Abstract
The recent coronavirus outbreak has changed the world’s economy and health sectors due to the high mortality and transmission rates. Because the development of new effective vaccines or treatments against the virus can take time, an urgent need exists for the rapid development and design of new drug candidates to combat this pathogen. Here, we obtained antiviral peptides obtained from the data repository of antimicrobial peptides (DRAMP) and screened their predicted tertiary structures for the ability to inhibit the main protease of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using multiple combinatorial docking programs, including PatchDock, FireDock, and ClusPro. The four best peptides, DRAMP00877, DRAMP02333, DRAMP02669, and DRAMP03804, had binding energies of −1125.3, −1084.5, −1005.2, and −924.2 Kcal/mol, respectively, as determined using ClusPro, and binding energies of −55.37, −50.96, −49.25, −54.81 Kcal/mol, respectively, as determined using FireDock, which were better binding energy values than observed for other peptide molecules. These peptides were found to bind with the active cavity of the SARS-CoV-2 main protease; at Glu166, Cys145, Asn142, Phe140, and Met165, in addition to the substrate-binding sites, Domain 2 and Domain 3, whereas fewer interactions were observed with Domain 1. The docking studies were further confirmed by a molecular dynamics simulation study, in which several descriptors, including the root-mean-square difference (RMSD), root-mean-square fluctuation (RMSF), solvent-accessible surface area (SASA), radius of gyration (Rg), and hydrogen bond formation, confirmed the stable nature of the peptide–main protease complexes. Toxicity and allergenicity studies confirmed the non-allergenic nature of the peptides. This present study suggests that these identified antiviral peptide molecules might inhibit the main protease of SARS-CoV-2, although further wet-lab experiments remain necessary to verify these findings.
Keywords: SARS-CoV-2, Peptide, Peptide–protein docking, Molecular dynamics, Lead identification
1. Introduction
The current coronavirus disease 2019 (COVID-19) pandemic has spread rapidly across the world, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which was first identified in December 2019 in Wuhan, Hubei Province, China (Huang et al., 2020, Wu et al., 2020b, Zu et al., 2020). COVID-19 has spread globally across 200 countries (Naqvi et al., 2020), and the most affected countries are the USA, Brazil, India, Russia, Colombia, Spain, the UK, Peru, Argentina, Mexico, France, and Italy. According to the most recent update, released on 2 July 2021 by the World Health Organization (WHO), 182,319,261 confirmed cases and 3,954,324 deaths have been recorded worldwide (https://covid19.who.int/). The consequences of COVID-19 appear to be exceedingly severe among people older than 55 years (Emergency and Team, 2020, Guan et al., 2020, Report, 2020, Uddin et al., 2020). By contrast, the fatality rate has been the lowest among individuals younger than 19 years, at 0–0.1%, and a low fatality rate has been observed among those aged 20–54 years, at 0.1–0.8%. The fatality rate increases gradually, ranging from 1.4% to 4.9% among those aged 55–74 years and reaching 4.3–10.5% among those aged 75–84 years, whereas those older than 85 years are associated with the highest fatality rate of 10.4–27.3% (Emergency and Team, 2020, Guan et al., 2020, Li et al., 2020, Report, 2020, Uddin et al., 2020). Individuals with delicate immune systems or chronic diseases, such as diabetes, kidney disease, liver disease, malignant tumors, and cardiovascular disease, are at the highest risk of COVID-19 and experience a higher mortality rate (Emergency and Team, 2020, Gao et al., 2020, Guan et al., 2020, Li et al., 2020, Report, 2020, Uddin et al., 2020).
Coronaviruses are single-stranded, positive-sense, enveloped ribonucleic acid viruses that are 60–140 nm in diameter and feature 9–12-nm long spike-like surface projections that manifest a crown or solar corona-like appearance (Singhal, 2020, Zu et al., 2020). Coronaviruses have been categorized into four genera, including α-, β-, γ-, and δ-coronaviruses, and SARS-CoV-2 belongs to the β-coronavirus genus, of the order Nidovirales, the family Coronaviridae, and the subfamily Coronavirinae (Liu et al., 2020, Naqvi et al., 2020, Wu et al., 2020a). Mammals can typically be infected by α- and β-coronaviruses, whereas avian species are typically infected by γ-coronaviruses, and both mammals and avians can be infected by δ-coronaviruses (Naqvi et al., 2020). SARS-CoV-2 represents the seventh coronavirus known to infect humans, including HCoV-229E, HKU-NL63, HCoV-OC43, HCoV-HKU1, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) (Chan et al., 2015, Chan et al., 2013, Chen et al., 2020, Liu et al., 2020, Wu et al., 2020a).
The SARS-CoV-2 genome contains 13–15 open reading frames (ORFs), which are straddled by the 3′-untranslated region (UTR) and the 5′-UTR (Lu et al., 2020, Mohammad et al., 2020). The ORF 1ab encodes the pp1a and pp1ab polyproteins, which are subsequently cleaved by the main protease (Mpro) and the papain-like protease (PLpro) to form 16 non-structural proteins (nsp1 to nsp16). Mpro, which is also known as 3CLpro, and PLpro play significant roles in the post-translational modification of the two replicase polyproteins, pp1a and pp1ab (Anand et al., 2003, Mohammad et al., 2020, Naqvi et al., 2020, Ton et al., 2020, Zhang et al., 2020), and are required to generate functional subunits. These abundant functional subunits are required for the flexing and packaging of newly formed virions and broadly assist in the viral replication process, which is necessary to propagate the infection (Du et al., 2004, Hegyi and Ziebuhr, 2002, Naqvi et al., 2020, Ullrich and Nitsche, 2020, Ziebuhr et al., 2000).
Mpro plays a central role in proteolysis, viral replication processes, transcriptional processes and is essential for the life cycle of the virus (Dai et al., 2020, Naqvi et al., 2020, Shamsi et al., 2020). Because Mpro is the key enzyme necessary for SARS-CoV-2 replication and propagation, its inhibition can block these processes, preventing viral spread. Therefore, Mpro is considered to be a more promising drug target than PLpro (Estrada, 2020, Khan et al., 2020, Zhang et al., 2020).
Some drugs have been examined explored for their ability to be repurposed for use in the treatment of COVID-19. Among these, remdesivir and favipiravir have been identified as RNA-dependent RNA polymerase (RdRp)-targeted drugs that exhibit antiviral activity against SARS-CoV-2. In addition, antimalarial drugs, including hydroxychloroquine and chloroquine, have been demonstrated to suppress SARS-CoV-2 replication (Kiplin Guy et al., 2020). Other antimalarial drugs, including halofantrine, doxycycline, mefloquine, lumefantrine, atovaquone, primaquine, and sulfonamides, have also been used to treat COVID-19 (Schlagenhauf et al., 2020). Typically, vaccines are required to successfully demonstrate efficacy and safety in three clinical trial phases (I, II, and III). As of 2 July 2021, according to the most recent update reported by the WHO, approximately 50, 37, and 32 vaccine candidates are currently undergoing Phase I, II, and III clinical trials, respectively (https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html). To date, several vaccines have been approved for use in different countries. On 31 December 2020, a COVID-19 vaccine named ‘BNT162b2/COMIRNATY Tozinameran (INN)’, manufactured by Pfizer, was granted an Emergency Use Listing (EUL) by the WHO. Subsequently, the WHO permitted the EUL use of two versions of the vaccine named ‘AZD1222’, manufactured by AstraZeneca and SKBio. In addition, the COVID-19 vaccine named ‘Covishield’ was manufactured by the Serum Institute of India, and the ‘mRNA-1273’ vaccine, manufactured by Moderna, became available near the end of February 2021. Two additional vaccine manufacturers, Sinopharm and Sinovac, launched two identically named vaccines, called ‘SARS-CoV-2 Vaccine,’ in early March. Additionally, on 12 March 2021, Janssen (Johnson & Johnson) developed the ‘Ad26.COV2.S’ vaccine. Besides, ‘Sputnik V,’ ‘Ad5-nCoV’, and ‘EpiVacCorona’ vaccines were developed by various manufacturers and have been issued EULs by the WHO (https://www.who.int/news-room/q-a-detail/coronavirus-disease-(covid-19)-; (Organización Mundial de la Salud, 2021)). Moreover, on March 15, China endorsed a vaccine for exigency use named “ZF2001” which has been made by two companies entitled- Anhui Zhifei Longcom and the Institute of Medical Biology at the Chinese Academy of Medical Sciences as copartners. As research continues, “NVX-CoV2373” is a vaccine developed by the company Novavax which is Maryland-based. Besides, the Beijing Institute of Biological Products manufactured the “BBIBP-CorV” vaccine which is authorized as exigence use by WHO on May 7. Nevertheless, the “CoronaVac” vaccine is developed by Sinovac Biotech, an unofficial Chinese company and on June 1 it gets permission for emergency use by WHO (https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html).
Peptides allocate themselves as fastidious, spiffing, benign signaling molecules that can able to affix to distinct cell surface receptors. Having these potential and underlying characteristics, peptides personate as novel therapeutics (Fosgerau and Hoffmann, 2015). As a drug, peptides propound sundry precedence comprehend with exuberant biological alacrity, bluffy specificity, and mangy toxicity (Bruckdorfer et al., 2005, Lien and Lowman, 2003). The comprehensive obstacle of being peptides therapeutics are petty active period due to impetuous renal clearance, scantiness of selective receptor subtype, minuscule half-life span. Peptides accommodate exorbitant affinity or specificity to a particular target and beneath toxic silhouette which makes assorted superiority over small molecules (Nestor, 2009, Sato et al., 2006). US Food and Drug Administration (FDA) endorse peptide therapeutics against the Hepatitis C virus in 2011 and also ratify as the first peptide drug of Human immunodeficiency virus (HIV) for clinical attestation. In addition, several peptides are undergoing phase trials against the Influenza virus, Hepatitis B virus, and Hepatitis D virus. Contrariwise, Chikungunya virus (CHIKV), Zika virus, and Dengue virus (DENV) have peptide therapeutics exhibiting antiviral activity (Agarwal and Gabrani, 2021).
With the aim of identifying plausible antiviral therapeutic agents to treat SARS-CoV-2, researchers all over the world have conducted virtual screening strategies that include molecular docking (Abdusalam and Murugaiyah, 2020, Abo-zeid et al., 2020, Cheke, 2020, Hagar et al., 2020, Joshi et al., 2020, Khaerunnisa et al., 2020, Nag et al., 2021, Rutwick Surya and Praveen, 2021) or molecular dynamics simulations combined with molecular docking (Al-Karmalawy et al., 2021, Chikhale et al., 2021, Das et al., 2020, Keretsu et al., 2020, Kumar et al., 2020a, Kumar et al., 2020b, Majumder and Mandal, 2020, Mhatre et al., 2020, Sepay et al., 2021, Teli et al., 2021). SARS-CoV-2 has exhibited almost 1516 nucleotide-level variations across various genome-wide annotations, and due to frequent adaptive mutations through which the viral genome evolves viral resistance, no effective or plausible antiviral treatment has yet been developed (Islam et al., 2020a, Xu et al., 2020). However, peptide-oriented molecular docking and molecular dynamics techniques can provide a rapid platform for reliable drug development in the race to combat SARS-CoV-2.
2. Materials and methods
2.1. Peptide screening and preparation
Potential antiviral peptides were retrieved from the data repository of antimicrobial peptides (DRAMP), (Kang et al., 2019) which contains 22,209 entries, including 5841 antimicrobial peptides. In our study, we only examined those peptide molecules with known antiviral activity. Based on antiviral activity, we screened 215 antiviral peptides from 5820 peptides. Additional screening procedures were executed according to the following parameters. First, we eliminated 62 antiviral peptides longer than 50 amino acids in length to facilitate the application of a new approach called PEP-FOLD 3.5 (Maupetit et al., 2010), which can accurately predict the structures of peptides containing from 5 to 50 amino acids. Two additional antiviral peptides with unknown amino acids in their sequences, denoted as ‘X,’ were also eliminated. Approximately 31 antiviral peptides were removed due to a lack of authentic information regarding their activity or function within the database. Another 16 antiviral peptides were removed due to a lack of any plausible antiviral activity or that presented with inefficient antiviral activities and were therefore not suitable for combating any viral disease. Consequently, among the initially identified 5841 antimicrobial peptides, we analyzed 104 antiviral peptides, each consisting of fewer than 50 amino acids and with evidence of antiviral activity, which were identified by an in-depth analysis of the database. The peptide sequence information was used to estimate the peptide structures using the PEP-FOLD 3.5 webserver. This webserver predicts the peptide structure using a Hidden Markov Model suboptimal sampling algorithm. The energy minimizations of the peptides were implemented in Avogadro software with the aid of MMFF94 force field, using the steepest descent algorithm and 500 steps.
2.2. Protein preparation
The three-dimensional structure of the SARS-CoV-2 Mpro (PDB ID: 6LU7) was retrieved from the Protein Data Bank (Protein Data Bank, 2019) with a resolution of 2.16 Å. The protein structure was initially prepared in Discovery Studio (San Diego: Accelrys Software Inc., 2012), where the water molecules and heteroatoms were removed. The cleaned protein structure was further minimized in YASARA tools by employing the AMBER14 force field (Land and Humble, 2018). The minimized protein structure was used for further molecular docking and dynamics studies.
2.3. Molecular docking
Peptide and protein docking was implemented as described in our previous study (Mahmud et al., 2021a). The protein structure of Mpro was input as the receptor molecule, and the peptides were used as the ligands in the PatchDock (Schneidman-Duhovny et al., 2005) webserver. The final clustering was selected on the basis of the root-mean-square deviation (RMSD) value. The top ten solutions were taken from PatchDock and further docked using FireDock (Mashiach et al., 2008) tools. The FireDock program optimizes lateral string conformations and rigid body formations to provide larger refinement. These web tools were employed to obtain rigid protein–peptide docking. The best protein–peptide complex was selected from among the top ten conformers based on energy scoring. All the 104 peptides were then further docked using the ClusPro program (Comeau et al., 2004) to obtain more accurate binding energy profiles from the corresponding peptide and protein complexes. Based on the combination of multiple docking programs, four peptide–protein complexes were selected for non-bonded structure analysis. The interaction analysis was conducted using PyMOL (DeLano, 2002) and Discovery Studio (San Diego: Accelrys Software Inc., 2012) software package.
2.4. Dynamics simulation
The docked peptide–protein molecules and their conformational variability were assessed through molecular dynamics simulations in YASARA dynamics software (Krieger et al., 2004). The AMBER14 force field (Maier et al., 2015) was used in this study, and the docked complexes were initially cleaned, along with hydrogen bond orientation and optimization for simulation. The cubic simulation box with a cell size of 110 × 110 × 110 Å3 was filled with water molecules, with a 0.9899 g/cm3 density, and the system was neutralized with 0.9% NaCl (Krieger et al., 2013). The TIP3P water model was used to solvate the complex. The acid dissociation constant value (pKa) was calculated for the amino acids in the complex. The SCWRL algorithm combined with hydrogen bond network optimization was applied to maintain the correct protonation state of each amino acid residue. The particle mesh Ewald method (Darden et al., 1993) was applied to calculate long-range electrostatic interactions, using a cutoff radius of 8 Å. The simulation cell box was 20 Å larger than the peptide–protein complex to allow for maximum free motion, and the periodic boundary condition was maintained (Krieger and Vriend, 2015). The energy of each simulated system was minimized by using the steepest descent gradient approaches (5000 cycles) via simulated annealing methods. An 80-ps position-restrained molecular dynamics simulation was performed for each system at constant pressure (1 atm) and temperature (310 K). Temperature coupling was performed using the Berendsen thermostat, with a temperature coupling constant of 0.1 ps, and a manometer method was used for pressure coupling, with a reference pressure of 1 atm. For the simulation study, a time step of 1.25 fs was used. The chemical bond lengths involving hydrogen bond atoms were fixed using the SHAKE algorithms (Brooks et al., 1983). After reaching an equilibrium state at 1 ns, the simulation was allowed to run for 250 ns, and the simulation trajectories were saved after every 100 ps. The simulation was conducted three times, and the average values from the trajectories were used to analyze the RMSD, root-mean-square fluctuation (RMSF), radius of gyration (Rg), solvent-accessible surface area (SASA), and hydrogen bonds (Bappy et al., 2020, Islam et al., 2020b, Khan et al., 2020, Swargiary et al., 2020).
Furthermore, the simulation snapshots were subjected to binding free energy calculations from MM-PBSA approaches by following equations (Mitra and Dash, 2018).
The YASARA macro was used to calculate the MM-PBSA binding energy where positive energy indicates the better bindings (Srinivasan and Rajasekaran, 2016).
2.5. Physiochemical properties
The physiochemical properties of the four best peptides were evaluated using ProtParam (Gasteiger et al., 2005) tools. This tool provides relevant properties, including molecular weight, net charge at pH 7, volume, peptide properties, stability, and charge.
2.6. Antigenicity and allergenicity prediction
The allergenicity and toxicity of the peptides were calculated using AllergenFP (Dimitrov et al., 2014b) and AllerTOP (Dimitrov et al., 2014a) webservers. The AllergenFP webserver predicts peptide allergenicity by employing a five E-descriptor-based fingerprinting method, whereas the AllerTOP webserver uses amino acid E-descriptors and k-nearest neighbor (kNN) machine learning approaches.
3. Results and discussion
3.1. Molecular docking
Molecular docking approaches can be used to identify the binding affinities of various ligands for the target protein structure to assist in the performance of rational drug design (Jones and Willett, 1995). This approach can contribute toward understanding the interaction dynamics and potential binding mechanisms, which can be applied for more rigorous inhibition (Liu et al., 2018). By combining several docking algorithms and programs, our approach provided enhanced structural accuracy and the precise calculation of binding interactions and affinities (Bartuzi et al., 2017). The peptide model structure derived from PEP-FOLD is presented in Fig. S1. The structures of 104 total peptides were modeled, and all 104 peptide structures were used to dock against the target Mpro enzyme. Howbeit, we selected the top ten elevated peptide molecules in conformity with the combined docking scores procured from PatchDock, ClusPro, and FireDock (Table 1 , Table S1). Furthermore, we opted top four plausible peptides amid the top ten peptide molecules predicated on the calculated binding energy for non-bonded structure analysis (Fig. 1 ). The DRAMP00877 peptide had the lowest FireDock global energy of −55.37 Kcal/mol and the lowest ClusPro docking energy (−1125.3 Kcal/mol). The binding energies for the DRAMP02333, DRAMP02669, and DRAMP03804 peptides in FireDock were −50.96 Kcal/mol, −49.25 Kcal/mol, and −54.81 Kcal/mol, respectively, whereas the binding energies in ClusPro were −1084.5 Kcal/mol, −1005.2 Kcal/mol, and −924.2 Kcal/mol.
Table 1.
The binding energy of the top 10 peptide molecules; DRAMP00877, DRAMP02333, DRAMP02669, DRAMP03804, DRAMP00832, DRAMP01366, DRAMP00837, DRAMP00876, DRAMP01671, DRAMP04504. The docking score from Firedock and Cluspro were tabulated where more negative energy indicates the favorable binding.
| DRAMP ID | Sequence | Firedock score (Global energy) | Cluspro Score |
|---|---|---|---|
| DRAMP00877 | GIPCGESCVWIPCISAALGCSCKNKVCYRN | −55.37 | −1125.3 |
| DRAMP02333 | FFRHLFRGAKAIFRGARQGWRAHKVVSRYRNRDVPETDNNQEEP | −50.96 | −1084.5 |
| DRAMP02669 | ACYCRIPACLAGERRYGTCIYQGRLWAFCC | −49.25 | −1005.2 |
| DRAMP03804 | VVCACRRALCLPRERRAGFCRIRGRIHPLCCRR | −54.81 | −924.2 |
| DRAMP00832 | GIPCAESCVWIPCTVTALLGCSCSNNVCYN | −51.84 | −888.9 |
| DRAMP01366 | GLLGLLGSVVSHVVPAIVGHF | −75.54 | −856.0 |
| DRAMP00837 | GIPCAESCVWIPCTVTALVGCSCSDKVCYN | −55.88 | −866.4 |
| DRAMP00876 | GLPVCGETCFTGTCYTNGCTCDPWPVCTRN | −49.08 | −871.4 |
| DRAMP01671 | ALWMTLLKKVLKAAAKALNAVLVGANA | −54.01 | −856.2 |
| DRAMP04504 | GILLNTLKGAAKNVAGVLLDKLKCKITGGC | −56.22 | −846.9 |
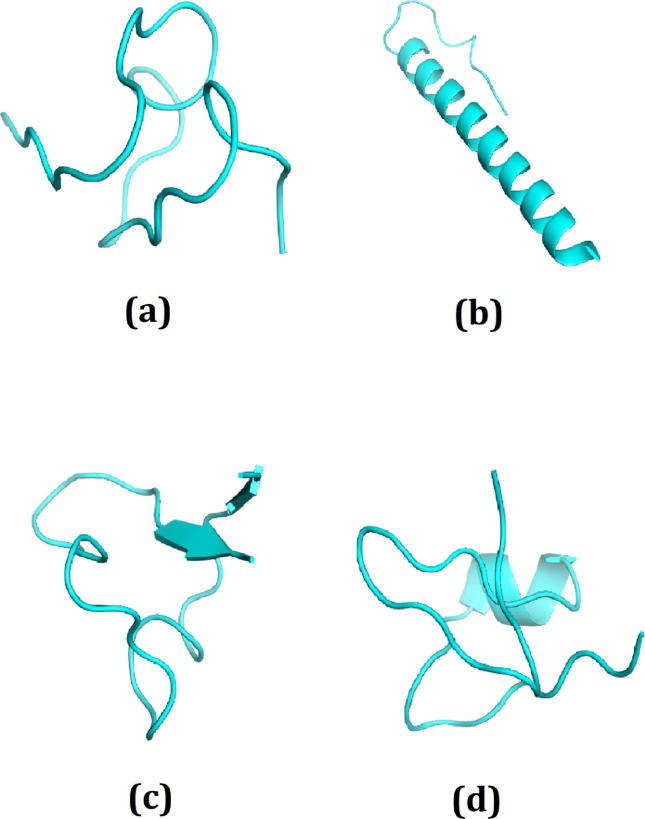
Fig. 1.
The best four peptide molecules based on binding affinity in docking program; (a) DRAMP00877, (b) DRAMP02333, (c) DRAMP02669, (d) DRAMP03804 peptide molecules.
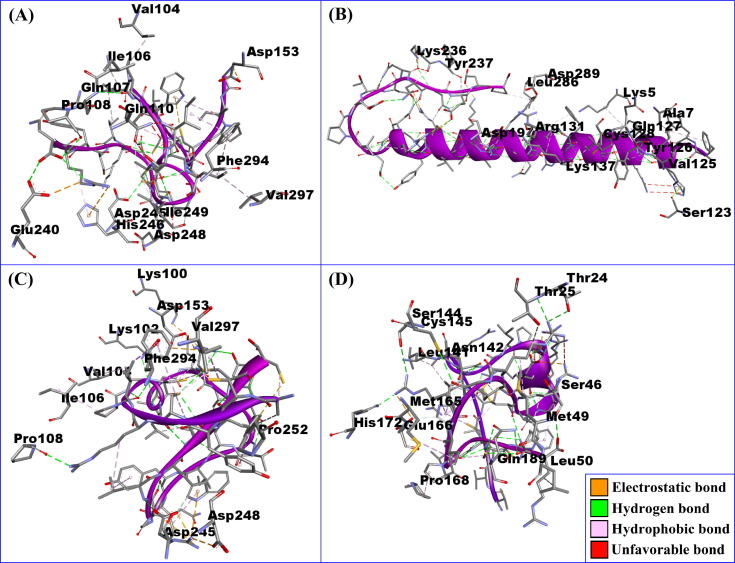
The non-bonded interactions between the peptide molecules and the SARS-CoV-2 Mpro are shown in Fig. 2 . The DRAMP00877 peptide and Mpro complex was stabilized by five hydrogen bonds at GLN107, ASP245, GLU240, ILE106, and PRO108, five hydrophobic bonds at VAL140, VAL297, HIS246, PHE294, and ILE249, two electrostatic bonds at ASP153 and GLU240 in addition to two unfavorable bonds at ASP248 and GLN110 (Fig. 2 and Table 2 ). The DRAMP02333 peptide and Mpro enzyme formed four hydrogen bonds at LYS137, LYS236, SER123, and TYR237, one electrostatic interaction at ASP197, and five hydrophobic bonds at ALA7, VAL125, CYS128, LEU286, and LYS5 together with four unfavorable bonds at TYR126, GLN127, ASP289, and ARG131 (Fig. 2 and Table 2). The DRAMP02669 peptide and Mpro complex formed five hydrophobic bonds at PHE294, VAL297, VAL104, ILE106 and PRO252, four hydrogen bonds at LYS100, LYS102, PRO108 and ASP245 and lastly four electrostatic bonds at LYS100, ASP153, ASP248 and ASP245. (Fig. 2 and Table 2). The DRAMP03804 peptide and Mpro complex created nine hydrogen bonds at ASN142, CYS145, GLN189, THR24, THR25, SER144, SER46, HIS172 and GLU166 and five hydrophobic bonds at MET165, Pro168, LEU141, Met49, and Leu50 and two electrostatic bonds at HIS172 and GLU166 (Fig. 2 and Table 2). The DRAMP03804 peptide binds in the active groove of Mpro, at Glu166, Cys145, Asn142, and Met165 (Jin et al., 2020). Binding to the active sites may lead to the possible inhibition of the target molecule (Daddam et al., 2020).
Fig. 2.
The non-bonded interaction of the DRAMP00877, DRAMP02333, DRAMP02669, DRAMP03804 peptides and main protease from SARS-CoV-2 at certain simulation times. Here, (A), (B), (C), (D), represents the binding interactions of the DRAMP00877, DRAMP02333, DRAMP02669, DRAMP03804 peptides and main protease complexes after 0 ns time respectively.
Table 2.
The binding interactions of the DRAMP00877, DRAMP02333, DRAMP02669, DRAMP03804 peptide and main protease from SARS-CoV-2 at certain simulation times. The snapshots were taken from 0 ns time.
| Peptide Name | Protein | Bond Distance(Å) | Interaction Category |
|---|---|---|---|
| DRAMP00877-0 ns | GLN107 | 2.95641 | Hydrogen Bond |
| ILE106 | 2.93253 | Hydrogen Bond | |
| PRO108 | 2.42821 | Hydrogen Bond | |
| VAL104 | 3.74178 | Hydrophobic Bond | |
| VAL297 | 4.52733 | Hydrophobic Bond | |
| HIS246 | 4.92969 | Hydrophobic Bond | |
| PHE294 | 4.01609 | Hydrophobic Bond | |
| ASP248 | 2.05342 | Unfavorable Bond | |
| GLN110 | 2.22731 | Unfavorable Bond | |
| ASP153 | 3.57136 | Electrostatic Bond | |
| GLU240 | 4.28431 | Hydrogen Bond;Electrostatic Bond | |
| ASP245 | 2.68394 | Hydrogen Bond | |
| ILE249 | 3.97462 | Hydrophobic Bond | |
| DRAMP02333-0 ns | LYS236 | 2.60219 | Hydrogen Bond |
| ASP197 | 4.11032 | Electrostatic Bond | |
| SER123 | 2.86314 | Hydrogen Bond | |
| ALA7 | 4.03287 | Hydrophobic Bond | |
| VAL125 | 4.47397 | Hydrophobic Bond | |
| CYS128 | 3.99697 | Hydrophobic Bond | |
| TYR126 | 2.28996 | Unfavorable Bond | |
| GLN127 | 2.20203 | Unfavorable Bond | |
| LYS137 | 3.78469 | Hydrogen Bond | |
| ASP289 | 2.24226 | Unfavorable Bond | |
| ARG131 | 2.83357 | Unfavorable Bond | |
| TYR237 | 4.83496 | Hydrogen Bond | |
| LEU286 | 2.35733 | Hydrophobic Bond | |
| LYS5 | 2.46289 | Hydrophobic Bond | |
| DRAMP02669-0 ns | LYS100 | 2.52983 | Hydrogen Bond;Electrostatic Bond |
| LYS102 | 3.08797 | Hydrogen Bond | |
| PHE294 | 5.00891 | Hydrophobic Bond | |
| ASP153 | 4.19372 | Electrostatic Bond | |
| VAL297 | 3.76384 | Hydrophobic Bond | |
| VAL104 | 2.94386 | Hydrophobic Bond | |
| ILE106 | 3.15834 | Hydrophobic Bond | |
| PRO108 | 2.88347 | Hydrogen Bond | |
| PRO252 | 3.43916 | Hydrophobic Bond | |
| ASP248 | 2.46233 | Electrostatic Bond | |
| ASP245 | 5.02135 | Hydrogen Bond;Electrostatic Bond | |
| DRAMP03804-0 ns | ASN142 | 3.28024 | Hydrogen Bond |
| CYS145 | 3.19041 | Hydrogen Bond | |
| GLN189 | 3.09088 | Hydrogen Bond | |
| MET165 | 5.24318 | Hydrophobic Bond | |
| PRO168 | 4.40674 | Hydrophobic Bond | |
| THR24 | 2.96432 | Hydrogen Bond | |
| THR25 | 3.57625 | Hydrogen Bond | |
| SER144 | 4.39765 | Hydrogen Bond | |
| LEU141 | 2.88614 | Hydrophobic Bond | |
| SER46 | 5.34029 | Hydrogen Bond | |
| MET49 | 4.17034 | Hydrophobic Bond | |
| HIS172 | 3.17295 | Hydrogen Bond;Electrostatic Bond | |
| GLU166 | 3.73191 | Hydrogen Bond;Electrostatic Bond | |
| LEU50 | 4.50938 | Hydrophobic Bond |
3.2. Molecular dynamics
The binding interaction and the conformational variations that occur during binding can be understood using a docking study to provide insight into the binding sites to a limited extent. Further changes in the ligand or the bound complex can be studied by varying the temperature and pressure in a molecular dynamics simulation, which can minimize the cost of performing such experiments in the laboratory (Arfin et al., 2018, Durrant and McCammon, 2011, Nair and Miners, 2014). A combined docking and molecular dynamics approach can be used to validate the docking-derived results (Gioia et al., 2017).
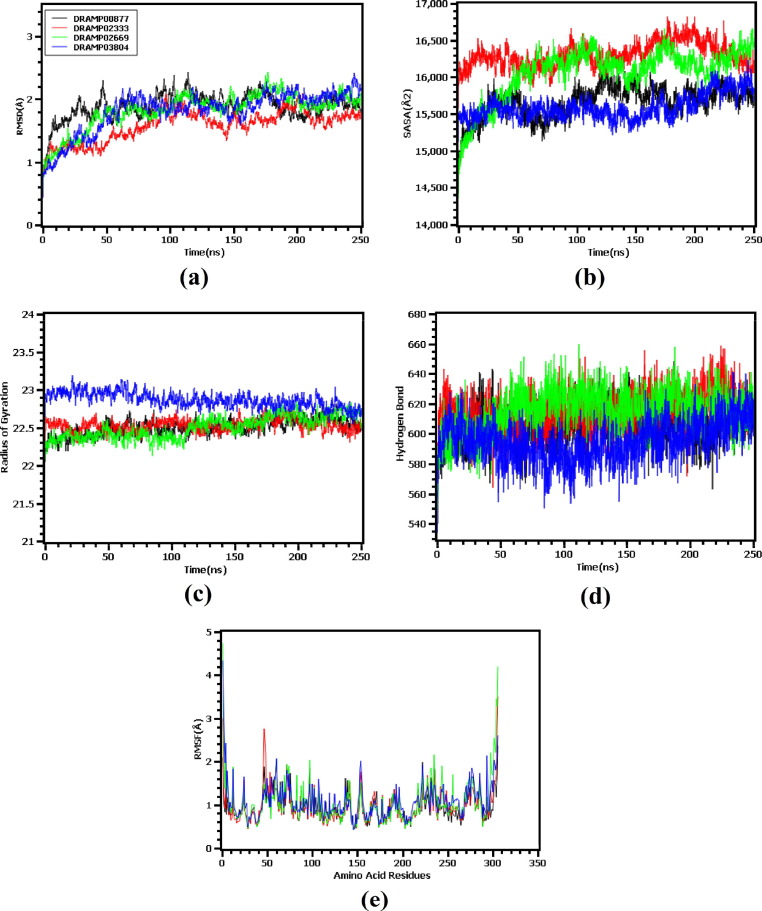
Molecular dynamics simulations were implemented for the peptide and SARS-CoV-2 Mpro complexes to confirm the structural rigidity and validate the docking outcomes for the complexes. The RMSD values of the C-alpha atoms were explored to understand the structural rigidity. Fig. 3 (a) demonstrated that the DRAMP00877, DRAMP02333, DRAMP02669, and DRAMP03804 complexes showed initial RMSD increases due to instability. The increase in RMSD was higher for the DRAMP00877 peptide complex than for the other complexes. The upward trend in RMSD was maintained until 40 ns, after which all four complexes achieved stability, which was maintained throughout the remainder of the simulation period. Although the DRAMP00877 complex had a higher RMSD trend during the initial phase, the RMSD profile decreased from 40 to 100 ns. The DRAMP02333 complex had a lower RMSD than the other complexes, indicating increased structural integrity. The average RMSD profile of the complexes formed with DRAMP00877, DRAMP02333, DRAMP02669, and DRAMP03804 were 1.88 Å, 1.58 Å, 1.82 Å, and 1.81 Å, respectively. The overall RMSD trend did not exceed 2.5 Å, which indicates overall structural stability as higher RMSD profiles for C-alpha atoms are indicators of low stability (Mahmud et al., 2021b).
Fig. 3.
The molecular dynamics simulation of the peptide and main protease complex, here (a) root mean square deviation of the c-alpha atoms, (b) solvent accessible surface area, (c) radius of gyration, (d) hydrogen bonding of the complexes, (e) root mean square fluctuation of the complexes to understand the flexibility of the amino acid residue.
The SASA values of the complexes were analyzed to evaluate changes in the surface of the SARS-CoV-2 Mpro in response to binding with the peptide molecules. The DRAMP00877, DRAMP02333, and DRAMP03804 complex had similar SASA trends throughout the entire simulation trajectory, indicating no change in protein volume. However, the DRAMP02669 complex showed an initial upward trend in SASA until 50 ns, indicating an expansion of the surface area following initial complex formation [Fig. 3 (b)]. The complex then stabilized and showed a trend similar to that of the DRAMP02333 peptide complex. The average SASA profiles of the four complexes were 15663.42 Å2, 16302.79 Å2, 16066.2Å2 and 15590.45 Å2 for DRAMP00877, DRAMP02333, DRAMP02669, and DRAMP03804, respectively. The SASA profiles of the peptide–protein complexes had similarly stable trends, with little fluctuation, indicating a lack of expansion or contraction for the protein complexes during the simulation time (Rakib et al., 2021).
The compactness of the protein complexes was evaluated using Rg, for which higher values indicate a more labile nature and a lower value indicates the firmness of the protein. The DRAMP02669 complex displayed fluctuations in the Rg value until 20 ns, indicating an initially loose packaging system for the protein [Fig. 3 (c)]. However, this complex maintained stability for the remainder of the simulation time. The DRAMP03804 complex had a larger Rg trend than all other complexes but was associated with a lower degree of deviation, demonstrating the achievement of a steady state. The comparative higher Rg of this peptide may be responsible for relatively more flexible state and mobile nature of the complexes. The average Rg values were 22.504, 22.527, 22.501, and 22.867 Å for DRAMP00877, DRAMP02333, DRAMP02669, and DRAMP03804, respectively. The higher Rg profile of the DRAMP03804 complex than other complexes may defines the more mobile nature and folding and unfolding of the protein complexes. In addition, hydrogen bond formation between the protein and peptide in complex plays a vital role in maintaining complex stability. Fig. 3 (d) demonstrated that the DRAMP02333 complex contained the highest number of hydrogen bonds throughout the entire simulation time. All of the evaluated peptide and Mpro complexes were stable, and high levels of fluctuation were generally absent for these complexes.
The RMSF of the peptide and Mpro complexes were explored to understand the flexibility across the amino acid region of Mpro. Fig. 3 (e) demonstrated that almost all residues had low RMSF, except Ser1 (helix-strand), Glu47 (beta-turn), Asp48 (beta-turn), Arg60 (helix-strand), Lys97 (beta-turn), Lys137 (beta-turn), Arg222 (beta-turn), Phe294 (helix-strand), Ser301 (beta-turn), Gly302 (beta-turn), Val303 (beta-turn), Thr304 (beta-turn), Phe305 (beta-turn), and Gln306 (beta-turn).
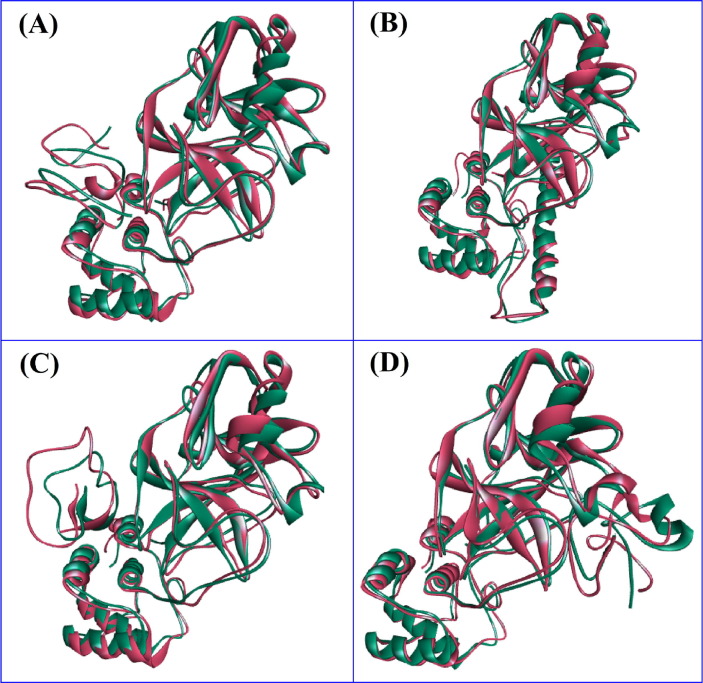
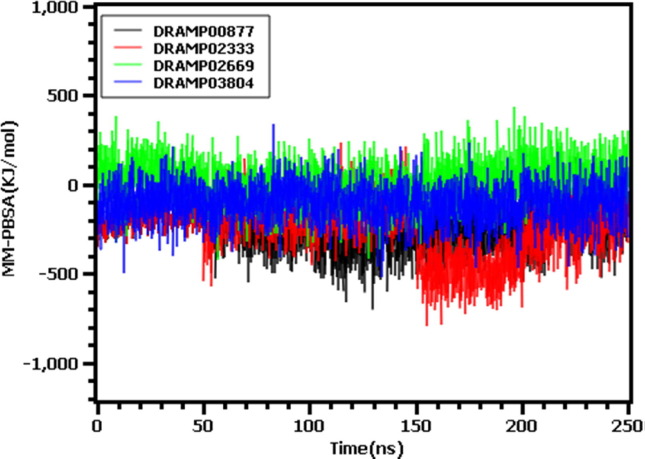
The pre- and post-molecular dynamics docked structures were superimposed to explore variations among these structures. The RMSD of the DRAMP00877, DRAMP02333, DRAMP02669, and DRAMP03804 peptides were found to be 2.076, 1.658. 1.793, and 1.778 Å, respectively, indicating limited flexibility among the structures (Fig. 4 ). Moreover, the binding free energy of the peptide-protein complexes were calculated from the MM-PBSA method (Fig. 5 ) where more positive score indicates the better binding of the complexes. The average free energy of the DRAMP00877, DRAMP02333, DRAMP02669, and DRAMP03804 peptide complexes were −245.612, −243.441, 19.51237, −51.5827KJ/mol respectively. The DRAMP02669 peptide complex exhibited more favorable binding than the other complexes.
Fig. 4.
The superimposition of the Pre and Post MD structure; (A) Pre and Post MD structure of DRAMP00877 peptide, (B) Pre and Post structure of DRAMP02333 peptide, (C) superimposed structure of DRAMP02669 peptide, (D) superimposed structure of DRAMP03804 peptide. The figure was analyzed in Discovery Studio software package.
Fig. 5.
The binding free energy of the top 4 peptide and protein complexes which were calculated by MM-PBSA methods.
The flexibility of the amino acid residues within the peptides was also assessed through RMSF analysis, and all four peptide molecules were found to display RMSF values below 2.5 Å, except Lys23 in the DRAMP00877 peptide; Tyr29, Asp33, and Val34 in the DRAMP02333 peptide; Glu13, Gly17, Thr18, Tyr21, and Gly22 in the DRAMP02669 peptide; and Val1, Val2, and Arg16 in the DRAMP03804 peptide. Lower RMSF values among the amino acid sequence indicate a less flexible and more stable nature of the peptide molecule (Fig. S6). The secondary structures of the peptide molecules were assessed to explore changes in the secondary structures, included the formation of α helices, turns, random coils, 310 helices, and pi helices (Fig. S7). The DRAMP00877 peptide displayed stable proportions of α-helices, β-sheets, turns, random coils, and pi helices, although some deviations were observed for 310 helices; however, 310 helices represented a lower percentage of secondary structure contents for the DRAMP00877 peptide. Random coils and helices were primarily noted for the DRAMP02333 peptide, which displayed a stable trend throughout the entire simulation times, with minor deviations in the proportions of 310 helices and turns. Random coils were primarily noted for the DRAMP02669 peptide, and stable profiles were observed for all secondary structures observed for this peptide throughout the simulation. Turns and random coils were primarily observed for the DRAMP03804 peptide, which featured stable trends, with some aberrations for α helix and 310 helix formation.
The SARS-CoV-2 Mpro consists of three domains; Domain 1, Domain 2, and Domain 3. Domain 1 contains residues 8–101; Domain 2 is formed by residues 102–184, which features an antiparallel β-barrel structure; and Domain 3 consists of residues 201–303 (Jin et al., 2020). Domain III (residues 201–303) comprises of five α- helices and is liable for the dimerization of enzymes exhibiting involvement in dimer formation (Chou et al., 2004, Kneller et al., 2020). In the Mpro dimerization, the troublesome the foreword of domain III has been adjusted by fragment elimination demonstrating that a decollated enzyme inexistent domain III stays as a monomer which is catalytically passive (Suárez and Díaz, 2020). Thus, Mpro constructs a functioning dimer via intermolecular interactivity, predominantly mesne the helical domains eliciting indispensable accosts of Domain III in the protease functionality (Kneller et al., 2020). The substrate-binding position is residing in the cleft interim domains I and II and the protomers, that cohere with every other via N-terminus residues 1–7, which are lying middle domains II and III with playing roles in the formation of the substrate-binding site. Four subsites namely; S1′, S1, S2, and S4 comprised the substrate-binding cleft (Mengist et al., 2021). The S1 subsite is formed with Phe140, Gly143, His163, His172, Cys145 and Glu166 while S2 comprised with Thr25, His41, and Cys145 amino acid residues; chiefly inlaid in electrostatic and hydrophobic interactions. The perfunctory subsites S3-S5 consist of Met49, His41, Met165, Glu166, and Gln189 amino acid residues. These shallow subsites can endure varied performance (Jin et al., 2020, Yang et al., 2005).
The binding interactions of the docked complexes were analyzed after taking snapshots at the 100, 200, and 250 ns time points during the simulation to understand interaction stability and deviations that occur across the simulation time. The DRAMP00877 peptide had several stable interactions at Domain 3 (Val 297, Phe294, His246, Glu240, Ile249, and Asp248), and equal stable interactions were found with Domain 2 (Gln107, Ile106, Pro108, Val104, Asp153, and Gln110) after 100 ns of simulation (Table S2, Fig. S2). Meanwhile, the DRAMP00877 peptide had the most stable interactions at Domain 3 (Phe294, Glu240, Ile249, His246, Asp248, and Asp245), and two stable interactions were observed with Domain 2 (Gln110 and Asp153) after 200 ns of simulation. Howbeit, the DRAMP00877 peptide had two stable interactions at Domain 2 (Gln110 and Asp153) and diverse stable interactions at Domain 3(Phe294, Ile249, Glu240, His246, and Asp245).
In case of the DRAMP02333 peptide molecule, there were diverse interactions in the simulation environment at Domain 3 (Lys236, Asn238, Leu287, Tyr237, Asp289 and Leu286) and four stable interactions at Domain 2 (Lys137, Cys128, Tyr126 and Ser123 in conjunction with an interaction at Domain 1 (Ala7). Domain2 and Domain 3 of Mpro are connected by a long loop region, and DRAMP02333 formed stable contacts with Asp197 within the loop region. Furthermore, the DRAMP02333 peptide molecule exhibited several interactions at Domain 2 (Val125, Gly124, Ser139, Lys137, Ser123, and Tyr126) and three stable interactions at Domain 3 (Asn238, Lys236, and Leu286) in tandem with an interaction at Domain 1 (Glu14 and Ala7) in the simulation environment after 200 ns of simulation (Table S3, Fig. S3), The DRAMP02333 constituted stable contacts with Asp197 within the loop region that connects Domain 2 and Domain 3 of Mpro. Howbeit, after 250 ns of simulation, the DRAMP02333 peptide molecule demonstrated various interactions in the simulation environment, including nine static interactions were noticed at Domain 2 (Val125, Lys137, Ser139, Gly124, Gly138, Tyr126, Pro122, Ser123 and Glu166) and six stable interactions at Domain 3 (Asn238, Lys236, Tyr237, Leu286, Leu287, and Met235) in conjunction with two rigid interactions at Domain 1 (Glu14, and Ala7). Here, the DRAMP02333 formed stable contacts with Thr199 within the loop region connecting Domain2 and Domain 3 of Mpro simulation.
For the DRAMP02669 peptide, sole one interaction audited at Domain 1 (Lys100), in addition with six interactions, which observed at Domain 2 (Lys102, Ile106, Val104, Gln110, Pro108, Asp153) and five interactions observed at Domain 3 (Phe294, Val297, Pro252, Asp248, Asp245) after the 100 ns simulation (Table S4, Fig. S4). Additionally, four interactions noticed at Domain 2 (Lys102, Val104, Gln107, Asp153), with five interactions observed at Domain 3 (Asp245, His246, Asp248, Phe294, Val297) after the 200 ns simulation. Besides, after the 250 ns simulation only three interactions were seen at Domain 2 (Lys102, Val104, Asp153), with notably seven interactions observed at Domain 3 (Pro252, His246, Phe294, Val297, Asp248, Ile249, Asp245).
There are six interactions observed for the DRAMP03804 peptide were formed with Domain 1 (His41, Ser46, Leu50, Thr25, Thr24, Met49), with six interactions observed at Domain 2 (Asn142, Met165, Pro168, Glu166, Phe140, Leu141) after the 100 ns simulation (Table S5, Fig. S5). Moreover, afterwards the 200 ns simulation, five interactions were observed at Domain 1 (Ser46, Met49, Leu50, Thr25, Thr24), including seven interactions observed at Domain 2 (Asn142, Met165, Pro168, Leu141, Phe140, Glu166, His172). Furthermore, only three interactions were observed at Domain 1 (Cys44, Ser46, Leu50), with six interactions observed at Domain 2 (Asn142, Glu166, Met165, Pro168, Leu141, His163) after the 250 ns simulation and this peptide constructed static contacts with GLN 189, ALA 191 among the loop region that appends Domain 2 and Domain 3 of Mpro. The interactions formed by the DRAMP03804 peptide primarily involve the active residues of the SARS-CoV-2 Mpro, which may interfere with the Mpro function. The co-crystalized ligand obtained with the Mpro structure binds in the substrate-binding pocket in extended conformations, and the inhibitor backbone atoms from an antiparallel sheet that interacts with residues 164–168 (Jin et al., 2020). Interestingly, the DRAMP03804 peptide molecules also featured three conserved interactions within these regions, at Met165, Pro168 and Glu166. It is cognizant that, DRAMP03804 peptide has Asn142, Gln189, Met165, Pro168, Leu141 and Glu166 residues that elicit resemblance with sundry of the abuzz residues of SARS-CoV-2 Mpro (Jin et al., 2020).
The peptide molecule DRAMP00877 interacted with the SARS-CoV-2 Mpro through conserved residues Phe294, His246, Glu240, Ile249, Asp153, and Gln110, whereas the DRAMP02333 peptide interaction involved Lys137, Tyr126, Ser123, Lys236, Ala7 and Leu286 throughout the simulation at 0 ns, 100 ns, 200 ns and 250 ns. Moreover, in all respects of the entire simulation trajectory, the amino acid residues Lys102, Phe294, Asp153, Val297, Val104, Asp248 and Asp245 of DRAMP02669 were involved in the interaction with SARS-CoV-2 Mpro and the Asn142, Gln189, Met165, Pro168, Leu141, Ser46, Glu166 and Leu50 residues contributed to the interaction between Mpro and DRAMP03804.
3.3. Physiochemical properties
We assessed the physicochemical properties of the best four peptide molecules (Table 4 ). The largest peptide contained 44 amino acids, whereas the smallest peptide only contained 30 amino acids. The molecular weights and theoretical isoelectric point (pI) were higher for the DRAMP02333 and DRAMP03804 peptides than for the other peptides. The peptides DRAMP00877, DRAMP02333, DRAMP02669, and DRAMP03804 contained 1,5,1, and 1 negatively charged residues, respectively. The total number of atoms included in the entire peptide molecule ranged from 400 to 600, except for DRAMP02333.
Table 4.
the physiochemical properties of the best four peptide molecules, the peptide properties were calculated from ProtParam webtools.
| DRAMPID | Number of amino acids | Molecular weight | Theoretical pI | Total number of negatively charged residues (Asp + Glu) | Total number of positively charged residues (Arg + Lys) | Total number of atoms |
|---|---|---|---|---|---|---|
| DRAMP00877 | 30 | 3175.78 | 8.33 | 1 | 3 | 433 |
| DRAMP02333 | 44 | 5313.94 | 11.23 | 5 | 10 | 738 |
| DRAMP02669 | 30 | 3448.09 | 8.68 | 1 | 4 | 466 |
| DRAMP03804 | 33 | 3897.79 | 11.40 | 1 | 10 | 551 |
3.4. Allergenicity and toxicity prediction
An allergenic antigen can trigger the activity of the Th2 response, causing B cells to produce IgE, which binds with the receptor molecule FcεRI and activates eosinophils. Activated eosinophils can lead to inflammation and tissue damage (Brock, 1995, Dimitrov et al., 2013, Huby et al., 2000). The AllerTOP tools predict allergens based on descriptors associated with amino acid properties and has been demonstrated to predict allergenicity with a high level of accuracy and robustness (Dimitrov et al., 2014a). The toxicity and allergenic profiles of the peptide molecules were evaluated using multiple web tools (Table 3 ). The DRAMP00877, DRAMP03804, and DRAMP02333 peptide molecules were non-allergic and were characterized as non-toxic, whereas the DRAMP02669 peptide was identified as a probable allergen in the AllerTOP tool, although it was reported as a non-allergen by the AllergenFP webserver.
Table 3.
The allergic and toxicity profile of the best peptide molecules where DRAMP02669 peptide was found as allergen in AllerTOP server.
| DRAMP ID |
Allergenicity prediction |
Toxicity prediction through ToxinPred | |
|---|---|---|---|
| AllerTOP v. 2.0 | AllergenFP v.1.0 | ||
| DRAMP00877 | PROBABLE NON-ALLERGEN | PROBABLE NON-ALLERGEN | Non-Toxin |
| DRAMP02333 | PROBABLE NON-ALLERGEN | PROBABLE NON-ALLERGEN | Non-Toxin |
| DRAMP02669 | PROBABLE ALLERGEN | PROBABLE NON-ALLERGEN | Non-Toxin |
| DRAMP03804 | PROBABLE NON-ALLERGEN | PROBABLE NON-ALLERGEN | Non-Toxin |
4. Conclusion
Antiviral peptides can serve as effective leads for the development of new therapeutic options against SARS-CoV-2. In this study, peptide molecules were tested against the SARS-CoV-2 Mpro, and the four best peptides were identified based on the binding affinities with SARS-CoV-2 Mpro. This study revealed that the best peptide molecules bind to the active amino acid residues in Mpro. The molecular dynamics simulation study confirmed the docked conformations and structural properties that were identified in the rigid state. In addition, allergenicity profiling confirmed the non-allergic properties of the peptide molecules selected in this current study. The present study may aid the development of effective drugs against this deadly virus.
Funding
The authors did not receive any external funding.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.arabjc.2021.103315.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Abdusalam A.A.A., Murugaiyah V. Identification of potential inhibitors of 3CL protease of SARS-CoV-2 from ZINC database by molecular docking-based virtual screening. Front. Mol. Biosci. 2020;7:1–11. doi: 10.3389/fmolb.2020.603037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abo-zeid Y., Ismail N.S., McLean G.R., Hamdy N.M. A molecular docking study repurposes FDA approved iron oxide nanoparticles to treat and control COVID-19 infection. Eur. J. Pharm. Sci. 2020;153 doi: 10.1016/j.ejps.2020.105465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal G., Gabrani R. Antiviral peptides: identification and validation. Int. J. Pept. Res. Ther. 2021;27:149–168. doi: 10.1007/s10989-020-10072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Karmalawy A.A., Alnajjar R., Dahab M., Metwaly A., Eissa I. Molecular docking and dynamics simulations reveal the potential of anti-HCV drugs to inhibit COVID-19 main protease. Pharm Sci. 2021 doi: 10.34172/PS.2021.3. [DOI] [Google Scholar]
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science (80-.) 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- Arfin M., Nasution F., Toepak E.P., Alkaff A.H. Flexible docking-based molecular dynamics simulation of natural product compounds and Ebola virus Nucleocapsid (EBOV NP): a computational approach to discover new drug for combating Ebola. BMC Bioinformatics. 2018;19 doi: 10.1186/s12859-018-2387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bappy S.S., Sultana S., Adhikari J., Mahmud S., Khan M.A., Kibria K.M.K., Rahman M.M., Shibly A.Z. Extensive immunoinformatics study for the prediction of novel peptide-based epitope vaccine with docking confirmation against envelope protein of Chikungunya virus: a computational biology approach. J. Biomol. Struct. Dyn. 2020;1–16 doi: 10.1080/07391102.2020.1726815. [DOI] [PubMed] [Google Scholar]
- Bartuzi D., Kaczor A.A., Targowska-Duda K.M., Matosiuk D. Recent advances and applications of molecular docking to g protein-coupled receptors. Molecules. 2017;22 doi: 10.3390/molecules22020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock J. Immunobiology: the immune system in health and disease. Immunol. Today. 1995;16:551–552. doi: 10.1016/0167-5699(95)80057-3. [DOI] [Google Scholar]
- Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. doi: 10.1002/jcc.540040211. [DOI] [Google Scholar]
- Bruckdorfer T., Marder O., Albericio F. From production of peptides in milligram amounts for research to multi-tons quantities for drugs of the future. Curr. Pharm. Biotechnol. 2005;5:29–43. doi: 10.2174/1389201043489620. [DOI] [PubMed] [Google Scholar]
- Chan J.F.W., Lau S.K.P., To K.K.W., Cheng V.C.C., Woo P.C.Y., Yue K.Y. Middle East Respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., To K.K.W., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheke R.S. The molecular docking study of potential drug candidates showing anti-COVID-19 activity by exploring of therapeutic targets of SARS-CoV-2. Eurasian J. Med. Oncol. 2020;4:185–195. doi: 10.14744/ejmo.2020.31503. [DOI] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhale R., Sinha S.K., Wanjari M., Gurav N.S., Ayyanar M., Prasad S., Khanal P., Dey Y.N., Patil R.B., Gurav S.S. Computational assessment of saikosaponins as adjuvant treatment for COVID-19: molecular docking, dynamics, and network pharmacology analysis. Mol. Divers. 2021 doi: 10.1007/s11030-021-10183-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.Y., Chang H.C., Hsu W.C., Lin T.Z., Lin C.H., Chang G.G. Quaternary structure of the severe acute respiratory syndrome (SARS) coronavirus main protease. Biochemistry. 2004 doi: 10.1021/bi0490237. [DOI] [PubMed] [Google Scholar]
- Comeau S.R., Gatchell D.W., Vajda S., Camacho C.J. ClusPro: An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 2004;20:45–50. doi: 10.1093/bioinformatics/btg371. [DOI] [PubMed] [Google Scholar]
- Daddam J.R., Sreenivasulu B., Peddanna K., Umamahesh K. Designing, docking and molecular dynamics simulation studies of novel cloperastine analogues as anti-allergic agents: Homology modeling and active site prediction for the human histamine H1 receptor. RSC Adv. 2020;10:4745–4754. doi: 10.1039/c9ra09245e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Rao Z., Zhang L.K., Xu Y., Yang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science (80-.). 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- Das P., Majumder R., Mandal M., Basak P. In-Silico approach for identification of effective and stable inhibitors for COVID-19 main protease (Mpro) from flavonoid based phytochemical constituents of Calendula officinalis. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1796799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002 [Google Scholar]
- Dimitrov I., Bangov I., Flower D.R., Doytchinova I. AllerTOP vol 2 - A server for in silico prediction of allergens. J. Mol. Model. 2014;20 doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- Dimitrov I., Flower D.R., Doytchinova I. AllerTOP - a server for in silico prediction of allergens. BMC Bioinformatics. 2013;14 doi: 10.1186/1471-2105-14-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov I., Naneva L., Doytchinova I., Bangov I. AllergenFP: Allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30:846–851. doi: 10.1093/bioinformatics/btt619. [DOI] [PubMed] [Google Scholar]
- Du Q.S., Wang S.Q., Zhu Y., Wei D.Q., Guo H., Sirois S., Chou K.C. Polyprotein cleavage mechanism of SARS CoV M pro and chemical modification of the octapeptide. Peptides. 2004;25:1857–1864. doi: 10.1016/j.peptides.2004.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant J.D., McCammon J.A. Molecular dynamics simulations and drug discovery. BMC Biol. 2011;9 doi: 10.1186/1741-7007-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emergency, C.-N., Team, C.M., 2020. Osong Public Health and Research Perspectives Early Epidemiological and Clinical Characteristics of 28 Cases of Coronavirus Disease in South Korea 11, 8–14. [DOI] [PMC free article] [PubMed]
- Estrada E. Topological analysis of SARS CoV-2 main protease. Chaos. 2020;30 doi: 10.1063/5.0013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosgerau K., Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discov. Today. 2015;20:122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:1–2. doi: 10.5582/BST.2020.01047. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. The proteomics protocols handbook. Proteomics Protoc. Handb. 2005;571–608 doi: 10.1385/1592598900. [DOI] [Google Scholar]
- Gioia D., Bertazzo M., Recanatini M., Masetti M., Cavalli A. Dynamic docking: a paradigm shift in computational drug discovery. Molecules. 2017;22 doi: 10.3390/molecules22112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J.L., Liang Z., Peng Y., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagar M., Ahmed H.A., Aljohani G., Alhaddad O.A. Investigation of some antiviral N-heterocycles as COVID 19 drug: molecular docking and DFT calculations. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21113922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83:595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huby R.D.J., Dearman R.J., Kimber I. Why are some proteins allergens? Toxicol. Sci. 2000;55:235–246. doi: 10.1093/toxsci/55.2.235. [DOI] [PubMed] [Google Scholar]
- Islam M.R., Hoque M.N., Rahman M.S., Alam A.S.M.R.U., Akther M., Puspo J.A., Akter S., Sultana M., Crandall K.A., Hossain M.A. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-70812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.S., Mahmud S., Sultana R., Dong W. Identification and in silico molecular modelling study of newly isolated Bacillus subtilis SI-18 strain against S9 protein of Rhizoctonia solani. Arab. J. Chem. 2020;13:8600–8612. doi: 10.1016/j.arabjc.2020.09.044. [DOI] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang Xinglou, You T., Liu Xiaoce, Yang Xiuna, Bai F., Liu H., Liu Xiang, Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jones G., Willett P. Docking small-molecule ligands into active sites. Curr. Opin. Biotechnol. 1995;6:652–656. doi: 10.1016/0958-1669(95)80107-3. [DOI] [PubMed] [Google Scholar]
- Joshi, T., Joshi, T., Sharma, P., Mathpal, S., Pundir, H., 2020. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking 4529–4536. [DOI] [PubMed]
- Kang X., Dong F., Shi C., Liu S., Sun J., Chen J., Li H., Xu H., Lao X., Zheng H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data. 2019 doi: 10.1038/s41597-019-0154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keretsu S., Bhujbal S.P., Cho S.J. Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation. Sci. Rep. 2020;10:1–14. doi: 10.1038/s41598-020-74468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S. Potential Inhibitor of COVID-19 main protease (M pro) from several medicinal plant compounds by molecular docking study. Preprints. 2020:1–14. [Google Scholar]
- Khan M.A., Mahmud S., Alam A.S.M.R.U., Rahman M.E., Ahmed F., Rahmatullah M. Comparative molecular investigation of the potential inhibitors against SARS-CoV-2 main protease: a molecular docking study. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1796813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiplin Guy R., DiPaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science (80-.). 2020;368:829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- Kneller D.W., Phillips G., O’Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:7–12. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, Elmar, G.V., Spronk, C., 2013. YASARA - Yet another scientific artificial reality application. YASARA.org.
- Krieger E., Darden T., Nabuurs S.B., Finkelstein A., Vriend G. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins Struct. Funct. Genet. 2004;57:678–683. doi: 10.1002/prot.20251. [DOI] [PubMed] [Google Scholar]
- Krieger E., Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36:996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Kumari K., Vishvakarma V.K., Jayaraj A., Kumar Dhiraj, Ramappa V.K., Patel R., Kumar V., Dass S.K., Chandra R., Singh P. Promising inhibitors of main protease of novel corona virus to prevent the spread of COVID-19 using docking and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1779131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Sharma P.P., Shankar U., Kumar D., Joshi S.K., Pena L., Durvasula R., Kumar A., Kempaiah P., Poonam, Rathi B. Discovery of new hydroxyethylamine analogs against 3CLproProtein target of SARS-CoV-2: molecular docking, molecular dynamics simulation, and structure-activity relationship studies. J. Chem. Inf. Model. 2020;60:5754–5770. doi: 10.1021/acs.jcim.0c00326. [DOI] [PubMed] [Google Scholar]
- Land H., Humble M.S. YASARA: a tool to obtain structural guidance in biocatalytic investigations. Methods Mol. Biol. 2018;1685:43–67. doi: 10.1007/978-1-4939-7366-8_4. [DOI] [PubMed] [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science (80-.) 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S., Lowman H.B. Therapeutic peptides. Trends Biotechnol. 2003;21:556–562. doi: 10.1016/j.tibtech.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Liu Y.C., Kuo R.L., Shih S.R. COVID-19: The first documented coronavirus pandemic in history. Biomed. J. 2020;2–6 doi: 10.1016/j.bj.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Liu Yujie, Zeng G., Shao B., Chen M., Li Z., Jiang Y., Liu Yang, Zhang Y., Zhong H. Application of molecular docking for the degradation of organic pollutants in the environmental remediation: a review. Chemosphere. 2018;203:139–150. doi: 10.1016/j.chemosphere.2018.03.179. [DOI] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud S., Paul G.K., Biswas S., Afrose S., Mita M.A., Hasan M., Saleh M. Prospective role of peptide-based antiviral therapy against the main protease of SARS-CoV-2. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.628585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud S., Uddin M.A.R., Paul G.K., Shimu M.S.S., Islam S., Rahman E., Islam A., Islam M.S., Promi M.M., Emran T.B., Saleh M.A. Virtual screening and molecular dynamics simulation study of plant-derived compounds to identify potential inhibitors of main protease from SARS-CoV-2. Brief. Bioinform. 2021 doi: 10.1093/bib/bbaa428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder R., Mandal M. Screening of plant-based natural compounds as a potential COVID-19 main protease inhibitor: an in silico docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1817787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H.J. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupetit J., Derreumaux P., Tufféry P. A fast method for large-scale de novo peptide and miniprotein structure prediction. J. Comput. Chem. 2010 doi: 10.1002/jcc.21365. [DOI] [PubMed] [Google Scholar]
- Mengist H.M., Dilnessa T., Jin T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021;9:1–19. doi: 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhatre, S., Naik, S., Patravale, V., 2020. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- 19. The COVID-19 resource centre is hosted on Elsevier Connect, the company’ s public news and information.
- Mitra S., Dash R. Structural dynamics and quantum mechanical aspects of shikonin derivatives as CREBBP bromodomain inhibitors. J. Mol. Graph. Model. 2018;83:42–52. doi: 10.1016/j.jmgm.2018.04.014. [DOI] [PubMed] [Google Scholar]
- Mohammad T., Shamsi A., Anwar S., Umair M., Hussain A., Rehman M.T., AlAjmi M.F., Islam A., Hassan M.I. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: towards the development of effective COVID-19 therapy. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Banerjee R., Chowdhury R.R., Krishnapura Venkatesh C. Phytochemicals as potential drug candidates for targeting SARS CoV 2 proteins, an in silico study. VirusDisease. 2021 doi: 10.1007/s13337-021-00654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair P.C., Miners J.O. Molecular dynamics simulations: from structure function relationships to drug discovery. Silico Pharmacol. 2014;2 doi: 10.1186/s40203-014-0004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta - Mol. Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor J., Jr. The medicinal chemistry of peptides. Curr. Med. Chem. 2009;16:4399–4418. doi: 10.2174/092986709789712907. [DOI] [PubMed] [Google Scholar]
- Organización Mundial de la Salud, 2021. Status of COVID-19 Vaccines within WHO EUL/PQ evaluation process (20 January 2021) 2.
- Protein Data Bank, 2019. RCSB PDB: Homepage. Rcsb Pdb.
- Rakib A., Nain Z., Sami S.A., Mahmud S., Islam A., Ahmed S., Siddiqui A.B.F., Babu S.M.O.F., Hossain P., Shahriar A., Nainu F., Emran T. Bin, Simal-Gandara J. A molecular modelling approach for identifying antiviral selenium-containing heterocyclic compounds that inhibit the main protease of SARS-CoV-2: an in silico investigation. Brief. Bioinform. 2021;22:1476–1498. doi: 10.1093/bib/bbab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Report, M.W., 2020. Bialek - 2020 Severe-outcomes-among-patients-with-coronavirus-disease-2019-COVID19--United-States-February-12march-16-20202020Morbidity-and-Mortality-Weekly-ReportOpen-Access.pdf 69, 343–346. [DOI] [PMC free article] [PubMed]
- Rutwick Surya U., Praveen N. A molecular docking study of SARS-CoV-2 main protease against phytochemicals of Boerhavia diffusa Linn. for novel COVID-19 drug discovery. VirusDisease. 2021 doi: 10.1007/s13337-021-00683-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Diego: Accelrys Software Inc., 2012. Discovery Studio Modeling Environment, Release 3.5 [WWW Document]. Accelrys Softw. Inc.
- Sato A.K., Viswanathan M., Kent R.B., Wood C.R. Therapeutic peptides: technological advances driving peptides into development. Curr. Opin. Biotechnol. 2006;17:638–642. doi: 10.1016/j.copbio.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Schlagenhauf P., Grobusch M.P., Maier J.D., Gautret P. Repurposing antimalarials and other drugs for COVID-19. Travel Med. Infect. Dis. 2020;34 doi: 10.1016/j.tmaid.2020.101658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005 doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepay N., Sekar A., Halder U.C., Alarifi A., Afzal M. Anti-COVID-19 terpenoid from marine sources: a docking, admet and molecular dynamics study. J. Mol. Struct. 2021;1228 doi: 10.1016/j.molstruc.2020.129433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamsi A., Mohammad T., Anwar S., AlAjmi M.F., Hussain A., Rehman M.T., Islam A., Hassan M.I. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020;40 doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan E., Rajasekaran R. Computational investigation of curcumin, a natural polyphenol that inhibits the destabilization and the aggregation of human SOD1 mutant (Ala4Val) RSC Adv. 2016;6:102744–102753. doi: 10.1039/c6ra21927f. [DOI] [Google Scholar]
- Suárez D., Díaz N. SARS-CoV-2 main protease: a molecular dynamics study. J. Chem. Inf. Model. 2020 doi: 10.1021/acs.jcim.0c00575. [DOI] [PubMed] [Google Scholar]
- Swargiary A., Mahmud S., Saleh M.A. Screening of phytochemicals as potent inhibitor of 3-chymotrypsin and papain-like proteases of SARS-CoV2: an in silico approach to combat COVID-19. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1835729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teli D.M., Shah M.B., Chhabria M.T. In silico screening of natural compounds as potential inhibitors of SARS-CoV-2 main protease and spike RBD: targets for COVID-19. Front. Mol. Biosci. 2021;7:1–25. doi: 10.3389/fmolb.2020.599079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol. Inform. 2020 doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M., Mustafa F., Rizvi T.A., Loney T., Al Suwaidi H., Al-Marzouqi A.H.H., Eldin A.K., Alsabeeha N., Adrian T.E., Stefanini C., Nowotny N., Alsheikh-Ali A., Senok A.C. SARS-CoV-2/COVID-19: Viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12:1–18. doi: 10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorganic Med. Chem. Lett. 2020;30 doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.C., Chen C.S., Chan Y.J. The outbreak of COVID-19: an overview. J. Chinese Med. Assoc. 2020;83:217–220. doi: 10.1097/JCMA.0000000000000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Kwok Y.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science (80-.) 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- Zu, Z.Y., Jiang, M. Di, Xu, P.P., Chen, W., Ni, Q.Q., Lu, G.M., Zhang, L.J., Hospital, J., Hospital, T., Zu, D., Zhang, L.J., Clinical, N., District, X., 2020. Pr e ss In Pr e. JAMA - J. Am. Med. Assoc. 2019, in press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.