Abstract
Background
Serological tests detecting severe acute respiratory syndrome coronavirus−2 (SARS-CoV-2) are widely used in seroprevalence studies and evaluating the efficacy of the vaccination program. Some of the widely used serological testing techniques are enzyme-linked immune-sorbent assay (ELISA), chemiluminescence immunoassay (CLIA), and lateral flow immunoassay (LFIA). However, these tests are plagued with low sensitivity or specificity, time-consuming, labor-intensive, and expensive. We developed a serological test implementing flow-through dot-blot assay (FT-DBA) for SARS-CoV-2 specific IgG detection, which provides enhanced sensitivity and specificity while being quick to perform and easy to use.
Methods
SARS-CoV-2 antigens were immobilized on nitrocellulose membrane to capture human IgG, which was then detected with anti-human IgG conjugated gold nanoparticle (hIgG-AuNP). A total of 181 samples were analyzed in-house. Within which 35 were further evaluated in US FDA-approved CLIA Elecsys SARS-CoV-2 assay. The positive panel consisted of RT-qPCR positive samples from patients with both <14 days and >14 days from the onset of clinical symptoms. The negative panel contained samples collected from the pre-pandemic era dengue patients and healthy donors during the pandemic. Moreover, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of FT-DBA were evaluated against RT-qPCR positive sera. However, the overall efficacies were assessed with sera that seroconverted against either nucleocapsid (NCP) or receptor-binding domain (RBD).
Results
In-house ELISA selected a total of 81 true seropositive and 100 seronegative samples. The sensitivity of samples with <14 days using FT-DBA was 94.7%, increasing to 100% for samples >14 days. The overall detection sensitivity and specificity were 98.8% and 98%, respectively, whereas the overall PPV and NPV were 99.6% and 99%. Moreover, comparative analysis between in-house ELISA assays and FT-DBA revealed clinical agreement of Cohen’s Kappa value of 0.944. The FT-DBA showed sensitivity and specificity of 100% when compared with commercial CLIA kits.
Conclusion
The assay can confirm past SARS-CoV-2 infection with high accuracy within 2 minutes compared to commercial CLIA or in-house ELISA. It can help track SARS-CoV-2 disease progression, population screening, and vaccination response. The ease of use of the assay without requiring any instruments while being semi-quantitative provides the avenue of its implementation in remote areas around the globe, where conventional serodiagnosis is not feasible.
Keywords: COVID-19, SARS-CoV-2, dot-blot immunoassay, AuNP, gold nanoparticle, serosurveillance, nucleocapsid, receptor binding domain
Plain Language Summary
Dot blot assay provided comparable sensitivity and specificity to SARS-CoV-2 NCP-IgG and SARS-CoV-2 RBD-IgG ELISA in both <14 days and >14 days.
The Dot-blot assay intensity scales directly correlated with the SARS-CoV-2 NCP-IgG and SARS-CoV-2 RBD-IgG antibody titers.
The flow-through dot-blot assay showed 100% similarity in sensitivity and specificity compared with Elecsys SARS-CoV-2 assay (NCP).
Introduction
A newly discovered coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) triggered the pneumonia outbreak in China’s Hubei province in December 2019.1 The World Health Organization (WHO) termed the infection COVID-19 (coronavirus disease 2019), which has now spread beyond China and has become a full-blown pandemic.2 To combat the virus’s spread, affected countries have adopted numerous public health measures such as isolation, quarantine, regional lockdown, social distancing, restriction on people’s movement, and limiting local and international traveling. Despite these preventive measures, the disease surges across countries, with more than 170 million confirmed cases with around 3.7 million deaths to date (June 9, 2021).3
SARS-CoV-2 has a higher transmission rate than the previous two coronaviruses: severe acute respiratory syndrome coronavirus (SARS-CoV) and middle east respiratory syndrome coronavirus (MERS-CoV).4 Ongoing research has emphasized finding therapeutic interventions and preventive vaccination, with limited success.5 Early diagnosis is critical for successfully containing this contagious outbreak.6 The US Food and Drug Administration (FDA) is issuing emergency use authorization (EUA) for different categories of in vitro diagnostic tests to increase testing capacity.7 While nucleic acid and antigen-based tests can detect active infection, low viral load, and variation in test sensitivity issues increase the risk of false-negative results, limiting their use.8,9 In addition, many COVID-19 victims are asymptomatic with a viral load lower than symptomatic individuals.10 These hinder these tests’ usefulness and make the epidemiological evaluation of the disease complex.11,12 Moreover, amidst mass COVID-19 vaccination, it is essential to monitor the antibody dynamics for COVID-19 containment.13,14
WHO recommends systemic serosurveys to determine the whole disease spectrum implemented by many countries around the world.13,15–19 Other than assessing risk and prevalence, serology testing is vital for contact tracing, detecting immune response against the virus, identifying potential plasma donors, in some instances to be used in adjunct with a molecular diagnosis, and evaluating the success of vaccination program in place.14,20–23
The widely used tests for serology-based diagnosis of SARS-CoV-2 include enzyme-linked immunosorbent assay (ELISA), automated chemiluminescence assay (CLIA), neutralization assay, and rapid immunoassay.20 Except for rapid tests, all assay systems require infrastructure, longer testing time, and qualified staff to conduct the tests and interpret the results, rendering these assays inadequate when a large number of testing are required immediately.24,25 These limitations call for an urgent need for easy and affordable rapid point-of-care testing (POCT). The present study reports developing such a rapid POCT antibody test, based on membrane immune-concentration flow-through principle, for SARS-CoV-2 specific IgG detection. There are numerous reports on rapid lateral flow immunochromatographic assay systems for SARS-CoV-2 antibody detection.7,26–29 However, LFIA systems have a higher possibility of the false-negative signal depending on the immobilized analyte concentration compared to the flow-through system.30,31
Here, we have developed a flow-through detection system with biofunctionalized colloidal gold nanoparticles (AuNP). Among numerous reporter molecules, AuNPs have wide applications due to their remarkable optical and physicochemical properties.32–35 AuNP-conjugated biomolecules are exceptional for their simplicity and high contrast visualization when coupled with a rapid immunoassay system.36–38
This report will detail how we developed and validated a rapid dot-blot serological assay to detect SARS-CoV-2 specific IgG in human serum using seropositive and seronegative samples, characterized by other available detection systems. Moreover, compared with nucleocapsid (NCP) or receptor-binding domain (RBD) ELISA specific for SARS-CoV-2, an additive sensitivity was observed in our assay system due to co-immobilization of both antigens to capture a wide range of antibodies.
Materials and Methods
Design Concept
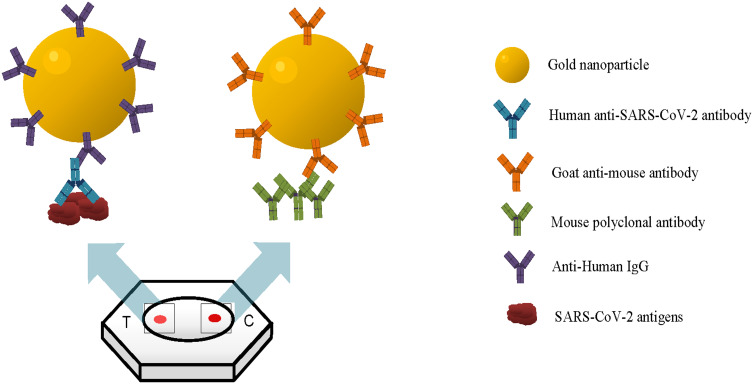
Rapid flow-through dot-blot immunoassay (FT-DBA) is a qualitative immunoassay to detect the presence of SARS-CoV-2 specific IgG antibodies in human serum. It utilizes the solid phase capture technique in a membrane immune-concentration flow-through system (Figure 1). The test device is a plastic cassette that contains a combo made up of an absorbent pad with nitrocellulose (N.C.) membrane with its active side on top, visible through a circular window. The test media (active side of the N.C. membrane) has two adjacent dots, designated T (test) and C (control) position to indicate test result and test kit validity, respectively. A mixture of SARS-CoV-2 antigens is immobilized on the test dot, and the control dot contains immobilized mouse IgG. Thus, the SARS-CoV-2 specific antibody, if present in the serum, is captured on the N.C. test dot, which becomes visible after the addition of AuNP conjugate.
Figure 1.
Design concept of the rapid flow-through dot-blot immunoassay (FT-DBA). The gold nanoparticle (AuNP) with antibody conjugated onto the surface can bind to the SARS-CoV-2 antigen. At the same time, the AuNP with goat anti-mouse within the same solution can bind to mouse polyclonal antibody, which is the control dot.
Reagents and Material
SARS-CoV and SARS-CoV-2 specific NCP, envelope (E), spike S1, spike S2, and RBD recombinant proteins were purchased from The Native Antigen (UK, Kidlington), M.P. Biomedicals (California, USA), Sino Biological (China), Fapon Biotech Inc. (China), and Creative Diagnostics (USA). Gold colloids (particle size: 10nm and 40nm) were purchased from Bhat Biotech Ltd. (India) and BBI Solutions (U.K.). PBS (Phosphate-buffered saline) tablets (pH 7.4), Tris-Buffered Saline (TBS) pH 7.2, glycerol, and sodium chloride (NaCl) were purchased from Thermo Fisher Scientific (USA). Cold-water gelatin was purchased from Sigma-Aldrich. Gold dilution and stabilization buffer (Bhat Biotech Ltd.), mouse IgG antibody (Fapon Biotech), goat-anti-mouse IgG (Fapon Biotech) and, goat-anti-human IgG (Fapon Biotech) were also purchased. N.C. membranes were purchased from Ken Biotech (China), Bhat Biotech Ltd. (India), and Sartorius (France) to determine the optimal support matrix for the immunoassay. Other materials (such as plastic cassettes) were purchased from Bhat Biotech (India) and Changzhou Dengfeng (China).
Optimization of AuNP Conjugate
Gold nanoparticles have already been accepted as a remarkable diagnostic tool worldwide.39 Therefore, the research team employed AuNP conjugated with anti-human IgG to detect SARS-CoV-2 specific IgG in human serum. Anti-human IgG-AuNP and anti-mouse IgG-AuNP conjugates were prepared according to the protocol described by Oliver C. et al.40 Briefly, two different sized (10 nm and 40 nm) gold colloids were evaluated and to determine the optimal concentration of both proteins for conjugation, aliquots of the anti-human IgG/anti-mouse IgG solutions (5 µg/mL, 10 µg/mL, 15 µg/mL and 20 µg/mL) in phosphate-buffered saline (PBS) were prepared. About 1 mL of gold colloid solution was added to each aliquot, and the tubes were incubated for 15 mins at room temperature. The minimum amount of anti-human IgG/anti-mouse IgG required to stabilize the conjugates was determined by assessing color change and agglomeration. The conjugates were then stabilized using 1% cold-water gelatin. Excess antibodies were removed using glycerol gradient, and the conjugates were dialyzed against TBS for 1 hr at room temperature. The final preparation was diluted with TBS and 1% cold-water gelatin and stored at 4 °C.
Selection of Control Samples for Assay Development
In this assay’s development and optimization, two SARS-CoV-2 positive and two negative control sera were utilized. Clinical symptoms, RT-qPCR confirmation, serostatus verification with in-house and commercial chemiluminescence assay (ROCHE, Elecsys Anti SARS-CoV-2) were considered control selection criteria.41 Besides, the antibody kinetics of positive control individuals were analyzed longitudinally to avoid spectrum bias.42
Specimen for Clinical Validation
The clinical performance of rapid FT-DBA has been evaluated with three panels of serum samples (n=181). Two sera panels comprise single and multiple collections of SARS-CoV-2 positive serum samples (n=81) from forty-five RT-qPCR confirmed individuals with clinical signs and symptoms of COVID-19. Panel 1 consists of twenty RT-qPCR positive samples that have been collected within two weeks from the onset of symptoms. Panel 2 (n=61) samples were also from RT-qPCR positive individuals with symptom onset of >14 days. Panel 3 samples were negative samples (n=100) collected during, i) pre-pandemic sera from healthy donors (n=40), ii) April to June 2020 from RT-qPCR negative individuals (n=36), and iii) pre-pandemic dengue-positive patients (n=24). The panels were characterized with SARS-CoV-2 IgG ELISA described by Sil et al against SARS-CoV-2 antigens: NCP and RBD.41–43 Comparative analyses were carried out with these samples, between in-house ELISA assays and the kit developed in this work. Moreover, according to FDA guidelines, the seropositive and seronegative samples based on in-house ELISA results were tested with the developed assay.
We analyzed 35 samples from these 181 in-house ELISA characterized sera by US FDA approved commercial Elecsys SARS-CoV-2 assay. Among these 35 samples, 15 (>21 days after being RT-qPCR positive) were previously identified as seropositive in both NCP-IgG and RBD-IgG ELISA, and 20 were seronegative NCP-IgG and RBD-IgG. All the samples were stored at −80 °C until further use.
Assay Development
At the development phase of the assay, each component and steps were optimized and screened. SARS-CoV-2 recombinant antigens: NCP, E, S1, S2, and RBD proteins were utilized as the potential capturing agent. Six different cocktail preparations (antigen dilution ranged from 1:10 to 1:800) from 16 antigens were analyzed. The combination generating the highest signal without cross-reaction was immobilized as a test dot. To avoid blocking the N.C. membrane, sample processing steps were optimized by diluting samples to 1:2, 1:4, and 1:8 in the commercial buffer (Bhat Biotech Ltd., India).
Assay Procedure
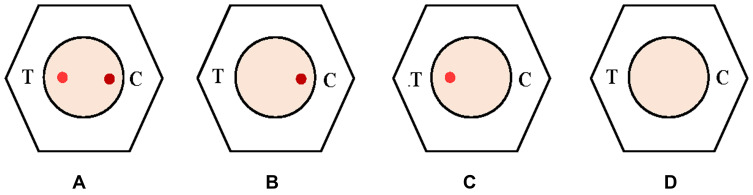
The untreated serum sample was thawed at 37 °C and processed further for the test procedure. During testing, the sample was diluted with 2–3 drops (50–75 µL) of dilution buffer. About 50 µL of diluted serum was then added to test media following two drops of (50µL) of wash buffer. The addition of one drop of AuNP conjugate mixture, followed by two drops of (50µL) of wash buffer, completed the test. Development of control dot attests to the fact that the device is working correctly and the presence or absence of test dot specifies positive or negative results. Results were interpreted as shown in Figure 2.
Figure 2.
Interpretation of the test result- (A) positive: both the dots are visible whereby implicating that the AuNP solution is in perfectly working condition; (B) negative: the presence of the only control dot signifies that there is an absence of anti-SARS-CoV-2 antibody while the AuNP is perfect working condition; (C) and (D) invalid test: the absence of control dot signifies that the AuNP is not in working condition, henceforth the test should not be interpreted but should be repeated.
Intensity Scale Generation
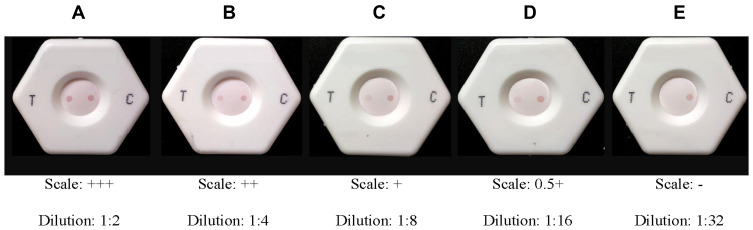
An intensity scale was developed for the semi-quantitative determination of the detection limit (LOD) of the rapid immunoassay. One positive control serum with a high antibody titer was selected based on reference ELISA value.41 Two-fold serial dilutions (1:2, 1:4, 1:8, 1:16, 1:32) of the chosen sera were prepared in standard negative serum and run through the assay. A five-point gradient scale was generated for semi-quantitative detection of IgG in the sample (Figure 3).
Figure 3.
Dot intensity and scale of measurement. (A) 3+: test dot is present at 8-fold dilution but may be absent or present in 16-fold dilution; (B) 2+: test dot is present at 4-fold dilution but absent at 8-fold dilution; (C) 1+: test dot is present at 2-fold dilution but absent at 4-fold dilution; (D) 0.5+: test dot is absent at 2-fold dilution; (E) test dot is absent. It should be ensured that the control dot is present in every experiment, or else the test would be void.
Internal Validation
Rapid dot-blot assay performance validation was designed to determine its clinical efficiency. The rapid assay’s performance was analyzed with the selected samples (n=181) and characterized seropositive and seronegative samples. The selected specimens’ serostatus was first evaluated with an established in-house ELISA test against SARS-CoV-2 recombinant NCP and RBD antigens.41–43 All the samples were then tested with the rapid dot-blot assay to assess their performance.
Co-Efficient of Variation
The coefficient of variation (CV) demonstrates test reproducibility and precision. The intra-assay and inter-assay variations were tested, with five replicates of two positive sera samples on the same day and in 15 different days for later. The coefficient of variation was determined using the following formula.
Coefficient of variation (CV)= (Standard Deviation/Mean) x 100%
Data Analysis
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV), and area under the curve (AUC) with 95% confidence interval were estimated to see the effectiveness of this rapid dot blot assay with in-house developed ELISA as well as FDA approved commercial kits. The calculation was done using a 2×2 table format with the formula shown in Table 1. A linear regression model was used to assess the mean difference of the Ratio (O.D./cut-off) with the different intensity scale. The analysis was performed with STATA 15 (StataCorp, L.P., College Station, Texas, USA), and GraphPad Prism 8.3 was used for graphical presentation.
Table 1.
Formula for Sensitivity, Specificity, Positive Predictive Value (PPV), Negative Predictive Value (NPV) Calculation
| Positive test result | True Positive (TP) | False Positive (FP) | PPV: TP/(TP+FP) |
| Negative test result | False Negative (FN) | True Negative (TN) | NPV: TN/(TN+FN) |
| Sensitivity: TP/(TP+FN) | Specificity: TN/(FP+TN) |
Ethical Approval
Human participants in this study were enrolled, maintaining the national research committee’s ethical standards and the 1964 Helsinki Declaration. All participants of this study were verbally explained in detail regarding the study’s aims, scopes, and purpose. Thereby, researchers obtained research participants’ written consent (approval) for their participation in this study, titled “AuNP Coupled Rapid Flow-Through Dot-Blot Immuno-Assay for Enhanced Detection of SARS-CoV-2 Specific Nucleocapsid and Receptor Binding Domain IgG”. The history of the participants was noted in a questionnaire before the collection of blood samples. The study was approved by the National Research Ethics Committee of Bangladesh (NREC), and the meeting was conducted by Bangladesh Medical Research Ethics Committee (BMRC) (https://www.bmrcbd.org/) [Reference No.: BMRC/NREC/2019-2022/1042]. The privacy of all participants in the study is ensured by not disclosing their information to the investigators.
Results
Screening and Optimization of Test Components
Test performance of rapid dot-blot mainly depends upon the appropriate combination of four major factors: coating agent concentration, nitrocellulose membrane blocking effect, sample processing, and sample volume. Different combinations of SARS-CoV and SARS-CoV-2 antigens were evaluated as capture antigens. Our results showed that the combo, which contained Escherichia coli derived recombinant envelope and S2 proteins, cross-reacted with negative controls (Data not shown). Nevertheless, the combination containing human cell line-derived NCP and RBD showed the best results with good sensitivity and specificity with the controls. Henceforth, a combo containing 1:10 dilution of NCP and RBD proteins (Sino Biologicals) was selected to be used as a test dot immobilized on the N.C. membrane.
Moreover, mouse IgG (Fapon) was immobilized as a control dot on the N.C. membrane. Direct use of sample without any processing blocked the N.C. membrane, which interfered with result interpretation. Sample processing steps were optimized to avoid blocking the N.C. membrane. Two-fold dilution was finalized among three dilutions, and 50 µL of diluted samples were used for testing. Moreover, AuNP conjugate prepared from 10 nm gold colloid gave a better resolving background than 40 nm. Thus, further evaluation conjugates of 10 nm gold colloids were chosen.
Semi-Quantitative LOD Determination
A two-fold serial diluted sample (P-1) was evaluated, and a scale was generated (Figure 3). Based on the results we found in our experiment, a range of 0.5 to 3 plus scale was considered positive for semi-quantitative differentiation of the dot-blot result. In contrast, negative was considered the absence of antibody.
Sample Selection and Characterization Through ELISA and Commercial CLIA Kit
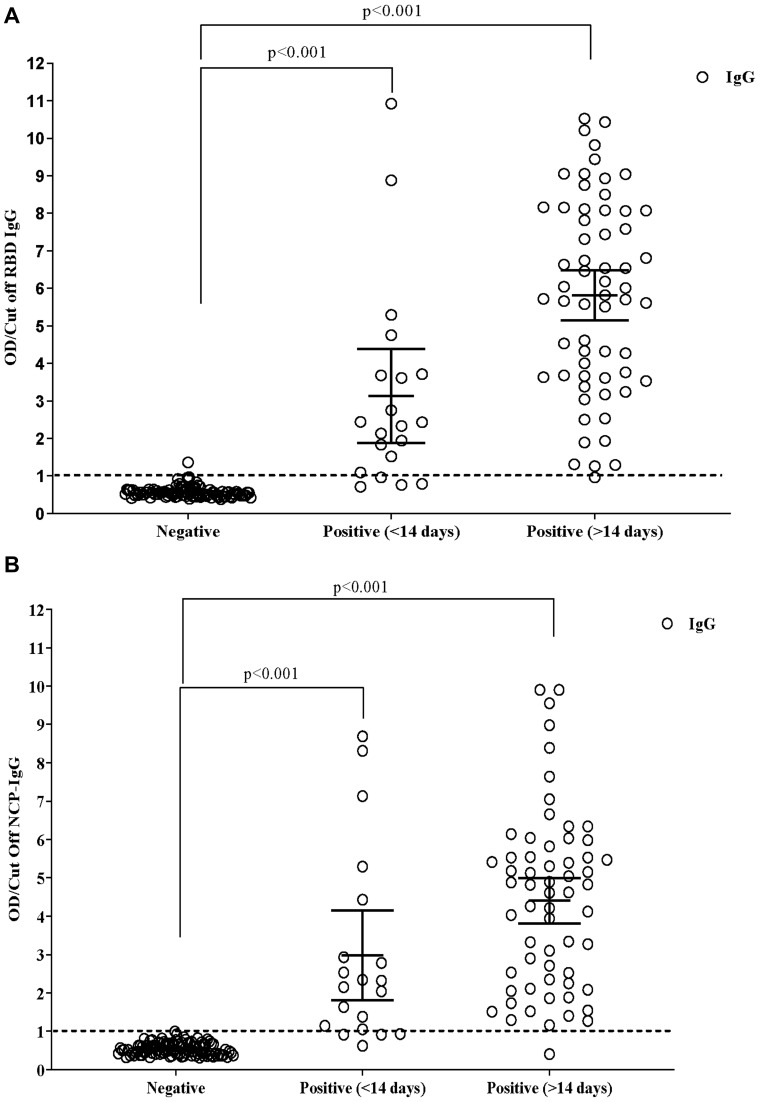
As a combination of NCP and RBD was used as capture antigen in the test platform, the serum samples of SARS-CoV-2 RT-qPCR confirmed patients (n=81), dengue positive patients (n=24), and healthy donors (n=76) were first characterized using IgG ELISA test against these two SARS-CoV-2 immunogens on previously developed in-house ELISA.41,43 Data analysis showed that the sera of COVID-19 infected individuals had a different antibody titer range (Figure 4). Three patterns of antibody response were observed among positive patients with blood collection in the first two weeks of infection: i) 16 patients mounted IgG response against both SARS-CoV-2 proteins and two were entirely negative, ii) two patients achieved only anti-NCP IgG, and iii) two patients mounted an only anti-RBD response. In the second group, all the patients are seropositive for both SARS-CoV-2 antigens except for two patients; one did not develop NCP-IgG, but RBD-IgG for other vice-versa result was observed (Figure 4). Only one sample from the dengue panel showed cross-reactivity to SARS-CoV-2 RBD protein (Figure 4A) but no reaction against NCP (Figure 4B). Other sera were negative for both SARS-CoV-2 antigens. Both the OD/cut-off of RBD (Figure 4A) and NCP (Figure 4B) of the positive cases at <14 and >14 days showed highly significant (p<0.001) with the participants who were negative against SARS-CoV-2. Besides this, the mean OD/cut-off between <14 (3.13±0.58; 2.97±0.53) and >14 days (5.81±0.33; 4.40±0.30) of RBD (p<0.001) and NCP (p=0.021), respectively also showed a significant difference.
Figure 4.
Mean difference in the positive (<14 and > 14 days) and negative samples in in-house ELISA to detect RBD (A) and NCP (B) specific IgG against SARS-CoV-2. The linear regression model was used to estimate the p-value, and the data were represented as mean with standard deviation (S.D.).
When 35 pre-characterized samples were tested in a commercial CLIA kit, they were found to be accurately characterized by our in-house ELISA, ie, fifteen NCP-IgG and RBD-IgG containing RT-qPCR positive samples were reactive in chemiluminescence assay. In contrast, the other twenty NCP-IgG and RBD-IgG negative samples were found non-reactive in tested CLIA (Table 2).
Table 2.
Comparison Between FT-DBA with FDA Approved Commercial Antibody Immunoassay
| Commercial Immunoassay (Elecsys SARS-CoV-2 Assay) | ||||||
|---|---|---|---|---|---|---|
| Positive | Negative | Total | Sensitivity, (95% CI) | Specificity, (95% CI) | ||
| FT-DBA | Positive | 15 | 0 | 15 | 100%(78.3%, 100%) | 100%(83.2%, 100%) |
| Negative | 0 | 20 | 20 | |||
| Total | 15 | 20 | 35 | |||
Note: Test agreement was evaluated by Kappa statistics.
Detection of Sensitivity and Specificity of the Rapid Dot-Blot Kit
The rapid dot blot assay’s performance efficiency was evaluated with the three chosen sample panels of sera (n=181). The assay detected SARS-CoV-2 infection in patients with symptoms less than 14 days with 85.0% (62.1%, 96.8%) sensitivity which increased to 100% (94.1%, 100.0%) after 14 days and the Cohen’s Kappa test agreement was 81% (Kappa=0.814; p<0.001) and 95% (Kappa=0.948; p<0.001), respectively (Table 3). At both the time point the detection specificity was 98.0% (93.0%, 99.8%). The PPV was 89.5% (95% CI; 66.9%, 98.7%) and 96.8% (95% CI; 89.0%, 100%) for <14 days and >14 days, respectively. NPV of the assays were 97.0% (95% CI; 91.6%, 99.4%) and 100% (95% CI; 96.3%, 100%) for both panels (<14 and >14 days) (Table 4). To check the specificity and cross-reactivity we have run 76 and 24 sera from healthy donors and dengue positive samples, respectively. Among them only 2 samples were misdiagnosed and the overall specificity was found 98.0% (95% CI: 93.0%, 99.8%) (Table 3). The overall sensitivity and specificity were calculated it was found to be 96.3% (95% CI; 89.6%, 99.2%) and 98%, (95% CI; 93.0%, 99.8%), respectively, with 97.5% PPV and 97.0% NPV (Tables 5 and 6). The overall test agreement was 94.4% (Kappa=0.944; p<0.001).
Table 3.
Comparison of AUC, Sensitivity, Specificity, and Kappa of Dot Blot with RBD-IgG and S1-IgG at Different Time Points with RT-PCR Positive and Negative Samples
| AUC (95% CI) | Sensitivity, % (95% CI) | Specificity (95% CI) | Kappa | p-value | |
|---|---|---|---|---|---|
| <14 Days | |||||
| NCP-IgG | 0.90(0.81, 0.99) | 80.0(56.3, 94.3) | 100(96.4, 100) | 0.800 | <0.001 |
| RBD-IgG | 0.90(0.81, 0.99) | 80.0(56.3, 94.3) | 99.0(94.6, 100) | 0.840 | <0.001 |
| Dot blot | 0.92(0.83, 1.00) | 85.0(62.1, 96.8) | 98.0(93.0, 99.8) | 0.845 | <0.001 |
| >14 days | |||||
| NCP-IgG | 0.99(0.98, 1.00) | 98.4(91.2, 100) | 100(94.1, 100.0) | 0.987 | <0.001 |
| RBD-IgG | 0.99(0.98, 1.00) | 98.4(91.2, 100) | 99.0(94.6, 100) | 0.974 | <0.001 |
| Dot blot | 0.99(0.98, 1.00) | 100(94.1, 100.0) | 98.0(93.0, 99.8) | 0.948 | <0.001 |
| Overall | |||||
| NCP-IgG | 0.96(0.93, 0.99) | 93.8(86.2, 98.0) | 100(96.4, 100) | 0.921 | <0.001 |
| RBD-IgG | 0.96(0.94, 0.99) | 93.8(86.2, 98.0) | 99.0(94.6, 100) | 0.933 | <0.001 |
| Dot blot | 0.97(0.95, 1.00) | 96.3(89.6, 99.2) | 98.0(93.0, 99.8) | 0.944 | <0.001 |
Table 4.
Positive and Negative Predicted Value and Test Agreement of the Assay Procedure of Rapid Dot Blot at Different Time Points
| Days | PPV, % (95% CI) | NPV, % (95% CI) |
|---|---|---|
| <14 days | 89.5(66.9, 98.7) | 97.0(91.6, 99.4) |
| >14 days | 96.8(89.0, 100) | 100(96.3, 100) |
| Overall | 97.5(91.3, 99.7) | 97.0(91.6, 99.4) |
Abbreviations: PPV, positive predictive value; NPV, negative predictive value.
Table 5.
Comparison of AUC, Sensitivity, Specificity, and Kappa of Dot-Blot with Seropositive or Seronegative Samples at Different Times
| AUC (95% CI) | Sensitivity, % (95% CI) | Specificity (95% CI) | Kappa | p-value | |
|---|---|---|---|---|---|
| <14 days | 0.97(0.92, 1.0) | 94.7(74, 99.9) | 98.0(93.0, 99.8) | 0.943 | <0.001 |
| >14 days | 0.99(0.98, 1.00) | 100(94.1, 100.0) | 98.0(93.0, 99.8) | 0.948 | <0.001 |
| Overall | 0.98(0.97, 1.0) | 98.8(93.3, 100) | 98.0(93.0, 99.8) | 0.946 | <0.001 |
Table 6.
Positive and Negative Predicted Value and Test Agreement of the Dot-Blot Assay with Characterized Seropositive and Seronegative Samples
| Days | PPV, % (95% CI) | NPV, % (95% CI) |
|---|---|---|
| <14 days | 94.4(72.7, 99.9) | 98.8(93.6, 100) |
| >14 days | 96.8(89.0, 100) | 100(96.3, 100) |
| Overall | 99.6(93.5, 100) | 99.0(94.5, 100) |
Abbreviations: PPV, positive predictive value; NPV, negative predictive value; 95% CI, 95% confidence interval.
Moreover, when the Dot-blot assay kit was evaluated with in-house ELISA (NCP and/ RBD) characterized seropositive and seronegative sera, the sensitivity and specificity in less than 14 days samples were 94.7% (95% CI; 74.0%, 99.9%) and 98.0% (95% CI; 93.0%, 99.8%), respectively, with 94.4% PPV and 98.8% NPV. As expected, the values increased for the samples collected more than 14 days of symptom onset, and sensitivity and specificity were 100% (95% CI; 94.1%, 100.0%) and 98.0% (95% CI; 93.0%, 99.8%), respectively. The PPV and NPV for this phase were 96.8% and 100%, respectively. The overall sensitivity and specificity were calculated and it was found to be 98.8% (95% CI; 93.3%, 100%) and 98% (95% CI, 93.0%, 99.8%), respectively, with 99.6% PPV and 99.0% NPV (Tables 5 and 6).
Comparison with Commercial CLIA Elecsys SARS-CoV-2 Assay
Interestingly, when thirty-five Elecsys SARS-CoV-2 assay characterized sera by evaluated in FT-DBA, fifteen sera found reactive in CLIA showed test dots. In contrast, the other twenty non-reactive sera did not produce any dot in the test region. This indicated the sensitivity and specificity of FT-DBA were 100% (95% CI; 78.3%, 100%) and 100% (95% CI; 83.2%, 100%) with a test agreement of 100% (Kappa 1.00; p<0.001), respectively when compared with the Elecsys SARS-CoV-2 assay (Table 2).
Dot-Blot Assay Can Semi-Quantitatively Reveal Antibody Titer
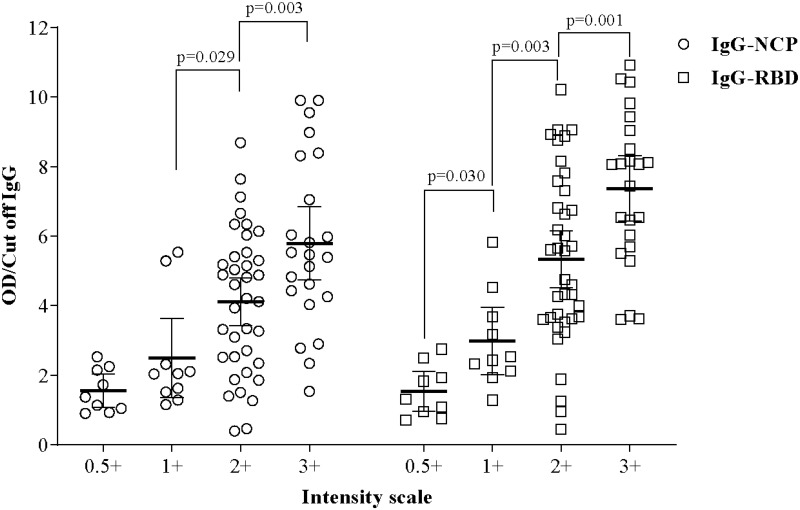
When compared with Reference ELISA values, it was revealed that the FT-DBA could detect SARS-CoV-2 specific IgG antibodies in human serum even when the OD/Cut-off ratio was meager. Linear regression model between the intensity scale and IgG cut-off of RBD and NCP showed significant difference.
The highest intensity 3+ had the most elevated IgG (Cut off) for RBD (7.37±2.18) and NCP (5.78±2.44), which showed significant differences with the intensity scale of 2+ (p= 0.001 and 0.003), respectively. Similarly, the intensity scale of 2+ showed significant differences with the intensity scale of 1+ of RBD and NCP (p=0.003 and 0.029), respectively. Whereas a significant difference (p=0.030) was noted in RBD between 1+ and 0.5+ scales. No such difference was found in NCP (Figure 5). Mean ELISA values in RBD and NCP corresponding to each intensity scale are listed in Supplementary Tables 1 and 2.
Figure 5.
Mean difference in RBD and NCP specific IgG (Cut off) in contrast with intensity scale. The linear regression model was used to estimate the p-value, and the data were shown as mean with a 95% confidence interval.
Co-Efficient Variation
Our analysis showed no intra-assay variation in the assay, but a 9.98% coefficient of variance was found in the inter-assay for the sera samples used.
Discussion
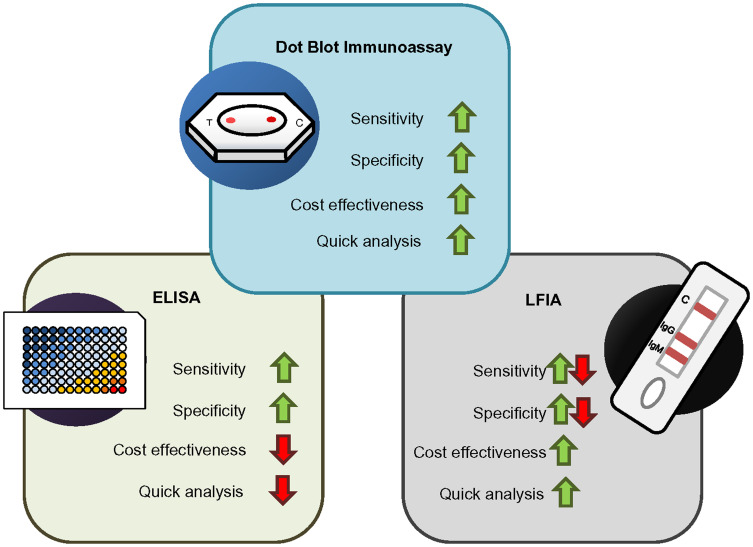
Rapid dot-blot has been a valuable tool for the diagnosis and epidemiological survey of various viral diseases.44–46 Currently, rapid SARS-CoV-2 antibody detection is primarily conducted through LFIA.27,28 Other techniques like surface-enhanced Raman spectroscopy (SERS), microfluidic immune-fluorescence assay, chromatographic digital immunoassays are evolving.47–49 However, the flow-through dot-blot-based approach is not yet widely available as COVID-19 immunoassay. This work includes developing and evaluating a rapid flow-through dot-blot assay (FT-DBA) to detect SARS-CoV-2 specific IgG in human serum. The Dot-blot principle provides a more reliable field-testing framework than LFIA. The latter has usual limitations, including mass transport limitation, binding kinetics of immunogen, and competitive inhibition of target analyte (Figure 6).31
Figure 6.
Comparison between dot-blot assay with ELISA and LFIA. The dot blot immunoassay has the advantage of higher sensitivity and specificity while being cost-effective and time-efficient.
The biofunctionalized theranostic agent can significantly improve targeting, imaging, and therapeutics in diagnostic and clinical settings. Carbon, gold, silver, lipid, nanoceria, bilirubin, and cerium oxide nanoparticles are widely used as theranostic agents.50–55 AuNP and silver nanoparticles (AgNP) are excellent theranostic agents, with a high ratio of area to the volume that allows the functionalization of these particles with various biomolecules.35,56 Moreover, the use of AuNP and enhancement of signal by Ag is reported to improve the sensitivity.56 Due to their multiple nanostructures, such as nanospheres, nanorods, nanosheets, nanotriangle, nanoshells, nanostars, nanocubes, gold nanoparticles, they have a wide range of clinical and diagnostics applications.50,57–61 Since our objective was to develop a rapid immunoassay that the naked eye can interpret, easy to maneuver, cost-effective, and widely available, we choose the AuNP in our assay.
For the role in viral pathogenesis and entry into a host cell, NCP and spike are considered the two most crucial target immunogens of SARS-CoV-2. The RBD of the spike is more advantageous for having the potential to induce neutralizing antibodies.62 Typically, an immune reaction to NCP evolves earlier than RBD, but exceptions have also been reported.43 In that context, for increased sensitivity in the acute phase of infection, an assay was designed with dual immune capture property instead of one, which might fail to identify a seroconverted person.63,64 Another issue considered was the possibility of cross-reaction with other alpha and beta-coronaviruses surrounding the use of the SARS-CoV-2 serological test. The two human coronaviruses SARS-CoV and MERS-CoV, tend to pose the greatest likelihood of cross-reaction. Due to the high level of sequence identity, SARS-CoV antigens are known to cross-neutralize SARS-CoV-2.1,65 However, since the SARS-CoV epidemic, it has been seventeen years, and specific antibody response against the virus has been reported to be diminished (90%).66 Therefore, a false-positive reaction at this point is an unlikely event. MERS-CoV is still active in the population along with the four endemic low pathogenic human CoVs (229E-CoV, NL63-CoV, OC43-CoV, and HKU1-CoV), so most humans might bear antibody against them.67,68 Nevertheless, their cross-reactivity against SARS-CoV-2 nucleoprotein and RBD protein is very low. Except for SARS-CoV, other human coronaviruses do not have any sequence resemblance to RBD and S1 domain of spike protein of SARS-CoV-2.69
Moreover, the assay was designed to take into account the dengue-endemic situation of Bangladesh.70 Since both diseases have common characteristics in the early phase and there has been a report of antigenic cross-reactivity between SARS-CoV-2 and dengue virus (DENV), there is a chance of misdiagnosis.71 To avoid the risk, 24 pre-pandemic dengue-positive sera were incorporated into the evaluation panel. Another strengthening point of the assay is that it was developed using SARS-CoV-2 positive sera, studied longitudinally for antibody response in one of our previous studies, eliminating any chance of spectrum bias.42,72
Laboratory evaluation revealed that when challenged with RT-qPCR positive or RT-qPCR positive and seropositive samples, the developed assay’s overall sensitivity was 96.3% (Table 3) for the former but increased to 98.8% (Table 5) for the latter group. The difference was that two RT-qPCR positive samples were never seroconverted in our study, as reported by others.73 A comparative study conducted between conventional ELISA and developed assay revealed a high correlation, which others can find.74 Dot intensity seemed to increase with corresponding ELISA value upon disease progression and antibody titer (Figure 5, Supplementary Tables 1 and 2). Statistical analysis revealed equivalent clinical agreement between the two techniques as well as with the gold-standard method of RT-qPCR, with a Cohen’s Kappa value of 0.84 (strong agreement) and 0.94 (robust agreement), respectively, in between <14 and >14 days (Table 3). A similar significant difference was observed between the results of two-time points in both assay techniques (Table 3). Moreover, a slightly increased sensitivity was observed in the dot assay compared to the two ELISA, which might have an additive effect of using two proteins instead of one (Table 3). 100% (95% CI; 78.3%, 100%) sensitivity and 100% (95% CI; 83.2%, 100%)
Elecsys SARS-CoV-2 assay (NCP) is a US FDA approved CLIA assay system, which qualitatively detects COVID-19 antibodies with high sensitivity of 99.5 (95% CI; 97.0–100.0) and specificity of 99.80 (95% CI; 99.69–99.88), when samples were RT-qPCR positive samples were tested >14 days of disease onset 75. Compared with other ELISA, CLIA, and LFIA, Elecsys SARS-CoV-2 assay has always been consistent with its performance.75–78 Moreover, researchers have employed this kit to identify the convalescent groups before vaccinations.79 We opted to use this assay to evaluate our FT-DBA kit compared to 100% similarity in sensitivity and specificity with thirty-five samples tested (Table 2). Our future endeavor will focus on using both NCP and S1 versions of Elecsys SARS-CoV-2 assay kits to evaluate our FT-DBA kit with a larger sample pool.
One of the limitations of our study is that it does not include infants and newborns, although recent studies have found that infants and newborns can also be infected with SARS-CoV-2.80,81 In addition, all the study participants were 18 years and above, who would provide informed consent without a guardian requirement.
Considering all these factors, the assay system elaborated in the present study can be regarded as a more feasible option for serosurveillance study than conventional ELISA, especially for low- and middle-income countries (LMICs), including Bangladesh. Moreover, with the ongoing vaccination programs, the serological test will be essential in addressing two fundamental issues: vaccine prioritization and monitoring of protective immunity development in post-vaccinated cohorts.
Conclusions
The developed kit uses AuNP technology to detect and determine the presence of SARS-CoV-2 antibodies in infected individuals. Compared to the in-house developed ELISA kit, the FT-DBA presents an overall sensitivity of 98% and a specificity of 98.8%. The specificity indicates that the kit can differentiate antibodies against SARS-CoV-2 antigens from those detecting against other coronaviruses antigens. Furthermore, the kit provides a reliable semi-quantitative result that can help determine the efficacy of the vaccination programs within the country. The convenient use and easy implementation mean that the developed system can be utilized in remote regions of the world with low healthcare penetration.
Recommendations
Implementing rapid antibody tests in low and middle-income countries (LMICs) would facilitate serostatus assessment after natural infection.
With the scarcity of vaccines in many LMICs, the antibody tests would provide a way to better implement the vaccination to those who have not been exposed or have low to no antibodies, thereby breaking the chain of SARS-CoV-2 transmission.
We have observed that flow-through immunoassay presented higher specificity with comparable sensitivity.
Implementation of RBD-specific antibody assay is necessary for observing the efficacy of vaccination.
Implementation of NCP-specific antibody assay will provide an insight into the previous infections. Simultaneously, the absence of NCP-specific antibodies and the presence of RBD-specific antibodies would correlate with the vaccination program’s efficacy in providing protection.
Acknowledgment
The authors of the current work appreciate and show their gratitude to Prof Mohammed S. Razzaque, MBBS, Ph.D. of Lake Erie College of Osteopathic Medicine (Pennsylvania, USA), for reading the manuscript and providing valuable suggestions.
Consent for Publication
All authors reviewed and approved the final version and have agreed to be accountable for all aspects of the work, including any issues related to accuracy or integrity.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the Journal to which the article has been submitted; and agreed to be accountable for all aspects of the work.
Disclosure
Mr Md. Ahsanul Haq reports a patent 10202006327W pending; Prof Dr Bijon Kumar Sil reports a patent 10202006327W pending; Dr Mohd Raeed Jamiruddin reports a patent 10202006327W pending to Intellectual Property Office of Singapore; Dr Nihad Adnan reports a patent 10202006327W pending; the authors report no other conflicts interest in this work. This research has the potential possibility for applying for patent rights. Mainul Haque declines not to be part of the patent.
References
- 1.Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Contini C, Di Nuzzo M, Barp N, et al. The novel zoonotic COVID-19 pandemic: an expected global health concern. J Infect Dev Countries. 2020;14(03):254–264. doi: 10.3855/jidc.12671 [DOI] [PubMed] [Google Scholar]
- 3.WHO. COVID-19 Dashboard. 2020; https://covid19.who.int/. Accessed June29, 2021.
- 4.Petersen E, Koopmans M, Go U, et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis. 2020;20(9):e238–e244. doi: 10.1016/S1473-3099(20)30484-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO. COVID-19 vaccines. 2021; https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines. Accessed June29, 2021.
- 6.Peeling RW, Wedderburn CJ, Garcia PJ, et al. Serology testing in the COVID-19 pandemic response. Lancet Infect Dis. 2020;20(9):e245–e249. doi: 10.1016/S1473-3099(20)30517-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.FDA. In Vitro Diagnostics EUAs. 2021; https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas. Accessed June29, 2021.
- 8.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false-negative rate of reverse transcriptase-polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med. 2020;173(4):262–267. doi: 10.7326/M20-1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinnes J, Deeks JJ, Adriano A, et al. Rapid, point‐of‐care antigen and molecular‐based tests for diagnosis of SARS‐CoV‐2 infection. Cochrane Database of Systematic Reviews. 2020;1(8):584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chau NVV, Thanh Lam V, Thanh Dung N, et al. The natural history and transmission potential of asymptomatic SARS-CoV-2 infection. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandhi M, Yokoe DS, Havlir DV. Asymptomatic Transmission, the Achilles’ Heel of Current Strategies to Control Covid-19. In: Mass Medical Soc; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahalingam S, Peter J, Xu Z, et al. Landscape of humoral immune responses against SARS-CoV-2 in patients with COVID-19 disease and the value of antibody testing. Heliyon. 2021;7(4):e06836. doi: 10.1016/j.heliyon.2021.e06836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Fu B, Chen L, Feng Y. Serological tests facilitate identification of asymptomatic SARS‐CoV‐2 infection in Wuhan, China. J Med Virol. 2020;92(10):1795–1796. doi: 10.1002/jmv.25904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Augustine R, Das S, Hasan A, et al. Rapid antibody-based COVID-19 mass surveillance: relevance, challenges, and prospects in a pandemic and Post-Pandemic world. J Clin Med. 2020;9(10):3372. doi: 10.3390/jcm9103372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollán M, Pérez-Gómez B, Pastor-Barriuso R, et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396(10250):535–544. doi: 10.1016/S0140-6736(20)31483-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar MS, Bhatnagar T, Manickam P, et al. National sero-surveillance to monitor the trend of SARS-CoV-2 infection transmission in India: protocol for community-based surveillance. Indian J Med Res. 2020;151(5):419. doi: 10.4103/ijmr.IJMR_1818_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havers FP, Reed C, Lim T, et al. Seroprevalence of antibodies to SARS-CoV-2 in 10 sites in the United States, March 23-May 12, 2020. JAMA Intern Med. 2020;180(12):1576–1586. doi: 10.1001/jamainternmed.2020.4130 [DOI] [PubMed] [Google Scholar]
- 18.Silveira MF, Barros AJ, Horta BL, et al. Population-based surveys of antibodies against SARS-CoV-2 in Southern Brazil. Nat Med. 2020;26(8):1196–1199. doi: 10.1038/s41591-020-0992-3 [DOI] [PubMed] [Google Scholar]
- 19.WHO. “Solidarity II” global serologic study for COVID-19. 2020; https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-2-global-serologic-study-for-covid-19. Accessed June29, 2021.
- 20.Espejo AP, Akgun Y, Al Mana AF, et al. Review of current advances in serologic testing for COVID-19. Am J Clin Pathol. 2020;154(3):293–304. doi: 10.1093/ajcp/aqaa112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Xiao M, Liu X, et al. Significance of serology testing to assist timely diagnosis of SARS-CoV-2 infections: implication from a family cluster. Emerg Microbes Infect. 2020;9(1):924–927. doi: 10.1080/22221751.2020.1752610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long Q-X, Liu B-Z, Deng H-J, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1 [DOI] [PubMed] [Google Scholar]
- 23.Krsak M, Johnson SC, Poeschla EM. COVID-19, Serosurveillance May Facilitate Return-to-Work Decisions. Am J Trop Med Hyg. 2020;102(6):1189–1190. doi: 10.4269/ajtmh.20-0302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubina R, Dziedzic A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10(6):434. doi: 10.3390/diagnostics10060434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallett SJ, Rayment M, Patel A, et al. Point-of-care serological assays for delayed SARS-CoV-2 case identification among healthcare workers in the U.K.: a prospective multicentre cohort study. Lancet Respir Med. 2020;8(9):885–894. doi: 10.1016/S2213-2600(20)30315-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C, Wen T, Shi FJ, Zeng XY, Jiao YJ. Rapid Detection of IgM Antibodies against the SARS-CoV-2 Virus via Colloidal Gold Nanoparticle-Based Lateral-Flow Assay. ACS Omega. 2020;5(21):12550–12556. doi: 10.1021/acsomega.0c01554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu JL, Tseng WP, Lin CH, et al. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J Infect. 2020;81(3):435–442. doi: 10.1016/j.jinf.2020.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAulay K, Bryan A, Greninger AL, et al. Retrospective clinical evaluation of 4 lateral flow assays for the detection of SARS-CoV-2 IgG. Diagn Microbiol Infect Dis. 2020;98(3):115161. doi: 10.1016/j.diagmicrobio.2020.115161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posthuma-Trumpie GA, Korf J, van Amerongen A. Lateral flow (Immuno) assay: its strengths, weaknesses, opportunities, and threats. A literature survey. Anal Bioanal Chem. 2009;393(2):569–582. doi: 10.1007/s00216-008-2287-2 [DOI] [PubMed] [Google Scholar]
- 31.Ross G, Salentijn GI, Nielen MW. A critical comparison between flow-through and lateral flow immunoassay formats for visual and smartphone-based multiplex allergen detection. Biosensors. 2019;9(4):143. doi: 10.3390/bios9040143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aldewachi H, Chalati T, Woodroofe MN, Bricklebank N, Sharrack B, Gardiner P. Gold nanoparticle-based colorimetric biosensors. Nanoscale. 2017;10(1):18–33. doi: 10.1039/C7NR06367A [DOI] [PubMed] [Google Scholar]
- 33.Liu G, Lu M, Huang X, Li T, Xu D. Application of Gold-Nanoparticle Colorimetric Sensing to Rapid Food Safety Screening. Sensors. 2018;18:12. doi: 10.3390/s18124166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang W, Wang J, Ding Y. Gold nanoparticle-conjugated nanomedicine: design, construction, and structure–efficacy relationship studies. J Mater Chem B. 2020;8(22):4813–4830. doi: 10.1039/C9TB02924A [DOI] [PubMed] [Google Scholar]
- 35.Pedrosa P, Baptista PV. Chapter 1 - Gold and Silver Nanoparticles for Diagnostics of Infection. In: Rai M, Kon K, editors. Nanotechnology in Diagnosis, Treatment, and Prophylaxis of Infectious Diseases. Boston: Academic Press; 2015:1–18. [Google Scholar]
- 36.Liu X, Xiang JJ, Tang Y, et al. Colloidal gold nanoparticle probe-based immunochromatographic assay for the rapid detection of chromium ions in water and serum samples. Anal Chim Acta. 2012;745:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan R, Jiang Y, Sun L, et al. Gold nanoparticle-based enhanced lateral flow immunoassay for detection of Cronobacter sakazakii in powdered infant formula. J Dairy Sci. 2018;101(5):3835–3843. doi: 10.3168/jds.2017-14265 [DOI] [PubMed] [Google Scholar]
- 38.Shen YSG, Shen G. Signal-Enhanced Lateral Flow Immunoassay with Dual Gold Nanoparticle Conjugates for the Detection of Hepatitis B Surface Antigen. ACS Omega. 2019;4(3):5083–5087. doi: 10.1021/acsomega.8b03593 [DOI] [Google Scholar]
- 39.Khandker SS, Shakil M, Hossen M. Gold Nanoparticles; Potential Nanotheranostic Agent in Breast Cancer: a Comprehensive Review with Systematic Search Strategy. Curr Drug Metab. 2020;21(8):579–598. doi: 10.2174/1389200221666200610173724 [DOI] [PubMed] [Google Scholar]
- 40.Oliver C. Conjugation of colloidal gold to proteins. In: Immunocytochemical Methods and Protocols. Springer; 2010:369–373. [DOI] [PubMed] [Google Scholar]
- 41.Sil BK, Jahan N, Haq MA, et al. Development and performance evaluation of a rapid in-house ELISA for retrospective serosurveillance of SARS-CoV-2. PLoS One. 2021;16(2):e0246346. doi: 10.1371/journal.pone.0246346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jamiruddin MR, Haq MATK, Kobatake E, et al. Longitudinal Antibody Dynamics Against Structural Proteins of SARS-CoV-2 in Three COVID-19 Patients Shows Concurrent Development of IgA, IgM, and IgG. J Inflamm Res. 2021:2497–2506. doi: 10.2147/JIR.S313188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sil BK, Adnan N, Oishee MJ, et al. Development and Evaluation of Two Rapid Indigenous IgG-ELISA immobilized with ACE-2 Binding Peptides for Detection Neutralizing Antibodies Against SARS-CoV-2. medRxiv. 2020. [Google Scholar]
- 44.Gentilomi G, Musiani M, Zerbini M, Gallinella G, Venturoli S, Manaresi E. Dot immunoperoxidase assay for detection of parvovirus B19 antigens in serum samples. J Clin Microbiol. 1997;35(6):1575–1578. doi: 10.1128/jcm.35.6.1575-1578.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akrami H, Hedayati A, Farshian M, Haqshenas G. Development of a recombinant protein-based dot-blot hybridization assay for the detection of antibody to chicken infectious bronchitis virus. Iran J Veterinary Res. 2013;14(4):350–353. [Google Scholar]
- 46.Tsukazaki T, Yoshida M, Namba H, Yamada M, Shimizu N, Nii S. Development of a dot blot neutralizing assay for HHV-6 and HHV-7 using specific monoclonal antibodies. J Virol Methods. 1998;73(2):141–149. doi: 10.1016/S0166-0934(98)00051-2 [DOI] [PubMed] [Google Scholar]
- 47.Oishee MJ, Ali T, Jahan N, et al. COVID-19 Pandemic: review of Contemporary and Forthcoming Detection Tools. Infect Drug Resist. 2021;14:1049–1082. doi: 10.2147/IDR.S289629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yadav S, Sadique MA, Ranjan P, et al. SERS Based Lateral Flow Immunoassay for Point-of-Care Detection of SARS-CoV-2 in Clinical Samples. ACS Applied Bio Materials. 2021;4(4):2974–2995. doi: 10.1021/acsabm.1c00102 [DOI] [PubMed] [Google Scholar]
- 49.Sharma B, Shahanshah MFH, Gupta S, Gupta V. Recent advances in the diagnosis of COVID-19: a bird’s eye view. Expert Rev Mol Diagn. 2021;1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tabish TA, Dey P, Mosca S, et al. Smart Gold Nanostructures for Light Mediated Cancer Theranostics: combining Optical Diagnostics with Photothermal Therapy. Adv Sci. 2020;7(15):1903441. doi: 10.1002/advs.201903441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loh KP, Ho D, Chiu GNC, Leong DT, Pastorin G, Chow EK. Clinical Applications of Carbon Nanomaterials in Diagnostics and Therapy. Adv Mater. 2018;30(47):e1802368. doi: 10.1002/adma.201802368 [DOI] [PubMed] [Google Scholar]
- 52.Khurana A, Allawadhi P, Khurana I, et al. Role of nanotechnology behind the success of mRNA vaccines for COVID-19. Nano Today. 2021;38:101142. doi: 10.1016/j.nantod.2021.101142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allawadhi P, Khurana A, Allwadhi S, Joshi K, Packirisamy G. Nanoceria as a possible agent for the management of COVID-19. Nano Today. 2020;35:100982. doi: 10.1016/j.nantod.2020.100982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khurana I, Allawadhi P, Khurana A, et al. Can bilirubin nanomedicine become a hope for the management of COVID-19? Med Hypotheses. 2021;149:110534. doi: 10.1016/j.mehy.2021.110534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niemiec SM, Hilton SA, Wallbank A, et al. Cerium oxide nanoparticle delivery of microRNA-146a for local treatment of acute lung injury. Nanomedicine. 2021;34:102388. doi: 10.1016/j.nano.2021.102388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu T, Chen C, Huang G, Yang X. Antibody modified-silver nanoparticles for colorimetric immuno sensing of Aβ(1–40/1–42) based on the interaction between β-amyloid and Cu2+. Sens Actuators B Chem. 2016;234:63–69. doi: 10.1016/j.snb.2016.04.159 [DOI] [Google Scholar]
- 57.Singh P, Pandit S, Mokkapati V, Garg A, Ravikumar V, Mijakovic I. Gold Nanoparticles in Diagnostics and Therapeutics for Human Cancer. Int J Mol Sci. 2018;19:7. doi: 10.3390/ijms19071979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Draz MS, Shafiee H. Applications of gold nanoparticles in virus detection. Theranostics. 2018;8(7):1985–2017. doi: 10.7150/thno.23856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsai TT, Huang CY, Chen CA, et al. Diagnosis of Tuberculosis Using Colorimetric Gold Nanoparticles on a Paper-Based Analytical Device. ACS Sens. 2017;2(9):1345–1354. doi: 10.1021/acssensors.7b00450 [DOI] [PubMed] [Google Scholar]
- 60.Nejati K, Dadashpour M, Gharibi T, Mellatyar H, Akbarzadeh A. Biomedical Applications of Functionalized Gold Nanoparticles: a Review. J Cluster Sci. 2021. doi: 10.1007/s10876-020-01955-9 [DOI] [Google Scholar]
- 61.Kang J, Yeom G, Jang H, Oh J, Park CJ, Kim MG. Development of Replication Protein A-Conjugated Gold Nanoparticles for Highly Sensitive Detection of Disease Biomarkers. Anal Chem. 2019;91(15):10001–10007. doi: 10.1021/acs.analchem.9b01827 [DOI] [PubMed] [Google Scholar]
- 62.He Y, Zhou Y, Liu S, et al. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem Biophys Res Commun. 2004;324(2):773–781. doi: 10.1016/j.bbrc.2004.09.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai X-F, Chen J, Hu J. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J Infect Dis. 2020;222(2):189–193. doi: 10.1093/infdis/jiaa243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian X, Li C, Huang A, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Em Microbes Infect. 2020;9(1):382–385. doi: 10.1080/22221751.2020.1729069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang F, Quan Y, Xin Z-T, et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186(12):7264–7268. doi: 10.4049/jimmunol.0903490 [DOI] [PubMed] [Google Scholar]
- 67.WHO. Middle East respiratory syndrome coronavirus (MERS-CoV) – the Kingdom of Saudi Arabia. 2020; https://www.who.int/csr/don/08-april-2020-mers-saudi-arabia/en/. Accessed June29, 2021.
- 68.Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Okba NMA, Muller MA, Li W, et al. Severe Acute Respiratory Syndrome Coronavirus 2-Specific Antibody Responses in Coronavirus Disease Patients. Emerg Infect Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rafi A, Mousumi AN, Ahmed R, Chowdhury RH, Wadood A, Hossain G. Dengue epidemic in a non-endemic zone of Bangladesh: clinical and laboratory profiles of patients. PLoS Negl Trop Dis. 2020;14(10):e0008567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harapan H, Ryan M, Yohan B, et al. Covid‐19 and dengue: double punches for dengue‐endemic countries in Asia. Rev Med Virol. 2020;e2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takahashi S, Greenhouse B, Rodríguez-Barraquer I. Are seroprevalence estimates for severe acute respiratory syndrome coronavirus 2 biased? J Infect Dis. 2020;222(11):1772–1775. doi: 10.1093/infdis/jiaa523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baron RC, Risch L, Weber M, et al. Frequency of serological non-responders and false-negative RT-PCR results in SARS-CoV-2 testing: a population-based study. Clin Chem Lab Med. 2020;58(12):2131–2140. doi: 10.1515/cclm-2020-0978 [DOI] [PubMed] [Google Scholar]
- 74.Ramachandran S, Singhal M, McKenzie KG, et al. A Rapid, Multiplexed, High-Throughput Flow-Through Membrane Immunoassay: a Convenient Alternative to ELISA. Diagnostics. 2013;3(2):244–260. doi: 10.3390/diagnostics3020244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peter Muench SJ, Wenderoth V, Ofenloch-Haehnle B, et al. Development and Validation of the elecsys anti-SARS-CoV-2 Immunoassay as a Highly specific tool for determining past exposure to SARS-CoV-2. J Clin Microbiol. 2020;58:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen SY, Lee YL, Lin YC, et al. Multicenter evaluation of two chemiluminescence and three lateral flow immunoassays for the diagnosis of COVID-19 and assessment of antibody dynamic responses to SARS-CoV-2 in Taiwan. Emerg Microbes Infect. 2020;9(1):2157–2168. doi: 10.1080/22221751.2020.1825016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Egger M, Bundschuh C, Wiesinger K, et al. Comparison of the Elecsys(R) Anti-SARS-CoV-2 immunoassay with the EDI enzyme-linked immunosorbent assays for the detection of SARS-CoV-2 antibodies in human plasma. Clin Chim Acta. 2020;509:18–21. doi: 10.1016/j.cca.2020.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Favresse J, Cadrobbi J, Eucher C, et al. Clinical performance of three fully automated anti-SARS-CoV-2 immunoassays targeting the nucleocapsid or spike proteins. J Med Virol. 2021;93(4):2262–2269. doi: 10.1002/jmv.26669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manisty C, Otter AD, Treibel TA, et al. Antibody response to first BNT162b2 dose in previously SARS-CoV-2-infected individuals. Lancet. 2021;397(10279):1057–1058. doi: 10.1016/S0140-6736(21)00501-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei M, Yuan J, Liu Y, Fu T, Yu X, Zhang ZJ. Novel Coronavirus Infection in Hospitalized Infants Under 1 Year of Age in China. JAMA. 2020;323(13):1313–1314. doi: 10.1001/jama.2020.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang ZJ, Yu XJ, Fu T, et al. Novel coronavirus infection in newborn babies aged <28 days in China. Eur Respir J. 2020;55:6. doi: 10.1183/13993003.00697-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]