Abstract
Scientific and consumer interest in healthy foods (also known as functional foods), nutraceuticals and cosmeceuticals has increased in the recent years, leading to an increased presence of these products in the market. However, the regulations across different countries that define the type of claims that may be made, and the degree of evidence required to support these claims, are rather inconsistent. Moreover, there is also controversy on the effectiveness and biological mode of action of many of these products, which should undergo an exhaustive approval process to guarantee the consumer rights. Computational approaches constitute invaluable tools to facilitate the discovery of bioactive molecules and provide biological plausibility on the mode of action of these products. Indeed, methodologies like QSAR, docking or molecular dynamics have been used in drug discovery protocols for decades and can now aid in the discovery of bioactive food components. Thanks to these approaches, it is possible to search for new functions in food constituents, which may be part of our daily diet, and help to prevent disorders like diabetes, hypercholesterolemia or obesity. In the present manuscript, computational studies applied to this field are reviewed to illustrate the potential of these approaches to guide the first screening steps and the mechanistic studies of nutraceutical, cosmeceutical and functional foods.
Graphical Abstract

Keywords: Health foods, Nutraceuticals, Cosmeceuticals, Machine learning, QSAR, Docking, Molecular dynamics
Introduction
In the last decade, the number of research papers on nutraceuticals, cosmeceuticals and functional foods has grown exponentially, with more than 11,000 manuscripts compiled in the Pubmed repository [1] during this time interval. Despite this increase, there is still a lot of controversy with these terms and their corresponding definitions.
Stephen DeFelice coined the term “nutraceutical” in the 1980s, as a combination of the words “nutrient” and “pharmaceutical”, which he defined as “a food (or part of a food) that provides medical or health benefits, including the prevention and/or treatment of a disease” [2]. Although this definition has evolved through the years, there is still no clear consensus over it, and a standard definition for this term is yet to be adopted. In general, the categorization and regulation of these products depends on their intended use and may vary across countries and regions. [3]. While the Food and Drug Administration (FDA) defines nutraceutical as “any substance that is a food or a part of a food and is able to induce medical and health benefits, including the prevention and treatment of disease” [4], there is no official definition for this term in the EU to date; nutraceutical claims submitted to the European Food Safety Authority (EFSA) are evaluated based on the Regulation of Nutritional and Health Claims (Regulation (EC) No 1924/2006) for possible authorization. However, this regulation does not cover medicinal applications in the EU [5]. A “functional food”, is defined by the FDA as a “nutrient consumed as part of a normal diet, delivering one or more active ingredients (that have physiological effects and may enhance health) within the food matrix” [4]. As is the case for nutraceuticals, this term has no legal definition in the EU; instead, one can refer to healthy foods or food constituents when health claims have been approved according to the EU Regulation [6].
The concept of cosmeceutical arises by merging the words “cosmetic” and “pharmaceutical”. Albert Kligman, defined a cosmeceutical as a “cosmetic product that exerts a pharmaceutical therapeutic benefit but not necessarily a biologic therapeutic benefit” [7]. Just as in the case of the nutraceutical concept, there is neither consensus on the definition of cosmeceuticals nor on the regulation of these kind of products.
Even though there is no consensus on the way to regulate and define these terms globally, they are widely used in labels to attract customers to commercialized products. The global nutraceutical market was worth USD 382.52 billion in 2020 and is expected to reach USD 722.49 billion by 2025 [8], with the United States as the leading country in nutraceutical production, followed by Europe, Asia, Latin America, Middle East and Africa [3]. Also, this market is expected to grow following the outbreak of the COVID-19, given potential benefits of these products on the immune system [9]. Moreover, the functional food market reached a worth of USD 177.41 billion and is projected to reach USD 275.77 billion by 2025 [10]. The cosmeceutical market peaked at USD 55.4 billion in 2020 and is projected to reach USD 70.0 billion by 2025 [11]. The main reason for the growth of the nutraceutical market worldwide is the trend to adopt healthier dietary habits by the population nowadays. Consumers skeptical or unsatisfied with the existent therapeutic approaches are seeking complementary products or alternatives that could help to prevent and/or improve the treatment of diseases [12].
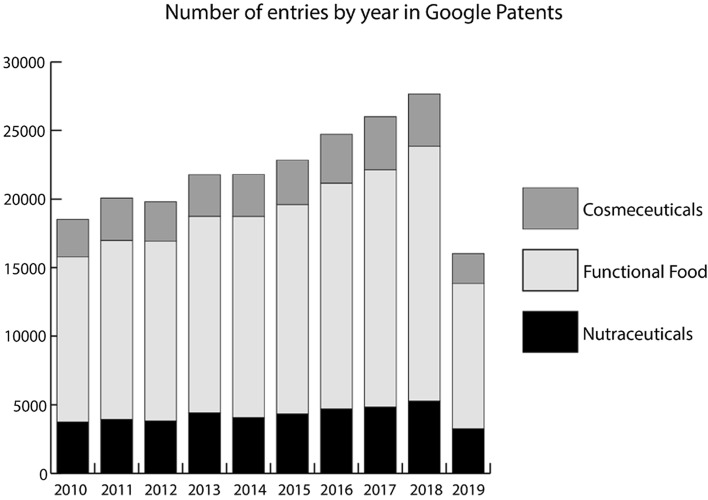
In the last ten years, the number of patents of these products has increased with the growth of the market. Figure 1 illustrates the trend in the number of entries indexed at Google Patents [13] during the period 2010–2019 worldwide. As it may be observed, the number of filed patents has grown to approximately 20,000 patents per year in the last decade. A drop in this positive trend was observed in 2019, probably due to the drop in fillings by China-based inventors amidst an overall shift in regulations there. Even then, an acceptable number of patents was filled [14].
Fig. 1.
Number of patents of every term by year on Google Patents
Several studies have demonstrated the global health benefit of consuming nutraceuticals, cosmeceuticals and functional foods, further stimulating research in this field to improve the quality of life of the society by promoting health and preventing disease. The main targets of nutraceuticals and functional foods are the antioxidant gastro-intestinal, reproductive and renal health-effects as well as the reduction of the risk of cardiovascular diseases, oral diseases, ostheoarthritis, diabetes or obesity [15, 16]. Furthermore, cosmeceuticals are intended not only to prevent the signs of aging, but also attenuate problems such as skin blemishes [17].
Nutraceuticals, cosmeceuticals and functional food have been studied in recent years, although the main challenge for this field continues to lie in the difficulty to substantiate with clinical evidences the efficacy of these compounds as well as to determine their mechanisms of action [3].
Computational techniques have been widely applied in the fields of chemistry and biology, particularly in the context of drug discovery applications in the pharmaceutical industry [18]. Many of the principles and tools employed in drug discovery may also be extrapolated to nutraceutical, cosmeceutical and functional food research, particularly in the identification of complementary leads and targets using molecular modelling, quantitative or qualitative “structure–activity” relationships ((Q)SAR), and pattern recognition methods [19]. Other possible applications of chemoinformatics and bioinformatics tools in the field of nutraceuticals, cosmeceutical and functional foods include the prediction of toxicity, activity and the elucidation of the corresponding mechanisms of action. The goal of this paper is to provide a comprehensive review on the use of computational techniques in the context of nutraceuticals, cosmeceutical and functional foods. Moreover, a prospective analysis of the impact of these techniques in this field will be provided.
Computational Methods
Chemical databases
Chemical databases have progressed over the past years from being simple repositories of chemicals, to playing an important role in chemoinformatic and bioinformatic applications [20]. A list of some of the most relevant chemical databases for computational purposes is provided in Table 1. These chemical databases allow for the querying of compounds following different criteria:
By structure allows for the query of structurally similar molecules or that share the same functional groups. Such a query is useful for paradigms where the bioactivity or target-ligand binding profile of a molecule is known, but structurally similar molecules with better bioactivity or binding profiles are desired. Also, this kind of search could be useful in ligand-based modeling
Table 1.
Chemical databases for nutraceutical, cosmeceutical and functional food research
| Database | Description | Organization | Website access |
|---|---|---|---|
| PubChem [22] | Open chemistry database with more than 100 million compounds. PubChem contains numerous small molecules but also larger molecules such as nucleotides, lipids, peptides and more | National Institutes of Health (NIH) | https://pubchem.ncbi.nlm.nih.gov |
| Zinc [23] | A free database of commercially available compounds with more than 230 million compounds | University of California | http://zinc.docking.org |
| ChEMBL [24] | A manually curated database of bioactive molecules with drug-like properties with 2 million compounds and their abstracted bioactivities | European Bioinformatics Institute | https://www.ebi.ac.uk/chembl/ |
| NCI [25] | A database with more than 275.000 small molecules for research in the field of cancer/AIDS | National Cancer Institute | https://cactus.nci.nih.gov/ncidb2.2/ |
| ChemDB [26] | Chemical database that contains about 5 million of small molecules and its physicochemical properties | Institute for Genomics and Bioinformatics from the University of California | http://cdb.ics.uci.edu |
| ChemSpider [27] | Free chemical structure database with over 67 million structures from hundreds of data sources | The Royal Society of Chemistry | http://www.chemspider.com |
| BindingDB [28] | A database with information from about 1 million binding data for more than 6000 protein targets and near 400.000 small molecules | Skaggs School of Pharmacy and Pharmaceutical Sciences at the University of California | https://www.bindingdb.org/bind/index.jsp |
| PDB-Bind [29] | A collection of more than 21.000 entries of binding affinities for protein–ligand complexes | Shanghai Institute of Organic Chemistry | http://www.pdbbind.org.cn |
| PDBeChem [30] | A database with more than 30.000 entries that contains small molecules, ligands and monomers searched in the Protein Data Bank | The European Bioinformatics Institute | https://www.ebi.ac.uk/pdbe-srv/pdbechem/ |
| KEGG [31] | A database that integrates genomic, chemical and systemic functional information | Kaneisha Laboratories | https://www.kegg.jp/kegg/ |
| HMDB [32] | The Human Metabolome Database contains detailed information about small molecules found in the human body | The Metabolomics Innovation Centre | https://hmdb.ca |
| SMPDB [33] | The Small Molecule Pathway Database with about 50.000 entries with pathway information about the human body | The Metabolomics Innovation Centre and DrugBank | https://smpdb.ca |
| BIAdb [34] | Database of benzylisoquinoline alkaloids with more than 800 entries | Indraprastha Institute of Information Technology | https://webs.iiitd.edu.in/raghava/biadb/index.html |
| DrugBank [35] | A database that combines chemical and pharmaceutical data with comprehensive drug target information. DrugBank contains more than 13.000 entries of which more than 130 are about nutraceutical molecules | Wishart Research Group | https://www.drugbank.ca |
| SuperNatural [36] | Freely available database of natural compounds with more than 300.000 entries | Charite University of Medicine | http://bioinf-applied.charite.de/supernatural_new/index.php |
| NPACT [37] | Curated database of plant derived natural compounds that exhibits anti-cancerous activity. Contains more than 1.500 entries | The Institute of Cytology and Preventive Oncology | https://webs.iiitd.edu.in/raghava/npact/index.html |
| TTD [38] | Therapeutic Target Database that provides information about the known therapeutic protein and nucleic acid targets and the corresponding drugs directed at each of these targets | Zhejiang University | http://db.idrblab.net/ttd/ |
| PharmGKB [39] | A pharmacogenomics database that encompasses clinical information of drug molecules with more than 600 entries | PharmGKB | https://www.pharmgkb.org |
| SuperDrug [40] | Database that contains about 2500 3D-structures of active compounds of marketed drugs | Charite University of Medicine | http://cheminfo.charite.de/superdrug2/index.html |
By target searches by target are an interesting option when the target which is studied is well-known. This type of query allows to obtain datasets of molecules with information related to a specific target.
By properties A search of molecules based on properties of interest may also be performed e.g. molecules whose hydrophobicity values have been reported may be compiled generating a dataset of compounds for this property to be used in posterior modeling studies.
In addition to the previously mentioned searches, there are other computational strategies that may be followed to find new active molecules structurally different from the existing ones for the cases when, for example, a molecule is shown to have activity, but also undesirable effects and as a result one needs to identify alternatives [21].
Chemical databases have become a fundamental tool in the first steps of computer aided molecular discovery (CAMD) research workflows. Due to the large number of molecules that they offer, it is often necessary that different query criteria and data mining strategies are applied to extract data with the necessary characteristics for each experiment in a more efficient way.
Chemoinformatic techniques
Chemoinformatics is a field of information technology that involves the use of computer techniques to collect, store, analyze and manipulate large sets of chemical data. This data is composed of chemical structures and requires special approaches to represent, store (i.e., using molecular descriptors or SMILES annotation) and retrieve them. Moreover, these techniques seek to establish clear relationships between structural patterns and the corresponding properties or activities [41, 42].
The application of chemoinformatic methods such as pharmacophore modelling, scaffold-hopping, read-across, similarity searching, as well as statistical and machine learning (ML) modeling, has contributed to optimizing and accelerating chemical research in various fields including toxicology, CAMD and biochemistry. Noteworthy among the chemoinformatics approaches employed in the context of nutraceuticals, cosmeceuticals, and functional foods are Quantitative Structure-Activity Relationship (QSAR) models.
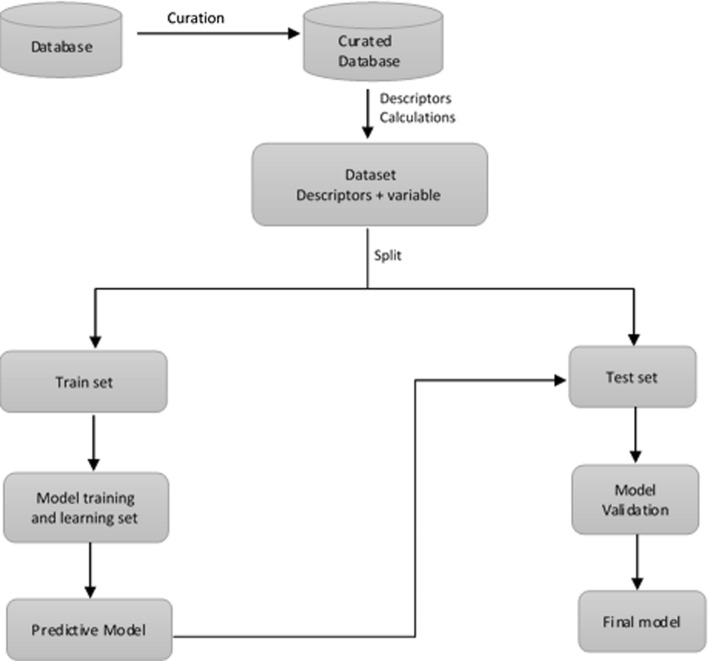
QSAR methodology, proposed over one hundred years ago by Crum Brown, is based on the idea that there exists a relationship between a chemical structure and its activity [43]. QSAR development involves the generation of a mathematical model relating the chemical structure of a dataset of molecules with a specific physicochemical, chemical or biological property. The development of these models requires the previous characterization of the molecules using numerical descriptors and the subsequent application of different statistical and ML tools to generate the algorithms that relate these descriptors with the studied parameter. After the development and validation of a QSAR model, it can be employed as a prediction tool for the property/activity of new molecules with known chemical structures [44, 45]. Figure 2 illustrates a general workflow for constructing QSAR models, which starts with the curation of the dataset of molecules to be employed. Next, a series of molecular descriptors are calculated for this dataset yielding an n × m data matrix, where n is the number of molecules and m the number of descriptors. The data matrix is split in train and test sets and the model training is performed over the training set using different techniques to yield a predictive model. This model is subsequently validated using the test set (also known as the external validation procedure).
Fig. 2.
QSAR general workflow
QSAR techniques may naturally be applied to the field of nutraceuticals, cosmeceuticals and functional foods to study the relationship between food components and a diversity of properties. Indeed this methodology has already been employed in several studies, allowing to translate experimental information on particular molecules into general knowledge on the corresponding biological or physicochemical mechanisms [46]. Here, the most relevant studies will be discussed.
The QSAR methodology has been employed to build multiple linear regression models for the antioxidant capacity of a series of flavonoids, hydrolyzates from various natural proteins (such as soybean or casein), tripeptides derived from β-lactoglobulin (β-LG), as well as peptides with sequences of up to 20 amino acids. Chemical structural interpretation of the flavonoid model revealed that the C-terminus is important for the antioxidant activity [47] and the bulky hydrophobic residues of this terminus were related to the antioxidant activity in three different free radical systems [48]. Moreover, the electronic and hydrogen-bonding properties of the amino acids in the sequences, as well as the steric properties at both terminus, were found to play an important role in the antioxidant activities of β-LG derived tripeptides [49].
QSAR models have been employed to evaluate some nutraceutical activities, such as the possible ACE inhibitory activity of food product components. To this end, models were developed for peptides obtained from plant derived inhibitors and posteriorly used for screening large libraries of peptides obtained from 15 major food commodities. Results showed that peptides from pork, beef and chicken, contained the highest number of potent inhibitory peptides, followed by proteins from egg, soybean and canola [50]. Specifically egg derived peptides were evaluated with the developed QSAR models to predict their ACE inhibitory activity. The obtained results revealed that eggs contained relatively large-chain peptides with this activity, some of them previously reported in the literature, which confirms the validity of the model [51]. Another QSAR model was developed with a dataset of milk-derived tripeptides with known ACE inhibitory activity. The built model (with an R2 of 0.845) was used for the screening of different milk-derived tripeptides with possible ACE inhibitory activity. Four tripeptides were selected from the results as a promissory candidates for this activity and further studied with in vivo experiments, which validated two of the tripeptides (i.e. IVP and VIP) [52].
ML approaches have also been employed in building QSAR models to study the properties of nutraceuticals, cosmeceuticals and functional foods. The main goal for ML is to optimize the performance of a model given an objective function and training dataset. These prediction models help to guide decisions on chemical compound prioritization in drug discovery efforts, predicting toxicity, etc. [53].
ML techniques generally comprise three main paradigms depending in how models are created and trained.
In the case where the model is developed using both input (e.g., molecule descriptors) and output (e.g., IC50) data, we refer to supervised learning. This is one of the most prominent paradigms and uses algorithms such as: support vector machine (SVM) which aims to identify the most optimum hyperplane that separates class instances, decision trees (DT)-employ a rule-based tree structure representation to label instances, or neural networks (NN) which aim to approximate underlying data relationship pattern through a process that mimics the functioning of the human brain, among others.
Contrary to this, if only input data is given then unsupervised learning is performed. ML algorithms attempt, in the context of unsupervised learning, to identify structure within the data (clustering) or reduce the dimensionality.
Finally, reinforcement learning is the part of ML that deals with decision-making, in which an agent learns good responses by modifying or acquiring new responses incrementally [54]. In this case, algorithms follow a sequential experience-driven learning paradigm in that for each step (or state) a mapping to certain actions is performed to maximize the cumulative reward, without the need of labeled data [55, 56].
We will now briefly overview of the most relevant case studies on the use of ML in nutraceuticals, cosmeceutical and functional foods QSAR modeling.
In recent years, ML approaches have been employed to build classification models for identifying antioxidants and made available using web servers. Firstly, the SeqSVM, a sequence-based classification model for the antioxidant activity of proteins extracted from the Universal Protein Resource (Uniprot) was built using the SVM algorithm. This model yielded an overall accuracy of 89.46% [57]. A year later, the AOPs-SVM model, a classifier of antioxidant proteins was developed. For this model, a library was obtained from Uniprot and 473 discrete features were extracted to construct the model, using the 473D feature extraction algorithm [58], based on the PSI-BLAST [59] and PSI-PRED [60] profiles. The built model yielded an average accuracy value of 94.2% and thus improved the performance of the previously obtained SeqSVM classifier. The AOPs-SVM model has been made available via an online platform (http://server.malab.cn/AOPs-SVM/index.jsp) [61].
On the other hand, ML algorithms have also been employed to model the antihypertensive activity. Dietary habits are crucial in the regulation of blood pressure and hypertension. In this sense, nutraceuticals combined with the diet could help to enhance the antihypertensive effect [62]. An in silico platform for antihypertensive peptides of different length has been developed by Kumar et al. Different databases such as AHTPDB [63] with peptides derived from different foods such as milk, eggs or fish were used for this work. For small peptides, a regression model based on the SVM algorithm was developed, yielding an R2 of 0.701 for dipeptides and 0.543 for tripeptides. In another study, classification models were built for tetrapeptides, pentapeptides, hexapeptides, medium peptides (7–12 aa’s) and large peptides (> 12 aa’s), and accuracy values of 76.67%, 72.04%, 77.39%, 82.61%, and 84.21%, were obtained, respectively. A web based platform was created for these models (http://crdd.osdd.net/raghava/ahtpin/) to help in predicting, screening and designing antihypertensive peptides with possible nutraceutical application [64]. Based on this study, another ML classification model was developed with the same dataset but, without considering the length of the peptides. This model yielded an accuracy of 84.73% and was subsequently implemented in a server, called PAAP (https://codes.bio/paap/) which allows to predict, screen or design peptides with antihypertensive activity [65].
ML approaches have been employed in the cosmetics field as well, particularly in finding anti-ageing compounds. A deep learning model was developed with a set of compounds with known antioxidant activity and subsequently employed to screen a database of natural compounds. A lead compound, called pep_RTE62G, was identified and subsequent in vitro and in vivo evaluations corroborated the in silico predictions, thus demonstrating the predictivity of the built model [66]. Furthermore, systemic toxicity of peptides employed in cosmeceutical products is a major concern. Models based on ML approaches using various properties of peptides in cosmeceutical field were developed for predicting their toxicity. The performance of dipeptide model was of 94.50% in terms of accuracy. Also, a hybrid model was developed combining the dipeptide model with information extracted on toxic motifs of peptides yielding an accuracy of approx. 90%. A webserver called ToxinPred (http://crdd.osdd.net/raghava/toxinpred/) has been developed with these models to facilitate the evaluation of the toxicity of peptides [67].
Molecular modelling (MM)
Molecular modelling comprises all theoretical and computational techniques employed to model or mimic the behavior of molecules in nature. These techniques are applied in diverse domains including drug discovery and development, computational chemistry, and materials science, among others, to study molecular systems of different sizes and complexity [68]. The MM techniques may be employed to elucidate reaction mechanisms and interaction modes involving molecular systems, and/or to predict macroscopic physicochemical, chemical or biological properties. These methods include mainly molecular docking and molecular dynamic simulations and are collectively denominated as structure-based computational methods. Each of these methods has proven to be useful in nutraceutical, cosmeceutical and functional food compounds research [69].
For these structure-based methods, good quality (high resolution) three-dimensional (3D) structures of molecules are deemed as a fundamental starting step for successful molecular modelling [70]. The most important techniques for obtaining these 3D structures are shown in Table 2.
Table 2.
Main techniques for obtaining the 3D structure of macromolecules
| Experimental | Technique | Definition |
| X-Ray crystallography | One of the most used technique for structure determination of proteins and macromolecules. A purified sample at high concentration is crystallized and exposed to an x-ray beam. Those results can be processed allowing to obtain the 3D structure of the molecule [71] | |
| Nuclear Magnetic Resonance Spectroscopy | Analytical chemistry technique that reveals the atomic structure of macromolecules in highly concentrated solutions based on the fact that certain atomic nuclei (such as H, 13C, 19F, 23Na, or 31P) are intrinsically magnetic [72]. Provides unique information about dynamics and interactions but the determination of the atomic structure is restricted to small complexes | |
| Cryo-Electron Microscopy | Based on the imaging of frozen-hydrated molecules by electron microscopy. Allows obtaining molecular resolution but in recent studies atomic resolution has been achieved [73] | |
| Computational | Homology modelling | Computational prediction method that allows to determine the protein 3D structure from its amino acid sequence based on 3D structure templates of proteins with similar sequences[74] |
Molecular docking
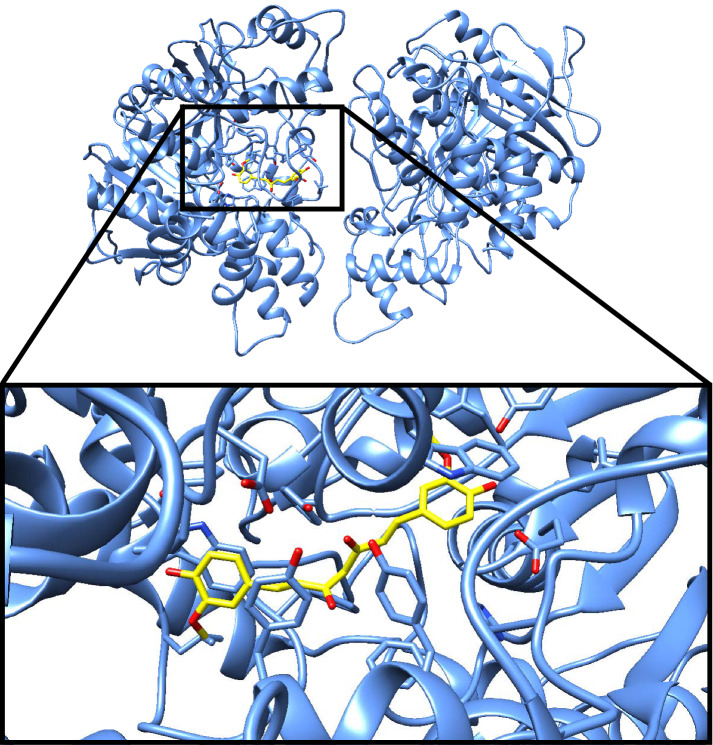
The goal of this technique is to predict the target-ligand binding affinity and possible binding modes, ultimately contributing to understanding molecular recognition process. Molecular docking includes structure-activity studies, finding potential leads by virtual screening, providing binding hypotheses and lead optimization [75]. Figure 3 shows an example of a docking pose of natural compound curcumin bonded to acetylcholinesterase, generated using Chimera [76].
Fig. 3.
Example of a docking pose result of the binding of curcumin in yellow with acetylcholinesterase(PDB ID: 6U3P) in blue
Molecular docking studies have been employed in the nutraceutical’s field, providing information on the first steps of nutraceutical research prior to the in vitro studies. Various molecular docking tools have been developed to date, employing specific algorithms and for different purposes. Some of the most known tools are provided in Table 3. We provide herein a review of the most relevant applications of molecular docking in the evaluation of the possible health benefits of nutraceuticals.
Table 3.
Different molecular docking tools and their algorithms
| Molecular Docking tool | Algorithm | Website |
|---|---|---|
| AutoDock [77] | Monte Carlo Simulated Annealing, Genetic Algorithm, and Lamarckian Genetic Algorithm | http://autodock.scripps.edu/ |
| Gold [78] | Genetic Algorithm | https://www.ccdc.cam.ac.uk/solutions/csd-discovery/components/gold/ |
| Glide [79] | In-house algorithm using different search criteria and refinement using the Monte Carlo method | https://www.schrodinger.com/products/glide |
| Haddock [80] | In-house algorithm that encodes information from identified or predicted protein interfaces in ambiguous interaction restraints to drive the docking process | https://wenmr.science.uu.nl/ |
| PyDock [81] | Fast protocol which uses electrostatics and desolvation energy to score docking poses generated with FFT-based algorithms | https://life.bsc.es/pid/pydock/ |
| SwissDock [82] | Based on the docking software EADock DSS | http://www.swissdock.ch/ |
| Rosetta [83] | Monte Carlo based multi-scale docking algorithm | https://www.rosettacommons.org/software |
| DOCK [84] | Based in a Geometric Matching Algorithm | http://dock.compbio.ucsf.edu/ |
| DockingServer [85] | Includes the PM6 semi-empirical method to AutoDock | https://www.dockingserver.com/web |
| Medusa Dock [86] | In-house algorithm using a stochastic rotamer library of ligands | https://dokhlab.med.psu.edu/cpi/#/MedusaDock |
The multidrug resistance-associated protein 2 (MRP2) is a transporter located in the membrane that acts as an important transporter of food product compounds with poor oral bioavailability. However, its mechanism of action is not well-known, and the corresponding crystal structure is yet to be solved. Fang et al. obtained the homology 3D structure which was subsequently used in a docking experiment of different flavonoids to understand the selectivity of the transporter. This study allowed to separate the flavonoids two in groups: flavonoids that seem to interact with the transporter and those that seem to not interact with the transporter. Further studies will be necessary but this analysis brings greater insight on the chemical-structural profiles responsible for the differences in the flavonoids bioavailability [87].
Legume-derived peptides are known to have diverse benefits in the organism as they present antioxidant and anti-inflammatory properties [88]. Among these, lentil-derived peptides seem to play a dual role presenting both antioxidant and Angiotensin Converting Enzyme (ACE) inhibitory properties. P. García-Mora et al. carried out a study aimed at identifying lentil-derived peptides with dual antioxidant and ACE inhibitory activity, in order to propose them as ingredients for functional foods. After a gastrointestinal digestion simulation, the selected peptides (ligands) and two structures of somatic ACEs (receptors) were prepared with Maestro [89], and induced fit docking studies performed using the Glide 5.7 [79] and Prime 3.0. [90] modules available in the Schrödinger suite. The obtained results showed the possible dual activity of lentil-derived peptides which could guide subsequent in vitro studies to assess this activity [91].
Also, molecular docking studies of tetrapeptides from Atlantic salmon with possible ACE inhibitory activity were performed, with the aim of elucidating their binding modes and determine the corresponding binding energies. For this study, the CHARMM-based program (CDOCKER) was employed. Ten conformational models were obtained but only the best ones were selected for further analysis to identify the implicated residues and the best tetrapeptide conformation. This study showed that residue Glu376 of ACE might play an important role in the interactions with peptides. The IC50 value of two potent ACE inhibitory peptides, PGAR and IGPR, were calculated as 0.598 ± 0.12 and 0.43 ± 0.09 mmol L−1, respectively. Although compared to the clinical hypertension drug lisinopril, both peptides showed lower ACE inhibitory activity, they still have potential for use in hypertension prevention [92].
Ursolic Acid (UA), a plant-derived nutraceutical compound, has been shown to regulate multiple proinflammatory transcription factors and inflammatory enzymes [93]. In a study aimed at identifying specific targets for this compound, the PharmMapper database was explored for potential targets [94] followed by molecular docking studies to select the most favorable ones, based on the corresponding scoring functions. An analysis of the interaction modes of the selected ursolic acid-target complexes allowed to identify the residues critical for the binding process. For this study, the Autodock [77] and MOE software were employed. The UA was observed to bind to Caspase 3 and the obtained results were used for posterior in vitro studies, which further validated this binding and allowed to hypothesize that the anti-inflammatory effect may be mediated by the MAPK signaling pathways [95].
Curcumin seems to have beneficial effects in Alzheimer, by inhibiting the formation of β-amyloid plaques related to this disease. This inhibition seems related to the interaction of curcumin with the acetylcholinesterase (AChE) receptor, which in turn affects the amyloid precursor protein. Molecular docking was employed by Sriraman et al. to evaluate the interaction between curcumin and eight different targets related to Alzheimer disease. The results revealed that that curcumin binds most favorably to AChE compared with the other studied targets [96].
Finally, molecular docking has been applied in the cosmetics sector, for example, to evaluate the possible interaction between tyrosinase and ginsenoside Re, a compound extracted from the roots of the ginseng plants. Tyrosinase plays an essential function in modulating the production of melanin, the primary protective barrier against ultraviolet damage and whose excessive production may result in severe skin ailments such as patches, ephelis and melasma [97]. The computational results of the performed docking study suggested that ginsenoside Re inhibits Tyrosinase and subsequent in vitro studies confirmed the predicted interaction profile. It was thus inferred that ginsenoside Re may be used as potential agent in cosmetics for the inhibition of melanogenesis [98].
Molecular dynamics (MD) simulation
This computational technique is employed to study the physical displacements of atoms and molecules based on Newtonian mechanics laws. To carry out MD simulations, molecular mechanics forcefields, defined as a set of equations that describe the dependence of the energy of a system on the coordinates of its particles and the interatomic potentials, are required [99, 100]. The impact of MD simulations in CAMD has grown dramatically in the last years. Using this technique, it is possible to predict the behavior of atoms in a molecular system over time based on a general model of the physics governing the inter-atomic interactions. These simulations could be very useful in studying important biomolecular processes such as ligand binding, protein folding or conformational changes. Also, such simulations can predict at an atomic level how these biomolecules will act with perturbations such as protonation, mutation or the addition or removal of ligands [101].
Food systems are complex and undergo lots of physical and chemical transformations during processing and storage. These transformations may be analyzed by MD simulations, providing key leads on the energetic and interaction profiles, as well as information on the conformational stability and/or preference [102].
MD simulations have been employed in the nutraceutical and cosmetical field to study the influence of pH and temperature on the corresponding chemical-biological profiles. For example, MD simulations have been employed to study the influence of temperature on the stability of gliadins, major antigenic wheat proteins reported to be responsible of food allergy and coeliac disease [103]. For this study, models of different gliadins were simulated at 25 ºC and 100 ºC until equilibrium was achieved, using the Gromacs platform [104]. This experiment showed that the exposure of linear epitopes and their location changed with temperature and this could affect the allergenicity of these proteins [105].
In another study, Stănciuc et al. applied MD simulations to explore how the conformational changes of β-LG, commonly used in the food industry due to its high nutritional value, contribute to its thermally induced behavior as a result of the chemical and/or physical processing in the food industry. For the MD simulation, the β-LG system was heated over a range of 30-90 ºC with equilibrations for every 10 ºC increment. The HyperChem software was employed for this study [106]. The obtained results indicated that at higher temperatures, β-LG structure was in a more flexible state that favored the hydrophobic exposure, allowing for a better understanding of the structure-function relationship [107]. Also, β-LG plays an important role as a carrier of flavonoids which could improve their bioavailability. The obtained results contributed to understanding the behavior of the studied complexes and how the ligands affect the conformational changes and the stability of β-LG, showing that quercitrin and rutin changed the conformation of β-LG, although the similarity of the atomic fluctuations profiles for β-LG and the β-LG –ligand complexes suggested that the structure of the ligand-binding site remained approximately rigid during the simulation [108].
MD simulations have been employed to study the solubility of nutraceutical and functional foods such as polyphenols, carbohydrates, or lipids. In this case, three polyphenols were employed to create different systems simulated using NAMD [91]. Hydration free energies were computed for each compound and the obtained results showed an exponential relationship between solubility and the hydration free energy [92]. Likewise, Zhang et al.sought to examine how the hydrothermal annealing treatment improves the low heat stability and crystal homogeneity of starch, particularly when mixed with other molecules [93]. Systems formed with cornstarch (CS) and fatty acids were simulated for 1 ns using Gromacs [104]. These results revealed that the total and potential energy were higher (in magnitude) for the lipids-CS complex when compared to CS alone, which led to the inference that more energy is necessary to damage the complex since it has more stability than CS alone [109].
MD simulations have also been employed to evaluate how the variation of the pH affects the physicochemical behavior of components in functional foods. For example, long chain fatty acids used as surfactants in the food industry may present varying aggregation behavior at different pH conditions. In a study by Benett et al. constant pH coarse-grained MD simulations were performed on systems of oleic acid (alone and in a bilayer) to assess the impact of different pH values using coarse-grained MD simulations. The following pH values were considered: 2.0, 5.5 and 9.0, respectively. The obtained results showed that the acidic behavior of oleic acid depend on the local chemical environment, observing an increment of the pKa and a large degree of anticooperativity in small micelles [110].
Conclusion/ expert opinion
Nutraceuticals, cosmeceuticals and functional foods combined with healthy lifestyles could aid in the preservation of health and the reduction of the risk of developing chronic diseases. Moreover, some of the bioactive molecules could be useful to reinforce or complement existing pharmacological therapies, extending their use beyond food boundaries. Despite the potential health benefits of these products and the steady market growth, important challenges remain in this field. One of the main challenges is to prove the specificity of their action during the premarket approval, through the understanding of the mechanism of action of the active molecules in the organism, just as is the case for drugs in the pharmaceutical sector.
Computational techniques can help with these challenges, especially in the first steps, when there is not so much knowledge on the active component(s), the molecular target or mode of action. The inclusion of machine learning approaches in the development phases constitutes an important breakthrough for this field. Indeed, ML methods have contributed to the development and refinement of predictive models, as well as in accelerating molecular docking and molecular dynamics simulations. The implementation of these techniques in studies related to this field not only do they contribute to generating more knowledge at a molecular level, but also allow for the analysis of bigger sets of molecules, for example, in screening of dataset repositories to select those that could possibly present the studied activity for posterior in vitro and in vivo studies. The field of ML has grown exponentially in the recent years as well as its role in CAMD paradigms. This growth is accompanied with an improvement and sophistication of the methods that contribute to better and more precise predictions.
Computational techniques could also help to elucidate the mechanisms of action of molecules of interest by providing information on the binding modes of molecules with macromolecular targets. By means of structural approaches such as molecular dynamics, it is possible to predict not only the binding of two molecules, but also how they interact with time. Also, it is possible to predict how a system of molecules will evolve following changes in parameters such as temperature or pressure, and how these modifications can affect the studied molecules. This can be useful for quality control during the shelf-life analysis as well as to understand and improve the production processes of these products.
Nonetheless, the use of computational techniques in nutraceutical, cosmeceutical and functional foods is still at an early stage when compared to the pharmaceutical field. Also, these methods have some limitations that need to be addressed in order to increase not only their presence but also their functionality in the field. As a case in point, QSAR modelling is not well established for mixtures, which means that QSAR models mostly analyse independent components in mixtures and thus do not contemplate possible synergic or antagonist effects. Also, techniques as molecular docking and molecular dynamic simulation depend on having quality 3D structural models of the studied molecules (ligands, lipids, proteins) or analogues.
Computational approaches could contribute to making the approval process of these products more robust and consolidate the knowledge on their mode of action, thus helping to support the effectiveness and safety of the products that are launched on the market. Also, the limitations that these technologies seem to have when applied to foods may be viewed as future challenges to be solved and opportunities to improve the usefulness of computational approaches in a broader context.
Acknowledgements
We gratefully acknowledged the financial support of the Agencia Valenciana de la Investigación (AVI, https://innoavi.es/en/) by its program Innodocto (Reference number INNTAL32/19/002). SJB acknowledges the funding assistance from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 893810.
Declaration
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.PubMed. https://pubmed.ncbi.nlm.nih.gov/. Accessed 10 Dec 2020
- 2.Kalra EK. Nutraceutical - definition and introduction. AAPS J. 2003;5:1–2. doi: 10.1208/ps050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daliu P, Santini A, Novellino E. A decade of nutraceutical patents: where are we now in 2018? Expert Opin Ther Pat. 2018;00:1–8. doi: 10.1080/13543776.2018.1552260. [DOI] [PubMed] [Google Scholar]
- 4.Santini A, Cammarata SM, Capone G, et al. Nutraceuticals: opening the debate for a regulatory framework. Br J Clin Pharmacol. 2018;84:659–672. doi: 10.1111/bcp.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bragazzi NL, Martini M, Saporita TC, et al (2017) Nutraceutical and functional food regulations in the European Union. In: Developing New Functional Food and Nutraceutical Products. Elsevier, 309–322
- 6.Serafini M, Stanzione A, Foddai S. Functional foods: traditional use and european legislation. Int J Food Sci Nutr. 2012;63:7–9. doi: 10.3109/09637486.2011.637488. [DOI] [PubMed] [Google Scholar]
- 7.Tsai TC, Hantash BM. Cosmeceutical agents : a comprehensive review of the literature. Clin Med Insights Dermatology. 2008;1:20. [Google Scholar]
- 8.Nutraceuticals Market Growth, Size, Trends and Forecast 2020 to 2025. https://www.marketdataforecast.com/market-reports/global-nutraceuticals-market. Accessed 1 Mar 2021
- 9.Global Nutraceuticals Market | Growth | Trends | Forecast (2020 - 2025). https://www.mordorintelligence.com/industry-reports/global-nutraceuticals-market-industry. Accessed 1 Mar 2021
- 10.Functional Food Market Growth, Size, Share and Forecast to 2025. https://www.marketdataforecast.com/market-reports/functional-food-market. Accessed 1 Mar 2021
- 11.Global Cosmeceutical Market Size & Share Report, 2019–2025. https://www.grandviewresearch.com/industry-analysis/cosmeceutical-market. Accessed 1 Mar 2021
- 12.Das L, Bhaumik E, Raychaudhuri U, Chakraborty R. Role of nutraceuticals in human health. J Food Sci Technol. 2012;49:173–183. doi: 10.1007/s13197-011-0269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Google Patents. https://patents.google.com/. Accessed 9 Dec 2020
- 14.Wipo (2010) World Intellectual Property Indicators 2010
- 15.Dutta S, Ali KM, Dash SK, Giri B. Role of Nutraceuticals on Health Promotion and Disease Prevention: a Review. J Drug Deliv Ther. 2018;8:4–10. doi: 10.22270/jddt.v8i4.1759. [DOI] [Google Scholar]
- 16.Zhao J. Nutraceuticals, nutritional therapy, phytonutrients, and phytotherapy for improvement of human health: a perspective on plant biotechnology application. Recent Pat Biotechnol. 2008;1:75–97. doi: 10.2174/187220807779813893. [DOI] [PubMed] [Google Scholar]
- 17.Lintner K, Mas-Chamberlin C, Mondon P, et al. Cosmeceuticals and active ingredients. Clin Dermatol. 2009;27:461–468. doi: 10.1016/j.clindermatol.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Peña-Castillo A, Méndez-Lucio O, Owen JR, et al. Chemoinformatics in food science. Appl Chemoinformatics. 2018 doi: 10.1002/9783527806539.ch10. [DOI] [Google Scholar]
- 19.Lagunin AA, Goel RK, Gawande DY, et al. Chemo- and bioinformatics resources for in silico drug discovery from medicinal plants beyond their traditional use: a critical review. Nat Prod Rep. 2014;31:1585–1611. doi: 10.1039/C4NP00068D. [DOI] [PubMed] [Google Scholar]
- 20.Miller MA. Chemical database techniques in drug discovery. Nat Rev Drug Discov. 2002;1:220–227. doi: 10.1038/nrd745. [DOI] [PubMed] [Google Scholar]
- 21.Ojeda-Montes MJ, Casanova-Martí À, Gimeno A, et al. Mining large databases to find new leads with low similarity to known actives: application to find new DPP-IV inhibitors. Future Med Chem. 2019;11:1387–1401. doi: 10.4155/fmc-2018-0597. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Chen J, Cheng T, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019 doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sterling T, Irwin JJ. ZINC 15 - Ligand discovery for everyone. J Chem Inf Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendez D, Gaulton A, Patrícia Bento A, 2019. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. [DOI] [PMC free article] [PubMed]
- 25.NCI/CADD Group Chemoinformatics Tools and User Services. https://cactus.nci.nih.gov/index.html. Accessed 10 Aug 2020
- 26.Chen JH, Linstead E, Swamidass SJ, et al. ChemDB update-full-text search and virtual chemical space. 2007;23:2348–2351. doi: 10.1093/bioinformatics/btm341. [DOI] [PubMed] [Google Scholar]
- 27.ChemSpider | Search and share chemistry. https://www.chemspider.com/. Accessed 10 Aug 2020
- 28.Gilson MK, Liu T, Baitaluk M, et al. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2015;44:1045–1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su M, Yang Q, Du Y, et al. Comparative assessment of scoring functions: the CASF-2016 update. J Chem Inf Model. 2019;59:895–913. doi: 10.1021/acs.jcim.8b00545. [DOI] [PubMed] [Google Scholar]
- 30.Dimitropoulos D, Ionides J, Henrick K (2006). Using MSDchem to Search the PDB Ligand Dictionary. In: Current Protocols in Bioinformatics. John Wiley & Sons, Inc., Hoboken, NJ, USA, 14.3.1–14.3.21 [DOI] [PubMed]
- 31.Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes [DOI] [PMC free article] [PubMed]
- 32.Wishart DS, Tzur D, Knox C, et al HMDB: the Human Metabolome Database. 10.1093/nar/gkl923 [DOI] [PMC free article] [PubMed]
- 33.Frolkis A, Knox C, Lim E, et al SMPDB: The Small Molecule Pathway Database. 10.1093/nar/gkp1002 [DOI] [PMC free article] [PubMed]
- 34.Singla D, Sharma A, Kaur J, et al. BIAdb: a curated database of benzylisoquinoline alkaloids. BMC Pharmacol. 2010;10:4. doi: 10.1186/1471-2210-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wishart DS, Feunang YD, Guo AC, et al (2018) DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:. 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed]
- 36.Banerjee P, Erehman J, Bj¨ B, et al. Super Natural II-a database of natural products. Nucleic Acids Res. 2014;43:935–939. doi: 10.1093/nar/gku886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangal M, Sagar P, Singh H, et al NPACT: Naturally Occurring Plant-based Anti-cancer Compound-Activity-Target database. 10.1093/nar/gks1047 [DOI] [PMC free article] [PubMed]
- 38.Wang Y, Zhang S, Li F, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2019;48:1031–1041. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whirl-Carrillo M, Mcdonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for. Pers Med. 2012 doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siramshetty VB, Eckert OA, Bj¨ B,, et al. SuperDRUG2: a one stop resource for approved/marketed drugs. Nucleic Acids Res. 2017;46:1137–1143. doi: 10.1093/nar/gkx1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J, Hagler A. Chemoinformatics and drug discovery. Molecules. 2002;7:566–600. doi: 10.3390/70800566. [DOI] [Google Scholar]
- 42.Brown F (2007) Introduction to Cheminformatics. 1–9
- 43.Brown AC, Fraser TR. V.—On the connection between chemical constitution and physiological action. part. i.—on the physiological action of the salts of the ammonium bases, derived from Strychnia, Brucia, Thebaia, Codeia, Morphia, and Nicotia. Trans R Soc Edinburgh. 1867;25:151–203. doi: 10.1017/S0080456800028155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halder AK, Moura AS, Cordeiro MNDS. QSAR modelling: a therapeutic patent review 2010-present. Expert Opin Ther Pat. 2018;28:467–476. doi: 10.1080/13543776.2018.1475560. [DOI] [PubMed] [Google Scholar]
- 45.Gozalbes R, Vicente de Julián-Ortiz J. Applications of Chemoinformatics in Predictive toxicology for regulatory purposes, especially in the context of the EU REACH legislation. Int J Quant Struct Relationships. 2017;3:1–24. doi: 10.4018/ijqspr.2018010101. [DOI] [Google Scholar]
- 46.Pripp AH, Isaksson T, Stepaniak L, et al. Quantitative structure activity relationship modelling of peptides and proteins as a tool in food science. Trends Food Sci Technol. 2005;16:484–494. doi: 10.1016/j.tifs.2005.07.003. [DOI] [Google Scholar]
- 47.Li Y-W, Li B, He J, Qian P. Structure-activity relationship study of antioxidative peptides by QSAR modeling: the amino acid next to C-terminus affects the activity. J Pept Sci. 2011;17:454–462. doi: 10.1002/psc.1345. [DOI] [PubMed] [Google Scholar]
- 48.Li YW, Li B. Characterization of structure-antioxidant activity relationship of peptides in free radical systems using QSAR models: Key sequence positions and their amino acid properties. J Theor Biol. 2013;318:29–43. doi: 10.1016/j.jtbi.2012.10.029. [DOI] [PubMed] [Google Scholar]
- 49.Tian M, Fang B, Jiang L, et al. Structure-activity relationship of a series of antioxidant tripeptides derived from β-Lactoglobulin using QSAR modeling. Dairy Sci Technol. 2015;95:451–463. doi: 10.1007/s13594-015-0226-5. [DOI] [Google Scholar]
- 50.Gu Y, Majumder K, Wu J. QSAR-aided in silico approach in evaluation of food proteins as precursors of ACE inhibitory peptides. Food Res Int. 2011;44:2465–2474. doi: 10.1016/j.foodres.2011.01.051. [DOI] [Google Scholar]
- 51.Majumder K, Wu J. A new approach for identification of novel antihypertensive peptides from egg proteins by QSAR and bioinformatics. Food Res Int. 2010;43:1371–1378. doi: 10.1016/j.foodres.2010.04.027. [DOI] [Google Scholar]
- 52.Jing P, Qian B, He Y, et al. Screening milk-derived antihypertensive peptides using quantitative structure activity relationship (QSAR) modelling and invitro/invivo studies on their bioactivity. Int Dairy J. 2014;35:95–101. doi: 10.1016/j.idairyj.2013.10.009. [DOI] [Google Scholar]
- 53.Angra S, Ahuja S (2017) Machine learning and its applications: A review. Proc 2017 Int Conf Big Data Anal Comput Intell ICBDACI 2017 57–60. 10.1109/ICBDACI.2017.8070809
- 54.François-lavet V, Henderson P, Islam R, et al. An Introduction to deep reinforcement learning. Found trends Mach Learn. 2018;II:1–140. doi: 10.1561/2200000071.Vincent. [DOI] [Google Scholar]
- 55.Strieth-Kalthoff F, Sandfort F, Segler M, Glorius F. Machine learning the ropes: principles, applications and directions in synthetic chemistry. Chem Soc Rev. 2020 doi: 10.1039/C9CS00786E. [DOI] [PubMed] [Google Scholar]
- 56.María PG-MMM-M, Martinez-Vilalluenga. ABRG-MEPJFC (2017). Identification, functional gastrointestinal stability and molecular docking studies of lentil peptides with dual antioxidant and angiotensin I converting enzyme inhibitory activities. In: Food Chemistry. 464–472 [DOI] [PubMed]
- 57.Xu L, Liang G, Shi S, Liao C. SeqSVM: a sequence-based support vector machine method for identifying antioxidant proteins. Int J Mol Sci. 2018;19:1773. doi: 10.3390/ijms19061773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei L, Liao M, Gao X, Zou Q. Enhanced protein fold prediction method through a novel feature extraction technique. IEEE Trans Nanobioscience. 2015;14:649–659. doi: 10.1109/TNB.2015.2450233. [DOI] [PubMed] [Google Scholar]
- 59.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 61.Meng C, Jin S, Wang L, et al. AOPs-SVM: a sequence-based classifier of antioxidant proteins using a support vector machine. Front Bioeng Biotechnol. 2019;7:224. doi: 10.3389/fbioe.2019.00224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Houston MC. Nutrition and nutraceutical supplements in the treatment of hypertension. Expert Rev Cardiovasc Ther. 2010;8:821–833. doi: 10.1586/erc.10.63. [DOI] [PubMed] [Google Scholar]
- 63.Kumar R, Chaudhary K, Sharma M, et al. AHTPDB: a comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic Acids Res. 2015;43:D956–D962. doi: 10.1093/nar/gku1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar R, Chaudhary K, Singh Chauhan J, et al. An in silico platform for predicting, screening and designing of antihypertensive peptides. Sci Rep. 2015;5:12512. doi: 10.1038/srep12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Win TS, Schaduangrat N, Prachayasittikul V, et al. PAAP: a web server for predicting antihypertensive activity of peptides. Future Med Chem. 2018;10:1749–1767. doi: 10.4155/fmc-2017-0300. [DOI] [PubMed] [Google Scholar]
- 66.Kennedy K, Cal R, Casey R, et al. The anti-ageing effects of a natural peptide discovered by artificial intelligence. Int J Cosmet Sci. 2020;42:388–398. doi: 10.1111/ics.12635. [DOI] [PubMed] [Google Scholar]
- 67.Diehl C. (2019). Peptides in cosmeceuticals. Ukr J Dermatology, Venerol Cosmetol 0:28–35. 10.30978/ujdvk2019-1-28
- 68.Intelligence M, Learning M (2013) Encyclopedia of Sciences and Religions
- 69.Tao X, Huang Y, Wang C, et al. Recent developments in molecular docking technology applied in food science: a review. Int J Food Sci Technol. 2020;55:33–45. doi: 10.1111/ijfs.14325. [DOI] [Google Scholar]
- 70.Schneider G, Fechner U. Computer-based de novo design of drug-like molecules. Nat Rev Drug Discov. 2005;4:649–663. doi: 10.1038/nrd1799. [DOI] [PubMed] [Google Scholar]
- 71.Smyth MS, Martin JHJ. x Ray crystallography. J Clin Pathol - Mol Pathol. 2000;53:8–14. doi: 10.1136/mp.53.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berg JM, Tymoczko JL, Stryer L (2002) Three-Dimensional Protein Structure Can Be Determined by NMR Spectroscopy and X-Ray Crystallography
- 73.Nakane T, Kotecha A, Sente A, et al. Single-particle cryo-EM at atomic resolution. Nature. 2020;587:152–156. doi: 10.1038/s41586-020-2829-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muhammed MT, Aki-Yalcin E. Homology modeling in drug discovery: overview, current applications, and future perspectives. Chem Biol Drug Des. 2019;93:12–20. doi: 10.1111/cbdd.13388. [DOI] [PubMed] [Google Scholar]
- 75.Schleinkofer K, Wang T, Wade RC. Molecular docking. Encycl Ref Genomics Proteomics Mol Med. 2006;443:1149–1153. doi: 10.1007/3-540-29623-9_3820. [DOI] [Google Scholar]
- 76.Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera - a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 77.Morris GM, Huey R, Lindstrom W, et al AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed]
- 78.Jones G, Willett P, Glen RC, et al. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 79.Friesner RA, Murphy RB, Repasky MP, et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 80.Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 81.Jiménez-García B, Pons C, Fernández-Recio J (2013) pyDockWEB: A web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. In: Bioinformatics. Bioinformatics, pp 1698–1699 [DOI] [PubMed]
- 82.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lyskov S, Gray JJ. The RosettaDock server for local protein-protein docking. Nucleic Acids Res. 2008;36:W233. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Allen WJ, Balius TE, Mukherjee S, et al. DOCK 6: Impact of new features and current docking performance. J Comput Chem. 2015;36:1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bikadi Z, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of autodock. J Cheminform. 2009;1:1–16. doi: 10.1186/1758-2946-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang J, Dokholyan NV. MedusaDock 2.0: efficient and accurate protein-ligand docking with constraints. J Chem Inf Model. 2019;59:2509–2515. doi: 10.1021/acs.jcim.8b00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fang Y, Cao W, Liang F, et al. Structure affinity relationship and docking studies of flavonoids as substrates of multidrug-resistant associated protein 2 (MRP2) in MDCK/MRP2 cells. Food Chem. 2019;291:101–109. doi: 10.1016/j.foodchem.2019.03.111. [DOI] [PubMed] [Google Scholar]
- 88.Garcia-Mora P, Frias J, Peñas E, et al. Simultaneous release of peptides and phenolics with antioxidant, ACE-inhibitory and anti-inflammatory activities from pinto bean (Phaseolus vulgaris L. var. pinto) proteins by subtilisins. J Funct Foods. 2015;18:319–332. doi: 10.1016/j.jff.2015.07.010. [DOI] [Google Scholar]
- 89.Maestro S (2020) Schrödinger Release 2020–2:
- 90.Schrödinger (2020) Schrödinger Release 2020–2: Prime
- 91.García-Mora P, Martín-Martínez M, Angeles Bonache M, et al. Identification, functional gastrointestinal stability and molecular docking studies of lentil peptides with dual antioxidant and angiotensin I converting enzyme inhibitory activities. Food Chem. 2017;221:464–472. doi: 10.1016/j.foodchem.2016.10.087. [DOI] [PubMed] [Google Scholar]
- 92.Yu Z, Chen Y, Zhao W, et al. Identification and molecular docking study of novel angiotensin-converting enzyme inhibitory peptides from Salmo salar using in silico methods. J Sci Food Agric. 2018;98:3907–3914. doi: 10.1002/jsfa.8908. [DOI] [PubMed] [Google Scholar]
- 93.Shanmugam MK, Dai X, Kumar AP, et al. Ursolic acid in cancer prevention and treatment: molecular targets, pharmacokinetics and clinical studies. Biochem Pharmacol. 2013;85:1579–1587. doi: 10.1016/j.bcp.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 94.Liu X, Ouyang S, Yu B, et al. PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 2010;38:W609–W614. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma X, Zhang Y, Wang Z, et al. Ursolic acid, a natural nutraceutical agent, targets caspase3 and alleviates inflammation-associated downstream signal transduction. Mol Nutr Food Res. 2017;61:3–8. doi: 10.1002/mnfr.201700332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Roy S, Sriraman S, Saha NG. Interaction of curcumin with different target proteins of Alzheimer’s disease: docking and MD simulation studies. Int J Comput Biol Drug Des. 2017;10:315. doi: 10.1504/ijcbdd.2017.10009070. [DOI] [Google Scholar]
- 97.Briganti S, Camera E, Picardo M. Chemical and instrumental approaches to treat hyperpigmentation. Pigment Cell Res. 2003;16:101–110. doi: 10.1034/j.1600-0749.2003.00029.x. [DOI] [PubMed] [Google Scholar]
- 98.Jiménez-pérez ZE, Kim Y, Castro-aceituno V, et al (2017) Jiménez et al ., Afr J Tradit Complement Altern Med ., ( 2017 ) 14 ( 5 ): 209–218 Department of Biotechnology and Ginseng Bank , 2 Department of Oriental Medicine Biotechnology , College of Life Sciences , Kyung Hee University , Yongin , Republic of Korea. 14:209–218
- 99.Chen G, Huang K, Miao M, et al. Molecular dynamics simulation for mechanism elucidation of food processing and safety: state of the art. Compr Rev Food Sci Food Saf. 2019;18:243–263. doi: 10.1111/1541-4337.12406. [DOI] [PubMed] [Google Scholar]
- 100.González MA. Force fields and molecular dynamics simulations. École thématique la Société Française la Neutron. 2011;12:169–200. doi: 10.1051/sfn/201112009. [DOI] [Google Scholar]
- 101.Hollingsworth SA, Dror RO. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Paramita VD, Kasapis S. Molecular dynamics of the diffusion of natural bioactive compounds from high-solid biopolymer matrices for the design of functional foods. Food Hydrocoll. 2019;88:301–319. doi: 10.1016/j.foodhyd.2018.09.007. [DOI] [Google Scholar]
- 103.Kahlenberg F, Sanchez D, Lachmann I, et al. Monoclonal antibody R5 for detection of putatively coeliac-toxic gliadin peptides. Eur Food Res Technol. 2006;222:78–82. doi: 10.1007/s00217-005-0100-4. [DOI] [Google Scholar]
- 104.Abraham MJ, Murtola T, Schulz R, et al. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 105.Stănciuc N, Banu I, Bolea C, et al. Structural and antigenic properties of thermally treated gluten proteins. Food Chem. 2018;267:43–51. doi: 10.1016/j.foodchem.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 106.Froimowitz M. HyperChem(TM): a software package for computational chemistry and molecular modeling. Biotechniques. 1993;14:1010–1013. [PubMed] [Google Scholar]
- 107.Stǎnciuc N, Aprodu I, Râpeanu G, Bahrim G. Fluorescence spectroscopy and molecular modeling investigations on the thermally induced structural changes of bovine β-lactoglobulin. Innov Food Sci Emerg Technol. 2012;15:50–56. doi: 10.1016/j.ifset.2012.03.001. [DOI] [Google Scholar]
- 108.Sahihi M, Heidari-Koholi Z, Bordbar A-K. The Interaction of polyphenol flavonoids with β-lactoglobulin: molecular docking and molecular dynamics simulation studies. J Macromol Sci Part B. 2012;51:2311–2323. doi: 10.1080/00222348.2012.672854. [DOI] [Google Scholar]
- 109.Zhang X, Wang YS, Chen HH. Effect of annealing temperature on morphology and physicochemical properties of cornstarch complexed with oleic acid and molecular dynamics simulation. Cereal Chem. 2019;96:668–677. doi: 10.1002/cche.10163. [DOI] [Google Scholar]
- 110.Bennett WFD, Chen AW, Donnini S, et al. Constant pH simulations with the coarse-grained MARTINI model — Application to oleic acid aggregates. Can J Chem. 2013;91:839–846. doi: 10.1139/cjc-2013-0010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Mendez D, Gaulton A, Patrícia Bento A, 2019. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. [DOI] [PMC free article] [PubMed]