Abstract
The mutant protein FOXL2C134W is expressed in at least 95% of adult-type ovarian granulosa cell tumors (AGCT) and is considered to be a driver of oncogenesis in this disease. However, the molecular mechanism by which FOXL2C134W contributes to tumorigenesis is not known. Here we show that mutant FOXL2C134W acquires the ability to bind SMAD4, forming a FOXL2C134W/SMAD4/SMAD2/3 complex that binds a novel hybrid DNA motif AGHCAHAA, unique to the FOXL2C134W mutant. This binding induced an enhancer-like chromatin state, leading to transcription of nearby genes, many of which are characteristic of epithelial-to-mesenchymal transition. FOXL2C134W also bound hybrid loci in primary AGCT. Ablation of SMAD4 or SMAD2/3 resulted in strong reduction of FOXL2C134W binding at hybrid sites and decreased expression of associated genes. Accordingly, inhibition of TGFβ mitigated the transcriptional effect of FOXL2C134W. Our results provide mechanistic insight into AGCT pathogenesis, identifying FOXL2C134W and its interaction with SMAD4 as potential therapeutic targets to this condition.
Graphical Abstract

Introduction
Granulosa cell tumors (GCTs) are sex cord stromal neoplasms representing approximately 5% of all ovarian cancers (1,2). GCTs are divided into two different groups based on their histological features and the age of onset. The juvenile GCTs (JGCTs) usually occur before puberty and represent approximately 4% of GCTs (3). The adult-type GCTs (AGCTs) are more prevalent, constituting 96% of GCTs and are most frequently diagnosed in perimenopausal women. AGCTs generally progress slowly, but recurrences 5–10 years after primary treatment are common (4).
The transcription factor FOXL2, is important for sex determination and differentiation during several stages of folliculogenesis (5) and for maintaining granulosa cell identity. FOXL2 is present in the female gonad from the initiation of ovarian determination and throughout reproductive life (6).
A mutation (c.402C>G) in the FOXL2 gene, giving rise to a C to W substitution (C134W) in the protein, is a hallmark of AGCTs (7–9). FOXL2C134W is therefore assumed to be a key driver of pathogenesis in this condition. The mutation is typically monoallelic (8,10) and is also found in the rare male version of GCTs and in some Sertoli-Leydig cell sex cord tumors (SLCTs) (11). Since FOXL2 mutations in AGCT are limited to one codon and always lead to a C134W substitution they are most likely gain-of-function mutations as seen for various kinases and oncogenes (12–14). The fact that mutant FOXL2C134W is found in practically all AGCTs, clearly indicates that the mutation is an early and necessary step in the development of AGCTs. Presently, however, the mechanism through which the FOXL2C134W mutant drives oncogenesis remains elusive.
A recent study (15) suggested that the C134W mutation negatively affects the stability of FOXL2. C134 is positioned in the extreme C-terminal part of the forkhead DNA binding domain, and is not directly involved in DNA binding. Therefore, it has been assumed that the C134W mutation would not affect DNA-binding (16). Blount et al. reported that wildtype FOXL2 binds directly to SMAD3 (but not SMAD2 or SMAD4) contributing to activin-mediated regulation of the FST gene (17). Others have reported that FOXL2 (both wildtype and C134W mutant) and SMAD3 can act synergistically in promoter transactivation studies in vitro, potentially in concert with GATA4 (18,19). Recent promoter transactivation studies have indicated that the C134W mutant interacts with SMAD3 and a FOX-binding DNA element upstream of the CYP19A gene to activate its promoter, whereas the wildtype form of FOXL2 was inactive (20).
Here we sought to define the role of the FOXL2C134W mutant in the pathogenicity of AGCT. We show, that FOXL2C134W acts as a gain-of function mutant acquiring the ability to bind SMAD4 thereby forming an aberrant FOXL2C134W/SMAD complex. This complex associates to with new sites in the genome that feature the hybrid DNA-motif AGHCAHAA to instill a transcriptional signature characterized by epithelial-mesechymal transition (EMT) and induction of various cytokines, oncogenes and factors involved in stemness. Importantly, the abnormal binding of FOXL2C134W to hybrid sites and its derived transcriptional effects observed in cell-lines could also be documented in primary AGCTs. Our results provide mechanistic insight into AGCT pathogenesis, explain why these cancers respond poorly to conventional chemotherapies but may respond to TGFβ-inhibition and provide the basis of new anti-cancer therapies to this condition; by targeting the interaction between mutant FOXL2 C134W and SMAD4 or by targeting FOXL2C134W directly.
Materials and Methods
A detailed description of materials and methods is provided in the Supplementary Information file S1.
Cell Culture.
KGN, HGrC1 and COV434 cells were cultured in DMEM-F12 supplemented with 10% FBS and 100U/ml penicillin and 100μg/ml streptomycin. 293FT, Flp-In T-Rex 293 (life technologies), TIG3 (from the Japanese Cancer Research Resources Bank, Tokyo, Japan), U2OS (ATCC) and HaCaT (Accegen) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with GlutaMaxTM (Gibco) supplemented with 10% heat-inactivated (v/v) fetal bovine serum (FBS) and 1X Pen-Strep (100μg/ml penicillin and 100μg/ml streptinomycin, Gibco). All cell lines used were tested and confirmed to be negative for mycoplasma. All cells were grown in radiation-sterilized dishes (Nunc) and kept in humidified incubators at 37°C with 5% CO2. Cells were passaged 1–4 times a week by detachment from the plate with 0.25% Trypsin-EDTA (1X, Gibco). KGN were established and supplied by dr. Toshihiko Yanase, HGrC1 cells were established and supplied by drs. Akira Iwase and Tomoko Nakamura. COV434 cells were obtained from Sigma-Aldrich (cat no.07071909).
Human AGCTs and Controls
Following approval by Memorial Sloan Kettering Cancer Center’s Institutional Review Board, fresh frozen tissue blocks of AGCTs and other sex cord stromal tumors were retrieved from the Biobank of the Department of Pathology at MSKCC. Written informed consents were obtained from each patient, and samples were anonymized prior to analysis. All cases were reviewed by an expert gynecologic pathologist (R.A.S.). Cases were classified as AGCT, n=5 or other sex cord stromal tumors, n=4 ( two Sertoli-Leydig cell tumors (SLCTs), and two juvenile granulosa cell tumors (JGCTs). FOXL2C134W mutational status was determined by Sanger sequencing as previously described (21). All AGCTs harbored the FOXL2C134W mutation, whereas the SLCTs and JGCTs did not. RNA was extracted from 5 AGCTs, 2 SLCTs, 2 JGCTs and subjected to RNA-sequencing as previously described (22). Fresh frozen tumor tissue from the same five AGCTs was homogenized, formaldehyde-fixed and subjected to ChIP-sequencing.
ChIP-Sequencing.
For ChIP-seq, the ChIP DNA was quantified using Qubit Fluorometer (Life Technologies). 0.5–5ng of ChIP DNA was used to make library using NEBNext Ultra DNA library kit for Illumina (New England Biolabs), according to the supplier’s instructions with AMPure XP beads (Beckman) for the size-selection step. Libraries were sequenced using an Illumina NextSeq 500 (75bp single-end).
RNA Sequencing.
500ng of total RNA was used for library preparation. Sequencing adaptors were added using the Truseq RNA library prep kit v2 (Illumina) according to the manufacturer’s instructions. Library quality was assessed by analysis on a Bioanalyzer 2100 and subsequently sequenced on Illumina HiSeq2000 (75-bp single-end). Fastq datasets were concatenated using the “concatenate” tool (Galaxy tool ID: Cat1, version1.0.0) and trimmed with Trimmomatic version 0.36.0. Sequencing data were aligned via RNAstar using hg38 (canonical) as reference genome and reads were counted using Htseq-count. Differential expression was analysed using Deseq2. Genes were called as differentially expressed for expression changes larger than 2-fold and an FDR ≤0.01. A minimum of three biological replicates of each sample were used.
Data and Software Availability
ChIP-seq and RNAseq data have been submitted to the NCBI Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) with accession numbers GSE138496 and PRJNA576673, for cell line data and tumor data respectively.
Results
FOXL2C134W Interacts with SMAD4
We hypothesized that the pathogenicity of the FOXL2C134W mutant could arise through changes in its interaction with protein partners or DNA. To test whether FOXL2C134W binds to a different repertoire of proteins than wildtype FOXL2, we purified proteins associated with wildtype (WT) FOXL2 or FOXL2C134W mutant (C134W) from FlipIn-293-TREX cells expressing Flag-HA tagged versions of FOXL2 or FOXL2C134W and determined the identity of the co-purified proteins by mass spectrometry.
Among the proteins enriched selectively after FOXL2C134W immunoprecipitation was SMAD4, an essential member of the TGFβ signaling pathways (Fig. 1A) (23,24). A list of proteins identified as interactors of FOXL2 and FOXL2C134W is provided in supplementary file S2. Western blotting confirmed the interaction between SMAD4 and FOXL2C134W (Fig. 1B). In contrast, no binding of SMAD4 was observed with wildtype FOXL2, indicating a unique interaction between FOXL2C134W and SMAD4 (Fig. 1A-B). To further validate the interaction, immunoprecipitation using Flag-antibody led to the presence of SMAD4 in immunoprecipitates from Flag-FOXL2C134W-expressing 293-TREX cells but not from Flag-FOXL2-expressing 293-TREX cells (Fig. 1C). A two-hybrid luciferase assay using U2OS reporter cell lines with pVP16-fused FOXL2 or FOXL2C134W and Gal4-fused SMAD4 showed almost 20-fold higher luciferase activity for FOXL2C134W (Fig. 1D) further supporting the selective interaction between FOXL2C134W and SMAD4. Performing co-immunoprecipitation with different truncated SMAD4 variants in 293-FT cells, we were able to map the interaction site for FOXL2C134W to the linker region of SMAD4 (Fig. 1E, Fig.S1A-B). Besides the acquired ability to bind to SMAD4, we found that FOXL2C134W is more prone to form homodimers than FOXL2 and heterotypic FOXL2-FOXL2C134W dimers also form more readily than FOXL2 homodimers (Fig.S1C).
Figure 1. The FOXL2C134W mutant binds SMAD4.
A. Quantification of nine peptides unique to SMAD4 by mass-spec of proteins immunoprecipitated with wild type or C134W mutant FOXL2 in 293 TREX cells. Experiments were performed in triplicates.
B. WB of proteins immunoprecipitated with Flag-HA-tagged FOXL2 wildtype or C134W mutant, respectively (same experiment as in A).
C. Validation of the interaction between FOXL2C134W and SMAD4. Co-immuno-precipitation (Co-IP) was performed with Flag antibodies in 293 TREX cells expressing Flag-tagged wildtype (WT) or C134W mutant (C134W) FOXL2.
D. Validation of the interaction between FOXL2C134W and SMAD4 using a reporter assay. Data show mean +/− SD of triplicates, p-values determined by a two-tailed parametric t-test are provided.
E. Mapping of the FOXL2C134W interacting site to the linker region of SMAD4 in 293 FT cells.
FOXL2C134W Exhibits a Unique Genome-wide Binding Profile
To investigate whether the amino acid substitution in mutant FOXL2 affects its binding to DNA, we mapped the genome-wide localization of wildtype and FOXL2C134W by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) in the human immortalized granulosa cell line HGrC1 (25). The cells carry the wildtype FOXL2 gene (25), but do not express detectable levels of FOXL2 mRNA and protein (Fig.S1D-E). Genomic binding profiles for both FOXL2 and FOXL2C134W were established from HGrC1 cells ectopically expressing FOXL2 or FOXL2C134W (Fig. 2A, three upper panels). In parallel, we performed ChIP-seq in the AGCT-derived cell line KGN (Fig. 2A, tracks 5–7 from the top) using three different antibodies to FOXL2. The KGN cell-line is derived from an AGCT patient expressing FOXL2C134W from one allele and wild type FOXL2 from the other (26) (Fig.S1F). Although the antibodies were unable to differentiate between FOXL2 and FOXL2C134W (Fig.S2A-D), comparison to the HGrC1 data-sets enabled us to readily distinguish between endogenous wildtype and C134W FOXL2-binding sites in KGN cells. Furthermore, we speculated that the selective interaction observed between FOXL2C134W and SMAD4 might influence the global binding pattern of mutant FOXL2C134W and that this could be regulated by TGFβ-signaling. Therefore genome-wide mappings and subsequent experiments were performed both in the presence and absence of TGFβ-stimulation (Fig. 2B).
Figure 2. A subset of FOXL2C134W mutant peaks occur at hybrid sites.
A. The distribution of C134W mutant and wildtype FOXL2 in HGrC1 cells transduced with empty vector (EV) wildtype (WT) or mutant (C134W) FOXL2 is shown at selected loci. ChIP-seq of endogenous FOXL2 was performed in KGN cells using three FOXL2 antibodies; two mouse monoclonal antibodies (mAb1, mAb2) and a goat polyclonal antibody (pAb). ChIP-seq of endogenous SMAD4 was perfomed in TGFβ-stimulated KGN cells. Black arrows indicate non-canonical (hybrid) FOXL2 peaks. y-axis values are fragments per kilobasepair per million (FPKM).
B. Experimental setup for ChIP-seq profiling of wildtype (WT) and C134W mutant FOXL2 in HGrC1 and KGN cells.
C. Top DNA motifs enriched at canonical FOXL2 sites and at non-canonical (hybrid) FOXL2C134W sites. The central enrichment of the top motifs identified in the FOXL2 dataset (solid line) and in a control dataset (broken line) are depicted with corresponding E-values. A schematic drawing illustrates the similarity between the top-motif enriched in the non-canonical FOXL2C134W dataset and the SMAD3/4 motif and canonical FOXL2 wildtype motif.
D. Ratiometric heatmap showing the weighted log2-fold difference in FOXL2 signal between FOXL2C134W and wildtype (WT) FOXL2 at FOXL2 binding sites in the absence (left) or presence (right) of TGFβ-stimulation. Signal intensities are shown as FPKM. Black triangles indicate peak center.
To characterize differences between the targets of FOXL2 wildtype and FOXL2C134W, peak-finding and clustering was performed on the four different FOXL2 ChIP datasets obtained in HGrC1 cells (FOXL2, FOXL2C134W, FOXL2+TGFβ, FOXL2C134W+TGFβ). This procedure generated a set of 21,516 peaks distributed on 10 clusters (0–9), (Fig.S2E-F). This also revealed that FOXL2C134W target sites could be divided in two main classes; 1) canonical target sites (n=17,264) overlapping with wildtype FOXL2 binding sites as exemplified by the SMAD3 and SMAD7 loci (Fig. 2A), and 2) some divergent non-canonical sites (n=4,252), unique to FOXL2C134W mutant (indicated by black arrows on Fig. 2A). The analysis also revealed a high consistency between endogenous FOXL2 binding sites detected in KGN cells and binding sites found in HGrC1 cells overexpressing FOXL2 in its wildtype or FOXL2C134W mutant forms (Fig. 2A). Thus, both canonical and non-canonical FOXL2 binding sites were present in KGN cells, reflecting the expression of both wild type and mutant FOXL2 (Fig. 2A). Moreover, the three different anti-FOXL2 antibodies employed gave very similar binding profiles in KGN cells (Fig. 2A, track 5–7 from the top). Compared to the low background seen in a ChIP-seq using an unspecific antibody (IgG, Fig. 2A, track 4 from the top), and in conjunction with antibody validation studies (Fig.S2A-D) this indicates a highly specific and consistent mapping of FOXL2 binding sites in the genome. Moreover, the very low background observed for the FOXL2 HGrC1 ChIP in the empty vector condition (EV, Fig. 2A top track) further confirmed the specificity of the FOXL2 ChIP. In the following experiments, FOXL2 ChIPs were performed using the polyclonal anti-FOXL2 antibody.
Non-canonical FOXL2C134W Binding Sites Feature a Potential FOXL2/SMAD Hybrid DNA-binding Motif
To identify putative DNA-binding motifs for the canonical FOXL2 and unusual non-canonical FOXL2C134W-specific peaks, respectively, we subjected the two peak-sets to de-novo motif analysis (27). The sequence WMAACA was highly enriched in both wildtype and mutant FOXL2 peak-sets and corresponds well to the consensus FOX-box binding sequence RYMAAMA (28). In contrast, the motif most highly enriched in the non-canonical FOXL2C134W peak-set was AGHCAHAA, (Fig. 2C). This motif has some similarities to the FOX-box motif (28), but also to the SMAD4/SMAD3 consensus motif, the so-called CAGA-box (29), potentially suggesting that it could be a FOXL2C134W/SMAD hybrid motif (Fig. 2C and Fig.S2G). In the following, this motif will be denoted the hybrid motif and the corresponding binding sites referred to as hybrid sites. Of note in this context, preliminary genome localization data indicated that SMAD4 appeared to be highly enriched at these hybrid sites only (Fig. 2A, bottom track).
The existence of canonical and hybrid sites for FOXL2C134W was also apparent on the global scale as reflected by the mutant to wildtype FOXL2 enrichment ratio (C134W/WT), which was low and high at canonical and hybrid sites, respectively, and seen both in absence or presence of TGFβ signaling (Fig. 2D). The occurrence of the hybrid sites was seen over a range of FOXL2 and FOXL2C134W expression levels and in several independent experiments (Fig.S3A-C).
SMAD4 and SMAD2/3 are Enriched Exclusively at Hybrid Sites by FOXL2C134W
Because of the increased ability of FOXL2C134W to interact with SMAD4 and its association with hybrid DNA binding sites, we speculated that FOXL2C134W, SMAD4 and associated SMAD2/3 would be co-recruited to the FOXL2C134W-specific hybrid sites. To investigate this, we mapped the genome-wide localization of FOXL2, SMAD4 and SMAD2/3 in HGrC1 cells ectopically expressing wildtype FOXL2 or FOXL2C134W in the absence or presence of TGFβ-stimulation. For comparison, ChIP-seq was also performed in the AGCT-derived cell-line KGN +/− TGFβ-stimulation.
SMAD4 and SMAD2/3 were found to be strongly enriched at FOXL2 binding sites where the FOXL2C134W occupancy exceeds that of wildtype FOXL2, e.g. putative hybrid sites, (Fig. 3A-B). In contrast, SMAD4 and SMAD2/3 enrichments were absent or negligible at FOXL2 binding sites where the wildtype signal was equal to or exceeded the C134W signal (canonical sites), (Fig. 3A-B). Interestingly, SMAD4 and SMAD2/3 were enriched at the same sites in KGN cells, which was stimulated by TGFβ (Fig. 3B). This effect was also immediately apparent at individual hybrid loci in both HGrC1 and KGN cells (Fig. 3C and Fig. 3D). Hence, whereas FOXL2 was unable to trigger SMAD4 and SMAD2/3 binding at canonical sites, the FOXL2C134W mutant led to recruitment of SMAD4 and SMAD2/3 at hybrid loci, which was strongly enhanced by TGFβ-stimulation (Fig. 3C). Strikingly, while strong FOXL2 occupancy was observed at some canonical sites in KGN cells (examples shown by open arrowheads, Fig. 3D), SMAD4 and SMAD2/3 were recruited exclusively to hybrid sites (marked by black arrowheads, Fig. 3D). This may seem surprising since both the C134W and wildtype forms of FOXL2 bind at these canonical sites, and because wildtype FOXL2 has been reported to interact with SMAD3 (17). The results are however perfectly consistent with our hypothesis that the non-canonical FOXL2C134W sites are hybrid sites requiring combined binding of FOXL2C134W, SMAD4 and SMAD2/3. The recruitment of endogenous SMAD4 exclusively to hybrid sites was also apparent on global scale (Fig. 3E) and was confirmed by ChIP-qPCR in both HGrC1 and KGN cells (Fig. 3F-G).
Figure 3. SMAD4 and SMAD2/3 are highly enriched at hybrid sites.
A. Pseudocolored 2D-histograms showing local SMAD4 and SMAD2/3 ChIP-seq signal in TGFβ-stimulated HGrC1 cells in relation to the log10 distance from each FOXL2 peak to the nearest TSS (x-axis) as well as the log10 ratio between C134W mutant and wildtype FOXL2 (y-axis). Coloring shows the average levels of SMAD4 and SMAD2/3 at local areas.
B. Pseudo-colored 2D-histograms showing local SMAD4 and SMAD2/3 ChIP-seq signal in TGFβ-stimulated KGN cells in relation to the log10 distance from each FOXL2 binding site to the nearest TSS (x-axis) as well as the log10 ratio between C134W mutant and wildtype FOXL2. Y-axis determined in HGrC1 cells).
C. ChIP-seq tracks of the ACVR1/ACVR1C locus in HGrC1 cells transduced with either wildtype (WT) or mutant (C134W) FOXL2 in the presence or absence of TGFβ-stimulation. Y-axis values are FPKM.
D. ChIP-seq tracks at various hybrid FOXL2 targets, in KGN cells (unstimulated or stimulated with TGFβ) and in HGrC1 cells transduced with wildtype (WT) or mutant (C134W) FOXL2. Hybrid peaks are indicated by black arrowheads. Selected canonical peaks are indicated by open arrowheads. Y-axis values are FPKM.
E. Ratiometric heatmap showing the weighted log2-fold difference in SMAD4 signal between FOXL2 C134W mutant and wildtype at FOXL2 binding sites in the absence or presence of TGFβ-stimulation. Signal intensities are shown as FPKM. Black triangles indicate peak center.
F. ChIP-qPCR using an unspecific antibody (IgG) and antibodies to FOXL2, SMAD4 and SMAD2/3-binding at various hybrid FOXL2 targets in HGrC1 cells transduced with empty vector (EV) wildtype (WT) or mutant (C134W) FOXL2 and after 24hrs TGFβ or mock-stimulation. Data are represented as % enrichment of input, mean ±SD, n=3, p-values determined by a two-tailed parametric t-test are provided.
G. ChIP-qPCR using an unspecific antibody (IgG) and antibodies to FOXL2, SMAD4 and SMAD2/3-binding at various hybrid FOXL2 targets in KGN cells after 24hrs TGFβ or mock-stimulation. Data are represented as % enrichment of input, mean ±SD, n=3, p-values determined by a two-tailed parametric t-test are provided.
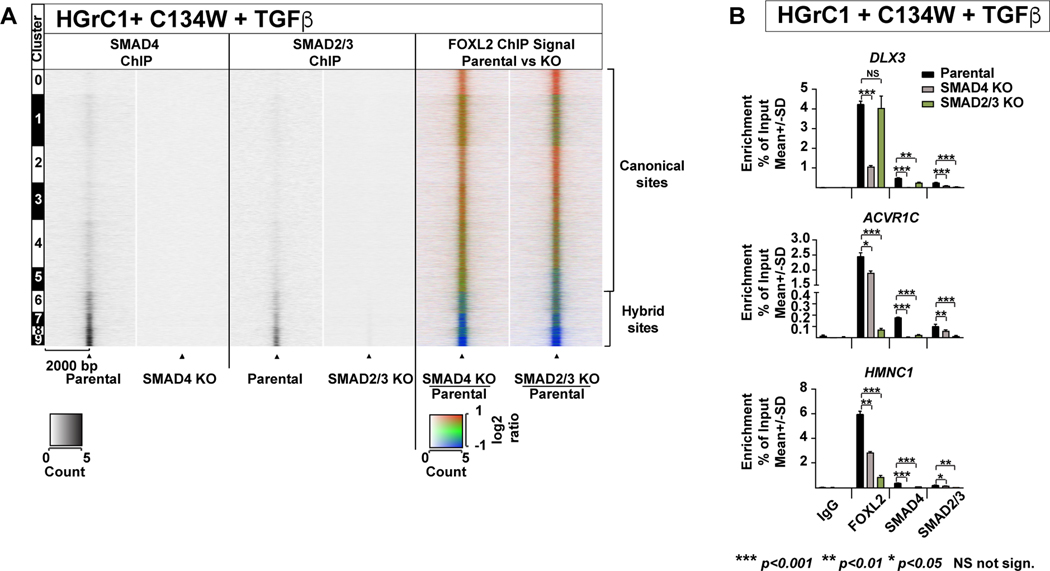
Binding of FOXL2C134W to Hybrid Sites is Dependent on SMAD4 and SMAD2/3
To test if FOXL2C134W binding at the putative hybrid sites is dependent on SMAD4 and SMAD2/3, we established SMAD4 and SMAD2/3 knockout HGrC1 cells. Next, we expressed FOXL2C134W in parental HGrC1 cells (wildtype for SMAD4 and SMAD2/3) and in HGrC1 SMAD4-KO and SMAD2/3-KO cells and performed ChIP-seq analysis for FOXL2, SMAD4 and SMAD2/3.
The knockout of SMAD4 or double knockout of SMAD2/3 strongly reduced FOXL2C134W binding to hybrid sites (Fig. 4A-B, Fig.S4A-E). In stark contrast, FOXL2C134W enrichment was either unaltered or increased at canonical loci in response to SMAD4 or SMAD2/3 knockout (Fig. 4A, the two pseudocolored panels to the right and Fig.S4B and E), indicating a re-distribution of the FOXL2C134W mutant from hybrid to canonical sites in the absence of SMAD4 and SMAD2/3. Of note, while SMAD4 deletion in general had little effect on SMAD2/3 binding to DNA, SMAD2/3 deletion led to a global decrease of SMAD4 binding to chromatin affecting both hybrid and canonical sites (Fig. S4E). Taken together, these results strongly support the hypothesis that putative hybrid FOXL2C134W sites are indeed “true” FOXL2C134W/SMAD hybrid DNA binding sites.
Figure 4. Depletion of SMAD4 or SMAD2/3 reduces the binding of FOXL2C134W to hybrid sites.
A. Parental HGrC1 cells (wildtype for SMAD4 and SMAD2/3) or HGrC1 cells KO for SMAD4 or double KO for SMAD2/3, all expressing FOXL2C134W were stimulated with TGFβ for 24hours and harvested for ChIP-seq. From left to right: 1. Heatmap showing the SMAD4 signal in parental HGrC1 and 2. SMAD4-KO HGrC1. 3. Heatmap showing the SMAD2/3 signal in parental HGrC1 and 4. SMAD4-2/3 KO HGrC1. 5. Ratiometric heatmap showing the weighted log2 fold difference in FOXL2 signal between the parental and SMAD4-KO HGrC1 cells and 6. between parental and SMAD2/3 knockout HGrC1 cells. Signal intensities are shown as FPKM. Black triangles indicate peak center.
B. ChIP-qPCR using an unspecific antibody (IgG) and antibodies to FOXL2, SMAD4 and SMAD2/3-binding at various hybrid FOXL2 targets in parental, SMAD4-KO or SMAD2/3-KO HGrC1 cells transduced with mutant FOXL2 and stimulated with TGFβ. Data are represented as % enrichment of input, mean ±SD, n=3, p-values determined by a two-tailed parametric t-test are provided.
FOXL2C134W Induces an Enhancer-like Chromatin State at Hybrid Sites
Having established that the FOXL2C134W/SMAD complex binds hybrid sites, we were intrigued by the idea that the FOXL2C134W mutant complex would regulate transcription by binding to promoters or enhancers at these locations or even leading to the formation of novel enhancer-like elements at these sites. To investigate this, we mapped H3K27 acetylation (H3K27ac) and H3K4 monomethylation (H3K4me1) sites; two histone modifications characteristic of enhancers and promoters, in KGN and HGrC1 cells.
Ectopic expression of FOXL2C134W in HGrC1 cells caused a strong induction of H3K27ac at hybrid sites (Fig.S5A), which was further enhanced by TGFβ-stimulation (Fig.S5A,C and D). In contrast, ectopic expression of wildtype FOXL2 (WT) either had little or no effect on H3K27ac accumulation at these sites (Fig.S5A,C,D). Moreover, a strong induction of H3K27ac was observed in KGN cells at hybrid sites in response to TGFβ-stimulation, most likely reflecting the effect of the endogenous FOXL2C134W mutant (Fig.S5B-C). This induction of H3K27ac was not observed at canonical FOXL2 sites (Fig.S5C). The ability of FOXL2C134W to induce H3K27ac was generally observed at hybrid DNA binding sites, and was confirmed by ChIP-qPCR on individual sites (Fig.S5C-D). Similarly, the FOXL2C134W mutant triggered formation of H3K4me1 at hybrid sites (Fig.S5C-D). Furthermore, the dip in histone mark (H3K27ac and H3K4me1) levels observed at the FOXL2 apex and most apparent at hybrid loci, (Fig.S5C) is an indication of nucleosome depletion and a further sign of transcriptional competence.
The requirement of SMAD4 and SMAD2/3 to attain a fully functional aberrant FOXL2C134W/SMAD complex, was underscored by the fact that depletion of SMAD4 or SMAD2/3 compromised the ability of FOXL2C134W to induce H3K27ac at hybrid loci (Fig.S5E-F).
Collectively, these experiments demonstrate that the FOXL2C134W/SMAD complex induces an enhancer-like chromatin state at FOXL2C134W/SMAD hybrid sites as defined by a high occupancy of H3K4me1 and H3K27ac.
FOXL2C134W Induces the Expression of Genes Associated with Hybrid DNA binding Sites
Since binding of FOXL2C134W to hybrid sites triggered the formation of an enhancer-like state at such areas, we speculated that FOXL2C134W would induce transcription of genes associated to such sites. To determine the transcriptional effects of ectopic expression of FOXL2 and FOXL2C134W in HGrC1 cells, we performed RNA-seq analysis. As shown in Figure 5A-B, genes associated to hybrid DNA binding sites were generally transcriptionally induced by FOXL2C134W as compared to wildtype FOXL2 and TGFβ-stimulation further enhanced their expression (Fig. 5A, two left panels). In KGN cells, genes associated with hybrid sites were also generally induced in response to TGFβ-stimulation, whereas genes associated with canonical FOXL2 sites were generally downregulated by FOXL2C134W (Fig. 5A, right panel).
Figure 5. Transcription is induced at genes associated with hybrid FOXL2C134W sites.
A. Pseudo-colored 2D-histograms showing the log10-fold transcriptional change of the nearest gene in relation to the log10 distance from each FOXL2 binding site to the nearest TSS (x-axis) as well as the log10 ratio between C134W mutant and wildtype (WT) FOXL2 (y-axis). Coloring shows the average transcriptional change (C134W vs WT) in HGrC1 cells in the absence or presence of TGFβ and the average transcriptional change (TGFβ vs Mock) in KGN cells.
B. Upper left panel: Histogram showing the percentage of canonical (Can) or hybrid (Hyb) FOXL2 peaks, associated with genes significantly (p<0.05) upregulated (Up) or downregulated (Down) in TGFβ stimulated HGrC1 cells transduced with C134W FOXL2 vs WT FOXL2.
Lower left panel: Box and whiskers plot showing the log2-fold transcriptional change (C134W vs WT) of genes associated with canonical (Can) or hybrid (Hyb) FOXL2 peaks in TGFβ stimulated HGrC1 cells transduced with C134W FOXL2 or WT FOXL2. Data are represented as median and 10–90% percentile.
Upper right panel: Histogram showing the percentage of canonical (Can) or hybrid (Hyb) FOXL2 peaks, associated with genes significantly (p<0.05) upregulated (Up) or downregulated (Down) respectively in TGFβ vs mock-stimulated KGN cells.
Lower right panel: Box and whiskers plot showing the log2-fold transcriptional change (TGFβ vs mock) of genes associated with canonical (Can) or hybrid (Hyb) FOXL2 peaks in TGFβ or mock -stimulated KGN cells. Data are represented as median and 10–90% percentile.
C. Quantitative real-time PCR of genes associated with hybrid FOXL2 sites in HGrC1 cells transduced with either an empty vector control (EV) wildtype FOXL2 (WT) or FOXL2C134W (C134W) after 24hrs TGFβ or mock-stimulation. Data are represented as fold change vs EV, mean ±SD, n=3, p-values determined by a two-tailed parametric t-test are provided.
D. Upper row: Enrichment of EMT gene sets in TGFβ-stimulated HGrC1 cells; C134W vs WT or C134W vs EV and in KGN cells (TGFβ vs mock) as revealed by GSEA.
Lower row: Enrichment of EMT gene sets in HGrC1 cells; C134W vs WT or C134W vs EV
The respective nominal p-values false discovery rate (FDR) q-values and family-wise error rate (FWER) p-values are provided.
Moreover, in both HGrC1 and KGN cells, the majority of genes near hybrid sites were transcriptionally induced (69% and 66% in HGrC1 cells and KGN cells, respectively, (Fig. 5B upper panels)). In contrast, genes close to canonical FOXL2 sites were more or less equally induced and repressed (Fig. 5B, upper panels). Furthermore, the transcriptional induction of genes associated with hybrid sites, in response to FOXL2C134W was significantly higher than for genes associated with canonical sites (Fig. 5B, lower panels).
FOXL2C134W Induces Genes involved in Cancer, Stemness and Epithelial-to-Mesemchymal Transition
Genes significantly upregulated by FOXL2C134W and associated with hybrid DNA binding sites included genes induced in ovarian cancer and or AGCT and genes encoding various cytokines, hormone receptors and oncoproteins, many of which not previously associated with FOXL2 function (Fig. 5C, Table S1). In contrast, genes significantly downregulated by FOXL2C134W expression were typically associated with canonical FOXL2 sites or were not direct targets of FOXL2 (wildtype or C134W mutant), (Table S1). Many genes involved in stemness, including the cancer stem cell (CSC) markers DCLK1, IL23A, CXCR4 and SOX4 were induced and found to be targets of FOXL2C134W. Among these, IL-23A has been reported to be highly expressed in ovarian CSCs and shown to maintain the tumor initiating capacity of ovarian CSCs in-vivo and mediate their self-renewal capability, presumably by activating the STAT3 and NF-κB signaling pathways (30). Also, CXCR4 has been shown to promote proliferation of ovarian CSC-like cells (31). The well-known ovarian CSC marker CD24 (32–34) is also strongly induced by FOXL2C134W, although FOXL2 does not appear to bind to its regulatory sequences. In addition to these CSC markers, some stem cell markers including PAX6, CXCL6, and POU2F2 were also induced by FOXL2C134Was was the stem cell reprogramming factor PBX1.
Using gene set enrichment analysis, we found that gene sets regulating epithelial-to-mesenchymal transition (EMT) were significantly enriched in HGrC1 cells expressing FOXL2C134W compared to both wildtype FOXL2 or empty vector control (Fig. 5D, upper row, left and middle panel). Similarly, EMT gene sets were enriched in KGN cells in response to TGFβ-stimulation (Fig. 5D, upper row, right panel). As examples of genes associated to hybrid peaks induced by FOXL2C134W that are known to promote EMT were IL6, CXCL1, MYO6 and FAS and the previously mentioned stemness markers CXCL6, DCLK1 and IL23A. Since TGFβ-stimulation per se may induce EMT, and given that the transcriptional effect seen in response to FOXL2C134W is strongly dependent upon SMAD4 and SMAD2/3 we cannot exclude the possibility that induction of EMT by FOXL2C134W may at least partially be caused by TGFβ-signalling. However, it is noteworthy that EMT gene sets were enriched also in response to FOXL2C134W alone (Fig. 5D, lower row).
Interestingly, many genes associated with canonical FOXL2 binding sites were found to be negatively regulated in response to FOXL2C134W. Some of these genes are negative regulators of EMT (genes marked in red, Table S1) and include DKK1 and PTX3.
Among the genes upregulated by FOXL2C134W were also the known markers of granulosa cell tumors AMH, ACVR1C, FST and PGR, ovarian cancer markers; CYP2S1 and GRIN2A as well as oncogenes and genes promoting metastases or other aspects of oncogenesis as MYB, PTGER2, HERC2, CYP24A1, IL6, among others (Table S1). Of further interest, various genes encoding members of the ABC-transporter pumps previously implicated in CSC-mediated drug-resistance (35) were induced by expression FOXL2C134W in HGrC1 cells, including ABCA1,3,4,5. Similarly, the stem cell reprogramming factor, PBX1 that has been reported to fulfill an essential role, in mediating chemoresistance in ovarian carcinomas (36) was induced by FOXL2C134W.
The Transcriptional Effects of FOXL2C134W are Dependent on SMAD4, SMAD2/3 and TGFβ Signaling
To test if the transcriptional effects of FOXL2C134W are dependent SMAD4 or SMAD2/3, we measured the transcriptional response to FOXL2C134W expression in parental HGrC1 cells and HGrC1 SMAD4 or SMAD2/3 knockout HGrC1 cells. As shown in Figures 6A-B, knockout of the SMAD proteins completely abolished the ability of FOXL2C134W to activate gene expression. Furthermore, TGFβ-inhibition strongly impaired the induction of target genes in response to FOXL2C134W in HGrC1 cells and of their expression in KGN cells (Fig. 6C-E).
Figure 6. Depletion of SMAD4, SMAD2/3 or TGFβ inhibition mitigates the transcriptional effect of the FOXL2C134W. FOXL2 is required for growth/survival of KGN cells.
A. RT-qPCR of selected genes associated with hybrid FOXL2 sites in parental or SMAD4-KO HGrC1 cells transduced with FOXL2C134W +/−TGFβ. Data are fold-change vs parental unstimulated HGrC1, mean±SD, n=3, p-values determined by a two-tailed parametric t-test are provided.
B. RT-qPCR of selected genes associated with hybrid FOXL2 sites in parental or SMAD2/3-KO HGrC1 cells transduced with FOXL2C134W and stimulated with TGFβ. Data are fold-change vs parental unstimulated HGrC1, mean±SD, n=3, p-values determined by a two-tailed parametric t-test are provided.
C. Effect of TGFβ-Inhibitors (TGFI) on canonical and hybrid FOXL2 targets as evaluated by RT-qPCR. Selected genes associated with hybrid FOXL2 sites in HGrC1 cells transduced with either an empty vector control (EV) wildtype FOXL2 (WT) or FOXL2C134W (C134W), +/−TGFβ. Data are fold-change vs EV, mean±SD, n=3, p-values determined by a two-tailed parametric t-test are provided.
D. Effect of TGFI on hybrid FOXL2 targets in HGrC1 cells by RT-qPCR. HGrC1 cells transduced as indicated, +/−TGFβ. Data are fold-change vs unstimulated (mock) EV, mean±SD, n=3, p-values determined by a two-tailed parametric t-test are provided.
E. Effect of a TGFI on selected hybrid FOXL2 targets in KGN cells as evaluated by RT qPCR. KGN cells +/−TGFβ in the presence or absence of TGFI. Data are fold-change vs mock, mean±SD, n=3, p-values determined by a two-tailed parametric t-test are provided.
F. KGN-Cas9-expressing cells were transduced with three different sgRNAs targeting FOXL2, an sgRNA targeting SMAD4, a non-targeting sgRNA as negative control (NT) or sgRNAs to the common essential genes PRMT5, HEATR1 and RPS19.The percentage of GFP-positive cells was measured 5 days after transduction (day 0). Graph shows percentage of GFP-positive cells relative to day 0.
FOXL2 is Required for Proliferation of KGN Cells
Our previous efforts to knockout FOXL2 in KGN cells were not successful, which may reflect the requirement for the oncogenic driver FOXL2C134W to support proliferation. To test this more rigorously, we stably expressed Cas9 in KGN cells and subsequently transduced them with lentiviral vectors expressing GFP and sgRNAs either targeting FOXL2, SMAD4, known essential genes (positive controls) or non-targeting sgRNAs (negative controls) and monitored GFP expression over time. GFP percentages dropped for cells containing sgRNAs targeting FOXL2 and for sgRNAs targeting common essential genes (PRMT5, HEATR1, MDM2 and RPS19), while staying stable in cells with non-targeting sgRNAs. GFP percentages were also decreased, although to a lesser extent, for an sgRNA targeting SMAD4 (Fig. 6F). Based on these results, we conclude that FOXL2 is required for the proliferation of KGN cells, which is consistent with a recent results reporting decreased proliferation of KGN cells depleted for FOXL2 (37).
FOXL2 is Enriched at Hybrid Binding Sites in AGCT Tissues
Having established that the binding of FOXL2C134W to hybrid DNA sites causes induction of nearby genes in cellular models of AGCT, we investigated whether similar effects could be observed in primary AGCT tumor tissues.
To address this, tumor tissues from five patients with AGCT, all carrying somatic FOXL2C134W mutations, and from four patients with other ovarian sex cord stromal tumors (SLCTs and JGCTs), wildtype for FOXL2 were assessed. RNA extracted from all AGCTs and SLCTs/JGCTs was subjected to RNA-seq analysis, and chromatin was prepared from the five AGCT samples and used for FOXL2 ChIP-seq analysis. ChIP failed for one of five AGCT samples. Although the ChIP-seq signals from AGCT tumor material were weak, when compared to cell-lines, our results show that a subset of FOXL2 DNA binding sites were significantly enriched in the different AGCT tumor samples (consensus targets, n=1,040), (Fig. 7A-C, Fig.S6A-C). Strikingly, the strongest and most consistent enrichment of FOXL2 was found at the hybrid DNA binding sites we previously identified in HGrC1 cells (Fig. 7A-C). Furthermore, we observed a significant overlap between target genes transcriptionally affected in primary AGCT tumors, HGrC1 cells expressing FOXL2C134W and in KGN cells in response to TGFβ-stimulation (Fig. 7D-F). Taken together, these results show that the binding of FOXL2C134W to hybrid sites and its derived transcriptional effects observed in cell lines are consistent with those present in primary AGCTs.
Figure 7. FOXL2 is enriched at hybrid binding sites in primary AGCT tissues.
A. FOXL2 ChIP-seq tracks at hybrid target genes in primary AGCT tissues, derived from 3 AGCT patients (AGCT1–3). HGrC1 cells transduced with WT or C134W FOXL2 and KGN cells are included for comparison. Y-axis values are FPKM.
B. Heatmap showing the signal in primary tumor tissue from 3 different AGCT patients at FOXL2 binding sites. FOXL2 binding sites were clustered into consensus and non-consensus peaks according to the overall FOXL2 signal in the four AGCT samples. Y-axis values are FPKM.
C. Parameter plot providing the corresponding log2 ratio of C134W to WT FOXL2 in TGFβ-stimulated HGrC1 cells in the non-consensus and consensus regions respectively (colors depict log2-fold ratio).
D. Histograms showing distribution of the log2-ratio of C134W to WT FOXL2 in TGFβ-stimulated HGrC1 cells in the non-consensus and consensus regions as percentage of total. Red shading shows regions with a log2-ratio>0. Expected and observed numbers of canonical and hybrid sites and corresponding Bonferroni-corrected p-values are given.
E. Table showing the Bonferroni-corrected p-values for a Γ-square test comparing the transcriptional changes observed in primary AGCT tissues to FOXL2 wildtype sex-cord stromal tumors (SLCTs and JGCTs) to transcriptional changes in 1) HGrC1 cells transduced with C134W vs WT FOXL2 2) TGFβ-stimulated HGrC1 cells transduced with C134W vs WT FOXL2. 3) KGN cells +TGFβ vs KGN cells-TGFβ.
F. Heatmap providing a selection of genes co-regulated in the three different conditions. The expression of listed genes was significantly changed (p<0.05, Benjamini-Hochberg corrected, p-value) in all three tests. The heatmap was sorted according to the log2-fold change observed in AGCT vs SCST. Coloring depicts log2-fold changes.
Discussion
This study demonstrates that FOXL2C134W acts as a gain-of-function mutation with the acquired ability to bind to SMAD4, enabling FOXL2C134W to bind to a newly defined FOXL2C134W/SMAD hybrid motif and to trigger the formation of an active enhancer signature and the induction of associated genes.
FOXL2C134W Induces Expression of Genes Associated with Oncogenesis
Our experiments show that FOXL2C134W induces oncogenes and various stemness factors including several prominent cancer stem cell markers and induces EMT. Also many genes characteristic of AGCT (AMH, ACVR1C, FST, PGR), ovarian cancer as well as genes promoting various aspects of oncogenesis and drug-resistance were induced by FOXL2C134Wsuggesting that the FOXL2C134W mutant may contribute to oncogenesis and drug-resistance in AGCTs.
Role of the TGFβ pathway in AGCTs
The transforming growth factor (TGF) superfamily affects differentiation and growth in a wide variety of physiological processes, including differentiation of granulosa cells (38). The TGF superfamily consists of several subgroups including TGFβs and Activins that signal through SMAD2 and 3, (AR-SMADs) and BMP receptors that signal through SMAD1,5,8 (BR-SMADs) (39,40). The AGCT-derived cell-line KGN can trigger the formation of metastatic cancers in immune-compromised mice (41). So far however, a FOXL2C134W-driven transgenic model of AGCT has not been described. In this context, it is noteworthy that the interaction-site of FOXL2 on SMAD4 is not very well conserved between mouse and human, potentially making it unique for humans. However, imbalances of AR-SMAD and BR-SMAD signaling pathways; e.g. decreased BR-SMAD signaling and increased AR-SMAD signaling, have been shown to cause granulosa-cell tumor-like pathologies in mice (42–44). A common feature of these experimental granulosa cell tumor models is the presence of a constitutively active AR-SMAD pathway as demonstrated by a strong immunoreactivity for nuclear phospho-SMAD2/3 (45,46). Our data demonstrate that TGFβ-signaling strongly enhances the effects of the FOXL2C134W mutant, contingent on its increased binding to SMAD4, and further suggests that the ability of AR-SMADs (SMAD2 and SMAD3) to enhance the transcriptional effects of the FOXL2C134W mutant is unique among the receptor SMADs. Thus, whereas activin-stimulation (that signals through SMAD2/3) also has an enhancing effect on the transcriptional response to FOXL2C134W, BMP-stimulation (signaling through SMAD1,5,8) fails to enhance this effect and partially seems to counteract it, pointing to different (opposed) effects of the BMP and TGFβ/Activin-signaling in granulosa cell biology (Fig.S6D-E). We speculate that FOXL2C134W may act to sequester SMAD4 from interacting with BR-SMADs or with other transcriptional co-factors. Thus, FOXL2C134W expression not only triggers the induction of genes at hybrid sites but may also in some cases contribute to the deregulation of genes associated to canonical FOXL2 sites. Similarly, various “classic” BMP targets including ID1 and ID2 are transcriptionally downregulated by the FOXL2C134W mutant (Fig.S6D). Hence FOXL2C134W may act to obstruct other signaling pathways dependent of SMAD4.
Potential Impact for the Treatment of AGCT patients
Although AGCTs display a unique histotype and clearly represent a distinct disease (7), patients with this disease are treated in a similar way as other ovarian cancers; with surgical tumor resection and combinations of platinum and taxane chemotherapy (47,48). Indeed, the current National Comprehensive Cancer Network (NCCN) guidelines for treatment of AGCTs recommend consideration of platinum-based chemotherapy in patients with high-risk and advanced/ recurrent disease (jnccn.org). This treatment regimen is optimized to target highly proliferative cells and may not be effective given the genomic stability and slow growth rate of these tumors, in particular in light of our results indicating transcriptional induction of factors involved in stemness, EMT and drug-resistance by FOXL2C134W. Indeed, these tumors are prone to late relapse (49) and recurrent disease is associated with poor response to chemotherapy and inferior survival (47,50). Thus, the tumor will relapse in 1 of 3 patients causing death of the majority of patients (51–53).
Importantly, our results show that the molecular and transcriptional effects precipitated by the FOXL2C134W mutant and documented in granulosa cell lines could be confirmed in primary tumor material from AGCT patients. Thus, FOXL2 binding sites in primary AGCTs were significantly enriched at hybrid-sites. Likewise, gene expression changes observed in cell lines in response to FOXL2C134W are significantly correlated to transcriptional changes detected in primary AGCTs when comparing to tumors wildtype for FOXL2 (SCST). Notably, our data show that TGFß/activin signaling may contribute to and enhance the transcriptional and potential oncogenic effects triggered by the FOXL2C134W mutant. Moreover, our experiments establish that TGFβ-inhibition dramatically antagonizes the transcriptional effect of the FOXL2C134W/SMAD complex, suggesting a potential benefit of TGFβ or activin-inhibitors in AGCT as documented in a very recent study (54). The potential tumor-suppressive role of TGFβ-signaling in some cancers (55–58) may obviously be a risk of such treatment.
The high frequency of FOXL2C134W mutations in AGCTs, in combination with our data showing that FOXL2C134W is required for the proliferation of AGCT cells, suggest that FOXL2C134W is an oncogenic driver and conceivably an excellent target for anti-cancer therapy in this disease. Wildtype FOXL2 is known to contribute to the lineage determination, differentiation and maturation of granulosa cells (59). Based on our results, we speculate that the ability of FOXL2C134W to induce various factors involved in EMT, oncogenesis and stemness may act to prevent the maturation and differentiation of granulosa cells contributing to AGCT oncogenesis.
The present study also clearly indicates that the acquired binding of the FOXL2C134W mutant to SMAD4 is key for its molecular and oncogenic effects and suggests the possibility of therapeutic targeting of the FOXL2C134W-SMAD4 interaction in AGCTs, or alternatively to target FOXL2C134W directly. Traditionally, transcription factors have not been viewed to be druggable, but recent advances in medicinal chemistry have opened the possibility of targeting classically non-druggable targets by novel approaches (60). A potential benefit of targeting FOXL2, as compared to components of the TGFβ-signaling pathway, is that the protein only is expressed in few tissues. Moreover, AGCT is typically manifests in post-menopausal women who in principle are devoid of functional granulosa cells. For this reason, few adverse events would be envisioned of pharmacological targeting of FOXL2. As mentioned previously, apart from the disease-defining FOXL2C134W mutant, mutations in other oncogenes or tumor suppressor genes in AGCTs are rare (7). Thus, FOXL2C134W may well be the main driving force of this disease. Seen in conjunction with our data implicating the FOXL2C134W mutant in the transcriptional induction of factors involved in EMT and cancer resistance, the pharmacological targeting of FOXL2 may offer the unprecedented potential of not only eliminating the driving oncogene, but may also contribute to prevent resistance to conventional therapies.
Supplementary Material
Significance.
FOXL2C134W highjacks SMAD4 and leads to the expression of genes involved in EMT, stemness, and oncogenesis in AGCT, making FOXL2C134W and the TGFβ pathway therapeutic targets in this condition.
Acknowledgements
We thank Bente Møller and Ulla Toftegård for expert technical assistance, members of the Helin laboratory for discussions and P. Selenica for sequence data handling. The work in the Helin laboratory was supported by the Danish Cancer Society (R167-A10877), the Danish National Research Foundation (DNRF82), the Independent Research Fund Denmark (6153-000005; 7016-00067; 8020-00044), The Neye Foundation, the Novo Nordisk Foundation (NNF; NNF16OC0023234), through a center grant from the NNF to the NNF Center for Stem Cell Biology (NNF17CC0027852), and through the Memorial Sloan Kettering Cancer Center Support Grant (NIH P30 CA008748). Proteomics research in the O.N. Jensen laboratory at SDU is supported by the Danish National Research Foundation (DNRF82), the VILLUM Foundation (VILLUM Center for Bioanalytical Sciences, grant no. 7292), and PRO-MS: Danish National Mass Spectrometry Platform for Functional Proteomics (grant no. 5072-00007B). BW was funded in part by Breast Cancer Research Foundation, Cycle for Survival and Stand Up To Cancer-American Association for Cancer Research Dream Team Translational Cancer Research Grant. Research reported in this publication was supported in part by a Cancer Center Support Grant of the NIH/NCI (Grant No. P30CA008748).
Footnotes
Conflict of Interests
Carol Aghajanian is a member of the advisory boards of Tesaro, Immunogen, Clovis and Eisai/Merck and a member of the steering committees of Genetech, AbbVie and Mateon Therapeutics. Carol Aghajanian has received Grants or personal fees from Tesaro, Immunogen, Clovis and Eisai/Merck Astra Zeneca, Genentech, AbbVie and Mateon Therapeutics. The remaining authors declare no competing interests.
References
- 1.Fox H, Agrawal K, Langley FA. A clinicopathologic study of 92 cases of granulosa cell tumor of the ovary with special reference to the factors influencing prognosis. Cancer 1975;35:231–41 [DOI] [PubMed] [Google Scholar]
- 2.Pectasides D, Pectasides E, Psyrri A. Granulosa cell tumor of the ovary. Cancer Treat Rev 2008;34:1–12 [DOI] [PubMed] [Google Scholar]
- 3.Kalfa N, Philibert P, Patte C, Thibaud E, Pienkowski C, Ecochard A, et al. [Juvenile granulosa-cell tumor: clinical and molecular expression]. Gynecol Obstet Fertil 2009;37:33–44 [DOI] [PubMed] [Google Scholar]
- 4.Bjorkholm E, Silfversward C. Prognostic factors in granulosa-cell tumors. Gynecol Oncol 1981;11:261–74 [DOI] [PubMed] [Google Scholar]
- 5.Bertho S, Pasquier J, Pan Q, Le Trionnaire G, Bobe J, Postlethwait JH, et al. Foxl2 and Its Relatives Are Evolutionary Conserved Players in Gonadal Sex Differentiation. Sex Dev 2016;10:111–29 [DOI] [PubMed] [Google Scholar]
- 6.Caburet S, Georges A, L’Hote D, Todeschini AL, Benayoun BA, Veitia RA. The transcription factor FOXL2: at the crossroads of ovarian physiology and pathology. Mol Cell Endocrinol 2012;356:55–64 [DOI] [PubMed] [Google Scholar]
- 7.Wang YK, Bashashati A, Anglesio MS, Cochrane DR, Grewal DS, Ha G, et al. Genomic consequences of aberrant DNA repair mechanisms stratify ovarian cancer histotypes. Nat Genet 2017;49:856–65 [DOI] [PubMed] [Google Scholar]
- 8.Shah SP, Kobel M, Senz J, Morin RD, Clarke BA, Wiegand KC, et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med 2009;360:2719–29 [DOI] [PubMed] [Google Scholar]
- 9.Al-Agha OM, Huwait HF, Chow C, Yang W, Senz J, Kalloger SE, et al. FOXL2 is a sensitive and specific marker for sex cord-stromal tumors of the ovary. Am J Surg Pathol 2011;35:484–94 [DOI] [PubMed] [Google Scholar]
- 10.Jamieson S, Butzow R, Andersson N, Alexiadis M, Unkila-Kallio L, Heikinheimo M, et al. The FOXL2 C134W mutation is characteristic of adult granulosa cell tumors of the ovary. Mod Pathol 2010;23:1477–85 [DOI] [PubMed] [Google Scholar]
- 11.Karnezis AN, Wang Y, Keul J, Tessier-Cloutier B, Magrill J, Kommoss S, et al. DICER1 and FOXL2 Mutation Status Correlates With Clinicopathologic Features in Ovarian Sertoli-Leydig Cell Tumors. Am J Surg Pathol 2019;43:628–38 [DOI] [PubMed] [Google Scholar]
- 12.. Levine AJ, Wu MC, Chang A, Silver A, Attiyeh EF, Lin J, et al. The spectrum of mutations at the p53 locus. Evidence for tissue-specific mutagenesis, selection of mutant alleles, and a “gain of function” phenotype. Ann N Y Acad Sci 1995;768:111–28 [DOI] [PubMed] [Google Scholar]
- 13.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr.,, Kinzler KW. Cancer genome landscapes. Science 2013;339:1546–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu K, Liu Q, Zhou Y, Tao C, Zhao Z, Sun J, et al. Oncogenes and tumor suppressor genes: comparative genomics and network perspectives. BMC Genomics 2015;16 Suppl 7:S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JH, Kim YH, Kim HM, Park HO, Ha NC, Kim TH, et al. FOXL2 posttranslational modifications mediated by GSK3beta determine the growth of granulosa cell tumours. Nat Commun 2014;5:2936. [DOI] [PubMed] [Google Scholar]
- 16.Rosario R, Cohen PA, Shelling AN. The role of FOXL2 in the pathogenesis of adult ovarian granulosa cell tumours. Gynecol Oncol 2014;133:382–7 [DOI] [PubMed] [Google Scholar]
- 17.Blount AL, Schmidt K, Justice NJ, Vale WW, Fischer WH, Bilezikjian LM. FoxL2 and Smad3 coordinately regulate follistatin gene transcription. J Biol Chem 2009;284:7631–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anttonen M, Pihlajoki M, Andersson N, Georges A, L’Hote D, Vattulainen S, et al. FOXL2, GATA4, and SMAD3 co-operatively modulate gene expression, cell viability and apoptosis in ovarian granulosa cell tumor cells. PLoS One 2014;9:e85545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benayoun BA, Anttonen M, L’Hote D, Bailly-Bechet M, Andersson N, Heikinheimo M, et al. Adult ovarian granulosa cell tumor transcriptomics: prevalence of FOXL2 target genes misregulation gives insights into the pathogenic mechanism of the p.Cys134Trp somatic mutation. Oncogene 2013;32:2739–46 [DOI] [PubMed] [Google Scholar]
- 20.Belli M, Iwata N, Nakamura T, Iwase A, Stupack D, Shimasaki S. FOXL2C134W-Induced CYP19 Expression via Cooperation With SMAD3 in HGrC1 Cells. Endocrinology 2018;159:1690–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conlon N, Schultheis AM, Piscuoglio S, Silva A, Guerra E, Tornos C, et al. A survey of DICER1 hotspot mutations in ovarian and testicular sex cord-stromal tumors. Mod Pathol 2015;28:1603–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pareja F, Lee JY, Brown DN, Piscuoglio S, Gularte-Merida R, Selenica P, et al. The Genomic Landscape of Mucinous Breast Cancer. J Natl Cancer Inst 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massague J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol 2000;1:169–78 [DOI] [PubMed] [Google Scholar]
- 24.Batlle E, Massague J. Transforming Growth Factor-beta Signaling in Immunity and Cancer. Immunity 2019;50:924–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayasula Iwase A, Kiyono T, Takikawa S, Goto M, Nakamura T, et al. Establishment of a human nonluteinized granulosa cell line that transitions from the gonadotropin-independent to the gonadotropin-dependent status. Endocrinology 2012;153:2851–60 [DOI] [PubMed] [Google Scholar]
- 26.Nishi Y, Yanase T, Mu Y, Oba K, Ichino I, Saito M, et al. Establishment and characterization of a steroidogenic human granulosa-like tumor cell line, KGN, that expresses functional follicle-stimulating hormone receptor. Endocrinology 2001;142:437–45 [DOI] [PubMed] [Google Scholar]
- 27.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 2011;27:1696–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol 2002;250:1–23 [DOI] [PubMed] [Google Scholar]
- 29.Dennler S, Huet S, Gauthier JM. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene 1999;18:1643–8 [DOI] [PubMed] [Google Scholar]
- 30.Wang D, Xiang T, Zhao Z, Lin K, Yin P, Jiang L, et al. Autocrine interleukin-23 promotes self-renewal of CD133+ ovarian cancer stem-like cells. Oncotarget 2016;7:76006–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Wang S, Bu S, Xu M, Lai D. Low-dose cisplatin-induced CXCR4 expression promotes proliferation of ovarian cancer stem-like cells. Acta Biochim Biophys Sin (Shanghai) 2016;48:282–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi YP, Shim HS, Gao MQ, Kang S, Cho NH. Molecular portraits of intratumoral heterogeneity in human ovarian cancer. Cancer Lett 2011;307:62–71 [DOI] [PubMed] [Google Scholar]
- 33.Gao MQ, Choi YP, Kang S, Youn JH, Cho NH. CD24+ cells from hierarchically organized ovarian cancer are enriched in cancer stem cells. Oncogene 2010;29:2672–80 [DOI] [PubMed] [Google Scholar]
- 34.Tarhriz V, Bandehpour M, Dastmalchi S, Ouladsahebmadarek E, Zarredar H, Eyvazi S. Overview of CD24 as a new molecular marker in ovarian cancer. J Cell Physiol 2019;234:2134–42 [DOI] [PubMed] [Google Scholar]
- 35.Hedditch EL, Gao B, Russell AJ, Lu Y, Emmanuel C, Beesley J, et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J Natl Cancer Inst 2014;106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung JG, Shih IM, Park JT, Gerry E, Kim TH, Ayhan A, et al. Ovarian Cancer Chemoresistance Relies on the Stem Cell Reprogramming Factor PBX1. Cancer Res 2016;76:6351–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang B, Zhang Y, Zhang W, Zhu Y, Yuan S. Deletion of FOXL2 by CRISPR promotes cell cycle G0/G1 restriction in KGN cells. Int J Mol Med 2019;43:567–74 [DOI] [PubMed] [Google Scholar]
- 38.Richards JS, Pangas SA. New insights into ovarian function. Handb Exp Pharmacol 2010:3–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyazono K, Kusanagi K, Inoue H. Divergence and convergence of TGF-beta/BMP signaling. J Cell Physiol 2001;187:265–76 [DOI] [PubMed] [Google Scholar]
- 40.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev 2005;19:2783–810 [DOI] [PubMed] [Google Scholar]
- 41.Imai M, Muraki M, Takamatsu K, Saito H, Seiki M, Takahashi Y. Spontaneous transformation of human granulosa cell tumours into an aggressive phenotype: a metastasis model cell line. BMC Cancer 2008;8:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matzuk MM, Finegold MJ, Mather JP, Krummen L, Lu H, Bradley A. Development of cancer cachexia-like syndrome and adrenal tumors in inhibin-deficient mice. Proc Natl Acad Sci U S A 1994;91:8817–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coerver KA, Woodruff TK, Finegold MJ, Mather J, Bradley A, Matzuk MM. Activin signaling through activin receptor type II causes the cachexia-like symptoms in inhibin-deficient mice. Mol Endocrinol 1996;10:534–43 [DOI] [PubMed] [Google Scholar]
- 44.Pangas SA, Li X, Umans L, Zwijsen A, Huylebroeck D, Gutierrez C, et al. Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol Cell Biol 2008;28:248–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, Ren YA, Pangas SA, Adams J, Zhou W, Castrillon DH, et al. FOXO1/3 and PTEN Depletion in Granulosa Cells Promotes Ovarian Granulosa Cell Tumor Development. Mol Endocrinol 2015;29:1006–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Middlebrook BS, Eldin K, Li X, Shivasankaran S, Pangas SA. Smad1-Smad5 ovarian conditional knockout mice develop a disease profile similar to the juvenile form of human granulosa cell tumors. Endocrinology 2009;150:5208–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mangili G, Ottolina J, Cormio G, Loizzi V, De Iaco P, Pellegrini DA, et al. Adjuvant chemotherapy does not improve disease-free survival in FIGO stage IC ovarian granulosa cell tumors: The MITO-9 study. Gynecol Oncol 2016;143:276–80 [DOI] [PubMed] [Google Scholar]
- 48.Khosla D, Dimri K, Pandey AK, Mahajan R, Trehan R. Ovarian granulosa cell tumor: clinical features, treatment, outcome, and prognostic factors. N Am J Med Sci 2014;6:133–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jamieson S, Fuller PJ. Molecular pathogenesis of granulosa cell tumors of the ovary. Endocr Rev 2012;33:109–44 [DOI] [PubMed] [Google Scholar]
- 50.van Meurs HS, Schuit E, Horlings HM, van der Velden J, van Driel WJ, Mol BW, et al. Development and internal validation of a prognostic model to predict recurrence free survival in patients with adult granulosa cell tumors of the ovary. Gynecol Oncol 2014;134:498–504 [DOI] [PubMed] [Google Scholar]
- 51.Cronje HS, Niemand I, Bam RH, Woodruff JD. Review of the granulosa-theca cell tumors from the emil Novak ovarian tumor registry. Am J Obstet Gynecol 1999;180:323–7 [DOI] [PubMed] [Google Scholar]
- 52.McConechy MK, Farkkila A, Horlings HM, Talhouk A, Unkila-Kallio L, van Meurs HS, et al. Molecularly Defined Adult Granulosa Cell Tumor of the Ovary: The Clinical Phenotype. J Natl Cancer Inst 2016;108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bryk S, Farkkila A, Butzow R, Leminen A, Heikinheimo M, Anttonen M, et al. Clinical characteristics and survival of patients with an adult-type ovarian granulosa cell tumor: a 56-year single-center experience. Int J Gynecol Cancer 2015;25:33–41 [DOI] [PubMed] [Google Scholar]
- 54.Tao JJ, Cangemi NA, Makker V, Cadoo KA, Liu JF, Rasco DW, et al. First-in-Human Phase I Study of the Activin A Inhibitor, STM 434, in Patients with Granulosa Cell Ovarian Cancer and Other Advanced Solid Tumors. Clin Cancer Res 2019;25:5458–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyaki M, Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem Biophys Res Commun 2003;306:799–804 [DOI] [PubMed] [Google Scholar]
- 56.Hilgers W, Song JJ, Haye M, Hruban RR, Kern SE, Fearon ER. Homozygous deletions inactivate DCC, but not MADH4/DPC4/SMAD4, in a subset of pancreatic and biliary cancers. Genes Chromosomes Cancer 2000;27:353–7 [PubMed] [Google Scholar]
- 57.Tarafa G, Villanueva A, Farre L, Rodriguez J, Musulen E, Reyes G, et al. DCC and SMAD4 alterations in human colorectal and pancreatic tumor dissemination. Oncogene 2000;19:546–55 [DOI] [PubMed] [Google Scholar]
- 58.Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res 2013;73:725–35 [DOI] [PubMed] [Google Scholar]
- 59.Bentsi-Barnes IK, Kuo FT, Barlow GM, Pisarska MD. Human forkhead L2 represses key genes in granulosa cell differentiation including aromatase, P450scc, and cyclin D2. Fertil Steril 2010;94:353–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheepstra M, Hekking KFW, van Hijfte L, Folmer RHA. Bivalent Ligands for Protein Degradation in Drug Discovery. Comput Struct Biotechnol J 2019;17:160–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and RNAseq data have been submitted to the NCBI Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) with accession numbers GSE138496 and PRJNA576673, for cell line data and tumor data respectively.