INTRODUCTION
The symptom of low back pain (LBP) is the fifth most frequent reason for physician office visits in the United States (US),[11] and the leading cause of years lived with disability.[18] It is the single most common site of pain among US adults.[17] Yet despite its prevalence and major societal impact, much remains unknown about the basic biology underlying this symptom. Identifying genetic risk factors for LBP is one possible route towards understanding the mechanisms of LBP and the development of new treatments.
LBP has a substantial genetic component, with broad-sense heritability estimates of 40% from classical twin studies.[54] We recently conducted the first two genome-wide association studies (GWAS) of self-reported LBP in the UK Biobank (UKB) and the Cohorts for Heart and Aging Research Genomic Epidemiology (CHARGE) consortium. These GWAS identified several genetic variants associated with LBP and demonstrated that genetic factors underpin the biopsychosocial model of LBP.[22; 54] These findings provide a starting point for genetic discovery into the mechanisms underlying LBP, yet other approaches may be needed to account for the heterogeneity of conditions associated with LBP. LBP is a symptom and not a diagnosis. Clinical spine specialists routinely “subgroup” patients with LBP into diagnostic categories, and offer different treatments depending on the subgroup implicated. The two most widely recognized diagnoses associated with LBP are lumbosacral radicular pain or radiculopathy (sometimes referred to as “lumbosacral radicular syndrome” [LSRS] in the research context[39]) and lumbar spinal stenosis (LSS).[26] While these two subgroups are generally accepted by clinical spine specialists and researchers, many other subgroups lack widely accepted diagnostic criteria and reliability in research studies.[25; 41] For this reason, much LBP research to date has focused on the management of those with “non-specific LBP”, in which potential peripheral nociceptive causes of LBP are not considered, beyond the exclusion of rare “red flag” conditions. Ignoring phenotypic heterogeneity related to LBP may decrease statistical power for genetic discovery through GWAS.[42] A different approach would be to target more specific diagnostic subgroups- called “endophenotypes” such as the well-recognized clinical syndromes of LSRS and LSS. While these diagnoses cannot be accurately identified by patient self-report, they can be identified by the diagnostic codes used by clinicians in routine clinical care, by leveraging the resources contained in existing genetic biobanks linked to longitudinal electronic health record (EHR) data. Genetic biobanks using EHR data make available patient/participants’ extended medical histories, and these longitudinal datasets may permit more refined pain phenotyping incorporating information about diagnostic subgroups.
In the current study, we leverage EHR biobanks for novel genetic discovery into the mechanisms underlying LBP and lumbar spinal disorders. The first aim was to identify novel genetic variants associated with LBP prompting health care utilization. The second aim was to identify novel genetic variants associated with LSRS and LSS. A third aim was to examine the external validity of variants previously identified in GWAS of self-reported LBP and lumbar spinal disorders, in the context of EHR biobanks.
METHODS
Overview
GWAS involve scanning large numbers of genetic markers across the genome in order to discover genetic variations associated with a trait or disease, using stringent correction for multiple statistical testing.[30] By examining genetic variation on a genome-wide basis, GWAS are often described as “agnostic” or “hypothesis-free” approaches,[34] and have largely proven to be more successful at producing replicable, generalizable findings for complex phenotypes as compared to candidate gene approaches.[19] We conducted a meta-analysis using two non-overlapping GWAS performed within the Electronic Medical Records and Genomics (eMERGE) network,[50] and the Geisinger Health system, followed by external validation of genome-wide significant variants in an independent sample from the UKB. eMERGE is a US network of medical centers with EHR data linked to biorepository samples and genomic data.[50] The network has existed for over 12 years, supported by successive phases of funding from NIH, primarily the National Human Genome Research Institute; the current study used data from eMERGE Phase 3 (eMERGE3). eMERGE3 involved nine non-pediatric study sites and associated medical centers (Columbia University Health Sciences, Geisinger Health, Partners Healthcare/Harvard University, Kaiser Permanente Washington/University of Washington (UW), Mayo Clinic, Marshfield Clinic, Mt. Sinai Health System, Northwestern University, and Vanderbilt University)[50]. A second GWAS was performed using data from Geisinger Health, excluding participants who were part of the eMERGE3 dataset so as to remove overlap of participants, and results from the two GWAS were meta-analyzed.
Phenotyping of LBP and lumbar spine disorders (discovery sample)
Cases and controls were defined using longitudinal EHR data and validated algorithms for International Classification of Diseases (ICD) Version 9 codes, (ICD-9) which have been widely used in research studies over the past 25 years.[9; 15; 16; 43] These algorithms were updated to include the International Classification of Diseases, version 10 (ICD-10).[35] Four EHR-defined phenotypes were studied: (1) Any LBP prompting health care utilization (the “LBP-HC” phenotype); (2) Lumbosacral radicular syndrome (the “LSRS” phenotype); (3) Lumbar spinal stenosis (the “LSS” phenotype), and (4) Any lumbar spine-associated neuropathic leg pain (the “LSRS/LSS phenotype”) (Supplemental File 1). To ensure a minimum time under observation for all participants, analyses were restricted to those age >18 years with at least 1 year of time in the dataset during which they used health care services (as reflected by ICD codes or Current Procedural Terminology codes). Following the “rule of two” to limit phenotype misclassification,[27] cases were defined as adults with 2 or more ICD-9 or ICD-10 codes indicating a phenotype, and controls were defined as adults with no codes indicating a phenotype (all codes included in Supplemental File 1). Adults with only 1 diagnostic code indicating a phenotype were omitted from the analysis (i.e., not included as cases or controls). The LBP-HC phenotype was intended to capture any episodes of LBP of sufficient severity to prompt health care utilization, with or without associated lower extremity neuropathic pain. It reflects non-specific LBP, without any assumptions regarding the possible underlying diagnostic subgroups involved. In contrast, the other phenotypes examined (LSRS, LSS, and LSRS/LSS), did rely on the distinctions made by the diagnostic codes used by clinicians during routine clinical care. We used a validated hierarchical EHR-based phenotyping algorithm for identifying LSRS and LSS;[43] associations between LSRS and LSS defined in this manner are associated with distinct trajectories of symptoms among those with LBP.[16] The LSRS phenotype includes the diagnoses of intervertebral disc displacement with lumbosacral radiculopathy and radicular pain, but may also include various other diagnoses that can result in a similar presentation, such as sciatic neuropathy, piriformis syndrome, sacroiliac joint pain, and others.[55] The LSS phenotype encompasses neurogenic claudication and other instances of symptoms in the setting of lumbar spinal stenosis.[55] Because the clinical syndromes of LSRS and LSS are related anatomically (e.g. any substantial disc herniation by definition causes at least some stenosis [narrowing] of the spinal canal) and in terms of their clinical presentations,[55] an LSRS/LSS phenotype was also defined where 2 or more ICD-9 or ICD-10 codes reflecting LSRS or LSS defined cases, and controls had no codes reflecting LSRS or LSS. No exclusions for red flag conditions were made in the phenotyping process, based on the documented very low frequency of red flags in clinical samples.[26; 35]
Genotyping (discovery sample)
We have previously reported genotyping methods for eMERGE3.[50] The eMERGE network conducted genotyping using Illumina and Affymetrix arrays in 83 batches from across the participating sites. Genotyping at Geisinger used the Illumina Global Screening Array-24 and Illumina OmniExpress arrays. Imputation of single nucleotide variants (SNVs) for both datasets was performed using guidelines from the Michigan Imputation Server,[13; 38] which relies on reference panels from the Haplotype Reference Consortium (HRC) release 1.1 genome build 37 (hg19).[44]. Human subjects approvals were provided by each participating eMERGE3 site and all participants completed informed consent.
Phenotyping and genotyping in UKB (validation sample)
Many proposed prognostic factors in LBP research do not replicate, or have inconsistent results between studies.[52; 56; 58] Replication in independent samples is a standard practice in GWAS that helps to assure validity and generalizability. Variants attaining genome-wide significance in the discovery phase were carried forward for external validation in UKB. UKB is a population-based prospective study involving more than 500,000 participants between the ages of 40 and 69 years, established to allow detailed studies of the genetic and nongenetic determinants of diseases of middle age and old age.[51] Although is evidence for a “healthy volunteer” selection bias in UKB, it is believed to have valid assessments of exposure-disease relationships that are widely generalizable to the general UK population.[23] In particular, UKB participants are similar to the general UK population in the prevalence of pain conditions and their association with sociodemographic and psychological factors.[40] Cases and controls were defined using ICD-10 diagnostic codes obtained during a hospitalization and medical conditions reported during an interview with a trained nurse.[4] Analyses were restricted to individuals who had at least one ICD-10 code of any type or one non-cancer diagnostic code recorded during the interview (Supplemental File 1); a minimum requirement of 2 diagnostic codes was not applied in UKB because the specificity of hospital-based diagnostic coding in UKB was expected to be higher than that of the outpatient- and hospital-based diagnostic coding used in eMERGE3/Geisinger. LSRS cases in UKB (LSRS-UKB) were participants who had 1 or more ICD-10 diagnostic codes indicating LSRS, or reported “sciatica” or “prolapsed disc/slipped disc” during the interview; cases were participants who had no ICD-10 codes indicating LSRS and no report of “sciatica” or “prolapsed disc/slipped disc” during the interview. Cases and controls for the LSS and LSS/LSRS phenotypes were defined using similar methods (Supplemental File 1). Due to substantial differences between eMERGE3/Geisinger and UKB in terms of the study settings, data collection, and phenotyping methods involved, replication in UKB was viewed as a challenging test of external validity using distinct yet related clinical phenotypes. Genotyping for UKB was conducted using the Affymetrix UK BiLEVE Axiom and UK Biobank Axiom arrays. Imputation of SNVs and indels was conducted by UKB (version 3), using the HRC, UK10K, and 1000 Genomes reference panels. Ethics approval was provided by the UK Biobank Research Ethics Committee (11/NW/0382). UKB data for this study was obtained under the project #18219.
Statistical analyses
Analyses of eMERGE3 and Geisinger (aims 1 and 2) were restricted to those of European ancestry (EA) in order to decrease genetic heterogeneity, according to the intersection of self-reported race and genetic ancestry determined by clustering of ancestry principle components.[50] Third-degree relatives and closer relatives were excluded from the analysis to account for interrelatedness. In the GWAS of eMERGE3 data, filters were applied for minor allele frequency [MAF] <0.005, imputation r2 <0.3, deviation from Hardy-Weinberg equilibrium (HWE) p-value < 10−6, genotyping call rate <0.98, and individual call rate <0.98. In the GWAS of Geisinger data, filters were applied for MAF <0.01, imputation r2 <0.3 or <0.4 depending on the array, deviation from HWE p-value < 10−15, genotyping call rate <0.90, and individual call rate <0.90. Logistic regression of imputed SNVs with an additive genotype model was conducted adjusting for sex, age, site-specific characteristics, and ancestry principal components 1 to 10 in PLINK 1.9.[8; 46] Analyses were not adjusted for body mass index due to the fact that elevated BMI may be a consequence of back pain. We harmonized GWAS results and conducted quality control using EasyQC, removing SNVs with MAF <0.01, imputation r2 <0.3, deviation from HWE p-value < 10−6, BETA>= 10, 0>=SE>=10 or SE=Inf, and MAC (expected minor allele count)<=100.[62]
The software METAL was used for meta-analysis.[61]. A p-value < 5 × 10−8 was used to define genome-wide statistical significance. Heritability was calculated using the linkage disequilibrium (LD) score regression (LDSR) method.[6] Liability scale heritability for LBP-HC was estimated assuming a lifetime prevalence of seeking health care for LBP in the general population of 48.7%, based on the product of estimates from the literature for the lifetime prevalence of an episode of LBP (84%)[7]and the proportion of LBP episodes that result in health care seeking (58%).[21] Similarly, liability scale heritabilities for LSRS, LSS, and LSRS/LSS were estimated assuming a lifetime prevalence in the general population of 26% for LSRS, [24] and 11% for LSS.[31] No population-based prevalence of LSRS/LSS was found in the literature, and a slightly higher prevalence than that for LSRS was assumed (29%). To quantify overall shared genetic factors between phenotypes, we calculated SNP-based genetic correlations (rg) between LBP-HC, LSRS, and LSS using LDSR, expecting such correlations to be substantial given that the LSRS and LSS phenotypes are subgroups within LBP-HC. We examined genome-wide significant SNVs using the University of Santa Cruz (UCSC) Genome Browser. We identified independent SNVs using visual inspection of LocusZoom plots[45] and, where applicable, examined associations conditional on the most significant variant at each locus using the conditional and joint analysis method implemented in the Genome-wide Complex Trait Analysis (GCTA) software package.[64] To add functional eQTL information to identified SNVs, we examined them in GTEx[12] and identified variants associated with differential expression of nearby genes.
In order to characterize potential shared genetic influences (pleiotropy) of genome-wide significant SNVs on other traits, we examined associations of the lead SNVs with other traits (p<1×10−5) using the PhenoScanner database version 2,[32; 49] and conducted a phenome-wide association study (PheWAS) of the lead SNVs in eMERGE3. Whereas GWAS systematically examine all variants associated with a phenotype, PheWAS systematically examine all phenotypes associated with a variant.[47] The PheWAS included all eMERGE3 EA participants and examined phenotypes defined by “phecodes”, a standardized system of curated ICD-9 and ICD-10 EHR-based phenotyping algorithms (Phecode map version 1.2) that have been widely used in prior genetic research and validated.[14; 60; 63] We examined associations between the lead SNVs and 890 phecodes with at least two ICD codes and greater than 200 cases defined in the eMERGE data, excluding spine-related phecodes due to overlap with the GWAS case definitions. We used a threshold of statistical significance for each lead SNV of p<5.6×10−5 (0.05/890). A list of all phecodes is provided in Supplemental File 1.
SNVs achieving genome-wide statistical significance in the meta-analysis of eMERGE3/Geisinger results were carried forward for replication in UKB EA participants.[20] One SNV associated with LSRS was carried forward using a replication significance threshold of p < 0.05 (0.05/1 SNV), and 1 SNV associated with LSS was carried forward using a replication significance threshold of p < 0.05 (0.05/1 SNV).
Analyses for aim 3 involved external validation of 3 SNVs that were previously associated with self-reported LBP in UKB (rs12310519 [SOX5], rs7814941 [CCDC26/GSDMC], and rs3180 [SPOCK2/ CHST3]),[22] in the current meta-GWAS of LBP-HCS conducted in eMERGE3/Geisinger, using a replication significance threshold p < 0.0167 [0.05/3 SNVs). Also studied was one SNV previously associated with lumbar discectomy for LSRS in Iceland ( rs7833174)[5], which was examined in the current meta-GWAS of LSRS conducted in eMERGE3/Geisinger (replication significance threshold p < 0.05 [0.05/1 SNV]).
RESULTS
GWAS of low back pain and lumbar spinal disorders in eMERGE3/Geisinger (discovery phase)
Characteristics of the study samples are provided in Table 1. The case phenotype prevalence in meta-analysis of eMERGE3/Geisinger was 48.8% for LBP-HC, 19.8% for LSRS, 7.9% for LSS, and 22.1% for LSRS/LSS. Histograms reflecting the underlying prevalence of different diagnostic codes for each phenotype are provided in Supplemental Figures S1–S4. Quantile-quantile plots did not demonstrate systematic deviations of GWAS association results from that expected by chance (Supplemental Figures S5–S8). Meta-GWAS results for the LBP-HC phenotype are presented as a Manhattan plot in Supplemental Figure S9. No SNVs were associated with LBP-HC at the genome-wide significant level (p<5 × 10−8). The genome wide SNV-based heritability of LBP-HC was 3.1% (±0.5%) on the observed scale and 4.9% (±0.8%) on the liability scale.
Table 1.
Characteristics of the study sample
| Meta-GWAS conducted in eMERGE3 and Geisinger (discovery)* | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Low back pain prompting health care utilization (LBP-HC) | Lumbosacral radicular syndrome (LSRS) | Lumbar spinal stenosis (LSS) | Lumbosacral radicular syndrome or lumbar spinal stenosis (LSRS/LSS) | ||||||
| Cases n=49,182 | Controls n=51,629 | Cases n=20,838 | Controls n=84,642 | Cases n=8,326 | Controls n=97,106 | Cases n=23,411 | Controls n=82,405 | ||
| Age | eMERGE3 | 63.4 (14.7) | 56.2 (16.7) | 64.5 (13.8) | 58.2 (16.5) | 69.3 (11.3) | 59.1 (16.3) | 65.3 (13.6) | 57.8 (16.5) |
| Geisinger | 63.1 (15.7) | 58.1 (17.7) | 64.6 (14.7) | 59.1 (17.4) | 72.5 (11.7) | 59.4 (16.9) | 65.3 (14.7) | 58.8 (17.3) | |
| Female Sex | eMERGE3 | 13,553 (57.1%) | 14,192 (51.2%) | 5,214 (56.7%) | 23,699 (53.3%) | 2,231 (49.8%) | 24,065 (52.4%) | 5,960 (55.1%) | 23,058 (53.5%) |
| Geisinger | 15,847 (62.3%) | 13,515 (56.5%) | 7,086 (60.9%) | 23,744 (59.1%) | 2,114 (55.0%) | 30,773 (60.1%) | 7,583 (60.2%) | 23,266 (59.2%) | |
| BMI (eMERGE) | eMERGE3 | 29.3 (6.3) | 28.6 (6.7) | 29.6 (6.1) | 28.8 (6.6) | 30.0 (5.9) | 28.9 (6.5) | 29.6 (6.1) | 28.8 (6.6) |
| Geisinger | 31.9 (7.6) | 30.8 (7.7) | 32.1 (7.4) | 31.0 (7.7) | 31.9 (6.7) | 31.2 (7.7) | 32.1 (7.4) | 31.0 (7.7) | |
n (%) or mean ± (SD) are presented; all participants were of European ancestry
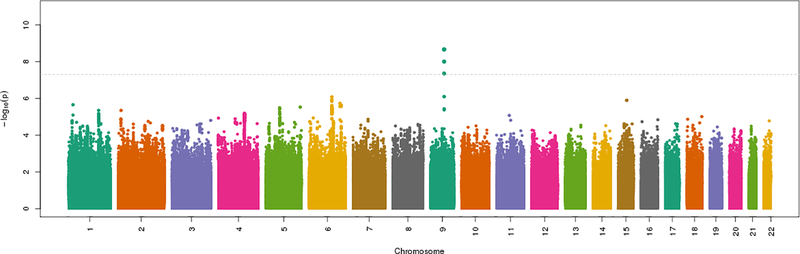
Results for the LSRS phenotype are presented as a Manhattan plot in Figure 1 and Table 2. The SNV-heritability of LSRS was 4.2% (±0.5%) on the observed scale and 9.4% (±1.0%) on the liability scale. Three SNVs at 1 locus were significantly associated with LSRS (Supplemental File 1). The lead SNV at this locus was rs146153280:C>G (OR=1.17 for the G allele, p=2.1 × 10−9, with G allele frequency 0.04), at an intergenic region on chromosome 9 between the UCSC-annotated genes NXNL2 (~402KB 5’ of variant) and C9orf47/S1PR3/SHC3 (~15KB 3’ of variant). The genes C9orf47, S1PR3 and SHC3 are closely located and/or partially overlapping with each other. The other two variants identified at this locus flank the lead SNV rs146153280, are also intergenic, are collinear with the lead SNV, and did not remain significant in analyses conditional on rs146153280. Locus Zoom plots for all genome wide significant results are presented in Supplemental Figures S10–12.
Figure 1. Manhattan plot of GWAS meta-analysis results for the LSRS phenotype*.
LSRS=lumbosacral radicular syndrome
Table 2.
Associations with Lumbar Spine Disorders in eMERGE3/Geisinger*
| Phenotype: Lumbosacral radicular syndrome (LSRS) | ||||||||||
| SNP rsID | Chr:PosA | Nearest Gene | Location | Effect allele | Other allele | EAF | OR | p-value | I2 | Het. p-value |
| rs146153280 | 9:91590608 | C9orf47 | Intergenic | G | C | 0.04 | 1.173 | 2.1 × 10−9 | 7.5 | 0.30 |
| Phenotype: Lumbar spinal stenosis (LSS) | ||||||||||
| rs13427243 | 2:69690187 | AAK1 | 3’ UTR | A | G | 0.40 | 1.099 | 4.3 × 10−8 | 0 | 0.87 |
| Phenotype: Lumbosacral radicular syndrome or lumbar spinal stenosis (LSRS/LSS) | ||||||||||
| rs146153280 | 9:91590608 | C9orf47 | Intergenic | G | C | 0.04 | 1.169 | 1.4 × 10−9 | 0 | 0.46 |
Chr:pos=chromosome-position, EAF=effect allele frequency eMERGE= Electronic Medical Records Genomics network, OR=odds ratio, het.=heterogeneity, 3’ UTR= untranslated region, ncRNA=non-coding RNA
Top variant at each locus meeting genome-wide significance level in analysis of eMERGE data (p<5.0×10−8)
Build GRCh37/hg19
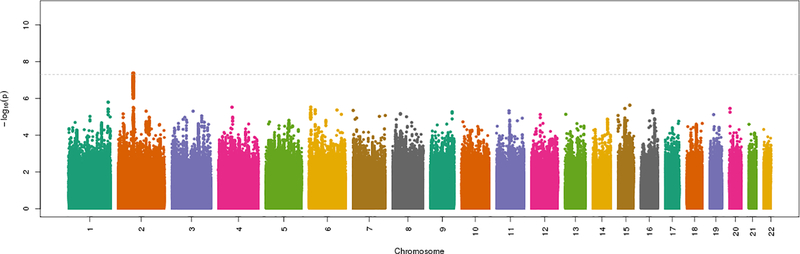
Results for the LSS phenotype are presented as a Manhattan plot in Figure 2 and Table 2. The SNV-heritability of LSS was 2.2% (±0.5%) on the observed scale and 8.0% (±2.0%) on the liability scale. Four SNVs at one locus were significantly associated with LSS (Supplemental File 1). The lead SNV at this locus was rs13427243:G>A (OR=1.10 for the A allele, p=4.3 × 10−8, allele A frequency 0.40), in the 5’ untranslated region (UTR) of AAK1 on chromosome 2. The haplotype defined by these four associated SNVs spans 131,465 bp and includes the genes GFPT1 and NFU1, in addition to AAK1. The other three variants at this locus were collinear and not significant in analyses conditional on rs13427243.
Figure 2. Manhattan plot of GWAS meta-analysis results for the LSS phenotype*.
LSS=lumbar spinal stenosis
Results for the LSRS/LSS phenotype are presented as a Manhattan plot in Supplemental Figure S13. The SNV-heritability of LSRS/LSS was 4.4% (±0.5%) on the observed scale and 9.3% (±1.0%) on the liability scale. This GWAS identified the same lead variant as in the GWAS of LSRS (rs146153280 on chromosome 9), which was also associated with LSRS/LSS rs146153280 (OR=1.17, p=1.4 × 10−9, Table 1) as well as two other variants that were not significant in analyses conditional on the lead variant (Supplemental File 1).
Genetic correlations (rg) with LBP were ~1.00 for LSRS, and 0.86 for LSS. The genetic correlation between LSRS and LSS was 0.67 (Supplemental File 1).
GTEx Functional expression characterization of associated variants
No significant eQTLs were reported for the lead SNV associated with LSRS and LSRS/LSS, rs146153280, in GTEx. The lead SNV associated with LSS, rs13427243, is an eQTL for GFPT1 and NFU1 genes in 20 tissues, including muscle and nerve (p-values between 7.4×10−5 to 1.8×10−20, Supplemental File 1). Violin plots demonstrating rs13427243-associated expression of GFPT1 and NFU1 are provided in Supplemental Figure S14.
Phenotype characterization of associated variants
The lead SNV associated with LSRS in chromosome 9 (rs146153280) was associated with the protein Semaphorin-4D and gene expression of C9orf47 traits included in the PhenoScanner database (Supplemental File 1). The lead SNV associated with LSS in chromosome 2 (rs13427243) was associated with gene expression (GFPT1, NFU1, ANXA4) and epigenetic (DNA methylations, histone modifications) traits in the PhenoScanner database (Supplemental File 1). PheWAS for the lead variants in these loci conducted within eMERGE3 revealed no associations with other phenotypes after correction for multiple statistical testing (Supplemental Figures S15 and S16).
External validation of lead variants, conducted in UKB (replication phase)
The phenotype prevalence for LSRS-UKB, LSS-UKB, LSRS/LSS-UKB was 5.8%, 1.2%, and 6.4% respectively, substantially lower than the corresponding prevalences in eMERGE/Geisinger (19.8%, 7.9%, and 22.1%) respectively. The lead variant rs146153280 that was associated with LSRS and LSRS/LSS in the discovery sample was not replicated in UKB (Supplemental File 1). However, the lead variant rs13427243 associated with LSS in the discovery sample was replicated in UKB (OR 1.11, p=5.4 × 10−5; Supplemental File 1).
External validation in eMERGE3/Geisinger of variants previously associated with LBP and LSRS
The variant rs12310519:C>T (SOX5) was significantly associated with LBP-HC in eMERGE/Geisinger (OR=1.05 for the T allele, p=0.011), with a very similar magnitude and direction effect to its previously-reported association with self-reported LBP in UKB (OR=1.05, Supplemental Table S2). In addition, rs7814941:G>A (CCDC26/GSDMC), was significantly associated with LBP-HC in eMERGE/Geisinger (OR=1.03 for A allele, p=0.005), with a similar magnitude and direction effect to its association with self-reported LBP in UKB (OR=1.04). The previously reported association of rs3180 (SPOCK2/CHST3) with self-reported LBP was not replicated in eMERGE/Geisinger, but had a similar magnitude and direction effect (OR=0.98, p=0.09; Supplemental Table S2) to that in UKB.
DISCUSSION
This GWAS leveraged EHR data from multiple health systems to identify genetic markers associated with LBP prompting health care utilization (LBP-HC) and two lumbar spine-related diagnoses (LSRS and LSS). Despite a large study sample of over 100,000 participants, no genome-wide significant associations were found with LBP-HC. However, we identified novel genetic associations with each of the two lumbar spine-related diagnoses (LSRS and LSS), despite lower numbers of cases and less statistical power compared to the LBP-HC phenotype. These included one locus associated with LSS and another associated with LSRS and a combined LSRS/LSS phenotype. In attempted validation using related yet distinct phenotypes drawn from a very different UK cohort, the lead variant rs13427243 associated with LSS was successfully replicated, while the lead variant rs146153280 associated with LSRS and LSRS/LSS was not.
Although a twin study reported substantial genetic contributions to imaging-detected LSS (heritability 67–81%)[3], no GWAS have found genome-wide significant associations with any LSS phenotype.[10] Our study identifies (lead variant rs13427243:G>A [OR for A=1.10, p=4.3 × 10−8]) and replicates (OR for A=1.11, p=5.4 × 10−5) in an independent sample a locus associated with LSS spanning the genes GFPT1, NFU1, and AAK1, which is linked to the expression of GFPT1 and NFU1 in multiple tissues. AAK1 was previously identified as a potential target for neuropathic pain emerging from a large screen of knockout mice to identify new pain targets.[37] Inhibitors of AAK1 have been tested for the treatment of neuropathic pain using animal models,[37], a phase 1 clinical trial for neuropathic pain was recently completed, [1] and a phase 2 clinical trial for diabetic peripheral neuropathic pain is currently underway.[59] Because LSS is a common lumbar spine-related cause of neuropathic pain, our findings of a genetic association with this locus reveals an unexpected convergence between the findings yielded by our agnostic GWAS approach in humans and those resulting from a drug development strategy using animal models. Symptoms attributed to LSS are often treated with drugs developed for peripheral neuropathic pain (such as gabapentin and pregabalin). If AAK1 inhibitors are shown effective and safe for the treatment of diabetic peripheral neuropathic pain in the future, the association of LSS with a locus that includes AAK1 raises the question of whether AAK1 inhibitors may have utility in the treatment of neuropathic pain due to LSS. Other genes at this locus have not previously been implicated in pain. GFPT1 encodes for glutamine-fructose-6-phosphate transaminase-1, a rate-limiting enzyme in the hexosamine biosynthetic pathway that is required for the glycosylation of proteins and lipids.[29] Variations in GFPT1 are associated with congenital myasthenic syndromes marked by weakness and sparing of the ocular and bulbar muscles. The gene NFU1 encodes a protein with an important role in iron-sulfur cluster biogenesis, and mutations in NFU1 have been associated with mitochondrial dysfunction syndromes.[2]
The strongest association found in this GWAS meta-analysis was with LSRS and the combined LSRS/LSS phenotype for the variant rs146153280 in an intergenic chromosome 9 region between NXNL2 and C9orf47. This locus did not replicate in UKB. This may be partially explained by the different phenotype definitions used in the replication sample, evidenced by the much lower prevalence of LSRS in UKB (5.8%) than in eMERGE3/Geisinger (19.8%). On the other hand, LSS in UKB also had a substantially lower prevalence (1.2%) than that in eMERGE/Geisinger (7.9%), yet this did not impede replication of rs13427243. These discordant results from replication efforts may reflect differences in how these phenotypes are captured by the hospital-based coding used in UKB, which would be expected to capture a larger proportion of true LSS cases (due to the poor natural history of LSS and the older mean age of individuals at risk, resulting in more hospitalizations and surgeries for LSS) than true LSRS cases (which has a favorable natural history and often does not require hospitalization or surgery).[53] We suggest attempted replication of the association of rs146153280 with LSRS using a similar phenotyping strategy to the discovery sample.
Our study replicated several previously reported genetic associations with self-reported LBP in the current meta-GWAS of EHR-defined LBP, one for rs12310519 in the SOX5 gene and a second for rs7814941 in an intergenic region near CCDC26/GSDMC.[22] The lead variants in these loci had similar magnitude and direction of effects in eMERGE/Geisinger as compared to those in the original setting of UKB. Another previously discovered locus associated with self-reported LBP (SPOCK2/CHST3)[22] did not surpass the significance threshold, but had a similar magnitude and direction of effect. Analyses of the LSRS phenotype in the current meta-GWAS also replicated a prior association of rs6651255 with lumbar disc herniation requiring decompression surgery in Icelanders,[5] but with a smaller magnitude effect in eMERGE/Geisinger (OR=0.95 for the C allele) as compared to Iceland (OR=0.81). This may be explained by the LSRS phenotype used in the Icelandic study representing surgical cases of LSRS only. By confirming an association of rs12310519 and rs7814941 with LBP-HC, and rs7814941 near CCDC26/GSDMC with LBP-HC, we demonstrate variants identified through GWAS of self-reported LBP also associate with LBP of a sufficient severity to warrant health care use- a rigorous test of external validity with a different, yet related, phenotype. Taken together, the findings of the current study support the utility of EHR-based genetic studies for discovery and replication of the genetic mechanisms underlying lumbar spinal disorders.
Overall, our findings suggest a greater yield for genetic discovery of individual variants from EHR-based phenotyping of specific lumbar spine-related diagnoses, such as LSRS and LSS, as compared to the commonly reported symptom of LBP, consistent with the generally accepted principle that better phenotyping yields more precise genetic discovery. This pattern may be unsurprising to clinical spine specialists, and corresponds to the clinical intuition that distinct subgroups of patients with LBP exist and warrant different diagnostic and treatment methods. For instance, lumbar decompression surgery is a reasonable treatment option in patients with intractable neuropathic leg pain or myotomal weakness attributed to LSRS or LSS,[33] but may be inappropriate for a patient with axial LBP without neuropathic symptoms/signs affecting lower extremities. At the same time, genetic correlations reflecting the genetic effects of all SNPs genome-wide implied essentially the same genetic influences on LSRS and LBP (rg~1.0), highly similar genetic influences on LSS and LBP-HC (rg=.86), and substantial shared genetic influences on LSRS and LSS (rg=.68). The magnitude of the genetic correlations between LSS and LBP or LSRS (different lumbar spine phenotypes) are comparable to those we have previously reported for different locations of musculoskeletal pain, such as neck pain and hip pain (rg=0.83) or neck pain and knee pain (rg=0.64), but shared genetic influences on LSRS and LBP appear even more tightly correlated. Taken together, the results of these GWAS findings for individual variants and genetic correlations reflecting all variants genome-wide are consistent with a common theme in contemporary pain genomics that shared genetic factors underlie diverse pain phenotypes, alongside genetic factors unique to specific pain phenotypes.[22; 54; 57]
There are limitations to our study. The diagnostic codes used in clinical practice for the syndromes of LSRS and LSS are applied differently between providers and health care systems. For instance, the codes we used to define LSRS or LSS might reflect imaging-detected pathoanatomic changes such as lumbar disc herniation or spinal canal stenosis, respectively, or could instead reflect the occurrence of the clinical syndromes that can be (but are not necessarily) associated with such imaging findings.[55] Therefore, it is all the more noteworthy that, despite this heterogeneity, more specific spine phenotyping resulted in greater yield in the current study. An important caveat to this study and all GWAS is that the specific variants identified usually do not reflect causal variants, but instead are likely to proxy another locus that confers disease risk. The replicated locus associated with LSS spanning the genes GFPT1, NFU1, and AAK1 was associated with functional eQTL expression in GFPT1 and NFU1, and involved multiple tissue types, highlighting the multiple functional effects that associated variants in this region may have. While significant associations with AAK1 expression were not found, this may be due to a variety of reasons such as the limited range of tissues in GTEx (which does not include the spinal canal morphologic characteristics that are central to LSS) or to the limited sample sizes available. Currently, while we know that variants in AAK1, GFPT1, and NFU1 are associated with LSS, we do not know the specific mechanisms of function involved- only that expression differences in neighboring genes suggest a functional effect in some manner for this region. While these results provide a starting point for genetic exploration into previously unknown mechanisms underlying LSS, further work is needed to elucidate the variants in this locus that are actually causal. While the effects of identified variants in the current study were small (e.g., a 10% increased odds of LSS associated with the A allele of rs13427243:G>A in the discovery phase, and an 11% increased odds of LSS in the replication phase), effect sizes are expected to increase with further studies that hone in on the causal variant(s) involved. As these analyses were performed in European-ancestry individuals, our findings may not be generalizable to other ancestries.
Future GWAS of lumbar spinal disorders may benefit from efforts to examine how EHR data can best be used to increase the phenotyping precision [48] and resulting power for discovery. Such studies may also benefit from alternatives to case-control approaches that take greater advantage of longitudinal data, such as time-to-event analytic methods.[28] Other distinct subgroups of LBP and spinal disorders based on pain severity, trajectories, and patterns over time have also been proposed,[16] and some have suggested that these subgroups may be more suitable targets for genetic discovery than the binary classifications typically applied to LBP and other spine phenotypes (e.g. absent vs. present, or chronic vs. acute).[36]
In conclusion, this first meta-GWAS study of lumbar spine-related phenotypes using longitudinal EHR data identified two novel associations with LSRS and LSS, the latter which was replicated in an independent dataset. The locus associated with LSS includes the gene AAK1, which has recently been suggested as a novel drug target for neuropathic pain, [1; 37; 59]. This raises questions about overlap between the mechanisms underlying spine-related neuropathic pain due to LSS and peripheral neuropathic pain, and draws attention to a candidate analgesic in the drug discovery pipeline. The study also replicated previously discovered associations with LBP for variants in SOX5 and CCDC26/GSDMC. EHR-based GWAS studies have utility for genetic discovery of the mechanisms underlying lumbar spinal disorders and pain.
Supplementary Material
ACKNOWLEDGEMENTS
Dr. Suri is a Staff Physician at the VA Puget Sound Health Care System in Seattle, Washington. Dr. Suri and this work were partially supported by VA Career Development Award # 1IK2RX001515 from the United States (U.S.) Department of Veterans Affairs Rehabilitation Research and Development Service. This work was also supported by the UW Clinical Learning, Evidence and Research (CLEAR) Center for Musculoskeletal Disorders funded by NIH/NIAMS P30AR072572. Dr. Aulchenko is supported by the Russian Ministry of Education and Science under the 5–100 Excellence Programme and by the Federal Agency of Scientific Organizations via the Institute of Cytology and Genetics (project 0259-2021-0009 / АААА-А17-117092070032-4). Dr. Tsepilov is supported by the Russian Foundation for Basic Research (project 19-015-00151) and by the Russian Ministry of Education and Science under the 5–100 Excellence Programme.
eMERGE3 was funded by the National Human Genome Research Institute, U01HG8657 and U01HG006375 (Kaiser Permanente Washington(formerly Group Health Cooperative)/University of Washington); U01HG8685, U01HG8676, and U01HG004424 (Brigham and Women’s Hospital/Partners Healthcare/Broad Institute); U01HG006378, U01HG8672, U01HG006385, and U01HG8701 (Vanderbilt University Medical Center); U01HG6379 and U01HG006379 (Mayo Clinic), U01HG8679 and U01HG006382 (Geisinger), U01HG8680 (Columbia University Health Sciences), U01HG8673 and U01HG006388 (Northwestern University), U01HG006389 (Marshfield Clinic), and U01HG006380 (Icahn School of Medicine at Mount Sinai). Other funding included 3U01HG008657–06S1 REV.The study was carried out under UK Biobank approved project #18219. We are grateful to participants from all cohorts as well as the studies’ clerical, clinical and support staff. The contents of this work do not represent the views of the U.S. Department of Veterans Affairs, the National Institutes of Health, or the United States Government.
Footnotes
CONFLICT OF INTEREST
YSA is a founder and co-owner of PolyOmica and PolyKnomics, private organizations that provide services, research, and development in the field of quantitative and statistical genetics and computational genomics. Other authors declare no conflicts of interest.
DATA AVAILABILITY
Summary statistics for GWAS have been deposited on zenodo.com under doi: 10.5281/zenodo.4265494. Data will be made available to researchers upon request.
REFERENCES
- [1].Agajanian MJ, Walker MP, Axtman AD, Ruela-de-Sousa RR, Serafin DS, Rabinowitz AD, Graham DM, Ryan MB, Tamir T, Nakamichi Y, Gammons MV, Bennett JM, Counago RM, Drewry DH, Elkins JM, Gileadi C, Gileadi O, Godoi PH, Kapadia N, Muller S, Santiago AS, Sorrell FJ, Wells CI, Fedorov O, Willson TM, Zuercher WJ, Major MB. WNT Activates the AAK1 Kinase to Promote Clathrin-Mediated Endocytosis of LRP6 and Establish a Negative Feedback Loop. Cell Rep 2019;26(1):79–93 e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ahting U, Mayr JA, Vanlander AV, Hardy SA, Santra S, Makowski C, Alston CL, Zimmermann FA, Abela L, Plecko B, Rohrbach M, Spranger S, Seneca S, Rolinski B, Hagendorff A, Hempel M, Sperl W, Meitinger T, Smet J, Taylor RW, Van Coster R, Freisinger P, Prokisch H, Haack TB. Clinical, biochemical, and genetic spectrum of seven patients with NFU1 deficiency. Front Genet 2015;6:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Battie MC, Ortega-Alonso A, Niemelainen R, Gill K, Levalahti E, Videman T, Kaprio J. Lumbar spinal stenosis is a highly genetic condition partly mediated by disc degeneration. Arthritis Rheumatol 2014;66(12):3505–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Biobank U Touchscreen Questionnaire, Vol. 2020, 2011. [Google Scholar]
- [5].Bjornsdottir G, Benonisdottir S, Sveinbjornsson G, Styrkarsdottir U, Thorleifsson G, Walters GB, Bjornsson A, Olafsson IH, Ulfarsson E, Vikingsson A, Hansdottir R, Karlsson KO, Rafnar T, Jonsdottir I, Frigge ML, Kong A, Oddsson A, Masson G, Magnusson OT, Gudbjartsson T, Stefansson H, Sulem P, Gudbjartsson D, Thorsteinsdottir U, Thorgeirsson TE, Stefansson K. Sequence variant at 8q24.21 associates with sciatica caused by lumbar disc herniation. Nat Commun 2017;8:14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C, Patterson N, Daly MJ, Price AL, Neale BM. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015;47(3):291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cassidy JD, Carroll LJ, Cote P. The Saskatchewan health and back pain survey. The prevalence of low back pain and related disability in Saskatchewan adults. Spine (Phila Pa 1976) 1998;23(17):1860–1866; discussion 1867. [DOI] [PubMed] [Google Scholar]
- [8].Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cherkin DC, Deyo RA, Volinn E, Loeser JD. Use of the International Classification of Diseases (ICD-9-CM) to identify hospitalizations for mechanical low back problems in administrative databases. Spine (Phila Pa 1976) 1992;17(7):817–825. [DOI] [PubMed] [Google Scholar]
- [10].Cheung JPY, Kao PYP, Sham P, Cheah KSE, Chan D, Cheung KMC, Samartzis D. Etiology of developmental spinal stenosis: A genome-wide association study. J Orthop Res 2018;36(4):1262–1268. [DOI] [PubMed] [Google Scholar]
- [11].Chou R, Shekelle P. Will this patient develop persistent disabling low back pain? Jama 2010;303(13):1295–1302. [DOI] [PubMed] [Google Scholar]
- [12].Consortium G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C. Next-generation genotype imputation service and methods. Nat Genet 2016;48(10):1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, Field JR, Pulley JM, Ramirez AH, Bowton E, Basford MA, Carrell DS, Peissig PL, Kho AN, Pacheco JA, Rasmussen LV, Crosslin DR, Crane PK, Pathak J, Bielinski SJ, Pendergrass SA, Xu H, Hindorff LA, Li R, Manolio TA, Chute CG, Chisholm RL, Larson EB, Jarvik GP, Brilliant MH, McCarty CA, Kullo IJ, Haines JL, Crawford DC, Masys DR, Roden DM. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol 2013;31(12):1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Deyo RA, Andersson G, Bombardier C, Cherkin DC, Keller RB, Lee CK, Liang MH, Lipscomb B, Shekelle P, Spratt KF, et al. Outcome measures for studying patients with low back pain. Spine 1994;19(18 Suppl):2032S–2036S. [DOI] [PubMed] [Google Scholar]
- [16].Deyo RA, Bryan M, Comstock BA, Turner JA, Heagerty P, Friedly J, Avins AL, Nedeljkovic SS, Nerenz DR, Jarvik JG. Trajectories of symptoms and function in older adults with low back disorders. Spine (Phila Pa 1976) 2015;40(17):1352–1362. [DOI] [PubMed] [Google Scholar]
- [17].Deyo RA, Mirza SK, Martin BI. Back pain prevalence and visit rates: estimates from U.S. national surveys, 2002. Spine; 2006;31(23):2724–2727. [DOI] [PubMed] [Google Scholar]
- [18].Dieleman JL, Baral R, Birger M, Bui AL, Bulchis A, Chapin A, Hamavid H, Horst C, Johnson EK, Joseph J, Lavado R, Lomsadze L, Reynolds A, Squires E, Campbell M, DeCenso B, Dicker D, Flaxman AD, Gabert R, Highfill T, Naghavi M, Nightingale N, Templin T, Tobias MI, Vos T, Murray CJ. US Spending on Personal Health Care and Public Health, 1996–2013. JAMA 2016;316(24):2627–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Duncan LE, Ostacher M, Ballon J. How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete. Neuropsychopharmacology 2019;44(9):1518–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Elgaeva EE, Tsepilov Y, Freidin MB, Williams FMK, Aulchenko Y, Suri P. ISSLS Prize in Clinical Science 2020. Examining causal effects of body mass index on back pain: a Mendelian randomization study. Eur Spine J 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ferreira ML, Machado G, Latimer J, Maher C, Ferreira PH, Smeets RJ. Factors defining care-seeking in low back pain--a meta-analysis of population based surveys. Eur J Pain 2010;14(7):747 e741–747. [DOI] [PubMed] [Google Scholar]
- [22].Freidin MB, Tsepilov YA, Palmer M, Karssen LC, Suri P, Aulchenko YS, Williams FM, Group CMW. Insight into the genetic architecture of back pain and its risk factors from a study of 509,000 individuals. Pain 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, Collins R, Allen NE. Comparison of Sociodemographic and Health-Related Characteristics of UK Biobank Participants With Those of the General Population. Am J Epidemiol 2017;186(9):1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Frymoyer JW, Pope MH, Clements JH, Wilder DG, MacPherson B, Ashikaga T. Risk factors in low-back pain. An epidemiological survey. J Bone Joint Surg Am 1983;65(2):213–218. [DOI] [PubMed] [Google Scholar]
- [25].Hancock MJ, Maher CG, Latimer J, Spindler MF, McAuley JH, Laslett M, Bogduk N. Systematic review of tests to identify the disc, SIJ or facet joint as the source of low back pain. Eur Spine J 2007;16(10):1539–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hartvigsen J, Hancock MJ, Kongsted A, Louw Q, Ferreira ML, Genevay S, Hoy D, Karppinen J, Pransky G, Sieper J, Smeets RJ, Underwood M, Lancet Low Back Pain Series Working G. What low back pain is and why we need to pay attention. Lancet 2018. [DOI] [PubMed] [Google Scholar]
- [27].Hebbring SJ. The challenges, advantages and future of phenome-wide association studies. Immunology 2014;141(2):157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hughey JJ, Rhoades SD, Fu DY, Bastarache L, Denny JC, Chen Q. Cox regression increases power to detect genotype-phenotype associations in genomic studies using the electronic health record. BMC Genomics 2019;20(1):805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Issop Y, Hathazi D, Khan MM, Rudolf R, Weis J, Spendiff S, Slater CR, Roos A, Lochmuller H. GFPT1 deficiency in muscle leads to myasthenia and myopathy in mice. Hum Mol Genet 2018;27(18):3218–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jannot AS, Ehret G, Perneger T. P < 5 × 10(−8) has emerged as a standard of statistical significance for genome-wide association studies. J Clin Epidemiol 2015;68(4):460–465. [DOI] [PubMed] [Google Scholar]
- [31].Jensen RK, Jensen TS, Koes B, Hartvigsen J. Prevalence of lumbar spinal stenosis in general and clinical populations: a systematic review and meta-analysis. Eur Spine J 2020;29(9):2143–2163. [DOI] [PubMed] [Google Scholar]
- [32].Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, Butterworth AS, Staley JR. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 2019;35(22):4851–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Katz JN, Harris MB. Clinical practice. Lumbar spinal stenosis. N Engl J Med 2008;358(8):818–825. [DOI] [PubMed] [Google Scholar]
- [34].Kitsios GD, Zintzaras E. Genome-wide association studies: hypothesis-”free” or “engaged”? Transl Res 2009;154(4):161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kneeman J, Battalio S, Korpak A, Rundell SD, Luo G, Cherkin D, Suri P. Predicting Persistent Disabling Low Back Pain in Veterans Health Administration Primary Care Using the STarT Back Tool. Accepted for publication, PM&R, 2020. [DOI] [PubMed] [Google Scholar]
- [36].Kongsted A, Kent P, Axen I, Downie AS, Dunn KM. What have we learned from ten years of trajectory research in low back pain? BMC Musculoskelet Disord 2016;17:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kostich W, Hamman BD, Li YW, Naidu S, Dandapani K, Feng J, Easton A, Bourin C, Baker K, Allen J, Savelieva K, Louis JV, Dokania M, Elavazhagan S, Vattikundala P, Sharma V, Das ML, Shankar G, Kumar A, Holenarsipur VK, Gulianello M, Molski T, Brown JM, Lewis M, Huang Y, Lu Y, Pieschl R, O’Malley K, Lippy J, Nouraldeen A, Lanthorn TH, Ye G, Wilson A, Balakrishnan A, Denton R, Grace JE, Lentz KA, Santone KS, Bi Y, Main A, Swaffield J, Carson K, Mandlekar S, Vikramadithyan RK, Nara SJ, Dzierba C, Bronson J, Macor JE, Zaczek R, Westphal R, Kiss L, Bristow L, Conway CM, Zambrowicz B, Albright CF. Inhibition of AAK1 Kinase as a Novel Therapeutic Approach to Treat Neuropathic Pain. J Pharmacol Exp Ther 2016;358(3):371–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Loh PR, Danecek P, Palamara PF, Fuchsberger C, Y AR, H KF, Schoenherr S, Forer L, McCarthy S, Abecasis GR, Durbin R, A LP. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet 2016;48(11):1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Luijsterburg PA, Verhagen AP, Ostelo RW, van Os TA, Peul WC, Koes BW. Effectiveness of conservative treatments for the lumbosacral radicular syndrome: a systematic review. Eur Spine J 2007;16(7):881–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Macfarlane GJ, Beasley M, Smith BH, Jones GT, Macfarlane TV. Can large surveys conducted on highly selected populations provide valid information on the epidemiology of common health conditions? An analysis of UK Biobank data on musculoskeletal pain. Br J Pain 2015;9(4):203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Maher C, Underwood M, Buchbinder R. Non-specific low back pain. Lancet 2017;389(10070):736–747. [DOI] [PubMed] [Google Scholar]
- [42].Manchia M, Cullis J, Turecki G, Rouleau GA, Uher R, Alda M. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS One 2013;8(10):e76295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Martin BI, Lurie JD, Tosteson AN, Deyo RA, Tosteson TD, Weinstein JN, Mirza SK. Indications for spine surgery: validation of an administrative coding algorithm to classify degenerative diagnoses. Spine (Phila Pa 1976) 2014;39(9):769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki AE, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PI, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R, Haplotype Reference C. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Robinson JR, Denny JC, Roden DM, Van Driest SL. Genome-wide and Phenome-wide Approaches to Understand Variable Drug Actions in Electronic Health Records. Clin Transl Sci 2018;11(2):112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shi X Harmonizing EHR Data via Automated Translation of Medical Concepts. In: Sentinel Innovation Center; editor, Vol. 2020, 2020. [Google Scholar]
- [49].Staley JR, Blackshaw J, Kamat MA, Ellis S, Surendran P, Sun BB, Paul DS, Freitag D, Burgess S, Danesh J, Young R, Butterworth AS. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics 2016;32(20):3207–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Stanaway IB, Hall TO, Rosenthal EA, Palmer M, Naranbhai V, Knevel R, Namjou-Khales B, Carroll RJ, Kiryluk K, Gordon AS, Linder J, Howell KM, Mapes BM, Lin FTJ, Joo YY, Hayes MG, Gharavi AG, Pendergrass SA, Ritchie MD, de Andrade M, Croteau-Chonka DC, Raychaudhuri S, Weiss ST, Lebo M, Amr SS, Carrell D, Larson EB, Chute CG, Rasmussen-Torvik LJ, Roy-Puckelwartz MJ, Sleiman P, Hakonarson H, Li R, Karlson EW, Peterson JF, Kullo IJ, Chisholm R, Denny JC, Jarvik GP, e MN, Crosslin DR. The eMERGE genotype set of 83,717 subjects imputed to ~40 million variants genome wide and association with the herpes zoster medical record phenotype. Genet Epidemiol 2019;43(1):63–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Suri P, Carlson MJ, Rainville J. Nonoperative Treatment for Lumbosacral Radiculopathy: What Factors Predict Treatment Failure? Clinical orthopaedics and related research, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Suri P, Hunter DJ, Jouve C, Hartigan C, Limke J, Pena E, Li L, Luz J, Rainville J. Nonsurgical treatment of lumbar disk herniation: are outcomes different in older adults? J Am Geriatr Soc 2011;59(3):423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Suri P, Palmer MR, Tsepilov YA, Freidin MB, Boer CG, Yau MS, Evans DS, Gelemanovic A, Bartz TM, Nethander M, Arbeeva L, Karssen L, Neogi T, Campbell A, Mellstrom D, Ohlsson C, Marshall LM, Orwoll E, Uitterlinden A, Rotter JI, Lauc G, Psaty BM, Karlsson MK, Lane NE, Jarvik GP, Polasek O, Hochberg M, Jordan JM, Van Meurs JBJ, Jackson R, Nielson CM, Mitchell BD, Smith BH, Hayward C, Smith NL, Aulchenko YS, Williams FMK. Genome-wide meta-analysis of 158,000 individuals of European ancestry identifies three loci associated with chronic back pain. PLoS Genet 2018;14(9):e1007601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Suri P, Rainville J, Kalichman L, Katz JN. Does this older adult with lower extremity pain have the clinical syndrome of lumbar spinal stenosis? JAMA 2010;304(23):2628–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Taylor JB, Goode AP, George SZ, Cook CE. Incidence and risk factors for first-time incident low back pain: a systematic review and meta-analysis. The spine journal : official journal of the North American Spine Society 2014;14(10):2299–2319. [DOI] [PubMed] [Google Scholar]
- [57].Tsepilov YA, Freidin MB, Shadrina AS, Sharapov SZ, Elgaeva EE, Zundert JV, Karssen Lcapital Es C, Suri P, Williams FMK, Aulchenko YS. Analysis of genetically independent phenotypes identifies shared genetic factors associated with chronic musculoskeletal pain conditions. Commun Biol 2020;3(1):329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Vlaeyen JWS, Maher CG, Wiech K, Van Zundert J, Meloto CB, Diatchenko L, Battie MC, Goossens M, Koes B, Linton SJ. Low back pain. Nat Rev Dis Primers 2018;4(1):52. [DOI] [PubMed] [Google Scholar]
- [59].Warner C Efficacy, Safety, and PK of LX9211 in Patients With Diabetic Peripheral Neuropathic Pain (RELIEF-DPN 1). clinicaltrials.gov, 2020. [Google Scholar]
- [60].Wei WQ, Bastarache LA, Carroll RJ, Marlo JE, Osterman TJ, Gamazon ER, Cox NJ, Roden DM, Denny JC. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One 2017;12(7):e0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Magi R, Ferreira T, Fall T, Graff M, Justice AE, Luan J, Gustafsson S, Randall JC, Vedantam S, Workalemahu T, Kilpelainen TO, Scherag A, Esko T, Kutalik Z, Heid IM, Loos RJ, Genetic Investigation of Anthropometric Traits C. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 2014;9(5):1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wu P, Gifford A, Meng X, Li X, Campbell H, Varley T, Zhao J, Carroll R, Bastarache L, Denny JC, Theodoratou E, Wei WQ. Mapping ICD-10 and ICD-10-CM Codes to Phecodes: Workflow Development and Initial Evaluation. JMIR Med Inform 2019;7(4):e14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yang J, Ferreira T, Morris AP, Medland SE, Genetic Investigation of ATC, Replication DIG, Meta-analysis C, Madden PA, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, Frayling TM, McCarthy MI, Hirschhorn JN, Goddard ME, Visscher PM. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44(4):369–375, S361–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics for GWAS have been deposited on zenodo.com under doi: 10.5281/zenodo.4265494. Data will be made available to researchers upon request.