Key Points
Neddylation is activated in mice and macrophages after MRSA infection.
Neddylation facilitates bactericidal ability via increasing macrophage ROS production.
Abstract
Neddylation, a posttranslational modification in which NEDD8 is covalently attached to target proteins, has emerged as an endogenous regulator of innate immunity. However, the role of neddylation in methicillin-resistant Staphylococcus aureus (MRSA) infection remains unknown. In this study, we found that neddylation was activated after MRSA infection in vivo and in vitro. Inhibition of neddylation with MLN4924 promoted injury of liver and kidneys in C57BL/6 mice with MRSA bloodstream infection and increased mortality. Blockade of neddylation, either pharmacologically (MLN4924, DI591) or through the use of Uba3 small interfering RNA, inhibited Cullin3 neddylation and promoted Nrf2 accumulation, thus reducing reactive oxygen species (ROS) induction and bacterial killing ability in mouse peritoneal macrophages. In summary, our findings suggest that activation of neddylation in macrophages plays a critical protective role against MRSA infection by increasing ROS production, partially by signaling through the NEDD8-Cullin3-Nrf2-ROS axis. Furthermore, our results may provide a new non-antibiotic treatment strategy for MRSA infection through targeting of neddylation.
Introduction
Staphylococcus aureus, one of the most common pathogens responsible for community-acquired infections and healthcare-related nosocomial infections in humans, has high morbidity and mortality (1). The extreme morbidity and mortality of S. aureus are due to the resistance of many S. aureus strains to a variety of antibiotics, such as penicillin, streptomycin, gentamicin, and vancomycin (2). With the overuse of antibiotics, methicillin-resistant S. aureus (MRSA) infection has increased, causing a significant public health problem (3, 4). It is estimated that more than 300 million premature deaths will occur due to antimicrobial resistance by the year 2050. Given the current dilemma of antibiotic resistance, the need to pursue new therapeutic targets in addition to antibiotics to improve non-antibiotic strategies for treating MRSA infection is both important and urgent.
Neural precursor cell-expressed developmentally downregulated 8 (NEDD8), a small ubiquitin-like protein, can be conjugated to substrate proteins in a process known as neddylation (5). Analogous to ubiquitination, neddylation is triggered by the activation of the NEDD8-activating enzyme (NAE) E1 (a heterodimer of APPBP1 and the catalytic subunit UBA3), the NEDD8-conjugating enzyme E2 (UBE2M or UBE2F), and NEDD8-E3 ligases, which are involved in many physiological and pathological processes, such as cell survival and differentiation (6), neurodegeneration (7), fibrosis (8), and cancer (9, 10). Accumulating evidence has recently suggested that neddylation participates in the pathogenesis of infection and inflammation (11–13). Activation of neddylation in T cells by Plasmodium during the blood stage of Plasmodium infection was critical for parasite control and improved survival in a mouse model of Plasmodium bloodstream infection (11). MLN4924, a neddylation inhibitor, abrogated the LPS-induced increase in the expression of cytokines, including IL-6 and TNF-α, by macrophages in vitro (12) and protected against LPS-induced acute kidney injury in mice (13). In addition, some pathogens, such as Burkholderia pseudomallei (14), Escherichia coli (15), and Chlamydia trachomatis (16), selectively deamidate NEDD8 in host cells to promote pathogen survival via consequent repression of the NF-κB pathway. Studies have also suggested that neddylation plays a protective role against the development of exogenous pathogen-induced infection.
Macrophages play a key role in antimicrobial immunity by recognizing, phagocytosing, and killing pathogens (17). Although macrophages are essential to defend bacteria, and the activation of neddylation plays an important role in exogenous pathogen invasion, whether and how the neddylation of macrophages is involved in defense against MRSA invasion are largely unknown. Herein, we investigated the effect of neddylation in macrophages on MRSA infection and its underlying mechanisms.
Materials and Methods
Bacterial preparation
S. aureus strain USA300 (ATCC-BAA-1556, Miaoling, Shanghai, China) was inoculated into Luria-Bertani medium, incubated for 8 h at 37°C, serially diluted in sterile PBS and cultured on solid Luria-Bertani plates. CFUs were counted after incubation at 37°C overnight. Then, the bacteria were pelleted by centrifugation, washed, and resuspended in PBS.
Animals and cell lines
Male C57BL/6 mice were housed in a specific pathogen-free facility in the Experimental Animal Center of Zhejiang University and used at 8 wk of age. The mouse experimental protocols were approved by the Ethics Committee for Animal Studies at Zhejiang University. NIH-3T3 cells were obtained from the American Type Culture Collection (Manassas, VA) and cultured in DMEM supplemented with 10% (v/v) FCS (Lonza, Basel, Switzerland).
Chemicals and reagents
MLN4924 (HY-70062), GSK2795039 (HY-18950), and ML385 (HY-100523) were purchased from MedChemExpress (Monmouth Junction, NJ). N-acetyl-l-cysteine (NAC, A9165) and MG132 (C2211) were purchased from Sigma-Aldrich (St. Louis, MO). CFSE (C1157) was purchased from Thermo Fisher Scientific (Carlsbad, CA, USA). DCFH-DA (S0033-1) was purchased from Beyotime Biotech (Shanghai, China).
Establishment of a mouse model of MRSA bloodstream infection
An MRSA-induced bloodstream infection model was established by intravenously injecting mice with S. aureus strain USA300 as previously described (18). The optimal dose of USA300 to be used in the study was determined by evaluating the survival rates of mice postinfection with strain USA300 at different dosages. As shown in Supplemental Fig. 1A, early death began on day 2 postinfection with strain USA300 at a dose of 1 × 108 CFUs, and the 7-d survival rate was 20% upon infection with strain USA300 at a dose of 5 × 107 CFUs. Thus, we chose a dose of 5 × 107 CFUs for the subsequent survival rate experiment, and a dose of 1 × 108 CFUs was used for the other in vivo experiments.
Detection of the bacterial load in different mouse organs
Liver, spleen, lung, and kidney tissues were collected at 24 h after MRSA infection and homogenized, and the homogenates were serially diluted in PBS, plated on solid Luria-Bertani plates, and then incubated under aerobic conditions for 16 h at 37°C. CFUs were counted, and the values are expressed as colony counts per gram of tissue.
Depletion of macrophages from mice
As described previously (19), we also used clodronate-encapsulated liposomes to deplete macrophages. In brief, mice were intravenously injected with 50 μl of clodronate-encapsulated liposomes or PBS liposomes (Clodronate Liposomes, Amsterdam, the Netherlands) 12 h before MLN4924 injection.
Isolation and purification of mouse peritoneal macrophages
Peritoneal macrophages (PMs) were prepared from mice as previously described (20). Briefly, 8-wk-old C57BL/6 mice were i.p. injected with 2 ml of 3% sterile thioglycolate medium (BD Biosciences, Sparks, MD). Three days later, PMs were extracted. To isolate and purify the PMs, each mouse was euthanized with 40 mg/kg pentobarbital sodium, and its abdomen was sprayed with 70% ethanol. The outer layer of the peritoneum was incised with scissors, and ice-cold RPMI 1640 medium was subsequently injected into the peritoneal cavity using a 20-ml syringe. After the peritoneum was gently massaged to dislodge any attached cells and suspend them in RPMI 1640 medium, the enterocoelic fluid was collected into a tube using a 20-ml syringe, kept on ice, and centrifuged at 250 × g at 4°C for 5 min. The supernatant was discarded, and the precipitate was suspended in RPMI 1640 medium supplemented with 10% FBS and 100 U/ml penicillin, after which the cells were counted using a hemocytometer. The cells were then added to 12- or 24-well tissue culture plates as needed to obtain a density of 5 × 105 cells/ml and cultured for 2 h at 37°C. Then, nonadherent cells were removed by gentle washing with PBS three times. The isolated macrophages were stained for CD11b and F4/80. The purity of the isolated macrophages (CD11b and F4/80 double-positive cells) was more than 90%, as analyzed by flow cytometry (Supplemental Fig. 1B), and the macrophages were prepared for in vitro experiments.
Tissue dissociation and cellular analysis of the liver, lungs, and spleen
For liver dissociation, mice were anesthetized and perfused through the hepatic portal vein with dissociation solution containing 80 µg/ml collagenase type IV (Sigma) and 50 µg/ml DNase (Sigma) at 37°C for 10 min. Then, the liver was collected and ground into a suspension using a 5-ml injection syringe. Lung tissues were collected, cut into small pieces, and incubated with dissociation solution containing 2 mg/ml collagenase type I (Sigma), 2 mg/ml collagenase type IV (Sigma), and 1 mg/ml DNase (Sigma) at 37°C for 2 h. Spleen tissues were directly ground into a suspension using a 5-ml injection syringe. Suspensions from the liver, lungs, and spleen were washed using PBS and dispersed through a 70-mm cell strainer. Single-cell suspensions were stained with the indicated Abs and analyzed with DxFLEX (Beckman Coulter, Brea, CA).
Analysis and isolation of macrophages from the mouse liver, lungs, and spleen
Single cells isolated from the liver, spleen, or lungs were stained with the indicated fluorescence-conjugated Abs for 20 min, washed, resuspended in PBS containing 1% FBS, and analyzed with a Cytoflex machine (Beckman Coulter). Flow cytometry data were plotted and quantified as the mean fluorescence intensity (MFI) using FlowJo software (Tree Star, Ashland, OR). The following fluorescence-conjugated Abs used for the experiment were purchased from BioLegend (San Diego, CA, USA): allophycocyanin-conjugated anti-mouse F4/80, FITC-conjugated anti-mouse F4/80, PE-conjugated anti-mouse CD11b, Pacific Blue (PB)–conjugated fixable viability dye, and allophycocyanin-conjugated anti-mouse CD45. To isolate macrophages from mouse livers and lungs, CD11b+F4/80+ macrophages were further sorted by a Beckman MoFlo Astrios EQ (Beckman Coulter).
Histopathology
The liver, lungs, and kidneys were dissected from individual mice, immediately fixed with 4% paraformaldehyde, and then subjected to H&E staining. At least three images of randomly selected microscopic fields were captured from each slide from each mouse.
Cytokine analysis
Mouse serum concentrations of the cytokines IL-6, IL-1β, and TNF-α were measured with an ELISA kit (BioLegend) according to the manufacturer’s instructions.
Immunofluorescence and confocal microscopy
PMs were incubated with or without MLN4924 (100 nM) for 6 h, after which MRSA was added at a multiplicity of infection (MOI) of 20 and incubated for another 6 h. The cells were washed and fixed in prechilled methanol for 10 min and then permeabilized with 0.1% Triton X-100 for another 10 min. After blocking with 5% BSA, the cells were incubated overnight at 4°C with an anti–NF erythroid-derived 2–like 2 (Nrf2) (Ab137550, Abcam, Cambridge, MA, 1:250) Ab. The primary Ab was detected using a DyLight 594–labeled secondary Ab (Ab96921, Abcam, 1:400). Nuclei were stained with DAPI (Invitrogen, Carlsbad, CA). Images of the cells and sections were captured with an Olympus confocal fluorescence microscope (Olympus, Tokyo, Japan).
Immunoblot analysis
A total of 20 µg of protein from the cell lysate was separated by SDS-PAGE on 10% gels and transferred onto polyvinylidene difluoride membranes. The membranes were blocked with 5% non-fat milk in PBS with Tween 20 (PBST) buffer and incubated with primary Abs overnight at 4°C. After washing, the bound Abs were detected with HRP-conjugated secondary Abs (goat anti-rabbit IgG, Beyotime Biotechnology, Shanghai, China, 1:5000) for 1 h and visualized using enhanced chemiluminescent reagents, followed by scanning with a Tanon 4500 gel imaging system. The following primary Abs were used: rabbit monoclonal anti-NEDD8 (Ab81264, Abcam, 1:2000), rabbit polyclonal anti-Nrf2 (Ab137550, Abcam, 1:2000), rabbit polyclonal anti-COPS5 (A1766, ABclonal, China, 1:1000), rabbit polyclonal anti-NADPH oxidase (NOX)2 (A1636, ABclonal, 1:1000), rabbit monoclonal anti-Cullin3 (Ab75851, Abcam, 1:1000), and rabbit polyclonal anti-p62 (Ab101266, Abcam, 1:1000). β-Actin (ET1701-80, HuaBio, China, 1:2000) was used as a sample loading control.
Real-time PCR
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. cDNA was synthesized using a cDNA synthesis kit (Takara, Dalian, Liaoning, China) following the manufacturer’s instructions. For mRNA detection, β-actin served as an internal control. Real-time PCR was conducted using SYBR Green (TaKaRa) with an Applied Biosystems 7500 real-time PCR system (Thermo Fisher Scientific). The primer sequences are listed in Table I.
Table I.
Primer sequences used
| Name | Primer Sequence (5′→3′) |
|---|---|
| β-Actin | F, GGCTGTATTCCCCTCCATCG; R, CCAGTTGGTAACAATGCCATGT |
| NEDD8 | F, GGAGCGAATCAAGGAGCGT; R, GGAACCACCTAGAATCTTGT |
| Cullin3 | F, AGCCGGAAGGACACCAAGA; R, GCTCCTCAAAACTAAGACCACTG |
| Nrf2 | F, TCTTGGAGTAAGTCGAGAAGTGT; R, GTTGAAACTGAGCGAAAAAGGC |
| CSN5 | F, TGGGTCTGATGCTAGGAAAGG; R, CTATGATACCACCCGATTGCATT |
| UBA3 | F, CCTTCACACACCCCGATTTC; R, GCCTTGGGTCTTCCGACATC |
| p62 | F, AGGATGGGGACTTGGTTGC; R, TCACAGATCACATTGGGGTGC |
| IL-6 | F, TAGTCCTTCCTACCCCAATTTCC; R, TTGGTCCTTAGCCACTCCTTC |
| IL-1β | F, GCAACTGTTCCTGAACTCAACT; R, ATCTTTTGGGGTCCGTCAACT |
| TNF-α | F, CAAACCACCAAGTGGAGGAG; R, GTGGGTGAGGAGCACGTAGT |
| Nox2 | F, GAATCAGCCTTAGTGTCACAGG; R, ATTCCGGTATGCGTCCAGC |
| GST | F, AGCTCACGCTATTCGGCTG; R, GCTCCAAGTATTCCACCTTCAGT |
| SOD1 | F, AACCAGTTGTGTTGTCAGGAC; R, CCACCATGTTTCTTAGAGTGAGG |
| CAT | F, AGCGACCAGATGAAGCAGTG; R, TCCGCTCTCTGTCAAAGTGTG |
F, forward; R, reverse.
Apoptosis assay
PMs were treated with or without MLN4924 for 6 h and then were or were not infected with MRSA at an MOI of 20 for an additional 6 h. Then, the cells were dissociated using 0.25% trypsin-EDTA and collected by centrifugation. Apoptosis was determined using an Annexin V-PI apoptosis detection kit (MultiSciences, Hangzhou, Zhejiang, China) and immediately analyzed by flow cytometry.
CFSE labeling, phagocytosis, and bacterial killing assay
MRSA was labeled with 5 μM CFSE (Invitrogen) at 37°C for 10 min according to the manufacturer’s instructions, washed three times with complete RPMI 1640 medium, and incubated with macrophages. For FACS-based phagocytosis, MLN4924- or DMSO-treated macrophages were infected with CFSE-labeled MRSA (MOI of 20) for 30 min, extensively washed with cold PBS three times, and measured immediately with FACS. For immunofluorescence measurement of the phagocytosis and killing abilities of the macrophages, 1 × 105 PMs were cultured in 24-well dishes and then infected with CFSE-labeled MRSA (MOI of 20) for 30 min, followed by washing with PBS three times. The infected cells were further cultured in sterile RPMI 1640 medium containing 2 µg/ml vancomycin (Sangon Biotech, Shanghai, China). At the indicated times, the macrophages were washed and fixed with prechilled methanol for 10 min at −20°C, washed again, and fixed with anti-fade mounting medium containing DAPI for image collection with an Olympus FV3000 confocal microscope (Olympus). For the in vitro bacterial killing assay, macrophages were incubated with MRSA (MOI of 20) for the indicated durations at 37°C. Then, the cells were collected and lysed in ddH20, and the lysates were diluted and spread onto solid Luria-Bertani plates. The CFUs were counted after incubation overnight at 37°C.
Analysis of reactive oxygen species (ROS) levels
To measure cellular ROS levels, cells (PMs or NIH-3T3 cells) were treated with or without 5 μM NAC (Sigma) as needed, followed by other indicated treatments (MLN4924 treatment, MRSA infection, or transfection). The culture medium was removed, and the cells were washed with PBS and incubated for 30 min at 37°C with dichloro-dihydro-fluorescein diacetate (DCFH-DA) at a final concentration of 10 μM in serum-free RPMI 1640 medium. The cells were washed with PBS, removed from the plates by 0.25% trypsin-EDTA, pelleted at 2000 rpm for 5 min, immediately resuspended in cold PBS, and analyzed by flow cytometry. Data analysis was performed using FlowJo software.
Construction of expression plasmids
The mouse Ube2m gene was amplified from mouse PMs by reverse transcription PCR and ligated into pcDNA3.1-Flag to construct a Flag-tagged expression plasmid. The mouse Dcn1 gene was amplified from RAW264.7 cells by RT-PCR and ligated into the PMX plasmid.
Plasmid and small interfering RNA (siRNA) transfection
NIH-3T3 cells were transfected with plasmids using JetPEI transfection reagent (Polyplus, Beijing, China). Mouse PMs were transfected with a scrambled negative control (NC) sequence or targeted siRNA using TransIT-TKO transfection reagent (Mirus Bio, Madison, WI) according to the manufacturer’s instructions. The siRNAs were synthesized by GenePharma (Shanghai, China). The following siRNA oligonucleotide sequences were used: Cullin3 siRNA, 5'-GGGUGCGAGAAGAUGUACUAAAUdTdT-3'; Nrf2 siRNA, 5'-CUUGAAGUCUUCAGCAUGUUAdTdT-3'; p62 siRNA, 5'-GAAUUCAUGUGAAGCCGAAdTdT-3'; Uba3 siRNA, 5'-CGACACUUUCUACCGACAA-UUCUCdTdT-3'; COP9 signalosome subunit 5 (Csn5) siRNA, 5'-GACUGAUGUCUCAGGUUAUUAdTdT-3'; NC siRNA, 5'-UUCUCCGAACGUGUCACGUdTdT-3'.
Generation of bone marrow-derived macrophages (BMDMs)
BMDMs were generated as previously described (21). Briefly, bone marrow mononuclear cells were prepared from C57BL/6J mouse (8 wk old) tibial and femur suspensions by depletion of RBCs and cultured in RPMI 1640 medium supplemented with 10% FBS, 1% penicillin/streptomycin, and 10 ng/ml recombinant murine GM-CSF (Millipore, Billerica, MA). For full differentiation of BMDMs, the cells were cultured for an additional 8 d with replacement of the above-mentioned medium every 2 d. All cells were CD11b+F4/80+ when analyzed by flow cytometry.
Macrophage infusion
For macrophage infusion experiments, mice were intravenously injected with 50 μl of clodronate-encapsulated liposomes. Twelve hours later, MLN4924 (100 nM) or DMSO-treated BMDMs (4 × 106 cells per mouse) were infused into the macrophage-depleted mice via their tail veins. Twelve hours after macrophage infusion, mice were injected with USA300 (5 × 107 CFUs per mouse for survival rate analysis; 1 × 108 CFUs per mouse for others).
Statistical analyses
All statistical analyses were performed with GraphPad Prism 8.0.2 software (GraphPad Software). The unpaired Student t test was used for comparisons between two groups. Kaplan-Meier curves of overall survival were compared using the log-rank test. A p value <0.05 indicated a statistically significant difference.
Results
Neddylation is activated in mice after MRSA bloodstream infection
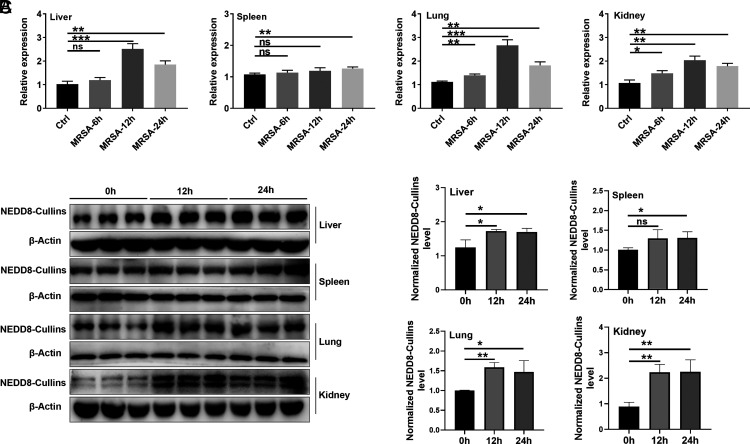
After MRSA infection, the mRNA levels of Nedd8 were increased at 6 h, peaked at 12 h, and declined at 24 h in lung and kidney tissues. In the liver, the mRNA level of Nedd8 increased at 12 and 24 h. In addition, a significant increase in Nedd8 mRNA levels in the spleen was also observed later, at 24 h after MRSA infection (Fig. 1A). Elevated NEDD8 levels can promote protein neddylation, which is reflected by the protein expression of NEDD8-conjugated Cullins (NEDD8-Cullins) (22) (Table I). Thus, we detected NEDD8-Cullins in these organs after MRSA infection. As expected, NEDD8-Cullins levels were obviously increased in the liver, lungs, and kidneys at 12 and 24 h after MRSA infection and in the spleen at 24 h after MRSA infection (Fig. 1B, 1C). These results suggest that MRSA infection activates neddylation in vivo.
FIGURE 1.
Neddylation is activated after MRSA-induced bloodstream infection. Mice were i.v. injected with S. aureus strain USA300 at a dose of 1 × 108 CFUs. Tissues were harvested at 0 (determined as control), 6, 12, and 24 h after USA300 injection (n = 3). (A) Time-dependent changes in relative Nedd8 mRNA expression in the liver, spleen, lung, and kidney tissues after USA300 infection were measured by real-time PCR. (B) NEDD8-Cullins in the liver, spleen, lung, and kidney tissues was observed by Western blot analysis. β-Actin was used as a sample loading control. (C) Quantification of β-actin–normalized NEDD8-Cullins protein levels [from (B)]. Representative results from three independent experiments are shown (mean and SD) (n = 3). ns, not significant. *p < 0.05, **p < 0.01, ***p < 0.001 (unpaired Student t test).
MLN4924 exacerbates MRSA bloodstream infection in mice
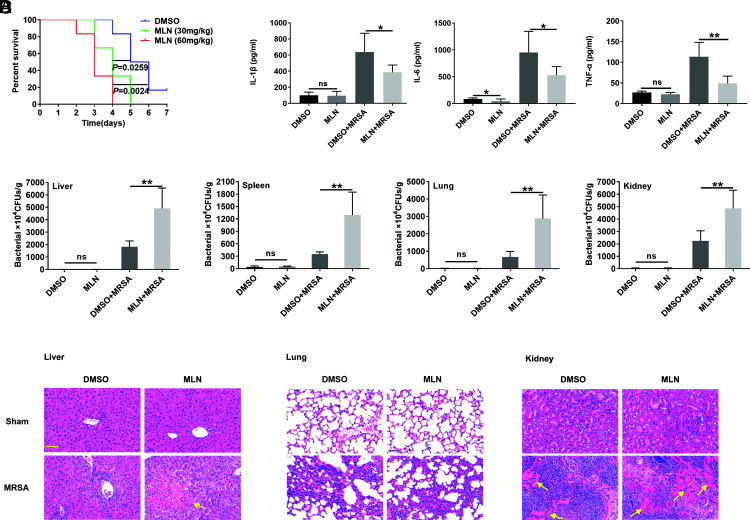
MLN4924, a well-known inhibitor of the neddylation E1 enzyme NAE1, is widely used to investigate the role of neddylation (23). NEDD8-Cullins levels in liver, spleen, lung, and kidney tissues were decreased at 12 h after i.p. injection of MLN4924 (30 mg/kg), although the decrease in NEDD8-Cullins levels in the liver was smaller (Supplemental Fig. 1C). MLN4924 alone did not impair liver and kidney functions (Supplemental Fig. 1D) and did not cause histopathological damage to the major organs (Supplemental Fig. 1E) or death (Supplemental Fig. 1F). Intraperitoneal administration of MLN4924 significantly decreased the survival rate after MRSA infection (Fig. 2A). MLN4924 treatment also inhibited the production of the serum inflammatory cytokines IL-1β, IL-6, and TNF-α at 24 h after MRSA infection (Fig. 2B), causing a notable increase in the bacterial burden (Fig. 2C) and increased tissue abscesses in liver and kidney tissues (Fig. 2D). These results suggest that neddylation probably has an inhibitory effect on MRSA infection.
FIGURE 2.
MLN4924 increases mouse susceptibility to USA300 sepsis. (A) Mice were i.p. injected with 30 or 60 mg/kg MLN4924 or the same volume of DMSO. Twelve hours later, 5 × 107 CFUs of USA300 were injected via the tail vein, and 7-d mortality was monitored. (B–D) After the administration of MLN4924 at a dose of 30 mg/kg for 12 h, 1 × 108 CFUs of USA300 was injected via the tail vein. Blood and organs were harvested 24 h after USA300 injection. Serum inflammatory cytokines (IL-1β, IL-6, and TNF-α) were measured by ELISA (B), the bacterial loads (CFUs) in the liver, lungs, spleen, and kidneys were measured (C), and inflammatory cell infiltration and abscess (yellow arrow) in the liver, lungs, and kidneys was visualized by H&E staining (D). Scale bars, 50 μm. Data are representative of three independent experiments (mean and SD) (n = 6). ns, not significant. *p < 0.05, **p < 0.01 (unpaired Student t test).
Macrophages play an essential role in MLN4924-mediated exacerbation of MRSA infection
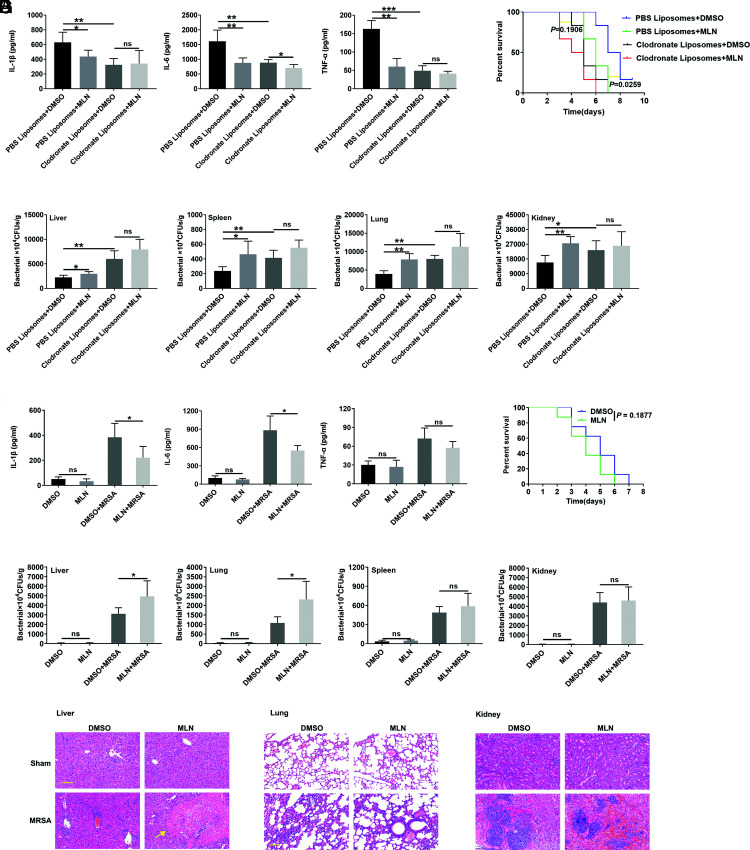
Because macrophages play an immediate role in defense against bacterial invasion by phagocytosing and killing pathogens (24), we wanted to determine the role of macrophages in the MLN4924-mediated exacerbation of MRSA infection. Thus, macrophage depletion experiments with clodronate-encapsulated liposomes were conducted as reported in a previous study (19). Macrophages could hardly be detected in liver, lung, and spleen tissues after clodronate-encapsulated liposome treatment (Supplemental Fig. 2A, 2B). Macrophage depletion resulted in a significant decrease in serum levels of the cytokines IL-1β, IL-6, and TNF-α and distinct increases in the MRSA load in liver, spleen, lung, and kidney tissues after MRSA bloodstream infection (Fig. 3A, 3B), indicating the critical role of macrophages in restraining MRSA infection. Except for a slight decrease in serum IL-6 levels in MLN4924-treated mice (Fig. 3A), the bacterial burden and survival rate of MLN4924-treated and control mice after MRSA infection were not different after macrophage depletion (Fig. 3B, 3C). In addition, we infused MLN4924- or DMSO-treated BMDMs into the macrophage-depleted mice via tail veins. Decreased inflammatory cytokines IL-1β and IL-6, increased bacteria burden in liver and lung, and increased abscess in liver could be detected in MLN4924-treated macrophage infusion mice (Fig. 3D–F). However, significant differences in inflammatory cytokine TNF-α, bacteria burden in spleen, and kidney and survival time were not observed (25, 26) (Fig. 3D–G). These results suggest that macrophages play an essential role in the effect of MLN4924.
FIGURE 3.
Macrophages play an essential role in the effect of MLN4924. (A and B) Twelve hours after their i.v. injection of 50 μl of clodronate-encapsulated liposomes, mice were i.p. administered MLN4924 (30 mg/kg). Twelve hours after MLN4924 admission, 1 × 108 CFUs of USA300 were injected into the mice through the tail vein. Lung, liver, kidney, and spleen tissues and blood were harvested 24 h later, and serum IL-1β, TNF-α, and IL-6 levels were measured by ELISA (A). The CFUs in the liver, lungs, spleen, and kidneys were measured (B) (n = 6). (C) Twelve hours after i.v. injection of 50 μl of clodronate-encapsulated liposomes, mice were i.p. administered MLN4924 (30 mg/kg). Twelve hours after MLN4924 admission, 5 × 107 CFUs of USA300 were injected into the mice through the tail vein, and the 10-d survival rate was recorded (n = 6). (D–G) MLN4924 (100 nM)- or DMSO-treated BMDMs (4 × 106 cells per mouse) were infused into the macrophage-depleted mice via tail veins. Twelve hours after macrophages infusion, USA300 (5 × 107 CFUs per mouse for survival rate analysis, 1 × 108 CFUs per mouse for others) was i.v. injected into the mice. Lung, liver, kidney, and spleen tissues and blood were harvested 24 h later, and serum IL-1β, TNF-α, and IL-6 levels were measured by ELISA (D). The CFUs in the liver, lungs, spleen, and kidneys were measured (E) (n = 5), inflammatory cell infiltration and abscess (yellow arrow) in the liver, lungs, and kidneys was visualized by H&E staining (F), and 8-d survival rate was recorded (G) (n = 8). Scale bars, 50 μm. Data are representative of three independent experiments (mean and SD). ns, not significant. *p < 0.05, **p < 0.01, ***p < 0.001 (unpaired Student t test).
MLN4924 inhibits MRSA clearance by macrophages in vitro
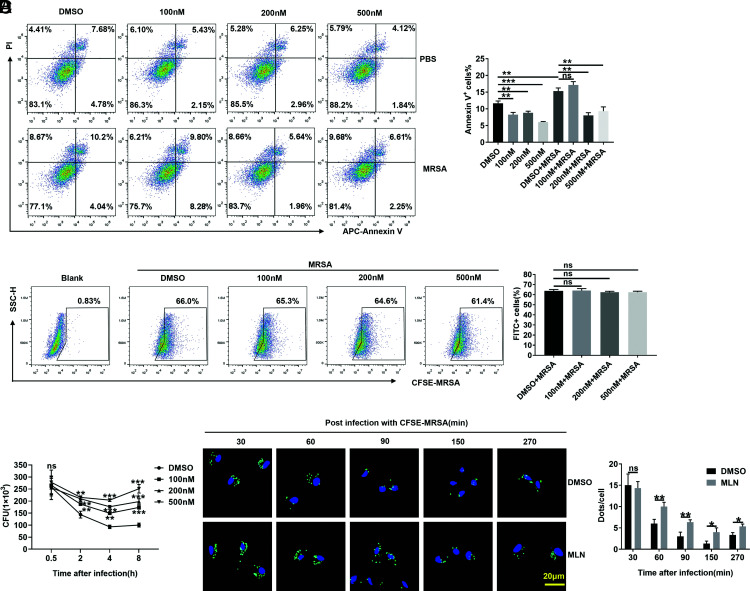
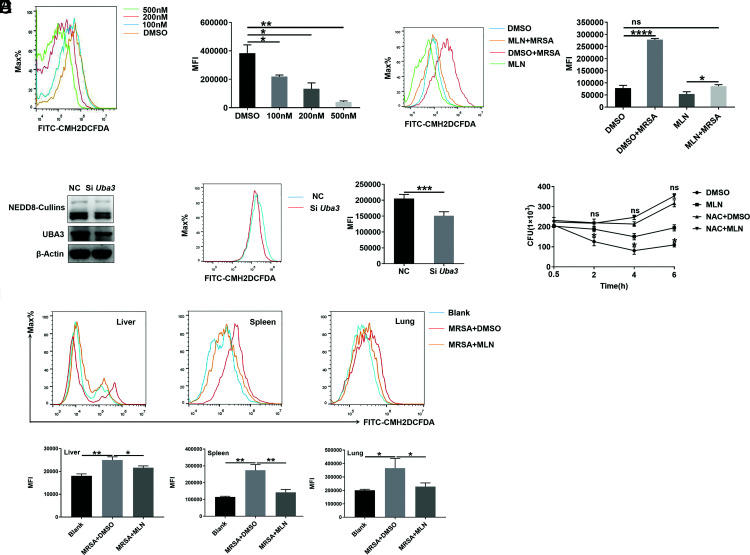
Then, the effects of MLN4924 on macrophages in vitro were examined, and we confirmed that MLN4924 effectively inhibited NEDD8-Cullins in PMs in vitro (Supplemental Fig. 2C). Because MLN4924 could induce the apoptosis of RAW264.7 cells (27), we investigated the effect of MLN4924 treatment on the apoptosis of PMs. We found that MLN4924 did not promote the apoptosis of PMs but rather inhibited apoptosis after MRSA infection (Fig. 4A). An in vivo experiment proved that MLN4924 did not affect the number of macrophages in the liver, spleen, or lungs of mice after MRSA infection (Supplemental Fig. 2D). Then, we hypothesized that MLN4924 might affect bacterial phagocytosis by PMs, but after coculture with PMs for 30 min, we found that MLN4924 did not affect the phagocytosis of strain USA300 (Fig. 4B). However, after 30 min of USA300 infection, when the infected PMs were further cultured, we found that MLN4924 treatment substantially increased the number of live intracellular USA300 bacteria in PMs postinfection (Fig. 4C). Furthermore, when macrophages were visualized by immunofluorescence, we found that MLN4924 treatment indeed inhibited macrophages from clearing USA300 (Fig. 4D). These results demonstrate that MLN4924 impaired MRSA clearance rather than macrophage-mediated engulfment of MRSA.
FIGURE 4.
MLN4924-treated macrophages are defective in USA300 killing. (A) PMs were treated with 100, 200, and 500 nM MLN4924 for 6 h, followed by infection with or without USA300 at an MOI of 20 for 6 h. Apoptotic cells were detected by flow cytometry (left) and statistically analyzed (right). (B) After treatment with 100, 200, and 500 nM MLN4924 for 6 h, PMs were infected with CFSE-labeled USA300 at an MOI of 20 for 30 min. CFSE-positive cells were analyzed by flow cytometry (left) and statistically analyzed (right). (C) After treatment with 100, 200, and 500 nM MLN4924 for 6 h, PMs were infected with CFSE-labeled USA300 at an MOI of 20 for 30 min. The infected cells were further cultured in sterile medium containing 2 µg/ml vancomycin, and CFUs were quantified at the indicated time points after USA300 infection. (D) PMs treated with MLN4924 (100 nM) or DMSO were infected with CFSE-USA300 at an MOI of 20 for 30 min and further cultured in sterile medium containing 2 µg/ml vancomycin. Then, the infected cells were washed, fixed, and stained with DAPI at the indicated times postinfection. CFSE-labeled USA300 (green) was visualized by fluorescence microscopy (left), and dots were analyzed (right). Scale bar, 20 μm. Data are representative of three independent experiments (mean and SD) (n = 3). ns, not significant. *p < 0.05, **p < 0.01, ***p < 0.001 (unpaired Student t test).
ROS induction in macrophages was defective after neddylation inhibition
ROS play a key role in bacterial killing by macrophages, especially in S. aureus infection (28–30). Hence, we explored whether ROS were related to the deficiency in MRSA killing by macrophages observed after MLN4924 treatment. We found that MLN4924 inhibited ROS production in a dose-dependent manner in PMs (Fig. 5A). At a concentration of 500 nM, MLN4924 completely abolished the ROS burst in PMs induced by MRSA infection (Fig. 5B). Only one neddylation E1, a heterodimer of UBA3 and NAE1, has been identified (31, 32). We also blocked neddylation by using Uba3 siRNA and then assessed the role of neddylation in ROS. Uba3 knockdown significantly decreased ROS levels in PMs (Fig. 5C, 5D). Thus, we wondered whether ROS were responsible for the defective antibacterial activity of macrophages upon MLN4924 treatment. NAC, a ROS scavenger (33), efficiently lowered the ROS level in PMs (Supplemental Fig. 3A). Pretreatment with NAC abrogated the difference in the bacterial clearance rate of PMs with and without MLN4924 treatment (Fig. 5E). Next, we investigated the effect of MLN4924 on ROS production in macrophages in mice after MRSA infection in vivo. Consistent with the in vitro results, MLN4924 treatment remarkably reduced ROS levels in macrophages in the liver, spleen, and lungs of mice after MRSA infection (Fig. 5F). Thus, these results indicate that MLN4924 impairs the antibacterial activity of macrophages mainly by reducing their production of ROS.
FIGURE 5.
Macrophages are defective in ROS induction after neddylation inhibition. (A) PMs were treated with 100, 200, and 500 nM MLN4924 for 6 h. Then, the ROS in PMs were analyzed by flow cytometry (left), and the MFI was statistically analyzed (right). (B) PMs were treated with 500 nM MLN4924 or DMSO for 6 h, followed by infection with or without USA300 at an MOI of 20 for 6 h. Then, the ROS in PMs were analyzed by flow cytometry (left), and the MFI values were statistically analyzed (right). (C and D) PMs were transfected with scrambled NC siRNA or Uba3 siRNA for 48 h, followed by USA300 infection at an MOI of 20 for 6 h. Then, NEDD8-Cullins and UBA3 levels were analyzed by Western blot analysis (C). ROS in the cells were analyzed by flow cytometry (left), and MFI values were statistically analyzed (right) (D). (E) PMs pretreated with 5 mM NAC for 0.5 h were treated with 100 nM MLN4924 for 6 h. Then, the cells were infected with USA300 at an MOI of 20, and CFUs were quantified at the indicated time points. (F) Mice were i.p. injected with 30 mg/kg MLN4924 or an equal volume of DMSO. Twelve hours later, the mice were infected with USA300 (1 × 108 CFUs) via tail vein injection. The ROS in macrophages of the liver, spleen, and lungs were analyzed by flow cytometry (top) and statistically analyzed (bottom). Data are representative of three independent experiments (mean and SD). ns, not significant. *p < 0.05, **p < 0.01, ***p < 0.001 (unpaired Student t test).
The MLN4924-mediated decrease in ROS production in macrophages occurred partially via the NEDD8-Cullin3-Nrf2 axis
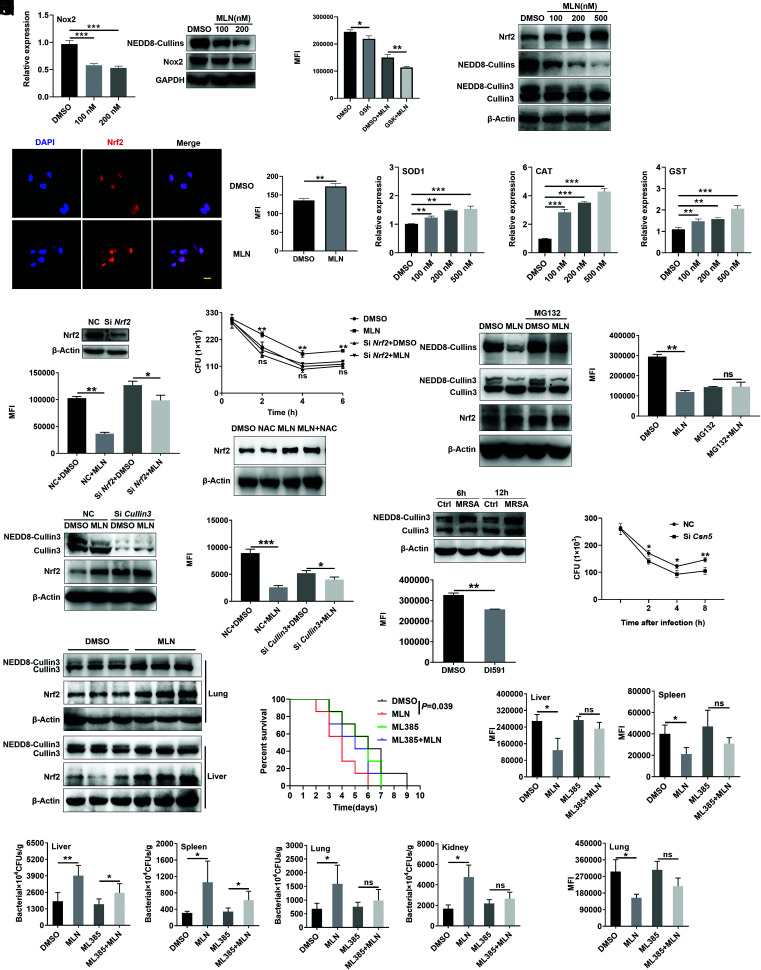
NOX is a major source of ROS production (34). We found that Nox2 mRNA and Nox2 protein levels slightly declined after MLN4924 treatment (Fig. 6A). However, the reduction in ROS in MLN4924-treated macrophages persisted when Nox2 inhibitor GSK2795039 existed (35) (Fig. 6B), implying that Nox2 had a limited effect on the MLN4924-induced decrease in ROS. Nrf2 plays a central role in regulating the expression of antioxidant proteins against oxidative damage triggered by injury and inflammation (36). In addition to downregulated NEDD8-Cullin levels, the Nrf2 protein levels in PMs were markedly increased after MLN4924 treatment (Fig. 6C), which was further confirmed by immunofluorescence staining, which showed enrichment of Nrf2 in the nuclei of PMs (Fig. 6D). We also found higher mRNA levels of Nrf2 target genes, including Sod1, Cat, and Gst, after MLN4924 treatment (Fig. 6E). Moreover, knockdown of Nrf2 reduced the inhibitory effect of MLN4924 on ROS production and bactericidal ability of PMs (Fig. 6F, 6G). After NAC treatment, increased Nrf2 in the MLN4924 group still existed (Fig. 6H). These results suggest that MLN4924 promotes Nrf2 accumulation, which inhibits the production of ROS in macrophages.
FIGURE 6.
MLN4924 decreases ROS partly via the NEDD8-Cullin3-Nrf2 pathway. (A) PMs were treated with 100 and 200 nM MLN4924 for 6 h, followed by USA300 infection at an MOI of 20 for an additional 6 h. The Nox2 level was analyzed by real-time PCR (right) and Western blotting (left). (B) PMs were pretreated with or without GSK2795039 (GSK, a Nox2 inhibitor) for 12 h, treated with or without 100 nM MLN4924 for 6 h, and stimulated with USA300 infection at an MOI of 20 for another 6 h. ROS in these cells were statistically analyzed by flow cytometry. (C–E) PMs were treated with 100, 200, and 500 nM MLN4924 for 6 h, followed by USA300 infection at an MOI of 20 for an additional 6 h. Nrf2, NEDD8-Cullins, and Cullin3 in these cells were detected by immunoblotting (C). The distribution of Nrf2 in PMs treated with 100 nM MLN4924 for 12 h was assessed by immunofluorescence (left) and statistical analysis (right) (D). Scale bar, 10 μm. Sod1, Cat and Gst mRNA levels were analyzed by real-time PCR (E). (F and G) PMs were transfected with NC or Nrf2 siRNA for 48 h, followed by treatment with 100 nM MLN4924 for 6 h. Then, the cells were infected with USA300 at an MOI of 20 for an additional 6 h. The Nrf2 levels were analyzed by Western blotting (top), and ROS in these cells were statistically analyzed by flow cytometry (bottom) (F). CFUs were quantified at the indicated time points after USA300 infection (G). (H) PMs pretreated with or without 5 mM NAC for 0.5 h, followed by 100 nM MLN4924 treatment for 6 h. Then, the cells were infected with USA300 at an MOI of 20 for 6 h. Nrf2 in PMs was detected by immunoblotting. (I and J) PMs were treated with MG132 (10 μM) for 2 h, followed by MLN4924 (100 nM) treatment for an additional 6 h. Then, the cells were incubated with USA300 at an MOI of 20 for an additional 6 h. The protein levels of NEDD8-Cullins, Nrf2, and Cullin3 were analyzed by Western blotting (I). ROS were analyzed by flow cytometry and statistically analyzed in (J). (K and L) PMs were transfected with NC or Cullin3 siRNA for 48 h, followed by 100 nM MLN4924 treatment for 6 h and infection with USA300 at an MOI of 20 for an additional 6 h. The Cullin3 and Nrf2 protein levels were analyzed by Western blot analysis (K). ROS in these cells were analyzed by flow cytometry and statistically analyzed in (L). (M) PMs were infected with USA300 at an MOI of 20 for 6 and 12 h. Then, the Cullin3 and NEDD8-Cullin3 proteins were observed by Western blotting. (N) After DI591 treatment for 12 h and infection with USA300 at an MOI of 20 for 6 h, the ROS in PMs were analyzed by flow cytometry. (O) PMs were transfected with NC or Csn5 siRNA for 48 h, followed by infection with USA300 at an MOI of 20 for 30 min, and CFUs were quantified at the indicated time points after USA300 infection. (P) Mice were i.p. injected with 30 mg/kg MLN4924 or an equal volume of DMSO. Twelve hours later, the mice were infected with USA300 (1 × 108 CFUs) via tail vein. CD11b+F4/80+ macrophages were sorted by flow cytometry 12 h after USA300 administration, and Cullin3 and Nrf2 protein levels were analyzed by Western blotting. (Q–S) Mice were i.p. injected with 30 mg/kg MLN4924 with or without 10 mg/kg ML385, and 12 h later, the mice were infected with USA300 via tail vein injection. The survival rate was recorded (n = 7) (Q), and ROS of macrophages in liver, lungs, spleen (n = 3) (R) and the bacterial loads in the liver, lungs, spleen, and kidneys were measured (n = 5) (S). Data are representative of three independent experiments with similar results. ns, not significant. *p < 0.05, **p < 0.01, ***p < 0.001 (unpaired Student t test).
Then, we wanted to determine how MLN4924 regulates Nrf2 in macrophages. In contrast to Nrf2 protein levels, Nrf2 mRNA levels were significantly decreased in MLN4924-treated PMs (Supplemental Fig. 3B). Nrf2 is degraded in a proteasome-dependent manner, and MG132, a proteasome inhibitor, increased the Nrf2 protein levels in PMs (37). Therefore, we hypothesized that MLN4924 suppressed Nrf2 degradation by proteasomes. In fact, MLN4924 did not increase Nrf2 protein levels in PMs after MG132 treatment (Fig. 6I). Furthermore, the decrease in ROS levels caused by MLN4924 was abolished after MG132 treatment (Fig. 6J). According to past research, p62 can competitively bind to Keap1, leading to the activation of Nrf2 (38). We found that although p62 protein levels were obviously increased in MLN4924-treated PMs (Supplemental Fig. 3C), MLN4924 could still reduce ROS levels in PMs after p62 knockdown (Supplemental Fig. 3D–F), which indicates that the decrease in ROS in MLN4924-treated macrophages was not due to increased p62.
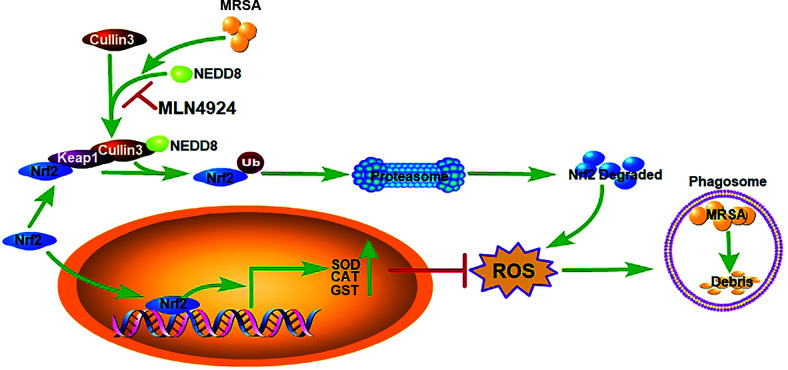
Nrf2 is also negatively regulated by the Cullin3-Keap1-E3 ubiquitin ligase complex, which targets Nrf2 for proteasome-mediated degradation (37). Since MLN4924 obviously inhibited Cullin3 neddylation (Fig. 6C), we wondered whether Cullin3 was responsible for the Nrf2-mediated decrease in ROS in macrophages after MLN4924 treatment. We found that Cullin3 knockdown abolished MLN4924-mediated Nrf2 accumulation in PMs (Fig. 6K). In addition, Cullin3 knockdown largely impaired the inhibitory effects of MLN4924 on the reduction in ROS levels in PMs (Fig. 6L). Furthermore, Cullin3 neddylation was obviously increased in PMs after MRSA infection in vitro (Fig. 6M). DI591, a specific inhibitor that interferes with conjugation between UBE2M (also known as UBC12) and DCN1 (39), also markedly inhibits Cullin3 neddylation (Supplemental Fig. 4A). Accordingly, DI591 treatment significantly inhibited ROS production in PMs after MRSA infection in vitro (Fig. 6N). UBE2M is an E2-conjugating enzyme, and DCN1 is an E3 ligase in Cullin3 neddylation (40–42). To further confirm the function of neddylated Cullin3 in ROS generation, we overexpressed E2-UBE2M (Supplemental Fig. 4B, 4C) or E3-DCN1 (Supplemental Fig. 4D, 4E) in NIH-3T3 cells and found that both could increase Cullin3 neddylation and ROS production. CSN5, an isopeptidase belonging to the COPS9 signalosome, is responsible for recycling NEDD8 from Cullin3-NEDD8 conjugation (43, 44). Silencing of CSN5 notably increased ROS levels (Supplemental Fig. 4F, 4G) and facilitated MRSA clearance in PMs (Fig. 6O). In addition, we found decreased neddylated Cullin3 and increased Nrf2 expression in liver and lung macrophages in mice after MLN4924 treatment (Fig. 6P). After using ML385, a pharmacological inhibitor of Nrf2 (45), the survival rate (Fig. 6Q) and ROS levels in macrophages of liver, spleen, and lungs (Fig. 6S) of the MLN4924 group were substantially improved and showed no significant difference compared with that of DMSO group. However, the difference in bacterial burdens between MLN4924 and DMSO was diminished in the liver, spleen, lungs, and kidneys, but it remained significantly higher in the MLN4924 group in the liver and spleen after cotreatment with ML385 (Fig. 6S). Collectively, these findings show that the inhibitory effect of MLN4924 on ROS production is partially dependent on the NEDD8-Cullin3-Nrf2 axis (Fig. 7).
FIGURE 7.
A proposed working model of the mechanism by which MLN4924 increased mouse susceptibility to MRSA infection. The neddylation of Cullin3 in macrophages was activated upon MRSA infection and abolished by MLN4924, which caused the accumulation of Nrf2 via the ubiquitin-proteasome pathway. Increased nuclear translocation of Nrf2 enhanced expression of Nrf2 target genes such as Sod1, Cat, and Gst, leading to the reduced generation of ROS and exacerbation of MRSA infection.
Discussion
In recent years, accumulating evidence has shown the important role of neddylation in the field of pathogen infection. The growth of influenza A virus in A549 cells was promoted in vitro via activation of the neddylation modification pathway (46), while MLN4924 protected mice against influenza A virus infection by inhibiting neddylation in vivo (47). Neddylation is required for Kaposi’s sarcoma-associated herpesvirus replication in cells, and MLN4924 kills Kaposi’s sarcoma-associated herpesvirus–infected lymphoma cells by inhibiting NF-κB activity (48). In addition, neddylation activation was effectively induced after blood-stage Plasmodium infection, which facilitated IFN-γ induction and the anti-Plasmodium T cell response in a mouse model (11). Enteropathogenic E. coli can selectively deamidate NEDD8, leading to decreased neddylation activation by abolishing the activity of Cullin-RING ubiquitin ligases, which is closely linked to enteropathogenic E. coli infection-induced cytopathic effects (14). In our study, MRSA infection promoted neddylation activation both in vivo and in vitro. Inhibition of neddylation activation by MLN4924 decreased the survival rate, exacerbated organ damage, and increased the bacterial burden. Our results show that neddylation activation plays a protective role against MRSA infection. Although it remains unclear whether neddylation is activated or inhibited and whether an altered neddylation status protects against or promotes infection with different pathogens, our study further confirmed the involvement of neddylation in the pathogenesis of exogenous infection.
Once neddylation was inhibited by MLN4924 in mice, a decreased survival rate, exacerbated organ damage, and an increased bacterial burden were observed, and the exacerbation of these phenomena via suppression of neddylation activation by MLN4924 disappeared when macrophages were depleted by clodronate liposomes. In addition, adoptively transferred MLN4924-treated BMDMs into macrophage-depleted mice will also aggravate MRSA infection in mice. These results suggest that macrophages play an essential role in the exacerbating effects of neddylation suppression on MRSA infection. In vitro, macrophages treated with MLN4924 were defective in MRSA killing but not phagocytosis, and this effect was related to a significant decrease in ROS levels in MLN4924-treated macrophages. In addition, increased neddylation activation induced in NIH-3T3 cells by the overexpression of Ube2m or Dcn1 and induced in macrophages by Csn5 interference facilitated ROS induction. Previous studies have found that abundant ROS production in macrophages was key to their ability to kill MRSA (49), while S. aureus exhibited long-term survival within the intracellular niche of macrophages due to a low level of ROS generation, facilitating its dissemination (50, 51). In addition, exogenous neutralization of ROS with NAC counteracted the MLN4924-induced decrease in MRSA clearance in vitro. Taken together, these results suggest that decreased ROS levels in macrophages after neddylation suppression can explain the increased bacterial burden observed in mice with MRSA infection after MLN4924 intervention and the subsequent serious organ damage and increased mortality.
The conjugation of NEDD8 with Cullin3 is vital for the ubiquitin ligase function of Cullin3-RING ubiquitin ligases (CRL3s) (37). Under normal conditions, Nrf2 is degraded by the proteasome through CRL3. When cells are faced with stress, the degradation of Nrf2 is retarded (52). Once it has accumulated and translocated into the nucleus, Nrf2, a powerful anti-redox transcription factor, promotes the transcription of its downstream antioxidant targets, including NADPH quinone dehydrogenase 1 (NQO1), haem oxygenase-1 (HO-1), and superoxide dismutase (SOD), leading to the inhibition of ROS induction (52). After MRSA was phagocytosed by macrophages, the neddylation of Cullin3 was further activated, which accelerated the degradation of Nrf2 and promoted ROS induction in macrophages. When the degradation of Nrf2 was reduced by MLN4924 during MRSA infection, the balance between Nrf2 and ROS was disrupted, which led to a significant decrease in ROS production. A mechanism schematic diagram for neddylation activation against MRSA infection is shown in (Fig. 7.
The Nrf2 inhibitor could not fully reverse the higher bacterial burdens in vivo after MLN4924 treatment, implying that other contributing factors may exist. We found that the expression of Nox2, which is the main source of ROS, slightly declined in macrophages after MLN4924 treatment in vitro and showed a limited effect on the MLN4924-mediated decrease in ROS in PMs, but its role in vivo deserved further exploration. In addition, Perforin-2 is a pore-forming protein required for the bactericidal activity of ROS in phagocytes (53). Recently, neddylation of Cullin-1 was reported to trigger a rapid redistribution of Perforin-2 and was important for its normal function (54). Thus, perforin-2 may participate in MLN4924-mediated ROS regulation, which needs further study.
Because of its good antitumor effect in vitro and in vivo, the neddylation inhibitor MLN4924 has been approved for phase II clinical trials as an antitumor drug for the treatment of human acute myeloid leukemia, non–small cell lung cancer, and mesothelioma (55–57). However, according to our research, caution should be used when MLN4924 is systematically applied in patients with MRSA infection. To the best of our knowledge, specific activators of neddylation are not currently available. The COP9 signalosome possesses metalloisopeptidase activity, which efficiently deneddylates NEDD8 from the Cullins family (58, 59). Recently, Klebsiella pneumoniae was found to prevent the neddylation of Cullin1 of the ubiquitin ligase E3-SCF-TrCP complex by inducing CSN5 expression (60). CSN5 is also an important regulator of parasite protein degradation and participates in the effect of zinc ditiocarb against Entamoeba histolytica (61). Our results proved that silencing the Csn5 gene facilitated ROS induction and increased bactericidal capacity in mouse PMs. Therefore, targets of the COP9 signalosome may be an efficient way to develop neddylation activators and therapeutic chemical agents for pathogens (62).
In summary, neddylation activation in macrophages plays a critical protective role against MRSA infection via ROS-mediated bactericidal effects, and this effect may be partially associated with a signaling pathway consisting of the NEDD8-Cullin3-Nrf2-ROS axis. These results provide a promising nonantibiotic strategy for treating MRSA infection by targeting neddylation activation.
Supplementary Material
This work was supported by National Key Research and Development Program of China Stem Cell and Translational Research (Stem Cell and Translational Research) Grant 2016YFA0501800 and Natural Science Foundation of Zhejiang Province and Grant LY19H150007 (to G.Z.), as well as by National Natural Science Foundations of China partial Grants 81971871 (to G.Z.) and 81901941 (to S.Z.).
The online version of this article contains supplemental material.
- BMDM
- bone marrow-derived macrophage
- CSN5
- COP9 signalosome subunit 5
- MFI
- mean fluorescence intensity
- MRSA
- methicillin-resistant S. aureus
- NAE
- NEDD8-activating enzyme
- NC
- negative control
- NEDD8
- neural precursor cell-expressed developmentally downregulated 8
- NEDD8-Cullins
- NEDD8-conjugated Cullins
- NOX
- NADPH oxidase
- Nrf2
- NF erythroid-derived 2–like 2
- ROS
- reactive oxygen species
- siRNA
- small interfering RNA
Disclosures
The authors have no financial conflicts of interest.
References
- 1. Chambers H. F., Deleo F. R.. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grundmann H., Aires-de-Sousa M., Boyce J., Tiemersma E.. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368: 874–885. [DOI] [PubMed] [Google Scholar]
- 3. Vikesland P., Garner E.. 2019. Differential drivers of antimicrobial resistance across the world. Acc. Chem. Res. 52: 916–924. [DOI] [PubMed] [Google Scholar]
- 4. Holmes A. H., Moore L. S., Sundsfjord A., Steinbakk M., Regmi S., Karkey A., Guerin P. J., Piddock L. J.. 2016. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387: 176–187. [DOI] [PubMed] [Google Scholar]
- 5. Kamitani T., Kito K., Nguyen H. P., Yeh E. T.. 1997. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 272: 28557–28562. [DOI] [PubMed] [Google Scholar]
- 6. Zhou X., Tan M., Nyati M. K., Zhao Y., Wang G., Sun Y.. 2016. Blockage of neddylation modification stimulates tumor sphere formation in vitro and stem cell differentiation and wound healing in vivo. Proc. Natl. Acad. Sci. USA 113: E2935–E2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vogl A. M., Brockmann M. M., Giusti S. A., Maccarrone G., Vercelli C. A., Bauder C. A., Richter J. S., Roselli F., Hafner A. S., Dedic N., et al. 2015. Neddylation inhibition impairs spine development, destabilizes synapses and deteriorates cognition. Nat. Neurosci. 18: 239–251. [DOI] [PubMed] [Google Scholar]
- 8. Zubiete-Franco I., Fernández-Tussy P., Barbier-Torres L., Simon J., Fernández-Ramos D., Lopitz-Otsoa F., Gutiérrez-de Juan V., de Davalillo S. L., Duce A. M., Iruzubieta P., et al. 2017. Deregulated neddylation in liver fibrosis. Hepatology 65: 694–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Enchev R. I., Schulman B. A., Peter M.. 2015. Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 16: 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Watson I. R., Irwin M. S., Ohh M.. 2011. NEDD8 pathways in cancer, sine quibus non. Cancer Cell 19: 168–176. [DOI] [PubMed] [Google Scholar]
- 11. Cheng Q., Liu J., Pei Y., Zhang Y., Zhou D.. 2018. Neddylation contributes to CD4+ T cell-mediated protective immunity against blood-stage Plasmodium infection. PLoS Pathog. 14: e1007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang F. M., Reyna S. M., Granados J. C., Wei S. J., Innis-Whitehouse W., Maffi S. K., Rodriguez E., Slaga T. J., Short J. D.. 2012. Inhibition of neddylation represses lipopolysaccharide-induced proinflammatory cytokine production in macrophage cells. J. Biol. Chem. 287: 35756–35767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fu Z., Liao W., Ma H., Wang Z., Jiang M., Feng X., Zhang W.. 2019. Inhibition of neddylation plays protective role in lipopolysaccharide-induced kidney damage through CRL-mediated NF-κB pathways. Am. J. Transl. Res. 11: 2830–2842. [PMC free article] [PubMed] [Google Scholar]
- 14. Cui J., Yao Q., Li S., Ding X., Lu Q., Mao H., Liu L., Zheng N., Chen S., Shao F.. 2010. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science 329: 1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Catic A., Misaghi S., Korbel G. A., Ploegh H. L.. 2007. ElaD, a deubiquitinating protease expressed by E. coli. PLoS One 2: e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Misaghi S., Balsara Z. R., Catic A., Spooner E., Ploegh H. L., Starnbach M. N.. 2006. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol. Microbiol. 61: 142–150. [DOI] [PubMed] [Google Scholar]
- 17. Wynn T. A., Chawla A., Pollard J. W.. 2013. Macrophage biology in development, homeostasis and disease. Nature 496: 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rom J. S., Atwood D. N., Beenken K. E., Meeker D. G., Loughran A. J., Spencer H. J., Lantz T. L., Smeltzer M. S.. 2017. Impact of Staphylococcus aureus regulatory mutations that modulate biofilm formation in the USA300 strain LAC on virulence in a murine bacteremia model. Virulence 8: 1776–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bader J. E., Enos R. T., Velázquez K. T., Carson M. S., Nagarkatti M., Nagarkatti P. S., Chatzistamou I., Davis J. M., Carson J. A., Robinson C. M., Murphy E. A.. 2018. Macrophage depletion using clodronate liposomes decreases tumorigenesis and alters gut microbiota in the AOM/DSS mouse model of colon cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 314: G22–G31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu J., Liu B., Mao W., Feng S., Yao Y., Bai F., Shen Y., Guleng A., Jirigala B., Cao J.. 2020. Prostaglandin E2 regulates activation of mouse peritoneal macrophages by Staphylococcus aureus through Toll-like receptor 2, Toll-like receptor 4, and NLRP3 inflammasome signaling. J. Innate Immun. 12: 154–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang P., Geng J.. 2019. Macrophage achieves self-protection against oxidative stress-induced ageing through the Mst-Nrf2 axis. Nat. Commun. 10: 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pan Z. Q., Kentsis A., Dias D. C., Yamoah K., Wu K.. 2004. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23: 1985–1997. [DOI] [PubMed] [Google Scholar]
- 23. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., et al. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458: 732–736. [DOI] [PubMed] [Google Scholar]
- 24. Cole J., Aberdein J., Jubrail J., Dockrell D. H.. 2014. The role of macrophages in the innate immune response to Streptococcus pneumoniae and Staphylococcus aureus: mechanisms and contrasts. Adv. Microb. Physiol. 65: 125–202. [DOI] [PubMed] [Google Scholar]
- 25. Thomas J. A., Pope C., Wojtacha D., Robson A. J., Gordon-Walker T. T., Hartland S., Ramachandran P., Van Deemter M., Hume D. A., Iredale J. P., Forbes S. J.. 2011. Macrophage therapy for murine liver fibrosis recruits host effector cells improving fibrosis, regeneration, and function. Hepatology 53: 2003–2015. [DOI] [PubMed] [Google Scholar]
- 26. Ma P. F., Gao C. C., Yi J., Zhao J. L., Liang S. Q., Zhao Y., Ye Y. C., Bai J., Zheng Q. J., Dou K. F., et al. 2017. Cytotherapy with M1-polarized macrophages ameliorates liver fibrosis by modulating immune microenvironment in mice. J. Hepatol. 67: 770–779. [DOI] [PubMed] [Google Scholar]
- 27. Li L., Liu B., Dong T., Lee H. W., Yu J., Zheng Y., Gao H., Zhang Y., Chu Y., Liu G., et al. 2013. Neddylation pathway regulates the proliferation and survival of macrophages. Biochem. Biophys. Res. Commun. 432: 494–498. [DOI] [PubMed] [Google Scholar]
- 28. Di Cara F., Sheshachalam A., Braverman N. E., Rachubinski R. A., Simmonds A. J.. 2018. Peroxisome-mediated metabolism is required for immune response to microbial infection. [Published erratum appears in 2081 Immunity 48: 832–833.] Immunity 48: 832–833. [DOI] [PubMed] [Google Scholar]
- 29. Fang F. C. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2: 820–832. [DOI] [PubMed] [Google Scholar]
- 30. Buvelot H., Posfay-Barbe K. M., Linder P., Schrenzel J., Krause K. H.. 2017. Staphylococcus aureus, phagocyte NADPH oxidase and chronic granulomatous disease. FEMS Microbiol. Rev. 41: 139–157. [DOI] [PubMed] [Google Scholar]
- 31. Liakopoulos D., Doenges G., Matuschewski K., Jentsch S.. 1998. A novel protein modification pathway related to the ubiquitin system. EMBO J. 17: 2208–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang D. T., Hunt H. W., Zhuang M., Ohi M. D., Holton J. M., Schulman B. A.. 2007. Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature 445: 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ravindran R., Loebbermann J., Nakaya H. I., Khan N., Ma H., Gama L., Machiah D. K., Lawson B., Hakimpour P., Wang Y. C., et al. 2016. The amino acid sensor GCN2 controls gut inflammation by inhibiting inflammasome activation. Nature 531: 523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bedard K., Krause K. H.. 2007. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol. Rev. 87: 245–313. [DOI] [PubMed] [Google Scholar]
- 35. Hirano K., Chen W. S., Chueng A. L., Dunne A. A., Seredenina T., Filippova A., Ramachandran S., Bridges A., Chaudry L., Pettman G., et al. 2015. Discovery of GSK2795039, a novel small molecule nadph oxidase 2 inhibitor. Antioxid. Redox Signal. 23: 358–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ahmed S. M., Luo L., Namani A., Wang X. J., Tang X.. 2017. Nrf2 signaling pathway: pivotal roles in inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 1863: 585–597. [DOI] [PubMed] [Google Scholar]
- 37. Villeneuve N. F., Lau A., Zhang D. D.. 2010. Regulation of the Nrf2-Keap1 antioxidant response by the ubiquitin proteasome system: an insight into Cullin-ring ubiquitin ligases. Antioxid. Redox Signal. 13: 1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Katsuragi Y., Ichimura Y., Komatsu M.. 2016. Regulation of the Keap1–Nrf2 pathway by p62/SQSTM1. Curr. Opin. Toxicol. 1: 54–61. [Google Scholar]
- 39. Zhou H., Lu J., Liu L., Bernard D., Yang C. Y., Fernandez-Salas E., Chinnaswamy K., Layton S., Stuckey J., Yu Q., et al. 2017. A potent small-molecule inhibitor of the DCN1-UBC12 interaction that selectively blocks cullin 3 neddylation. Nat. Commun. 8: 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou W., Ma L., Ding L., Guo Q., He Z., Yang J., Qiao H., Li L., Yang J., Yu S., et al. 2019. Potent 5-cyano-6-phenyl-pyrimidin-based derivatives targeting DCN1-UBE2M interaction. J. Med. Chem. 62: 5382–5403. [DOI] [PubMed] [Google Scholar]
- 41. Zhou H., Zhou W., Zhou B., Liu L., Chern T.-R., Chinnaswamy K., Lu J., Bernard D., Yang C.-Y., Li S., et al. 2018. High-affinity peptidomimetic inhibitors of the DCN1-UBC12 protein-protein interaction. J. Med. Chem. 61: 1934–1950. [DOI] [PubMed] [Google Scholar]
- 42. Kurz T., Chou Y. C., Willems A. R., Meyer-Schaller N., Hecht M. L., Tyers M., Peter M., Sicheri F.. 2008. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell 29: 23–35. [DOI] [PubMed] [Google Scholar]
- 43. Cavadini S., Fischer E. S., Bunker R. D., Potenza A., Lingaraju G. M., Goldie K. N., Mohamed W. I., Faty M., Petzold G., Beckwith R. E., et al. 2016. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 531: 598–603. [DOI] [PubMed] [Google Scholar]
- 44. Wu J.-T., Lin H.-C., Hu Y.-C., Chien C.-T.. 2005. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat. Cell Biol. 7: 1014–1020. [DOI] [PubMed] [Google Scholar]
- 45. Singh A., Venkannagari S., Oh K. H., Zhang Y. Q., Rohde J. M., Liu L., Nimmagadda S., Sudini K., Brimacombe K. R., Gajghate S., et al. 2016. Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem. Biol. 11: 3214–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang T., Ye Z., Yang X., Qin Y., Hu Y., Tong X., Lai W., Ye X.. 2017. NEDDylation of PB2 reduces its stability and blocks the replication of influenza A virus. Sci. Rep. 7: 43691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun H., Yao W., Wang K., Qian Y., Chen H., Jung Y. S.. 2018. Inhibition of neddylation pathway represses influenza virus replication and pro-inflammatory responses. Virology 514: 230–239. [DOI] [PubMed] [Google Scholar]
- 48. Hughes D. J., Wood J. J., Jackson B. R., Baquero-Pérez B., Whitehouse A.. 2015. NEDDylation is essential for Kaposi’s sarcoma-associated herpesvirus latency and lytic reactivation and represents a novel anti-KSHV target. PLoS Pathog. 11: e1004771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abuaita B. H., Schultz T. L., O’Riordan M. X.. 2018. Mitochondria-derived vesicles deliver antimicrobial reactive oxygen species to control phagosome-localized Staphylococcus aureus. Cell Host Microbe 24: 625–636.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Surewaard B. G., Deniset J. F., Zemp F. J., Amrein M., Otto M., Conly J., Omri A., Yates R. M., Kubes P.. 2016. Identification and treatment of the Staphylococcus aureus reservoir in vivo. [Published erratum appears in 2016 J. Exp. Med. 213: 3087.] J. Exp. Med. 213: 1141–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krause K., Daily K., Estfanous S., Hamilton K., Badr A., Abu Khweek A., Hegazi R., Anne M. N., Klamer B., Zhang X., et al. 2019. Caspase-11 counteracts mitochondrial ROS-mediated clearance of Staphylococcus aureus in macrophages. EMBO Rep. 20: e48109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tonelli C., Chio I. I. C., Tuveson D. A.. 2018. Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 29: 1727–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McCormack R. M., de Armas L. R., Shiratsuchi M., Fiorentino D. G., Olsson M. L., Lichtenheld M. G., Morales A., Lyapichev K., Gonzalez L. E., Strbo N., et al. 2015. Perforin-2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria. Elife 4: e06508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McCormack R. M., Lyapichev K., Olsson M. L., Podack E. R., Munson G. P.. 2015. Enteric pathogens deploy cell cycle inhibiting factors to block the bactericidal activity of Perforin-2. eLife 4: e06505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Swords R. T., Erba H. P., DeAngelo D. J., Bixby D. L., Altman J. K., Maris M., Hua Z., Blakemore S. J., Faessel H., Sedarati F., et al. 2015. Pevonedistat (MLN4924), a First-in-Class NEDD8-activating enzyme inhibitor, in patients with acute myeloid leukaemia and myelodysplastic syndromes: a phase 1 study. Br. J. Haematol. 169: 534–543. [DOI] [PubMed] [Google Scholar]
- 56. Swords R. T., Coutre S., Maris M. B., Zeidner J. F.. 2018. Pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, combined with azacitidine in patients with AML. Blood 131: 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shah J. J., Jakubowiak A. J., O’Connor O. A., Orlowski R. Z., Harvey R. D., Smith M. R., Lebovic D., Diefenbach C., Kelly K., Hua Z., et al. 2016. Phase I study of the novel investigational NEDD8-activating enzyme inhibitor pevonedistat (MLN4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin. Cancer Res. 22: 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. von Arnim A. G. 2003. On again-off again: COP9 signalosome turns the key on protein degradation. Curr. Opin. Plant Biol. 6: 520–529. [DOI] [PubMed] [Google Scholar]
- 59. Wei N., Deng X. W.. 2003. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19: 261–286. [DOI] [PubMed] [Google Scholar]
- 60. Sá-Pessoa J., Przybyszewska K., Vasconcelos F. N., Dumigan A., Frank C. G., Hobley L.. 2020. Klebsiella pneumoniae reduces SUMOylation to limit host defense responses. mBio 11: e01733-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ghosh S., Farr L., Singh A.. 2020. COP9 signalosome is an essential and druggable parasite target that regulates protein degradation. PLoS Pathog. 16: e1008952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schlierf A., Altmann E., Quancard J., Jefferson A. B., Assenberg R., Renatus M., Jones M., Hassiepen U., Schaefer M., Kiffe M., et al. 2016. Targeted inhibition of the COP9 signalosome for treatment of cancer. Nat. Commun. 7: 13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.