Abstract
Acute basophilic leukaemia (ABL) is a rare subtype of acute myeloid leukaemia (AML); therefore, few data are available about its biology. Herein, we analysed two ABL patients using flow cytometry and next‐generation sequencing (NGS). Two cell populations were detected by flow cytometry in both patients. In Case no. 1, blasts (CD34+, CD203c−, CD117+, CD123dim+) and basophils (CD34−, CD203c+, CD117±, CD123+) were identified, both of which were found by NGS to harbour the 17p deletion and have loss of heterozygosity of TP53. In Case no. 2, blasts (CD33+, CD34+, CD123−) and basophils (CD33+, CD34+, CD123+) were identified. NGS detected NPM1 mutations in either blasts or basophils, and TET2 in both. These data suggest an overlap of the mutational landscape of ABL and AML, including TP53 and TET2 mutations. Moreover, additional mutations or epigenetic factors may contribute for the differentiation into basophilic blasts.
Keywords: acute basophilic leukaemia, gemtuzumab ozogamicin, next‐generation sequencing
1. INTRODUCTION
Acute basophilic leukaemia (ABL) is a rare subtype of acute myeloid leukaemia (AML) recognized by the 2016 World Health Organization (WHO) classification. The proposed diagnostic criteria for ABL are as follows: (a) myeloblasts + metachromatic blasts (>20%) and basophils (>40%) of nucleated bone marrow or peripheral blood cells; and (b) persistent hyperbasophilia. 1 However, the molecular cytogenetic data available on ABL are scarce. Indeed, it remains unclear whether ABL shares a similar mutational profile with AML. Moreover, there is no established treatment strategy for ABL, highlighting the need for a better understanding of this malignancy.
Here, we diagnosed two patients with ABL based on morphology and cytology. In addition, bone marrow samples were analysed using next‐generation sequencing (NGS). This report will add new information about the mutational landscape of this rare entity.
2. CASE DESCRIPTION
2.1. Case no. 1
The patient was a 59‐year‐old Japanese man with a history of cardiac arrhythmia. One month before presentation to our institution, he was diagnosed with pneumonia and treated with tazobactam/piperacillin (TAZ/PIPC) and teicoplanin at another hospital. Three weeks before presentation, bone marrow aspiration was performed for the evaluation of leukopenia, which revealed an increased number of blasts and immature basophils. Two weeks before presentation, he was diagnosed with acute myocardial infarction (AMI), for which emergent percutaneous coronary intervention was performed. After AMI treatment, he was referred to our hospital for ABL treatment.
Blood examination on admission to our hospital showed a leucocyte count of 0.52 × 109/L (10% blasts and 6% immature basophils). Bone marrow aspiration revealed a further increase in blasts (38.2%) and immature basophils (33.8%; Figure 1A), but no dysplasia. Toluidine blue staining was positive for immature basophils (Figure 1B). Electron microscopic examination demonstrated basophilic granules (Figure 1C). Flow cytometry detected two populations, namely blasts (CD34+, CD203c‐, CD117+, CD123dim+) and basophils (CD34‐, CD203c+, CD117±, CD123+; Figure 1D). Blasts showed basophilic differentiation. Although basophils in the bone marrow were less than 40%, his condition was consistent with ABL, based on morphology, flow cytometry and electron microscopy findings. G‐band karyotyping from the bone marrow showed a complex karyotype (42, XY, −1, −5, −7, add(7)(q11.2), −8, add(9)(p11), −10, −11, −14, der(17)t(11;17)(q13;p11.2), −18, +4mar [4/20 cells] / 43, idem, +mar [7/20 cells]/46, XY [9/20 cells]). PCR results were negative for DEK‐NUP214, and major, minor and micro‐BCR‐ABL1. NGS detected TP53 mutation (R175H; Figure 1E), 17p deletion and loss of heterozygosity of TP53.
FIGURE 1.
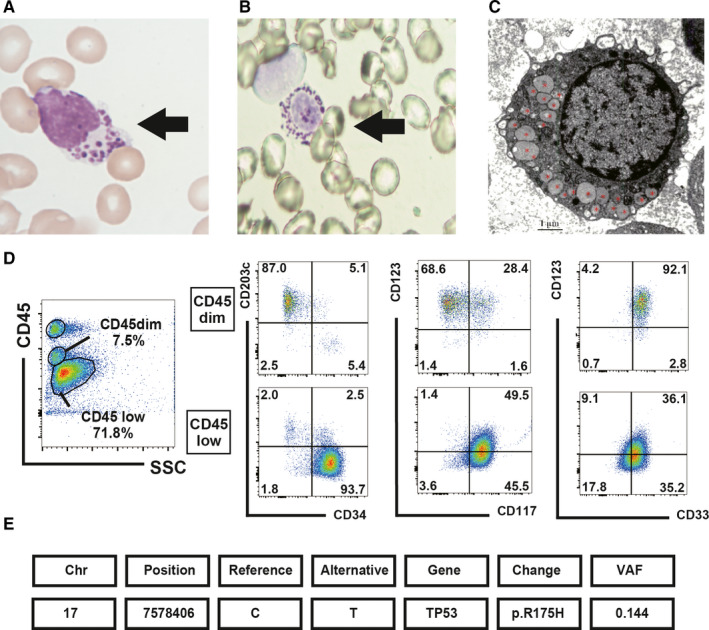
Clinical analysis of peripheral blood and bone marrow revealed an increase in blasts and basophils with TP53 mutation (Case no. 1). A, May‐Giemsa staining and B, toluidine blue staining of basophils (black arrow) in the peripheral blood (May‐Giemsa and toluidine blue staining; oil immersion lens, original magnification, ×1000). C, Electron microscopy image of basophils in the peripheral blood (red asterisks represent basophilic granules). D, Flow cytometry analysis of the bone marrow. Two tumour populations were detected: basophils (CD45dim+, CD34‐, CD203c+, CD117±, CD123+) and blasts (CD45 low, CD34+, CD203c‐, CD117+, CD123dim+), which differentiated into basophils. E, Next‐generation sequencing data
He was administered dual histamine blockade (H1 and H2) and prednisolone to prevent symptoms due to excessive release of histamine. 1 Induction therapy with DNR/AraC (daunorubicin 50 mg/m2 on days 1‐5 and cytarabine 100 mg/m2 on days 1‐7) was ineffective. Six weeks later, re‐induction therapy with MEC (mitoxantrone 8 mg/m2, etoposide 100 mg/m2 and cytarabine 1 g/m2) was administered. However, it resulted in significant inflammation and temporal cognitive impairment, possibly due to tumour lysis syndrome on day 2. Therefore, it was discontinued. He was not tolerant to high‐dose conventional chemotherapy. Four weeks later, gemtuzumab ozogamicin (GO; 10 mg/body) was administered, because the tumour cells express CD33. Following administration of GO, he developed an infusion reaction and was agitated again. He did not respond to any chemotherapeutic regimens and passed away 4 months after the initial diagnosis.
2.2. Case no. 2
A 72‐year‐old Japanese man with a medical history of mitral stenosis and benign prostatic hyperplasia was referred to our institute for high‐grade fever. Two weeks before presentation, he had fever and diarrhoea, which resolved simultaneously. On admission, he had high‐grade fever and hypotension. He was admitted to our hospital for suspected urosepsis as the urinalysis showed bacteriuria.
Blood examination showed a leucocyte count of 2.62 × 109/L (3% blasts and 27% immature basophils; Figure 2A) and thrombocytopenia (57 × 109/L). Bone marrow aspiration revealed increased percentage of blasts (57.9%) and immature basophils (20.0%), without any dysplasia. Flow cytometry detected basophils (CD33+, CD45low, CD123+; Figure 2B). He was diagnosed with ABL. G‐band karyotyping showed normal karyotype (46, XY). PCR for major and minor BCR‐ABL1 was negative. NGS revealed NPM1 and TET2 mutations (Figure 2C).
FIGURE 2.
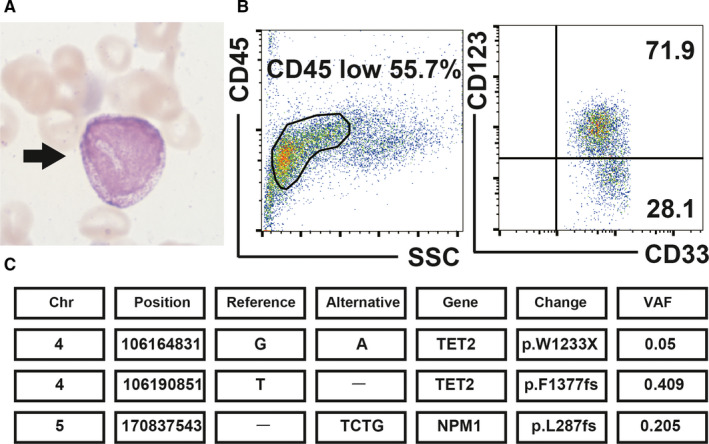
Clinical analysis of bone marrow showed an increase in blasts and basophils with TET2 and NPM1 mutations (Case no. 2). A, Basophilic granules (black arrow) were present in the bone marrow (May‐Giemsa stain, oil immersion lens, original magnification×1000). B, Flow cytometry analysis of the bone marrow: tumour cells were divided into two populations, blasts (CD33+, CD45dim+ and CD123‐) and basophils (CD33+, CD45dim+ and CD123+). C, Next‐generation sequencing data
For urosepsis treatment, he received meropenem and vancomycin. As hypotension persisted even after recovery from sepsis, he was administered vasopressor during induction chemotherapy. We used histamine blockers and prednisolone similar to that in Case no. 1. After induction therapy with DNR/AraC (daunorubicin 40 mg/m2 day 5‐7 and cytarabine 80 mg/m2 day 1‐7), hypotension was resolved and haematological CR was achieved at day 33. However, soon after, he had a relapse of the disease. He was refractory to subsequent chemotherapeutic regimens including mitoxantrone (MIT) monotherapy and MIT/AraC. He died 8 months after initial diagnosis.
3. METHODS
Data from bone marrow samples were collected using BD FACSCalibur & FACSLyric (BD Biosciences) and analysed using FlowJo version 10.
For targeted sequencing, 200 ng of genomic DNA from bone marrow was enriched for the target regions by liquid‐phase hybridization using the SureSelect custom kit (Agilent Technologies®). We used an in‐house panel designed to examine 390 genes that are implicated in myeloid malignancies and bone marrow failure using SureDesign (Agilent Technologies®). The purified library was subjected to high‐throughput sequencing analysis with Illumina HiSeq 2500 using 125 bp pair‐end mode. Sequencing reads were aligned to the human genome reference (hg19) using Burrows‐Wheeler Aligner (version 0.7.8) with default parameter settings, and annotation was conducted with Genomon pipeline 2.6.2, as previously reported. 2 Mapping errors were resolved by visual inspection on the Integrative Genomics Viewer browser.
4. RESULTS AND DISCUSSION
In the present study, we presented two cases of ABL. We considered differential diagnosis of secondary cause of acute basophilic leukaemia such as blast phase of CML, AML with t(8;21) and basophilia, AML with t(6;9)(p23;q34), monocytic/monoblastic AML with basophilia, acute promyelocytic leukaemia (APL), mast cell leukaemia (MCL), MPN or MDS with basophilia. Secondary ABL was excluded based on the results of bone marrow examination, G‐banding of karyotype and detection of leukaemia fusion gene by RT‐PCR. Although a bone marrow basophil count of less than 40% in our cases did not fulfil the proposed criteria by Valent P et al 1 there is no consensus regarding the number of basophilic blasts. Hence, we diagnosed these patients with ABL based on multiple confirmation of basophils via morphological, cytological and electron microscopic analysis. With dual histamine blockade, we were able to suppress the symptoms caused by the excessive histamine release, including shock, systemic anaphylaxis, urticaria, peptic ulceration and gastrointestinal bleeding. 3 Multiple chemotherapeutic regimens including GO therapy were also used for Case no. 1, but none were efficacious. These cases illustrate the challenges in treating ABL.
Some reports have described cytogenetic abnormalities in ABL, such as t(X;6)(p11;q23), 4 del(8), t(10;11)(p13;q21), del(16)(q22), t(16;17)(q22;21), 5 t(2;6)(q23?4;p22?3), as well as del(12)(p11). 6 However, no recurrent mutations in ABL were identified except t(X;6)(p11;q23). Case no. 1 had a complex karyotype with 17p11 translocation, whereas Case no. 2 had a normal karyotype.
To understand the underlying pathogenesis of ABL, we performed NGS in these cases. In Case #1, a TP53 mutation (R175H) at a variant allelic frequency (VAF) of 14.4%, chromosome 17p deletion and loss of heterozygosity of TP53 were identified. Bone marrow (38.2% of blasts and 33.8% of basophils) and G‐banding data (55% of cells harboured the 17p11 translocation) suggested that both blasts and basophils had translocation of 17p11. Considering that tumour cells with 17p10‐12 translocation led to loss of TP53, 7 we suspected that both cell types lacked TP53 activity. Moreover, either blasts or basophils had a point mutation in TP53 (R175H). In Case no. 2, two TET2 mutations at VAF of 40.9% and 5% and a NPM1 mutation at a VAF of 20.5% were identified. Both basophils and blasts seemed to carry TET2 mutations. However, we could not determine the population harbouring the NPM1 mutation.
Some molecular pathways may influence the phenotype and development of ABL. In four male infants with ABL, t(X;6)(p11;q23) and c‐MYB–GATA1 mutations were reported. These were the first recurrent mutations of ABL 4 and promoted basophilic differentiation by induction of IL‐33, NGF, IL1RL1 and NTRK1 expression. 8 c‐Myb was stimulated by p16INK4a. 9 In Case #2, NPM1 through interaction with p16INK4a may influence c‐Myb. 10 C/EBPα up‐regulation may be also associated with the development of ABL. TET2 encodes an epigenetic modifier, which is known to act as an upstream regulator of mast cell and basophil lineage commitment. The absence of TET2 causes up‐regulation of C/EBPα (basophil‐specific genes). 11 Therefore, we hypothesized that an additional NPM1 mutation, along with a TET2 mutation, could act synergistically to drive the leukemic transformation. 12 TET2 mutation combined with the epigenetic modification of C/EBPα may have modulated downstream molecules to induce basophil differentiation. As the relationship between TET2, NPM1, p53 and c‐MYB–GATA1 has not been described yet, further studies are needed to reveal the molecular link.
In conclusion, we reported two ABL cases using NGS analysis. Although mutations in these cases (TP53, NPM1 and TET2) are common in AML, they have not been previously reported in ABL. This suggests an overlap of the mutational profiles of ABL and AML. Case no. 1 had a complex karyotype with TP53 loss, whereas Case no. 2 had a normal karyotype with NPM1 and TET2 mutations, which suggests that epigenetic factors may promote differentiation into basophilic blasts. As cytogenetic and mutations of ABL are heterogeneous, further research is necessary for a better understanding on the factors that encourage basophilic differentiation in ABL.
CONFLICT OF INTEREST
The authors declare no conflicts of interest associated with the present study.
AUTHOR CONTRIBUTIONS
Takuya Shimizu: Conceptualization (lead); Writing‐original draft (lead). Tadakazu Kondo: Conceptualization (supporting); Project administration (lead); Supervision (lead); Writing‐review & editing (lead). Yasuhito Nannya: Formal analysis (lead); Methodology (lead); Project administration (supporting); Supervision (supporting); Writing‐review & editing (supporting). Mizuki Watanabe: Methodology (supporting); Supervision (supporting); Writing‐review & editing (supporting). Toshio Kitawaki: Formal analysis (supporting); Writing‐review & editing (supporting). Takero Shindo: Project administration (supporting); Supervision (supporting); Writing‐review & editing (supporting). Masakatsu Hishizawa: Supervision (supporting); Writing‐review & editing (supporting). Kouhei Yamashita: Supervision (supporting); Writing‐review & editing (supporting). Seishi Ogawa: Formal analysis (supporting); Funding acquisition (lead); Methodology (supporting); Project administration (supporting); Supervision (supporting); Writing‐review & editing (supporting). Akifumi Takaori‐Kondo: Project administration (lead); Supervision (lead); Writing‐review & editing (supporting).
ETHICAL APPROVAL
Informed consent was obtained for using the biospecimens for research, in compliance with the Declaration of Helsinki under the protocols approved by the Ethics Committee and Institutional Review Board of Kyoto University Hospital (Ref. number: G697).
ACKNOWLEDGEMENTS
The authors thank the patients and their families, all the medical staff members involved in treating this patient and Dr Ohno and Daiki Shimomura from Tenri Hospital for electron microscopy analysis (Case no. 1).
Shimizu T, Kondo T, Nannya Y, et al. Next‐generation sequencing in two cases of de novo acute basophilic leukaemia. J Cell Mol Med. 2021;25:7095–7099. 10.1111/jcmm.16591
Funding information
This work was supported by JSPS KAKENHI under Grant Numbers JP15H05909 and JP26221308, by Ministry of Education, Culture, Sports, Science and Technology under Grant Number hp160219 and by AMED under Grant Number JP18ck0106250h0002 and JP20cm0106501h0005 to SO.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Valent P, Sotlar K, Blatt K, et al. Proposed diagnostic criteria and classification of basophilic leukemias and related disorders. Leukemia. 2017;31:788‐797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yoshizato T, Nannya Y, Atsuta Y, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129:2347‐2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kritharis A, Brody J, Koduru P, et al. Acute basophilic leukemia associated with loss of gene ETV6 and protean complications. J Clin Oncol. 2011;29:e623‐e626. [DOI] [PubMed] [Google Scholar]
- 4. Quelen C, Lippert E, Struski S, et al. Identification of a transforming MYB‐GATA1 fusion gene in acute basophilic leukemia: a new entity in male infants. Blood. 2011;117:5719‐5722. [DOI] [PubMed] [Google Scholar]
- 5. Shvidel L, Shaft D, Stark B, et al. Acute basophilic leukaemia: eight unsuspected new cases diagnosed by electron microscopy. Br J Haematol. 2003;120:774‐778. [DOI] [PubMed] [Google Scholar]
- 6. Giagounidis AAN, Hildebrandt B, Heinsch M, et al. Acute basophilic leukemia. Eur J Haematol. 2001;67:72‐76. [DOI] [PubMed] [Google Scholar]
- 7. Finka SR, Smoleya SA, Stockero KJ, et al. Loss of TP53 is due to rearrangements involving chromosome region 17p10~p12 in chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2006;167:177‐181. [DOI] [PubMed] [Google Scholar]
- 8. Ducassou S, Prouzet‐Mauléon V, Deau MC, et al. MYB–GATA1 fusion promotes basophilic leukaemia: involvement of interleukin‐33 and nerve growth factor receptors. J Pathol. 2017;242:347‐357. [DOI] [PubMed] [Google Scholar]
- 9. Lei W, Liu F, Ness SA. Positive and negative regulation of c‐Myb by cyclin D1, cyclin‐dependent kinases, and p27 Kip1. Blood. 2005;105:3855‐3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitrea DM, Kriwacki RW. On the relationship status for Arf and. NPM1–it's complicated. FEBS J. 2018;285(5):828‐831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palam L, Mali R, Ramdas B, et al. Loss of epigenetic regulator TET2 and oncogenic KIT regulate myeloid cell transformation via PI3K pathway. JCI Insight. 2018;3:e94679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patel JL, Schumacher JA, Frizzell K, et al. Coexisting and cooperating mutations in NPM1‐mutated acute myeloid leukemia. Leuk Res. 2017;56:7‐12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


