Highlights
-
•
An AI-enabled risk staging method, MRS, is developed using easy-to-acquire parameters.
-
•
Genomic tests cannot be performed owing to economical or geographical constraints.
-
•
MRS does not use cytogenetic abnormalities for risk stage prediction unlike RISS.
-
•
K-adaptive partitioning (KAP) used to find new thresholds for the parameters.
Keywords: Machine learning, Risk stratification of multiple myeloma, J48 decision tree, BIRCH clustering, Hazard ratios, Hematological malignancy
Abstract
Introduction
: An efficient readily employable risk prognostication method is desirable for MM in settings where genomics tests cannot be performed owing to geographical/economical constraints. In this work, a new Modified Risk Staging (MRS) has been proposed for newly diagnosed Multiple Myeloma (NDMM) that exploits six easy-to-acquire clinical parameters i.e. age, albumin, β2-microglobulin (β2M), calcium, estimated glomerular filtration rate (eGFR) and hemoglobin.
Materials and Methods
: MRS was designed using a training cohort of 716 NDMM patients of our inhouse MM Indian (MMIn) cohort and validated on MMIn (n=354) cohort and MMRF (n=900) cohort. K-adaptive partitioning (KAP) was used to find new thresholds for the parameters. Risk staging rules, obtained via training a J48 classifier, were used to build MRS.
Results
: New thresholds were identified for albumin (3.6 g/dL), β2M (4.8 mg/L), calcium (11.13 mg/dL), eGFR (48.1 mL/min), and hemoglobin (12.3 g/dL) using KAP on the MMIn dataset. On the MMIn dataset, MRS outperformed ISS for OS prediction in terms of C-index, hazard ratios, and its corresponding p-values, but performs comparable in prediction of PFS. On both MMIn and MMRF datasets, MRS performed better than RISS in terms of C-index and p-values. A simple online tool was also designed to allow automated calculation of MRS based on the values of the parameters.
Discussion
: Our proposed ML-derived yet simple staging system, MRS, although does not employ genetic features, outperforms RISS as confirmed by better separability in KM survival curves and higher values of C-index on both MMIn and MMRF datasets.
Funding
: Grant: BT/MED/30/SP11006/2015 (Department of Biotechnology, Govt. of India), Grant: DST/ICPS/CPS-Individual/2018/279(G) (Department of Science and Technology, Govt. of India), UGC-Senior Research Fellowship.
Introduction
Staging of disease in oncology practice has been a useful tool for risk stratification as it helps in identifying patients requiring intense therapy upfront and/or a higher monitoring frequency during the follow-up periods. The first staging system for multiple myeloma (MM) was proposed by Salmon and Durie in 1975 that divided patients into three risk categories with differential overall survival [1]. Subsequently, in 2005 an International staging system (ISS) based on two simple laboratory parameters of serum albumin and beta2-microglobulin (β2M) was proposed by Greipp PR and colleagues [2]. Serum albumin reflected the normalcy of the protein compartment and serum β2M reflected the tumor burden. With the development of novel agents such as immunomodulators (IMIDs) and proteasome inhibitors (PSI) for treatment of MM, the landscape of responses and survival changed drastically [3,4]. In addition, the advances in molecular biology allowed investigators to look closely at the genomic changes in MM and, especially, in subgroups of patients with poor outcome. This led to the inclusion of cytogenetic aberrations into the staging system used for MM and thereby, emerged the Revised-ISS (RISS) [5]. The survival data used for developing RISS consisted predominantly of patients who were treated with immunomodulatory agents. As the new class of drugs, i.e., the PSI made their way into the treatment of MM, some of the cytogenetic aberrations like t(4;14) included in the RISS seem to lose their poor prognostic impact [6]. From an academic and research perspective, it is desirable to characterize subset of patients with poor clinical outcome to develop effective therapies but in clinical practice, it is desirable to have a staging system that is based on clinical and laboratory parameters that are easily accessible in healthcare setting across the globe.
In recent times, data analytics including advanced machine learning methods are being used to extract valuable information from medical records. Machine learning algorithms have been shown to be useful in devising risk stratification system in type-2 diabetic patients by Ricci and colleagues [7], in cardiovascular disorders by Ahuja and Schaar [8], in prostate cancer by Varghese [9], in resected gastric cancer[10] and in nasopharyngeal carcinoma [11]. In this study, we used machine learning methods to develop a new risk stratification system, namely, Modified Risk Staging (MRS) for MM using six easy-to-acquire laboratory parameters: albumin, β2M, calcium, eGFR, hemoglobin along with age. The model was developed on a training dataset of patients with newly diagnosed multiple myeloma (NDMM) and validated on two test datasets. Rigorous comparison of the proposed risk staging model with ISS and RISS was undertaken to check its efficacy on the predictions of progression free survival (PFS) and overall survival (OS).
Methods
Study population
The computerized database search on June 28, 2019 with keyword ‘ICD C90’ returned 1675 entries of patients registered at the Institute Rotary Cancer centre, All India Institute of Medical Sciences (AIIMS). A total of 253 patients had plasma cell dyscrasia other than MM, 132 patients were lost to follow up after a single visit (n=111) or before first response could be assessed (n=21), and 121 patients’ records had inadequate clinical and/or laboratory parameters. Patients who died within 16 weeks of diagnosis were labelled as early deaths (n=99) and were excluded from the staging algorithms. Remaining cohort of 1070 Indian patients of MM, referred to as the MMIn cohort, was evaluated in this study (Supplementary Fig. S1). An independent cohort of 900 patients of MM enrolled in Multiple Myeloma Research Foundation (MMRF) repository, for which the clinical and laboratory data is available publicly, was used for validation.
Clinical and laboratory characteristics
The clinical, laboratory, and radiological data was obtained from the medical case files. A subset of patients, for whom the molecular data (n=627) was available, were assigned RISS as described previously [12]. Treatment response was assessed as per the International uniform response criteria for multiple myeloma [13]. Progression free survival (PFS) was calculated from the date of diagnosis until progression or death. Overall survival (OS) was calculated from the date of diagnosis until death due to any cause or was censored at last follow-up. Clinical and laboratory features of the patients are given in Supplementary Table S1.
Design strategy
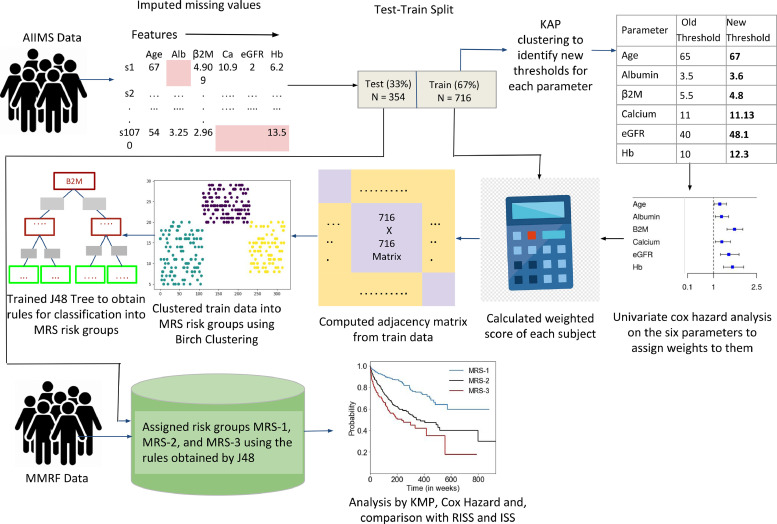
Patients (n=1070) in MMIn were randomly split in the ratio of 67:33 as training (n=716) and test cohorts (n=354). The test cohort did not have any missing value. In the training cohort, 41 patients (5.7% of 716 patients) had one or two missing values that were imputed with the median value of the parameters. The training data was used to develop the proposed staging system called Modified Risk Staging (MRS) and the test dataset was used to evaluate the correctness of the MRS. The staging system was then validated on MMRF data. No missing imputation was applied on the test cohort or the MMRF dataset. Complete MRS design strategy, shown in Fig. 1, is explained below.
Fig. 1.
Workflow for the development of Modified Risk staging (MRS) system for Multiple Myeloma.
Initially seven parameters, i.e., albumin, β2M, calcium, eGFR, hemoglobin, lactate dehydrogenase (LDH), and age were evaluated for designing MRS. β2M and LDH levels are reflective of tumor burden and serum albumin, hemoglobin, calcium and creatinine are reflective of the bone and renal homeostasis. eGFR was calculated from creatinine concentration using MDRD eGFR equation [14]. LDH values were brought to a common scale by multiplying each entry by 280 and dividing it by the upper limit of LDH provided for that particular entry. For each parameter, patients were initially divided into high-risk and low-risk groups using the well-established cut-offs of these parameters. Established thresholds for albumin and β2M are derived from ISS [2] and for eGFR, calcium, hemoglobin are derived from revised IMWG criteria [15]. Log-rank test on the Kaplan-Meier curves yielded significant p-values for all the parameters except LDH which was, therefore, not used further (Table 1). Next, K-adaptive partitioning (KAP) [16] algorithm was used to find new threshold values for the six parameters. KAP was performed on the training patients’ parameters yielding two threshold values for each parameter, one from PFS and the other from OS analysis. The threshold with lower p-value of the two was chosen as the new cut-off for each parameter.
Table 1.
Comparison of established and proposed cut-offs for laboratory parameters for stratification of patients for progression free survival (PFS) and overall survival (OS) in training data (n=716 patients) using Kaplan Meier analysis and the weights assigned to the laboratory parameters for calculation of score.
| PFS |
OS |
||||||
|---|---|---|---|---|---|---|---|
| Parameter | Established Threshold value | Proposed Threshold value | p-value with established threshold | p-value with proposed threshold | p-value with established threshold | p-value with proposed threshold | Weights assigned after univariate Cox Hazard analysis |
| Age | >65 | >67 | 0.19 | 0.016 | 8.7e-4 | 1.86e-5 | 2.85 |
| Albumin | ≤ 3.5 | ≤ 3.6 | 0.59 | 0.089 | 0.06 | 2.7e-3 | 1 |
| β2M | ≥ 5.5 | ≥ 4.8 | 5.41e-6 | 5.81e-6 | 5.6e-6 | 8.78e-8 | 2.85 |
| Calcium | ≥ 11 | ≥ 11.13 | 0.011 | 6.5e-3 | 0.02 | 0.029 | 1.07 |
| eGFR | ≤ 40 | ≤ 48.1 | 0.03 | 0.012 | 7.9e-3 | 1.2e-3 | 1.14 |
| Hb | ≤ 10 | ≤ 12.3 | 0.012 | 4.88e-4 | 0.027 | 1.2e-4 | 4.0 |
| LDH | < 280 | < 95 | 0.66 | 0.96 | 0.52 | 0.43 | – |
*Most significant p-values under each category are highlighted in bold.
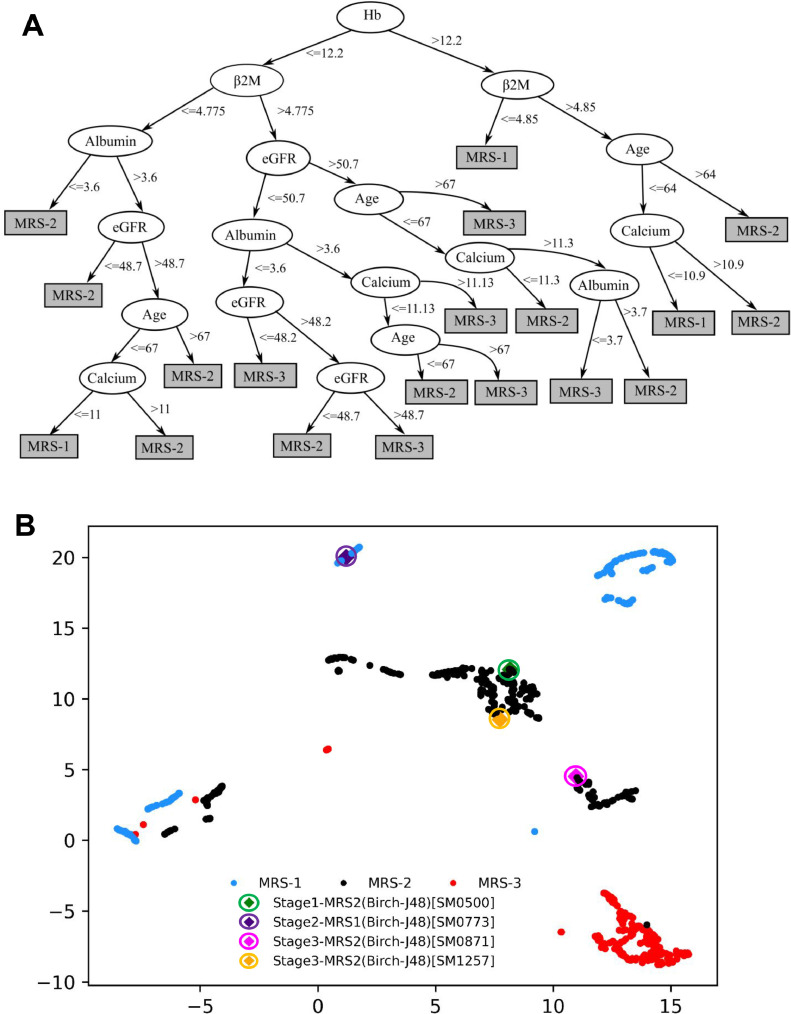
For the cumulative integration of parameters into risk staging, weights were assigned to each parameter using their respective hazard ratios (HR) for PFS and OS obtained from the univariate Cox-proportional hazard test on the training data (Supplementary Table S2). For each parameter, the highest of the two HR values obtained from PFS and OS was chosen and normalized using ‘minmax’ scaling in the range of 1 to 4. The scaled HR values were assigned as the respective weights of each of the parameters (Table 1). This captures the relative impact of each parameter on the patients’ survival. Next, a new score for each patient was calculated by adding the weights of all those parameters that had values (in the respective patient) beyond the threshold defined for the high-risk group. These patient scores were used to compute an adjacency matrix of 716 rows and 716 columns (columns are features), where each row corresponds to one patient and each entry in the row is the absolute difference between the score of that patient with each of the 716 patients including self. BIRCH (Balanced Iterative Reducing and Clustering using Hierarchies) Clustering, an unsupervised ML method, was applied on the adjacency matrix to cluster the patients of the training dataset into three risk groups [17]. Each cluster of patients was assigned one label: Stage-1 (low-risk), Stage-2 (intermediate-risk), or Stage-3 (high-risk). Initially assigned risk stages via BIRCH clustering on training patients were used as ground truth labels. BIRCH is an unsupervised clustering algorithm that works on the entire data. It does not provide rules that can be employed on any prospective subject to determine its risk stage. Hence, there was a need to obtain rules of risk staging. At the same time, a supervised classifier cannot be trained initially, because there is no risk stage-label. A novel methodology is employed, wherein the risk stage labels provided by BIRCH were used as ground truth class labels (risk stages) on the training data to train a J48 classifier (a rule-based supervised decision-tree classifier). The trained J48 classifier provided the rules in terms of laboratory parameters and age for the identification of risk groups, labeled as MRS-1 (low risk), MRS-2 (intermediate-risk), and MRS-3 (high-risk) (Fig. 2). The risk stage assigned by the J48 tree was considered the actual risk class for each patient. All the patients in the test dataset were also assigned to one of the MRS groups using the J48 rules. These MRS groups were then analyzed for OS and PFS, and compared with those obtained with the ISS and RISS.
Fig. 2.
A- Hierarchical rule based tree structure to assign data samples to MRS-1, MRS-2 and MRS-3 groups. Parameters: Age: Age; Alb: Albumin; β2M: beta2-macroglobulin; Ca: Calcium; eGFR: estimated glomerular filtration rate and Hb: hemoglobin. B- UMAP scatter plot of training data depicting the three labels identified by J48 classifier rules and the four mismatched patients (highlighted in circles).
Results
Clinical and laboratory characteristics of myeloma patients
The baseline demographic and laboratory features of patients are given in Supplementary Table S1; the training and the test cohorts were comparable in baseline demographic, laboratory, and clinical parameters (Supplementary Table S1). All the patients received novel agents (IMIDs:thalidomide or lenalidomide and/or PSI i.e. bortezomib) either as primary or maintenance therapy and dexamethasone. 56.5% of patients received triplet regimen. With a median follow up period of 166 weeks (range: 14–961 weeks), 626 patients had progressed and 372 died; the median PFS and OS of the entire cohort was 117 weeks and 166 weeks, respectively.
Patients in the training cohort (n=716) were initially stratified into high-risk and low-risk groups based on the new thresholds deduced by KAP algorithm that yielded better separability (lower p-values) between the two groups (Table 1). Weights assignment to each parameter on a scale of 1 to 4 led to the highest weight of 4 to hemoglobin and 2.85 to β2M and age, indicating them to be the important prognostic factors for risk stratification (Table 1). The trained J48 classifier yielded the rules for the risk stage assignment (Fig. 2A) as well as ten-fold cross-validation accuracy of 96.5% and the weighted-average ROC area of 97.5%. The patients were assigned labels, MRS-1 (Low-risk), MRS-2 (Intermediate-risk) and MRS-3 (High-risk) using the rules obtained from the J48 classifier. An online version of the MRS calculator (Supplementary Fig. S2) has also been developed. It calculates the risk stage of the patient based on the values of the six parameters, age, albumin, β2M, calcium, eGFR and hemoglobin. It also displays median PFS and OS in weeks for the patient depending on his assigned risk group. Median PFS and OS have been calculated on the combined data (n=1970 patients) of MMIn and MMRF cohort.
Results on the training (n=716), test (n=354) and complete MMIn cohort (n=1070)
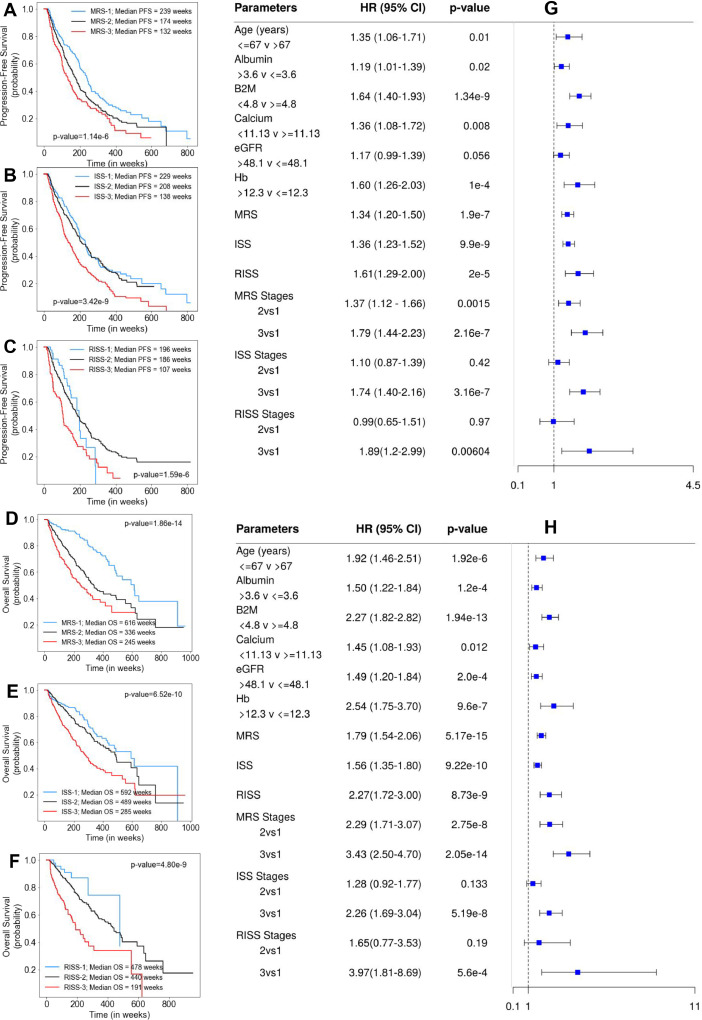
Largest proportion of training cohort (n=716) were assigned to MRS-2 (n=332, 46.36%), followed by MRS-3 (n=199, 27.80%) and MRS-1 (n=185, 25.84%). Results of the median PFS on MRS groups (p=6.28e-6) and ISS groups (p=1.25e-5) as well as of median OS on MRS groups (p=8.15e-10) and ISS groups (p=2.03e-5) show better performance of MRS than ISS (lower p-values; Supplementary Table S3). Similar findings were obtained on the test cohort (n=354; Supplementary Table S3). Univariate Cox analysis of the entire patient cohort (n=1070, Supplementary Table S4), revealed increased risk of progression and mortality for age>67 years, albumin≤3.6, β2M≥4.8, calcium≥11.13, eGFR≤48.1 and hemoglobin≤12.3. Using MRS, the largest proportion of patients were placed in MRS-2 (n=511, 47.76%) followed by MRS-1 (n=281, 26.26%) and MRS-3 (n=278, 25.98%). KM survival analysis of MRS groups indicated statistically significant difference in PFS between MRS-1 and MRS-2 groups (p=0.0012) and between MRS-2 and MRS-3 groups (p=0.0055). For ISS, the difference was significant between ISS-2 and ISS-3 groups (p=1.118e-6), but not between ISS-1 and ISS-2 groups (p=0.46). For RISS, there was statistically significant difference between RISS-2 and RISS-3 (p=5.6e-7) but not between RISS-1 and RISS-2 (p=0.96). KM survival analysis of MRS groups further revealed statistically significant difference in OS between MRS-1 and MRS-2 groups (p=5.9e-9) and between MRS-2 and MRS-3 groups (p=0.001). For ISS, the difference in OS was significant between ISS-2 and ISS-3 groups (p=3.12e-6) but not between ISS-1 and ISS-2 groups (p=0.118) and. For RISS, there was statistical difference in OS between RISS-2 and RISS-3 groups (p=8.32e-9), but was not significant between RISS-1 and RISS-2 groups (p=0.2) (Fig. 3). Results of multivariate Cox hazards model are also shown in Supplementary Table S5.
Fig. 3.
A, B, C- Progression-Free Survival in patients with MM from MMIn cohort (n=1070) stratified by the proposed MRS (n=1070), ISS (n=1070) and RISS (n=627), respectively. D, E, F- Overall Survival in patients with MM from MMIn cohort (n=1070) stratified by the proposed MRS (n=1070), ISS (n=1070) and RISS (n=627), respectively. G, H- Cox Hazard Analysis of PFS and OS. Univariate analysis of parameters- Age, Albumin, β2M, Calcium, eGFR, Hb and different staging methods-MRS, ISS and RISS. Multivariate analysis of different groups of MRS, ISS and RISS.
The C-Statistic computed on MRS and ISS demonstrate slightly better performance of MRS than ISS with respect to PFS and OS. C-Statistic for MRS was 0.57 (HR=1.34, 95% CI=1.20–1.5, p=1.9e-7) for PFS and 0.63 (HR=1.79, 95% CI=1.54–2.06, p=5.17e-15) for OS as compared to 0.57 (HR=1.36, 95% CI=1.23–1.52, p=9.9e-9) and 0.60 (HR=1.56, 95% CI=1.35–1.8, p=9.22e-10) for ISS (Fig. 3). RISS was available for only 627 patients, hence, MRS stages were determined separately for these patients. C-statistic was better for MRS with values of 0.57 (HR=1.38, 95% CI=1.17–1.62, p=9e-5) for PFS and 0.63 (HR=1.90, 95% CI=1.53–2.35, p=6.3e-9) for OS as compared to 0.56 (HR=1.61, 95% CI=1.29–2.00, p=2e-5) and 0.60 (HR=2.27, 95% CI=1.72–3.00, p = 8.73e-9) for RISS (Fig. 3).
Results on the MMRF cohort
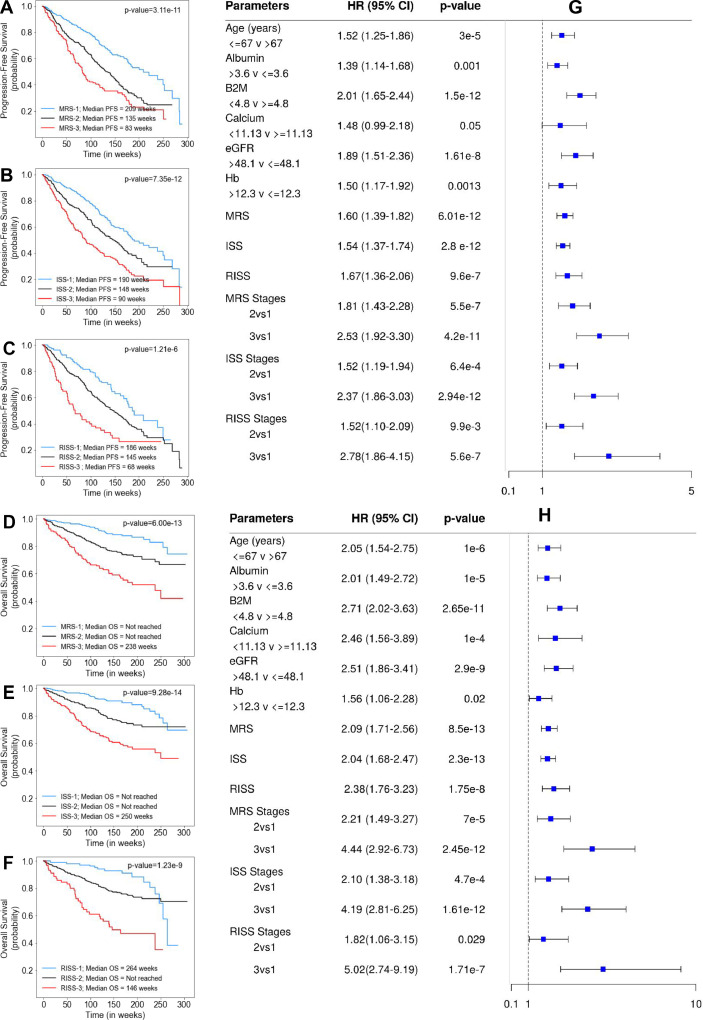
MRS was further evaluated by comparing it with ISS and RISS using the MMRF dataset. ISS was available for 900 patients and RISS was available for 703 patients. In MMRF cohort, majority of the patients were placed in MRS-2 (n=405, 45%) followed by MRS-1 (n=348, 38.67%) and MRS-3 (n=147, 16.33%). In the univariate Cox hazard analysis of the MMRF data, risk of progression and mortality was increased for age>67 years, β2M≥4.8, albumin≤3.6, hemoglobin≤12.3, eGFR≤48.1 and calcium≥11.13 (Supplementary Table S4). Results of the median PFS on MRS groups (p=3.11e-11), ISS groups (p=7.35e-12), and RISS groups (p=1.21e-6) as well as of median OS on MRS groups (p=6.00e-13), ISS groups (p=9.28e-14), and RISS groups (p=1.23e-9) show comparable performance of MRS than ISS and RISS (comparable p-values; Supplementary Table S3; Fig. 4). The risk of progression and that of mortality was increased for MRS 2vs1, MRS 3vs1, ISS 2vs1, ISS 3vs1, RISS 2vs1, and RISS 3vs1 (Fig. 4). The C-Statistic for MRS in MMRF data is 0.60 (HR=1.60, 95% CI=1.39–1.82, p=6.01e-12) for PFS and 0.65 (HR=2.09, 95% CI=1.71–2.56, p=8.50e-13) for OS as compared to 0.61 (HR=1.54, 95% CI=1.37–1.74, p=2.8e-12) and 0.667 (HR=2.04, 95% CI=1.68–2.47, p=2.3e-13) for ISS; 0.58 (HR=1.67, 95% CI=1.36–2.06, p=9.60e-7) for RISS and 0.62 (HR=2.38, 95% CI=1.76–3.23, p=1.75e-8) for RISS, respectively. Results of multivariate Cox hazards model are also shown in Supplementary Table S6. The 5-year OS for the complete MMIn data (n=1070) was 85.82% for MRS-1, 61.73% for MRS-2 and 48.78% for MRS-3 (Table 2). The difference in the percentages of the 5-year OS and median OS for different risk groups indicated that the groups were significant. A similar stratification was achieved when the MRS model was applied on the MMRF test dataset. The 5-year OS for MMRF data was 79.06% for MRS-1, 66.66% for MRS-2 and 41.91% which is quite comparable to that obtained in the MMIn data.
Fig. 4.
A, B, C- Progression-Free Survival in patients with MM from MMRF cohort (n=900) stratified by the proposed MRS (n=900), ISS (n=900) and RISS (n=703), respectively. D, E, F- Overall Survival in patients with MM from MMRF cohort (n=900) stratified by the proposed MRS (n=900), ISS (n=900) and RISS (n=703), respectively. G, H- Cox Hazard analysis of PFS and OS. Univariate analysis of parameters- Age, Albumin, β2M, Calcium, eGFR, Hb and different staging methods- MRS, ISS and RISS. Multivariate analysis of different groups of MRS, ISS and RISS.
Table 2.
Prediction of Progression-free survival and overall survival (in%) for MRS, ISS and RISS at 1, 2, 3, 4 and 5 years in MMIn (n=1070) and MMRF datasets (n=900).
| MMIn dataset | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Year | MRS (n = 1070) |
ISS (n = 1070) |
RISS (n = 627) |
|||||||
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | ||
| PFS | 1 | 89.78 | 85.73 | 77.11 | 89.64 | 88.48 | 80.24 | 91.11 | 86.76 | 68.24 |
| 2 | 77.00 | 71.45 | 58.46 | 80.33 | 73.89 | 62.68 | 84.15 | 72.39 | 53.41 | |
| 3 | 69.13 | 54.94 | 43.02 | 68.50 | 62.48 | 46.22 | 62.62 | 58.07 | 36.87 | |
| 4 | 55.66 | 40.28 | 33.60 | 53.22 | 49.98 | 33.48 | 33.46 | 45.28 | 27.44 | |
| 5 | 43.04 | 33.30 | 27.48 | 40.25 | 41.67 | 27.22 | 26.77 | 36.66 | 18.29 | |
| OS | 1 | 96.75 | 93.19 | 86.74 | 93.64 | 95.88 | 89.94 | 97.78 | 95.56 | 82.27 |
| 2 | 94.39 | 84.18 | 71.89 | 90.10 | 88.82 | 78.01 | 93.28 | 87.91 | 70.40 | |
| 3 | 91.72 | 76.52 | 63.62 | 87.84 | 82.63 | 69.45 | 90.76 | 81.80 | 59.32 | |
| 4 | 90.58 | 68.34 | 55.29 | 85.82 | 76.00 | 61.30 | 86.98 | 73.61 | 49.03 | |
| 5 | 85.82 | 61.73 | 48.78 | 80.94 | 71.91 | 52.79 | 86.98 | 68.65 | 40.39 | |
| MMRF dataset | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Year | MRS (n = 900) |
ISS (n = 900) |
RISS (n = 703) |
|||||||
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | ||
| PFS | 1 | 88.95 | 77.64 | 69.84 | 89.95 | 79.44 | 69.08 | 90.32 | 80.49 | 62.03 |
| 2 | 77.56 | 59.90 | 42.20 | 77.11 | 61.86 | 46.24 | 79.57 | 61.73 | 38.13 | |
| 3 | 62.82 | 42.17 | 35.10 | 59.75 | 48.21 | 33.6 | 62.95 | 47.73 | 28.99 | |
| 4 | 51.49 | 28.09 | 23.16 | 47.7 | 32.95 | 22.35 | 46.41 | 33.74 | 26.36 | |
| 5 | 35.61 | 24.66 | 13.89 | 34.81 | 29.27 | 14.37 | 27.69 | 24.96 | 26.36 | |
| OS | 1 | 96.93 | 90.90 | 82.81 | 96.76 | 91.61 | 85.29 | 98.07 | 91.17 | 83.21 |
| 2 | 93.73 | 82.69 | 66.44 | 93.91 | 85.24 | 68.65 | 96.11 | 83.81 | 61.09 | |
| 3 | 88.43 | 76.18 | 59.20 | 90.73 | 77.16 | 60.79 | 92.86 | 77.28 | 49.78 | |
| 4 | 86.56 | 72.64 | 51.88 | 88.21 | 73.39 | 55.8 | 88.33 | 73.52 | 47.02 | |
| 5 | 79.06 | 66.66 | 41.91 | 74.73 | 71.92 | 48.93 | 57.48 | 70.49 | 35.27 | |
For MMRF data, 5-year OS was 57.48% for RISS-1, 70.49% for RISS-2 and 35.27% for RISS-3 (Table 2) which suggested some anomaly since RISS-1 should have a higher OS as compared to RISS-2. This anomaly may be because of assigning a much larger number of patients to RISS-2 having greater OS time as compared to RISS-1.
Discussion
The advent of immunomodulatory drugs and PSI has considerably improved treatment outcomes in MM and hence, the current risk stratifications based on ISS and RISS need to be relooked at. ISS is a simple model based on two laboratory parameters of serum-albumin and β2M, but is largely based on data from patients treated in the pre-IMID era and is not very informative on PFS in NDMM patients [2]. Since the length of PFS is an important predictor of long-term OS, a better model to assess PFS is desirable [12]. The RISS for MM takes into consideration the molecular abnormalities and is based on data from patients treated with either IMIDS or PI, or both and is informative on PFS as well as OS [5]. However, many molecular aberrations such as 1q gain and chromothripsis that adversely affect outcome in MM have been overlooked and t(4;14) included in RISS has lost significance in patients treated with triplet regimens [6,18]. The preferred frontline treatment for MM is triplet regimen consisting of an IMID, PI and steroid regardless of molecular aberrations or risk stratification. The targeted therapy in MM is reserved for patients with RAS and BRAF mutations in progressive disease and relapsed refractory setting, and the upfront molecular characterization adds to the cost of healthcare in clinical settings. It is, thus, desirable to develop simple risk staging models for MM to enable judicious use of healthcare resources reserving the molecular analysis for patients who progress or relapse on frontline therapy and in setting of clinical trials when a targeted therapy is intended to be used.
Performance of MRS as compared to RISS and ISS
On the MMIn dataset, the proposed MRS performed better than ISS in prediction of OS in terms of C-index, HR, and p-values, while the performance in PFS was comparable. The performance of MRS was superior to RISS in terms of C-index and p-values. On the MMRF dataset, the performance of MRS was superior to RISS in terms of C-index and p-values, but was comparable to ISS. The performance on MMRF dataset indicated that there may be nuances with respect to ethnicity and race as the population of MMIn and MMRF dataset is different in terms of ethnicity and race. The algorithm was trained on MMIn dataset using the KAP-proposed thresholds on MMIn dataset while the developed MRS model was only tested on the MMRF dataset.
Specific characteristics of different staging systems
Levels of albumin and β2M are significant in ISS staging method, while LDH along with albumin and β2M are significant in RISS staging method (Supplementary Table S8). LDH was not observed to be significant in our preliminary findings and was, therefore, excluded from MRS staging. eGFR although significant came lower in the tree. The possible reason could be that renal dysfunction gets reversed in a significant number of patients of MM treated with novel agents as opposed to alkylating agents used in the past to treat MM. Age remained a significant parameter of outcome in the new staging as well. In MRS, none of the deranged parameters individually dominated the underlying risk of death. For example, when the level of hemoglobin was less than 12.2 and β2M was greater than 4.775, then for eGFR levels less than 50.7 and albumin and calcium levels greater than 3.6 and 11.13 respectively, the patient was placed in the MRS-3 (High risk) group. However, even if the eGFR level was greater than 50.7 but age was greater than 67, the patient was still placed in MRS-3. Hence, it was evident that poor outcome was associated with a combination of abnormally high or low levels of multiple prognostic factors. Thus, MRS staging does not rely on a single parameter but takes into consideration multiple parameters that are associated with hemodynamic systems as a whole.
In the MMRF dataset, 703 patients out of 900 had RISS labels. 91 out of these 703 patients (12.9%) were labeled as RISS-3. In these 91 patients, 43 were labeled as MRS-3, 43 as MRS-2 and 5 were labeled as MRS-1. The median OS for these 5 patients was 70 weeks and no death event was observed in any of these patients. In fact, there was no disease progression in these 5 patients in the first year of diagnosis. Hence, the staging provided by MRS is more accurate as it positioned these patients in the low-risk stage (MRS-1) contrary to the high-risk stage (RISS-3) provided by the RISS scheme. Similarly, 239 patients out of 900 (26.56%) patients were stratified as ISS-3. Out of these 239 patients, 125 were labeled MRS-3, 105 were labeled MRS-2 and 9 were labeled MRS-1. Median OS of these 9 patients was 70 weeks and no death event was observed in these patients. Further, there was no disease progression in the first year of diagnosis. MRS correctly placed the patients in a low-risk group as compared to ISS. Thus, it can be deduced that MRS helps in better filtering of the patients compared to RISS and ISS. Further, there were overall 147 patients (16.33%) that were stratified as MRS-3 in MMRF dataset. None of the 147 patients was present in the low-risk stages of ISS and RISS, thereby establishing the efficacy of MRS staging in identification of high risk patients.
Hierarchical rules in J48 tree and mismatched labels between BIRCH and J48 classifier
Hemoglobin and β2M were observed to be the most important poor prognostic factors in MRS staging followed by others. Hemoglobin had the greatest weight assigned based on hazard ratios obtained from the univariate Cox hazard analysis. It was present at the first level (highest node) for classification in the J48 tree, thus confirming the high prognostic value of β2M on PFS and OS. It was observed that no leaf node of the decision ended in MRS-3 stage for hemoglobin values greater than the threshold 12.2, while five leaf nodes ended in MRS-3 stage for β2M values lower than 12.2, thereby, indicating that the patients with higher disease load have lower hemoglobin. J48 tree utilized the values of hemoglobin at continuous scale and provided a single cut-off for hemoglobin as 12.2 in the decision rules (Fig. 2A) which is quite close to the proposed threshold value of 12.3 obtained via KAP and hence, justified the choice of our new threshold for hemoglobin. Lower levels of hemoglobin were predictors of poor outcome and were associated with high-risk patients, as evident from the hierarchical rules obtained from J48 classifier. Similarly, β2M levels of 4.775 or lower were associated with either low or intermediate risk as observed in the J48 classifier rules. J48 tree provided two cut-offs for β2M, 4.775 and 4.85 in the decision rules. These cut-offs were quite close to the proposed threshold for β2M, 4.8 and justified the choice of our new threshold for β2M. Apart from hemoglobin and β2M, the hierarchical rules in the J48 tree for other parameters also exhibited values comparable to the proposed thresholds.
The J48 assigned risk group label of four patients (0.5%) of the training set were found to have a mismatch with that assigned by the unsupervised BIRCH clustering. Training data was visualized using UMAP scatter plot. UMAP (Uniform Manifold Approximation and Projection) is a technique of dimensionality reduction mostly used for visualization of high dimensional data. It is evident from the plot that SM0500 (green), SM0773 (violet), SM0871 (magenta), and SM1257 (yellow) were assigned labels different from the labels obtained from BIRCH clustering. Further, there is a possibility of overfitting due to a smaller number of training samples (n=716), which was addressed by performing pruning on the J48 tree. Results in terms of p-values and hazard ratios in the test set of MMIn dataset and in the overall cohort of MMRF data is further suggestive of the reduced possibility of overfitting.
Sensitivity analysis of the J48 decision tree classifier
We first trained the J48 decision tree classifier using all the variables and observed the 10-fold cross validation accuracy. As already discussed, the ground truth risk stages were obtained via BIRCH clustering. We then trained the J48 classifier under the same settings six times, each time excluding one of the variables and observed their classification accuracy. Highest classification accuracy was 96.5% when all the parameters were used for training. Classification accuracy was least affected by absence of calcium. However, classification accuracy decreased drastically (Supplementary Table S7) in absence of one of the variables- albumin, β2M, Hb, eGFR and age (ordered in rank from the highest impact to the lowest impact).
Conclusion
Overall, this work presents a new reliable and inexpensive staging system, namely, MRS that utilizes easily acquirable laboratory parameters. It is valuable for the settings where genomic tests cannot be performed owing to economical and/or geographical constraints. The thresholds, proposed by this study, of laboratory parameters via KAP produce distinct PFS and OS patterns that are quantified by minimum p-value with better separation of MRS groups compared with those obtained with established thresholds and hence, can be adopted. 10-fold cross validation classification accuracy and ROC area confirm that our hierarchical stratification model can correctly classify patients into different risk groups. Application of machine learning techniques in MRS has led to better prediction of the survival outcome and identified different risk groups with distinct characteristics. The study recommends training of machine learning models on larger datasets because that can provide efficient upfront prognostication that may be useful in selection of therapy of appropriate intensity especially in high-risk MM patients. Further, the performance on MMRF dataset indicates that there may be nuances with respect to ethnicity and race. MRS model is trained on MMIn dataset using the new cut-offs proposed via KAP and is only tested on the MMRF dataset. Both the dataset belong to populations of different ethnicity and race. Therefore, the impact of ethnicity and race on risk staging ML models can be explored in future.
Data availability/calculator availability
An online version of the MRS calculator (https://github.com/AkankshaFarswan/MRS_Calculator) (Supplementary Fig S2) is also available which provides the risk stage of the patient based on the values of the six parameters-age, albumin, β2M, calcium, eGFR and hemoglobin.
Author contributions statement
Akanksha Farswan: Methodology, Software, Formal analysis, Investigation, Writing- Original draft preparation. Anubha Gupta: Methodology, Investigation, Validation, Writing- Original draft preparation, Resources, Project management, Supervision. Ritu Gupta: Conceptualization, Investigation, Validation, Resources, Writing- Original draft preparation, Project management, Supervision. Saswati Hazra: Formal analysis, Writing- Original draft preparation. Sadaf Khan: Resources. Lalit Kumar: Resources. Atul Sharma: Resources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
This work was supported by grant from Department of Biotechnology, Govt. of India [Grant: BT/MED/30/SP11006/2015] and Department of Science and Technology, Govt. of India [Grant: DST/ICPS/CPS-Individual/2018/279(G)]. Akanksha Farswan would like to thank the University Grants Commission, Govt. of India for the UGC-Senior Research Fellowship. Authors acknowledge MMRF and dbGaP (Project #18964) for providing the dataset. These data were generated as part of the Multiple Myeloma Research Foundation Personalized Medicine Initiative. Authors would also like to thank centre of Excellence in Healthcare, IIIT-Delhi for support in their research.
Role of funding source
The funding bodies had no role in study design, data collection, data analysis, data interpretation or writing of the report. The corresponding authors had full access to all the data used in the study and had final responsibility for the decision to submit for publication.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101157.
Contributor Information
Anubha Gupta, Email: anubha@iiitd.ac.in.
Ritu Gupta, Email: drritu.laboncology@aiims.edu.
Appendix. Supplementary materials
References
- 1.Durie B.G., Salmon S.E. A clinical staging system for multiple myeloma correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer. 1975;36(3):842–854. doi: 10.1002/1097-0142(197509)36:3<842::aid-cncr2820360303>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 2.Greipp P.R., Miguel J.S., Durie B.G. International staging system for multiple myeloma. J. Clin. Oncol. 2005;23(15):3412–3420. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 3.Dimopoulos M.A., Delimpasi S., Katodritou E. Significant improvement in the survival of patients with multiple myeloma presenting with severe renal impairment after the introduction of novel agents. Ann. Oncol. 2014;25(1):195–200. doi: 10.1093/annonc/mdt483. [DOI] [PubMed] [Google Scholar]
- 4.Fouquet G., Pegourie B., Macro M. Safe and prolonged survival with long-term exposure to pomalidomide in relapsed/refractory myeloma. Ann. Oncol. 2016;27(5):902–907. doi: 10.1093/annonc/mdw017. [DOI] [PubMed] [Google Scholar]
- 5.Palumbo A., Avet-Loiseau H., Oliva S. Revised international staging system for multiple myeloma: a report from International Myeloma Working Group. J. Clin. Oncol. 2015;33(26):2863. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avet-Loiseau H., Leleu X., Roussel M. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p) J. Clin. Oncol. 2010;28(30):4630–4634. doi: 10.1200/JCO.2010.28.3945. [DOI] [PubMed] [Google Scholar]
- 7.Beatrice Ricci, van der Schaar M., Yoon J. Machine learning techniques for risk stratification of non-ST-elevation acute coronary syndrome: the role of diabetes and age. Circulation. 2017;136(suppl_1):A15892. [Google Scholar]
- 8.K. Ahuja, M. van der Schaar, Risk-stratify: confident stratification of patients based on risk. arXiv preprint arXiv:1811.00753, 2018.
- 9.Varghese B., Chen F., Hwang D. Objective risk stratification of prostate cancer using machine learning and radiomics applied to multiparametric magnetic resonance images. Sci. Rep. 2019;9(1):1570. doi: 10.1038/s41598-018-38381-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bria E., De Manzoni G., Beghelli S. A clinical–biological risk stratification model for resected gastric cancer: prognostic impact of Her2, Fhit, and APC expression status. Ann. Oncol. 2013;24(3):693–701. doi: 10.1093/annonc/mds506. [DOI] [PubMed] [Google Scholar]
- 11.Hui E.P., Li W.F., Ma B.B. Integrating post-radiotherapy plasma Epstein-Barr virus DNA and TNM stage for risk stratification of nasopharyngeal carcinoma to adjuvant therapy. Ann. Oncol. 2020;31(6):769–779. doi: 10.1016/j.annonc.2020.03.289. [DOI] [PubMed] [Google Scholar]
- 12.Gupta R., Kaur G., Kumar L. Nucleic acid based risk assessment and staging for clinical practice in multiple myeloma. Ann. Hematol. 2018;97(12):2447–2454. doi: 10.1007/s00277-018-3457-8. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S., Paiva B., Anderson K.C. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17(8):e328–e346. doi: 10.1016/S1470-2045(16)30206-6. [DOI] [PubMed] [Google Scholar]
- 14.Florkowski C.M., Chew-Harris J.S. Methods of estimating GFR–different equations including CKD-EPI. Clin. Biochem. Rev. 2011;32(2):75. [PMC free article] [PubMed] [Google Scholar]
- 15.Rajkumar S.V. Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2016;91(7):719–734. doi: 10.1002/ajh.24402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S.H. Eo, H.J. Kang, S.M. Hong, H. Cho, K-adaptive partitioning for survival data, with an application to cancer staging. arXiv preprint arXiv:1306.4615, 2013.
- 17.Goswami C., Poonia S., Kumar L. Staging system to predict the risk of relapse in multiple myeloma patients undergoing autologous stem cell transplantation. Front. Oncol. July 2019;9:633. doi: 10.3389/fonc.2019.00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur G., Gupta R., Mathur N. Clinical impact of chromothriptic complex chromosomal rearrangements in newly diagnosed multiple myeloma. Leuk. Res. 2019;76:58–64. doi: 10.1016/j.leukres.2018.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
An online version of the MRS calculator (https://github.com/AkankshaFarswan/MRS_Calculator) (Supplementary Fig S2) is also available which provides the risk stage of the patient based on the values of the six parameters-age, albumin, β2M, calcium, eGFR and hemoglobin.