Abstract
Background
De-regulated host response to severe coronavirus disease 2019 (COVID-19), directly referring to the concept of sepsis-associated immunological dysregulation, seems to be a strong signature of severe COVID-19. Myeloid cells phenotyping is well recognized to diagnose critical illness-induced immunodepression in sepsis and has not been well characterized in COVID-19. The aim of this study is to review phenotypic characteristics of myeloid cells and evaluate their relations with the occurrence of secondary infection and mortality in patients with COVID-19 admitted in an intensive care unit.
Methods
Retrospective analysis of the circulating myeloid cells phenotypes of adult COVID-19 critically ill patients. Phenotyping circulating immune cells was performed by flow cytometry daily for routine analysis and twice weekly for lymphocytes and monocytes subpopulations analysis, as well as monocyte human leukocyte antigen (mHLA)-DR expression.
Results
Out of the 29 critically ill adult patients with severe COVID-19 analyzed, 12 (41.4%) developed secondary infection and six patients died during their stay. Monocyte HLA-DR kinetics was significantly different between patients developing secondary infection and those without, respectively, at day 5–7 and 8–10 following admission. The monocytes myeloid-derived suppressor cells to total monocytes ratio was associated with 28- and 60-day mortality. Those myeloid characteristics suggest three phenotypes: hyperactivated monocyte/macrophage is significantly associated with mortality, whereas persistent immunodepression is associated with secondary infection occurrence compared to transient immunodepression.
Conclusions
Myeloid phenotypes of critically ill COVID-19 patients may be associated with development of secondary infection, 28- and 60-day mortality.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13613-021-00896-4.
Keywords: COVID-19, Sepsis-induced immunodepression, Secondary infection, mHLA-DR, Myeloid-derived suppressor cells
Introduction
Critically ill patients with COVID-19 are now recognized to develop frequently secondary bacterial infection with an incidence ranging between 14 and 25% [1, 2]. Among the contributing factors for secondary infections, prolonged ICU stay is well established, but environmental conditions are known to greatly influence its development. The host response to acute and/or prolonged aggression is well recognized to participate in critical illness-induced immunodepression and its clinical consequences [3]. Both innate and adaptive immune signatures characterize this condition, but myeloid function and phenotypic profiles are the most robust predictors of acquisition of secondary infections and mortality [4].
Monocytes have the capacity to detect aggression and danger signals, to trigger inflammation and to initiate resolution of inflammation. This extreme functional plasticity is closely associated with myeloid phenotypes throughout myelopoiesis and maturational process modifications. Various inflammatory/infectious signals (e.g., calprotectin, Interleukin (IL)-6, Toll Like Receptors (TLR) agonists) are identified to reorient and promote release from the bone marrow of immature myeloid cells that suppress both innate and adaptive immune response [5]. Those cells, named myeloid-derived suppressor cells (MDSC), are now well identified to participate in the host response to infection by blocking T lymphocyte and natural killer cells proliferation, and production of high amount of immunosuppressive cytokine such as IL-10. In addition, the resulting pro-/anti-inflammatory function of monocytes is globally reflected by the level of surface expression of human leukocyte antigen (HLA)-DR [6] which is a highly predictive of secondary infection development and mortality in patients with sepsis [7, 8]. Increasing evidence suggests a central role of myeloid cells and of severe immunosuppression in the pathogenesis of severe COVID-19 [9–11].
Hereby, we aimed at defining the myeloid cells phenotypes during the first 2 weeks following ICU admission of critically ill COVID-19 and explore their relations with development of secondary infections, 28- and 60-day mortality.
Methods
All critically ill adult patients with confirmed COVID-19 (positive RT-PCR for SARS-CoV-2 and suggestive chest CT-scan) admitted to the Pediatric ICU of the Bicêtre Hospital, AP-HP Paris Saclay University, between March and April 2020 were included. Due to the shortage of adult ICU beds, 16 PICU beds were converted to admit adult COVID-19 critically ill patients and staffed with the PICU professionals reinforced by professionals coming from preserved French regions [12]. A senior adult intensivist (JLT) completed the team. All patients were prospectively included in the CLOVIS cohort study (ClinicalTrial.gov Identifier: NCT04544878). The study (French study classification: MR004) was approved by the local IRB and French Data Protection Authority (CNIL registration number: 2219981) waiving the need of written consent. All patients or relatives received information on the study and could refuse to participate at any time. Clinical characteristics including age, gender, comorbidities, occurrence of organ failure, infective complications and related microbiological documentation, length of stay, 28- and 60-day mortality were obtained. Biological characteristics included on admission ferritin, brain natriuretic peptide, troponin T, complement factors, immunoglobulins and sub-classes, daily standard blood workup, and twice weekly non-specific inflammatory markers (e.g., C-reactive protein, procalcitonin), circulating cell phenotyping (complete cell count, T cells and subtypes, B cells, natural killer cells, monocytes and subtypes, neutrophils and subtypes) and monocyte histocompatibility leukocyte antigen (HLA)-DR expression measurement (anti-HLA-DR/anti-monocyte Quantibrite assay, BD Biosciences, San Jose, USA). Data were reported at admission, and at four periods within the first 2 weeks: days 1–4, days 5–7, days 8–10 and days 11–14. Circulating cell phenotyping was performed using ethylenediaminetetraacetic acid (EDTA) anticoagulated blood. Detailed antibodies used for myeloid phenotyping are reported elsewhere (see Additional file 1: Table S1). Monocyte subsets analysis was based on HLA-DR and CD11b expression in CD19−CD14+CD15− cells. Among these, monocytes myeloid-derived suppressor cells (M-MDSC) were identified as CD11b+ HLA-DR−. Thresholds selected for HLA-DR “low-immunosuppressed” group was set at < 15,000 antibody per cell (AB/C) and “high-elevated” group > 30,000 AB/C according to recent published data in COVID-19 critically ill patients [11]. Standard immunophenotyping was performed for lymphocytes T and subclass, NK cells, lymphocytes B, neutrophils. The primary endpoint was the occurrence of secondary infection during the ICU stay as defined by IDSA definitions [13, 14]. The secondary endpoint was 28- and 60-day mortality.
Data are described as number (%) and median (interquartile range (IQR) for categorical and continuous variables, respectively). Comparison of baseline characteristics between patients with a blood sample was done using the Fisher’s exact test or the Chi-square test, as appropriate. The Wilcoxon Mann–Whitney U test was used to determine differences between immunological variables at each time point. We measured the discrimination of M-MDSC to total monocytes ratio using the area under the receiver operating characteristics (AUROC) curve. The best threshold was obtained with the calculation of sensitivity, specificity, positive and negative predictive values and the Youden’s index (Y = sensitivity + specificity − 1). A p-value of less than 0.05 was considered statistically significant. All analyses were performed using GraphPad Prism, v 8.4.2 (GraphPad Software, LLC).
Results
From March 26 to April 15, 2020, 32 critically ill adult patients with suspected COVID-19 were admitted to the Pediatric ICU of the Bicêtre Hospital, AP-HP Paris Saclay University. Out of the 32 patients, one died within the first 6 h following admission and two ultimately had negative SARS-Cov-2 PCR and were definitively considered as non-COVID-19 patients (Fig. 1). Twenty-nine patients were analyzed and their characteristics displayed in Table 1. Secondary infection occurred in 12 patients (41.4%) and six patients (20.7%) died in the ICU. Length of stay in the ICU ranged between 7 and 28 days. At the end of the ICU stay, all survivors were transferred to an adult stepdown unit or wards of our hospital. Five patients were transferred to a medical unit with ventilator rehabilitation facilities. Secondary infections were primarily ventilator-associated pneumonia including pulmonary abscess (n = 2) and Aspergillus spp pneumonia (n = 2) (see Additional file 1: Table S2). On univariate analysis, age, SAPS II, comorbidities and mechanical ventilation were significantly different between patients developing or not secondary infection (Table 1). No biologic markers but NK cells count, monocytes count and M-MDSC to total monocytes ratio significantly differed between both groups.
Fig. 1.
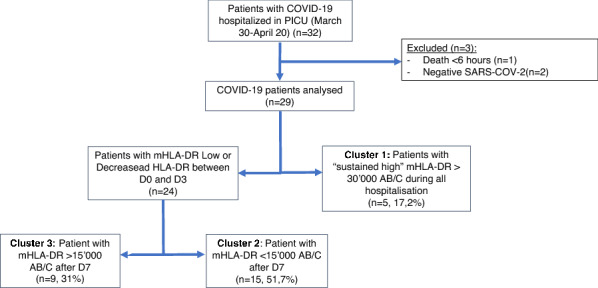
Flowchart
Table 1.
Patients characteristics
| No secondary infection (n = 17) | Secondary infection (n = 12) | p value | |
|---|---|---|---|
| Age (years) | 52 (47–54) | 64 (62–66) | 0.005 |
| BMI (kg/m2) | 28.5 (26.3–31.4) | 29.3 (27.4–33) | 0.44 |
| Gender male | 10 (58.8) | 10 (83.3) | 0.16 |
| SOFA score | |||
| Day 1 | 7 (4–11) | 10 (7–14) | 0.095 |
| Day 3 | 7 (4–10) | 11 (7–14.5) | 0.024 |
| Day 7 | 6 (4.5–7) | 11 (8–14.5) | 0.013 |
| Day 10 | 3 (3–6) | 11 (9–13.5) | 0.02 |
| SAPS II score | |||
| Day 1 | 45 (27–58) | 59 (54–67) | 0.03 |
| Day 3 | 48 (24–55) | 62 (58–64) | 0.04 |
| Day 7 | 40 (25–49) | 60(58–69) | 0.02 |
| Day 10 | 23 (13–35) | 65 (56–70) | 0.001 |
| Comorbidities | |||
| Cardiologic | 5 (29.4) | 10 (83.3) | 0.004 |
| Hypertension | 5 (100) | 9 (90) | – |
| Hypertrophic cardiomyopathy | 1 (20) | 0 | – |
| Peripheral artery occlusive disease | 1 (20) | 1 (10) | – |
| Ischemic cardiopathy | 0 | 2 (20) | – |
| Pneumologic | 2 (11.8) | 1(8.3) | 0.77 |
| Neurologic | 2 (11.8) | 3 (25) | 0.35 |
| Immunologic | 3 (17.6) | 1 (8.3) | 0.47 |
| Metabolic | 7 (41.2) | 8 (66.7) | 0.18 |
| Supportive care | |||
| Norepinephrine | 5 (29.4) | 5 (41.7) | 0.49 |
| Mechanical ventilation | 12 (70.6) | 12 (100) | 0.04 |
| CRRT | 3 (17.6) | 5 (41.7) | 0.154 |
| Immunotherapy | 1 (5.8) | 2 (33.3) | 0.55 |
| Laboratory values at admission | |||
| Leucocytes (/mm3) | 7520 (5970–9330) | 11,910 (7160–14,750) | 0.082 |
| Neutrophils (/mm3) | 6420 (4880–8310) | 10,010 (6238–12,225) | 0.09 |
| Immature neutrophils (%) | 4 (2.57–7.54) | 4.9 (2.5–6.1) | 0.98 |
| Lymphocytes (/mm3) | 800 (600–1191) | 1235 (663–1442) | 0.143 |
| CD4 (/mm3) | 330 (222.5–526.5) | 333 (240.5–481.5) | 0.917 |
| CD8 (/mm3) | 167 (114.5–257) | 346 (163–399.5) | 0.102 |
| NK (/mm3) | 74 (63.25 -109.25) | 127 (95–249) | 0.046 |
| B (/mm3) | 136 (97.5–279) | 132 (100–333.5) | 0.827 |
| Monocytes (/mm3)a | 198.7 (81.5–284) | 328.6 (170.9–453.8) | 0.03 |
| mHLA-DR (AB/C) | 17,737 (10,060–32,503) | 11,877 (7039–22,702) | 0.3 |
| M-MDSC (%) | 3.8 (0.95–13.96) | 10.25 (4.14–29.32) | 0.04 |
| Immunoglobulin G (g/L) | 9.87 (7.6–11.35) | 10.8 (8.71–13.80) | 0.364 |
| Immunoglobulin M (g/L) | 1.22 (0.7–1.39) | 0.81 (0.63–1.30) | 0.427 |
| Immunoglobulin A (g/L) | 2.73 (2.21–3.3) | 2.49 (2.21–5.02) | 0.584 |
| C3 (g/L) | 1.28 (1.05–1.43) | 1.4 (1.15–1.45) | 0.568 |
| C4 (g/L) | 0.27 (0.23–0.36) | 0.29 (0.22–0.34) | 0.765 |
| CH50 (U/mL) | 70 (56.5–73.3) | 70 (57–73) | 0.943 |
| Platelets (× 103/mm3) | 271(182–340) | 444(263–503.7) | 0.097 |
| Fibrinogen (mg/dL) | 7.3 (6.5–8.1) | 8.1 (7–8.3) | 0.28 |
| d-dimers (g/dL) | 2145 (1100–3595) | 2859 (1612–2870) | 0.29 |
| Ferritin (μg/L) | 1223 (818.25–1378.5) | 1691 (303–4406.5) | 0.287 |
| Creatinine (μmol/L) | 67 (50–109) | 88.5 (65.8–145) | 0.25 |
| Total bilirubin (μmol/L) | 9 (6–12) | 11 (5.75–21.5) | 0.73 |
| PCT (ng/mL) | 0.64 (0.34–2.77) | 0.71(0.36–3.22) | 0.9 |
| CRP (mg/L) | 131.5 (89.7–210.5) | 188(98.5–265) | 0.34 |
| Outcome | |||
| 28-day mortality | 3 (17.6) | 3 (25) | 0.63 |
| 60-day mortality | 5 (29.4%) | 6 (50) | 0.26 |
| PICU length of stay | 10 (7–15) | 26 (21.5–28) | 0.0001 |
Values are expressed as median (IQR) or number (%)
BMI, body mass index; SOFA, Sequential Organ Failure Assessment; SAPS, Simplified Acute Physiological Score; CRRT, continuous renal replacement therapy; CD, cluster of differentiation; mHLA-DR, monocyte histocompatibility leucocyte antigen-DR; M-MDSC, monocyte myeloid-derived suppressor cell (CD11b+ HLA-DR−), C, complement; CH50, total complement activity; PCT, procalcitonin; CRP, C-reactive protein; PICU, pediatric intensive care unit
aTotal monocytes: CD19− CD14+ CD15−, monocytes subtypes based on CD11b and HLA-DR
Five patients had persistently high mHLA-DR throughout their ICU stay. In the remaining 24 patients, mHLA-DR at admission was similar, but patients with secondary infection had a persistently low mHLA-DR level throughout the first 2 weeks, whereas those without secondary infection significantly increased their mHLA-DR within the first 5–7 days after admission (median range increase from day 1–4: 14,826 to 25,355 AB/C at day 5–7 and 14,826 to 27,082 at day 8–10 AB/C) (Fig. 2a). Interestingly, no significant change in mHLA-DR occurs in both groups between days 5 to 10, suggesting that most of the myeloid response is set in the first days following admission (see Additional file 1: Figure S1). This significant deviation of mHLA-DR between patients developing or not secondary infection occurred 1 to 3 days before infection diagnosis was made. No significant difference in mHLA-DR was found between survivors and deceased ones during the study (Fig. 2b). No statistical difference in the percentage of M-MDSC to total circulating monocytes between patients with or without secondary infection was observed (Fig. 3a). M-MDSC proportion to total circulating monocytes was associated with 60-day survival (AUROC curve = 0.70, 95% CI 0.39 to 1) with M-MDSC to total monocytes being significantly lower at admission in patients alive at day 60 (n = 10/29, p = 0.044) (Fig. 3b). A cut-off value of 21% (Youden index = 0.54) was associated with a sensitivity of 67% and specificity of 87%.
Fig. 2.
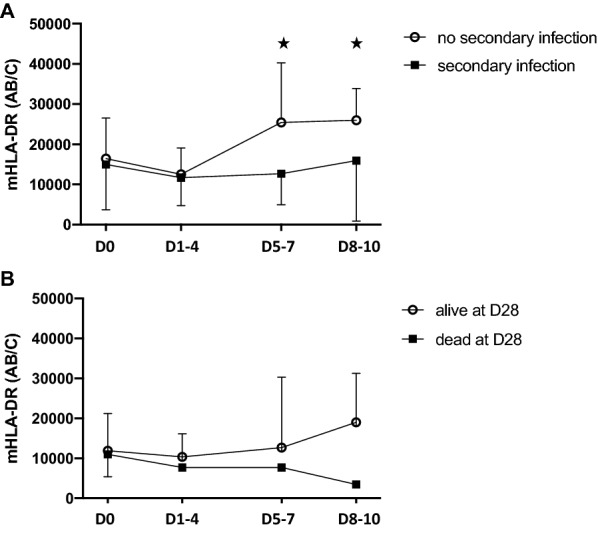
Monocyte HLA-DR kinetics. Monocyte HLA-DR antibody per cell surface expression from admission to day 8–10 in patients with secondary infection (a) or survivors at day 28 (b). *p < 0.05
Fig. 3.
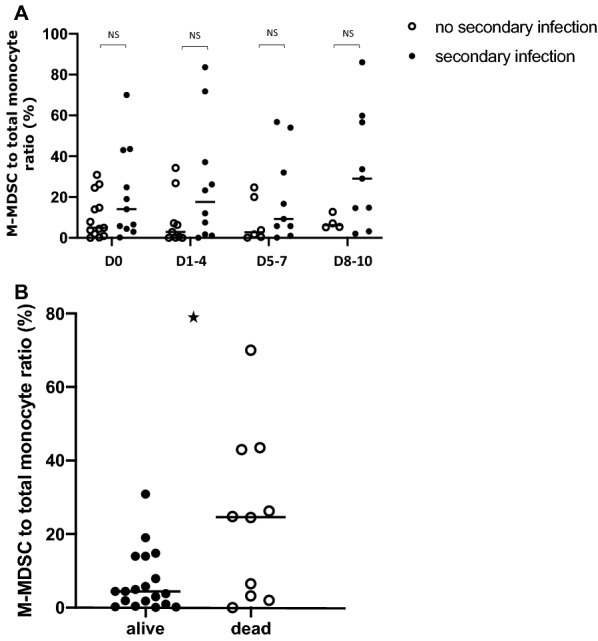
Monocyte myeloid-derived suppressor cells kinetics. Monocyte myeloid-derived suppressor cells (M-MDSC) kinetics from admission to day 8–10 in patients with secondary infection (a) or survivors at day 28 (b). **p < 0.005
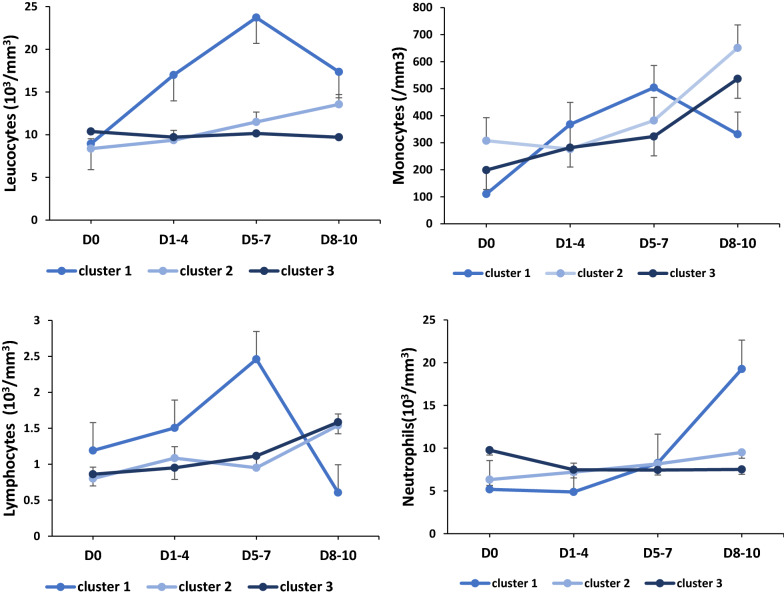
Circulating myeloid cell phenotyping suggested three different groups of patients according to the evolution of the mHLA-DR rate: patients with persistently high mHLA-DR > 30,000 AB/C during the study (cluster 1), patients with persistently low mHLA-DR < 15,000 AB/C after day 5–7 (cluster 2) and patients with rising mHLA-DR > 15,000 AB/C after day 5–7 (cluster 3) (Table 2). On admission neither clinical nor biochemical parameters, but circulating cell phenotypes, differentiated the three phenotypes. Although on admission leukocytes count were similar (Fig. 4), patients from cluster 1 showed a non-significant trend toward early increased in leukocytes (Wilcoxon sign rank test, p = 0.06). Since patient from cluster 1 always displayed no to low M-MDSC (range: 0 to 3.19%), analysis of M-MDSC proportion to total circulating monocytes, without cluster 1 patients, was significantly associated with 28 days mortality (p = 0.003, AUROC curve = 0.94, 95% CI 0.84 to 1; cut-off value 18%, Youden Index = 0.85). The three phenotypes were associated with clinical outcomes: patients from cluster 1 displayed a hyperactivated monocytes/macrophage (HAMM) phenotype associated with the highest mortality, whereas those from cluster 2 and 3 displaying either prolonged immunodepression (PID), and transient immunodepression (TID) were differentially associated with occurrence of secondary infection (Table 3). Interestingly, the development of a secondary infection among the M-MDSC to total monocytes ratio > 18% group only occurred in patients with PID.
Table 2.
Severe COVID-19 myeloid phenotypes at admission
| Cluster 1 (HAMM, n = 8) | Cluster 2 (PID, n = 18) | Cluster 3 (TID, n = 9) | p | |
|---|---|---|---|---|
| Age (years) | 44 (43–57) | 63 (59–67) | 55 (53–59) | 0.04 |
| BMI (kg/m2) | 27.6 (26.3–31.5) | 29.4 (26.7–31.8) | 29.2 (26.4–31.4) | 0.94 |
| Gender male | 3 (60%) | 12 (66%) | 5 (55%) | 0.07 |
| SOFA score | 10 (2–14) | 7 (6.5–12) | 8 (6–12) | 0.96 |
| SAPS II score | 57 (22–59) | 55 (48–65) | 54 (45–58) | 0.63 |
| Laboratory values | ||||
| Leucocytes (/mm3) | 8940 (7520–15,600) | 8370 (6800–11,260) | 10,380 (5970–12,740) | 0.74 |
| Neutrophils (/mm3) | 5180 (4770–8310) | 6320 (5525–11,415) | 9770 (7960–11,080) | 0.66 |
| Immature neutrophils (%) | 7.3 (4–23) | 3.3 (2.7–6.7) | 4.6 (1.1–7.2) | 0.39 |
| Lymphocytes (/mm3) | 1191 (970–1370) | 800 (620–1340) | 860 (780–1220) | 0.87 |
| CD4 (/mm3) | 438 (308–670) | 396 (230–529) | 312 (226–339) | 0.44 |
| CD8 (/mm3) | 246 (74–356) | 268 (110–376) | 213 (162–243) | 0.93 |
| NK (/mm3) | 69 (63–92) | 102 (65–167) | 111 (91.5–193.5) | 0.24 |
| B (/mm3) | 301 (42–313) | 132 (122–283) | 127 (89–201) | 0.80 |
| Monocytes (/mm3)a | 110.2 (51.6–284) | 307.4 (169.4–350.3) | 198.7 (81.5–270.3) | 0.2 |
| mHLA-DR (AB/C) | 46,340 (41,550–48,017) | 10,060 (6329–16,281) | 15,646 (10,873–28,568) | 0.002 |
| M-MDSC (%) | 1.8 (0.4–1.87) | 13.9 (5.4–27.8) | 3 (0.9–14.8) | 0.009 |
| CD11b+ HLA-DR+ | 96.2 (95.8–97.95) | 82.2 (69.0–94.6) | 96.9 (85.2–14.8) | 0.01 |
| CD11− HLA-DR+ | 0.37 (0–1.8) | 0 (0–0.3) | 0 (0–0) | 0.28 |
| LNR (%) | 20.3 (11–26.4) | 12.6 (9.2–17.4) | 10.7 (9.8–17.1) | 0.45 |
| LMR (%) | 9.1 (3.4–10.8) | 3.5 (2.6–4.3) | 3.1 (2.2–5.2) | 0.2 |
| Immunoglobulin G (g/L) | 7.66 (7.54–10.10) | 9.93 (8.02–12.5) | 11.5 (9.80–14.30) | 0.30 |
| Immunoglobulin M (g/L) | 1.21 (0.90–1.34) | 0.81 (0.63–1.24) | 1.3 (0.90–1.55) | 0.36 |
| Immunoglobulin A (g/L) | 3.40 (1.94–4.32) | 2.63 (2.21–3.71) | 2.80(2.20–3.30) | 0.99 |
| C3 (g/L) | 1.32 (1.09–1.53) | 1.28 (1.09–1.43) | 1.39 (1.23–1.44) | 0.73 |
| C4 (g/L) | 0.25 (0.23–0.25) | 0.24 (0.16–0.35) | 0.29 (0.28–0.36) | 0.40 |
| CH50 (U/mL) | 68 (63–69) | 64 (52.5–73) | 79 (72–83) | 0.16 |
| Platelets (× 103/mm3) | 340 (333–387) | 308 (237–482) | 242 (182–312) | 0.73 |
| Fibrinogen (mg/dL) | 8.1 (6.5–8.1) | 7.2 (6.5–8.1) | 8.1 (6.7–8.3) | 0.81 |
| d-dimers (g/dL) | 2875 (2027.5–3370) | 1620 (1480–3510) | 2610 (1880–4000) | 0.86 |
| Ferritin (μg/L) | 507 (418–919) | 1416 (768–1730) | 1313 (729–1675) | 0.30 |
| Creatinine (μmol/L) | 67 (42–172) | 68 (57–121) | 85 (60–109) | 0.84 |
| Total bilirubin (μmol/L) | 5 (5–6) | 11 (6–17) | 10 (8–21) | 0.22 |
| PCT (ng/mL) | 0.57 (0.47–2.95) | 0.54 (0.19–1.74) | 0.88 (0.64–2.77) | 0.29 |
| CRP (mg/L) | 99 (74.5–117) | 163 (85–229) | 206.5 (149.8–233) | 0.33 |
| Outcomes | ||||
| 28-day mortality | 2 (40%) | 2(13%) | 2 (22%) | 0.36 |
| 60-day mortality | 2 (40%) | 5 (33%) | 3 (33%) | 0.24 |
| PICU length of stay | 7 (6–10) | 20 (11–24) | 15 (11–23.5) | 0.30 |
Values are expressed as median (IQR) or number (%)
BMI, body mass index; SOFA, Sequential Organ Failure Assessment; CD, cluster of differentiation; LNR, lymphocytes-to-neutrophils ratio; LMR, lymphocytes-to-monocytes ratio; mHLA-DR, monocyte histocompatibility leucocyte antigen-DR; M-MDSC, monocyte myeloid-derived suppressor cell (CD11b+HLA-DR−), C, complement; CH50, total complement activity; PCT, procalcitonin; CRP, C-reactive protein; PICU, pediatric intensive care unit
aTotal monocytes: CD19− CD14+ CD15−, monocytes subtypes based on CD11b and HLA-DR
Fig. 4.
Circulating cells kinetics in severe COVID-19 myeloid phenotypes
Table 3.
Myeloid phenotypes diagnostic criteria in severe COVID-19 and targeted investigation and therapy
| Hyper-activated monocytes/macrophage (HAMM) | Transient immune-depression (TID) | Persistent immune-depression (PID) | |
|---|---|---|---|
| Dynamic diagnostic criteria | |||
| M-MDSC day 0a,b | < 5% | ||
| mHLA-DR day 0a | > 30,000 AB/C | < 30,000 AB/C | |
| mHLA-DR day 5–7a | > 15,000 AB/C | < 15,000 AB/C | |
| Targeted investigation | Screen for MAS-like: myelogram | Track VAP | |
| Targeted therapy | If MAS-like, consider immunotherapy (e.g. Anti-IL6) | Consider empiric antibiotics by day 5–7 | |
M-MDSC, monocytes myeloid-derived suppressor cell to total monocytes ratio defined as CD19− CD14+ CD15−CD11b+HLA-DR− cells/CD19− CD14+ CD15− cells; mHLA-DR, monocytes Human leukocyte antigen-DR; AB/C, antibody per cell; MAS, macrophage activation syndrome; VAP, ventilator-associated pneumonia
aFollowing ICU admission
bM-MDSC > 18% is associated with 28-day mortality in PID and TID, with PID systematically associated with secondary infection
Although few patients were included in therapeutic institutional immunotherapy study [15], independently to the myeloid phenotypes, one patient with HAMM and confirmed macrophage activation was treated with compassionate anti-IL6 therapy (tocilizumab) and successfully weaned from ECMO within 48 h and fully recovered.
Discussion
This pilot study identified two prototypic myeloid characteristics associated in critically ill COVID-19 adult patients with the occurrence of secondary infection and mortality. Monocyte HLA-DR kinetics and presence of M-MDSC, both illustrating myeloid antigen presentation and immune cell suppressive function, showed specific profiles in severe COVID-19.
In contrast to published series suggesting a systematic decrease or low (< 15,000AB/C) mHLA-DR in severe COVID-19 [11], we identified three distinct mHLA-DR kinetics, suggestive of three clusters of patients (Fig. 1). Analysis of mHLA-DR kinetics has proven to be an accurate characterization of the immunological adaptation during critical illnesses. Although in COVID-19, mHLA-DR level seems not as low as what is seen in septic shock (< 8000 AB/C), in our study, profile kinetics seen in cluster 2 (PID) and cluster 3 (TID) were differentially associated with the development of secondary infection, which is in line with published experience in sepsis [16]. A decrease in mHLA-DR in severe COVID-19 was shown in both adults and children [17–21]. Spinetti et al. showed that critically ill patients initially admitted in general wards and secondarily transferred to the ICU, had a higher mHLA-DR than those directly admitted to the ICU [17]. Similarly, Moratto et al. showed that COVID-19 hospitalized patients progressing to critical illness had lower mHLA-DR level [18]. In a cohort of 157 COVID-19 patients, Wang et al. showed that the 62 patients who died had a significantly decreased mHLA-DR after 10 days [19]. Interestingly, preliminary report in children suggested that mHLA-DR was strongly decreased in children with severe COVID-19 multisystem inflammatory syndrome in children, but could not evidence an association with mortality [20]. In our study, mHLA-DR kinetics between deceased and survivors were not significantly different, although this result could have been hampered by the limited follow-up period.
In contrast to published experience, we identified a third cluster of patients, with persistently high mHLA-DR, suggestive of protracted monocyte activation phenotype. This mHLA-DR kinetic, although less frequent, was also identified in one series [17]. In our cohort, those patients had severe systemic inflammation, high ferritin, elevated fibrinogen and lymphopenia and in one patient, confirmed macrophage activation on bone marrow analysis reinforcing a potentially “hyperactivated monocyte/macrophage” (HAMM) phenotype. Use of mHLA-DR to identify critically ill patients with macrophage activation syndrome was recently suggested and may prove to be an effective screening criterion in patients with severe COVID-19 and suspected HAMM mimicking macrophage activation syndrome [22]. This observation further completed earlier observations suggesting that COVID-19 may be associated with macrophage activation-like syndrome [23–25].
Although not unique, impairment of myeloid response is increasingly recognized in COVID-19. Relative disappearance of non-classical CD14lowCD16high monocytes, seems to be characteristics of SARS-CoV-2 infection, not seen in other viral infections, and may be associated with severity [9, 26]. As shown, a second characteristic seen in severe COVID-19 is the decrease in HLA-DR expression on CD14high monocytes that could be associated with altered responsiveness of circulating monocyte to TLRs bacterial agonists [27]. There is currently significant arguments to suggest that emergency myelopoiesis occurs in severe COVID-19. Silvin et al. showed massive amounts of calprotectin and immature myeloid precursors raising the hypothesis of a probable expansion of myeloid-derived suppressor cells during SARS-Cov-2 infection [5, 9]. Hereby, we demonstrated the massive presence of M-MDSC in critically ill COVID-19 patients and its association with mortality. Few preliminary data suggest the presence of myeloid-derived suppressor cells and their role in severe COVID-19 and its pathophysiology [28–30]. Bordoni et al. demonstrated that patients with severe COVID-19 admitted in ICU had decreased frequency of lymphocyte T and natural killer, which paralleled expansion of MDSC and high level of cytokines [31]. In complement to our results, Sacchi et al. showed that polymorphonuclear (PMN)-MDSC percentage > 54% was associated with mortality in severe COVID-19, heralding the importance of MDSC in severe COVID-19 [30]. Although accumulation of immature myeloid cells is a recognized severity marker of sepsis since the early 1970s, the role of MDSC in critically ill septic patient’s pathophysiology has been poorly studied [5, 32]. Myeloid-derived suppressor cells are bone marrow heterogeneous myeloid immature precursors that may in certain pathological conditions, such as cancer, sepsis, and autoimmune diseases, partially interrupt their differentiation and expend. Two types of myeloid-derived suppressor cells are identified whether originating from PMN or monocytes precursors. MDSC induces lymphocytes T and natural killer cells apoptosis, inhibits T cells proliferation, induces expansion of T regulatory cells, and produces immunosuppressive cytokines such as IL-10 [33, 34]. Both high proportion of PMN- and M-MDSC were shown to be associated with development of secondary infections and early mortality [35–38]. Waeckel et al. in a series of secondary gated M-MDSC-like cells (CD14+HLA-DRlow, neither CD15 nor CD33 staining) in septic patients, identified a cut-off value of > 9% significantly associated with 28-day mortality and occurrence of secondary infections [35]. Altogether, there is convergent data suggesting that MDSC are central in the pathophysiology of sepsis, but its regulation, especially in regard to the temporal and spatial organization, seems to be essential in severe COVID-19 [9, 27].
Our study has several limitations. Inherent to the design, this study remains a preliminary single-center observation in need of further validation. The limited number of patients in the three clusters and their association with outcome warrant further confirmation. Nevertheless, the phenotypic characterization of the three clusters and their reciprocal differences are clear enough to suggest association with specific clinical courses and may be of some utility to select patients for innovative therapy [21, 25, 39].
To conclude, our data suggest that mHLA-DR kinetics allows to identify three myeloid phenotypes in critically ill COVID-19 adult patients potentially associated with specific infective paths: hyperactivated monocyte/macrophage, transient immunodepression, and persistent immunodepression phenotypes.
Supplementary Information
Additional file 1: Figure S1. Monocyte HLA-DR variation in patients with or without secondary infection. Table S1. Circulating cells phenotyping antibodies clones. Table S2. Site of secondary infection and associated pathogens.
Acknowledgements
The COVID-19 Immune Suppression (CLOVIS) study group: Simon Barreault, Mélissa Beggaz, Ramy Charbel, Caroline Claude, Zoé Demailly, Philippe Durand, Gaspard Gerschenfeld, Jessica Giraldi, Matteo Guerra, Manon Hily, Martin Journaux, Christopher Lai, Pauline Leroux, Clémence Marais, Zied Merchaoui, Jordi Miatello, Clarisse Niçaise, Jean-Eudes Piloquet, Melissa Ren, Marie Simbozel, Nada Semaan, Jean-Louis Teboul, Pierre Tissieres, Brendan Travert.
Abbreviations
- COVID-19
Coronavirus disease 2019
- mHLA-DR
Monocyte human leukocyte antigen-DR
- HAMM
Hyperactivated monocytes/macrophage
- PID
Prolonged immunodepression
- TID
Transient immunodepression
- SARS-CoV-2
Respiratory distress syndrome due to coronavirus 2
- CI
Confidence interval
- ICU
Intensive care unit
- IL−
Interleukin−
- TLR
Toll like Receptor
- M-/PMN-MDSC
Monocyte-/polymorphonuclear-myeloid-derived suppressor cell
- IRB
Institutional board review
- EDTA
Ethylenediaminetetraacetic acid
- CD
Cluster of differentiation
- NK
Natural killer
- IDSA
Infection Disease Society of America
- AUROC
Area under the receiver operating characteristics
- AB/C
AntiBody per cell
- ECMO
Extracorporeal membrane oxygenation
Authors’ contributions
Design of the work: JM, PT. Acquisition, analysis, or interpretation of data for the work: CM, NS, CC, JM, PT. Drafting the manuscript: CM, JM, PT. Revising it critically for important intellectual content: all authors. All authors approved the final version submitted for publication and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Funding
No funding.
Availability of data and materials
All materials are available upon request to Pr. Pierre Tissières at pierre.tissieres@aphp.fr.
Declarations
Ethics approval and consent to participate
The study (French study classification: MR004) was approved by the local IRB and French Data Protection Authority (CNIL Registration number: 2219981) waiving the need of written consent. The study was registered at ClinicalTrial.gov Identifier: NCT04544878.
Consent for publication
All authors agreed for publication of this study.
Competing interests
Pr. Tissieres reports research grants from Merieux Foundation and Chiesi Inc, outside the submitted work, and consulting fee for Baxter, Bristol Myers Squibb, bioMerieux, Faron, Chiesi, Inotrem. All other authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pierre Tissières, Email: pierre.tissieres@aphp.fr.
for The COVID-19 Immune Suppression (CLOVIS) Study Group:
Simon Barreault, Mélissa Beggaz, Ramy Charbel, Caroline Claude, Zoé Demailly, Philippe Durand, Gaspard Gerschenfeld, Jessica Giraldi, Matteo Guerra, Manon Hily, Martin Journaux, Christopher Lai, Pauline Leroux, Clémence Marais, Zied Merchaoui, Jordi Miatello, Clarisse Niçaise, Jean-Eudes Piloquet, Melissa Ren, Marie Simbozel, Nada Semaan, Jean-Louis Teboul, Pierre Tissieres, and Brendan Travert
References
- 1.Lansbury L, Lim B, Baskaran V, Lim WS. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giacobbe DR, Battaglini D, Ball L, Brunetti I, Bruzzone B, Codda G, Crea F, De Maria A, Dentone C, Di Biagio A, Icardi G, Magnasco L, Marchese A, Mikulska M, Orsi A, Patroniti N, Robba C, Signori A, Taramasso L, Vena A, Pelosi P, Bassetti M. Bloodstream infections in critically ill patients with COVID-19. Eur J Clin Investig. 2020 doi: 10.1111/eci.13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 2013;13(12):862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drewry AM, Ablordeppey EA, Murray ET, Beiter ER, Walton AH, Hall MW, Hotchkiss RS. Comparison of monocyte human leukocyte antigen-DR expression and stimulated tumor necrosis factor alpha production as outcome predictors in severe sepsis: a prospective observational study. Crit Care. 2016;20(1):334. doi: 10.1186/s13054-016-1505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ost M, Singh A, Peschel A, Mehling R, Rieber N, Hartl D. Myeloid-derived suppressor cells in bacterial infections. Front Cell Infect Microbiol. 2016;6:37. doi: 10.3389/fcimb.2016.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venet F, Demaret J, Gossez M, Monneret G. Myeloid cells in sepsis-acquired immunodeficiency. Ann N Y Acad Sci. 2020 doi: 10.1111/nyas.14333. [DOI] [PubMed] [Google Scholar]
- 7.Monneret G, Lepape A, Voirin N, Bohé J, Venet F, Debard AL, Thizy H, Bienvenu J, Gueyffier F, Vanhems P. Persisting low monocyte human leukocyte antigen-DR expression predicts mortality in septic shock. Intensive Care Med. 2006;32(8):1175–1183. doi: 10.1007/s00134-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 8.Landelle C, Lepape A, Voirin N, Tognet E, Venet F, Bohé J, Vanhems P, Monneret G. Low monocyte human leukocyte antigen-DR is independently associated with nosocomial infections after septic shock. Intensive Care Med. 2010;36(11):1859–1866. doi: 10.1007/s00134-010-1962-x. [DOI] [PubMed] [Google Scholar]
- 9.Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L, Almire C, Hénon C, Kosmider O, Droin N, Rameau P, Catelain C, Alfaro A, Dussiau C, Friedrich C, Sourdeau E, Marin N, Szwebel TA, Cantin D, Mouthon L, Borderie D, Deloger M, Bredel D, Mouraud S, Drubay D, Andrieu M, Lhonneur AS, Saada V, Stoclin A, Willekens C, Pommeret F, Griscelli F, Ng LG, Zhang Z, Bost P, Amit I, Barlesi F, Marabelle A, Pène F, Gachot B, André F, Zitvogel L, Ginhoux F, Fontenay M, Solary E. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182(6):1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Remy KE, Mazer M, Striker DA, Ellebedy AH, Walton AH, Unsinger J, Blood TM, Mudd PA, Yi DJ, Mannion DA, Osborne DF, Martin RS, Anand NJ, Bosanquet JP, Blood J, Drewry AM, Caldwell CC, Turnbull IR, Brakenridge SC, Moldwawer LL, Hotchkiss RS. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5(17):e140329. doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benlyamani I, Venet F, Coudereau R, Gossez M, Monneret G. Monocyte HLA-DR measurement by flow cytometry in COVID-19 patients: an interim review. Cytometry A. 2020;97(12):1217–1221. doi: 10.1002/cyto.a.24249. [DOI] [PubMed] [Google Scholar]
- 12.Geslain G, Miatello J, Barreault S, de Melo C, Durand P, Deho A, Naudin J, Zitvogel F, Tissieres P, Dauger S, Guilbert AS, Levy M. Paediatric intensive care unit mutation to adult unit during the initial Covid-19 wave: does it make sense? Minerva Anestesiol. 2020 doi: 10.23736/S0375-9393.20.14935-6. [DOI] [PubMed] [Google Scholar]
- 13.O'Grady NP, Alexander M, Burns LA, Dellinger EP, Garland J, Heard SO, Lipsett PA, Masur H, Mermel LA, Pearson ML, Raad II, Randolph AG, Rupp ME, Saint S, Healthcare Infection Control Practices Advisory Committee Guidelines for the prevention of intravascular catheter-related infections. Am J Infect Control. 2011;39(4 Suppl 1):S1–34. doi: 10.1016/j.ajic.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O'Grady NP, Bartlett JG, Carratalà J, El Solh AA, Ewig S, Fey PD, File TM Jr, Restrepo MI, Roberts JA, Waterer GW, Cruse P, Knight SL, Brozek JL. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the infectious diseases society of America and the American Thoracic Society. Clin Infect Dis. 2016;63(5): e61–e111. 10.1093/cid/ciw353. (Epub 2016 Jul 14). Erratum in: Clin Infect Dis. 2017;64(9):1298. Erratum in: Clin Infect Dis. 2017;65(8):1435. Erratum in: Clin Infect Dis. 2017;65(12):2161. [DOI] [PMC free article] [PubMed]
- 15.Hermine O, Mariette X, Tharaux PL, Resche-Rigon M, Porcher R, Ravaud P, CORIMUNO-19 Collaborative Group Effect of tocilizumab vs usual care in adults hospitalized with COVID-19 and moderate or severe pneumonia: a randomized clinical trial. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venet F, Lukaszewicz AC, Payen D, Hotchkiss R, Monneret G. Monitoring the immune response in sepsis: a rational approach to administration of immunoadjuvant therapies. Curr Opin Immunol. 2013;25(4):477–483. doi: 10.1016/j.coi.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spinetti T, Hirzel C, Fux M, Walti LN, Schober P, Stueber F, Luedi MM, Schefold JC. Reduced monocytic human leukocyte antigen-DR expression indicates immunosuppression in critically ill COVID-19 patients. Anesth Analg. 2020;131(4):993–999. doi: 10.1213/ANE.0000000000005044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moratto D, Chiarini M, Giustini V, Serana F, Magro P, Roccaro AM, Imberti L, Castelli F, Notarangelo LD, Quiros-Roldan E. Flow cytometry identifies risk factors and dynamic changes in patients with COVID-19. J Clin Immunol. 2020;40(7):970–973. doi: 10.1007/s10875-020-00806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Hou H, Yao Y, Wu S, Huang M, Ran X, Zhou H, Liu Z, Sun Z. Systemically comparing host immunity between survived and deceased COVID-19 patients. Cell Mol Immunol. 2020;17(8):875–877. doi: 10.1038/s41423-020-0483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter MJ, Fish M, Jennings A, Doores KJ, Wellman P, Seow J, Acors S, Graham C, Timms E, Kenny J, Neil S, Malim MH, Tibby SM, Shankar-Hari M. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat Med. 2020;26(11):1701–1707. doi: 10.1038/s41591-020-1054-6. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen LS, Ait Hamou Z, Gastli N, Chapuis N, Pène F. Potential role for interferon gamma in the treatment of recurrent ventilator-acquired pneumonia in patients with COVID-19: a hypothesis. Intensive Care Med. 2021;47(5):619–621. doi: 10.1007/s00134-021-06377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Remy S, Gossez M, Belot A, Hayman J, Portefaix A, Venet F, Javouhey E, Monneret G. Massive increase in monocyte HLA-DR expression can be used to discriminate between septic shock and hemophagocytic lymphohistiocytosis-induced shock. Crit Care. 2018;22(1):213. doi: 10.1186/s13054-018-2146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prilutskiy A, Kritselis M, Shevtsov A, Yambayev I, Vadlamudi C, Zhao Q, Kataria Y, Sarosiek SR, Lerner A, Sloan JM, Quillen K, Burks EJ. SARS-CoV-2 infection-associated hemophagocytic lymphohistiocytosis. Am J Clin Pathol. 2020;154(4):466–474. doi: 10.1093/ajcp/aqaa124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, Antonakos N, Damoraki G, Gkavogianni T, Adami ME, Katsaounou P, Ntaganou M, Kyriakopoulou M, Dimopoulos G, Koutsodimitropoulos I, Velissaris D, Koufargyris P, Karageorgos A, Katrini K, Lekakis V, Lupse M, Kotsaki A, Renieris G, Theodoulou D, Panou V, Koukaki E, Koulouris N, Gogos C, Koutsoukou A. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimopoulos G, de Mast Q, Markou N, Theodorakopoulou M, Komnos A, Mouktaroudi M, Netea MG, Spyridopoulos T, Verheggen RJ, Hoogerwerf J, Lachana A, van de Veerdonk FL, Giamarellos-Bourboulis EJ. Favorable anakinra responses in severe Covid-19 patients with secondary hemophagocytic lymphohistiocytosis. Cell Host Microbe. 2020;28(1):117–123.e1. doi: 10.1016/j.chom.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatti A, Radrizzani D, Viganò P, Mazzone A, Brando B. Decrease of non-classical and intermediate monocyte subsets in severe acute SARS-CoV-2 infection. Cytometry A. 2020;97(9):887–890. doi: 10.1002/cyto.a.24188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, Sigal N, Feng Y, Bristow L, Tak-Yin Tsang O, Wagh D, Coller J, Pellegrini KL, Kazmin D, Alaaeddine G, Leung WS, Chan JMC, Chik TSH, Choi CYC, Huerta C, Paine McCullough M, Lv H, Anderson E, Edupuganti S, Upadhyay AA, Bosinger SE, Maecker HT, Khatri P, Rouphael N, Peiris M, Pulendran B. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369(6508):1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reyes M, Filbin MR, Bhattacharyya RP, Sonny A, Mehta A, Billman K, Kays KR, Pinilla-Vera M, Benson ME, Cosimi LA, Hung DT, Levy BD, Villani AC, Sade-Feldman M, Baron RM, Goldberg MB, Blainey PC, Hacohen N. Induction of a regulatory myeloid program in bacterial sepsis and severe COVID-19. bioRxiv. 2020:2020.09.02.280180. 10.1101/2020.09.02.280180. (Preprint).
- 29.Thompson E, Cascino K, Ordonez A, Zhou W, Vaghasia A, Hamacher-Brady A, Brady N, Sun IH, Wang R, Rosenberg A, Delanoy M, Rothman RE, Fenstermacher K, Sauer L, Shaw-Saliba K, Bloch EM, Redd A, Tobian AA, Horton M, Smith K, Pekosz A, D'Alessio F, Yegnasubramanian S, Ji H, Cox AL, Powell JD. Mitochondrial induced T cell apoptosis and aberrant myeloid metabolic programs define distinct immune cell subsets during acute and recovered SARS-CoV-2 infection. medRxiv. 2020:2020.09.10.20186064. 10.1101/2020.09.10.20186064. (Preprint).
- 30.Sacchi A, Grassi G, Bordoni V, Lorenzini P, Cimini E, Casetti R, Tartaglia E, Marchioni L, Petrosillo N, Palmieri F, D'Offizi G, Notari S, Tempestilli M, Capobianchi MR, Nicastri E, Maeurer M, Zumla A, Locatelli F, Antinori A, Ippolito G, Agrati C. Early expansion of myeloid-derived suppressor cells inhibits SARS-CoV-2 specific T-cell response and may predict fatal COVID-19 outcome. Cell Death Dis. 2020;11(10):921. doi: 10.1038/s41419-020-03125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bordoni V, Sacchi A, Cimini E, Notari S, Grassi G, Tartaglia E, Casetti R, Giancola L, Bevilacqua N, Maeurer M, Zumla A, Locatelli F, De Benedetti F, Palmieri F, Marchioni L, Capobianchi MR, D’Offizi G, Petrosillo N, Antinori A, Nicastri E, Ippolito G, Agrati C. An inflammatory profile correlates with decreased frequency of cytotoxic cells in COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schrijver IT, Théroude C, Roger T. Myeloid-derived suppressor cells in sepsis. Front Immunol. 2019;10:327. doi: 10.3389/fimmu.2019.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, Rodriguez PC, Sica A, Umansky V, Vonderheide RH, Gabrilovich DI. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waeckel L, Venet F, Gossez M, Monard C, Rimmelé T, Monneret G. Delayed persistence of elevated monocytic MDSC associates with deleterious outcomes in septic shock: a retrospective cohort study. Crit Care. 2020;24(1):132. doi: 10.1186/s13054-020-02857-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhel F, Azzaoui I, Grégoire M, Pangault C, Dulong J, Tadié JM, Gacouin A, Camus C, Cynober L, Fest T, Le Tulzo Y, Roussel M, Tarte K. Early expansion of circulating granulocytic myeloid-derived suppressor cells predicts development of nosocomial infections in patients with sepsis. Am J Respir Crit Care Med. 2017;196(3):315–327. doi: 10.1164/rccm.201606-1143OC. [DOI] [PubMed] [Google Scholar]
- 37.Mathias B, Delmas AL, Ozrazgat-Baslanti T, Vanzant EL, Szpila BE, Mohr AM, Moore FA, Brakenridge SC, Brumback BA, Moldawer LL, Efron PA, the Sepsis, Critical Illness Research Center Investigators Human myeloid-derived suppressor cells are associated with chronic immune suppression after severe sepsis/septic shock. Ann Surg. 2017;265(4):827–834. doi: 10.1097/SLA.0000000000001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janols H, Bergenfelz C, Allaoui R, Larsson AM, Rydén L, Björnsson S, Janciauskiene S, Wullt M, Bredberg A, Leandersson K. A high frequency of MDSCs in sepsis patients, with the granulocytic subtype dominating in gram-positive cases. J Leukoc Biol. 2014;96(5):685–693. doi: 10.1189/jlb.5HI0214-074R. [DOI] [PubMed] [Google Scholar]
- 39.Coopersmith CM, Antonelli M, Bauer SR, Deutschman CS, Evans LE, Ferrer R, Hellman J, Jog S, Kesecioglu J, Kissoon N, Martin-Loeches I, Nunnally ME, Prescott HC, Rhodes A, Talmor D, Tissieres P, De Backer D. Surviving sepsis campaign: research priorities for COVID-19 in critical illness. Crit Care Med. 2021;49(4):598–622. doi: 10.1097/CCM.0000000000004895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Monocyte HLA-DR variation in patients with or without secondary infection. Table S1. Circulating cells phenotyping antibodies clones. Table S2. Site of secondary infection and associated pathogens.
Data Availability Statement
All materials are available upon request to Pr. Pierre Tissières at pierre.tissieres@aphp.fr.