Graphical abstract

Keywords: antibody, mutation, variant, deep learning, clinical trial
Abstract
The ongoing massive vaccination and the development of effective intervention offer the long-awaited hope to end the global rage of the COVID-19 pandemic. However, the rapidly growing SARS-CoV-2 variants might compromise existing vaccines and monoclonal antibody (mAb) therapies. Although there are valuable experimental studies about the potential threats from emerging variants, the results are limited to a handful of mutations and Eli Lilly and Regeneron mAbs. The potential threats from frequently occurring mutations on the SARS-CoV-2 spike (S) protein receptor-binding domain (RBD) to many mAbs in clinical trials are largely unknown. We fill the gap by developing a topology-based deep learning strategy that is validated with tens of thousands of experimental data points. We analyze 796,759 genome isolates from patients to identify 606 non-degenerate RBD mutations and investigate their impacts on 16 mAbs in clinical trials. Our findings, which are highly consistent with existing experimental results about Alpha, Beta, Gamma, Delta, Epsilon, and Kappa variants shed light on potential threats of 100 most observed mutations to mAbs not only from Eli Lilly and Regeneron but also from Celltrion and Rockefeller University that are in clinical trials. We unveil, for the first time, that high-frequency mutations R346K/S, N439K, G446V, L455F, V483F/A, F486L, F490L/S, Q493L, and S494P might compromise some of mAbs in clinical trials. Our study gives rise to a general perspective about how mutations will affect current vaccines.
Introduction
Since the first positive cases of coronavirus disease, 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was reported in late December 2019, over 2.5 million lives have been taken away in the COVID-19 pandemic up to March 10, 2021. The developments of vaccines and antibody therapies are the most significant scientific accomplishments that offer essential hope to win the battle against COVID-19. Nonetheless, the emerging SARS-CoV-2 variants signal a major threat to existing vaccines and antibody drugs.
SARS-CoV-2 is a novel -coronavirus, which is an enveloped, unsegmented positive-sense single-strand ribonucleic acid (RNA) virus. It gains entry into the host cell through the binding of its spike (S) protein receptor-binding domain (RBD) to the host angiotensin-converting enzyme 2 (ACE2) receptor, primed by host transmembrane protease, serine 2 (TMPRSS2).1 According to epidemiological and biochemical studies, the binding free energy (BFE) between the S protein and ACE2 is proportional to the infectivity of different SARS-CoV-2 variants in the host cells.2, 3 Intrinsically, the mutation-induced BFE changes () of S protein and ACE2 complex provide a method to measure the infectivity changes of a SARS-CoV-2 variant compared to the first SARS-CoV-2 strain that deposited to GenBank (Access number: NC 045512.2).4 Specifically, a positive mutation-induced BFE change of S and ACE2 indicates that this mutation would strengthen the infectivity of SARS-CoV-2, while a negative mutation-induced BFE change reveals the possibility of the weakening transmissible and infectious. Therefore, one can predict the impact of SARS-CoV-2 RBD variants on infectivity by estimating their BFE changes.4, 5, 6
Moreover, the binding of S protein and ACE2 will trigger the host adaptive immune system to produce antibodies against the invading virus.7, 8 As illustrated in Fig. 1 , antibodies are secreted by a type of white blood cell called B cell (mainly by plasma B cells or memory B cells). An antibody can either attach to the surface of B cell (called B-cell receptor (BCR)) or exist in the blood plasma in a solute form. An antibody can be generated in three different ways: (1) Once SARS-CoV-2 invades the host cell, the adaptive immune system will be triggered, and the B cells will generate and secrete antibodies. (2) In antibody therapies, antibodies are initially generated from patient immune response and T-cell pathway inhibitors,7 which are called antibody drugs.8 Most COVID-19 antibody drugs primarily target S protein. (3) The vaccine is designed to stimulate an effective host immune response, which is another way to make B cells secrete antibodies.9 At this stage, various vaccines, including two mRNA vaccines designed by Pfizer-BioNTech and Moderna, have been granted authorization for emergency use in many countries, aiming to give our human cells instructions to make a harmless S protein piece to initiate the immune response actively. Although COVID-19 vaccines are the gamechanger, S protein mutations might weaken the binding between the SARS-CoV-2 S protein and antibodies and thus, reduce the efficiency and efficacy of the existing vaccines and antibody therapies.10
Figure 1.
SARS-CoV-2 S protein antibodies are secreted by B cells in aiming to compete with the host ACE2 for binding to the S protein RBD.
Although SARS-CoV-2 has a higher fidelity in the replication process which benefits from its genetic proofreading mechanism regulated by the non-structural protein 14 (NSP14) and RNA-dependent RNA polymerase (RdRp),11, 12 over 5,000 unique mutations has been found on SARS-CoV-2 S protein,5 which raises the question that how these mutations on S protein will affect the existing vaccines and antibody drugs. Antibody resistance of SARS-CoV-2 variants Alpha (B.1.1.7) and Beta (B.1.351) was reported.10 Mutation E484K on S protein RBD may help SARS-CoV-2 slip past the host immune defenses, is broadly founded in the Beta (a.k.a 20H/501Y.V2) variant 13 and the Gamma (P.1) (a.k.a 20 J/501Y.V3) variant.14 The ongoing evaluation of susceptibility of variants in subjects treated with the antibody-drug bamlanivimab shows that E484K substitution in Alpha, Beta, and Iota (B.1.526) variants have reduced susceptibility to bamlanivimab.15 Moreover, the K417N+E484K+N501Y substitutions in Beta and Gamma variants have also reduced susceptibility to bamlanivimab.15 Specifically, a 50% increment in the transmission of the Beta variant is estimated.16 Both Beta and Gamma variants cause negative effects on the neutralization by emergency use authorization (EUA) monoclonal antibody therapeutics,17, 18 and the moderate reductions in neutralizing activity were observed by using convalescent and post-vaccination sera.19 Furthermore, the Epsilon (B.1.427/B.1.429) variant carries an L452R mutation on the S protein RBD, which approximately increases 20% of the transmissibility of SARS-CoV-2,19 and has a mild negative impact on neutralization by some EUA therapeutics according to the Food and Drug Administration (FDA) report.15, 20 Notably, by using convalescent and post-vaccination sera, moderate reductions in neutralizing activity of L452R were observed.19
However, the determination of whether a mutation will reduce susceptibility to the existing antibodies and antibody drugs from wet laboratory experiments is time-consuming. Current experimental studies are restricted to only a small fraction of known RBD mutations that have been observed. There is no reliable measurement about whether a mutation will evade a vaccine because none knows how many different antibodies will be created from the vaccination of the general population of different races, genders, ages, and health conditions. Based on the molecular mechanism of SARS-CoV-2 infectivity, antibody, and vaccine, one can quantitatively estimate mutation impacts on SARS-CoV-2 infectivity and an antibody-drug through computing mutation-induced BFE changes of the S protein-ACE2 complex and the S protein-antibody complex, respectively. Using machine learning models to predict protein-protein interaction binding free energies can efficiently deliver consistent results.21, 22, 23 However, applying a machine learning model in practical studies requires validation with experimental data. In our earlier work, we proposed a TopNetTree model to predict the RBD-induced binding free energy (BFE) changes of S protein with ACE2 and 106 antibodies,5, 24 where we also illustrated the validation on experimental data 25, 26, 27, 28, 29. We showed that RBD mutation N501Y could significantly strengthen SARS-CoV-2 infectivity,5 which is consistent with experiment.16 Our results indicated that K417N, E484K, and L452R are all antibody-escape and infectivity-strengthening mutations, which are consistent with the findings from many experimental labs.10, 24, 5 Among them, mutation L452R in the Epsilon variant can significantly increase infectivity.5 We found that the T478K mutation in variant B.1.1.222, which has a rapid growth rate in Mexico, has the highest value of predicted BFE changes among high-frequency mutations.24, 5 Our prediction is confirmed from a report that mutation T478K is spreading at an alarming speed.30 Containiingg both T478K and L452R, the Delta variant is about four times more infectious than the original virus. We also predicted 1149 most likely, 1912 likely, and 625 unlikely receptor-binding domain (RBD) mutations.6 Currently, all known RBD mutations were correctly predicted as the most likely ones in our work.6, 5 Most recently, we have analyzed 506,768 SARS-CoV-2 genome isolates from patients and found that essentially all of 100 most observed RBD mutations have favorable predicted BFE changes, which provides a population-level confirmation of the reliability of our predictions. 24
The objective of this work is to reveal the mutational threats to 16 antibody drug candidates that are either in clinical trials or associated with clinical trial antibodies, as shown in Fig. 3. To this end, we analyze 796,759 complete SARS-CoV-2 genome sequences isolated from patients to identify 27,960 unique single mutations up to May 24, 2021 (see our Mutation Tracker https://users.math.msu.edu/users/weig/SARS-CoV-2_Mutation_Tracker.html).1 Among them, 606 non-degenerate mutations are found on the S protein RBD of SARS-CoV-2. We develop an algebraic topology-based deep learning model to estimate the mutation-induced BFE changes. Our study of antibody-drug candidates is invaluable and complementary to experimental results in the following senses. First, our machine learning and deep learning models validated with tens of thousands of experimental data points, including SARS-CoV-2 related deep mutations, are reliable as confirmed by emerging experimental data on various SARS-CoV-2 variants. Second, many fast-growing RBD mutations around the world pose imminent threats to existing and future vaccines and antibody therapies. The current experimental capability lags behind the rapidly growing RBD mutations. For example, there is no experimental study about the rapidly increasing B.1.1.222 variant. Our approach helps close the gap by combining genotyping and mutation-induced BFE change analysis. This work provides a threat analysis of all 606 existing RBD mutations. However, our emphasis is given to the 100 most observed RBD mutations. Third, current experiments in the literature are limited to two EUA monoclonal antibody therapeutics from Regeneron 31 and Eli Lilly.8 We extend our analysis to many other antibody therapeutic candidates that are in various stages of clinical trials, such as those from Celltrion 32 and the Rockefeller University. Finally, we introduce an interactive website, “SARS-CoV-2 Mutation Analyzer” (https://weilab.math.msu.edu/MutationAnalyzer/), to rank the worldwide frequency, BFE change, and antibody disruption of all observed mutations.
Figure 3.
3D structure superposition of 16 antibodies and ACE2 on the S protein RBD. (a) CT-P59 (7CM4),32 REGN10933 (6XDG),31 LY-CoV016 (CB6) (7C01).33 (b) LY-CoV488 (7KMH),34 LY-CoV481 (7KMI),34 C102 (7K8M),35 C105 (6XCM).36 (c) LY-CoV555 (7KMG),34 C002 (7K8T),36 C104 (7K8U),36 C119 (7K8W),36 C121 (7K8X),36 C144 (7K90).36 (d) REGN10987 (6XDG),31 C110 (7K8V),36 C135 (7K8Z).36
Results
Analysis of observed S protein RBD mutations
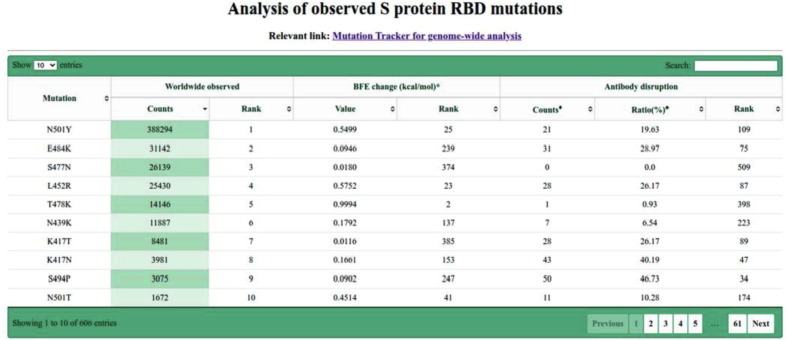
We first construct an interactive website, “SARS-CoV-2 Mutation Analyzer” (https://weilab.math.msu.edu/MutationAnalyzer/) to present a summary of 606 observed RBD mutations as shown in Fig. 2 . The interactive website allows one to choose different display options. Note that infectious mutation N501Y in Alpha, Beta, and Gamma variant has been observed 388,294 times worldwide. Mutations E484K, S477N, and L452R have been found over 25,000. Among them, E484K and L452R are vaccine-escape mutations.24 In particular, mutation L452R, which is in Delta, Epsilon, and Kappa variants, is as infectious as N501Y and as antibody disruptive as E484K. Mutation T478K in variant B.1.1.222 is the most infectious one among frequently observed mutations. Therefore, all significant variants have at least one infectious mutation.
Figure 2.
Analysis of observed S protein RBD mutations. Here, “BFE change” refers to the binding free energy change for the S protein and human ACE2 complex induced by a single-site S protein RBD mutation. A negative BFE change weakens the binding between S protein and ACE2, while a positive BFE change strengthens the binding between S protein and ACE2, giving rise to a more infectious variant.5 “Counts” of antibody disruption give the number of antibodies and S protein complexes disrupted by a specific mutation. We consider an antibody and S protein complex to be disrupted if its binding affinity is reduced by more than 0.3 kcal/mol.24 “Ratio” shows the ratio of disrupted antibody and S protein complexes out of 106 known complexes.24
Antibodies in clinical trials
In this work, we study 16 antibodies, including 5 antibodies in phase 3 clinical trials or EUA, and 2 antibodies in phase 1 clinical trials. The rest of the antibodies are closely related to those in clinical trials. For the 5 antibodies in phase 3 clinical trials or EUA, there are two antibody combination treatments, casirivimab/imdevimab (REGN10933/REGN10987), and bamlanivimab/etesevimab (LY-CoV555/LY-CoV016 (CB6)), and one single antibody treatment, regdanvimab (CT-P59) from Celltrion. C135 and C144 are two antibodies from the Rockefeller University in phase 1 clinical trials. The rest antibodies are C102, C105, C002, C104, C110, C119, C121, LY-CoV481, and LY-CoV488. Most of the antibodies are isolated or derived from COVID-19 human neutralizing antibodies,33, 32, 34, 35, 36 while REGN10933 and REGN10987 are derived from the treatments for Ebola – one from humanized mice and one from a convalescent patient.31 According to the literature,33, 31, 34 antibodies REGN10933, REGN10987, LY-CoV555, and LY-CoV016 were optimized through fluorescence-activated cell sorters.
In Fig. 3 , we align 16 three-dimensional (3D) antibody structures with ACE2. Fig. 3(a) and (b) show 7 antibodies that directly compete with ACE2 on the binding domain. Three clinical-trial antibodies, namely CT-P59, REGN10933, and LY-CoV016, can be found in Fig. 3(a). Fig. 3(c) shows 6 antibodies whose binding domains partially overlap with that of ACE2. Among them, LY-CoV555 and C144 are in clinical trials. Fig. 3(d) shows 3 antibodies that partially share their binding domains with ACE2. Antibodies REGN10987 and C135 do not compete with ACE2 directly and thus, they can be complements of other antibodies.
Impacts of SARS-CoV-2 on antibody efficacy and infectivity
SARS-CoV-2 variants with specific genetic markers are correlated to BFE changes on the RBD, degrade the neutralization by antibody treatments, or antibodies of the self immune system, and increase the difficulty of virus diagnostic or transmissible prediction. Especially, the mutations that enhance transmissibility and weaken antibody neutralization should be prioritized in the investigation. In Fig. 4 , we illustrate RBD mutations involved in the Alpha, Beta, Gamma, Delta, Epsilon, Kappa, and B.1.1.222 variants. In this figure, each RBD residue is colored by the maximum mutation-induced BFE change on the S protein-ACE2 complex from 19 possible mutations. One can notice that all the seven mutations in plethora variants have positive BFE changes that enhance the binding of S protein RBD and ACE2, and consequently, the infectivity of SARS-CoV-2.
Figure 4.
3D structure of human ACE2 (hACE2) and RBD (PDB 6M0J)27. Color on the RBD structure indicates the BFE changes induced by mutations, where blue means binding strengthening and red means weakening.
Fig. 5 illustrates SARS-CoV-2 S protein RBD mutation-induced BFE changes to the complexes of S protein with antibodies or ACE2. Here, we only consider 100 most observed mutations that have been observed with most times, and a similar study for all known RBD mutations is presented in the Supporting information. Note that there is a strong correlation between the positive predicted mutation-induced BFE changes and the observed mutation frequencies. For a given mutation, if its BFE changes for antibodies are very negative value while for ACE2 very positive, then this mutation has a combined antibody-escape and infectivity-strengthening effort. Therefore, one can observe that mutations, R346K/S, K417N, L452R, E484K/Q, F486L, F490L/S, S494P, and N501Y, have this effect, while R346K/S and N501Y induce a relatively moderate weakening effect to most antibodies.
Figure 5.
Illustration of the BFE changes of the complexes of S protein and antibodies or ACE2, induced by RBD mutations with frequencies being greater than 10. Positive changes strengthen the binding while negative changes weaken the binding. Here, only mutations that occurred on the relevant random coil of the S protein RBD are considered. The Grey color indicates that PDB structures do not involve specific residues.
Fig. 6 shows the BFE changes induced by seven RBD mutations (K417N, K417T, L452R, T478K, E484K, E484Q, and N501Y) for the S protein complexes with antibodies and ACE2. First of all, it is noted that all RBD mutations give positive BFE changes for binding to ACE2, leading to more infectious variants. Additionally, the magnitude of BFE changes on each mutation is correlated to the distance to antibodies. Therefore, antibodies having more overlap with ACE2 are impacted more significantly by mutations. For example, according to their 3D alignment in Fig. 3, LY-CoV016, CT-P59, REGN10933, C102, C105, LY-CoV481, and LY-CoV488 that are directly competing with ACE2 have large BFE changes in five mutations. Antibodies that partially overlap with ACE2 in terms of binding domain, i.e., C002, C104, C119, C121, C144, and LY-CoV555, have only a few significant BFE changes. Antibodies C110, C135, and REGN10987, which bind to the other side of the RBD, have very mild changes in all the mutations.
Figure 6.
BFE changes induced by new SARS-CoV-2 mutations, K417N, K417T, L452R, T478K, E484K, E484Q, and N501Y. C110∗ and C135∗: no results due to incomplete PDB structure.
More specifically, mutation T478K whose frequency has risen exponentially since early 2021 in Mexico (B.1.1.222), induces a very large positive BFE change in the ACE2-S protein complex. This could explain why T478K is a fast-growing mutation although it might not affect the binding of antibodies to the S protein. As for three variants from Alpha, Beta, and Gamma, they share the same mutation, N501Y, while the Alpha variant is the only one that contains one mutation on RBD and Beta and Gamma variants contain other mutations K417N/T and E484K. Meanwhile, the experimental results show that most antibodies demonstrate neutralizing capability against the Alpha variant.17, 18, 37 Interestingly, as reported by European Medicines Agency,38 regdanvimab (CT-P59) shows neutralizing ability against the Alpha variant. These results are highly consistent with the small positive BFE changes of N501Y on antibodies in Fig. 6. For the key substitution, L452R, of Epsilon, regdanvimab (CT-P59), and bamlanivimab (LY-CoV555) have large negative BFE changes. Here, we define large negative BFE changes as BFE change values are less than −0.5 kcal/mol. In the FDA report of bamlanivimab (LY-CoV555) and etesevimab (LY-CoV016),17, 18 the mutation L452R has a large fold reduction in susceptibility of single bamlanivimab and mild fold reduction of the combination of bamlanivimab and etesevimab. Lastly, we study the Beta and Gamma variants, which share the same mutations E484K and N501Y but are different in K417N/T. For antibodies in EUA, REGN10987 has mild changes on mutations, K417N/T and E484K, while REGN10933 and LY-CoV016 respond with large negative changes and LY-CoV555 has a significant negative change on E484K. Our predictions for the Beta and Gamma variants are in excellent agreement with experimental data.37, 39
Mutation impacts on antibodies in clinical trials
In this section, we study five antibodies in clinical trials or emergency use authorization. Two antibodies of Regeneron Pharmaceuticals, casirivimab and imdevimab, are studies together, followed by other three antibodies in phase 3, regdanvimab, bamlanivimab, and etesevimab. Two antibodies in phase 1 are discussed in the end as well. We emphasize 100 most observed RBD mutations and denoted them as high-frequency mutations. A complete study of all known RBD mutations is given in the Supporting information.
Regeneron Aatibodies REGN10933 and REGN10987 (aka Casirivimab and Imdevimab)
Regeneron’s Casirivimab and Imdevimab antibody cocktail against SARS-CoV-2 is the first combination therapy, which receives an FDA emergency use authorization. As the only one in the clinical trial antibodies that have the 3D structure of two antibodies binding to the RBD, we first study the BFE changes of them as an antibody combination. We examine the BFE changes induced by RDB mutations whose frequencies are greater than 10 in Fig. 7 of the antibody cocktail, REGN10933 and REGN10987, binding to the S protein RBD. The single antibody analysis is provided in the Supporting information. Notably, mutations K417T, N439K, G446V, E484K, and F486L lead to large negative BFE changes. For positive BFE changes, it is good to see that there are high-frequency mutations, which indicates that this antibody combination potentially prevents the new variants of SARS-CoV-2, especially for variants with mutations L452R, S477N, and K501Y. However, some mutations with negative BFE changes have a very large magnitude, indicating that the antibody combination of REGN10933 and REGN10987 was an immune product optimized for the original un-mutated S protein. In general, parts of the mutations on the S protein weaken the REGN10933+REGN10987 binding and make the antibodies less competitive to ACE2. This cocktail is prone to Beta and Gamma variants (K417N/T, E484K) but remains effective for Alpha and Epsilon variants (L452R and K501Y) (see Fig. 8 ).
Figure 7.
Illustration of SARS-CoV-2 mutation-induced binding free energy changes for the complexes of S protein and antibodies REGN10933 and REGN10987 (PDB 6XDG).31 Here, mutations K417T, N439K, G446V, E484K, and F486L could potentially disrupt the binding of antibodies and S protein RBD.
Figure 8.
The binding complex of S protein RBD and REGN10933+REGN10987 (PDB 6XDG).31
Additionally, these two antibodies can be studied separately, as shown in Figure 9, Figure 10 . By comparing the stand-alone BFE predictions to those in Fig. 7, it can be concluded that antibody REGN10933 plays the main role in the antibody neutralization, while the antibody REGN10987 is a complement for two reasons. First, the antibody REGN10933 shares the same disrupted mutations with the combination and has larger BFE changes on those mutations. Secondly, the BFE changes for REGN10987 are mild, and most of them are positive values. According to the 3D alignment, the antibody REGN10987 does not directly compete with ACE2. Lastly, in the comparison, one can notice that the magnitude of BFE changes is smaller on the mutations for the combination. This indicates a more stable binding of the antibody combination.
Figure 9.
Illustration of SARS-CoV-2 mutation-induced binding free energy changes for the complexes of S protein and antibody REGN10933 (PDB 6XDG)31. Here, mutations K417T, N439K, G446V, E484K/Q, and F486L could potentially disrupt the binding of antibody and S protein RBD.
Figure 10.
Illustration of SARS-CoV-2 mutation-induced binding free energy changes for the complexes of S protein and antibody REGN10987 (PDB 6XDG).
Eli Lilly antibodies LY-CoV555 and LY-CoV016 (aka Bamlanivimab and Etesevimab)
Bamlanivimab (LY-CoV555) was first developed as a single antibody therapy for the treatment of mild to moderate COVID-19 illness. However, it is not distributed alone due to the SARS-CoV-2 variant resistance and is used as an antibody combination with Etesevimab (LY-CoV016 (CB6)). Here, we first examine Bamlanivimab’s response to S protein RBD mutations followed by the discussion of Etesevimab.
In the BFE changes prediction of LY-CoV555 (PDB 7KMG) as shown in Fig. 12 , most mutations have mild changes, while mutations L452R, V483F/A, E484K/Q, F486L, F490L/S, and S494P have large negative BFE changes. For positive BFE changes, the largest value is only 0.75 kcal/mol and the average of positive BFE changes is 0.16 kcal/mol. However, many mutations with negative BFE changes have very large magnitudes, such that 7 mutations having binding free energy less than −2 kcal/mol, and the least value is −4.1 kcal/mol for E484K. This could indicate that antibody LY-CoV555 was an immune product optimized with respect to the original un-mutated S protein. In general, the mutations on S protein weaken the LY-CoV555 binding to S protein and make it less competitive with ACE2 as most mutations strengthen the S protein and ACE2 binding. The Beta variant (E484K) and Epsilon variant (L452R) have a strong antibody-escape effect.
Figure 12.
Illustration of SARS-CoV-2 RBD mutation-induced binding free energy changes for the complexes of S protein and antibody LY-CoV555 (PDB 7KMG). Here, mutations L452R, V483F/A, E484K/Q, F486L, F490L/S, Q493K/R, and S494P could potentially disrupt the binding of antibodies and S protein RBD.
In Fig. 13 , we illustrate the mutation-induced BFE changes for antibody LY-CoV016 (PDB 7C01), which directly competes with ACE2. One can notice that K417T/N, A475S, and N501Y, have large negative BFE changes, and three of them belong to SARS-CoV-2 variants. The rest mutations have a small magnitude of changes. There are no large positive BFE changes. Antibody LY-CoV016 is isolated from peripheral blood mononuclear cells of patients convalescing from COVID-19 at the early stage and optimized based on an early version of the SARS-CoV-2 virus.
Figure 13.
Illustration of SARS-CoV-2 mutation-induced binding free energy changes for the complexes of S protein and antibody LY-CoV016 (PDB 7C01). Here, mutations K417T/N, A475S, and N501Y could potentially disrupt the binding of antibody and S protein RBD.
In the 3D structure superposition of Fig. 11 right chart, antibodies LY-CoV555 and LY-CoV016 share a partial binding domain with ACE2. Therefore, they are not only competing with ACE2 but also with each other. Comparing the BFE change prediction on both LY-CoV555 and LY-CoV016, one can note that two antibodies respond to S protein RBD mutations differently and thus are complementary. We deduce that the combined antibody will enhance the single antibody neutralization.
Figure 11.
The binding complexes of S protein RBD with Left: LY-CoV555 (PDB 7KMG)34 and Middle: LY-CoV016 (7C01)33. Right: a crash at the interface between two antibodies.
Celltrion antibody CT-P59
Regdanvimab (CT-P59) has been approved for emergency use treatment in South Korea and is under review by European Medicines Agency (EMA) (see Fig. 14). We present the BFE changes in Fig. 15 . Antibody CT-P59 shares a similar binding domain with ACE2 and is a potent candidate for the direct neutralization of SARS-CoV-2. Most mutations induce small changes in the binding free energy, while mutations Y449H, L452R, L455F, E484K, F490L/S, Q493K/R, and S494P induce large negative BFE changes. This indicates antibody CT-P59 has an antibody-escape effect for many variants, including the Beta variant (B.1.351 with E484K) and the Epsilon variant (with L452R). It is noticed that CT-P59 has a large positive BFE change for mutation N501Y, indicating CT-P59 can neutralize the SARS-CoV-2 Alpha variant (B.1.17) .
Figure 14.
The binding complex of S protein and CT-P59 (PDB 7CM4)32.
Figure 15.
Illustration of SARS-CoV-2 mutation-induced binding free energy changes for the complexes of S protein and antibody CT-P59 (PDB 7CM4).32 Here, mutations Y449H, L452R, L455F, E484K, F490L/S, Q493K/R, and S494P could potentially disrupt the binding of antibodies and S protein RBD.
Rockefeller University antibodies C135 and C144
Lastly, we study C135 and C144, another antibody combination treatment currently on phase 1 study. Due to fact that there is no 3D structure of C135 and C144 binding to RBD simultaneously, we present their BFE change predictions based on PDB 7K8Z and 7K90, separately (see Fig. 16).
Figure 16.
The binding complexes of S protein and antibodies C135 (PDB 7K8Z)36 and C144 (PDB 7K90)36.
In the BFE change calculation of antibody C135 on Fig. 17, most mutations have mild BFE changes, while two mutations, R346K/S, induce large negative BFE changes, and three mutations, N440K, N450K, and P499H, lead to positive BFE changes greater than 0.5 kcal/mol. Notably, C135 is not an antibody that directly competes with ACE2 in terms of the binding domain. For mutations of emergent variants, K417T/N, L452N, and N501Y, they all have small BFE changes. With mild changes of most mutations, the antibody C135 could be a complement for other antibodies that are directly competing with ACE2 on the binding domain.
Figure 17.
Illustration of SARS-CoV-2 mutation-induced binding free energy changes for the complexes of S protein and antibody C135 (PDB 7K8Z).36 Here, mutations R346K/S could potentially disrupt the binding of antibodies and S protein RBD.
The last antibody is C144 on Fig. 18, which shares a part of the binding domain with ACE2. It is obvious that except for mutations E484K/Q, the rest mutations induce mild BFE changes. As the mutation E484K is part of the Gamma and Beta variants, this antibody treatment could have antibody-escape effects. However, since most mutations lead to mild BFE changes and mutations K417N/T, L452R, T478K, and N501Y render mild positive BFE changes, this antibody can have neutralizing efficacy for many emerging variants, such as the Alpha, Epsilon, and B.1.1.222 variants.
Figure 18.
Illustration of SARS-CoV-2 mutation-induced binding free energy changes for the complexes of S protein and antibody C144 (PDB 7K90). Here, mutations E484K/Q could potentially disrupt the binding of antibodies and S protein RBD.
Discussion and Conclusion
There are emerging variants spreading worldwide, which increase the virus transmissibility, reduce the neutralization of antibodies, and degrade the efficacy of antibody treatments or vaccines. The S protein plays the most important role in leading the virus to access the host cell. The RBD of S protein directly contacts ACE2, and its substitutions induced by variants can significantly weaken its binding with original antibodies either created from current vaccines or induced through existing antibody therapies. RBD mutations that enhance the RBD binding to ACE2 and weaken the RBD binding to many antibodies pose potential threats to vaccines and antibody therapies. Figure S1 in the Supporting information provides a detailed analysis of the 606 RBD mutations to 16 antibodies.
Alpha B.1.1.7 lineage The Alpha variant has one mutation, N501Y, on the RBD and was detected from the COVID-19 pandemic in the United Kingdom 40, which increases viral transmission 16 and severity based on hospitalizations.41 However, for antibodies in clinical trials, it has a minor impact on neutralization in terms of BFE changes based on our predictions. Similar findings for the B.1.1.7 lineage have been reported for experimental neutralization by EUA therapeutics 42, 17, 18 and for other antibodies.10
Beta B.1.351 lineage The Beta variant is different from the Gamma variant only for one RBD mutation, i.e., K417N and is first reported in South Africa 40. We can claim a similar statement but moderate impacts on all the clinical trial antibodies. The same pattern can be found in the CDC report 17, 18 and the literature.10
Gamma P.1 lineage The Gamma variant has three RBD mutations K417T, E484K, and N501Y, which was reported by the National Institute of Infectious Diseases, Japan, in people who had travel experience of Brazil 14. According to our BFE predictions, casirivimab (REGN10933) is moderately influenced by K417N and E484K on neutralization, while for imdevimab (RENG10987), the mutation impact is less significant. Regdanvimab (CT-P59) could still maintain its neutralizing capability. Bamlanivimab (LY-CoV555) shows a large fold reduction in susceptibility on mutation E484K in our prediction, which is consistent with a CDC report,18 while the combination of bamlanivimab and etesevimab gives a better response to P.1 lineage.17 We hypothesize that LY-CoV016 competes with LY-CoV555 and preserve its neutralization capacity with E484K. The combination of bamlanivimab and etesevimab has a large BFE reduction from P.1 lineage, which indicates that K417T has a negative impact on the binding.
Delta B.1.617.2 lineage This variant has two RBD mutations L452R and T478K detected in India.40 These two mutations have two different types of impacts on the neutralization for antibody therapies. While L452R will have a native impact on the neutralization for regdanvimab (CT-P59) and bamlanivimab (LY-CoV555) and a mild impact on others, the mutation T478K has a positive impact on almost all the known antibodies but strongly strengthens the binding of S protein and ACE2.
Epsilon B.1.427/429 lineage For the Epsilon variant, mutation L452R has a negative impact on the neutralization for regdanvimab, but a minimal impact on the neutralization by the two antibody combinations. L452R reduces the capacity of bamlanivimab, which can be shown by the prediction and the CDC report.18 Interestingly, the fact of a small impact on the antibody combination, bamlanivimab and etesevimab, shown by the prediction and report 17 indicates that etesevimab dominants the binding process. The Epsilon variant was first detected in California, US.40
Iota B.1.526 lineage The Iota variant is studied by only considering E484K and had spread rapidly in New York, US40. It reduces the neutralization of REGN10933, C144, and LY-CoV555. Based on our predictions, the impact on REGN10933 can be reduced if REGN10987 is also used in the treatment as well.
Kappa B.1.617.1 lineage The Kappa variant has two RBD mutations L452R and E484Q, and was first detected in India.40 It has mutation L452R shared with Delta and Epsilon variants. The Kappa variant has a mutation E484Q, which is different from the rest. According to the BFE prediction, mutation E484Q has a negative impact on antibodies REGN10933, C144, and LY-CoV555. Similar to E484K, the Regeneron antibody combination can reduce Kappa variant’s negative impact on REGN10933.
B.1.1.222 lineage The B.1.1.222 variant involves RBD mutation T478K and has a larger positive BFE change on the binding of ACE2 and RBD. However, it has minor effects on existing antibodies.
Fig. 19 illustrates two comparisons of experimental data and our BFE change predictions. The left chart is the comparison of natural log of experimental escape fraction 28 and our BFE change predictions. It is seen that BFE change predictions have a high correlation, i.e., , to the natural log of escape fraction. Especially, for variants significantly escaping antibodies (with escape fraction close to 1), the BFE predictions have large negative changes. The second comparison on the right chart is about virus infectivity changes induced by mutations. The experimental pseudovirus infection changes induced by N501Y or L452R in reference to D614G were reported in relative luciferase units.19 These results are compared with our predicted BFE changes of the RBD and ACE2 complex for N501Y or L452R. It is seen that two results match extremely well, suggesting that SARS-CoV-2 infectivity is mainly determined by the RBD and ACE2 binding.
Figure 19.
(a) Comparison of the natural log of experimental escape fraction 28 and predicted BFE changes for various RBD mutations associated with major antibody therapeutic candidates. The escape fraction is from 0 to 1 and is the descriptor of a given mutation that escapes antibody binding with the S protein. The natural logarithm is taken according to the equation of the enrichment ratio used in deep mutational scanning raw data. The Pearson correlation of natural logs of escape fractions and BFE changes is 0.80. (b) Comparison of relative luciferase units 19 for pseudovirus infection changes and predicted BFE changes of ACE2 and S protein complex induced by mutations L452R and N501Y.
Fig. 20 gives an overall comparison of experimental and predicted patterns of variant impacts on major antibody drug candidates. There is an excellent agreement between our predictions and various experimental data, except for a minor discrepancy. Specifically, our prediction shows a potentially twofold reduction in binding strength for LY-CoV016 from B.1.1.7 due to N501Y (see Fig. 6), while the experiment records little change 10 , which is the only difference between a large number of experimental reports and our predictions.
Figure 20.
Comparison of experimental (Exp.) pattern and predicted (Pred.) pattern of the impact of SARS-CoV-2 variants on major antibody therapeutic candidates. Light green indicates mild or no change in neutralization; pink indicates significant reduction in neutralization; grey indicates no available data. RBD mutations in various variants: B.1.526: E484K; B.1.1.7: N501Y; B.1.427: L452R; P.1: K417T+E484K+N501Y; B.1.351: K417N+E484K+N501Y. The BFE changes are accumulated for multi-mutation predictions. Data resource: REGN10933,10, 39, 43 REGN10987,10, 39, 43 REGN cocktail,10 LY-CoV016,10,?,? C135,44, 10 C144,44 and LY-CoV555.18, 10, 43
Conclusion In summary, the Eli Lilly antibody therapies bamlanivimab and etesevimab are likely compromised by known emerging variants and other high-frequency mutations V483F/A, E484Q/V/A/G/D, F486L, F490L/V/S, Q493L, and S494P. For Regeneron antibody therapies casirivimab and imdevimab, while there is no experimental data regarding K417T from variant P.1, our predictions indicate that there is a potential compromise from mutation K417T. Additionally, Regeneron antibodies are prone to high-frequency mutations N439K, G446V, E484G, and F486L. The Celltrion antibody therapy regdanvimab (i.e., CT-P59) is predicted to be compromised by variants P.1, B1.351, B.1.427, and B.1.526, although there is no experimental data now. It can also be weakened by high-frequency mutations L455F, E484A, F490L/S, and S494P/L. Rockefeller University antibody C135 can be evaded by high-frequency mutations R346K/S. The antibody C144 from Rockefeller University is prone to variants P.1, B1.351, and B.1.526, while the experiment has only confirmed the adversarial impact of variant B.1.526. Additionally, it can be compromised by high-frequency mutations E484Q/A. In the Supporting information, we further identify that low-frequency RBD mutations V401I/L, I402V, E406G, Q409L, I410V, D420A/G, N422S, N448D, N450D, Y453F, F456L, Y473F, E484Q/A/G/D, G485S/R/C/V, F486L/V/C, F490I/L/V/Y/S, S393A/L, N501I, and Y508S have potential to become future vaccine or antibody escape variants. These mutations are predicted to enhance the RBD binding to ACE2 while weaken the binding between RBD and most antibodies.
Methods
Genome sequence data and pre-processing
Complete SARS-CoV-2 genome sequences are available from the GISAID database.40 In this work, a total of 796,759 complete SARS-CoV-2 genome sequences with high coverage and exact collection date are downloaded from the GISAID database 40 (https://www.gisaid.org/) as of May 24, 2021. We take the first complete SARS-CoV-2 genome from the GenBank (NC_045512.2) as the reference genome,45 and the multiple sequence alignment is applied by the Clustal Omega 46, 47 with default parameters, which results in 27,960 single nucleotide polymorphism profiles. On the S protein RBD, i.e., residues 329 to 530, a total of 606 non-degenerate mutations are found. Among them, 100 mutations have been observed more than 40 times.
Machine learning datasets
Datasets are important to train accurate machine learning models. Both the BFE changes and enrichment ratios describe the effects on the binding affinity of protein-protein interactions. Therefore, integrating both kinds of datasets can improve the prediction accuracy. Especially, due to the urgency of COVID-19, the BFE changes of SARS-CoV-2 data are rarely reported, while the enrichment ratio data via high-throughput deep mutations are relatively easy to obtain. The most important dataset that provides the information for binding free energy changes upon mutations is the SKEMPI 2.0 dataset.48 The SKEMPI 2.0 is an updated version of the SKEMPI database, which contains new mutations and data from other three databases: AB-Bind,49 PROXiMATE,50 and dbMPIKT.51 There are 7,085 elements, including single- and multi-point mutations in SKEMPI 2.0. 4,169 variants in 319 different protein complexes are filtered as single-point mutations are used for our TopNetTree model training. Moreover, SARS-CoV-2 related datasets are also included to improve the prediction accuracy after a label transformation. They are all deep mutation enrichment ratio data, mutational scanning data of ACE2 binding to the receptor-binding domain (RBD) of the S protein,25 mutational scanning data of RBD binding to ACE2,26, 27 and mutational scanning data of RBD binding to CTC-445.2 and of CTC-445.2 binding to the RBD.27 Note that our training datasets used in the validation do not include the test dataset, which is a mutational scanning data of RBD binding to ACE2.
Feature generation of PPIs
Algebraic topology 52, 53 has had tremendous success in describing biochemical and biophysical properties.54 Element-specific and site-specific persistent homology can effectively simplify the structural complexity of protein-protein complex and extract the abstract properties of the vital biological information in PPIs.21, 6 The algebraic topological analysis on PPIs is constructed based on a series of atom subsets of complex structures, which are atoms of the mutation sites, , atoms in the neighborhood of the mutation site within a cut-off distance , antibody atoms within r of the binding site, , antigen atoms within r of the binding site, , and atoms in the system that has atoms of element type of {C, N, O}, . Additionally, a bipartition graph is introduced to describe the antibody and antigen in PPIs. Then, molecular atoms construct point clouds for simplicial complex, which is a finite collection of sets of linear combinations of points. We apply the Vietoris-Rips (VR) complex for dimension 0 topology, and alpha complex for point cloud of dimensions 1 and 2 topology.54 Overall, element-specific and site-specific persistent homology is devised to capture the multiscale topological information over different scales along a filtration 52 and is important for our machine learning predictions.
Simplex and simplicial complex
Given a set of independent points in , the convex combination is a point , where and . The convex hull of U is the collection of convex combinations of U, and a k-simplex is the convex hull of independent points U. For example, a 0-simplex is a point, a 1-simplex is an edge, a 2-simplex is a triangle, and a 3-simplex is a tetrahedron. A proper m-face of the k-simplex is a subset of the vertices of a k-simplex with vertices forms a convex hull in a lower dimension and . The boundary of a k-simplex is defined as a sum of all its –faces as
| (1) |
where is a convex hull formed by vertices of excluding . A simplicial complex denotes by K is a collection of finitely many simplices forms a simplicial complex. Thus, faces of any simplex in K are also simplices in K, and intersections of any 2 simplices are only faces of both or an empty set. A k-simplex is in Vietoris–Rips complex if and only if for and is in alpha complex if and only if .
Homology
For a simplicial complex K, a k-chain of K is a formal sum of the k-simplices in K defined as , where is the k-simplices and is coefficients. can be in different fields such as , and . Typically, is chosen to be , which is and forms an Abelian group . Then, the boundary operator can be extended to a k-chain as
| (2) |
such that and satisfies , follows from that boundaries are boundaryless. The chain complex is defined as a sequence of complexes by boundary maps is called a chain complex
| (3) |
The k-homology group is the quotient group defined by taking k-cycle group module of k-boundary group as
| (4) |
where is the k-homology group, and k-cycle group and the k-boundary group are the subgroups of defined as,
| (5) |
The Betti numbers are defined by the ranks of kth homology group as . reflects the number of connected components, reflects the number of loops, and reflects the number of cavities.
Filtration and persistent homology
A filtration of a topology space K is a nested sequence of K such that
| (6) |
Then, a sequence of chain complexes and a homology sequence are constructed on the filtration. The pth persistent of kth homology group of are defined as
| (7) |
and the Betti numbers . These persistent Betti numbers are applied to represent topological fingerprints.
Auxiliary features
Features of topological invariants are not enough to reflect the whole picture of PPIs. Importantly, chemical and physical information, including surface areas, partial charges, Coulomb interactions, van der Waals interaction, electrostatic solvation free energy, mutation site neighborhood amino acid composition, pKa shifts, and secondary structure, is added as auxiliary features to improve the predictive power of the machine learning model.5
Machine learning and deep learning algorithms
We illustrate the construction of a topology-based network (TopNet) model for the BFE change prediction of protein-protein interactions (PPIs) on SARS-CoV-2 studying. These approaches have been widely applied in studying protein-ligand and protein-protein binding free energy predictions.6, 5 Firstly, one ensemble method, gradient boosting decision tree (GBDT), is studied as baseline in comparison to deep neural network methods. The ensemble methods naturally handle correlation between descriptors and are robust to redundant features. Therefore, they usually do not depend on a sophisticated feature selection procedure and a complicated grid search of hyper-parameters. The implemented GBDT is a function from the scikit-learn package (version 0.22.2.post1).55 The number of estimators and the learning is optimized for ensemble methods as 20000 and 0.01, respectively. For each set, 10 runs (with different random seeds) were done and the average result is reported in this work. Considering a large number of features, the maximum number of features to consider is set to the square root of the given descriptor length for GBDT methods to accelerate the training process. The parameter setting shows that the performance of the average of sufficient runs is decent.
A neural network is a network of neurons that maps an input feature layer to an output layer. The neural network simulates a biological brain that solves problems with numerous neuron units by backpropagation to update weights on each layer. To reveal the facts of input features at different levels and abstract more properties, one can construct more layers and more neurons in each layer, which is known as a deep neural network. Optimization methods for feedforward neural networks and dropout methods are applied to prevent overfitting. In 10-fold cross-validations, the neural network model has a slightly better performance than the GBDT model, where Pearson correlations for these algorithms are 0.864 and 0.838 and root mean square errors are 1.019 kcal/mol and 1.063 kcal/mol, respectively. Thus, we applied the deep neural network for predictions, validation, and comparison.
Optimization
To train feedforward neural networks, backpropagation is applied where the loss function is evaluated at the output layer and is propagated backward through the network to update the model’s weights and bias. As the calculation of gradient required, one popular approach is the stochastic gradient descent (SGD) method with momentum, which estimates a small portion of training data and applies the idea of exponentially weighted averages. Thus, the momentum term can accelerate the convergence of the algorithm. A popular way to implement the SGD with momentum is given as
| (8) |
where is the parameters in the network, is the objective function, is the learning rate, and are the input and target of the training set, and is a scalar coefficient for the momentum term.
Dropout
Fully connected layers possess a large number of degrees of freedom. This can easily cause an overfitting issue, while the dropout technique is an easy way of preventing network overfitting.56 In the training process, hidden units are randomly set zero values to their connected neurons in the next layer. Suppose that a percentage of neurons at a certain layer is chosen to be dropped during training. The number of computed neurons of this layer is equal to the neuron number multiplied by a coefficient such as 1-p, where p is the dropout rate. Then, in the testing process, the output of these layers is computed by randomly dropouts the same rate of neurons, to approximate the network in each training step.
Validation
Deep learning algorithms
A deep neural network (DNN) consists of multi-layers of neurons. In the output layer, the single neuron gets full connections with the last hidden layer and calculates predictions. Notice that the network is constructed for mutation-induced BFE changes, one should preserve the consistency of all labels. An optimizer is used to minimize the following loss function:
| (9) |
where N is the number of samples, f is a function of the feature vector parametrized by a weight vector W and bias term b, and represents a penalty constant.
Here, we present a validation of our BFE change prediction for mutations on S protein RBD compared to the experimental deep mutational enrichment data.27 Fig. 21 presents a comparison between experimental deep mutational enrichment data and BFE change predictions on SARS-CoV-2 RBD binding to ACE2. In the heatmap of Fig. 21, both BFE changes and enrichment ratios describe the affinity changes of the S protein RBD-ACE2 complex induced by mutations. It is obvious that the predicted BFE changes are highly correlated to the enrichment ratio data. Pearson correlation is 0.70. It should be noticed that the deep mutational scanning data from different labs might vary dramatically due to different experimental conditions. For example, the RBD deep mutational scanning data of the SARS-CoV-2 RBD binding to ACE2 reported by two teams 26, 27 have a relatively small Pearson correlation of 0.666.
Figure 21.
A comparison between experimental RBD deep mutation enrichment data and predicted BFE changes for SARS-CoV-2 RBD binding to ACE2 (6M0J).27Top left: deep mutational scanning heatmap showing the average effect on the enrichment for single-site mutants of RBD when assayed by yeast display for binding to the S protein RBD.27Right: RBD colored by average enrichment at each residue position bound to the S protein RBD. Bottom left: machine learning predicted BFE changes for single-site mutants of the S protein RBD.
Data and Model Availability
The SARS-CoV-2 single nucleotide polymorphism data in the world is available at Mutation Tracker. The analysis of RBD mutations is available at Mutation Analyzer. The machine learning training datasets and the trained machine learning model are available at TopNetmAb. The related training process is described in Supporting information.
CRediT authorship contribution statement
Jiahui Chen: Methodology, Software, Visualization, Writing - original draft. Kaifu Gao: Data curation, Writing - review & editing. Rui Wang: Data curation, visualization, Writing - original draft. Guo-Wei Wei: Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported in part by NIH grant GM126189, NSF grants DMS-2052983, DMS-1761320, and IIS-1900473, NASA grant 80NSSC21M0023, Michigan Economic Development Corporation, MSU Foundation, Bristol-Myers Squibb 65109, and Pfizer. The authors thank The IBM TJ Watson Research Center, The COVID-19 High Performance Computing Consortium, NVIDIA, and MSU HPCC for computational assistance. RW thanks Dr. Changchuan Yin for useful discussion.
Edited by Michael Sternberg
Footnotes
GISAID found in early June of 2021 that 14 SARS-CoV-2 records submitted by three labs (Israel Institute for Biological Research, the Institute of Virology at Hannover Medical School, and Laboratoire de Biotechnologie) were wrong, which leads to a significant number of artificial low-frequency single nucleotide polymorphisms (SNPs) in worldwide research publications.
The supporting information is available for S1 BFE changes for the complexes of S protein RBD binding to antibodies or ACE2 induced by 606 RBD mutations and S2 Machine learning models. Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jmb.2021.167155.
Supplementary Data
The following are the Supplementary data to this article:
References
- 1.Hoffmann Markus, Kleine-Weber Hannah, Schroeder Simon, Krüger Nadine, Herrler Tanja, Erichsen Sandra, Schiergens Tobias S., Herrler Georg, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiu-Xia Qu., Hao Pei, Song Xi-Jun, Jiang Si-Ming, Liu Yan-Xia, Wang Pei-Gang, Rao Xi, Song Huai-Dong, et al. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280(33):29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Rui, Chen Jiahui, Gao Kaifu, Hozumi Yuta, Yin Changchuan, Wei Guo-Wei. Analysis of sars-cov-2 mutations in the united states suggests presence of four substrains and novel variants. Commun. Biol. 2021;4(1):1–14. doi: 10.1038/s42003-021-01754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Wendong, Shi Zhengli, Yu Meng, Ren Wuze, Smith Craig, Epstein Jonathan H., Wang Hanzhong, Crameri Gary, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310(5748) doi: 10.1126/science.1118391. 676–679. [DOI] [PubMed] [Google Scholar]
- 5.Chen Jiahui, Gao Kaifu, Wang Rui, Wei Guowei. Prediction and mitigation of mutation threats to covid-19 vaccines and antibody therapies. Chem. Sci. 2021;12 (20):6929–6948. doi: 10.1039/d1sc01203g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Jiahui, Wang Rui, Wang Menglun, Wei Guo-Wei. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020;432:5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Jiahui, Gao Kaifu, Wang Rui, Nguyen Duc Duy, Wei Guo-Wei. Review of covid-19 antibody therapies. Ann. Rev. Biophys. 2020;50 doi: 10.1146/annurev-biophys-062920-063711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Peter, Nirula Ajay, Heller Barry, Gottlieb Robert L., Boscia Joseph, Morris Jason, Huhn Gregory, Cardona Jose, et al. SARS-COV-2 neutralizing antibody LY-COV555 in outpatients with covid-19. New Engl. J. Med. 2021;384(3):229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amanat Fatima, Krammer Florian. SARS-COV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Pengfei, Nair Manoj S., Liu Lihong, Iketani Sho, Luo Yang, Guo Yicheng, Wang Maple, Yu Jian, et al. Antibody resistance of sars-cov-2 variants b. 1.351 and b. 1.1. 7. Nature. 2021;10 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 11.Ferron François, Subissi Lorenzo, Morais Ana Theresa Silveira De, Le Nhung Thi Tuyet, Sevajol Marion, Gluais Laure, Decroly Etienne, Vonrhein Clemens, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. 2018;115(2):E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sevajol Marion, Subissi Lorenzo, Decroly Etienne, Canard Bruno, Imbert Isabelle. Insights into RNA synthesis, capping, and proofreading mechanisms of SARS-coronavirus. Virus Res. 2014;194:90–99. doi: 10.1016/j.virusres.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mwenda Mulenga, Saasa Ngonda, Sinyange Nyambe, Busby George, Chipimo Peter J., Hendry Jason, Kapona Otridah, Yingst Samuel, et al. Detection of b. 1.351 SARS-CoV-2 variant strain–zambia, december 2020. MMWR Morb. Mortal Wkly Rep. 2021. 2020;70:280–282. doi: 10.15585/mmwr.mm7008e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faria Nuno R., Claro Ingra Morales, Candido Darlan, Franco LA Moyses, Andrade Pamela S., Coletti Thais M., Silva Camila A.M., Sales Flavia C., et al. Genomic characterisation of an emergent sars-cov-2 lineage in manaus: preliminary findings. Virological. 2021 [Google Scholar]
- 15.Emergency Use Authorization (EUA) for bamlanivimab 700mg IV Center for Drug Evaluation and Research (CDER) Memorandum on Fact Sheet Update (2021).
- 16.Davies Nicholas G., Abbott Sam, Barnard Rosanna C., Jarvis Christopher I., Kucharski Adam J., Munday James, Pearson Carl A.B., Russell Timothy W., et al. Estimated transmissibility and severity of novel sars-cov-2 variant of concern 20*20*12/01 in England. MedRxiv. 2021 2020–12. [Google Scholar]
- 17.Fact Sheet For Health Care Providers Emergency Use Authorization (Eua) Of Bamlanivimab And Etesevimab 02092021 (fda.gov).
- 18.Fact Sheet For Health Care Providers Emergency Use Authorization (EUA) Of REGEN-COV (fda.gov).
- 19.Deng Xianding, Garcia-Knight Miguel A, Khalid Mir M., Servellita Venice, Wang Candace, Morris Mary Kate, Sotomayor-Gonzalez Alicia, Glasner Dustin R., et al. Transmission, infectivity, and antibody neutralization of an emerging sars-cov-2 variant in california carrying a l452r spike protein mutation. medRxiv. 2021 [Google Scholar]
- 20.Emergency Use Authorization (EUA) for bamlanivimab 700 mg and etesevimab 1400 mg IV Center for Drug Evaluation and Research (CDER) Memorandum on Fact Sheet Update (2021).
- 21.Wang Menglun, Cang Zixuan, Wei Guo-Wei. A topology-based network tree for the prediction of protein–protein binding affinity changes following mutation. Nature Mach. Intell. 2020;2(2):116–123. doi: 10.1038/s42256-020-0149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Ning, Chen Yuting, Lu Haoyu, Zhao Feiyang, Alvarez Roberto Vera, Goncearenco Alexander, Panchenko Anna R., Li Minghui. Mutabind2: predicting the impacts of single and multiple mutations on protein-protein interactions. Iscience. 2020;23(3):100939. doi: 10.1016/j.isci.2020.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodrigues Carlos H.M., Myung Yoochan, Pires Douglas E.V., Ascher David B. MCSM-PPI2: predicting the effects of mutations on protein–protein interactions. Nucl. Acids Res. 2019;47(W1):W338–W344. doi: 10.1093/nar/gkz383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Rui, Chen Jiahui, Gao Kaifu, Wei Guo-Wei. Vaccine-escape and fast-growing mutations in the united kingdom, the united states, singapore, spain, india, and other covid-19-devastated countries. Genomics. 2021;113 (4):2158–2170. doi: 10.1016/j.ygeno.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan Kui K., Dorosky Danielle, Sharma Preeti, Abbasi Shawn A., Dye John M., Kranz David M., Herbert Andrew S., Procko Erik. Engineering human ace2 to optimize binding to the spike protein of sars coronavirus 2. Science. 2020;369(6508):1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starr Tyler N., Greaney Allison J., Hilton Sarah K., Ellis Daniel, Crawford Katharine H.D., Dingens Adam S., Navarro Mary Jane, Bowen John E., et al. Deep mutational scanning of SARS-COV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linsky Thomas W., Vergara Renan, Codina Nuria, Nelson Jorgen W., Walker Matthew J., Su Wen, Barnes Christopher O., Hsiang Tien-Ying, et al. De novo design of potent and resilient hACE2 decoys to neutralize SARS-COV-2. Science. 2020;370(6521):1208–1214. doi: 10.1126/science.abe0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starr Tyler N., Greaney Allison J., Addetia Amin, Hannon William W., Choudhary Manish C., Dingens Adam S., Li Jonathan Z., Bloom Jesse D. Prospective mapping of viral mutations that escape antibodies used to treat covid-19. Science. 2021;371(6531):850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greaney Allison J., Starr Tyler N., Gilchuk Pavlo, Zost Seth J., Binshtein Elad, Loes Andrea N., Hilton Sarah K., Huddleston John, et al. Complete mapping of mutations to the SARS-COV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giacomo Simone Di, Mercatelli Daniele, Rakhimov Amir, Giorgi Federico M. Preliminary report on SARS-COV-2 spike mutation t478k. bioRxiv. 2021 doi: 10.1002/jmv.27062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen Johanna, Baum Alina, Pascal Kristen E., Russo Vincenzo, Giordano Stephanie, Wloga Elzbieta, Fulton Benjamin O., Yan Ying, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369(6506):1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim Cheolmin, Ryu Dong-Kyun, Lee Jihun, Kim Young-Il, Seo Ji-Min, Kim Yeon-Gil, Jeong Jae-Hee, Kim Minsoo, et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-COV-2 spike protein. Nature Commun. 2021;12(1):1–10. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Rui, Shan Chao, Duan Xiaomin, Chen Zhihai, Liu Peipei, Song Jinwen, Song Tao, Bi Xiaoshan, et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020:1–8. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 34.Jones Bryan E., Brown-Augsburger Patricia L., Corbett Kizzmekia S., Westerndorf Kathryn, Davies Julian, Cujec Thomas P., Wiethoff Christopher M., Blackbourne Jamie L., et al. LY-COV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-COV-2 infection. BioRxiv. 2020 [Google Scholar]
- 35.Barnes Christopher O., Jette Claudia A., Abernathy Morgan E., Dam Kim-Marie A., Esswein Shannon R., Gristick Harry B., Malyutin Andrey G., Sharaf Naima G., et al. SARS-COV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020:1–6. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnes Christopher O., West Anthony P., Huey-Tubman Kathryn, Hoffmann Magnus A.G., Sharaf Naima G., Hoffman Pauline R., Koranda Nicholas, Gristick Harry B., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. bioRxiv. 2020 doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Pengfei, Liu Lihong, Iketani Sho, Luo Yang, Guo Yicheng, Wang Maple, Yu Jian, Zhang Baoshan, et al. Increased resistance of SARS-COV-2 variants b. 1.351 and b. 1.1. 7 to antibody neutralization. BioRxiv. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 38.EMA review of regdanvimab for COVID-19 to support national decisions on early use (ema.europa.eu).
- 39.Wang Pengfei, Wang Maple, Yu Jian, Cerutti Gabriele, Nair Manoj S., Huang Yaoxing, Kwong Peter D., Shapiro Lawrence, et al. Increased resistance of SARS-COV-2 variant p. 1 to antibody neutralization. bioRxiv. 2021 doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shu Yuelong, McCauley John. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.NERVTAG note on B.1.1.7 severity for SAGE.
- 42.Emary, Katherine R. W., Golubchik, Tanya, Aley, Parvinder K., Ariani, Cristina V., Angus, Brian John, Bibi, Sagida, Blane, Beth, Bonsall, David, et al. (2021). Efficacy of chadox1 NCOV-19 (azd1222) vaccine against SARS-COV-2 voc 202012/01 (b. 1.1. 7).
- 43.Planas Delphine, Veyer David, Baidaliuk Artem, Staropoli Isabelle, Guivel-Benhassine Florence, Rajah Maaran, Planchais Cyril, Porrot Francoise, et al. Reduced sensitivity of infectious SARS-COV-2 variant b. 1.617. 2 to monoclonal antibodies and sera from convalescent and vaccinated individuals. bioRxiv. 2021 [Google Scholar]
- 44.Weisblum Yiska, Schmidt Fabian, Zhang Fengwen, DaSilva Justin, Poston Daniel, Lorenzi Julio C.C., Muecksch Frauke, Rutkowska Magdalena, et al. Escape from neutralizing antibodies by SARS-COV-2 Spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Fan, Su Zhao, Yu Bin, Chen Yan-Mei, Wen Wang, Song Zhi-Gang, Hu Yi, Tao Zhao-Wu, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers Fabian, Higgins Desmond G. Multiple sequence alignment methods. Springer; 2014. Clustal omega, accurate alignment of very large numbers of sequences; pp. 105–116. [DOI] [PubMed] [Google Scholar]
- 47.Yin Changchuan. Genotyping coronavirus SARS-COV-2: methods and implications. Genomics. 2020;112(5):3588–3596. doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jankauskaitė Justina, Jiménez-García Brian, Dapkūnas Justas, Fernández-Recio Juan, Moal Iain H. SKEMPI 2.0: an updated benchmark of changes in protein–protein binding energy, kinetics and thermodynamics upon mutation. Bioinformatics. 2019;35(3):462–469. doi: 10.1093/bioinformatics/bty635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sirin Sarah, Apgar James R., Bennett Eric M., Keating Amy E. AB-Bind: antibody binding mutational database for computational affinity predictions. Protein Sci. 2016;25(2):393–409. doi: 10.1002/pro.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jemimah Sherlyn, Yugandhar K., Michael Gromiha M. Proximate: a database of mutant protein–protein complex thermodynamics and kinetics. Bioinformatics. 2017;33(17):2787–2788. doi: 10.1093/bioinformatics/btx312. [DOI] [PubMed] [Google Scholar]
- 51.Liu Quanya, Chen Peng, Wang Bing, Zhang Jun, Li Jinyan. dbmpikt: a database of kinetic and thermodynamic mutant protein interactions. BMC Bioinformatics. 2018;19(1):1–7. doi: 10.1186/s12859-018-2493-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlsson Gunnar. Topology and data. Bull. Am. Math. Soc. 2009;46(2):255–308. [Google Scholar]
- 53.Edelsbrunner Herbert, Letscher David, Zomorodian Afra. Proceedings 41st annual symposium on foundations of computer science. IEEE; 2000. Topological persistence and simplification; pp. 454–463. [Google Scholar]
- 54.Xia Kelin, Wei Guo-Wei. Persistent homology analysis of protein structure, flexibility, and folding. Int. J. Numer. Methods Biomed. Eng. 2014;30(8):814–844. doi: 10.1002/cnm.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pedregosa Fabian, Varoquaux Gaël, Gramfort Alexandre, Michel Vincent, Thirion Bertrand, Grisel Olivier, Blondel Mathieu, Prettenhofer Peter, et al. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 56.Srivastava Nitish, Hinton Geoffrey, Krizhevsky Alex, Sutskever Ilya, Salakhutdinov Ruslan. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15(1):1929–1958. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.