Summary
During mitosis in animal cells, the centrosome acts as a microtubule organizing center (MTOC) to assemble the mitotic spindle. MTOC function at the centrosome is driven by proteins within the pericentriolar material (PCM), however the molecular complexity of the PCM makes it difficult to differentiate the proteins required for MTOC activity from other centrosomal functions. We used the natural spatial separation of PCM proteins during mitotic exit to identify a minimal module of proteins required for centrosomal MTOC function in C. elegans. Using tissue-specific degradation, we show that SPD-5, the functional homologue of CDK5RAP2, is essential for embryonic mitosis, while SPD-2/CEP192 and PCMD-1, which are essential in the one-cell embryo, are dispensable. Surprisingly, although the centriole is known to be degraded in the ciliated sensory neurons in C. elegans1-3, we find evidence for “centriole-less PCM” at the base of cilia and use this structure as a minimal testbed to dissect centrosomal MTOC function. Super-resolution imaging revealed that this PCM inserts inside the lumen of the ciliary axoneme and directly nucleates the assembly of dendritic microtubules towards the cell body. Tissue-specific degradation in ciliated sensory neurons revealed a role for SPD-5 and the conserved microtubule nucleator γ-TuRC, but not SPD-2 or PCMD-1, in MTOC function at centriole-less PCM. This MTOC function was in the absence of regulation by mitotic kinases, highlighting the intrinsic ability of these proteins to drive microtubule growth and organization and further supporting a model that SPD-5 is the primary driver of MTOC function at the PCM.
Graphical Abstract
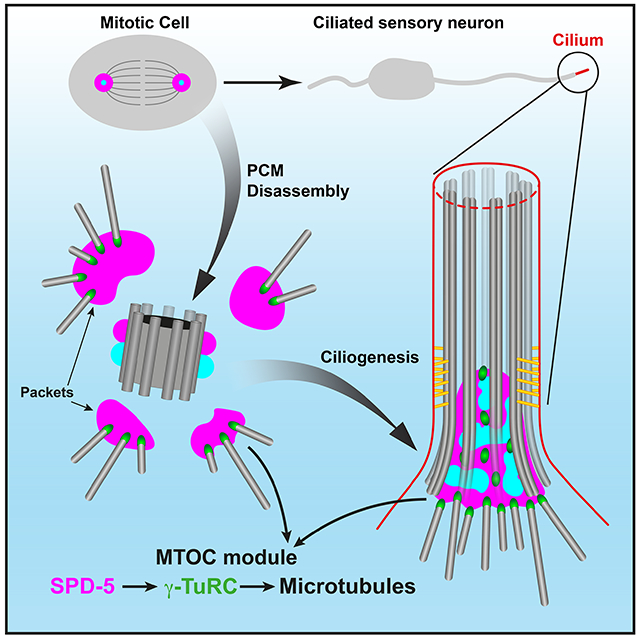
eTOC blurb:
Magescas et al. use tissue-specific degradation to investigate the role of PCM proteins in the microtubule organizing capacity of the centrosome in C. elegans. They identify a novel MTOC at the base of cilia in sensory neurons and reveal a central role for SPD-5/~CDK5RAP2 in MTOC function at this centriole-less PCM and at the mitotic centrosome.
Results
Microtubules perform numerous cellular functions, including chromosome segregation and intracellular transport, that require spatial organization imparted by cellular sites called microtubule organizing centers (MTOCs). The best-studied MTOC is the centrosome, a non-membrane bound organelle composed of two barrel-shaped microtubule-based centrioles surrounded by a layered network of pericentriolar material4-6 (PCM). The MTOC activity of the centrosome cycles with the cell cycle, peaking during mitosis when the PCM expands due to regulation by mitotic kinases to recruit microtubules.
The molecular complexity of the PCM and its regulation has obscured the identity of proteins that function in microtubule organization per se rather than other aspects of centrosome biology. In C. elegans, the PCM is relatively simple and composed of two main scaffolding proteins, SPD-2/CEP192 and SPD-5 (the functional homologue of CDK5RAP2), and a recently discovered coiled-coil protein PCMD-17. These proteins have largely been characterized in the zygote where they are interdependently recruited to the centrosome and, together with AIR-1/Aurora-A, localize the conserved microtubule nucleation complex γ-TuRC8-14. Despite the simplicity of C. elegans PCM, the direct intramolecular interactions that ultimately build and anchor microtubules are largely unknown. We therefore aimed to understand which PCM proteins impart the MTOC function of the centrosome in multiple cellular contexts.
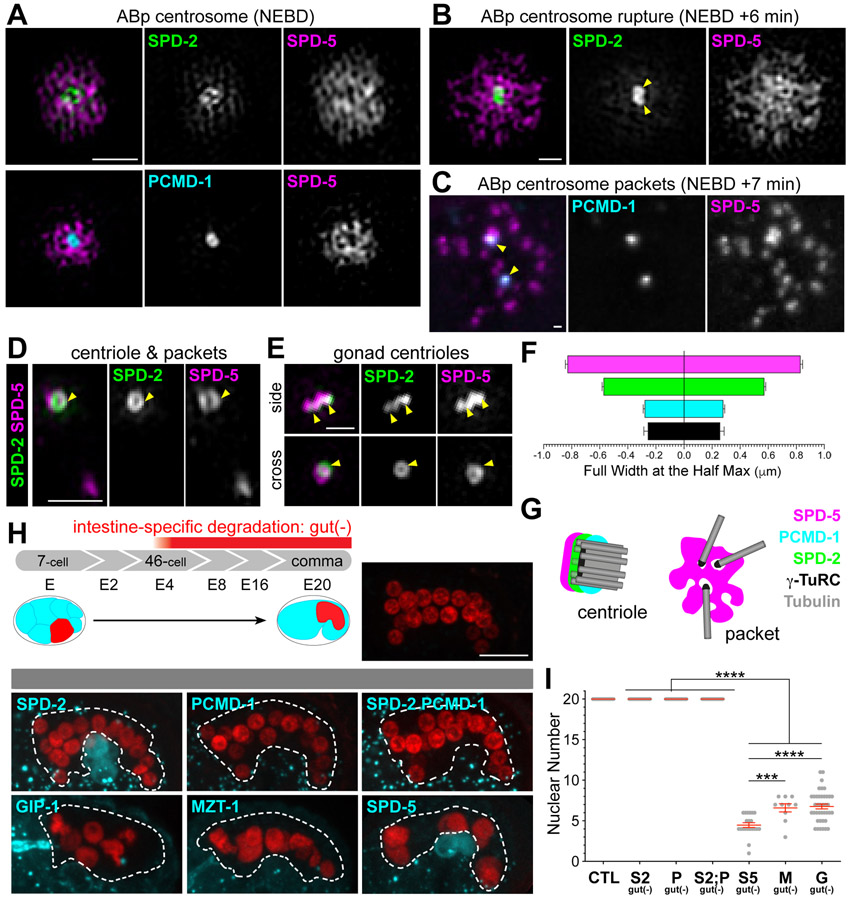
Our recent analysis in early dividing embryonic cells found evidence for distinct subcomplexes within the C. elegans PCM5, only some of which are spatially associated with MTOC function. In particular, we found that the PCM is organized with an ‘inner’ sphere that localizes all PCM proteins and an ‘outer’ sphere that colocalizes SPD-5 and proteins like γ-TuRC required for microtubule growth and assembly. These subcomplexes are not only spatially separated but also appear to be functionally distinct as MTOC function segregates with outer sphere proteins during PCM disassembly in mitotic exit: proteins in the outer sphere rupture into “packets” which co-localize with dynamic microtubules. Thus, SPD-5 is the central PCM component in the outer sphere and in packets5 (Figure 1A-G). 3-Dimensional Structured Illumination Microscopy (3D-SIM) imaging of PCM revealed that SPD-2 and PCMD-1 were concentrated more centrally (Figure 1A-C and 1F) while SPD-5 formed a broad, perforated matrix. Neither SPD-2 nor PCMD-1 localized in packets during disassembly5 (Figure 1C and 1D). During packet formation, SPD-2 localization was restricted to two toroids that we assume surround the centrioles as the localization resembled that of the centrioles in the C. elegans gonad that have little associated PCM15 (Figure 1D and 1E). In addition to localizing to packets, SPD-5 also localized as a toroid, however, the SPD-2 and SPD-5 toroids appeared to be offset and stacked (Figure 1D and 1E).
Figure 1. MTOC function at the centrosome is driven by SPD-5 and not SPD-2 or PCMD-1.
(A-E) 3D-SIM (A-B, D-E) or confocal (C) imaging of PCM proteins at indicated stages with centrioles (yellow arrowheads) and packets (white arrowheads) indicated. Scale bar= 1 μm. (F) Average width of pixel intensity profile for each protein at ABp centrosome at nuclear envelope breakdown: SPD-5: 0.83±0.01μm, n= 18; SPD-2: 0.57±0.01μm, n= 21; PCMD-1: 0.26±0.01μm, n= 12; SAS-4: 0.25±0.03μm, n= 19. (G) Cartoon representing the separation of function of PCM proteins, with MTOC function specifically associated with packets. (H) Top: Cartoon depicting the development of the C. elegans embryonic intestine (left) relative to the E4-stage onset of elt-2p promoter expression. Embryonic intestines have 20 cells by comma stage, which can be visualized using an intestine-specific histone::mCherry as indicated in control embryo. Bottom: Intestine-specific ZIF-1 expression (elt-2p::zif-1) effectively degrades indicated ZF:GFP tagged proteins (cyan). Asterisk indicates germ cells. Scale bar= 10μm. (I) Analysis of intestinal nuclear number at comma stage: control (‘CTL’): mean= 20±0.0 S.D. nuclei, n=20 embryos; SPD-2gut(−) (‘S2 gut (−)’): mean= 20.0±0.0 S.D. nuclei, n=20 embryos; PCMD-1gut(−) (‘P gut(−)’): mean= 20±0.0 S.D. nuclei, n = 20 embryos; [SPD-2;PCMD-1]gut(−) (‘S2;P gut(−)’): mean= 20±0.0 S.D. nuclei, n = 20 embryos; SPD-5gut(−) (‘S5 gut(−)’): mean= 4.5±1.3 S.D. nuclei, n= 21 embryos; MZT-1gut(−) (‘M gut(−)’): mean= 6.6±1.6 S.D. nuclei, n= 10 embryos; GIP-1gut(−) (‘G gut(−)’): mean= 6.8±1.9 S.D. nuclei, n= 40 embryos. Tukey’s multiple comparisons test, *** p-value=0.0008, **** p-value<0.0001.
See also Figure S1
Post-zygotic mitosis in the embryo requires SPD-5, but not SPD-2 or PCMD-1
Based on our localization studies, we hypothesized that MTOC function at the centrosome is controlled by a SPD-5-dependent module that functions independently of SPD-2 and PCMD-1. To test this hypothesis, we depleted essential centrosome components in embryonic cell divisions using a tissue-specific degradation system to deplete endogenous SPD-5, SPD-2, PCMD-1 and γ-TuRC components GIP-1/GCP3 and MZT-1/Mozart1 from the 4-cell stage embryonic intestine as observed by the loss of GFP signal from the endogenously tagged protein12,16,17 (‘gut(−)’, Figure 1H and Figure S1A). Intestinal cells all derive from the E-blastomere through successive rounds of divisions leading to an invariant 20-cell intestine in control embryos (Figure 1H and 1I). γ-TuRC depletion resulted in significantly fewer intestinal cell nuclei12 (Figure 1H, 1I, and Figure S1B). Degradation of SPD-5 resulted in an even greater reduction of intestinal cell nuclei (Figure 1H, 1I and Figure S1B), perhaps due to a more direct effect on spindle formation. Surprisingly, degradation of SPD-2, PCMD-1, or both proteins together had no impact on intestinal nuclear number, suggesting no perturbation in spindle formation or centriole duplication (Figure 1H, 1I and Figure S1B). We further explored a role for SPD-2, PCMD-1, and SPD-5 in other embryonic contexts, removing each protein from the early embryonic through morphogenesis-stage divisions (Figure S1). As in the intestine, depletion of SPD-5 led to dramatic embryonic defects while loss of SPD-2 and/or PCMD-1 did not (Figure S1C, S1D and S1F). In contrast to their strong requirement in the C. elegans zygote (Figure S1E), these results suggest that SPD-2 and PCMD-1 are not required for centrosome function past the third round of embryonic mitosis.
SPD-5 is maintained at the base of C. elegans cilia
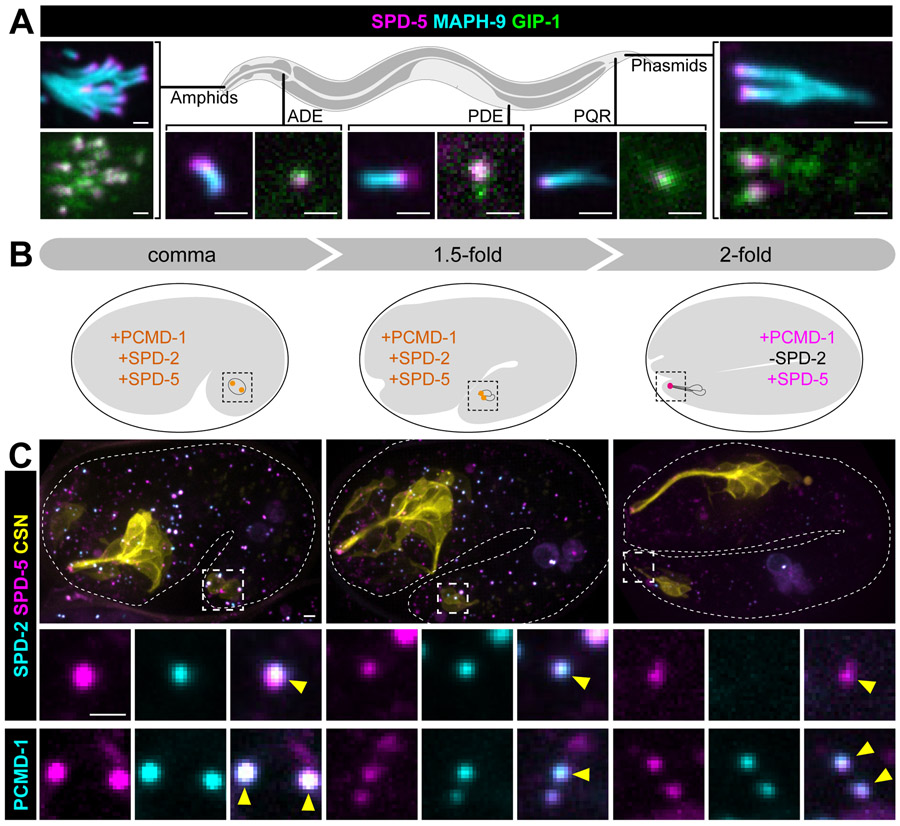
Specific roles for PCM proteins or subcomplexes during mitosis are difficult to distill as mitotic PCM is rapidly evolving and under heavy regulation18. Thus, we sought to find interphase PCM that retains MTOC function to probe the role of distinct PCM proteins and/or subcomplexes. In many differentiated cells types, MTOC function is reassigned to a non-centrosomal site as part of the normal process of differentiation19. Cells with cilia provide an exception to this general phenomenon as the basal body derived from the centrosome often retains MTOC activity20,21. In C. elegans, only a subset of sensory neurons are ciliated and previous work found that after migration of the centriole to the cell surface to template the growth of the cilium, the centriole is degraded beginning with the loss of SPD-2 and followed by the removal of centriole cartwheel components such as SAS-4/CPAP1,2,22. As we would have expected the PCM to be removed with the centriole, we were therefore surprised to find endogenous SPD-5 localized to the ciliary base (CB) of all ciliated sensory neurons in adult worms alongside GIP-1 (Figure 2A). Analysis of endogenous centriole and PCM components in the phasmid (Figure 2B and 2C) and amphid neurons (Figure S2A and 2B) revealed that SPD-5, PCMD-1, and GIP-1 are maintained at the CB through the entire process of ciliogenesis despite the previously described2 loss of SPD-2 and SAS-4. This pattern of localization suggests that the CB in C. elegans is composed of centriole-less PCM.
Figure 2. SPD-5 is maintained at the ciliary base while other centriolar and PCM protein are lost.
(A) Localization of SPD-5 (magenta) and GIP-1 (green) in indicated ciliated sensory neurons relative to the axoneme (MAPH-9, cyan) in adult worms. (B) Cartoon depicting the stages of phasmid ciliogenesis in the embryo. The phasmid neuronal precursor cell divides from comma to the 1.5-fold stage of the embryo. Dendrite extension and centriole (orange) migration occurs from the 1.5 to 2-fold stage and centrioles (magenta) subsequently lose SPD-2 by the 2-fold stage. (C) Localization of endogenously tagged indicated proteins in phasmid neurons (yellow: ‘CSN’: maph-9p::myr::BFP) of live embryos. Magnified views of centrioles in the phasmid neurons shown below (yellow arrowheads). Scale bars = 1μm.
See also Figure S2.
A unique MTOC organization at the ciliary base
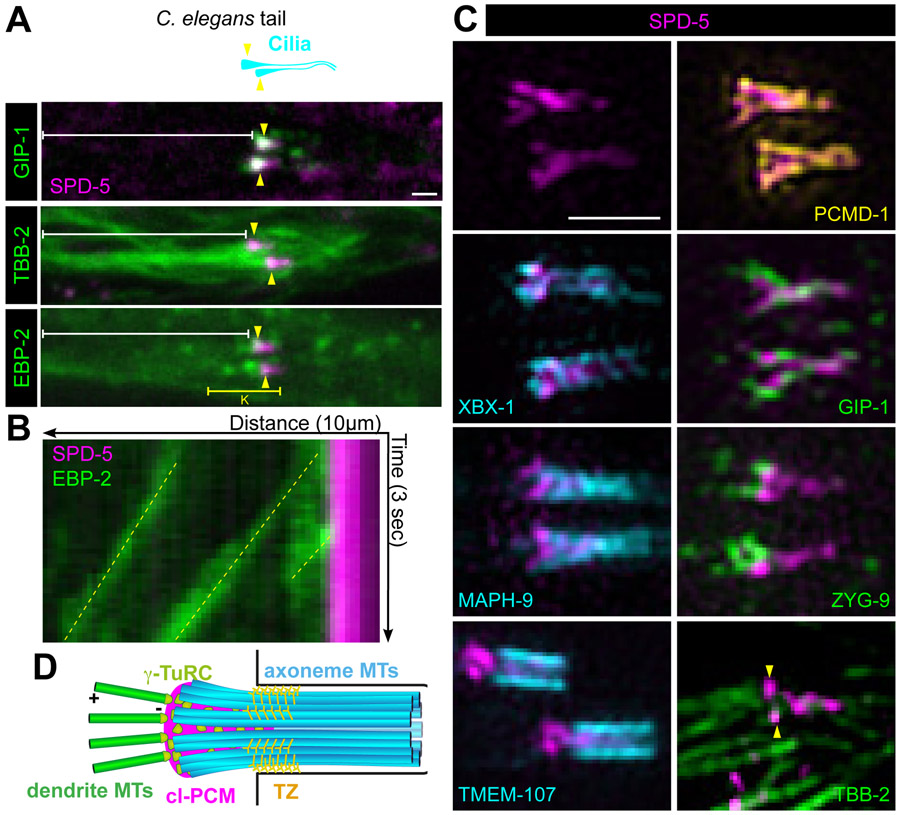
The CB has previously been suggested to function as an MTOC; γ-tubulin localizes near the CB and EBP-2/EB3 comets have been shown in the dendrite moving away from the cilium toward the cell body, however the structure imparting MTOC function was unclear from previous analysis10,23. To determine if the centriole-less PCM at the CB acts as an MTOC, we characterized the CB structure and microtubule organization in the adult phasmid neurons, a more isolated pair of ciliated sensory neurons. Localization of endogenous TBB-2/β-tubulin, GIP-1 and EBP-2 suggested that dynamic microtubules emanate directly from the CB; GIP-1 colocalized with SPD-5 at the CB and EBP-2 comets appeared to originate directly from the SPD-5 containing region (Figure 3A and 3B). In contrast to their localization to mitotic PCM, AIR-1/Aurora A and PLK-1/Plk1 did not localize to the centriole-less PCM at the CB (Figure S2C and S2D).
Figure 3. Centriole-less PCM at the ciliary base serves as a MTOC.
(A) Cartoon (top) and images from live adult worms (bottom) of the C. elegans phasmid cilia (cyan) at the tip of the dendrite (white bracket) with ciliary base indicated (yellow arrowhead). (B) Kymograph of yellow bracketed region in A of EBP-2 comets (green, yellow dotted line) traveling from the ciliary base (SPD-5, magenta) toward the cell body. (C) 3D-SIM analysis of SPD-5 (magenta) localization at the ciliary base in phasmids cilia relative to PCMD-1 (yellow), axonemal (cyan), and microtubule-related (green) proteins; Yellow arrowheads indicate centriole-less PCM and white arrowheads indicate cytoplasmic microtubules. (D) Cartoon summarizing the organization of the ciliary base as revealed by 3D-SIM. Axonemal MTs (axMTs, cyan), transition zone (TZ), centriole-less PCM (cl-PCM, magenta), and dendrite microtubules (dendrite MTs, green) are indicated. Scale bars = 1μm.
See also Figure S2.
We further characterized the CB structure using 3D-SIM (Figure 3C), revealing that SPD-5 localizes in a ‘T’-shape, projecting inside the luminal space of the axoneme: SPD-5 localizes inside the barrel of the axoneme (XBX-1/DNC2L1 and MAPH-9/MAP9) and transition zone (TMEM-107/TMEM107), together with PCMD-1, consistent with previous anecdotal reports of PCMD-1 localization in ciliated sensory neurons7. GIP-1 appeared to localize heterogeneously around and intermingled with the SPD-5 T-shape structure. The microtubule associated protein ZYG-9/chTOG was restricted to the dendritic space juxtaposed to SPD-5. Importantly, individual dendritic microtubules appeared to directly connect to the SPD-5 structure (Figure 3C, arrowheads, TBB-2). These observations revealed that microtubules are organized from a T-shaped centriole-less PCM and extend from the CB toward the cell body likely stabilized in the dendritic spaces by microtubule associated proteins such as ZYG-9. These data also suggest that the CB provides a minimal testbed to isolate and study roles of PCM proteins in MTOC function divorced from their other centrosomal functions.
SPD-5 based centriole-less PCM drives MTOC function at the CB and ciliogenesis
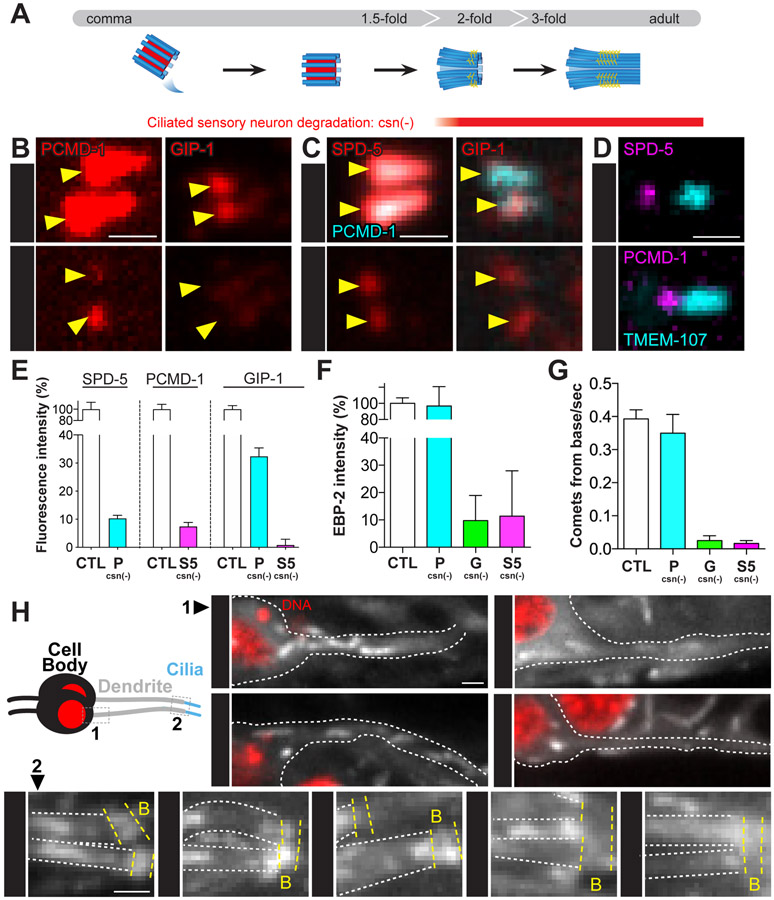
To test whether PCM components localize interdependently at the CB and have a role in MTOC function, we degraded endogenous proteins specifically in ciliated sensory neurons following centriole migration and subsequent docking at the membrane at the onset of ciliogenesis and transition zone establishment (‘csn(−),’ Figure 4A and Figure S3A). To avoid perturbation to cell division or centriolar migration, we used the osm-6 promoter to drive degradation in the early stages of ciliogenesis (Figure S3A and S3B). Removal of GIP-1 had no impact on the localization of either PCMD-1 or SPD-5 (Figure S3C). SPD-5 degradation caused a reduction of PCMD-1 and the complete loss of GIP-1 from the CB (Figure 4B, 4D and 4E), while PCMD-1 degradation resulted in the incomplete loss of either SPD-5 or GIP-1 (Figure 4C-4E) from the onset of degradation (Figure S3A and S3B). Similar observations were obtained using a putative null allele of pcmd-17 (pcmd-1(syb975), Figure S3D and S3E). Intriguingly, the populations of SPD-5 or PCMD-1 that remained following depletion of either PCMD-1 or SPD-5, respectively, were spatially distinct with the SPD-5 pool localizing closer to the dendrite and the PCMD-1 pool localizing closer to the axoneme (Figure 4D). These data suggest that SPD-5 and PCMD-1 are present at the CB in different subcomplexes and that only a fraction of the centriole-less PCM is a complex of both SPD-5 and PCMD-1.
Figure 4. SPD-5 drives MTOC function at the ciliary base and ciliogenesis.
(A) Cartoon of ciliogenesis in phasmid neurons with centriolar microtubules (blue), internal centriole structures (red), and transition zone fibers (yellow) indicated. The osm-6 promoter drives ciliated sensory neurons specific degradation (csn(−)). (B-D) Localization of indicated proteins at the ciliary base (yellow arrowhead) of adult phasmid cilia following SPD-5 or PCMD-1 depletion. (E) Analysis of fluorescence intensity of indicated proteins following degradation of SPD-5 or PCMD-1. SPD-5: control (‘CTL’) = 100±12.45%, n= 10 worms; PCMD-1csn(−) (‘P csn(−)’) = 10.32±1.06%, n= 14 worms; PCMD-1: control = 100±8.83%, n= 11 worms; SPD-5csn(−) (‘S5 csn(−)’) = 7.45±1.39%, n= 12 worms; GIP-1: control = 100%±6.41, n= 10 worms; PCMD-1csn(−) = 32.36±3.01%, n= 10; SPD-5csn(−) = 0.82±2.09%, n= 10 worms. Values are mean ± standard error of mean. (F) EBP-2 fluorescence intensity near the ciliary base relative to control: control = 100± 6.87%, n= 10 worms; PCMD-1csn(−) = 96.85±23.8%, n= 10 worms; GIP-1csn(−) (‘G csn(−)’) = 9.74±9.25%, n= 10 worms; SPD-5 csn(−) = 11.35±16.6%, n= 10 worms. (G) EBP-2 comets emanating from ciliary base: control = 0.40±0.03 comets/sec., n= 10 worms; PCMD-1csn(−) = 0.35±0.06 comets/sec., n= 10 worms; GIP-1csn(−) = 0.03±0.01 comets/sec., n= 10 worms; SPD-5csn(−) = 0.02±0.01 comets/sec., n= 10 worms. (H) Cartoon of phasmid neuron with region imaged along the dendrite (‘1’) and at the ciliary base (‘2’) indicated. 1: 12-second time-projection of EBP-2 comets in the dendrite moving toward the cell body (red, DNA). 2: 3-second time-projection of EBP-2 comets along the dendrites (outlined by white dashed lines, ‘D’) near the ciliary base (yellow dashed lines, ‘B’). Scale bars = 1μm.
The presence of distinct subcomplexes of protein at the CB suggested that, like at the centrosome, these populations could also be functionally distinct. Furthermore, the partial loss of SPD-5 and γ-TuRC following PCMD-1 degradation suggested that PCMD-1 localizes a specific subcomplex of SPD-5 that is not required for MTOC function. To test the role of these subcomplexes in MTOC function at the CB, we monitored the movement of EBP-2 comets from the CB and within the dendrite following degradation. In control animals, we observed an enrichment of EBP-2 at the CB with comets projecting from the CB toward the cell body (Figure 4F-H and Figure S3F; Video S1). Upon degradation of SPD-5 or GIP-1, we observed a loss of both EBP-2 enrichment and comets, while PCMD-1 degradation or mutation did not affect either (Figure 4F-H and Figure S3F; Video S1). Together, these results indicate that γ-TuRC is recruited by a SPD-5 dependent subcomplex of the centriole-less PCM to drive MTOC function in the dendrite.
Finally, we tested whether centriole-less PCM is required for ciliogenesis. SPD-5 but not PCMD-1 depletion led to variable defects in ciliary structure ranging from shorter to absent phasmid cilia, as visualized by an endogenously tagged dynein component XBX-1 or the cilia-specific β-tubulin isoform TBB-4 (Figure S3G and S3H). The phenotypic variability is likely due to variation in the timing of degradation but is consistent with a role for SPD-5 and the centriole-less PCM in establishing and/or maintaining the ciliary axoneme.
Discussion
Here, using multiple cellular contexts, we found evidence for a SPD-5-based module that controls MTOC function within the PCM. This claim is supported by three lines of evidence. First, SPD-5 is associated with dynamic microtubules and microtubule associated proteins in sub-PCM fragments that are spatially distinct from other PCM proteins such as SPD-2 and PCMD-1. Second, SPD-5, but not SPD-2 or PCMD-1, is essential for mitosis throughout development. Third, SPD-5 is associated with centriole-less PCM at the CB, a structure we argue represents the “pure” MTOC function of the centrosome. As loss of both γ-TuRC and SPD-5 from ciliated sensory neurons inhibited microtubule growth from the CB, we propose that SPD-5 alone recruits γ-TuRC to build and localize microtubules. As the SPD-5 functional homologues Centrosomin and CDK5RAP2 can directly interact with ɣ-TuRC, SPD-5 might serve as a platform to directly recruit γ-TuRC. Extensive studies have shown that the ability of the centrosome to expand its PCM is controlled by phosphorylation by mitotic kinases24. However, we did not observe AIR-1 or PLK-1 at the CB, indicative of the ability of SPD-5 to recruit microtubule regulators independent of this canonical mitotic regulation and making the CB a unique platform to study MTOC regulation.
The fact that SPD-2 and PCMD-1 are dispensable for later embryonic cell divisions is in sharp contrast to their essential function in the first cell division of the C. elegans zygote and in the germ cells. Not only are both proteins required for centrosome maturation, but SPD-2/DSpd-2/Cep192 also plays a conserved role in centriole duplication13. Thus, either centrosome regulation is divergent across cell types or we are unable to effectively deplete these proteins in later somatic cells due to limitations of our degradation system. Although we attempted to control for these limitations (Figure S1), it is possible that SPD-2 or PCMD-1 beyond the detection limit of GFP perdured or that yet undiscovered isoforms of either protein are left untagged by our endogenous degron insertion. Despite these limitations, our work is in agreement with reports of phenotypes observed for spd-225 and pcmd-1 temperature sensitive mutants7, suggesting that the essential nature of SPD-2 and PCMD-1 may be due to their specific function in the germline and zygote and speculate that this requirement is due to the unique nature of these developmental contexts. For example, the first mitosis of the embryo requires the reactivation of the heavily repressed sperm centrosome26 and mitotic centrosomes in the adult germline are localized to a stem cell niche. Thus, SPD-2 and PCMD-1 may act as a tissue-specific catalyst for PCM assembly as has been suggested from in vitro studies27. Although the centrosome in other embryonic divisions is inactive as an MTOC in interphase, especially once embryonic cell cycles achieve gap phases28,29, a residual shell of PCM in interphase could serve the same purpose4,6,30.
We found that SPD-5 and PCMD-1 are maintained at the base of C. elegans cilia despite the previously reported absence of centrioles and SPD-2. This centriole-less PCM could be a stabilized remnant of PCM packets, however since PCMD-1 does not localize to packets, other mechanisms such as protection from dephosphorylation by mitotic phosphatases5 or proteasome-dependent degradation likely maintain proteins at the CB. Alternatively, or in addition, SPD-5 and PCMD-1 could be maintained through continued expression. Interestingly, SPD-5 contains an X-box motif in its promoter region, a conserved signature of genes regulated by ciliogenic transcription factors and indicating continued expression of spd-5 specifically in ciliated sensory neurons31,32. We hypothesize that the combination of targeted protein degradation and transcriptional regulation drive the specific loss or maintenance of centrosomal proteins.
We described a novel organization of PCM proteins, independent of the centriole, where SPD-5 drives MTOC function. Intriguingly, similar patterns of modification to the centriole structure have been observed at the base of flagella in human sperm, including the loss of CEP192/SPD-2, and the maintenance of CDK5RAP2/~SPD-5. This structure is also competent to serve as an MTOC in mitotic extract suggesting conservation of the mechanisms described here33. Together, our studies reveal the innate ability of SPD-5 to recruit microtubule regulators which likely underlies the ability of the centrosome to function as an MTOC.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jessica Lynn Feldman (feldmanj@stanford.edu).
Materials Availability:
Strains and plasmids are available upon request.
Data and Code Availability
This published article includes all datasets generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains were maintained at 20°C unless otherwise specified and cultured on NGM plates seeded with E. coli OP5034. For strains that were maintained with a balancer, phenotypes were analyzed on non-balanced worms 2 to 3 generations off of the balancer.
METHOD DETAILS
CRISPR/Cas9 Editing
Endogenously tagged proteins used in this study were generated using the CRISPR Self Excising Cassette (SEC) method35. Cas9 and sgRNAs were delivered using a single plasmid based on pDD162, in which the corresponding sgRNA was incorporated using the Q5® Site directed mutagenesis kit (sgRNA sequences used for generating the alleles can be found in Table S1). Repair templates for each corresponding edit were generated by adding two homology arms flanking the SEC with the appropriate knock-in fluorescent protein. Homology arms were generated by amplifying the region close to the desired insertion site (oligonucleotides used to amplify these genomic regions can be found in Table S1). The repair vector was digested to remove the CcDB cassettes flanking the CRISPR insert and self-excising cassette, and the digested vector and homology arms were assembled using a HiFi DNA assembly reaction (New England BioLabs) accordingly to the manufacturer recommendation. DNA mixtures (sgRNA and Cas9 containing plasmid and repair template) were injected into young adults, and CRISPR edited worms were selected by treatment with hygromycin followed by visual inspection for appropriate expression and localization35. CRISPR edited worm lines were then back crossed at least two times, homozygosed and maintained under standard conditions.
Degradation
Protein degradation strategies were performed using the ZF/ZIF-1 degradation system12,16. Genes of interest were tagged with a ZF-degron and GFP (‘zf::gfp’), using CRISPR/Cas9 to insert a zf::gfp tag into each locus. zf::gfp-tagged alleles were generated and maintained in a zif-1(gk117) null mutant background so ZF:GFP targets would not be depleted by endogenous ZIF-1 expression12. ZIF-1 was then expressed transgenically in the desired tissue from a single-copy transgene inserted into the genome that has been generated and validated in other conditions12,16,17. Degradation conditions were created by crossing worms carrying zf::gfp-tagged alleles with those carrying the appropriate ZIF-1 expressing transgene and balancer chromosome when necessary. In all cases, worms or embryos analyzed for experiments were isolated from parents at least 2 generations away from the balancer. It is of note that all strains were able, due to the nature of the tissue specific degradation, to be maintained away from the balancer with the exception of zf::gfp::spd-5; cdc-42::ZIF-1 where all heterozygous cross progeny were embryonic lethal. Unless otherwise indicated, <25 embryos were analyzed in each condition.
RNAi treatment
RNAi treatment was performed by feeding L4 worms with HT115 bacteria transformed with GFP::3xFlag(RNAi) or LacZ(RNAi) plasmids36. The GFP::3xFlag(RNAi) plasmid was generated by amplifying the GFP::3xFlag present in the CRISPR repair vector and inserting it into the RNAi vector L4440. RNAi plates (NGM supplemented with IPTG and Ampicillin)36 were seeded with bacterial culture and grown 48 hours at room temperature protected from light. L4 stage worms were grown on RNAi plates at 25°C for 48 to 72 hours.
Assessment of embryonic lethality
To assess embryonic viability, 7 young adult worms of each genotype were singled onto 60mm NGM petri dish plates and allowed to lay for 4 hours at 20 °C, and then adults were removed and eggs were counted. After 3 days at 20 °C, the number of surviving worms present on each plate was counted, and viability for each plate was calculated as the total number of L4s and adults divided by the number of eggs. To generate SPD-5gut(−), SPD-5zif-1(+), and SPD-5soma(−) embryos, the appropriate genotype was generated by balancing spd-5 with the hT2 balancer. SPD-5gut(−) embryos showed a maternal effect phenotype and thus were analyzed from spd-5:zf:gfp homozygous mothers. In contrast, either heterozygous SPD-5Zif-1(+) or SPD-5soma(−) embryos balanced with hT2 were lethal. Embryonic lethality assessment for the GFP RNAi experiment was performed as has described above with the exception that the young adults selected were generated on GFP RNAi plates, then singled on GFP RNAi plates and allowed to lay for 4 hours at 25 °C.
Neuron and ciliary positioning
To track the ciliogenesis of phasmids neurons, we generated a plasmid expressing TagBFP2 with a N-terminal myristoylation signal (MGSSKSKPK) under the control of the promoter from the gene encoding MAPH-9 (2kb upstream of the ATG), a ciliated sensory neuron specific protein. This plasmid has been used to generate a stable extrachromosomal array that was then crossed into desired genotypes. In order to compare the localization of PCM protein after degradation of desired proteins, we generated a plasmid expressing a C-terminal TagBFP2-tagged TMEM-107 under the control of its own promoter (559 bp upstream of the ATG)37. The plasmid was injected into desired genotypes with an injection marker (pRF4, roller marker) and young adult Rol hermaphrodites were analyzed 5 days after injection.
Image acquisition
Worms were mounted on a pad (5% agarose dissolved in M9) immersed in 1mM Levamisol sandwiched between a microscope slide and no. 1.5 coverslip. The same technique was applied without Levamisole for mounting embryos. Spinning-disk confocal images were acquired on a Nikon Ti-E inverted microscope (Nikon Instruments) equipped with a 1.5x magnifying lens, a Yokogawa X1 confocal spinning disk head, and an Andor Ixon Ultra back thinned EM-CCD camera (Andor), all controlled by NIS Elements software (Nikon). Images were obtained using a 60x (NA= 1.4) or 100x Oil Plan Apochromat objective (NA= 1.45). Z-stacks were acquired using a 0.2 μm step. For EBP-2 comet imaging, images were acquired as a stream of single plane of images focused on the ciliary base or cell cortex of adult phasmids neurons, with a 100ms exposure. Structured illumination microscopy images were acquired on an OMX BLAZE V4 microscope (GE) in the Stanford Cell Science Imaging Facility equipped with a U-PLANAPO 100x (NA= 1.4) objective and Photometrics Evolve 512 EM-CCD cameras, all controlled by DeltaVision software. Structured Illumination images were generated using SoftWoRx software. Images were adjusted for brightness and contrast using ImageJ software. Unless otherwise indicated, >25 embryos were analyzed for each experiment.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical details of experiments can be found in the corresponding figure legends. Graphs and statistical analysis were performed using R and Prism (GraphPad software, La Jolla, Ca, USA).
To measure the space occupied by the PCM proteins at the centrosome (Figure 1F), stack images (30 images at 0.5 μm z-steps) centered around the centrosome of the corresponding endogenously GFP-tagged protein were taken in the ABp cell of the 4-cell embryo at nuclear envelope break down. To create a standardized cropped centrosome stack, the following method was followed: 1) a 30 μm wide square region of interest (ROI) was manually defined around the ABp centrosome and a substack of 15 z-slices was created from this centrosome ROI; 2) the z-slice from this substack containing the maximum intensity was then used to find the xy centroid of the centrosome. This z-slice was thresholded using the half max intensity. Using this threshold, we defined the centroid value by using the Analyze Particle tool (ImageJ); 3) xy coordinates obtained through this method were used to create a 15 μm wide substack of ± 7 slices around the max intensity containing slice centered on those coordinates; 4) a PCM intensity profile was then measured from a 10μm line centered on the xyz centroid of the signal. Profiles of each centrosome were analyzed separately to measure the width of the profile at the half max intensity. Measurements were then compiled and graphed using R software. Only the centrosome closest to the coverslip was analyzed to maintain consistency in imaging5.
To determine nuclear number in embryonic intestines (Figure 1I), comma-stage embryos of the desired genotype were collected, mounted on a 3% agarose pad, imaged on a spinning disk confocal microscope, and nuclei were manually counted.
To measure PCM intensity (Figure 4E, Figure S3B and Figure S3E), fluorescence intensity was measured in ImageJ by defining a square image stack 20 pixels wide x 11, 0.2 μm z-slices deep around the ciliary base of the two phasmid neurons of young adult hermaphrodites (Figure 4E and Figure S3E) or 1.5-fold stage embryos (Figure S3B). Another stack of the exact same dimensions was generated in the neighboring tissue. Both stacks were sum projected and the phasmid intensity was measured by subtracting the total intensity of the neighboring sum projection from the total intensity of the phasmids sum projection. This measurement was performed for the closest phasmid pair relative to the coverslip and only one pair was measured per worm.
To measure EBP-2 intensity (Figure 4F and Figure 4G), fluorescence intensity was measured in ImageJ by defining a round Region of Interest of 10 pixels in diameter around the phasmid in the single plane of young adult hermaphrodites. Another ROI of the exact same dimensions was generated in the neighboring tissue. The base of cilia intensity was measured by subtracting the intensity of the neighboring tissue from the intensity of the base of cilia.
Supplementary Material
EBP-2 comets upon ciliated sensory neuron specific degradation of PCM proteins, related to Figure 4.
Yellow circle represents the position of the ciliary base. Contour of the phasmids are delimited by the yellow dashed line.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli OP50 standard food | CGC | N/A |
| E. coli HT115 RNAi bacteria | CGC | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Levamisole hydrochloride | Sigma-Aldrich | Cat#31742 |
| Ampicillin Sodium Salt | Thermo Fisher Scientific | BP1760 |
| Isopropyl-β-D-thiogalactopyranoside (IPTG) | Thermo Fisher Scientific | BP175510 |
| Critical Commercial Assays | ||
| Q5® Site-Directed Mutagenesis kit | New England BioLabs | Cat#E0554S |
| NEBuilder® HiFi DNA Assembly | New England BioLabs | Cat#E5520S |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain JLF425: spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I; spd-2(wow60[spd-2:: gfp::3xflag]) I | 5 | N/A |
| C. elegans: Strain JLF429: spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I; sas-4(wow32[zf::gfp::3xflag::sas-4]), zif-1(gk117) III | 5 | N/A |
| C. elegans: Strain JLF776: spd-5(wow36[tagrfp-t::3xmyc::spd-5]), pcmd-1(wow109[gfp::zf::3xflag::pcmd-1]) I; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain OD2768: ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; unc-119(ed3) III | 18 | N/A |
| C. elegans: Strain JLF788: gip-1(wow5[gfp::zf::3xflag::gip-1]) III; ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)]II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF789: spd-2(wow61[spd-2::zf::gfp::3xflag]) I; ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF790: pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]) I; ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF791: mzt-1(wow23[zf::gfp::3xflag::mzt-1]); ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF792: spd-5(wow53[zf::gfp::3xflag::spd-5]) I; ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF787: spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I; maph-9(wow95[zf::gfp::3xflag::maph-9]) IV; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF777: spd-5(wow53[zf::gfp::3xflag::spd-5]) I; gip-1(wow25[tagRFP-T::3xmyc::gip-1]), zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF771: spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I; tbb-2(tj26[gfp::tbb-2]) | This paper | N/A |
| C. elegans: Strain JLF428: spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I; ebp-2(wow47[ebp-2::gfp::3xflag]) II | 5 | N/A |
| C. elegans: Strain JLF792: spd-5(wow53[zf::gfp::3xflag::spd-5]) I; zif-1(gk117) III; xbx-1(cas502[xbx-1::tagrfp-t::3xmyc]) V | This paper | N/A |
| C. elegans: Strain JLF793: spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I; oqEx500[tmem-107::GFP + unc-122::DsRed] | This paper /CGC | N/A |
| C. elegans: Strain JLF794: spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I; zyg-9(wow12[zf::gfp::3xflag::zyg-9]) II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain TMD119: pcmd-1(syb486[gfp::pcmd-1]) I | 7 | N/A |
| C. elegans: Strain JLF795: pcmd-1(syb486[gfp::pcmd-1]), spd-5(wow53[zf::gfp::3xflag::spd-5]) I; ltSi915[Posm-6::ZIF-1::operon- linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF796: spd-5(wow53[zf::gfp::3xflag::spd-5]) I; ltSi915[Posm-6::ZIF-1::operon- linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; gip-1(wow25[tagRFP-T::3xmyc::gip-1]), zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF797: spd-5(wow36[tagrfp-t::3xmyc::spd-5]), pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]) I; ltSi915[Posm-6::ZIF-1::operon- linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF775: pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]) I; gip-1(wow25[tagRFP-T::3xmyc::gip-1]), zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF798: pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]) I; ltSi915[Posm-6::ZIF-1::operon- linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; gip-1(wow25[tagRFP-T::3xmyc::gip-1]), zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF799: spd-5(wow53[zf::gfp::3xflag::spd-5]) I; ebp-2(wow47[ebp-2::gfp::3xflag]), ltSi915[Posm-6::ZIF-1::operon- linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF800: ebp-2(wow47[ebp-2::gfp::3xflag]), ltSi915[Posm-6::ZIF-1::operon- linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; gip-1(wow5[zf::gfp::3xflag::gip-1]), zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF801: pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]) I; ebp-2(wow47[ebp-2::gfp::3xflag]), ltSi915[Posm-6::ZIF-1::operon- linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF802: spd-5(wow53[zf::gfp::3xflag::spd-5]) I; ltSi915[Posm-6::ZIF-1::operon- linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III; xbx-1(cas502[xbx-1::tagrfp-t::3xmyc]) V | This paper | N/A |
| C. elegans: Strain JLF803: pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]) I; ltSi915[Posm-6::ZIF-1::operon- linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III; xbx-1(cas502[xbx-1::tagrfp-t::3xmyc]) V | This paper | N/A |
| C. elegans: Strain JLF136: air-1(wow14[air-1::zf::gfp:3xflag]); zif-1(gk117) III | 12 | N/A |
| C. elegans: Strain OD2425 : plk-1(it17[plk-1::sgfp]loxp) III | 39 | N/A |
| C. elegans: Strain JLF943: spd-2(wow61[spd-2::zf::gfp::3xflag]), pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]) I; ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF944: spd-2(wow61[spd-2::zf::gfp::3xflag]) I; xnIs520 [Pcdc-42::ZIF-1, Pcdc-42::mCherry]; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF945: pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]) I; xnIs520 [Pcdc-42::ZIF-1, Pcdc-42::mCherry]; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF946: spd-2(wow61[spd-2::zf::gfp::3xflag]), pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]) I; xnIs520 [Pcdc-42::ZIF-1, Pcdc-42::mCherry]; zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF947: spd-2(wow61[spd-2::zf::gfp::3xflag]), spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I | This paper | N/A |
| C. elegans: Strain JLF948: pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]), spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I | This paper | N/A |
| C. elegans: Strain JLF949: spd-2(wow61[spd-2::zf::gfp::3xflag]), pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]), spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I | This paper | N/A |
| C. elegans: Strain JLF950: spd-5(wow36[tagrfp-t::3xmyc::spd-5]), spd-2(wow60[spd-2:: gfp::3xflag]) I; wowEx161[Pmaph-9::myr::tagBFP2, Punc-122:RFP-T] | This paper | N/A |
| C. elegans: Strain JLF951: pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]), spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I; zif-1(gk117) III; wowEx161[Pmaph-9::myr::tagBFP2, Punc-122:RFP-T] | This paper | N/A |
| C. elegans: Strain JLF952: pcmd-1(wow109[zf::gfp::3xflag::pcmd-1]), spd-5(wow36[tagrfp-t::3xmyc::spd-5]) I; ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III; wowEx161[Pmaph-9::myr::tagBFP2, Punc-122:RFP-T] | This paper | N/A |
| C. elegans: Strain JLF929: tbb-4(wow150[tbb-4::tagrfp-t::3xmyc]) X | This paper | N/A |
| C. elegans: Strain JLF953: spd-5(wow53[zf::gfp::3xflag::spd-5]) I; zif-1(gk117) III; tbb-4(wow150[tbb-4::tagrfp-t::3xmyc]) X | This paper | N/A |
| C. elegans: Strain JLF954: pcmd-1(syb975)/hT2 I; tbb-4(wow150[tbb-4::tagrfp-t::3xmyc]) X | This paper | N/A |
| C. elegans: Strain JLF9555: pcmd-1(syb975)/hT2 I; ebp-2(wow47[ebp-2::gfp::3xflag]) II | This paper | N/A |
| C. elegans: Strain JLF956: spd-5(wow53[zf::gfp::3xflag::spd-5]) I; zif-1(gk117) III; wowEx161[Pmaph-9::myr::tagBFP2, Punc-122:RFP-T] | This paper | N/A |
| C. elegans: Strain JLF957: spd-5(wow53[zf::gfp::3xflag::spd-5]) I; ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; zif-1(gk117) III; wowEx161[Pmaph-9::myr::tagBFP2, Punc-122:RFP-T] | This paper | N/A |
| C. elegans: Strain JLF958: spd-5(wow52[gfp::3xflag::spd-5]) I; ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; gip-1(wow5[gfp::zf::3xflag::gip-1]), zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF959: pcmd-1(syb486[gfp::pcmd-1]) I; ltSi910[Pelt-2::vhhGFP4::ZIF-1::operon-linker::mCherry::histone::tbb-2_3'UTR; cb-unc-119(+)] II; gip-1(wow5[gfp::zf::3xflag::gip-1]), zif-1(gk117) III | This paper | N/A |
| C. elegans: Strain JLF960: pcmd-1(syb975), spd-5(wow36[tagrfp-t::3xmyc::spd-5])/hT2 I | This paper | N/A |
| C. elegans: Strain JLF961: pcmd-1(syb975)/hT2 I; gip-1(wow5[gfp::zf::3xflag::gip-1]), zif-1(gk117) III | This paper | N/A |
| Oligonucleotides | ||
| CRISPR targeting sequence (+PAM): ZF::GFP::3xflag::PCMD-1: GTCGTATTCCACCTCCATag(tgg) | This paper | N/A |
| CRISPR targeting sequence (+PAM): ZF::GFP::3xflag::MAPH-9: ccaacatgttccagtgaaAT(GGG) | This paper | N/A |
| CRISPR targeting sequence (+PAM): TBB-4::TagRFP-T::3xflag: TGATGAGCACGATCAAGATG(TGG) | This paper | N/A |
| Primers for CRISPR edits, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pJM54: GFP::3xflag RNAi vector | This paper | N/A |
| Plasmid: L4440: RNAi empty vector | Addgene | Cat#1654 |
| Plasmid: pJM74: Pmaph-9::myr::tagBFP2::unc-54UTR | This paper | N/A |
| Plasmid: pJM75: Ptmem-107::tmem-107::tagBFP2::unc-54UTR | This paper | N/A |
| Plasmid: pDD162: Peft-3::Cas9 + Empty sgRNA | Addgene | Cat#47549 |
| Plasmid: pJF250: ZF::GFP::3xFlag Empty repair template | 12 | N/A |
| Plasmid: pDD286: TagRFP-T::3xMyc Empty repair template | Addgene | Cat#70684 |
| Plasmid: pJM52: Peft-3::Cas9 + pcmd-1 sgRNA | This paper | N/A |
| Plasmid: pJM13: Peft-3::Cas9 + spd-5 sgRNA | 5 | N/A |
| Plasmid: pJM31 Peft-3::Cas9 + spd-2 sgRNA | 5 | N/A |
| Plasmid: pMT02 Peft-3::Cas9 + maph-9 sgRNA | This paper | N/A |
| Plasmid: pJM56: Peft-3::Cas9 + tbb-4 sgRNA | This paper | N/A |
| Plasmid: pJM53: ZF::GFP::3xFlag pcmd-1 repair template | This paper | N/A |
| Plasmid: pJM30: ZF::GFP::3xFlag spd-2 repair template | This paper | N/A |
| Plasmid: pJM17: ZF::GFP::3xFlag spd-5 repair template | This paper | N/A |
| Plasmid: pMT03: ZF::GFP::3xFlag maph-9 repair template | This paper | N/A |
| Plasmid: pMT04: tagRFP-T::3xMyc tbb-4 repair template | This paper | N/A |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| Prism9 | GraphPad | https://www.graphpad.com |
| NIS elements | Nikon Instruments | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
Highlights:
SPD-5 is physically and functionally central to the MTOC function of the centrosome
SPD-5 and PCMD-1 are maintained at the ciliary base in the absence of a centriole
Centriole-less pericentriolar material at the base of C. elegans cilia is an MTOC
A module of SPD-5 and γ-TuRC drives MTOC function at the ciliary base
Acknowledgements
We thank Karen Oegema, Tamara Mikeladze-Dvali, Dan Dickinson, and Bob Goldstein for providing strains and CRISPR advice and protocols. We also thank Kang Shen, Piali Sengupta, and members of the Feldman lab for helpful discussions about the manuscript. Some of the nematode strains used in this work were provided by the Caenorhabditis Genetic Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by an NIH New Innovator Award DP2GM119136-01 and R01GM136902 awarded to J.L.F. J.M. is supported by an American Heart Postdoctoral Fellowship. The project described was also supported, in part, by Award Number 1S10OD01227601 from the National Center for Research Resources (NCRR). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCRR or the National Institutes of Health.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li W, Yi P, Zhu Z, Zhang X, Li W, and Ou G (2017). Centriole translocation and degeneration during ciliogenesis in Caenorhabditis elegans neurons. The EMBO Journal 36, 2553–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serwas D, Su TY, Roessler M, Wang S, and Dammermann A (2017). Centrioles initiate cilia assembly but are dispensable for maturation and maintenance in C. elegans. The Journal of Cell Biology 216, 1659–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nechipurenko IV, Olivier-Mason A, Kazatskaya A, Kennedy J, McLachlan IG, Heiman MG, Blacque OE, and Sengupta P (2016). A Conserved Role for Girdin in Basal Body Positioning and Ciliogenesis. Developmental Cell 38, 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, and Agard DA (2012). Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nature Cell Biology 14, 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magescas J, Zonka JC, and Feldman JL (2019). A two-step mechanism for the inactivation of microtubule organizing center function at the centrosome. Elife 8, 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawo S, Hasegan M, Gupta GD, and Pelletier L (2012). Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nature Cell Biology 14, 1148–1158. [DOI] [PubMed] [Google Scholar]
- 7.Erpf AC, Stenzel L, Memar N, Antoniolli M, Osepashvili M, Schnabel R, Conradt B, and Mikeladze-Dvali T (2019). PCMD-1 Organizes Centrosome Matrix Assembly in C. elegans. Curr. Biol [DOI] [PubMed] [Google Scholar]
- 8.Lin T-C, Neuner A, and Schiebel E (2015). Targeting of γ-tubulin complexes to microtubule organizing centers: conservation and divergence. Trends in Cell Biology 25, 296–307. [DOI] [PubMed] [Google Scholar]
- 9.Oakley BR, Paolillo V, and Zheng Y (2015). γ-Tubulin complexes in microtubule nucleation and beyond. Molecular Biology of the Cell 26, 2957–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bobinnec Y, Fukuda M, and Nishida E (2000). Identification and characterization of Caenorhabditis elegans gamma-tubulin in dividing cells and differentiated tissues. Journal of Cell Science 113 Pt 21, 3747–3759. [DOI] [PubMed] [Google Scholar]
- 11.Hannak E, Oegema K, Kirkham M, Gönczy P, Habermann B, and Hyman AA (2002). The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. The Journal of Cell Biology 157, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sallee MD, Zonka JC, Skokan TD, Raftrey BC, and Feldman JL (2018). Tissue-specific degradation of essential centrosome components reveals distinct microtubule populations at microtubule organizing centers. PLoS Biol 16, e2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, and O'Connell KF (2004). Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Developmental Cell 6, 511–523. [DOI] [PubMed] [Google Scholar]
- 14.Hamill DR, Severson AF, Carter JC, and Bowerman B (2002). Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Developmental Cell 3, 673–684. [DOI] [PubMed] [Google Scholar]
- 15.Mikeladze-Dvali T, von Tobel L, Strnad P, Knott G, Leonhardt H, Schermelleh L, and Gonczy P (2012). Analysis of centriole elimination during C. elegans oogenesis. Development 139, 1670–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armenti ST, Lohmer LL, Sherwood DR, and Nance J (2014). Repurposing an endogenous degradation system for rapid and targeted depletion of C. elegans proteins. Development 141, 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Tang NH, Lara-Gonzalez P, Zhao Z, Cheerambathur DK, Prevo B, Chisholm AD, Desai A, and Oegema K (2017). A toolkit for GFP-mediated tissue-specific protein degradation in C. elegans. Development (Cambridge, England) 144, 2694–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo MY, Jang W, and Rhee K (2015). Integrity of the Pericentriolar Material Is Essential for Maintaining Centriole Association during M Phase. PLoS ONE 10, e0138905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez AD, and Feldman JL (2016). Microtubule-organizing centers: from the centrosome to non-centrosomal sites. Current Opinion in Cell Biology 44, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jana SC, Mendonça S, Machado P, Werner S, Rocha J, Pereira A, Maiato H, and Bettencourt-Dias M (2018). Differential regulation of transition zone and centriole proteins contributes to ciliary base diversity. Nature Cell Biology 20, 928–941. [DOI] [PubMed] [Google Scholar]
- 21.Clare DK, Magescas J, Piolot T, Dumoux M, Vesque C, Pichard E, Dang T, Duvauchelle B, Poirier F, and Delacour D (2014). Basal foot MTOC organizes pillar MTs required for coordination of beating cilia. Nat Commun 5, 4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nechipurenko IV, Berciu C, Sengupta P, and Nicastro D (2017). Centriolar remodeling underlies basal body maturation during ciliogenesis in Caenorhabditis elegans. Elife 6, R816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harterink M, Edwards SL, de Haan B, Yau KW, van den Heuvel S, Kapitein LC, Miller KG, and Hoogenraad CC (2018). Local microtubule organization promotes cargo transport in C. elegans dendrites. Journal of Cell Science 131, jcs223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, Jiang Q, and Zhang C (2014). The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. Journal of Cell Science 127, 4111–4122. [DOI] [PubMed] [Google Scholar]
- 25.O'Rourke SM, Carter C, Carter L, Christensen SN, Jones MP, Nash B, Price MH, Turnbull DW, Garner AR, Hamill DR, et al. (2011). A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS ONE 6, e16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McNally KLP, Fabritius AS, Ellefson ML, Flynn JR, Milan JA, and McNally FJ (2012). Kinesin-1 prevents capture of the oocyte meiotic spindle by the sperm aster. Developmental Cell 22, 788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, and Hyman AA (2017). The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 169, 1066–1077.e10. [DOI] [PubMed] [Google Scholar]
- 28.Feldman JL, and Priess JR (2012). A role for the centrosome and PAR-3 in the hand-off of MTOC function during epithelial polarization. Curr. Biol 22, 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang R, and Feldman JL (2015). SPD-2/CEP192 and CDK Are Limiting for Microtubule-Organizing Center Function at the Centrosome. Current Biology 25, 1924–1931. [DOI] [PubMed] [Google Scholar]
- 30.Pimenta-Marques A, Bento I, Lopes CAM, Duarte P, Jana SC, and Bettencourt-Dias M (2016). A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science 353, aaf4866–aaf4866. [DOI] [PubMed] [Google Scholar]
- 31.Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, et al. (2005). Functional genomics of the cilium, a sensory organelle. Current Biology 15, 935–941. [DOI] [PubMed] [Google Scholar]
- 32.Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, and Swoboda P (2005). Analysis of xbx genes in C. elegans. Development 132, 1923–1934. [DOI] [PubMed] [Google Scholar]
- 33.Fishman EL, Jo K, Nguyen QPH, Kong D, Royfman R, Cekic AR, Khanal S, Miller AL, Simerly C, Schatten G, et al. (2018). Author Correction: A novel atypical sperm centriole is functional during human fertilization. Nat Commun 9, 2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickinson DJ, Pani AM, Heppert JK, Higgins CD, and Goldstein B (2015). Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics 200, 1035–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahringer J (2006). Reverse genetics. WormBook. [Google Scholar]
- 37.Lambacher NJ, Bruel A-L, van Dam TJP, Szymanska K, Slaats GG, Kuhns S, McManus GJ, Kennedy JE, Gaff K, Wu KM, et al. (2016). TMEM107 recruits ciliopathy proteins to subdomains of the ciliary transition zone and causes Joubert syndrome. Nature Cell Biology 18, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
EBP-2 comets upon ciliated sensory neuron specific degradation of PCM proteins, related to Figure 4.
Yellow circle represents the position of the ciliary base. Contour of the phasmids are delimited by the yellow dashed line.
Data Availability Statement
This published article includes all datasets generated or analyzed during this study.