Abstract
Upregulation of the PI3K/AKT/mTOR pathway has been implicated in glioma-related epileptogenesis. In this retrospective analysis, epilepsy characteristics and response to treatment were evaluated in patients with gliomas harboring somatic mutation variants in PIK3CA. A cohort of 134 patients with 150 PIK3CA variants was extracted from previously validated databases. Patients with the hotspot H1047R, R88Q, E542K, and G118D variants comprised a subset (n=41) for epilepsy phenotyping. In multivariate analysis, the presence of H1047R (n=15) was associated with worse seizure control (p=0.026). These results support preclinical findings and suggest that glioma PIK3CA variation may have promise as a biomarker for epilepsy severity and response to treatment.
Keywords: Glioma, Focal seizure, Epilepsy, Somatic mutation, PIK3CA
1. Introduction
Seizures are commonly seen in association with gliomas.1,2 Seizure prevalence varies by glioma subtype, suggesting variable mechanisms of glioma-related epileptogenesis. Given its role in both gliomagenesis and epileptogenesis, overactivation of the PI3K/AKT/mTOR pathway has been implicated as one possible mechanism.3,4 Recent preclinical evidence further suggests that activating mutations in PIK3CA, the PI3K catalytic subunit, may have differing effects on peritumoral hyperexcitability based on the specific variant involved, although clinical evidence in humans is lacking.5
Predictive biomarkers of peritumoral hyperexcitability are necessary to improve seizure risk stratification in glioma patients, and may help guide treatment decisions. Furthermore, it is uncertain whether tumor biomarkers may also predict clinical response to anti-epileptic drugs (AEDs). In this retrospective analysis, we utilized the recently described Dana-Farber Cancer Institute (DFCI) Neuro-Oncology Cohort with clinical and sequencing data to identify and study 134 glioma patients harboring PIK3CA variants,6 and characterized the electrophysiologic and clinical phenotypes associated with four commonly observed variants (H1047R, R88Q, E542K, G118D).
2. Material and methods
2.1. Patient Selection
Patients treated at DFCI (Boston, MA) with histologically confirmed glioma (WHO Grades II-IV) and OncoPanel testing revealing any PIK3CA somatic mutation between 2011-2019 were included to comprise an initial dataset. Overall we utilized data from 1600 glioma patients in the DFCI Neuro-Oncology Cohort (dataset publicly available from DFCI-AACR GENIE cBioPortal or by contacting K.L.L.).6 Case records were retrieved using standardized queries. The majority (93%) of records were retrieved from the previously validated oncology database (OncDRS Project #117304) covering cases between 2011-2016.7 The remainder were retrieved from the Research Patient Data Registry, a quality-controlled centralized electronic health record data repository,8 followed by manual review to confirm the diagnosis and OncoPanel results.
A subset for phenotyping was defined containing patients with one of the PIK3CA variants occurring with a count of ≥10 in the initial dataset. Cases with >1 PIK3CA variant or additional variants in PIK3C2B, PIK3R1, or MTOR were then excluded to limit biasing the genotype/phenotype association (Figure 1A).
Figure 1.
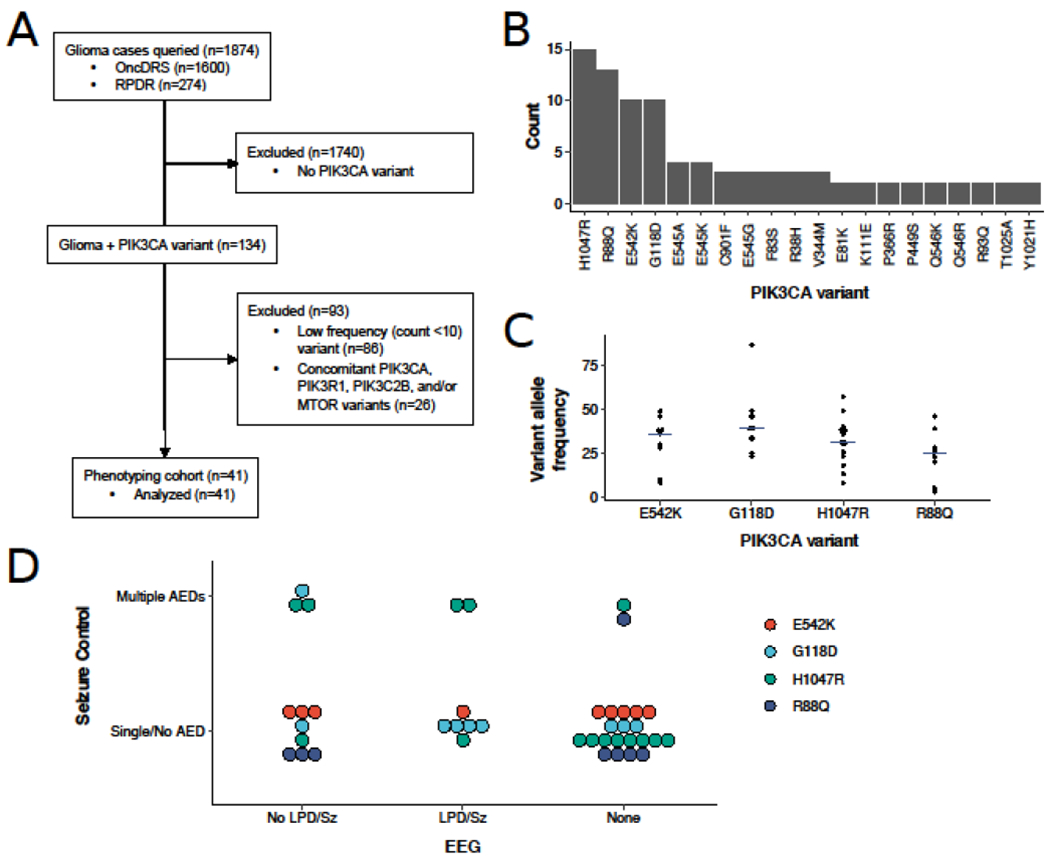
PIK3CA genotype-phenotype correlation. (A) Flowchart of sample determination for analysis. (B) Histogram of unique PIK3CA variants. Variants with a single occurrence are not shown. (C) Variant allele frequencies for each PIK3CA variant included in the phenotyping cohort. Crossbars indicate the median. (D) Distribution of patients with the PIK3CA variants H1047R, R88Q, E542K, and G118D stratified by EEG features and seizure control. AED, anti-epileptic drug; GBM, glioblastoma multiforme; LPD, lateralized periodic discharges; OncDRS, Oncology Data Retrieval System; RPDR, Research Patient Data Registry; Sz, seizure; WHO, World Health Organization.
All protocols for Human Subjects Research were approved by the Institutional Review Board at DFCI with waiver of informed consent (Protocol #20-345).
2.2. Glioma Genetic Sequencing
Tumor sequencing was performed using the OncoPanel genomic assay as previously described.7,9,10 In formalin-fixed paraffin-embedded tissue specimens with sufficient tumor density, DNA was extracted and fragmented for library preparation and PCR enrichment, hybrid capture with the OncoPanel DNA bait set, and Illumina sequencing, followed by single-nucleotide mutation analysis, copy number analysis, and structural variant analysis. Only tumor somatic mutation variants were included in this study.
2.3. Electrophysiology Analysis
EEG features were determined by review of all electrophysiology reports in the medical record and confirmation by visual waveform review when available. The presence of epileptiform discharges, periodic discharges, and seizures were recorded. Periodic discharges were defined according to American Clinical Neurophysiology Society Standardized Critical Care EEG Terminology.11
2.4. Seizure Outcomes
Pre-operative and post-operative seizures were determined by review of the medical record and defined with respect to the initial glioma resection. To evaluate the primary outcome of clinical response to AEDs, each patient was categorized as requiring ≤1 versus >1 AED to control seizures at any given time after glioma resection. AEDs were determined by review of medication reconciliation lists, EEG reports (when available), and clinician notes at timepoints pre-operatively, post-operatively, at time of EEG (if applicable), and at most recent follow-up. AEDs documented for non-seizure indications were excluded.
2.5. Definition of Variables
Temporal/insular lobe tumor localization and cortical T2/FLAIR involvement were defined by the brain MRI at diagnosis. The presence of gross total resection at initial diagnosis was determined by review of the radiology report of the first post-operative gadolinium-enhanced MRI and documentation by the treating neuro-oncologist. IDH1/2 mutation status was determined by review of immunohistochemistry and OncoPanel sequencing.
2.6. Statistical Analysis
Univariate comparisons between categorical and continuous variables were performed using the Fisher Exact Test and Mann-Whitney U-test (two groups) or Kruskal-Wallis H-test (multiple groups), respectively. Variables with p<0.2 in univariate analysis were included in multivariate analysis using a multiple linear regression model. All tests were two-tailed with a significance threshold of 0.05. Analysis was performed using R 4.0.3 (R Foundation for Statistical Computing, Vienna, Austria).
3. Results
3.1. Distribution of Glioma PIK3CA Variants
There were 134 patients with glioma and PIK3CA somatic mutation variants identified. Of these, 93 (69%) had WHO Grade IV IDH wildtype glioma (92 glioblastoma, 1 diffuse midline glioma), 39 (29%) had WHO Grade II-IV gliomas with IDH1 R132H mutation (9 glioblastoma, 3 astrocytoma, and 27 oligodendroglioma), and 2 (1.5%) had IDH1 wildtype astrocytoma. OncoPanel testing revealed a total of 150 PIK3CA variants, of which 81 were unique. The four most common known pathogenic hotspot variants H1047R, R88Q, E542K, and G118D accounted for 48/150 (32%) of the total variants observed, occurring in 48/134 (36%) of the patients (Figure 1B). No differences in allele frequencies were observed between variants (Figure 1C; p=0.20). In contrast, 61 different PIK3CA variants of rare recurrence or unknown significance were observed only once across the cohort. Among the 134 cases, 18 (13%) also had variants in PIK3C2B, PIK3R1, and/or MTOR.
3.2. Demographic and Clinical Features of Common PIK3CA Variants
Clinical-electrophysiological data were analyzed for the 41 patients harboring a single PIK3CA variant in H1047R, R88Q, E542K, or G118D, without any additional mutations in PIK3C2B, PIK3R1, or MTOR. Seven (17%) had exclusively pre-operative seizure, 12 (29%) had exclusively post-operative seizures, 10 (24%) had both pre-operative and post-operative seizures, and 12 (29%) had neither. WHO Grade IV gliomas were present in 11/15 cases with H1047R, 7/8 with R88Q, 6/9 with E542K, and 9/9 with G118D. Patient demographics, oncologic characteristics, and epilepsy details are shown in Table 1.
Table 1.
Demographics and clinical characteristics of patients with H1047R, R88Q, E542K, and G118D variants
| N=41 (%) | |
|---|---|
| Age at diagnosis (median, range) | 57 (18–81) |
| Sex (female) | 19 (46.3) |
| Histology | |
| Glioblastoma | 33 (80.5) |
| Anaplastic astrocytoma | 2 (4.9) |
| Anaplastic oligodendroglioma | 5 (12.1) |
| Oligodendroglioma | 1 (2.4) |
| IDH1 mutated * | 9 (22.0) |
| Gross total resection | 16 (39.0) |
| Temporal/insular lobe localization | 14 (34.1) |
| Cortical involvement † | 37 (92.5) |
| Pre-operative seizure | 17 (41.4) |
| Post-operative seizure | 22 (53.7) |
| EEG performed | 19 (46.3) |
| EEG with epileptiform discharges †† | 11 (57.9) |
| EEG with electrographic seizure †† | 2 (10.5) |
| EEG with periodic discharges and/or seizure †† | 8 (42.1) |
All IDH1 mutations were R132H; there were no IDH2 mutations
Percentage reported out of 40 cases with pre-operative MRI available
Percentages are reported as % of cases in which EEG performed
3.3. Electrophysiologic Characteristics of PIK3CA Variants
EEGs were performed on 19/41 (46%) of patients, 68% of which were completed post-operatively. Periodic discharges and/or electrographic seizures were seen in 4/6 cases with G118D variant, 3/6 with H1047R, 1/4 with E542K, and 0/3 with R88Q (Figure 1D).
3.4. Treatment Response by PIK3CA Variant
Patients stratified by seizure control and electrophysiologic features are shown in Figure 1D. In univariate analysis, the association of multiple AEDs with age (p=0.039), gross total resection (OR 1.21, 95% CI 0.15-8.44, p=0.69), temporal/insular lobe tumor localization (OR 1.55, 95% CI 0.19-11.02, p=0.67), IDH1 mutation status (OR 3.37, 95% CI 0.39-26.54, p=0.17), and PIK3CA H1047R variant (OR 5.71, 95% CI 0.78-69.59, p=0.079) were tested. In multivariate analysis, the presence of H1047R variant (p=0.026), but not age (p=0.11) or IDH1 mutation (p=0.75), was significantly associated with worse seizure control.
4. Discussion
In this analysis, we characterized the epilepsy phenotype associated with four glioma PIK3CA somatic mutation variants. We found that patients with the H1047R variant were more likely to require multiple AEDs to control seizures compared to patients with other PIK3CA variants. These results suggest that glioma PIK3CA variation may have promise as a predictive biomarker for epilepsy severity and response to treatment.
The results support preclinical findings implicating PIK3CA variation in glioma-related epileptogenesis. Recent work using a mouse glioblastoma model demonstrated that specific variants in PIK3CA may promote electrophysiologic hyperexcitability and seizures. In this model, tumors with C420R and H1047R variants were associated with greater epileptiform activity on EEG, behavioral seizures, and an imbalance between excitatory and inhibitory synaptogenesis compared to R88Q.5 In particular, the H1047R variant promoted hyperexcitability in a cell-autonomous manner, although the mechanism has yet to be fully elucidated.5 These data provide mechanistic evidence to support the validity of the clinical effect observed in this cohort. Each of the four PIK3CA variants analyzed are established gain of function mutations that lead to increased downstream phosphorylation and activation of AKT.12–14 Given the involvement of the PI3K/AKT/mTOR pathway in the pathogenesis of various brain malformations,15 somatic PIK3CA variation may represent a therapeutic target across various epilepsy syndromes.
Although the sample size for each individual PIK3CA variant was small, the size of the total cohort was relatively large. To our knowledge, prior studies of PIK3CA mutations in human glioblastoma have reported total sample sizes ranging up to 22 individuals.16–18 In contrast, this analysis involved 134 individuals harboring PIK3CA variants including 15 with H1047R mutations, allowing for focused clinical correlation to specific somatic mutations. Given the variable influence on peritumoral hyperexcitability and underlying mechanisms, it was necessary to analyze each PIK3CA variant independently at the cost of statistical power. Nevertheless, we found clinically meaningful trends that may motivate future large-scale efforts to confirm the observed associations, as well as improve outcome prediction and individualized treatment strategies for patients with glioma-related epilepsy. In addition to other epilepsy risk factors including age, tumor subtype and localization, and extent of resection, it is expected that multiple somatic variants influence peritumoral hyperexcitability and the manifestation of seizures. While the variants described here may apply to only a small proportion of glioma-related epilepsy, the increasingly widespread use of massively parallel tumor sequencing will aid in developing models to detect and integrate polygenic effects.
This study has limitations. Determination of seizure control, seizure type, and AEDs through retrospective analysis is subject to error. We attempted to minimize bias by cross-referencing different data sources at multiple serial timepoints. EEG data was available in under half of cases at variable timepoints and likely obtained on a subset according to clinical indications, which may have introduced bias.
5. Conclusions
Glioma somatic mutations in PIK3CA may variably impact peritumoral hyperexcitability. In this study, the H1047R variant was associated with more difficult to control seizures. PIK3CA variants should be considered in future predictive models assessing post-operative seizure risk in patients with glioma.
Highlights.
The distribution of PIK3CA variants detected in 134 glioma patients was evaluated
Epilepsy phenotype was analyzed for H1047R, R88Q, E542K, and G118D variants
Periodic discharges on EEG were common with H1047R and G118D variants
The H1047R variant was associated with more difficult to control seizures
Acknowledgments
Funding:
This work was supported by the National Cancer Institute [NCI P50CA165962-06A1 Sub-Project 5140] (S.T.); and the Brigham and Women’s Hospital A.J. Trustey Epilepsy Research Fund (S.T.).
Declarations of Interest:
S.T. has received research support from Eisai. K.L.L. is a founder and board member of Travera LLC, a consultant for BMS, Integragen, and Rarecyte, and has received research support from BMS, Lilly, Novartis, Amgen, and Deciphera. J.W.L. has served as a consultant for Biogen, performed contract work for Bioserenity and Teladoc, and is the co-founder and scientific advisor for Soterya, Inc. W.P. and E.L. have no declarations of interest.
Abbreviations
- AED
anti-epileptic drug
- EEG
electroencephalogram
- WHO
World Health Organization
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pallud J, Audureau E, Blonski M, et al. Epileptic seizures in diffuse low-grade gliomas in adults. Brain . 2014;137(2):449–462. doi: 10.1093/brain/awt345 [DOI] [PubMed] [Google Scholar]
- 2.Englot DJ, Berger MS, Chang EF, Garcia PA. Characteristics and treatment of seizures in patients with high-grade glioma: a review. Neurosurg Clin N Am. 2012;23(2):227–235, vii–viii. doi: 10.1016/j.nec.2012.01.009 [DOI] [PubMed] [Google Scholar]
- 3.Huberfeld G, Vecht CJ. Seizures and gliomas––towards a single therapeutic approach. Nat Rev Neurol. 2016;12(4):204–216. doi: 10.1038/nrneurol.2016.26 [DOI] [PubMed] [Google Scholar]
- 4.Yuan Y, Xiang W, Yanhui L, et al. Activation of the mTOR signaling pathway in peritumoral tissues can cause glioma-associated seizures. Neurol Sci. 2017;38(1):61–66. doi: 10.1007/s10072-016-2706 [DOI] [PubMed] [Google Scholar]
- 5.Yu K, Lin C-CJ, Hatcher A, et al. PIK3CA variants selectively initiate brain hyperactivity during gliomagenesis. Nature. Published online January 29, 2020:1–6. doi: 10.1038/s41586-020-1952-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Touat M, Li YY, Boynton AN, et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature. 2020;580(7804):517–523. doi: 10.1038/s41586-020-2209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orechia J, Pathak A, Shi Y, et al. OncDRS: An integrative clinical and genomic data platform for enabling translational research and precision medicine. Appl Transl Genom. 2015;6:18–25. doi: 10.1016/j.atg.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Estiri H, Stephens KA, Klann JG, Murphy SN. Exploring completeness in clinical data research networks with DQe-c.J Am Med Inform Assoc. 2018;25(1):17–24. doi: 10.1093/jamia/ocx109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagle N, Berger MF, Davis MJ, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2(1):82–93. doi: 10.1158/2159-8290.CD-11-0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia EP, Minkovsky A, Jia Y, et al. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med. 2017;141(6):751–758. doi: 10.5858/arpa.2016-0527-OA [DOI] [PubMed] [Google Scholar]
- 11.Hirsch LJ, LaRoche SM, Gaspard N, et al. American Clinical Neurophysiology Society’s Standardized Critical Care EEG Terminology: 2012 version. J Clin Neurophysiol. 2013;30(1):1–27. doi: 10.1097/WNP.0b013e3182784729 [DOI] [PubMed] [Google Scholar]
- 12.Oda K, Okada J, Timmerman L, et al. PIK3CA Cooperates with Other Phosphatidylinositol 3’-Kinase Pathway Mutations to Effect Oncogenic Transformation. Cancer Res. 2008;68(19):8127–8136. doi: 10.1158/0008-5472.CAN-08-0755 [DOI] [PubMed] [Google Scholar]
- 13.Orloff MS, He X, Peterson C, et al. Germline PIK3CA and AKT1 mutations in Cowden and Cowden-like syndromes. Am J Hum Genet. 2013;92(1):76–80. doi: 10.1016/j.ajhg.2012.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dogruluk T, Tsang YH, Espitia M, et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res. 2015;75(24):5341–5354. doi: 10.1158/0008-5472.CAN-15-1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain. 2015;138(6):1613–1628. doi: 10.1093/brain/awv045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka S, Batchelor TT, Iafrate AJ, et al. PIK3CA activating mutations are associated with more disseminated disease at presentation and earlier recurrence in glioblastoma. Acta Neuropathologica Communications. 2019;7(1):66. doi: 10.1186/s40478-019-0720-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallia GL, Rand V, Siu I-M, et al. PIK3CA Gene Mutations in Pediatric and Adult Glioblastoma Multiforme. Mol Cancer Res. 2006;4(10):709–714. doi: 10.1158/1541-7786.MCR-06-0172 [DOI] [PubMed] [Google Scholar]
- 18.Lee J-K, Wang J, Sa JK, et al. Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49(4):594–599. doi: 10.1038/ng.3806 [DOI] [PMC free article] [PubMed] [Google Scholar]


