Abstract
Aberrant DNA methylation is considered to play a critical role in the chemoresistance of epithelial ovarian cancer (EOC). In this study, we explored the relationship between hypermethylation of the Mahogunin Ring Finger 1 (MGRN1) gene promoter and primary chemoresistance and clinical outcomes in high-grade serous ovarian cancer (HGSOC) patients. The MALDI-TOF mass spectrometry assays revealed a strong association between hypermethylation of the MGRN1 upstream region and platinum resistance in HGSOC patients. Spearman’s correlation analysis revealed a significantly negative connection between the methylation level of MGRN1 and its expression in HGSOC. In vitro analysis demonstrated that knockdown of MGRN1 reduced the sensitivity of cells to cisplatin and that expression of EGR1 was significantly decreased in SKOV3 cells with low levels of MGRN1 expression. Similarly, EGR1 mRNA expression was lower in platinum-resistant HGSOC patients and was positively correlated with MGRN1 mRNA expression. Kaplan-Meier analyses showed that high methylation of the MGRN1 promoter region and low expression of MGRN1 were associated with worse survival of HGSOC patients. In multivariable models, low MGRN1 expression was an independent factor predicting poor outcome. Furthermore, low expression of EGR1 was also been confirmed to be significantly related to the poor prognosis of HGSOC patients by Kaplan-Meier. The hypermethylation of the MGRN1 promoter region and low expression of MGRN1 were associated with platinum resistance and poor outcomes in HGSOC patients, probably by altering EGR1 expression.
Keywords: MGRN1, HGSOC, methylation, platinum resistance, prognosis
Introduction
Due to the lack of initial symptoms and sensitive screening methods, approximately 70% of women with epithelial ovarian cancer (EOC) are diagnosed at an advanced stage of disease (1, 2); EOC is the most lethal gynecologic malignancy in China (3, 4). Currently, the treatment strategy for patients with advanced EOC is platinum-based chemotherapy following primary debulking surgery (5–7). Although the majority of EOC patients respond well to first-line chemotherapy, most of these patients relapse and develop platinum resistance within 2 years (7, 8). In addition, nearly 20% of patients do not respond at the beginning of chemotherapy (9–11). Therefore, chemotherapy resistance has become an important cause of high mortality among EOC patients.
Resistance to chemotherapeutics, including intrinsic and acquired resistance, is based on highly complex and individually variable biological mechanisms (12). Abnormal methylation of DNA has been considered to play an important role during the development of acquired chemoresistance in EOC patients (13). However, there are currently very few studies about the effect of DNA methylation on the development of intrinsic resistance in EOC patients. In our previous study, reduced representation bisulfite sequencing (RRBS) analysis showed that the promoter region of the Mahogunin Ring Finger 1 (MGRN1) gene had abnormal hypermethylation in high-grade serous ovarian cancer (HGSOC) patients with platinum resistance (14). MGRN1 is an intracellular C3HC4 RING finger domain protein that exhibits E3 ubiquitin ligase activity and plays critical roles in the control of protein degradation (15). Ubiquitin-mediated proteolysis has played a crucial role in controlling protein level homeostasis and regulating the cell cycle, cell proliferation, apoptosis and DNA damage responses, which are involved in tumorigenesis, tumor development, prognosis and drug resistance (16). However, the role of MGRN1 in tumorigenesis, tumor progression, and drug responses is not currently well understood. A study by Dugué et al. suggests that hypomethylation of MGRN1 CpG sites in peripheral blood DNA is associated with the development of sporadic and familiar breast cancer (17).
Based on the results of RRBS, this study investigated the role of hypermethylation of the MGRN1 upstream region in platinum resistance in HGSOC patients. First, we examined the effects of MGRN1 methylation status and expression in ovarian tumor samples on the prognosis of HGSOC patients. Furthermore, we also investigated the possible role and mechanism of decreased MGRN1 expression in ovarian cancer cells in the response to cisplatin in vitro. To the best of our knowledge, this is the first study to investigate the role of the methylation status of the MGRN1 promoter region in platinum resistance in HGSOC patients.
Materials and Methods
Tissue Samples
A total of 96 HGSOC tissues were obtained from the Department of Gynecology at the Fourth Hospital, Hebei Medical University, China (November 2011–June 2015). The detailed clinicopathological features of the patients are summarized in Table 1 . The informed consent of each subject was obtained, and this study was approved by the Medical Ethics Committee of the Fourth Affiliated Hospital of Hebei Medical University. Based on the platinum-free interval (PFI), all the study participants were divided into a platinum-sensitive group (n=55) and a platinum-resistant group (n=41). PFI of less than 6 months is widely used to clinically define platinum-resistant disease, whereas a PFI greater than 6 months is often used to define platinum-sensitive disease (18). The participants were regularly followed-up for 5 years. Overall survival (OS) and progression-free survival (PFS) were used to assess the survival status of the patients.
Table 1.
Clinical characteristics of 96 HGSOC patients.
| Characteristics | Stage | Patients (n) | Recurrence | p | Survival | p |
|---|---|---|---|---|---|---|
| HR (95% CI) | HR (95% CI) | |||||
| Age | <50 Years | 34 | Reference | 0.71 | Reference | 0.32 |
| ≥50 Years | 62 | 1.12 (0.74–1.98) | 1.12 (0.87–3.11) | |||
| FIGO stage | I-II | 21 | Reference | Reference | ||
| III-IV | 75 | 8.73 (1.25–30.15) | 0.03 | 6.66 (1.13–20.12) | 0.02 | |
| Grade | 1 | 24 | Reference | Reference | ||
| 2 | 42 | 2.95 (1.75–6.95) | 0.05 | 1.40 (0.37–4.18) | 0.30 | |
| 3 | 30 | 5.40 (1.92–16.01) | 0.01 | 6.40 (0.47–13.90) | 0.02 | |
| tumor residual size |
0 | 25 | Reference | Reference | ||
| <1cm | 48 | 5.38 (1.95–11.93) | 0.01 | 5.59 (0.37–12.80) | 0.02 | |
| >1cm | 23 | 4.09 (1.75–8.99) | <0.01 | 4.34 (1.35–7.68) | <0.01 | |
| MGRN1 | High expression | 35 | Reference | Reference | ||
| Expression | Low expression | 61 | 1.48 (0.77–2.83) | 0.24 | 3.12 (1.31–7.11) | 0.01 |
| EGR1 | High expression | 22 | Reference | Reference | ||
| Expression | Low expression | 74 | 1.39 (0.81-2.97) | 0.03 | 1.66 (0.73-3.21) | 0.01 |
Genomic DNA Extraction and MALDI-TOF Mass Spectrometry
Of the 96 HGSOC samples, high-quality DNA from 26 HGSOC tissue samples was isolated using the Wizard Genomic DNA Purification Kit (Promega, Madison, Wisconsin), as described by the manufacturers. MALDI-TOF mass spectrometry (Sequenom, San Diego, California, U.S.) was used to detect the methylation level of the MGRN1 promoter region. This experiment was conducted at CapitalBio Co., Ltd. (Beijing, China). PCR primers were designed using Methprimer (http://www.urogene.org/methprimer). For each reverse primer, an additional T7 promoter tag for in vivo transcription was added, whereas a 10 m tag on the forward primer was used to adjust melting temperature differences. Mass spectra were obtained via MassARRAY Compact MALDI-TOF (Sequenom). The resultant methylation calls were analysed with EpiTyper software v1.0 (Sequenom) to generate quantitative results for each CpG site or an aggregate of multiple CpG sites.
RNA Extraction and Quantitative Real-Time Reverse Transcriptase-PCR (RT-qPCR)
Total RNA was isolated from 96 HGSOC tissue samples using the TRIzol-chloroform extraction method (Generay Biotech Co., Ltd., Shanghai, China), as described by the manufacturers. The total cDNA was reverse-transcribed using the Revert Aid First-Strand cDNA Synthesis Kit (Thermo Scientific, USA). The specific primers for the target genes that were used in RT-qPCR were designed using Primer Premier 5.0 and produced by Sangon Biotech Co., Ltd. (Shanghai, China). GAPDH was used as the housekeeping gene. The primer sequences were as follows: MGRN1 forward, 5’-TACAAAGACGATGCCGACAG-3’; MGRN1 reverse, 5’-GCCTGGCAGTAGATGGTGAT-3’; GAPDH forward, 5’-AATCCC ATCACCATCTTCCA-3’; and GAPDH reverse, 5’-TGGACTCCACGA CGTACTCA-3’. The reactions were run with the QuantiNova TMSYBR® Green PCR Kit (Qiagen, Hilden, Germany) in an Mx3005P instrument. The comparative quantification of each target gene was performed based on the cycle threshold (Ct) and normalized to GAPDH using the 2-ΔCt method.
MGRN1 Immunohistochemical (IHC) Study of the Clinical Samples
Of the 96 HGSOC samples, 52 paraffin-embedded HGSOC tissue samples collected in the pathology department of the Fourth Hospital of Hebei Medical University were used for immunohistochemical (IHC) staining of MGRN1. MGRN1 immunostaining was performed using a primary antibody, namely rabbit antihuman MGRN1 (RNF156, 1:500 dilution; Proteintech, China). Briefly, 4-μm thick sections were dewaxed in xylene and dehydrated through a graded series of ethanol. After blocking endogenous peroxidase and nonspecific binding, the sections were incubated overnight at 4°C with primary antibody and then with biotinylated secondary antibody and streptavidin-peroxidase complex. After the sections were washed in PBS, they were incubated with DAB reagent and counterstained with haematoxylin. Negative control sections were incubated with PBS instead of primary antibody. The sections were independently examined by two pathologists, who were blinded to the clinicopathological information. The immunoreactivity of MGRN1 was considered to be positive in tumor cells showing cytoplasmic staining without nuclear staining. The immunohistochemical staining was evaluated using a previously reported scoring method (19). If the final score was ≥ 4, the tumor was considered to have high MGRN1 expression, whereas a score < 4 indicated low MGRN1 expression.
Cell Culture
The human serous ovarian cancer SKOV3 cell line was purchased from the iCell Bioscience Inc. (Shanghai, China). The SKOV3 cell line was cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco; Thermo Fisher Scientific, Inc.) The medium was always supplemented with 10% (w/v) fetal bovine serum, 100 U penicillin, and 100 µg/L streptomycin (Gibco; Thermo Fisher Scientific, Inc.). The cells were maintained in a 95% humidified and 5% CO2 atmosphere at 37°C. Each vitro experiments were repeat three times biologically.
Stable Cell Lines
MGRN1 expression plasmids and lentiviral packaging reagents and shRNA were purchased from Genecopoeia Inc. (MD, USA). The designed three target sequences in the MGRN1 gene were 5′-GGAAACTACTTTGCTTCGCAC-3′ (shRNAa); 5′-GCGTGTTTCCAGTAGTCATC C-3′ (shRNAb) and 5′- GGCATTGAGAACAAGAACAAC-3′ (shRNAc). The most effective construct, recombinant plasmid inserted with MGRN1 gene shRNA expression vector shRNAa was selected for the study. A random sequence of shRNA (shNC) was used as the negative control. Transfection of the SKOV3 cell line was performed according to the manufacturer’s protocol. In brief, SKOV3 cells were seeded in a six-well plate at a density of 4×105 cells/mL in a volume of 2mL/well. When the SKOV3 cells reached 70–80% confluence, they were transfected with shRNA. Forty-eight hours later, culture medium containing 1 μg/ml puromycin (Genecopoeia) was used for selection for one week. The surviving cells were reseeded in fresh culture medium. Then, MGRN1 gene expression was observed under a fluorescence microscope, and the cells were subjected to RT-qPCR analysis to confirm the downregulation of MGRN1.
Detection of Changes in MGRN1 via Western Blotting
Proteins were isolated using RIPA lysis buffer. Total protein was extracted and BCA protein assay kit (Thermo) was used to quantify protein, followed by SDS-PAGE electrophoresis. The protein separated by electrophoresis was electro-transferred onto PVDF membranes, and blocked with 5% skim milk. Then, 1: 1000 rabbit anti-human MGRN1 antibody(RNF156, Proteintech, China) was added into the protein, and the solution was kept at 4°C overnight. The membranes were washed with TBST buffer 3 times (10 min each time). Anti-rabbit IgG was used as the secondary antibody (a dilution of 1: 5000; Proteintech, China), and the solution was cultured for 1h at 37°C. The membranes were washed with TBST for 3 times (10 min each time), and then with TBS for 10min. The antigen-antibody reaction was visualized by detection with Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE, USA), and β-actin (ab8226, Abcam, Cambridge, UK) served as internal reference.
Cell Viability Assays
The cells were inoculated in 96-well microplates in medium containing 10% fetal bovine serum and penicillin/streptomycin. After overnight incubation, the cells were treated with cisplatin (Pfizer), returned to the incubator for 24 h, and then analyzed. Cell Counting Kit-8 (CCK-8) was used to measure cell activity. Ten microliters of CCK-8 was added to each well and incubated for 3 h (37°C; 5% carbon dioxide). Then, the absorbance was measured at 492 nm with a microplate reader. Each vitro experiments were repeat three times biologically.
Apoptosis Assays
The Annexin V Apoptosis Detection kit I (BD Biosciences, Franklin Lakes, NJ, USA) was used to analyze the apoptosis of the SKOV3 cells. Briefly, the concentration of ovarian cancer cells at logarithmic growth phase was adjusted to 3×105/mL, and the cells were inoculated into 6-well plates at 2 ml/well. The cells were collected after 24 h of cisplatin treatment. After washing with PBS two times, the cells were resuspended in 100 μL of 1 × binding buffer and subsequently incubated with 5 μL of Annexin V staining solution at room temperature for 30 min in the dark. Then, 400 μL of 1 × binding buffer was added, and the cell apoptosis rate of each group was determined by flow cytometry.
RNA Sequencing
This experiment was conducted at Differential Gene Technology Co., Ltd. (Anhui, China). EdgeR was used to identify the top ten enriched annotation terms among the differentially expressed genes (1.5-fold in either direction, P<0.05) between the SKOV3 sh-NC group and the SKOV3 sh-MGRN1 group.
Statistical Analysis
The statistical analyses were performed using SPSS 21.0 (Chicago, IL, USA). The Wilcoxon rank sum test was used to compare the methylation level and mRNA expression of MGRN1 between the two groups. The χ2 test was used to compare the protein expression of MGRN1 in each group. Spearman correlation analysis was performed to analyze the correlation between MGRN1 expression and methylation status. Kaplan–Meier analysis and the Cox proportional hazard model were used to analyze the relationship between the methylation level and mRNA expression of MGRN1 and the prognosis of HGSOC. A t test was used to analyze the cell activity and apoptosis data.
Results
MGRN1Promoter Methylation Levels in the Platinum-Resistant Group and Platinum-Sensitive Group
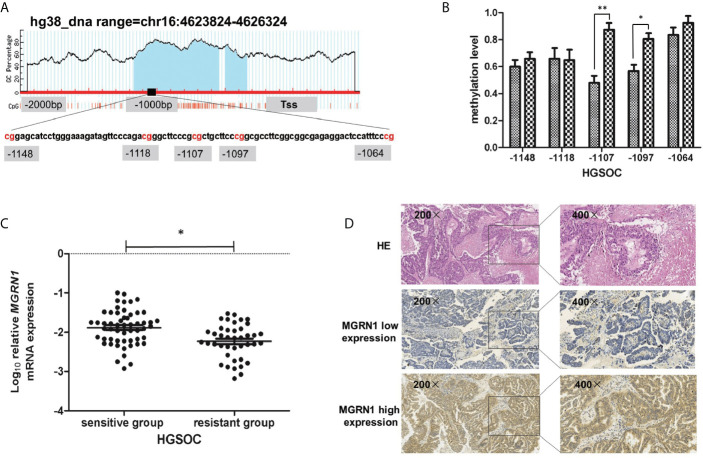
In our previous study, we used the RRBS assay to compare the differences in the genome-wide methylation patterns between 8 platinum-resistant epithelial ovarian cancer patients and 8 platinum-sensitive epithelial ovarian cancer patients. The results showed that a region from -1148 to -1064 within the promoter of MGRN1 was significantly hypermethylated in the platinum-resistant group compared to the platinum-sensitive group ( Figure 1A ). To further confirm the results of the RRBS assay, MALDI-TOF mass spectrometry was used to examine the methylation levels of this region in 12 platinum-resistant HGSOC patients and 14 platinum-sensitive HGSOC patients. In the present study, we tested the methylation levels of five CpG sites (-1148, -1118, -1107, -1097 and -1064 from the transcription start site) within this region. The analysis revealed that methylation levels of two CpG sites (-1107 and -1097) were significantly higher in the tumor tissues of platinum-resistant HGSOC patients than in those of platinum-sensitive HGSOC patients (P=0.01, 0.04, Figure 1B ).
Figure 1.
The methylation level of MGRN1 promoter was associated with platinum-resistant in HGSOC patients. (A) Schematic diagram of the abnormal methylation region (-1148 to -1064 upstream) in the promoter region of the MGRN1 gene. (B) The methylation of -1107 and -1097 CpG site were significantly hypermethylated in platinum-resistant HGSOC tissues compared with platinum-sensitive HGSOC tissues by MALDI-TOF Sequenom MassARRAY. (C) The mRNA expression of MGRN1 was lower in platinum-resistant HGSOC patients than platinum-sensitive HGSOC patients. (D) MGRN1 protein expression in HGSOC tissues. *P < 0.05, **P < 0.01.
MGRN1 Expression in the Platinum-Resistant Group and Platinum-Sensitive Group
RT-qPCR was used to determine the mRNA levels of MGRN1 in the tumor tissues from 41 platinum-resistant HGSOC patients and 55 platinum-sensitive HGSOC patients. The results showed that the mRNA level of MGRN1 in platinum-resistant HGSOC patients was 1.20-fold lower than that in platinum-sensitive HGSOC patients (P=0.01, Figure 1C ). Furthermore, IHC analysis was conducted to examine the protein expression of MGRN1 in 25 platinum-resistant HGSOC patients and 27 platinum-sensitive HGSOC patients. The analysis results showed that the frequency of positive MGRN1 expression in platinum-resistant HGSOC patients was significantly lower than that in platinum-sensitive HGSOC patients (P=0.02, Table 2 ). IHC staining showed that the MGRN1 protein was mainly expressed in the cytoplasm of HGSOC tissues ( Figure 1D ).
Table 2.
Comparison of MGRN1 protein expression between platinum-resistant HGSOC tissues and platinum-sensitive HGSOC tissues.
| MGRN1 expression | Resistant group n (%) | Sensitive group n (%) | P |
|---|---|---|---|
| High | 15(60.0) | 23(85.2) | 0.02 |
| Low | 10(40.0) | 4(14.8) |
Association Between MGRN1 mRNA Expression and Its Methylation Levels in HGSOC
Spearman’s correlation analysis showed that MGRN1 mRNA expression was significantly negatively correlated with the methylation level of the MGRN1 promoter region (average of -1107/-1097 CpGs: r=-.511, P=0.01). The results indicated that the hypermethylation of the MGRN1 promoter may be responsible for the downregulation of MGRN1 mRNA expression in HGSOC tissues.
Silencing of MGRN1 Expression in Serous Ovarian Cancer Cells by shRNA
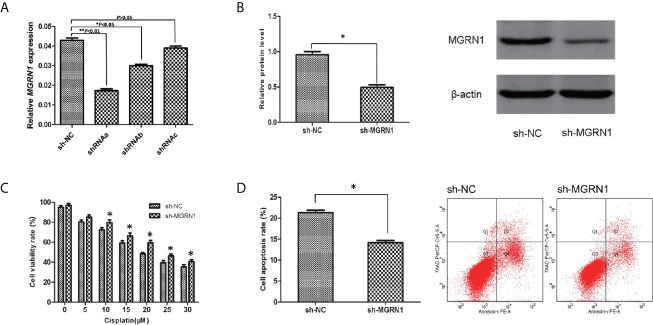
To investigate the role of MGRN1 expression in the sensitivity of serous ovarian cancer cells to cisplatin, SKOV3 cells were transfected with shRNAa-MGRN1, shRNAb-MGRN1, shRNAc-MGRN1 plasmid or shNC plasmid, respectively. After transfection for 48 hours, the expression of MGRN1 was confirmed by RT-qPCR. As shown in Figure 2A , shRNAa-MGRN1 could effectively decrease MGRN1 expression in SKOV3 cells, as compared with shNC groups. We also confirmed the expression of MGRN1 in shRNAa-MGRN1 group by Western blot. Therefore, we established MGRN1 stable knockdown cell lines using shRNAa-MGRN1 (P<0.05, Figures 2A, B ).
Figure 2.
The alteration of the sensitivity to cisplatin after a knockdown of MGRN1 expression in SKOV3 cells. (A) RT-qPCR assay showed the reduced expression of MGRN1 in shRNAa-MGRN1, shRNAb-MGRN1 and shRNAc-MGRN1 cells compared to shNC cells. (B) Western blot assay showed the reduced expression of MGRN1 in shRNAa-MGRN1 cells compared to shNC cells. (C) CCK-8 assays showed a significant increase in the proliferation rates in the shRNA-MGRN1 cells compared with the shNC cells after cisplatin treatment at several concentrations for 24h. (D) Flow cytometry showed that the apoptosis rate in the shRNA-MGRN1 cells was significantly lower than that in the shNC cells after exposure to cisplatin at the 10μM concentration for 24h. *P < 0.05, **P < 0.01. All the experiments were performed in triplicate.
Effect of MGRN1 Knockdown on the Cellular Response to Cisplatin
CCK-8 assays were used to compare the cell proliferation of the shRNA-MGRN1 group and shNC group. The proliferation rate was significantly higher in the shRNA-MGRN1 group than in the shNC group after treatment with cisplatin at several concentrations for 24 h (P<0.05, Figure 2C ). In addition, flow cytometry analysis demonstrated that the percentage of apoptotic cells in the shRNA-MGRN1 group was significantly lower than that in the shNC group after exposure to 10 μM cisplatin (P =0.03, Figure 2D ).
RNA-Seq Analysis Reveals That EGR1 Expression Is Differentially Regulated by MGRN1 in Ovarian Cancer Cells
To gain a better understanding of the differential regulation of transcription between the shNC transfection group and shRNA-MGRN1 transfection group, we performed an RNA-seq analysis of the total RNA harvested from the shNC group and shRNA-MGRN1 group. The top ten annotated, protein-coding genes that were differentially regulated in the shNC group compared to the shRNA-MGRN1 group are shown in Table 3 (P<0.05). Of these genes, EGR1 was the most differentially expressed, with 4.26-fold lower expression in the shRNA-MGRN1 group than in the shNC group (P=8.65E-07). EGR1 is a key gene involved in regulating cell proliferation and apoptosis in a variety of cancer tissues, and knockdown of EGR1 has been shown to promote resistance to cisplatin. Thus, we further validated the mRNA levels of EGR1 in cells and tissues by quantitative reverse-transcription PCR (RT-qPCR). The results showed that EGR1 mRNA expression was reduced by 72% in the SKOV3 shRNA-MGRN1 group compared with the SKOV3 shNC group (P<0.01, Figure 3A ). The mRNA level of EGR1 in 41 platinum-resistant HGSOC patients was 1.15-fold lower than that in 55 platinum-sensitive HGSOC patients (P=0.02, Figure 3B ). Spearman’s correlation analysis showed that MGRN1 mRNA expression was significantly positively correlated with EGR1 mRNA expression (r=-.379, P=0.01). Next, we used publicly available ovarian cancer data from The Cancer Genome Atlas (TCGA) series to validate our results. MGRN1 mRNA expression was also strongly associated with EGR1 mRNA expression in the ovarian cancer population (P<0.01).
Table 3.
Genes differentially expressed between shRNA-MGRN1 group and shNC group.
| Gene symbol | Description | Fold change | p |
|---|---|---|---|
| FOSB | FosB Proto-Oncogene, AP-1 Transcription Factor Subunit | -6.51 | 5.00E-08 |
| EGR1 | Early Growth Response 1 | -4.26 | 8.65E-07 |
| NGFR | Nerve Growth Factor Receptor | -5.53 | 0.000002 |
| ADM | Adrenomedullin | -3.25 | 0.00009 |
| KRT5 | Keratin 5 | -4.44 | 0.00010 |
| HK2 | Hexokinase 2 | -3.23 | 0.00011 |
| CES1P2 | Carboxylesterase 1 Pseudogene 2 | 8.9 | 0.00015 |
| DEPP1 | DEPP1 Autophagy Regulator | -4.26 | 0.00024 |
| STC1 | Stanniocalcin 1 | -3.6 | 0.00026 |
| EDIL3 | EGF Like Repeats And Discoidin Domains 3 | -4.71 | 0.00033 |
Figure 3.
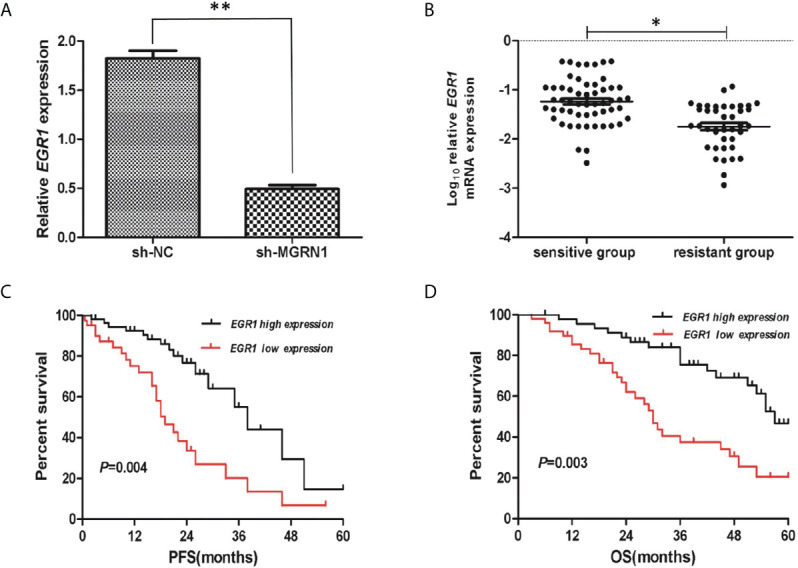
The low mRNA expression of EGR1 was associated with platinum-resistant in HGSOC patients. (A) RT-qPCR showed the reduced expression of EGR1 in sh-MGRN1 cells compared to shNC cells. (B) The mRNA expression of EGR1 in platinum-resistant HGSOC patients and platinum-sensitive HGSOC patients. (C, D) Kaplan-Meier analysis of PFS and OS according to the EGR1 mRNA expression in HGSOC patients. *P < 0.05, **P < 0.01.
Hypermethylation and Low Expression of MGRN1 Correlate With Poor Prognosis of HGSOC Patients
A total of 26 HGSOC patients were divided into hypermethylation and hypomethylation groups based on the median value of MGRN1 methylation. Kaplan-Meier survival analysis revealed that the MGRN1 hypermethylation group exhibited lower PFS and OS of HGSOC patients compared to the MGRN1 hypomethylation group (P=0.04, Figure 4A ; P=0.03, Figure 4B ). Next, we investigated the correlation between MGRN1 mRNA expression and the clinical outcomes of HGSOC patients. A total of 96 HGSOC patients were divided into low- and high-expression groups based on the median value of MGRN1 mRNA expression. Kaplan-Meier analysis showed that compared with high MGRN1 expression, low MGRN1 expression was associated with significantly lower PFS and OS of HGSOC patients (P=0.02, Figure 4C ; P=0.01, Figure 4D ). After adjusting for other prognostic factors (age, stage, grade and tumor residual size), low MGRN1 expression was also significantly associated with shorter OS ( Table 1 , P=0.01), demonstrating that MGRN1 expression was an independent predictor of poorer clinical outcomes in HGSOC patients. Furthermore, Kaplan-Meier analysis demonstrated that the low EGR1 expression group was associated with worse prognosis compared to the high EGR1 expression group (P<0.01, Figures 3C, D ). Multivariable analysis showed that EGR1 expression was an independent predictor of worse clinical outcomes in HGSOC patients ( Table 1 , P=0.03, 0.01).
Figure 4.
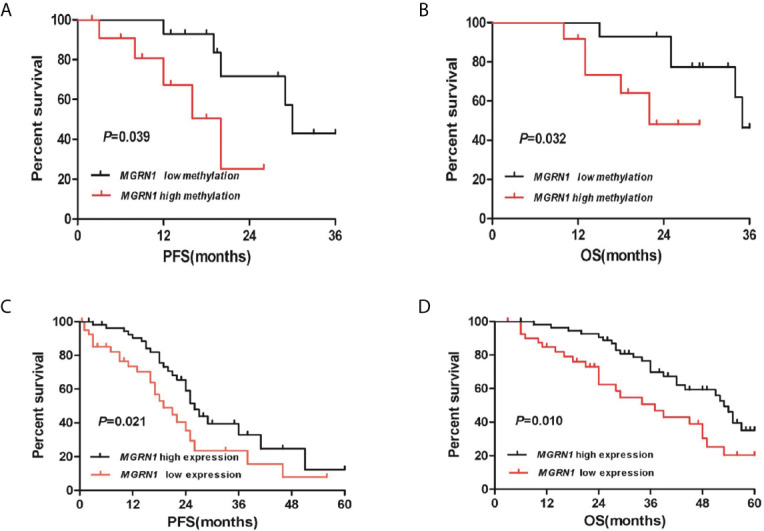
The high methylation of MGRN1 and its low mRNA expression are associated with poor survival in HGSOC patients. (A, B) Kaplan-Meier analysis of PFS and OS according to the MGRN1 methylation level in 26 HGSOC patients. (C, D) Kaplan-Meier analysis of PFS and OS according to the MGRN1 mRNA expression in 96 HGSOC patients.
Discussion
In this study, we confirmed that MGRN1 gene promoter hypermethylation is associated with platinum resistance in patients with HGSOC based on the following findings: 1) the upstream region of MGRN1 was significantly hypermethylated in the cancer tissues of platinum-resistant patients with HGSOC, 2) the lower expression of MGRN1 due to hypermethylation of the upstream region was associated with clinical outcomes of patients with HGSOC, 3) knockdown of MGRN1 expression could desensitize SKOV3 ovarian cancer cells to cisplatin, 4) knockdown of MGRN1 expression in SKOV3 cells could significantly reduce EGR1 mRNA expression, which significantly correlated with the treatment outcomes in cisplatin-treated cancer patients, and 5) EGR1 expression in the cancer tissues of platinum-resistant patients was significantly lower than that of platinum-sensitive patients and was related to the clinical prognosis of patients with HGSOC.
Based on the results of previous RRBS analysis, we found that the methylation level of the MGRN1 upstream region (-1148 to -1064) was significantly higher in the platinum-resistant HGSOC patients. Mass spectrometry further analysis showed that hypermethylation of the MGRN1 promoter region was associated with platinum resistance in HGSOC patients. We also discovered that the expression levels of MGRN1 mRNA and protein in platinum-resistant HGSOC patients were significantly lower than those in platinum-sensitive HGSOC patients. Correlation analysis indicated that the methylation level of the MGRN1 promoter region was associated with MGRN1 mRNA expression. Furthermore, knockdown of MGRN1 expression could increase proliferation and decrease apoptosis in SKOV3 cells challenged with cisplatin. These findings suggested that lower MGRN1 expression due to hypermethylation of its promoter region might induce platinum resistance in HGSOC.
MGRN1, an E3 ubiquitin ligase of the Really Interesting New Gene (RING) finger family, is involved in many biological and cellular mechanisms (20). However, there is no study of the role of MGRN1 in chemotherapy resistance in cancer patients to date. In the current study, microarray analysis of total RNA showed that knockdown of MGRN1 expression in SKOV3 cells resulted in significant downregulation of multiple genes, including early growth response protein 1 (EGR1). EGR1 is a transcription factor that can be induced by a variety of stimuli or stressors, including growth factors, hormones, ionizing radiation, and chemotherapy drugs (21–23), and plays essential roles in cell proliferation and apoptosis (24–26). Knockdown of EGR1 expression can decrease cisplatin-induced apoptosis in a variety of cancer cells (27–29), while overexpression of this gene sensitizes ovarian cancer cells to cisplatin-induced apoptosis (28). He et al. found that EGR1 expression levels were significantly higher in ovarian cancer tissues with low ERCC1 expression than in ovarian cancer tissues with high ERCC1 expression, suggesting that EGR1 expression is positively correlated with potential cisplatin-sensitive ovarian cancer, since ERCC1 is widely accepted as a biomarker of platinum resistance (28). In our study, it was also observed that the expression of EGR1 in HGSOC patients with platinum resistance was significantly downregulated and was positively correlated with the expression of MGRN1. Although the mechanism by which MGRN1 regulates EGR1 is still unclear, a strong association between MGRN1 mRNA expression and EGR1 mRNA expression was noted in the TCGA ovarian cancer dataset ( Additional File 1 and Figure S1 ). Therefore, we speculate that MGRN1 may affect the platinum resistance of ovarian cancer by regulating the expression of EGR1. Of course, the molecular mechanism by which MGRN1 regulates EGR1 requires further study.
More importantly, Kaplan-Meier analyses showed that hypermethylation of the MGRN1 promoter region was associated with worse survival of HGSOC patients in this study. Further, multivariable analysis also indicated that HGSOC patients with lower MGRN1 mRNA expression have a worse prognosis than those with higher MGRN1 mRNA expression, which suggested the prognostic value of MGRN1 methylation for HGSOC patients due to the negative correlation between MGRN1 methylation and its expression. Here, we also confirmed that patients with low EGR1 mRNA expression had significantly shorter PFS and OS than those with high EGR1 mRNA expression. Of note, data derived from TCGA showed that the expression of EGR1 significantly correlated with the treatment outcomes in cisplatin-treated cancer patients (30). Therefore, EGR1 expression may be a useful marker for predicting the clinical prognosis of HGSOC patients.
In summary, our study demonstrated that the hypermethylation of MGRN1 is an independent marker of poor prognosis in HGSOC patients and may also be predictive of platinum resistance in HGSOC patients. Considering that DNA methylation may be used as a molecular marker for ovarian cancer chemotherapy, we believe that our findings warrant confirmation in a larger patient cohort and could facilitate patient selection for chemotherapy.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the institute Medical Ethics Committee of Hebei Medical University, Fourth Hospital.
Author Contributions
Administrative support: X-FL, SK, and YL. Provision of study materials or patients: X-FL and H-YS. Collection and assembly of data: X-FL and TH. Data analysis and interpretation: X-FL, H-BZ, and Y-JT. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly acknowledge several doctors in the Department of Obstetrics and Gynecology, the Fourth Affiliated Hospital of Hebei Medical University, China, for their assistance in recruiting the study subjects.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.659254/full#supplementary-material
The association between MGRN1 mRNA expression and EGR1 mRNA expression in the TCGA ovarian cancer dataset.
References
- 1. Sato S, Itamochi H. Neoadjuvant Chemotherapy in Advanced Ovarian Cancer: Latest Results and Place in Therapy. Ther Adv Med Oncol (2014) 6(6):293–304. 10.1177/1758834014544891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buono NL, Morone S, Parrotta R, Giacomino A, Funaro A. Ectoenzymes in Epithelial Ovarian Carcinoma: Potential Diagnostic Markers and Therapeutic Targets: Intech (2012). 10.5772/28428 [DOI] [Google Scholar]
- 3. Jia J, Wang Z, Cai J, Zhang Y. PMS2 Expression in Epithelial Ovarian Cancer is Posttranslationally Regulated by Akt and Essential for Platinum-Induced Apoptosis. Tumor Biol (2016) 37(3):1–11. 10.1007/s13277-015-4143-2 [DOI] [PubMed] [Google Scholar]
- 4. Chen J, Wang L, Matyunina LV, Hill CG, McDonald JF. Overexpression of miR-429 Induces Mesenchymal-to-Epithelial Transition (MET) in Metastatic Ovarian Cancer Cells. Gynecol Oncol (2011) 121(1):200–5. 10.1016/j.ygyno.2010.12.339 [DOI] [PubMed] [Google Scholar]
- 5. Borley J, Wilhelm-Benartzi C, Brown R, Ghaem-Maghami S. Does Tumour Biology Determine Surgical Success in the Treatment of Epithelial Ovarian Cancer? A Systematic Literature Review. Br J Cancer (2012) 107(7):1069–74. 10.1038/bjc.2012.376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leary A, Cowan R, Chi D, Kehoe S, Nankivell M. Primary Surgery or Neoadjuvant Chemotherapy in Advanced Ovarian Cancer: The Debate Continues…. Am Soc Clin Oncol Educ Book (2016) 35:153–62. 10.14694/EDBK_160624 [DOI] [PubMed] [Google Scholar]
- 7. Mueller JJ, Zhou QC, Iasonos A, O’Cearbhaill RE, Alvi FA, Haraki AE, et al. Neoadjuvant Chemotherapy and Primary Debulking Surgery Utilization for Advanced-Stage Ovarian Cancer at a Comprehensive Cancer Center. Gynecol Oncol (2016) 140(3):436–42. 10.1016/j.ygyno.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rochet N, Kieser M, Sterzing F, Krause S, Lindel K, Harms W, et al. Phase II Study Evaluating Consolidation Whole Abdominal Intensity-Modulated Radiotherapy (IMRT) in Patients With Advanced Ovarian Cancer Stage FIGO Iii - The Ovar-IMRT-02 Study. BMC Cancer (2011) 11(1):1–7. 10.1186/1471-2407-11-41. 11, 1(2011-01-28). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mantia-Smaldone GM, Edwards RP, Vlad AM. Targeted Treatment of Recurrent Platinum-Resistant Ovarian Cancer: Current and Emerging Therapies. Cancer Manage Res (2011) 3(default):25–38. 10.2147/CMR.S8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siddiqui GK, Maclean AB, Elmasry K, Fong AWT, Morris RW, Rashid M, et al. Immunohistochemical Expression of VEGF Predicts Response to Platinum Based Chemotherapy in Patients With Epithelial Ovarian Cancer. Angiogenesis (2011) 14(2):155–61. 10.1007/s10456-010-9199-4 [DOI] [PubMed] [Google Scholar]
- 11. Colombo PE, Manoir SD, Orsett B, Rui BG, Theillet C. Ovarian Carcinoma Patient Derived Xenografts Reproduce Their Tumor of Origin and Preserve an Oligoclonal Structure. Oncotarget (2015) 6(29):28327–40. 10.18632/oncotarget.5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu Y, Zhu Y, Ma Y, Fei L, Zhu B. Genomic Insights Into Intrinsic and Acquired Drug Resistance Mechanisms in Achromobacter Xylosoxidans. Antimicrobial Agents Chemother (2015) 59(2):1152. 10.1128/aac.04260-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilting RH, Dannenberg JH. Epigenetic Mechanisms in Tumorigenesis, Tumor Cell Heterogeneity and Drug Resistance. Drug Resistance Updates (2012) 15(1-2):21–38. 10.1016/j.drup.2012.01.008 [DOI] [PubMed] [Google Scholar]
- 14. Hua T, Kang S, Li X, Tian Y, Li Y. DNA Methylome Profiling Identifies Novel Methylated Genes in Epithelial Ovarian Cancer Patients With Platinum Resistance. J Obstetr Gynaecol Res (2021) 47(3):1031–9. 10.1111/jog.14634 [DOI] [PubMed] [Google Scholar]
- 15. Phan LK, Feng L, Leduc CA, Chung WK, Leibel RL. The Mouse Mahoganoid Coat Color Mutation Disrupts a Novel C3HC4 RING Domain Protein. J Clin Invest (2002) 110(10):1449–59. 10.1172/JCI16131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang L, Chen J, Huang X, Zhang E, He J, Cai Z. Novel Insights Into E3 Ubiquitin Ligase in Cancer Chemoresistance. Am J Med Sci (2017) 355(4):368–76. 10.1016/j.amjms.2017.12.012. S0002962917306900. [DOI] [PubMed] [Google Scholar]
- 17. Dugué P-A, Milne RL, Southey MC. A Prospective Study of Peripheral Blood DNA Methylation at RPTOR, MGRN1 and RAPSN and Risk of Breast Cancer. Breast Cancer Res Treat (2017) 161(1):181–3. 10.1007/s10549-016-4032-4 [DOI] [PubMed] [Google Scholar]
- 18. Bookman MA, Tyczynski JE, Espirito JL, Wilson TW, Fernandes AW. Impact of Primary Platinum-Free Interval and BRCA1/2 Mutation Status on Treatment and Survival in Patients With Recurrent Ovarian Cancer. Gynecol Oncol (2017) S0090825817307928. 10.1016/j.ygyno.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 19. Liu YB, Mei Y, Tian ZW, Long J, Luo CH, Zhou HH. Downregulation of RIF1 Enhances Sensitivity to Platinum-Based Chemotherapy in Epithelial Ovarian Cancer (EOC) by Regulating Nucleotide Excision Repair (Ner) Pathway. Cell Physiol Biochem (2018) 46(5):1971–84. 10.1159/000489418 [DOI] [PubMed] [Google Scholar]
- 20. Upadhyay A, Amanullah A, Chhangani D, Mishra R, Prasad A, Mishra A. Mahogunin Ring Finger-1 (MGRN1), a Multifaceted Ubiquitin Ligase: Recent Unraveling of Neurobiological Mechanisms. Mol Neurobiol (2016) 53(7):4484–96. 10.1007/s12035-015-9379-8 [DOI] [PubMed] [Google Scholar]
- 21. Gregg J, Fraizer G. Transcriptional Regulation of EGR1 by EGF and the ERK Signaling Pathway in Prostate Cancer Cells. Genes Cancer (2011) 2(9):900–9. 10.1177/1947601911431885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim HR, Kim YS, Yoon JA, Lyu SW, Shin H, Lim HJ, et al. Egr1 is Rapidly and Transiently Induced by Estrogen and Bisphenol A Via Activation of Nuclear Estrogen Receptor-Dependent ERK1/2 Pathway in the Uterus. Reprod Toxicol (2014) 50:60–7. 10.1016/j.reprotox.2014.10.010 [DOI] [PubMed] [Google Scholar]
- 23. Ko H, Kim JM, Kim S, Shim SH, Ha CH, Chang HI. Induction of Apoptosis by Genipin Inhibits Cell Proliferation in AGS Human Gastric Cancer Cells Via Egr1/p21 Signaling Pathway. Bioorg Med Chem Lett (2015) 25(19):4191–6. 10.1016/j.bmcl.2015.08.005 [DOI] [PubMed] [Google Scholar]
- 24. Gabriella MA, Giulia P, Daniela T, Gerardo LR, Elisa B, Laura M, et al. Coordinated Sumoylation and Ubiquitination Modulate EGF Induced EGR1 Expression and Stability. PLoS One (2011) 6(10):e25676. 10.1371/journal.pone.0025676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ning ZC, Shi JD, Zhou Y, Xiao-Ling DU, Zhang J. Egr1 Promotes Proliferation in Prostatic Hyperplasia Epithelial Bph-1 Cells. Chin J Biochem Mol Biol (2013) 29(8):751–8. 10.13865/j.cnki.cjbmb.2013.08.003 [DOI] [Google Scholar]
- 26. Wang L, Sun H, Wang X, Hou N, Zhao L, Tong D, et al. EGR1 Mediates miR-203a Suppress the Hepatocellular Carcinoma Cells Progression by Targeting HOXD3 Through EGFR Signaling Pathway. Oncotarget (2016) 7(29):45302–16. 10.18632/oncotarget.9605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao. Groβ and its Downstream Effector EGR1 Regulate Cisplatin-Induced Apoptosis in WHCO1 Cells. Oncol Rep (2011) 25(4):1031–7. 10.3892/or.2011.1163 [DOI] [PubMed] [Google Scholar]
- 28. He J, Yu JJ, Xu Q, Wang L, Zheng JZ, Liu LZ, et al. Downregulation of ATG14 by EGR1-MIR152 Sensitizes Ovarian Cancer Cells to Cisplatin-Induced Apoptosis by Inhibiting Cyto-Protective Autophagy. Autophagy (2015) 11(2):373–84. 10.1080/15548627.2015.1009781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parra E. Increased Expression of p21Waf1/Cip1 and JNK With Costimulation of Prostate Cancer Cell Activation by a Sirna Egr-1 Inhibitor. Oncol Rep (2013) 30(2):911–6. 10.3892/or.2013.2503 [DOI] [PubMed] [Google Scholar]
- 30. Dilruba S, Grondana A, Schiedel AC, Ueno NT, Kalayda GV. Non-Phosphorylatable PEA-15 Sensitises SKOV-3 Ovarian Cancer Cells to Cisplatin. Cells (2020) 9(2):515. 10.3390/cells9020515 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The association between MGRN1 mRNA expression and EGR1 mRNA expression in the TCGA ovarian cancer dataset.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.