Abstract
Alcoholic liver disease (ALD) and nonalcoholic fatty liver disease (NAFLD) are typically asymptomatic and slow-progressing but potentially fatal diseases that are common causes of liver cirrhosis and related complications. Exosomes are nano-sized extracellular vesicles that have been linked to various intercellular communication processes and can carry biological materials reflecting the state and severity of disease. In this study, shotgun proteomic analysis of the protein expression profiles of extracellular vesicles, including exosomes and microvesicles, enriched from human serum samples of 24 patients diagnosed with various fatty liver diseases was performed using liquid chromatography tandem mass spectrometry (LC-MS/MS) followed by protein identification and label-free quantification using the MaxQuant platform. A total of 329 proteins, including 190 previously reported exosome-specific proteins, were identified from four types of liver disease, where significant differences in protein expression were found in apolipoproteins, immunoglobulins, and other previously reported markers of liver disease. Principal component analysis of 61 proteins identified from MaxQuant analysis of the LC-MS/MS data provided a confident differentiation between ALD and NAFLD.
Keywords: extracellular vesicles, exosomes, liver disease, LC-MS/MS, proteomics
Introduction
Liver disease refers to any disorder of the liver, including cirrhosis, hepatitis, liver cancer, and other metabolic and genetic liver disorders. Among liver diseases, nonalcoholic fatty liver disease (NAFLD), alcoholic hepatitis (AH), drug-induced liver injury (DI), and viral infection are the major causes of cirrhosis and liver complications. NAFLD, a relatively asymptomatic disease, is characterized by an excessive accumulation of fat (steatosis) in the liver without a history of heavy alcohol consumption. NAFLD consists of two clinical entities: simple steatosis (SS), accounting for 80–90% of NAFLD cases, and nonalcoholic steatohepatitis (NASH), accounting for 10–20% of cases [1]. NASH can develop into liver fibrosis and cirrhosis and is associated with high liver-related risk and cardiovascular mortality. NAFLD, AH, and DI are often diagnosed from abnormalities in liver function tests, where the disease is further diagnosed by a thorough examination of the patient’s background information, such as excess alcohol usage, medication history, obesity, or diabetes [2]. Fatty liver, also known as hepatic steatosis, can be diagnosed in an abdominal ultrasound, which is non-invasive and cost effective, but has low sensitivity and is highly subjective. A definitive diagnosis of NASH and liver fibrosis can only be achieved by liver biopsy; however, since the ratio of SS to NASH is relatively high, the use of such an invasive and costly method is considered unnecessary [1,3]. Thus, a minimally invasive method for detection of liver diseases from patient serum is needed at this time.
Extracellular vesicles (EVs) comprise various small particles released by most living cells. EVs have been intensely studied in recent years [4,5] because of their role in intercellular communication. EVs derived from mammalian cells often contain materials from the original parent cells, including but not limited to proteins, lipids, and genetic materials such as DNA and RNA [4,6]. These materials are transferred and exchanged between cells through the secretion and reception of EVs, which defines the role of EVs in cellular signaling. EVs are typically classified into three groups, exosomes, microvesicles (MVs), and apoptotic bodies, based on the origin, size, and features of the vesicle [7]. MVs mostly range from 100 to 1000 nm in diameter [5]. MVs primarily originate from platelets and are released by outward budding and fission of the plasma membrane [8,9]. Exosomes are 30–100-nm-sized intraluminal vesicles formed by inward budding of the endosomal membrane, and are released to the extracellular environment upon fusion of multivesicular bodies with the plasma membrane [4,10,11]. Both MVs and exosomes are involved in many biological processes such as the immune response, angiogenesis, aging, and apoptosis. Especially, exosomes are considered to play a critical biological role in cancer development and progression [7,12–15].
Previously, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS)-based characterization of intact low-molecular-weight proteins of exosomes was introduced as a tool for biomarker discovery and classification of multiple states of liver diseases [16]. The peaks identified in MALDI-MS analysis of exosomes from liver disease patients indicated the presence of haptoglobin alpha-1 chain and multiple C3a protein fragments with different intensity ratios to the exosome marker protein platelet factor 4 (PLF4). However, this approach has certain limitations for full proteome characterization of exosomes due to sample complexity, limited m/z range, and relatively low resolution of the instrument. Shotgun proteomics is the gold standard of mass spectrometry-based proteomic investigation [17,18], where whole proteins in a complex mixture are investigated using liquid chromatography-tandem mass spectrometry (LC-MS/MS) of peptides derived from proteolytic digestion of intact proteins. Shotgun proteomics can provide reliable, accurate, and high-throughput data that can be qualitatively and quantitatively analyzed using proteomic software.
In the current study we have assessed the potential of differentiating various types of liver diseases such as NASH, SS, AH, and DI using LC-MS/MS shotgun proteomic analysis of extracellular vesicles, including exosomes and microvesicles, obtained from human serum, with confirmed clinical diagnosis results as a reference. Label-free quantification (LFQ) of the LC-MS/MS data was performed to obtain spectral protein intensities for statistical analysis, and the current methodology based on proteomic analysis of extracellular vesicles enriched from patient sera was proposed as a novel tool for classification of liver disease.
Experimental
Sample collection
The extracellular vesicles were enriched from blood serum of 24 patients (around 1 mL each) with different types of liver disease. The sample pool included patients with NAFLD consisting of SS (n = 5) and NASH (n = 8); AH (n = 4); DI (n = 5); and non-specific hepatitis (NS; n = 2). The disease type of each patient was confirmed by biopsy and other diagnosis methods. The detailed information and the summary of background profiles (age, gender, and history of background diseases) as well as clinical blood screening results of the patients are provided in Supplementary Table S1 and Figure S1, respectively.
Written informed consent was obtained from all patients to use their sera for experiments and publication. Sample collection and experiments were approved by the institutional review board of Korea University Guro Hospital (KUGH16089–001). Blood samples were collected in accordance with hospital protocol, and the samples of sera were separated using centrifugation and stored at −20°C prior to extracellular vesicle isolation.
Extracellular vesicle isolation and enrichment
For extracellular vesicle isolation, the serum samples were pretreated by centrifugation at 2000 × g for 30 min followed by centrifugation at 12,000 × g for 45 min and filtered through a 0.2 μm membrane. Extracellular vesicles were then isolated and enriched from serum samples using a centrifugation method with multiple cycles [16,19]. Samples were subjected to five cycles of centrifugation at 40,000 × g at 4°C; the first cycle was 120 min and the remaining four cycles were 75 min. After each centrifugation cycle, the supernatant was removed (~ 980 μL) and the pellet was reconstituted with 1 mL of 0.01 M phosphate-buffered saline (PBS). All chemicals in the present study were obtained from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise specified. The final pellet after the fifth centrifugation step was reconstituted with 50 μL of PBS buffer and stored prior to further analysis. Field emission-scanning electron microscopy (FE-SEM) and MALDI-MS analyses were performed to confirm the presence of extracellular vesicles in the enriched pellets [16].
Lysis and protein digestion
Lysis of extracellular vesicles and digestion of extracellular vesicle proteins prior to LC-MS/MS analysis were performed using the filter-aided sample preparation method as described previously [19]. Briefly, the extracellular vesicle samples (10 μL) were incubated with 2× RIPA lysis buffer in a 1:1 ratio at 4°C for 30 min. The lysed extracts were then reduced with 100 mM dithiothreitol for 10 min at 70°C, filtered with a 30 kDa molecular weight cut-off centrifugal spin filter, and alkylated with 50 mM iodoacetamide solution in urea buffer. The samples were washed twice with 8 M urea solution and three times with 50 mM ammonium bicarbonate. Following tryptic digestion overnight at 37°C, the samples were desalted using a Pierce C18 Spin Column (Thermo Scientific, Waltham, MA, USA) prior to LC-MS/MS analysis.
LC-MS/MS analysis and data interpretation
The tryptically digested peptides were redissolved in 10 μL of the loading solution (water with 0.1% formic acid). Each sample (5 μL) was injected and analyzed in triplicate with a nanoflow LC-MS/MS system consisting of an Easy nLC 1000 (Thermo Fisher Scientific, Waltham, MA, USA) and LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) equipped with a nano-electrospray source. The peptides were first loaded onto a trap column (C18, 75 µm × 20 mm, 3 µm; Thermo Fisher Scientific), which was washed with the loading solution and switched in-line with an analytical column (EASY-Spray, 75 µm × 500 mm PepMap RSLC C18 2 µm; Thermo Fisher Scientific). The mobile phases A and B were composed of 0 and 100% acetonitrile containing 0.1% formic acid, respectively. The peptides were eluted from the column at a flow rate of 250 nL/min using a 120 min gradient. The LC gradient began with 5% B and was ramped to 25% B for 76 min, 40% B for 20 min, 95% B for 1 min, and remained at 95% B for 8 min. Finally, it was ramped to 5% B for another 5 min. The column was re-equilibrated with 5% B for 10 min before the next run. Quality control procedures for the LC-MS/MS experiments were performed by injecting a quality control standard (Pierce™ HeLa Protein Digest Standard, ThermoFisher Scientific) and a tryptic digest of human serum (Sigma-Aldrich), as well as blank injections after every 3–5 runs.
The eluting peptides were analyzed with an Orbitrap analyzer with full scan spectra acquired between m/z 400 and 2000 at a resolution of 120,000 followed by data-dependent higher energy collisional dissociation MS/MS acquisition for the top 12 most abundant ions at 27% normalized collision energy scans using a linear ion trap analyzer. The dynamic exclusion duration was set as 60 s. The voltage applied to produce an electrospray was +2.1 kV.
The MS/MS data were subjected to LFQ analysis using the MaxQuant platform (v1.6.7.0) [20]. Database search of the MS/MS data was performed in MaxQuant using the Andromeda search engine against the 20,659 reviewed proteins of the Homo sapiens taxonomy catalogued in the UniProt database (UP000005640; September 30, 20192020) and 245 commonly found contaminants. Digestion mode was set to Trypsin/P specificity, with a fixed carbamidomethyl modification of cysteine, and variable modifications of protein N-terminal acetylation and methionine oxidation. Protein and peptide identifications were achieved at a false discovery rate (FDR) of 0.01, minimum number of peptides required for protein identification at 1, maximum missed cleavages at 2, minimum score for modified peptides at 40, and no lower limit for unmodified peptides.
Visualizations of the LFQ data were performed using the Perseus platform to find correlations between disease types. Volcano plots were reconstructed from Perseus data using Microsoft Excel. Venn diagrams were illustrated using the InteractiVenn tool [21]. Multivariate analysis was visualized using SIMCA (Umetrics, Umea, Sweden). Pathway analysis of the shortlisted proteins was performed using Reactome (https://reactome.org).
Results and discussion
Extracellular vesicle confirmation using FE-SEM and MALDI-MS analyses
Confirmation of the presence of extracellular vesicles enriched from blood serum of 24 patients was performed using FE-SEM and MALDI-MS analyses, as shown in Figure 1 and Figure 2, respectively. The SEM images show that the diameter of enriched extracellular vesicles is in the range of 57–155 nm with an average diameter of 106 ± 30 nm. The size of the extracellular vesicles was slightly different between each sample probably due to varying dehydration states during sample preparation [22,23]; however, the size differences of extracellular vesicles between disease groups were not significant (p < 0.05) as shown in Supplementary Figure S2. The MALDI-MS analyses of extracellular vesicles from the 24 patients in the low m/z region confirmed in most samples the presence of an exosome marker protein PLF4 at m/z 7766 and in the same samples haptoglobin alpha-1 chain at m/z 9178 and C3a protein fragments at m/z 8127 and m/z 8932. Pretreatment and enrichment of extracellular vesicles by multiple cycles of centrifugation resulted in extracellular vesicle pellets of high purity as well as reduced risk of contaminations from high-abundance serum proteins such as albumin, as reported in previous work [16,19]. Therefore, the purity of extracellular vesicle pellets enriched using the described method was considered adequate for LC-MS/MS analysis.
Figure 1.
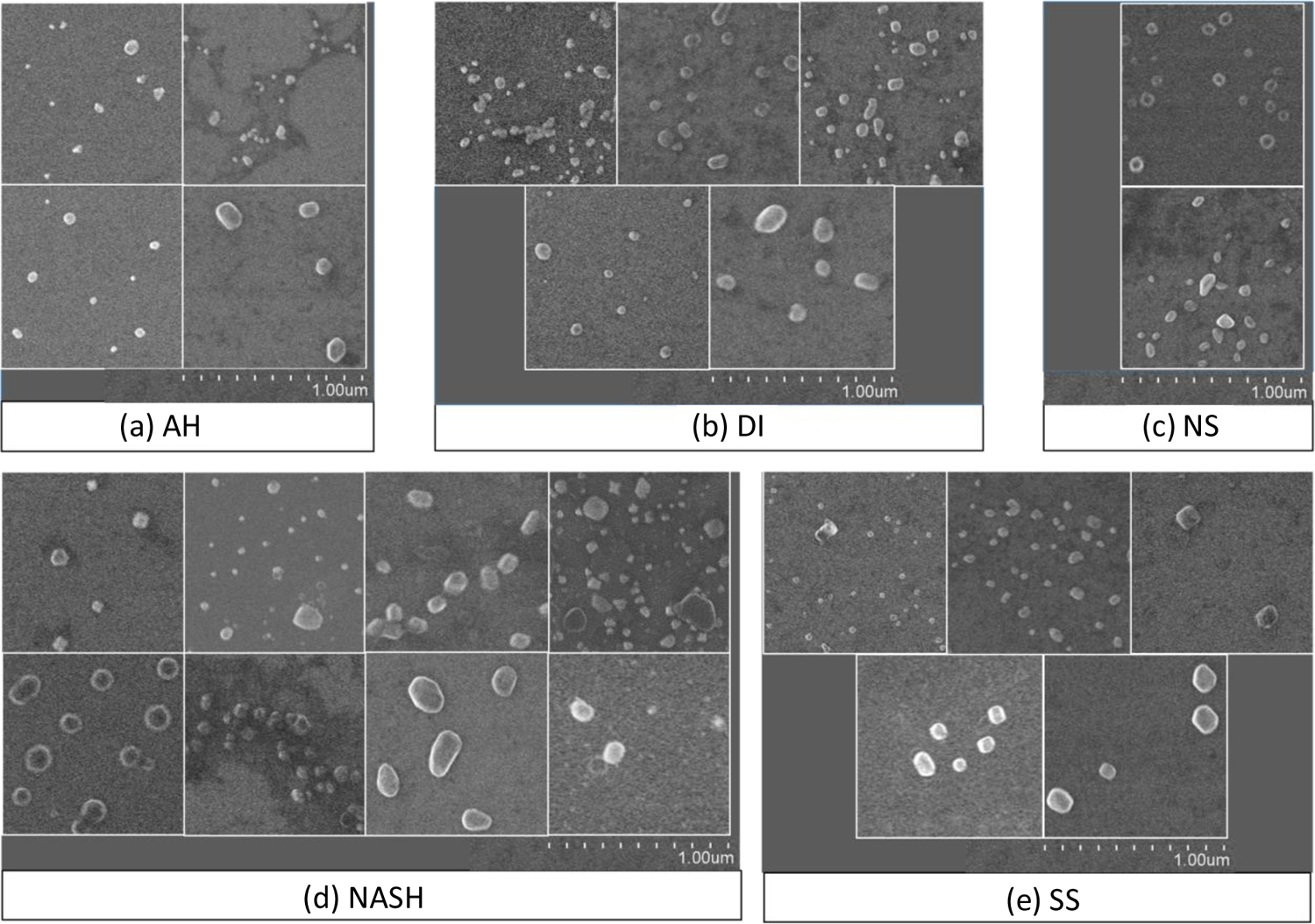
SEM images of extracellular vesicles enriched from the serum of 24 patients. Each panel represents 1.0 μm2 area of the original images, with the magnification level of 40,000.
Figure 2.
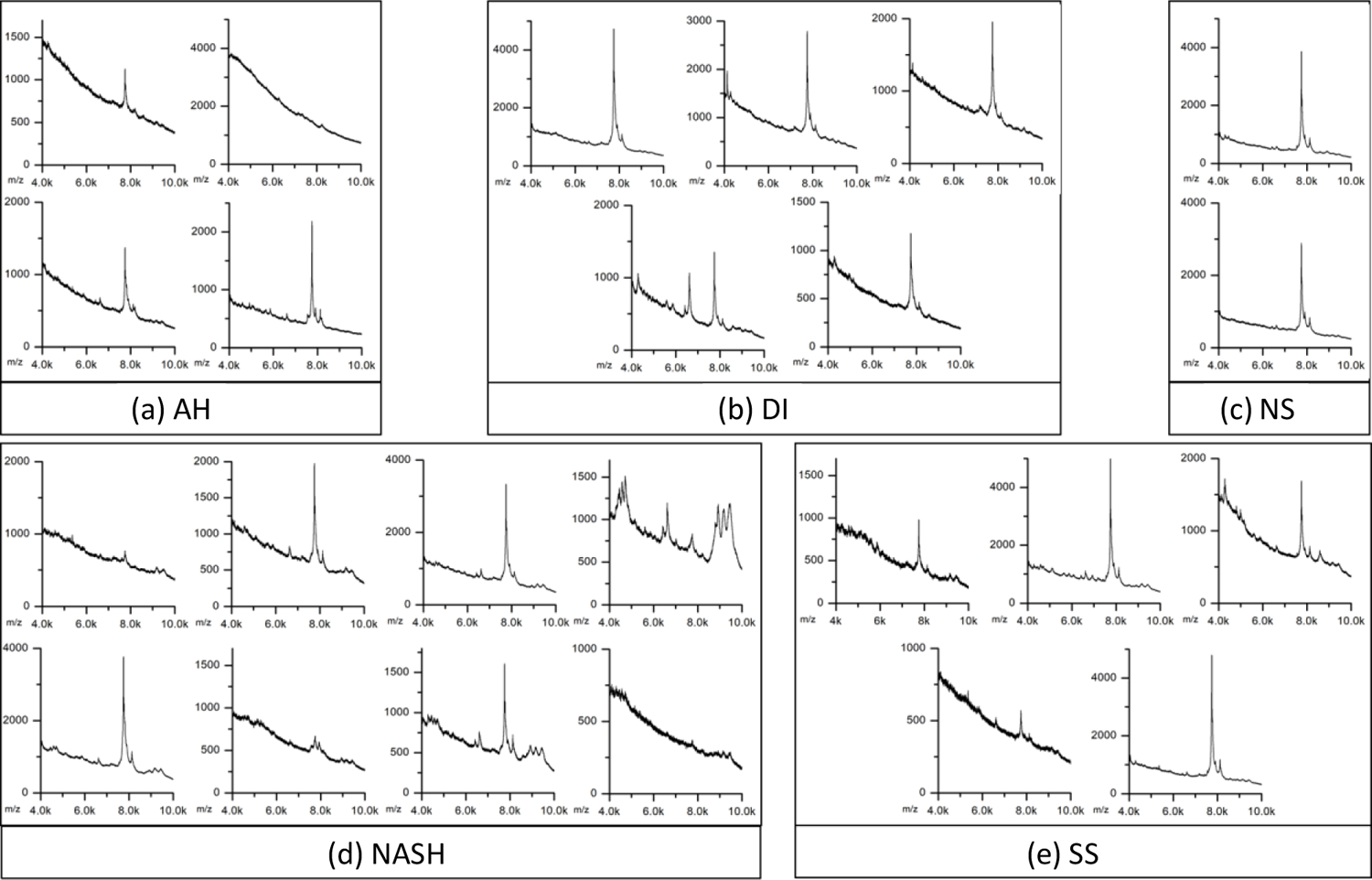
MALDI mass spectra of extracellular vesicles enriched from the serum of 24 patients. Each panel represents the spectrum in the range of m/z 4,000 – 10,000.
Protein identification from LC-MS/MS data
From the sample pool of all 24 patients, MaxQuant analysis of the LC-MS/MS data successfully identified 2577 peptides and 329 proteins. These proteins were then mapped to 329 unique gene name IDs, of which 190 proteins (57.8%) were exosomal markers annotated by the ExoCarta database. Commonly identified exosomal proteins, such as tetraspanins (CD63), alpha-enolase (ENO1), heat-shock proteins (HSPA2 and HSPA1A), and galectin-binding proteins (LGALS3BP and LGALS7B), were frequently identified in the current analysis. In addition, the exosomal proteins previously identified using MALDI-MS analysis (PLF4, haptoglobin, and complement C3) were also commonly identified. PLF4 has previously been reported as an exosome marker protein [24,25].
All identified proteins and peptides are provided in Supplementary Table S2 and Table S3, respectively. The number of proteins and exosome-derived proteins identified in this study were reasonable compared to similar previously reported data [26,27] considering the starting amount of serum (approximately 1.0 mL).
The relative abundance of protein profiles for each patient is provided as log2 LFQ intensity values in Supplementary Table S4 to provide an insight on the distribution of the differential protein expression across different groups of patients at the individual patient level. The quantitative comparison and statistical significance are also provided in Supplementary Table S4, showing the detailed statistical data with cutoff conditions of the statistical analysis. No clear correlation was observed between the size of the extracellular vesicles and the relative abundance of protein profiles.
Diagnosis-dependent classification of the identified proteins in each disease group was visualized using a Venn diagram (Figure 3). The diagram shows that an average of 210 proteins were found in each group, and 147 proteins were found in all the specified disease groups (AH, DI, SS, and NASH). Overall, the number of commonly expressed proteins between disease groups was sufficient (approximately 70%) for comparison. The discovery of both common and disease-specific proteins indicates that further quantifications of the LC-MS/MS data may reveal correlations between different states of liver disease.
Figure 3.
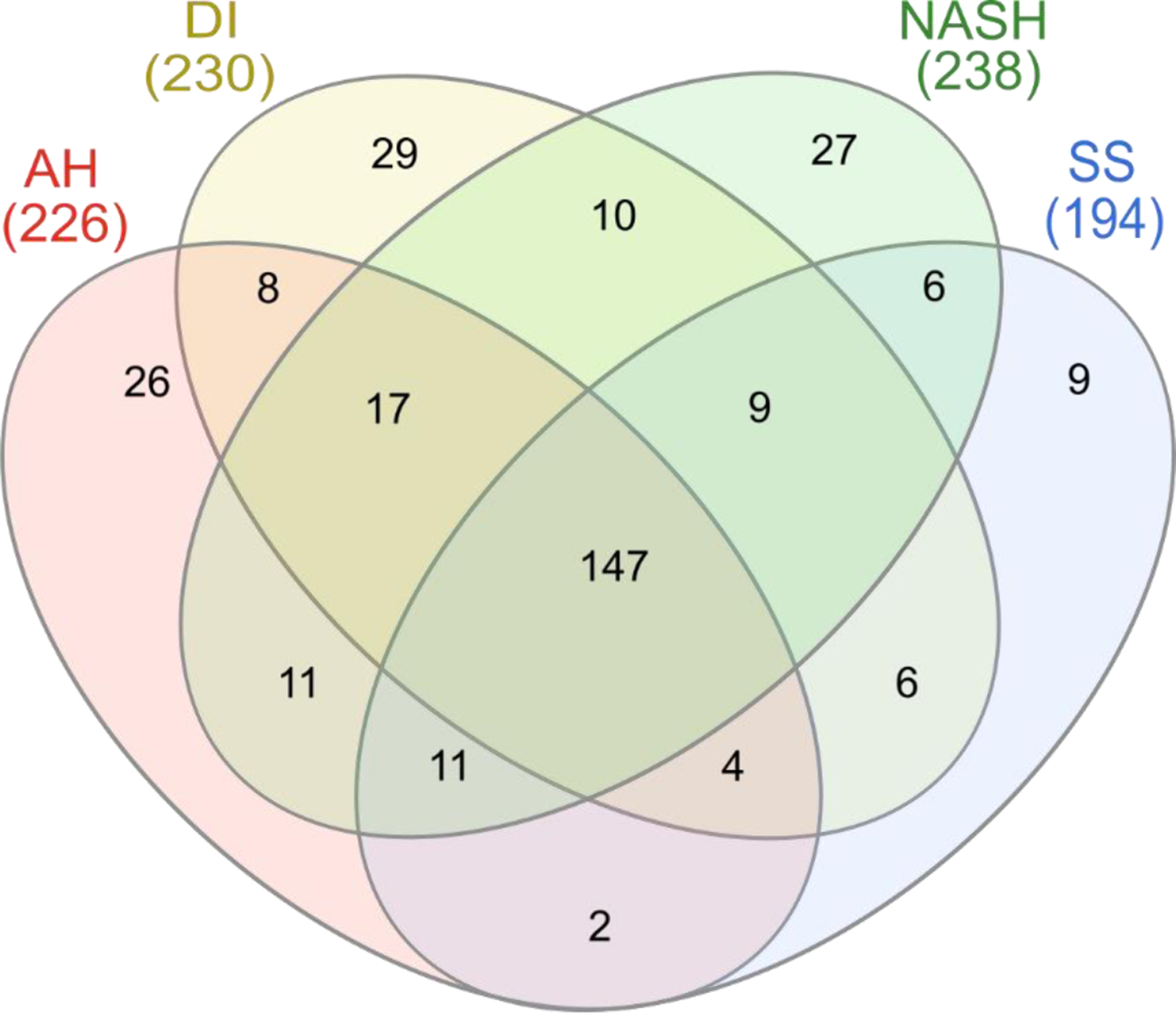
Venn diagram showing the distribution of identified proteins in each disease type. The overlapped region shows that 147 proteins are expressed in all disease types; 92 of these are indexed in the ExoCarta database.
Differential protein expression of extracellular vesicles between two liver disease types
Volcano plots of differentially expressed proteins between two selected disease groups were generated by plotting log2 (fold change) versus ‒log10 (p-value) to assess the significant differences in protein expression in extracellular vesicles enriched from serum samples (Figure 4). The p-values and the FDR values were calculated using a two-tailed Student’s t-test of each pair and co-validated by 250 randomizations with the Perseus program. Proteins with the FDR values less than 0.05 and absolute value of log2 (fold change) higher than 1 were considered significantly different, representing trends of differential expression between the two compared groups of liver disease.
Figure 4.
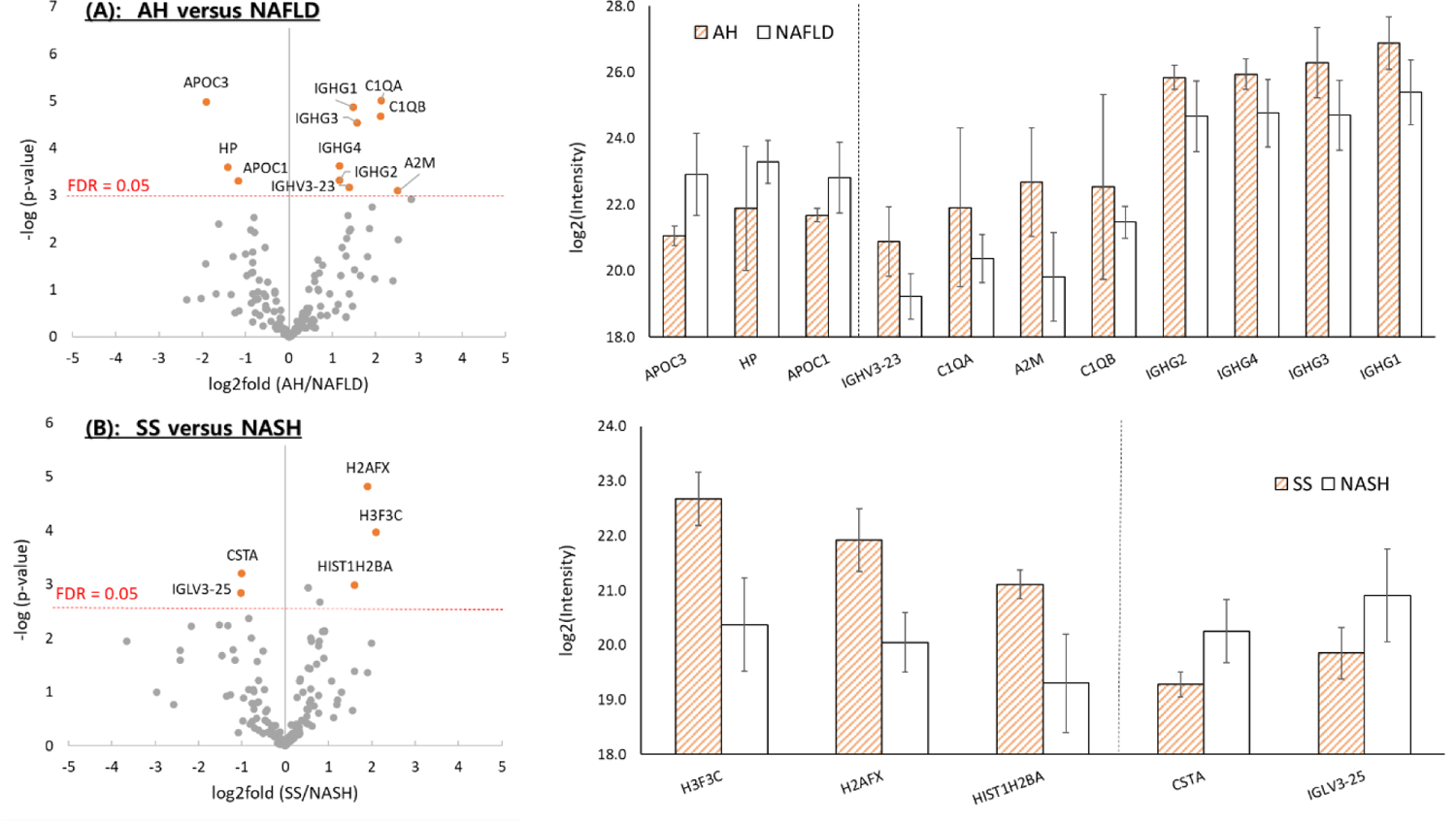
Volcano plots and log2 (fold change) comparisons showing the differences in extracellular vesicle protein expression between (A) AH and NAFLD, and (B) SS and NASH. Only statistically significant protein differences (FDR = 0.05 limit) were displayed in the log2 (fold change) comparisons.
Table 1 shows the list of differentially expressed proteins between compared disease types, along with protein information. AH and NAFLD, both of which describe an inflammatory phase of liver disease, are differentiated based on alcohol consumption history. Therefore, elevated expression of inflammatory markers in extracellular vesicles from patients with AH and NAFLD is expected. The volcano plot comparing AH and NAFLD (including both SS and NASH) is shown in Figure 4A. Among the 192 commonly expressed proteins, eleven proteins were considered significantly expressed (FDR < 0.05), with eight proteins (IGHG1, IGHG2, IGHG3, IGHG4, IGHV3–23, C1QA, C1QB, and A2M) expressed at a higher level in AH patients and three proteins (APOC1, APOC3, and HP) expressed at a higher level in NAFLD patients. The highest differences in protein expression were observed in the level of A2M for AH patients, and APOC3 for NAFLD patients [FDR < 0.05, and log2 (fold change) > 2]. Since NAFLD is significantly more difficult to reverse than AH or DI [28], APOC3, the most confidently identified NAFLD protein, could have a significant meaning as a potential biomarker. In comparison with previously reported literature, the extracellular vesicle protein expressions were observed to be in correlation with those from blood serum or plasma. Thabut et al. observed the increase in A2M as well as decrease in HP in blood serum analysis of patients diagnosed with alcoholic steatohepatitis, and included the two proteins as biomarkers for alcoholic steatohepatitis in their patented diagnostic panel (AshTest) [29]. APOC3 was previously reported to be linked to lipoprotein metabolism in NAFLD [30]. It has already been reported that the increased concentration of APOC3 is related to NAFLD [31,32]. Since APOC3 inhibits the activity of lipoprotein lipase which hydrolyzes triglycerides to generate free fatty acids, an increased expression of APOC3 leads to significantly increased triglyceride accumulation, resulting in increased steatosis in NAFLD [33–35]. Previous studies have shown that apolipoprotein C3 overexpressing mice are susceptible to hepatic steatosis [31,32].
Table 1.
Description of the proteins differentially expressed in the different types of liver disease.
| A. AH (4 patients) in comparison to NAFLD (13 patients) | ||||||
|---|---|---|---|---|---|---|
| # | Uniprot Accession | Gene name | Peptide count [Unique] | log2(AH/NAFLD) | p-value | Description |
| 1 | P02656 | APOC3 | 3[3] | −1.90 | < 0.0001 | Apolipoprotein C-III |
| 2 | P00738 | HP | 15 [7] | −1.40 | 0.0002 | Haptoglobin |
| 3 | P02654 | APOC1 | 3 [3] | −1.16 | 0.0005 | Apolipoprotein C-I |
| 4 | P01764 | IGHV3–23 | 2 [1] | 1.39 | 0.0006 | Immunoglobulin heavy variable 3–23 |
| 5 | P02745 | C1QA | 4 [4] | 2.13 | < 0.0001 | Complement C1q subcomponent subunit A |
| 6 | P01023 | A2M | 37 [34] | 2.52 | 0.0008 | Alpha-2-macroglobulin |
| 7 | P02746 | C1QB | 6 [6] | 2.12 | < 0.0001 | Complement C1q subcomponent subunit B |
| 8 | P01859 | IGHG2 | 9 [4] | 1.17 | 0.0005 | Immunoglobulin heavy constant gamma 2 |
| 9 | P01861 | IGHG4 | 8 [2] | 1.18 | 0.0002 | Immunoglobulin heavy constant gamma 4 |
| 10 | P01860 | IGHG3 | 13 [6] | 1.58 | < 0.0001 | Immunoglobulin heavy constant gamma 3 |
| 11 | P01857 | IGHG1 | 13 [6] | 1.48 | < 0.0001 | Immunoglobulin heavy constant gamma 1 |
| B. NASH (8 patients) in comparison to SS (5 patients) | ||||||
| # | Uniprot Accession | Gene name | Peptide count [Unique] | log2(SS/NASH) | p-value | Description |
| 1 | Q6NXT2 | H3F3C | 4 [4] | 2.11 | 0.0001 | Histone H3 3C |
| 2 | P16104 | H2AFX | 6 [6] | 1.91 | < 0.0001 | Histone H2AX |
| 3 | Q96A08 | HIST1H2BA | 4 [4] | 1.60 | 0.0010 | Histone H2B type 1-A |
| 4 | P01040 | CSTA | 3 [3] | −1.00 | 0.0006 | Cystatin-A |
| 5 | P01717 | IGLV3–25 | 4 [1] | −1.02 | 0.0015 | Immunoglobulin lambda variable 3–25 |
Significance: The current investigation revealed the difference among various types of liver disease using LC-MS/MS of exosomes enriched from human serum samples of 24 patients where the most significantly up-regulation proteins were alpha-2-macroglobulin for alcoholic hepatitis and apolipoprotein C3 for nonalcoholic fatty liver disease.
Another comparison was conducted in the NAFLD category, between SS with no or minimal inflammation and no fibrosis, and NASH with inflammation and fibrosis, which can lead to cirrhosis, liver failure, and hepatocellular carcinoma (Figure 4B). Three proteins in the histone family including H2AFX, H3F3C, and HIST1H2BA were expressed at a significantly higher level (FDR < 0.05) in SS patients, while two proteins (CSTA and IGLV3–25) were significantly expressed in NASH patients. The circulating histone levels were reported to significantly increase in patients with acute liver failure [36]. The protein CSTA (Cystatin A, also known as Stefin A) was previously indicated as a potential biomarker for liver diseases where an elevated serum level of Stefin A was observed in patients with liver diseases compared to healthy subjects [37]. In addition, overweight and obese patients with type 2 diabetes showed high stefin A serum concentrations [38]. These results suggest that the protein expressions of serum extracellular vesicles are related to those found in other media such as serum or plasma. Therefore, proteomic profiling of extracellular vesicles could become an assisting method for finding potential disease-related markers.
Multivariate statistical analysis
The results of the differential protein expression analysis of different types of liver disease as shown in the previous section of ‘Differential protein expression of extracellular vesicles between two liver disease types’ were used for multivariate statistical analysis to identify possible correlations among proteins of the three disease types (AH, SS, and NASH). Proteins with −log (p-value) ≥ 1.5 (or p-value < 0.0316), where the p-values were obtained from the t-test of log2 LFQ intensity values, were used as variables in multivariate data analysis. The relatively high p-value cutoff was used to include enough proteins for the multivariate analysis while removing proteins not related to the differentiation. Proteins found in multiple groups were merged into unique single entities. This filter process resulted in 61 unique proteins from extracellular vesicles of 22 different patients (Supplementary Table S2).
The principal component analysis (PCA) score plot and the loading plot of the above filtered data are displayed in Figure 5. After applying a dimension reduction algorithm, 52.9% of the total variance was explained in the first two components of the model, and up to 65% was explained with the third principal component. Detailed results are included in Supplementary Figure S3. In summary, the 2D score plot of the model revealed that data points from AH patients were clearly resolved from the NAFLD disease types (SS and NASH). Data points from SS also appeared to be grouped with high consistency, but not clearly resolved from the data points of NASH, indicating a high similarity in protein expression of extracellular vesicles in these two forms of NAFLD compared with AH. Data points from DI patients were also partially separated from AH and NAFLD disease types. Therefore, we confirmed that the pattern of protein expressions by serum-enriched extracellular vesicles analyzed by LC-MS/MS has a potential to differentiate these liver diseases. However, the relatively small sample size of each disease group potentially limit the classification ability and overall prediction power of the model. Cross-validation of the model using random sampling or external validation based on independent large datasets could reduce false associations and data overfitting. By increasing the sample pool size and incorporation of automatic cloud computing, this approach could result in the development of an assistance method for diagnosis and characterization of various liver diseases.
Figure 5.
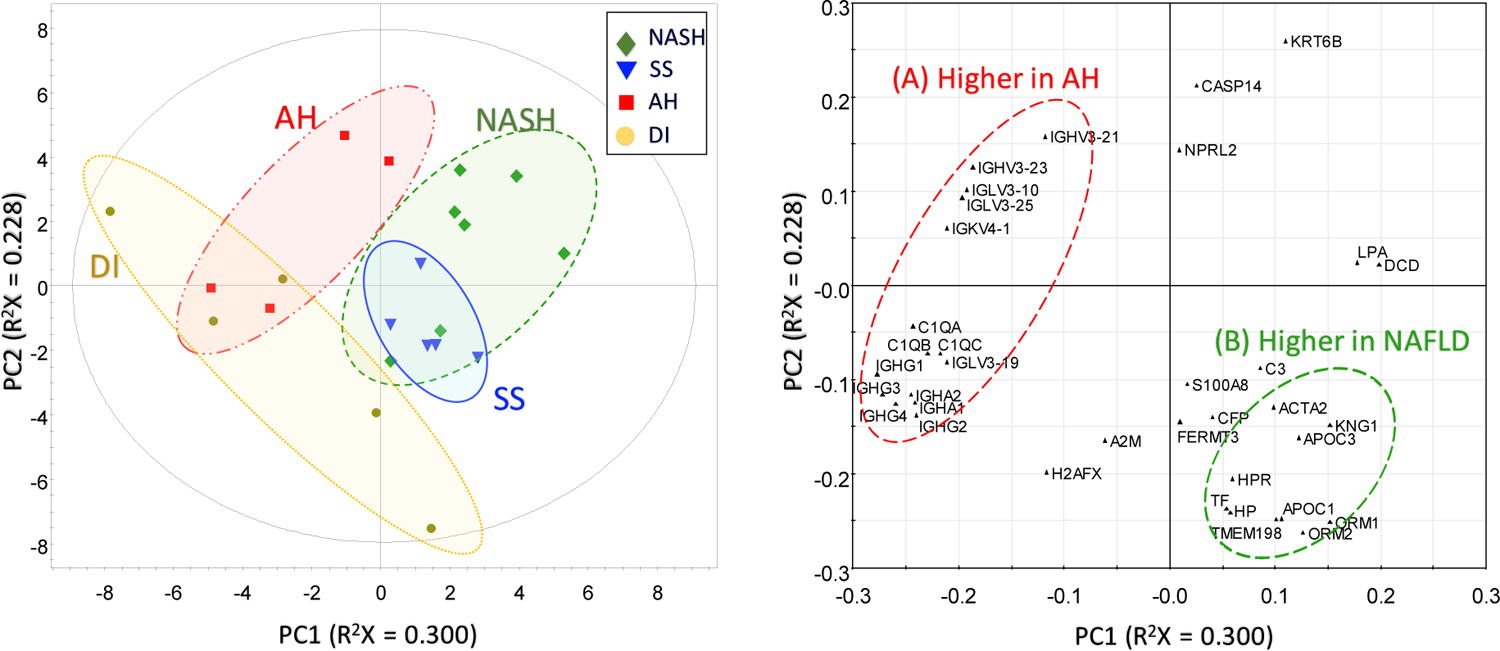
PCA plot (left) and loading plot (right) from multivariate analysis of the filtered dataset, showing classification of the disease types (AH, NASH, and SS).
The corresponding loading plot of the first two principal components provided insight into the relationship between expression levels of different protein groups and its contribution to the classification ability of the multivariate analysis. In the loading plot of Figure 5, a cluster (A) consisting of immunoglobulin proteins (IGHV, IGLV, IGKV, and IGHG) and complement C1q subcomponents (C1QA, C1QB, and C1QC) is located in the opposite vector direction of the cluster (B) consisting of apolipoproteins (APOC1 and APOC3) and lipocalins (ORM1 and ORM2). This distribution indicates that the immunoglobulin proteins and complement C1q proteins are present in higher levels in AH patient samples, while apolipoproteins and lipocalins are highly expressed in NAFLD patients. These results are highly correlated with the results of the volcano plots of differentially expressed proteins, where overexpression of immunoglobulin proteins and complement C1q proteins and decreased expression of apolipoproteins and haptoglobin are major factors in the characterization and classification of alcoholic-related liver diseases from nonalcoholic liver diseases. Therefore, we validated the possibility of using multivariate data analysis for characterization of liver disease groups by LC-MS/MS analysis.
Pathway analysis
NAFLD encompasses a spectrum of obesity-associated liver diseases from SS to NASH. The pathogenesis of NAFLD involves a complex mixture of individual factors such as diet, obesity, and other environmental and genetic factors [39]. In this study, pathway analysis was performed on the 61 proteins potentially related to the liver diseases obtained for multivariate analysis. The results of the pathway analysis are shown in Supplementary Figure S4 and Supplementary Table S5. Supplementary Table S5 shows a list of the 15 most significant pathways for the 61 proteins; a large proportion of the proteins are related to the immune system and complement cascade. The three most relevant pathways were antibody-mediated complement activation, regulation of complement cascade, and initial triggering of complement. The complement system plays an important role in the protection of tissue against infection and disposal of damaged tissue. There is evidence that a heavy alcohol intake increases the hepatic deposition of complement C1, C3q, C8, and C9; it also increases inflammation and steatosis [40]. However, further studies are required to determine whether the presence of C1q, C1r, C1s, C3, C4, and C6 in our investigation of extracellular vesicles is directly related to the origin and cause of liver diseases.
Conclusions
Differential and multivariate analyses of LC-MS/MS data obtained from extracellular vesicles from sera of patients with different liver disease types demonstrated that extracellular vesicle protein expression was significantly related to the liver disease types. Differences in the expression of C1q, C3 complement, and apolipoproteins, as well as previously identified disease markers, were found to be important factors in the classification of disease types. Our results suggest that exosome analysis may be a useful tool for the rapid diagnosis of liver diseases with the potential for a minimally invasive liquid biopsy.
Supplementary Material
Highlights.
Exosomes were enriched from human serum samples with fatty liver diseases
LC-MS/MS analyses of digested exosomal proteins were performed in triplicate
A total of 329 proteins were identified from the enriched exosomes
Confident differentiation was obtained between alcoholic liver disease and nonalcoholic fatty liver disease
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1D1A1B02008854 and 2019M3E5D1A01068997), supported by Chungnam National University (2019–2020) and by the National Cancer Institute under grant 1R01 CA160254 (DML).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hashimoto E, Taniai M, Tokushige K, Characteristics and diagnosis of NAFLD/NASH, J. Gastroenterol. Hepatol 28 (2013) 64–70. 10.1111/jgh.12271. [DOI] [PubMed] [Google Scholar]
- [2].Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ, The diagnosis and management of non-alcoholic fatty liver disease: Practice guideline by the American Gastroenterological Association, American Association for the Study of Liver Diseases, and American College of Gastroenterology, Gastroenterology 142 (2012) 1592–1609. 10.1053/j.gastro.2012.04.001. [DOI] [PubMed] [Google Scholar]
- [3].Dyson JK, Anstee QM, McPherson S, Non-alcoholic fatty liver disease: a practical approach to diagnosis and staging, Frontline Gastroenterol 5 (2014) 211 LP–218. 10.1136/flgastro-2013-100403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Van Niel G, D’Angelo G, Raposo G, Shedding light on the cell biology of extracellular vesicles, Nat. Rev. Mol. Cell Biol 19 (2018) 213–228. 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- [5].Doyle LM, Wang MZ, Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis, Cells 8 (2019) 727. 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yoon YJ, Kim OY, Gho YS, Extracellular vesicles as emerging intercellular communicasomes, BMB Rep 47 (2014) 531–539. 10.5483/BMBRep.2014.47.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Whiteside TL, Tumor-Derived Exosomes and Their Role in Cancer Progression, Adv. Clin. Chem 74 (2016) 103–141. 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C, Microvesicles: Mediators of extracellular communication during cancer progression, J. Cell Sci 123 (2010) 1603–1611. 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cocucci E, Racchetti G, Meldolesi J, Shedding microvesicles: artefacts no more, Trends Cell Biol (2009). 10.1016/j.tcb.2008.11.003. [DOI] [PubMed]
- [10].Schorey JS, Cheng Y, Singh PP, Smith VL, Exosomes and other extracellular vesicles in host-pathogen interactions, EMBO Rep 16 (2015) 24–43. 10.15252/embr.201439363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Raposo G, Stoorvogel W, Extracellular vesicles: Exosomes, microvesicles, and friends, J. Cell Biol 200 (2013) 373–383. 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Whiteside TL, The potential of tumor-derived exosomes for noninvasive cancer monitoring, Expert Rev. Mol. Diagn 15 (2015) 1293–1310. 10.1586/14737159.2015.1071666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Vlassov AV, Magdaleno S, Setterquist R, Conrad R, Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials, Biochim. Biophys. Acta - Gen. Subj 1820 (2012) 940–948. 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- [14].Simpson RJ, Lim JWE, Moritz RL, Mathivanan S, Exosomes: Proteomic insights and diagnostic potential, Expert Rev. Proteomics 6 (2009) 267–283. 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- [15].Koga K, Matsumoto K, Akiyoshi T, Kubo M, Yamanaka N, Tasaki A, Nakashima H, Nakamura M, Kuroki S, Tanaka M, Katano M, Purification, characterization and biological significance of tumor-derived exosomes, Anticancer Res 25 (2005) 3703–3707. [PubMed] [Google Scholar]
- [16].Nguyen H-Q, Lee D, Kim Y, Paek M, Kim M, Jang K-S, Oh J, Lee Y-S, Yeon JE, Lubman DM, Kim J, Platelet Factor 4 as a Novel Exosome Marker in MALDI-MS Analysis of Exosomes from Human Serum, Anal. Chem 91 (2019) 13297–13305. 10.1021/acs.analchem.9b04198. [DOI] [PubMed] [Google Scholar]
- [17].Vildhede A, Nguyen C, Erickson BK, Kunz RC, Jones R, Kimoto E, Bourbonais F, Rodrigues AD, Varma MVS, Comparison of Proteomic Quantification Approaches for Hepatic Drug Transporters: Multiplexed Global Quantitation Correlates with Targeted Proteomic Quantitation, Drug Metab. Dispos 46 (2018) 692 LP–696. 10.1124/dmd.117.079285. [DOI] [PubMed] [Google Scholar]
- [18].Han X, Aslanian A, Yates JR, Mass spectrometry for proteomics, Curr. Opin. Chem. Biol 12 (2008) 483–490. 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kim J, Tan Z, Lubman DM, Exosome enrichment of human serum using multiple cycles of centrifugation, Electrophoresis 36 (2015) 2017–2026. 10.1002/elps.201500131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cox J, Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification, Nat. Biotechnol 26 (2008) 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- [21].Heberle H, Meirelles VG, da Silva FR, Telles GP, Minghim R, InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams, BMC Bioinformatics 16 (2015) 1–7. 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chernyshev VS, Rachamadugu R, Tseng YH, Belnap DM, Jia Y, Branch KJ, Butterfield AE, Pease LF, Bernard PS, Skliar M, Size and shape characterization of hydrated and desiccated exosomes, Anal. Bioanal. Chem 407 (2015) 3285–3301. 10.1007/s00216-015-8535-3. [DOI] [PubMed] [Google Scholar]
- [23].Chuo ST-Y, Chien JC-Y, Lai CP-K, Imaging extracellular vesicles: current and emerging methods, J. Biomed. Sci 25 (2018) 91. 10.1186/s12929-018-0494-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nguyen H-Q, Lee D, Kim Y, Paek M, Kim M, Jang K-S, Oh J, Lee Y-S, Yeon JE, Lubman DM, Kim J, Platelet Factor 4 as a Novel Exosome Marker in MALDI-MS Analysis of Exosomes from Human Serum, Anal. Chem 91 (2019). 10.1021/acs.analchem.9b04198. [DOI] [PubMed] [Google Scholar]
- [25].An M, Lohse I, Tan Z, Zhu J, Wu J, Kurapati H, Morgan MA, Lawrence TS, Cuneo KC, Lubman DM, Quantitative Proteomic Analysis of Serum Exosomes from Patients with Locally Advanced Pancreatic Cancer Undergoing Chemoradiotherapy, J. Proteome Res 16 (2017) 1763–1772. 10.1021/acs.jproteome.7b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Duijvesz D, Burnum-Johnson KE, Gritsenko MA, Hoogland AM, Vredenbregt-van den Berg MS, Willemsen R, Luider T, Paša-Tolić L, Jenster G, Proteomic Profiling of Exosomes Leads to the Identification of Novel Biomarkers for Prostate Cancer, PLoS One 8 (2014) e82589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang C, Guo W. -b., Zhang W. -s., Bian J, Yang J. -k., Zhou Q. -z., Chen M. -k., Peng W, Qi T, Wang C. -y., Liu C. -d., Comprehensive proteomics analysis of exosomes derived from human seminal plasma, Andrology 5 (2017) 1007–1015. 10.1111/andr.12412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].George J, Anstee Q, Ratziu V, Sanyal A, NAFLD: The evolving landscape, J. Hepatol 68 (2018) 227–229. 10.1016/j.jhep.2017.11.016. [DOI] [PubMed] [Google Scholar]
- [29].Thabut D, Naveau S, Charlotte F, Massard J, Ratziu V, Imbert-Bismut F, Cazals-Hatem D, Abella A, Messous D, Beuzen F, Munteanu M, Taieb J, Moreau R, Lebrec D, Poynard T, The diagnostic value of biomarkers (AshTest) for the prediction of alcoholic steato-hepatitis in patients with chronic alcoholic liver disease, J. Hepatol 44 (2006) 1175–1185. 10.1016/j.jhep.2006.02.010. [DOI] [PubMed] [Google Scholar]
- [30].Jiang ZG, Robson SC, Yao Z, Lipoprotein metabolism in nonalcoholic fatty liver disease, J. Biomed. Res 27 (2013) 1–13. 10.7555/JBR.27.20120077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lee H-Y, Birkenfeld AL, Jornayvaz FR, Jurczak MJ, Kanda S, Popov V, Frederick DW, Zhang D, Guigni B, Bharadwaj KG, Choi CS, Goldberg IJ, Park J-H, Petersen KF, Samuel VT, Shulman GI, Apolipoprotein CIII overexpressing mice are predisposed to diet-induced hepatic steatosis and hepatic insulin resistance, Hepatology 54 (2011) 1650–1660. 10.1002/hep.24571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Paiva AA, Raposo HF, Wanschel ACBA, Nardelli TR, Oliveira HCF, Apolipoprotein CIII Overexpression-Induced Hypertriglyceridemia Increases Nonalcoholic Fatty Liver Disease in Association with Inflammation and Cell Death, Oxid. Med. Cell. Longev 2017 (2017). 10.1155/2017/1838679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Duseja A, Aggarwal R, APOC3 and PNPLA3 in non-alcoholic fatty liver disease: Need to clear the air, J. Gastroenterol. Hepatol 27 (2012) 848–851. 10.1111/j.1440-1746.2012.07103.x. [DOI] [PubMed] [Google Scholar]
- [34].Ibrahim SH, Kohli R, Gores GJ, Mechanisms of lipotoxicity in NAFLD and clinical implications, J. Pediatr. Gastroenterol. Nutr 53 (2011) 131–140. 10.1097/MPG.0b013e31822578db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Petersen KF, Dufour S, Hariri A, Nelson-Williams C, Foo JN, Zhang X-M, Dziura J, Lifton RP, Shulman GI, Apolipoprotein C3 Gene Variants in Nonalcoholic Fatty Liver Disease, N. Engl. J. Med 362 (2010) 1082–1089. 10.1056/NEJMoa0907295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wen Z, Lei Z, Yao L, Jiang P, Gu T, Ren F, Liu Y, Gou C, Li X, Wen T, Circulating histones are major mediators of systemic inflammation and cellular injury in patients with acute liver failure, Cell Death Dis (2016) e2391. 10.1038/cddis.2016.303. [DOI] [PMC free article] [PubMed]
- [37].G. Tumminello FM; Pizzolanti G; Gebbia N; Leto, Stefin A and cathepsin B and L circulating serum levels in patients with malignant or non-malignant liver diseases: A preliminary report, Med. Sci. Res 23 (1995) 741–742. [Google Scholar]
- [38].Kirpich IA, Gobejishvili LN, Homme MB, Waigel S, Cave M, Arteel G, Barve SS, McClain CJ, Deaciuc IV, Integrated hepatic transcriptome and proteome analysis of mice with high-fat diet-induced nonalcoholic fatty liver disease, J. Nutr. Biochem 22 (2011) 38–45. 10.1016/j.jnutbio.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Arab JP, Arrese M, Trauner M, Recent Insights into the Pathogenesis of Nonalcoholic Fatty Liver Disease, Annu. Rev. Pathol. Mech. Dis 13 (2018) 321–350. 10.1146/annurev-pathol-020117-043617. [DOI] [PubMed] [Google Scholar]
- [40].Bykov IL, Väkevä A, Järveläinen HA, Meri S, Lindros KO, Protective function of complement against alcohol-induced rat liver damage, Int. Immunopharmacol 4 (2004) 1445–1454. 10.1016/j.intimp.2004.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


