Abstract
Context
Broad genomic analyses among thyroid histologies have been described from relatively small cohorts.
Objective
Investigate the molecular findings across a large, real-world cohort of thyroid fine-needle aspiration (FNA) samples.
Design
Retrospective analysis of RNA sequencing data files.
Setting
Clinical Laboratory Improvement Amendments laboratory performing Afirma Genomic Sequencing Classifier (GSC) and Xpression Atlas (XA) testing.
Participants
A total of 50 644 consecutive Bethesda III-VI nodules.
Intervention
None.
Main Outcome Measures
Molecular test results.
Results
Of 48 952 Bethesda III/IV FNAs studied, 66% were benign by Afirma GSC. The prevalence of BRAF V600E was 2% among all Bethesda III/IV FNAs and 76% among Bethesda VI FNAs. Fusions involving NTRK, RET, BRAF, and ALK were most prevalent in Bethesda V (10%), and 130 different gene partners were identified. Among small consecutive Bethesda III/IV sample cohorts with one of these fusions and available surgical pathology excision data, the positive predictive value of an NTRK or RET fusion for carcinoma or noninvasive follicular thyroid neoplasm with papillary-like nuclear features was >95%, whereas for BRAF and ALK fusions it was 81% and 67%, respectively. At least 1 genomic alteration was identified by the expanded Afirma XA panel in 70% of medullary thyroid carcinoma classifier–positive FNAs, 44% of Bethesda III or IV Afirma GSC suspicious FNAs, 64% of Bethesda V FNAs, and 87% of Bethesda VI FNAs.
Conclusions
This large study demonstrates that almost one-half of Bethesda III/IV Afirma GSC suspicious and most Bethesda V/VI nodules had at least 1 genomic variant or fusion identified, which may optimize personalized treatment decisions.
Keywords: molecular diagnostics, indeterminate cytology, thyroid nodule, thyroid cancer, personalized healthcare, variant detection
Within the past decade, it was common practice for most cytologically indeterminate thyroid nodules to undergo surgery given their 20% to 30% risk of malignancy (1). Since then, guidelines have increasingly included molecular testing as a diagnostic adjunct toward management of cytologically indeterminate thyroid nodules, ruling out cancer with a high negative predictive value test and reducing the need for diagnostic hemithyroidectomy and identifying genomic alterations that are highly predictive of malignancy (2-4). Additionally, novel therapies have emerged that target specific genomic alterations to treat refractory thyroid cancer and offer the potential for increased efficacy with more tolerable side effects.
The Afirma Genomic Sequencing Classifier (GSC) is used to rule out malignancy and reclassify cytologically indeterminate (Bethesda III or IV) nodules to molecularly benign or suspicious (5). The original Afirma Xpression Atlas (XA) panel reported on 761 genomic variants and 130 fusion pairs from 511 genes (6). In March 2020, the panel was expanded to include 905 genomic variants and 235 fusion pairs from 593 genes (7). The Afirma XA intended-use population includes cytologically indeterminate nodules with an Afirma GSC suspicious result, those that are cytologically suspicious for malignancy or malignant (Bethesda V or VI, respectively), and extrathyroidal masses that are known or suspected to be thyroid cancer metastases (such as suspicious lymph nodes) (6, 7).
To date, broad genomic analyses among malignant and benign thyroid histologies have typically been reported from cohorts of up to several hundred samples (8-13). Here, we report Afirma GSC and XA data from 50 644 Bethesda III-VI real-world clinical samples consecutively resulted in the Veracyte Clinical Laboratory Improvement Amendments (CLIA) laboratory. These novel data shed light on the landscape and potential clinical utility available from the preoperative genomic analysis of Bethesda III-VI thyroid nodules.
Materials and Methods
The Afirma GSC and XA results of 50 644 Bethesda III-VI nodules consecutively determined in the Veracyte CLIA laboratory from August 2017 through September 2019 were analyzed. The median patient age was 58 years and 78% were female, with a median nodule size of 2.1 cm. These fine-needle aspiration (FNA) samples were clinically categorized according to the Bethesda System for Reporting Thyroid Cytopathology by the cytopathologists of record (either local [67%] or Thyroid Cytopathology Partners [33%], Austin, TX). Samples were submitted by 5786 physicians from 2141 practice sites. Molecular testing was performed using RNA sequencing as previously described (5, 6, 14-16). Testing that was not performed clinically was done via analysis of the original RNA sequencing data file. No FNA samples were physically reprocessed. Blinded to clinical outcomes status, these samples were reanalyzed using the expanded Afirma XA pipeline. Although outside of the Afirma GSC intended use, deidentified Bethesda V and VI samples were investigated with Afirma GSC for research purposes via analysis of the original RNA sequencing data file. Conversely, the outside-of-intended-use Afirma GSC benign samples were not analyzed via the Afirma XA pipeline (Afirma XA results among a smaller cohort of Afirma GSC benign samples was previously reported (6)). Samples without Bethesda classification were excluded. Gene pairs are listed alphabetically.
For simplicity, we report percentages in the text to the nearest whole digit for values ≥1%, and to 2 decimal places for values <1%. Statistical analyses of paired nominal data with a dichotomous trait were performed by McNemar’s χ 2 test via R version 3.2.3.
Investigations were conducted with an institutional review board (IRB) exemption to use delinked and anonymized clinical specimens (Copernicus Group IRB, Cary, NC). Copernicus Group IRB approval (study number: 1255781) was used for investigations that linked the laboratory specimen to the treatment outcome, including the surgical histopathologic diagnosis.
Results
Prevalence of GSC benign results and parathyroid or medullary thyroid carcinoma classifiers
A total of 66% of 48 952 cytologically indeterminate (Bethesda III or IV) thyroid nodules were classified as GSC benign (Table 1). The prevalence of BRAF V600E Classifier positive samples was 2%, 3%, 41%, and 76% for Bethesda categories III-VI, respectively (Table 1). One hundred and twenty medullary thyroid carcinoma (MTC) classifier positives were identified among all indeterminate nodules (0.25%) and comprised 0.72% of GSC suspicious indeterminate nodules. They were more common in Bethesda IV (0.57%) than among Bethesda III (0.17%). The parathyroid classifier was positive in 278 (0.57%) indeterminate nodules. See Table 1 for data for each Bethesda category.
Table 1.
Prevalence of Afirma GSC classifier results among consecutive resulted Bethesda III-VI samples from the Veracyte CLIA laboratory
| Afirma GSC | Cytopathology Result | Sum | |||
|---|---|---|---|---|---|
| AUS/FLUS (III) | FN/SFN (IV) | SFM (V) | Malignant (VI) | ||
| GSC benign | 26900 | 5180 | 125 | 32 | 32 237 |
| BRAF V600E | 818 | 316 | 340 | 652 | 2126 |
| MTC | 66 | 54 | 23 | 9 | 152 |
| CCDC6/RET or NCOA4/RET | 64 | 30 | 34 | 21 | 149 |
| Other suspiciousa | 11 408 | 3838 | 314 | 140 | 15 700 |
| Parathyroid | 208 | 70 | 1 | 1 | 280 |
| Sum | 39 464 | 9488 | 837 | 855 | 50 644 |
| % GSC benign | 68.16 | 54.60 | 14.93 | 3.74 | |
| % BRAF V600E | 2.07 | 3.33 | 40.62 | 76.26 | |
| % MTC | 0.17 | 0.57 | 2.75 | 1.05 | |
| % Parathyroid | 0.53 | 0.74 | 0.12 | 0.12 | |
Lines under “Afirma GSC” indicate results positive for that specific result.
Abbreviations: AUS/FLUS (Bethesda III), atypical of unknown significance/follicular lesion of unknown significance; Bethesda VI, malignant; Bethesda V, suspicious for malignancy; FN/SFN (Bethesda IV), follicular neoplasm/suspicious for follicular neoplasm; GSC, Genomic Sequencing Classifier; MTC, medullary thyroid carcinoma.
a Indicates GSC suspicious without being positive for BRAF V600E, MTC classifier, CCDC6/RET (RET/PTC1) or NCOA4/RET (RET/PTC3) fusions. Afirma GSC is not routinely performed in Bethesda V/VI, but results are shown here for completeness. The Afirma parathyroid, BRAF, and MTC classifiers are routinely performed with the Afirma Xpression Atlas in Bethesda V/VI samples.
Overall prevalence of variants and fusions per Bethesda category
Among the Afirma XA Intended Use population (Afirma GSC suspicious Bethesda III/IV nodules [n = 16 594] and Bethesda V/VI nodules [n = 1692]), 466 unique alteration profiles were identified at the level of the protein sequence or fusion partner, including no alterations, single alterations, and combinations of multiple alterations. In this cohort of 18 286 Afirma XA samples, 46% had precisely 1 alteration identified (Table 2). More than 1 alteration was seen in 1% of samples, including 5 samples with 3 variants, 6 samples with 2 fusions, and 1 sample with 2 variants plus a fusion.
Table 2.
Frequency of genomic variants and/or gene fusions among FNA samples from Afirma GSC suspicious Bethesda III/IV nodules and Bethesda V/VI nodules
| No. of alterations identified | Expanded XA panel fusions | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | |||
| BRAF V600E Classifier + expanded XA panel variants | 0 | 9681 | 1154 | 6 | |
| 1 | 7190 | 30 | 0 | ||
| 2 | 219 | 1 | 0 | ||
| 3 | 5 | 0 | 0 | ||
| XA intended use cohort | 18286 | ||||
Among Afirma Xpression Atlas (XA) intended use cohort (n = 18 286) (Afirma GSC suspicious Bethesda III/IV nodules [n = 16 594] and Bethesda V/VI nodules [n = 1692]), 45.6% of samples (7190 + 1154) had precisely 1 alteration (variant or fusion) identified. More than 1 alteration was seen in 1.4% of the target population (219 + 5 + 30 + 1 + 6). Six samples had 2 fusions identified.
Abbreviations: FNA, fine-needle aspiration; GSC, Genomic Sequencing Classifier.
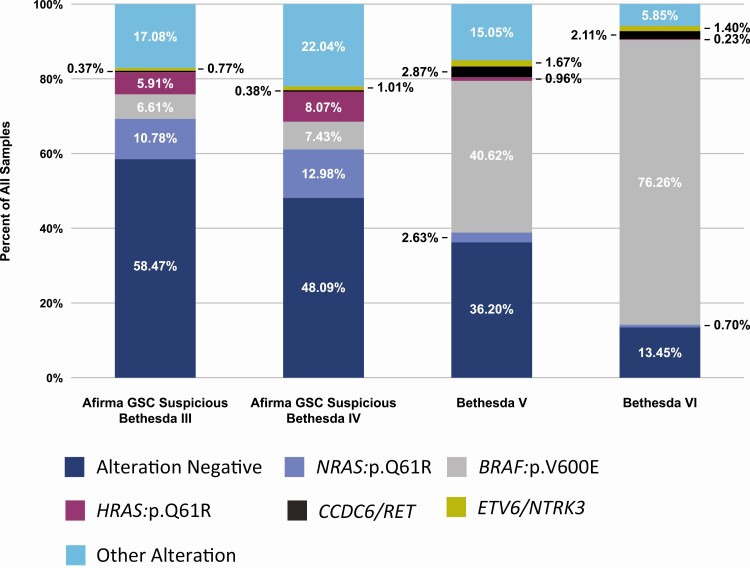
Among all 16 594 GSC suspicious, cytologically indeterminate thyroid nodules (shown in Table 1), 42% had at least 1 alteration identified on the original Afirma XA panel. This percentage increased to 44% on the expanded Afirma XA panel with 7331 samples having at least 1 alteration. In total, 7566 alterations were found among these 7331 samples. The most common alterations among these 16 594 samples were NRAS p.Q61R (11%, n = 1882), BRAF p.V600E (7%, n = 1132), and HRAS p.Q61R (6%, n = 1072), accounting for 54% (4086/7566) of all identified alterations (Fig. 1 and Supplemental Table 1) (17).
Figure 1.
Percent of alterations among Afirma Xpression Atlas Intended Use samples: Afirma GSC Suspicious Bethesda III (n = 12,356), Afirma GSC Suspicious Bethesda IV (n = 4,238), Bethesda V (n = 837), and Bethesda VI (n = 855). The three most frequent alterations in each Bethesda category were identified. The frequency of these five alterations among each Bethesda category is shown. All other alterations are shown as “Other Alteration”.  = Alteration Negative;
= Alteration Negative;  = NRAS:p.Q61R;
= NRAS:p.Q61R;  = BRAF:p.V600E;
= BRAF:p.V600E;  = HRAS:p.Q61R;
= HRAS:p.Q61R;  = CCDC6/RET;
= CCDC6/RET;  = ETV6/NTRK3;
= ETV6/NTRK3;  = Other Alteration.
= Other Alteration.
Among 837 Bethesda V nodules, 534 (64%) contained at least 1 variant or fusion. In total, 543 alterations were identified. The most common alterations were BRAF p.V600E (41%, n = 340), CCDC6/RET (3%, n = 24), and NRAS p.Q61R (3%, n = 22), accounting for 71% (386/543) of all alterations (Fig. 1 and Supplemental Table 1) (17).
Among 855 Bethesda VI nodules, 740 (87%) contained at least 1 variant or fusion. In total, 763 alterations were identified. The most common alterations were BRAF p.V600E (76%, n = 652), CCDC6/RET (2%, n = 18), and ETV6/NTRK3 (1%, n = 12), accounting for 89% (682/763) of all identified alterations (Fig. 1 and Supplemental Table 1) (17).
Prevalence of ALK, BRAF, NTRK, and RET fusions per Bethesda category
ALK, BRAF, NTRK1/3, and RET fusions were not identified among the 32 358 Bethesda III/IV Afirma GSC benign or parathyroid classifier–positive samples. Among 16 594 Bethesda III/IV GSC suspicious FNAs, 2% (n = 367) were positive for ALK, BRAF, RET, or NTRK1/3 fusions from the original Afirma XA version. When the panel was expanded to include 104 additional ALK, BRAF, NTRK1/3, and RET fusion combinations, the percentage of positive nodules increased to 3% (n = 529), P < 0.001. Among the 1692 Bethesda V/VI FNAs, the number of nodules positive for ALK, BRAF, NTRK1/3, and RET fusions increased from 6% (n = 100) to 8% (n = 135), P < 0.001. Among these combined cohorts of Bethesda III/IV Afirma GSC suspicious and Bethesda V/VI, the most common gene fusions included NTRK3 (1%), RET (1%), BRAF (0.89%), ALK (0.30%), and NTRK1 (0.15%). In total, 132 unique ALK, BRAF, NTRK1/3, or RET gene partner combinations were identified.
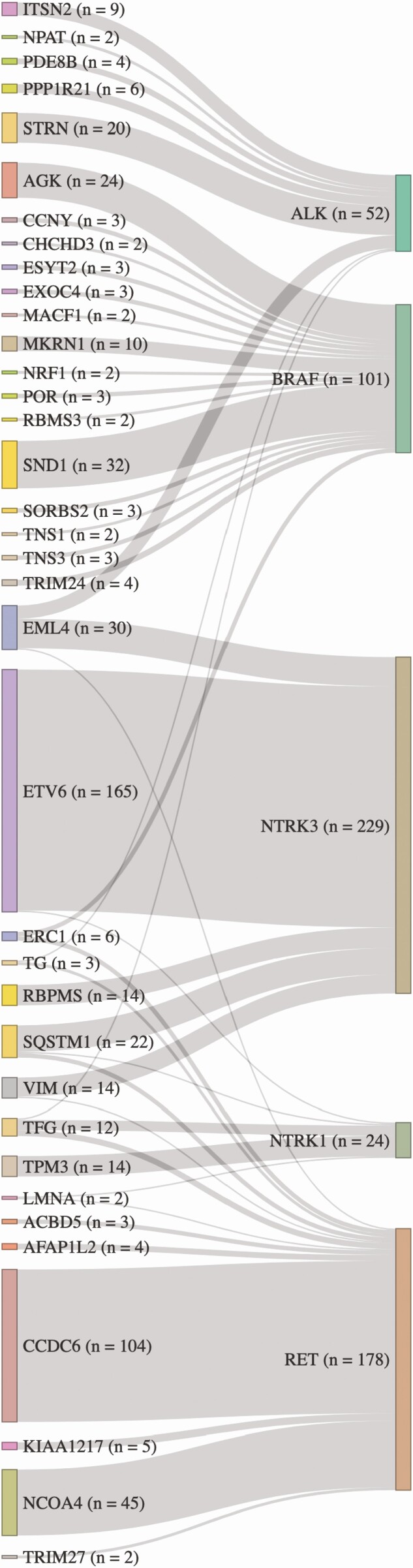
The most common fusion observed for each of the 5 studied receptor tyrosine kinase genes on the expanded panel was: ETV6/NTRK3 (72% of NTRK3 fusions; 64% of all NTRK fusions), CCDC6/RET (RET/PTC1, 55% of RET), BRAF/SND1 (20% of BRAF), and ALK/STRN (37% of ALK), and NTRK1/TPM3 (50% of NTRK1) (Fig. 2). BRAF showed the highest diversity of fusions, with 80 gene partners. Different gene partners with RET, ALK, NTRK1, and NTRK3 numbered 25, 11, 9, and 5, respectively.
Figure 2.
Sankey diagram of ALK, BRAF, NTRK, and RET fusions when the partner gene was identified in at least two samples. On the left are partner genes, and on the right are the key kinase genes. The thicknesses of the vertical bars denote the number of times each partner was observed. The thicknesses of the connections reveal the number of times each partner is fused to the key kinase gene. Several partners fused with more than one key kinase gene (EMLA4, ERC1, TG, SQSTM1, VIM, TFG, and LMNA).
Among all 50 644 nodule samples, a rise in ALK, BRAF, NTRK, and RET fusion prevalence was seen across the Bethesda categories and peaked in Bethesda V (Table 3). ALK fusions were not observed in Bethesda VI specimens.
Table 3.
Number and prevalence of selected RTK gene fusions among 50 644 thyroid FNA samples by Bethesda category
| RTK gene | AUS/FLUS (III) | FN/SFN (IV) | SFM (V) | Malignant (VI) |
|---|---|---|---|---|
| ALK | 29 (0.07%) | 22 (0.23%) | 3 (0.36%) | 0 (0.00%) |
| BRAF | 88 (0.22%) | 47 (0.50%) | 15 (1.79%) | 13 (1.52%) |
| NTRK1 | 16 (0.04%) | 8 (0.08%) | 2 (0.24%) | 2 (0.23%) |
| NTRK3 | 126 (0.32%) | 67 (0.71%) | 21 (2.51%) | 15 (1.75%) |
| RET | 86 (0.22%) | 40 (0.42%) | 40 (4.78%) | 24 (2.81%) |
| Any | 345 (0.87%) | 184 (1.94%) | 81 (9.68%) | 54 (6.32%) |
Table 1 reports the total number of samples in each Bethesda category and the Bethesda category abbreviations.
Abbreviation: RTK, receptor tyrosine kinase.
Positive predictive value of ALK, BRAF, NTRK, and RET fusions in Bethesda III/IV thyroid fine-needle aspirates
Consecutive cohorts of Bethesda III/IV nodules with ALK, BRAF, NTRK, or RET fusions (other than RET/PTC1 and RET/PTC3) that were submitted to Veracyte for molecular analysis were identified. Local surgical pathology diagnoses were sought with IRB approval. Only 1 nodule per patient was included.
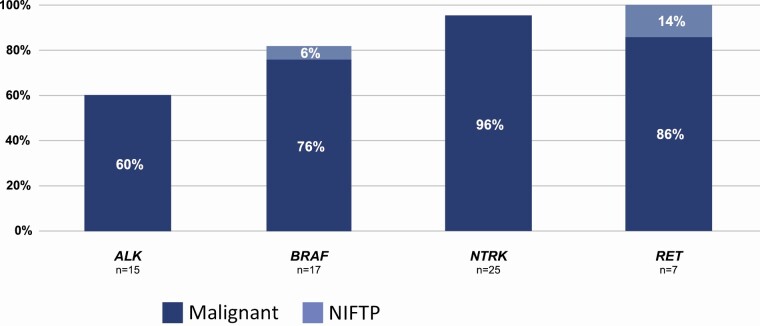
Excisional surgical pathologic diagnoses were available for 64 thyroid nodules from 64 patients. No sample had a concurrent variant. Fifteen ALK fusion partners included 10 STRN and 5 EML4. Nine (60%) were in malignant tumors (Fig. 3), whereas 6 (4 STRN and 2 EML4) were identified in adenomatoid or hyperplastic nodules (positive predictive value [PPV] = 60%). One of these adenomatoid nodules harbored both an STRN fusion partner and an additional MRPS16/TTC18 gene fusion (Table 2 shows the frequency of finding 2 fusions in an FNA sample). Seventeen BRAF fusion partners included 5 AGK, 6 SND1, 2 CCNY, 2 MKRN1, 1 POR, and 1 MACF1. Final pathologic diagnoses included 13 carcinomas (76%), 1 noninvasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP) (AGK), and 3 follicular adenomas (1 each AGK, SND1, and MKRN1) (PPV = 76%). Twenty-five NTRK fusion partners included 20 ETV6, 3 TPM3, and 2 RBPMS. Their excisional diagnoses included 24 carcinomas (96%) and 1 (ETV6) hyperplastic nodule (PPV = 96%). Seven RET fusion partners included 3 ERC1, 1 TRIM33, 1 AKAP13, 1 PRKAR1A, and 1 FKBP15. Excisional pathologic diagnoses included 6 carcinomas (86%) and 1 NIFTP (ERC1) (PPV = 100%). The specific pathological diagnoses are shown in Table 4.
Figure 3.
Percent of consecutive Bethesda III/IV nodules with ALK, BRAF, NTRK, or RET fusions (other than RET/PTC1 and RET/PTC3) demonstrating carcinoma or Non-Invasive Follicular Thyroid neoplasm with Papillary-like nuclear features (NIFTP) on local surgical histopathology (positive predictive value).  = Malignant;
= Malignant;  = NIFTP.
= NIFTP.
Table 4.
Local histopathological diagnosis of consecutive Bethesda III/IV nodules with ALK, BRAF, NTRK, or RET fusions (other than CCDC6/RET [RET/PTC1] or NCOA4/RET [RET/PTC3]) with available local surgical pathology diagnoses
| Fusion kinase domain | Local histology |
|---|---|
|
ALK
n = 15 |
• PTC 33% |
| • Hyperplasia 27% | |
| • Follicular variant PTC 13% | |
| • Adenomatoid nodule 13% | |
| • Follicular thyroid cancer 7% | |
| • Unspecified thyroid malignancy 7% | |
|
BRAF
n = 17 |
• PTC 53% |
| • Follicular variant PTC 23% | |
| • Follicular adenoma 18% | |
| • NIFTP 6% | |
|
NTRK
n = 25 |
• PTC 56% |
| • Follicular variant PTC 40% | |
| • Hyperplasia 4% | |
|
RET (not CCDC6/RET or NCOA4/RET) n = 7 |
• PTC 57% |
| • Follicular variant PTC 29% | |
| • NIFTP 14% |
Abbreviations: NIFTP, noninvasive follicular thyroid neoplasm with papillary-like features; PTC, papillary thyroid cancer.
Genomic landscape of FNAs positive for MTC
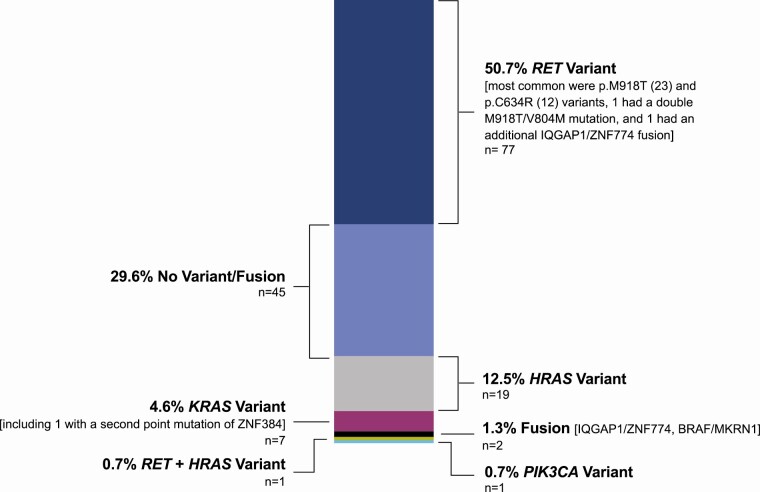
Examination of 50 644 Bethesda III-VI FNAs revealed 152 MTC classifier-positive cases (Table 1). Of them, 51% harbored a RET variant (including samples with other additional alterations), 5% contained a KRAS variant (including samples with other additional alterations), 13% included an HRAS variant, 0.66% had a BRAF fusion, 0.66% demonstrated other fusions, and 30% held no variant/fusion (Fig. 4 and Supplemental Table 2) (17). Among samples with more than 1 alteration was 1 sample with 2 RET variants (RET p.M918T, RET p.V804M) and 1 sample with a RET and HRAS variant (HRAS p.Q61L, RET p.C620Y).
Figure 4.
Variants and fusions identified in 152 MTC Classifier positive FNA samples using the expanded Afirma Xpression Atlas.
Discussion
Here, we report Afirma GSC and XA data from 50 644 Bethesda III-VI real-world thyroid nodules consecutively resulted over a 2-year time frame in the Veracyte CLIA laboratory. At least 1 alteration was identified by Afirma XA among 44% of Afirma GSC suspicious Bethesda III/IV FNAs, 64% of Bethesda V FNAs, and 87% of Bethesda VI FNAs. Although 466 unique combinations of Afirma XA results were found, the list of the 3 most common individual alterations in each Bethesda category (III-VI) identified only 5 unique alterations (BRAF:p.V600E, CCDC6/RET, ETV6/NRK3, HRAS:p.Q61R, and NRAS:pQ61R) (Fig. 1). Their PPVs among Bethesda III/IV nodules range from 38% to 100% (18). Though uncertainty exists regarding the PPVs of most individual variants and fusions among Bethesda III and IV nodules (18), alterations of BRAF:p.V600E, HRAS, and NRAS, and fusions of ALK, BRAF, NTRK1/3, and RET are known or suspected to convey a PPV of ≥50%. These potentially PPV-raising alterations were present among 31% and 39% of Afirma GSC Suspicious Bethesda III and IV FNAs, respectively.
Two-thirds of the 48 952 cytologically indeterminate nodules were reported as molecularly benign (Afirma GSC benign). GSC benign results were seen in 68% of Bethesda III samples and 55% of Bethesda IV samples (Table 1). In the proper clinical context, this rate of a benign molecular result, combined with its high negative predictive value (5), supports the opportunity to avoid diagnostic surgery among many thyroid nodules with cytologically indeterminate results (2-4). Low rates of surgical resection among GSC benign nodules have been reported in multiple independent experiences (19-26), including nodules with oncocytic (Hürthle cell) dominant cytology (27). Although the Afirma GSC test is not clinically performed on cytologically malignant nodules, the low rate of Afirma GSC benign results (3.7%, Table 1) seen here among deidentified Bethesda VI (cytologically malignant) samples supports the high sensitivity for cancer observed during clinical validation (5).
Suspected thyroid nodules that are enlarged parathyroid glands (within or adjacent to the thyroid) are a challenge for both ultrasonography and cytopathology (28). Nearly three-quarters of parathyroid classifier–positive samples were called Bethesda III, and almost all of the remainder was called Bethesda IV (Table 1). Overall, the Afirma parathyroid classifier was positive in 0.57% of cytologically indeterminate nodules. Parathyroid adenoma, parathyroid hyperplasia, and the rare parathyroid carcinoma should be considered in the differential diagnosis of a positive result. Diagnosis of MTC is also a cytological challenge (29, 30). The Afirma MTC classifier was positive in 0.25% of cytologically indeterminate nodules, and higher prevalence was seen among Bethesda categories V and VI (Table 1).
The Afirma BRAF V600E classifier result is reported for all samples tested for Afirma GSC or XA. The Afirma BRAF V600E Classifier was the dominant finding among Bethesda VI nodules (Table 1 and Fig. 1). It has been previously well documented that BRAF V600E variants are much less common among Bethesda III or IV nodules (31). Here again, it was uncommon among all Bethesda III and IV nodules (2%, Table 1). A positive Afirma BRAF V600E classifier result was present in only 7% of Afirma GSC suspicious Bethesda III/IV nodules. These data demonstrate that samples confidently diagnosed as papillary thyroid carcinoma (PTC) on cytopathologic examination are highly enriched for BRAF p.V600E-altered neoplasms. It seems that the “textbook” Bethesda VI FNA sample is a BRAF p.V600E-altered PTC. In contrast, the Bethesda V category showed the highest prevalence of ALK, BRAF, NTRK, and RET fusions (Table 3), suggesting that prevalence of genomic alterations across Bethesda categories may reflect both prevalence of malignancy and a relationship between genotype and cytological appearance. One may postulate that these gene fusions are associated with morphological changes that are less definitive for malignancy than are the morphological changes associated with the BRAF p.V600E variant. Alternatively, it may be that cytopathologists more strongly recognize the morphological changes of the BRAF p.V600E variant as the malignant standard. Regardless, substantial differences in the molecular profiles of Bethesda V and VI samples are present (Tables 1 and 3; Supplemental Table 1) (17), including higher rates of RAS variants among Bethesda V samples (Table 5).
Table 5.
Prevalence of RAS variants among 50 644 thyroid FNA samples by Bethesda category
| RAS gene | AUS/FLUS (III) | FN/SFN (IV) | SFM (V) | Malignant (VI) |
|---|---|---|---|---|
| HRAS variant | 1156 (2.93%) | 495 (5.22%) | 12 (1.43%) | 4 (0.47%) |
| KRAS variant | 314 (0.80%) | 129 (1.36%) | 12 (1.43%) | 6 (0.70%) |
| NRAS variant | 1712 (4.34%) | 681 (7.18%) | 29 (3.46%) | 7 (0.82%) |
| Any RAS variant | 3179 (8.06%) | 1304 (13.74%) | 53 (6.33%) | 17 (1.99%) |
Some samples contained more than 1 RAS variant. Table 1 reports the total number of samples in each Bethesda category and the Bethesda category abbreviations.
Abbreviation: FNA, fine-needle aspiration.
The predictive value of malignancy for ALK, BRAF, NTRK, and RET fusions (other than CCDC6/RET and NCOA4/RET) among Bethesda III/IV nodules is unknown (18). We studied their PPV according to local histopathologic diagnoses. NTRK and RET fusions (other than CCDC6/RET and NCOA4/RET) among Bethesda III/IV nodules were associated with malignancy in 28 of 30 nodules. Risk of malignancy was lower among nodules with ALK (67%) or BRAF (75%) fusions (Fig. 3). We found it notable that 2 nodules with BRAF or RET fusions, expected to be BRAFV600E-like (12, 32), were diagnosed as NIFTP. Additionally, 4 nodules were reported as hyperplastic despite harboring fusions expected to drive neoplasia (2 ALK/STRN, 1 ALK/EML4, 1 ETV6/NTRK3), although the pathologic diagnoses were not subsequently rereviewed by a blinded endocrine pathologist. These findings highlight the histological gray zone differentiating hyperplasia from clonal neoplasia based on light microscopy alone. With a modest sample size, our findings demonstrate histopathologic risk of malignancy associated with several receptor tyrosine kinase fusions among nodules with indeterminate cytopathology. Future studies with expert histopathologists may provide additional comparative insight, as will long-term clinical outcomes associating fusion partners with biological outcomes.
Receptor tyrosine kinase fusions are being targeted by small molecule inhibitors to treat late-stage cancers, including advanced thyroid cancer (33). Inhibitors targeting malignancies with NTRK fusions and RET alterations have received pan- or multicancer US Food and Drug Administration approval (34-37). Additionally, having these molecular data up front, even for Bethesda V/VI nodules, may provide insight for surgical management along with preplanned therapeutic options for cancers with the potential for recurrence or aggressive behavior (38, 39). Here, we investigated the prevalence of ALK, BRAF, NTRK, and RET fusions among 50 644 consecutive Bethesda III-VI nodule FNA samples submitted to the Veracyte CLIA laboratory. Positive cases were detected in all Bethesda categories and peaked near 10% in FNA samples cytologically suspicious for malignancy (Bethesda V, Table 3). Prior studies have shown that mutations in known driver genes are typically concordant between the primary tumor and distant metastatic deposits, suggesting that molecular knowledge from the primary tumor, or the most accessible metastatic deposit, may be concordant with less accessible metastatic deposits (40-42).
The genomic landscape of MTC has been studied by others using surgical samples (43, 44). Here, we report that an alteration was identified in 70% of MTC classifier–positive samples via a less invasive FNA sample (Fig. 4). The alterations we identified were primarily in RET, a target of several US Food and Drug Administration–approved and investigational therapies (33, 37, 45). Afirma XA does not differentiate germline from somatic alterations. Although family history, physical examination, and Afirma XA result may suggest a sporadic or familial origin of MTC, germline RET protooncogene testing (46) is recommended for all MTC cases regardless of the Afirma XA result.
Our study has several limitations, including that we did not correlate the surgical pathology outcome with most of the FNA samples described here. We did report surgical histology, performed locally, for small consecutive cohorts of Bethesda III/IV nodules with alterations of specific interest. There was no central blinded histopathologic review by experienced endocrine pathologists.
Another limitation is potential selection bias, especially among Bethesda V and VI samples. It is likely that a smaller fraction of Bethesda V and VI samples was submitted for molecular testing as they represented only 3% of the study cohort. This greater selectivity may increase selection bias. For example, we previously reported the incidence of the first generation Afirma (GEC) MTC classifier positivity among small cohorts of consecutive Bethesda V and VI samples and did not limit testing to those where molecular testing was clinically requested. We did that to minimize selection bias, and a progressive rise in test positivity across the Bethesda III-VI categories was seen (47). Here, however, we investigated only samples submitted to Veracyte for molecular testing and we saw a peak in MTC classifier positivity in the Bethesda V category (Table 1). It is possible that the difference between these observations is the result of selection bias.
Strengths of our study include whole exome RNA sequencing and variant/fusion reporting on a large cohort of thyroid nodule samples consecutively submitted in real-world practice by treating physicians. We report genomic classifiers, variants, and fusions across the entire cohort, thus avoiding selection bias when some of tests may not have been requested by the submitting physician.
Conclusions
The Afirma GSC and XA assays have been analytically and clinically validated. In the current characterization of the molecular landscape of real-world Bethesda III-VI nodules, we found that two-thirds of Bethesda III/IV nodules resulted Afirma GSC benign. A genomic alteration was identified by Afirma XA in almost one-half of those that resulted Afirma GSC suspicious, most commonly a variant of NRAS or HRAS. Alterations likely associated with an increased PPV were identified in about one-third of Afirma GSC suspicious samples. Conversely, Bethesda V/VI nodules were dominated by the high PPV BRAF V600E variant. Fusions involving ALK, BRAF, NTRK1, NTRK3, or RET were more common among Bethesda V/VI nodules. Almost 70% of MTC classifier–positive samples had an alteration identified by Afirma XA, most commonly variants of RET, KRAS, or HRAS. Molecular findings are increasingly being associated with tumor behaviors ranging from benignity to malignancy, neoplastic patterns of metastases, extrathyroidal spread, and responses to targeted oncological treatment. We speculate that knowledge of these molecular findings from preoperative thyroid nodule FNA specimens and thyroid cancer metastases may advance personalized treatment decisions.
Acknowledgments
Financial Support : The authors received no specific funding for this work.
Glossary
Abbreviations
- CLIA
Clinical Laboratory Improvement Amendments
- FNA
fine-needle aspiration
- GSC
Genomic Sequencing Classifier
- IRB
institutional review board
- MTC
medullary thyroid carcinoma
- NIFTP
noninvasive follicular thyroid neoplasm with papillary-like nuclear features
- PPV
positive predictive value
- PTC
papillary thyroid carcinoma
- XA
Xpression Atlas.
Additional Information
Disclosures : B.C.S., C.D., J.F.K., M.J.L., P.S., S.A., and S.P.W. report no competing financial interests. M.E.Z. reports personal fees from Eli Lilly and grant support from Eli Lilly, GeneproDx, and Merck. M.I.H. reports personal fees from Blueprint Medicine, Eli Lilly, Loxo Oncology, and Veracyte, and research support from Eli Lilly. L.J.W. reports personal fees from Bayer, Blueprint Medicine, Eisai, Eli Lilly, Genentech, Loxo Oncology, and Merck. P.W.L. reports personal fees from Veracyte. S.G.W. reports personal fees from Bayer. G.C.K., J.E.B., R.T.K., and Y.H. are Veracyte employees and equity owners. G.C.K. holds relevant patents.
Data Availability
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in References.
References
- 1. American Thyroid Association Guidelines Taskforce on Thyroid N, Differentiated Thyroid C, Cooper DS, Doherty GM, Haugen BR, et al. . Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2009;19(11):1167-1214. [DOI] [PubMed] [Google Scholar]
- 2. Patel KN, Yip L, Lubitz CC, et al. . The American Association of Endocrine Surgeons guidelines for the definitive surgical management of thyroid disease in adults. Annals of surgery. 2020;271(3):e21-e93. [DOI] [PubMed] [Google Scholar]
- 3. NCCN Clinical Practice Guidelines in Oncology. Thyroid carcinoma. NCCN Clinical Practice Guidelines in Oncology. National Comprehensive Cancer Network; 2021. Version 1.2021. [Google Scholar]
- 4. Haugen BR, Alexander EK, Bible KC, et al. . 2015 American Thyroid Association management guidelines for adult patients with thyroid nodules and differentiated thyroid cancer: The American Thyroid Association guidelines task force on thyroid nodules and differentiated thyroid cancer. Thyroid. 2016;26(1):1-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Patel KN, Angell TE, Babiarz J, et al. . Performance of a genomic sequencing classifier for the preoperative diagnosis of cytologically indeterminate thyroid nodules. JAMA Surg. 2018;153(9):817-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Angell TE, Wirth LJ, Cabanillas ME, et al. . Analytical and clinical validation of expressed variants and fusions from the whole transcriptome of thyroid FNA Samples. Front Endocrinol (Lausanne). 2019;10:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krane JF, Cibas ES, Endo M, et al. . The Afirma Xpression Atlas for thyroid nodules and thyroid cancer metastases: insights to inform clinical decision-making from a fine-needle aspiration sample. Cancer Cytopathol. 2020;128(7):452-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu B, Fuchs T, Dogan S, et al. . Dissecting anaplastic thyroid carcinoma: a comprehensive clinical, histologic, immunophenotypic, and molecular study of 360 cases. Thyroid. 2020;30(10):1505-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ciampi R, Romei C, Ramone T, et al. . Genetic landscape of somatic mutations in a large cohort of sporadic medullary thyroid carcinomas studied by next-generation targeted sequencing. Iscience. 2019;20:324-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pozdeyev N, Gay LM, Sokol ES, et al. . Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res. 2018;24(13):3059-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang J, Cai W, Feng D, et al. . Genetic landscape of papillary thyroid carcinoma in the Chinese population. J Pathol. 2018;244(2):215-226. [DOI] [PubMed] [Google Scholar]
- 12. Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye L, Zhou X, Huang F, et al. . The genetic landscape of benign thyroid nodules revealed by whole exome and transcriptome sequencing. Nat Commun. 2017;8:15533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hao Y, Choi Y, Babiarz JE, et al. . Analytical verification performance of afirma genomic sequencing classifier in the diagnosis of cytologically indeterminate thyroid nodules. Front Endocrinol (Lausanne). 2019;10:438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Randolph G, Angell TE, Babiarz J, et al. . Clinical validation of the afirma genomic sequencing classifier for medullary thyroid cancer (Clinical Oral Abstract 29). Thyroid. 2017;27(S1):A105. [Google Scholar]
- 16. Sosa J, Angell TE, Barbiarz J, et al. . Clinical validation of the afirma genomic sequencing parathyroid classifier (poster 168). Thyroid. 2017;27(S1):A50-A51. [Google Scholar]
- 17. Hu MI, Waguespack SG, Dosiou C, et al. . Data from: Supplemental Tables of: Afirma Genomic Sequencing Classifier & Xpression Atlas Molecular Findings in Consecutive Bethesda III-VI Thyroid Nodules. 2021. Deposited 2 April 2021. Doi: 10.6084/m9.figshare.14367905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goldner WS, Angell TE, McAdoo SL, et al. . Molecular variants and their risks for malignancy in cytologically indeterminate thyroid nodules. Thyroid. 2019;29(11):1594-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harrell RM, Eyerly-Webb SA, Golding AC, Edwards CM, Bimston DN. Statistical comparison of Afirma GSC and Afirma GEC outcomes in a community endocrine surgical practice: early findings. Endocr Pract. 2019;25(2):161-164. [DOI] [PubMed] [Google Scholar]
- 20. Angell TE, Heller HT, Cibas ES, et al. . Independent comparison of the Afirma genomic sequencing classifier and gene expression classifier for cytologically indeterminate thyroid nodules. Thyroid. 2019;29(5):650-656. [DOI] [PubMed] [Google Scholar]
- 21. Endo M, Nabhan F, Porter K, et al. . Afirma gene sequencing classifier compared with gene expression classifier in indeterminate thyroid nodules. Thyroid. 2019;29(8):1115-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei S, Veloski C, Sharda P, Ehya H. Performance of the Afirma genomic sequencing classifier versus gene expression classifier: an institutional experience. Cancer Cytopathol. 2019;127(11):720-724. [DOI] [PubMed] [Google Scholar]
- 23. San Martin VT, Lawrence L, Bena J, et al. . Real-world comparison of afirma GEC and GSC for the assessment of cytologically indeterminate thyroid nodules. J Clin Endocrinol Metab. 2020;105(3):e428-e435. [DOI] [PubMed] [Google Scholar]
- 24. Andrioli M, Carocci S, Alessandrini S, et al. . Testing for afirma in thyroid nodules with high-risk indeterminate cytology (TIR3B): first Italian experience. Endocr Pathol. 2020;31(1):46-51. [DOI] [PubMed] [Google Scholar]
- 25. Livhits MJ, Zhu CY, Kuo EJ, et al. . Effectiveness of molecular testing techniques for diagnosis of indeterminate thyroid nodules: a randomized clinical trial. JAMA Oncol. 2021;7(1):70-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geng Y, Aguilar-Jakthong JS, Moatamed NA. Comparison of Afirma gene expression classifier with gene sequencing classifier in indeterminate thyroid nodules: a single-institutional experience. Cytopathology. 2021;32(2):187-191. [DOI] [PubMed] [Google Scholar]
- 27. Endo M, Nabhan F, Angell TE, et al. . Letter to the editor: use of molecular diagnostic tests in thyroid nodules with hürthle cell-dominant cytology. Thyroid. 2020;30(9):1390-1392. [DOI] [PubMed] [Google Scholar]
- 28. Domingo RP, Ogden LL, Been LC, Kennedy GC, Traweek ST. Identification of parathyroid tissue in thyroid fine-needle aspiration: a combined approach using cytology, immunohistochemical, and molecular methods. Diagn Cytopathol. 2017;45(6):526-532. [DOI] [PubMed] [Google Scholar]
- 29. Trimboli P, Treglia G, Guidobaldi L, et al. . Detection rate of FNA cytology in medullary thyroid carcinoma: a meta-analysis. Clin Endocrinol (Oxf). 2015;82(2):280-285. [DOI] [PubMed] [Google Scholar]
- 30. Workman AD, Soylu S, Kamani D, et al. . Limitations of preoperative cytology for medullary thyroid cancer: proposal for improved preoperative diagnosis for optimal initial medullary thyroid carcinoma specific surgery. Head Neck. 2021;43(3):920-927. [DOI] [PubMed] [Google Scholar]
- 31. Kloos RT, Reynolds JD, Walsh PS, et al. . Does addition of BRAF V600E mutation testing modify sensitivity or specificity of the Afirma gene expression classifier in cytologically indeterminate thyroid nodules? J Clin Endocrinol Metab. 2013;98(4): E761-E768. [DOI] [PubMed] [Google Scholar]
- 32. Yoo SK, Lee S, Kim SJ, et al. . Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. Plos Genet. 2016;12(8):e1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. San Roman Gil M, Pozas J, Molina-Cerrillo J, et al. . Current and future role of tyrosine kinases inhibition in thyroid cancer: from biology to therapy. Int J Mol Sci. 2020;21(14):4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Drilon A, Laetsch TW, Kummar S, et al. . Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Entrectinib OK’d for cancers with NTRK fusions, NSCLC. Cancer Discov. 2019;9(10):OF2. [DOI] [PubMed] [Google Scholar]
- 36. Markham A. Selpercatinib: first approval. Drugs. 2020;80(11):1119-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wirth LJ, Sherman E, Robinson B, et al. . Efficacy of selpercatinib in RET-altered thyroid cancers. N Engl J Med. 2020;383(9):825-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chu YH, Wirth LJ, Farahani AA, et al. . Clinicopathologic features of kinase fusion-related thyroid carcinomas: an integrative analysis with molecular characterization. Mod Pathol. 2020;33(12):2458-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chu YH, Dias-Santagata D, Farahani AA, et al. . Clinicopathologic and molecular characterization of NTRK-rearranged thyroid carcinoma (NRTC). Mod Pathol. 2020;33(11):2186-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sohn SY, Park WY, Shin HT, et al. . Highly concordant key genetic alterations in primary tumors and matched distant metastases in differentiated thyroid cancer. Thyroid. 2016;26(5):672-682. [DOI] [PubMed] [Google Scholar]
- 41. Masoodi T, Siraj AK, Siraj S, et al. . Whole-exome sequencing of matched primary and metastatic papillary thyroid cancer. Thyroid. 2020;30(1):42-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jung CK, Jung SH, Jeon S, et al. . Risk stratification using a novel genetic classifier including PLEKHS1 promoter mutations for differentiated thyroid cancer with distant metastasis. Thyroid. 2020;30(11):1589-1600. [DOI] [PubMed] [Google Scholar]
- 43. Agrawal N, Jiao Y, Sausen M, et al. . Exomic sequencing of medullary thyroid cancer reveals dominant and mutually exclusive oncogenic mutations in RET and RAS. J Clin Endocrinol Metab. 2013;98(2):E364-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moura MM, Cavaco BM, Leite V. RAS proto-oncogene in medullary thyroid carcinoma. Endocr Relat Cancer. 2015;22(5): R235-R252. [DOI] [PubMed] [Google Scholar]
- 45. FDA approves selpercatinib; pralsetinib may soon follow. Cancer Discov. 2020;10(7):OF1. [DOI] [PubMed] [Google Scholar]
- 46. Wells SA Jr, Asa SL, Dralle H, et al. ; American Thyroid Association Guidelines Task Force on Medullary Thyroid Carcinoma . Revised American Thyroid Association guidelines for the management of medullary thyroid carcinoma. Thyroid. 2015;25(6):567-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kloos RT, Monroe RJ, Traweek ST, Lanman RB, Kennedy GC. A genomic alternative to identify medullary thyroid cancer preoperatively in thyroid nodules with indeterminate cytology. Thyroid. 2016;26(6):785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in References.