative analysis of coiled-coil prediction methods. J. Struct. Biol. 155, 140-145. 10.1016/j.jsb.2006.03.009 [DOI] [PubMed] [Google Scholar]
- Gruss, O. J., Wittmann, M., Yokoyama, H., Pepperkok, R., Kufer, T., Silljé, H., Karsenti, E., Mattaj, I. W. and Vernos, I. (2002). Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4, 871-879. 10.1038/ncb870 [DOI] [PubMed] [Google Scholar]
- Hackney, D. D. and Stock, M. F. (2000). Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat. Cell Biol. 2, 257-260. 10.1038/35010525 [DOI] [PubMed] [Google Scholar]
- Hammond, J. W., Cai, D., Blasius, T. L., Li, Z., Jiang, Y., Jih, G. T., Meyhofer, E. and Verhey, K. J. (2009). Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 7, e72. 10.1371/journal.pbio.1000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, J. W., Blasius, T. L., Soppina, V., Cai, D. and Verhey, K. J. (2010). Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J. Cell Biol. 189, 1013-1025. 10.1083/jcb.201001057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, M., Subramanian, R., Bangs, F., Omelchenko, T., Liem, K. F., Jr, Kapoor, T. M. and Anderson, K. V. (2014). The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 16, 663-672. 10.1038/ncb2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa, N., Noda, Y., Tanaka, Y. and Niwa, S. (2009). Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682-696. 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- Hu, Y., Wu, M. Z., Gu, N. J., Xu, H. T., Li, Q. C. and Wu, G. P. (2020). Human papillomavirus 16 (HPV 16) E6 but not E7 inhibits the antitumor activity of LKB1 in lung cancer cells by downregulating the expression of KIF7. Thorac. Cancer 11, 3175-3180. 10.1111/1759-7714.13640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.-F. and Banker, G. (2012). The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic 13, 549-564. 10.1111/j.1600-0854.2011.01325.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, B. and Allingham, J. S. (2020). These motors were made for walking. Protein Sci. 29, 1707-1723. 10.1002/pro.3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo, L., Yue, Y., Ren, J., Yu, J., Liu, J., Yu, Y., Ye, F., Xu, T., Zhang, M. and Feng, W. (2012). The CC1-FHA tandem as a central hub for controlling the dimerization and activation of kinesin-3 KIF1A. Structure 20, 1550-1561. 10.1016/j.str.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Ibisler, A., Hehr, U., Barth, A., Koch, M., Epplen, J. T. and Hoffjan, S. (2015). Novel KIF7 mutation in a Tunisian boy with acrocallosal syndrome: case report and review of the literature. Mol. Syndromol. 6, 173-180. 10.1159/000439414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi, M., Endres, N. F., Gennerich, A. and Vale, R. D. (2006). Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J. Cell Biol. 174, 931-937. 10.1083/jcb.200605179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, S., Mani, N., Wilson-Kubalek, E. M., Ku, P.-I., Milligan, R. A. and Subramanian, R. (2019). Interplay between the kinesin and tubulin mechanochemical cycles underlies microtubule tip tracking by the non-motile ciliary kinesin Kif7. Dev. Cell 49, 711-730.e8. 10.1016/j.devcel.2019.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaer, K., Yuksel, Z., Ichkou, A., Calisir, C. and Attié-Bitach, T. (2015). A novel KIF7 mutation in two affected siblings with acrocallosal syndrome. Clin. Dysmorphol. 24, 61-64. 10.1097/MCD.0000000000000080 [DOI] [PubMed] [Google Scholar]
- Kelliher, M. T., Yue, Y., Ng, A., Kamiyama, D., Huang, B., Verhey, K. J. and Wildonger, J. (2018). Autoinhibition of kinesin-1 is essential to the dendrite-specific localization of Golgi outposts. J. Cell Biol. 217, 2531-2547. 10.1083/jcb.201708096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, C.-I., Barbarulo, A., Solanki, A., Saldaña, J. I. and Crompton, T. (2017). The kinesin motor protein Kif7 is required for T-cell development and normal MHC expression on thymic epithelial cells (TEC) in the thymus. Oncotarget 8, 24163-24176. 10.18632/oncotarget.15241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.-R., Shin, H., Choi, J., Ko, J., Kim, S., Lee, H. W., Kim, K., Rho, S.-H., Lee, J. H., Song, H.-E.et al. (2004). An intramolecular interaction between the FHA domain and a coiled coil negatively regulates the kinesin motor KIF1A. EMBO J. 23, 1506-1515. 10.1038/sj.emboj.7600164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S. A., Ivanov, I. E., Lee, G.-H. and Cowan, N. J. (1989). Organization of microtubules in dendrites and axons is determined by a short hydrophobic zipper in microtubule-associated proteins MAP2 and tau. Nature 342, 498-505. 10.1038/342498a0 [DOI] [PubMed] [Google Scholar]
- Li, Z. J., Nieuwenhuis, E., Nien, W., Zhang, X., Zhang, J., Puviindran, V., Wainwright, B. J., Kim, P. C. W. and Hui, C. C. (2012). Kif7 regulates Gli2 through Sufu-dependent and -independent functions during skin development and tumorigenesis. Development 139, 4152-4161. 10.1242/dev.081190 [DOI] [PubMed] [Google Scholar]
- Liang, S., Shi, X., Yu, C., Shao, X., Zhou, H., Li, X., Chang, C., Lai, K. S., Ma, J. and Zhang, R. (2020). Identification of novel candidate genes in heterotaxy syndrome patients with congenital heart diseases by whole exome sequencing. Biochim. Biophys. Acta Mol. Basis Dis. 1866, 165906. 10.1016/j.bbadis.2020.165906 [DOI] [PubMed] [Google Scholar]
- Liem, K. F., Jr, He, M., Ocbina, P. J. R. and Anderson, K. V. (2009). Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA 106, 13377-13382. 10.1073/pnas.0906944106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. C., Couzens, A. L., Deshwar, A. R., Ld, B. M.-C., Zhang, X., Puviindran, V., Scott, I. C., Gingras, A.-C., Hui, C.-C. and Angers, S. (2014). The PPFIA1-PP2A protein complex promotes trafficking of Kif7 to the ciliary tip and Hedgehog signaling. Sci. Signal. 7, ra117. 10.1126/scisignal.2005608 [DOI] [PubMed] [Google Scholar]
- Lupas, A. (1996). Prediction and analysis of coiled-coil structures. Methods Enzymol. 266, 513-525. 10.1016/S0076-6879(96)66032-7 [DOI] [PubMed] [Google Scholar]
- Mann, B. J. and Wadsworth, P. (2019). Kinesin-5 regulation and function in mitosis. Trends Cell Biol. 29, 66-79. 10.1016/j.tcb.2018.08.004 [DOI] [PubMed] [Google Scholar]
- Masson, D. and Kreis, T. E. (1993). Identification and molecular characterization of E-MAP-115, a novel microtubule-associated protein predominantly expressed in epithelial cells. J. Cell Biol. 123, 357-371. 10.1083/jcb.123.2.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya, A. K., Ben, J., Zhao, Z., Lee, R. T. H., Niah, W., Ng, A. S. M., Iyu, A., Yu, W., Elworthy, S., Van Eeden, F. J. M.et al. (2013). Positive and negative regulation of Gli activity by Kif7 in the zebrafish embryo. PLoS Genet. 9, e1003955. 10.1371/journal.pgen.1003955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirvis, M., Stearns, T. and James Nelson, W. (2018). Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem. J. 475, 2329-2353. 10.1042/BCJ20170453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moua, P., Fullerton, D., Serbus, L. R., Warrior, R. and Saxton, W. M. (2011). Kinesin-1 tail autoregulation and microtubule-binding regions function in saltatory transport but not ooplasmic streaming. Development 138, 1087-1092. 10.1242/dev.048645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niceta, M., Dentici, M. L., Ciolfi, A., Marini, R., Barresi, S., Lepri, F. R., Novelli, A., Bertini, E., Cappa, M., Digilio, M. C.et al. (2020). Co-occurrence of mutations in KIF7 and KIAA0556 in Joubert syndrome with ocular coloboma, pituitary malformation and growth hormone deficiency: a case report and literature review. BMC Pediatr. 20, 120. 10.1186/s12887-020-2019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, S., Lipton, D. M., Morikawa, M., Zhao, C., Hirokawa, N., Lu, H. and Shen, K. (2016). Autoinhibition of a neuronal kinesin UNC-104/KIF1A regulates the size and density of synapses. Cell Rep. 16, 2129-2141. 10.1016/j.celrep.2016.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putoux, A., Thomas, S., Coene, K. L. M., Davis, E. E., Alanay, Y., Ogur, G., Uz, E., Buzas, D., Gomes, C., Patrier, S.et al. (2011). KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat. Genet. 43, 601-606. 10.1038/ng.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putoux, A., Nampoothiri, S., Laurent, N., Cormier-Daire, V., Beales, P. L., Schinzel, A., Bartholdi, D., Alby, C., Thomas, S., Elkhartoufi, N.et al. (2012). Novel KIF7 mutations extend the phenotypic spectrum of acrocallosal syndrome. J. Med. Genet. 49, 713-720. 10.1136/jmedgenet-2012-101016 [DOI] [PubMed] [Google Scholar]
- Putoux, A., Alqahtani, A., Pinson, L., Paulussen, A. D. C., Michel, J., Besson, A., Mazoyer, S., Borg, I., Nampoothiri, S., Vasiljevic, A.et al. (2016). Refining the phenotypical and mutational spectrum of Taybi-Linder syndrome. Clin. Genet. 90, 550-555. 10.1111/cge.12781 [DOI] [PubMed] [Google Scholar]
- Putoux, A., Baas, D., Paschaki, M., Morlé, L., Maire, C., Attié-Bitach, T., Thomas, S. and Durand, B. (2019). Altered GLI3 and FGF8 signaling underlies acrocallosal syndrome phenotypes in Kif7 depleted mice. Hum. Mol. Genet. 28, 877-887. 10.1093/hmg/ddy392 [DOI] [PubMed] [Google Scholar]
- Reid, T. A., Schuster, B. M., Mann, B. J., Balchand, S. K., Plooster, M., Mcclellan, M., Coombes, C. E., Wadsworth, P. and Gardner, M. K. (2016). Suppression of microtubule assembly kinetics by the mitotic protein TPX2. J. Cell Sci. 129, 1319-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, J., Wang, S., Chen, H., Wang, W., Huo, L. and Feng, W. (2018). Coiled-coil 1-mediated fastening of the neck and motor domains for kinesin-3 autoinhibition. Proc. Natl. Acad. Sci. USA 115, E11933-E11942. 10.1073/pnas.1811209115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu, J., Cade, N. I. and Surrey, T. (2015). Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol. 17, 1422-1434. 10.1038/ncb3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez, I. and Dynlacht, B. D. (2016). Cilium assembly and disassembly. Nat. Cell Biol. 18, 711-717. 10.1038/ncb3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, N., Zwetsloot, A. J., Bachmann, A., Roth, D., Hussain, H., Brandt, J., Kaverina, I. and Straube, A. (2019). PTPN21 and Hook3 relieve KIF1C autoinhibition and activate intracellular transport. Nat. Commun. 10, 2693. 10.1038/s41467-019-10644-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. K., Pandey, H., Al-Bassam, J. and Gheber, L. (2018). Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles. Cell. Mol. Life Sci. 75, 1757-1771. 10.1007/s00018-018-2754-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S., Soundara Rajan, D., Gaesser, J., Wen-Ya Lo, C. and Panigrahy, A. (2019). Olfactory bulb and olfactory tract abnormalities in acrocallosal syndrome and Greig cephalopolysyndactyly syndrome. Pediatr. Radiol. 49, 1368-1373. 10.1007/s00247-019-04480-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhune, E. A., Cuevas, M. T., Monley, A. M., Wethey, C. I., Chen, X., Cattel, M. V., Bayrak, M. N., Bland, M. R., Sutphin, B., Devon Trahan, G.et al. (2020). Mutations in KIF7 implicated in idiopathic scoliosis in humans and axial curvatures in zebrafish. Hum. Mutat. 42, 392-407. 10.1002/humu.24162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunovic, S., Barañano, K. W., Barkovich, J. A., Strober, J. B., Jamal, L. and Slavotinek, A. M. (2015). Novel KIF7 missense substitutions in two patients presenting with multiple malformations and features of acrocallosal syndrome. Am. J. Med. Genet. A 167A, 2767-2776. 10.1002/ajmg.a.37249 [DOI] [PubMed] [Google Scholar]
- Van Der Vaart, B., Van Riel, W. E., Doodhi, H., Kevenaar, J. T., Katrukha, E. A., Gumy, L., Bouchet, B. P., Grigoriev, I., Spangler, S. A., Yu, K. L.et al. (2013). CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell 27, 145-160. 10.1016/j.devcel.2013.09.010 [DOI] [PubMed] [Google Scholar]
- Van Riel, W. E., Rai, A., Bianchi, S., Katrukha, E. A., Liu, Q., Heck, A. J. R., Hoogenraad, C. C., Steinmetz, M. O., Kapitein, L. C. and Akhmanova, A. (2017). Kinesin-4 KIF21B is a potent microtubule pausing factor. eLife 6, e24746. 10.7554/eLife.24746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey, K. J. and Hammond, J. W. (2009). Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10, 765-777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Verhey, K. J., Lizotte, D. L., Abramson, T., Barenboim, L., Schnapp, B. J. and Rapoport, T. A. (1998). Light chain-dependent regulation of Kinesin's interaction with microtubules. J. Cell Biol. 143, 1053-1066. 10.1083/jcb.143.4.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, D. M., Shalev, S. A., Simpson, M. A., Morgan, N. V., Gelman-Kohan, Z., Chemke, J., Trembath, R. C. and Maher, E. R. (2013). Acrocallosal syndrome: identification of a novel KIF7 mutation and evidence for oligogenic inheritance. Eur. J. Med. Genet. 56, 39-42. 10.1016/j.ejmg.2012.10.004 [DOI] [PubMed] [Google Scholar]
- Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M. and Barton, G. J. (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189-1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek, M., Bechstedt, S., Chaaban, S. and Brouhard, G. J. (2015). Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell Biol. 17, 907-916. 10.1038/ncb3188 [DOI] [PubMed] [Google Scholar]
- Williams, L. S., Ganguly, S., Loiseau, P., Ng, B. F. and Palacios, I. M. (2014). The auto-inhibitory domain and ATP-independent microtubule-binding region of Kinesin heavy chain are major functional domains for transport in the Drosophila germline. Development 141, 176-186. 10.1242/dev.097592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, C. W., Nguyen, C. T., Chen, M.-H., Yang, J.-H., Gacayan, R., Huang, J., Chen, J.-N. and Chuang, P.-T. (2009). Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature 459, 98-102. 10.1038/nature07883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, K. Y., Liu, J. and Chan, K. W. (2017). KIF7 attenuates prostate tumor growth through LKB1-mediated AKT inhibition. Oncotarget 8, 54558-54571. 10.18632/oncotarget.17421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K. H., Hanada, T. and Chishti, A. H. (2007). The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry 46, 10039-10045. 10.1021/bi701169w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Y., Blasius, T. L., Zhang, S., Jariwala, S., Walker, B., Grant, B. J., Cochran, J. C. and Verhey, K. J. (2018). Altered chemomechanical coupling causes impaired motility of the kinesin-4 motors KIF27 and KIF7. J. Cell Biol. 217, 1319-1334. 10.1083/jcb.201708179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.