Abstract
Long non-coding RNA (lncRNA) TP53 target 1 (TP53TG1) is known to be strongly associated with tumor and cancer progression. However, its expression profile, unique role, and regulatory pathways in retinoblastoma (RB) are not known. Here, we revealed a large expression of TP53TG1 in RB tissues and cell lines. Conversely, we showed marked suppression of cell proliferation, migration, and invasion in TP53TG1 knocked down RB cells. Mechanistically, we established that TP53TG1 directly interacted with microRNA (miR)-33b in RB cells. Furthermore, TP53TG1 transcripts were found to be inversely correlated with miR-33b in RB tissues. We also showed that miR-33b suppression partly reversed the TP53TG1 knockdown mediated effects on tumor biology. Finally, TP53TG1 was shown to modulate the levels of SHC Binding and Spindle Associated 1 (SHCBP1), a direct target of miR-33b in RB cells. Based on the above data, we propose that TP53TG1 regulates RB progression via its modulation of the miR-33b/SHCBP1 pathway.
Keywords: long non-coding RNA, TP53TG1, SHCBP1, miR-33b, retinoblastoma
Introduction
Retinoblastoma (RB) is a highly prevalent eye cancer affecting children 1 . Recent advancements in early tumor detection and treatment have enhanced the chances of survival in children with RB 2 . But, in many cases, RB survivors experience vision loss due to metastasis and resistant-chemotherapy or radiotherapy 2,3 . Given the detrimental effects of RB, it is crucial to explore the underlying molecular pathways of RB initiation and metastasis in order to develop targeted therapeutic drugs against this detrimental disease.
Using high-throughput transcriptome analyses and genome-wide assays, over 90% of total mammalian genome was found to encode numerous short or long noncoding RNAs (lncRNAs) 4,5 . MicroRNAs (miRNAs) are 19-25 nucleotides long noncoding (nc) RNA molecules that modulate the expression of protein-coding genes by marking them for degradation 6 . LncRNAs, on the other hand, are >200 nucleotide long no-coding transcripts 7 that are biologically active in that they regulate multiple physiological and pathological systems, including cellular multipotency, differentiation, apoptosis, X chromosome imprinting, epigenetic regulation, alternative splicing, RNA decay, and tumorigenesis 8,9 . As a result, multiple lncRNAs were shown to be dysregulated in cancerous cells, suggesting a role as tumor modulators or a potential role as tumor biomarkers 1,10,11 . Recently, lncRNAs were also reported to be involved in RB growth and metastasis 12 –18 . In one study, overexpression of a novel lncRNA transcript, RBAT1 resulted in the significant elevation of E2F3 oncogene expression via the recruitment of the transcription factor HNPNPL 15 . Other studies also suggested that lncRNA can act as an miRNA sponge to inhibit or block the transcription of tumor suppressor genes and accelerate the progression of RB 19,20 . In fact, the lncRNA ILF3-AS1 was highly expressed in both RB tissues and cell lines and was reported to promote RB growth and metastasis via the miR-132-3p/SMAD2 pathway 21 .
In addition, we also focused on a unique p53-induced lncRNA TP53TG1. A previous study demonstrated that TP53TG1 (GenBank Accession Number NM_007233), a long intergenic non-protein-coding RNA with 751 nt in length, was revealed to exert a tumor-suppressor feature in human cancer 22 . Another report showed that TP53TG1 under glucose deprivation may promote cell proliferation and migration by influencing the expression of glucose metabolism related genes in glioma 23 . And TP53TG1 was reported to be associated with radiosensitivity 24 . The role of TP53TG1 had also been studied in lung cancer and nasopharyngeal carcinoma 25,26 , but its expression profile and regulatory pathways are not known in RB. In this study, we have examined the role of lncRNA in the progression of RB, TP53TG1. Our aim was to provide a better understanding of the TP53TG1 mediated signaling pathway that regulates RB disease progression for the development of highly effective and target specific RB therapeutics.
Materials and Methods
Human Tissue Samples
70 retinoblastoma and 70 healthy retinal specimens were obtained from surgery, between January 2013 and January 2015, at the First Affiliated Hospital of Xi’an Jiaotong University and The First Hospital of Qingdao University, China. The average age of the tissue donors were 3.12 years (range, 6 months to 12 years). The tumor samples were assessed by two separate pathologists and assigned a grade, according to the 7th edition of the American Joint Committee on Cancer guidelines 27 . The healthy retinal samples were collected from the ruptured globes of children, averaging 16.3 years of age. Both tumor and healthy specimens were flash frozen shortly after harvest and maintained at −80°C until further analysis. The RB patients were followed up post surgery and the overall survival (OS) rates were recorded.
RB Cell Culture and Transfection
Both the RB cell lines (SO-RB50, WERI-Rb1, Y79 and RBL-13) and the healthy human retinal epithelial cell line (ARPE-19) were acquired from the American Type Culture Collection (Shanghai, China). They were maintained in RPMI-1640 media (Gibco, Carlsbad, CA) with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA) at 37°C in a humidified environment of 95% air and 5% CO2. siRNAs targeting TP53TG1 (si-TP53TG1), negative control siRNA (si-NC), miR-33b mimics, NC mimics (miR-NC), miR-33b inhibitors, and corresponding NC (miR-NC) were acquired from GenePharma (Shanghai, China). Transient transfections were performed in RB cells using lipofectamine 3000 (Invitrogen, Carlsbad, CA), following manufacturers’ protocols.
RNA Isolation and Quantitative Real-Time PCR (qPCR)
Both tissue samples and cultured cells were used to extract total RNA using RNAiso (TaKaRa, Japan), following manufacturer’s protocols. Next, 1 µg of the RNA was reverse-transcribed into cDNA with miRNA Reverse Transcription Kit (RiboBio, China) and miR-33b levels were measured using the miRNA kit (RiboBio, China) and normalized using the endogenous U6 control. To verify our findings and to examine alterations in the transcription of relevant genes, 2 µg of total RNA was reverse transcribed using a PrimeScript first-strand cDNA synthesis kit (Takara, Japan) and qPCR was executed using SYBR® Premix Ex Taq™ II (Takara, Japan) following manufacturer’s guidelines. Supplementary Table 1 lists the primers used in this study. ABI 7900 cycler (Applied Biosystems, Foster City, CA) was employed for the qPCR amplification and GAPDH was employed as the endogenous control. The relative expression of relevant genes were determined with the 2−ΔΔCt method.
Cell Proliferation (CP) Using CCK-8 Analysis
Transfected Y79 cells were trypsinized and re-plated in 96-well plates (5000 cells/ well). Cell growth was measured with a cell counting kit-8 kit (CCK-8; Dojindo Laboratories), based on manufacturer’s guidelines. CP calculations were performed using an enzyme-based immunosorbent assay reader (Thermo Labsystems) at 450 nm.
CP, Cell Migration (CM), Cell Invasion (CI), and the EdU Assay
For colony forming estimation, low concentration cells (1000 cells/plate) were plated and allowed to form into colonies. Media, nourishing the cells, was changed once in 6 days. Upon colony formation, 0.1% crystal violet (Beyotime, China) stain was introduced to the cells and >50 cell colonies were counted and recorded.
To analyze CM capability, the wound healing assay was employed. Briefly, cells were cultured in a 6-well plate and wounded by dragging a pipette tip over the surface. Following this, the cells were PBS-washed to remove debris. Wound images were taken at the time of wounding and at 24 hours after the wounding.
The CM data was further verified with transwell inserts (8-µm pore filter, 24-well cell clusters; Millipore, Corp., Bedford, MA), following manufacturer’s guidelines. Briefly, cells were transfected for 48 h before they were trypsinized and plated (2 × 105 cells/well) on the top chamber with RPMI 1640 media and 2% FBS. The bottom chamber contained only 500 µl complete media and 10% FBS. Next, CM was allowed to occur from the top to the bottom chamber over a period of 24 hours in a humidified environment 37°C in with 5% CO2. The cells from the top chamber were removed, whereas those on the bottom chamber were fixed with 4% paraformaldehyde at room temperature for 30 min before a 10 min counterstain with 0.1% crystal violet. Lastly, the migrated cells were then counted using 5 random, non-overlapping fields per well using the software ImageJ (Media Cybernetics, Bethesda, MD, USA). CI assays was conducted similar to the transwell assay; only, the top chamber was precoated with Matrigel (BD Biosciences, San Jose, CA), and 1 × 105 cells were introduced to each top chamber.
To conduct the EdU assay, cells were plated in 24-well plates and incorporated with si-TP53TG1 or si-NC. Following this, 40 μM EdU was introduced to the cells for 2 h before fixing, permeabilizing, and EdU staining, following manufacturer’s guidelines. The nuclei staining was done with DAPI (Sigma Aldrich, Louis, MO). And the CP quantification was performed using a fluorosescent microscope (Celenas; Nikon, Japan). The EdU incorporation rate was assessed with ImageJ.
Western Blot Analysis
The Beyotime protein extraction kit was employed for the extraction of total protein from cultured cells. Protein measurement was completed with BCA kit (Qiagen, Valencia, CA). 30 µg of total protein was used to perform western blot analysis using 10% sodium dodecyl sulfate-polyacrylamide gel and polyvinylidene difluoride (PVDF) membranes (Millipore, Corp., Bedford, MA). The membrane was then blocked with 5% skim milk at 37°C for 2 h, and incubated with human SHCBP1 primary antibody (1:500, Abcam, Cambridge, MA) and the endogenous control GAPDH (1:1000, CST.) overnight at 4°C. Membranes were then exposed to the corresponding secondary horseradishperoxidase-conjugated antibody (Abcam, Cambridge, MA) for 1 h at 37°C. Finally, protein quantification was performed by analyzing enhanced chemiluminescence signals (Amersham Biosciences, Arlington Heights, IL) using Image Lab 2.0 (Bio-Rad Laboratories, Hercules, CA).
Dual-Luciferase Reporter Assay
MiRBase (http://www.mirbase.org), TargetScan ( http://www.targetscan.org /mamm_31/), and starBase (http://starbase.sysu.edu.cn/index.php) were used to characterize potential targets of miR-33b. The 3’-UTR of SHCBP1 transcript was found to have potential complementary sequence, thereby increasing its likelihood of being a target for miR-33b. Therefore, RB cells were transfected for 48 h with luciferase vectors pGL6-miR (Beyotime, China), a luciferase vector with the wild-type (WT) target gene’s 3’-UTR and one with a mutant-type target gene’s 3’-UTR (MUT) for TP53TG1 or SHCBP1, along with miR-33b mimics or miR-NC, via Lipofectamine 3000 Reagent (Invitrogen, Carlsbad, CA). Finally, the luciferase luminescence was evaluated with the Dual-Luciferase Reporter Assay System (Promega, WI), using manufacturer’s guidelines, and the firefly luciferase activity was normalized to the control Renilla luciferase activity.
Formation of Tumor in Nude Mice
Upon sh-TP53TG1 or sh-NC transfection, Y79 cells (5 × 106) were re-suspended in 150 µl of culture media, in the absence of FBS, and injected into the subcutaneous layer of the right flank of 4-week-old BALB/c nude mice, acquired from the Slac Laboratory Animal (Shanghai, China). The animals were kept in a sterile room under a 12-h light-dark cycle with open access to food and water. Each implantation group consisted of 6 mice. The tumor volume (TV) was determined as follows: TV = (width × width × length)/2, as previous described. The animals were executed with 1% pentobarbital sodium (50 mg/kg, Sigma Aldrich, Louis, MO), decapitated, and the tumors were harvested and weighed 4 weeks post cell implantation. We followed the strict guidelines of the First Affiliated Hospital of Xi’an Jiaotong University for the Care and Use of Animals in all animal studies.
Statistical Analysis
We employed SPSS 19.0 (SPSS) and GraphPad Prism 6 (GraphPad, La Jolla, CA) for all statistical analysis. The chi-square test was employed to determine the correlation between RB patients’ clinical pathological features and the level of TP53TG1 in their cancerous tissues. Statistical significance (P < .05) between groups was analyzed using t-test or one-way analysis of variance (ANOVA). Potential correlations between TP53TG1 and miR-33b or SHCBP1 expressions were evaluated with Spearman’s correlation. Lastly, statistical significance was represented by asterisks.
Results
High TP53TG1 Expression is Associated with RB Progression
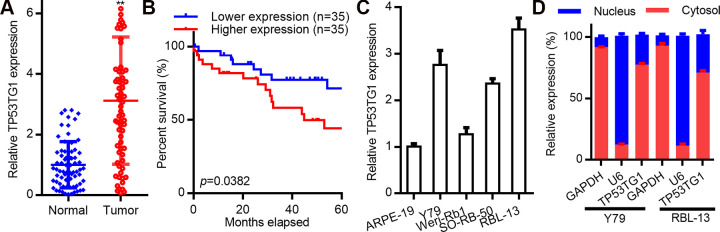
To determine a possible correlation between TP53TG1 and RB disease progression, the mRNA expression of TP53TG1 was analyzed in RB (n = 70) and normal retina tissues (n = 70). TP53TG1 levels were remarkably high in RB tissues, as opposed to healthy retina (Fig. 1A). To further analyze the relationship between TP53TG1 and RB, TP53TG1 expression was evaluated against the clinical data from RB patients. We found that patients with elevated TP53TG1 levels had shorter OS time, as opposed to patients with low TP53TG1 levels (P < .01, Fig. 1B). Similarly, TP53TG1 levels were also upregulated in other RB cell lines, namely Y79 and RBL-13, relative to healthy retinal cells (Fig. 1C) and were mostly located in the cytoplasm (Fig. 1D). Taken together, these data suggests that TP53TG1 upregulation is closely linked to the progression of RB.
Figure 1.
LncRNA TP53TG1 is highly expressed in retinoblastoma (RB) and accelerates disease progression. Relative expression of TP53TG1 in RB tissue (n = 70) and in normal retinal samples (n = 70). (B) Kaplan–Meier curves of RB survivors (n = 70) evaluated using the logrank test. (C) Relative TP53TG1 levels in human retinal epithelial cells (ARPE-19) and in RB cell lines (including SO-RB50, Weri-Rb1, Y79 and RBL-13). (D) Cellular localization of TP53TG1 in Y79 and RBL-13 cells. **P < .01.
TP53TG1 Knockdown Inhibited Cell Proliferation (CP)
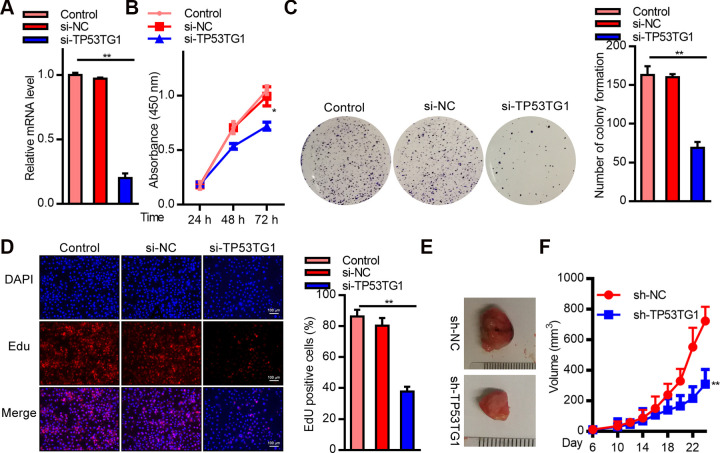
To explore the role of TP53TG1 in RB cells, we conducted a TP53TG1 knockdown using TP53TG1 specific siRNA. As shown in Fig. 2A, TP53TG1 levels were markedly reduced after treatment with siRNA. The CCK8 assay, assessing cell viability, revealed low cell survival (Fig. 2B). Similarly, the number of colonies reduced significantly in the TP53TG1 siRNA treated Y79 cells, as compared to those treated with si-NC (Fig. 2C). Moreover, the EdU assay demonstrated markedly reduced EdU positive cells, suggesting vastly reduced CP in the TP53TG1 siRNA treated cells, as opposed to si-NC treated controls (Fig. 2D). Additionally, we examined the TP53TG1 mediated regulation of tumor growth on nude mice xenografts. TP53TG1 was silenced in Y79 cells using recombinant plasmid carrying specific shRNA before implantation into nude mice. Our results demonstrated that TP53TG1-silenced Y79 cells were slow to form tumors and produced smaller sized tumors, as opposed to the tumors originating from sh-NC incorporated cells (Fig. 2E, F). In summary, these results strongly suggest that TP53TG1 silencing suppressed RB cell proliferation.
Figure 2.
SiRNA mediated TP53TG1 knockdown markedly inhibited RB cell proliferation (CP) in vitro and in vivo. (A) Relative TP53TG1 expression in Y79 cells after TP53TG1 knockdown. (B) CP, via CCK8 assay, in Y79 cells after TP53TG1 knockdown. (C) Colony forming assay after TP53TG1 knockdown. (D) EdU assay, assessing CP, after TP53TG1 knockdown. (E) Formation of tumors in xenograft nude mice after implantation of Y79 cells incorporated with sh-TP53TG1 or sh-NC. (F) Tumor volume analysis after implantation of Y79 cells incorporated with sh-TP53TG1 or sh-NC. **P < .01 vs. si-NC; **P < .01 vs. sh-NC.
TP53TG1 knockdown Suppresses Cell Migration (CM) and Cell Invasion (CI) of RB Cells
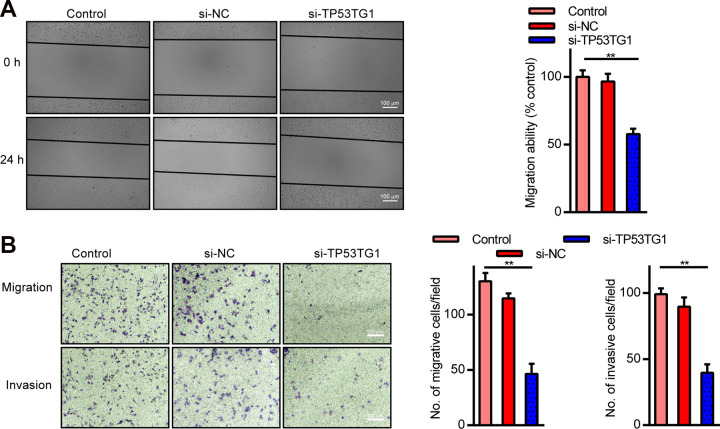
To characterize the potential regulatory mechanisms whereby TP53TG1 modulated tumor progression in RB, TP53TG1 was knocked down in Y79 cells and the CM and CI properties were assessed. TP53TG1 knockdown was shown to greatly inhibit CM, as evidenced by the wound healing assay (Fig. 3A). This result was further verified using the transwell assay that demonstrated that TP53TG1 knockdown significantly reduced both migratory and invasive behavior of Y79 cells (Fig. 3B). Taken together, this suggests that TP53TG1 knockdown can effectively inhibit both CM and CI in RB cells.
Figure 3.
TP53TG1 knockdown markedly suppressed cell migration (CM) and invasion (CI). (A) RB CM in TP53TG1 knocked down cells verses control after wounding. (B and C) RB CM and CI patterns in TP53TG1 knocked down cells verses control, using the transwell assay. **P < .01 vs. si-NC.
TP53TG1 Competes with Other Endogenous RNA in its Interaction with miR-33b
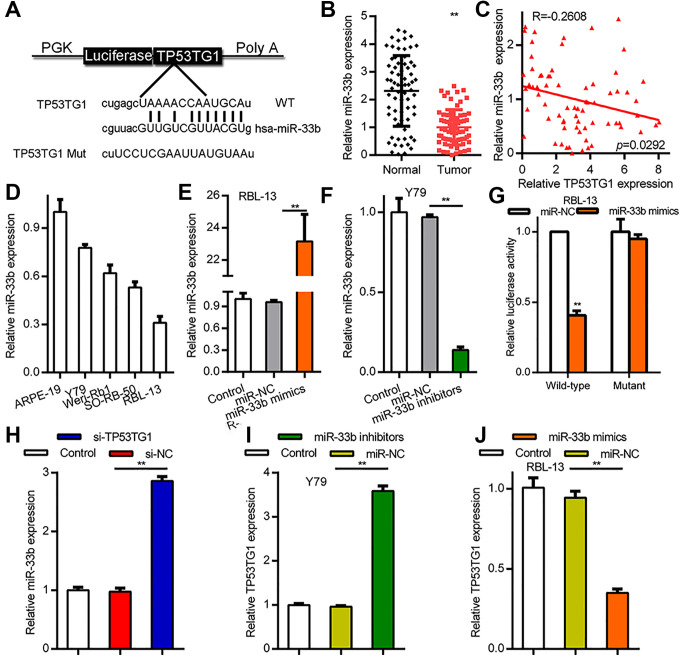
Multiple studies have reported that one of function of lncRNAs is to competitively interact with miRNAs to prevent miRNA-mediated degradation of target transcripts. As such, miRNA candidates that could possibly interact with TP53TG1 and, therefore, be a target of TP53TG1 were searched using miRBase, TargetScan and starBase. The predicted miR-33b binding site within TP53TG1 is shown in Fig. 4A. Comparing between RB tissues and normal retinal tissues, miR-33b showed a marked reduction in RB tissues, but not in normal retina (Fig. 4B). These data were further confirmed by the pearson correlation analysis which demonstrated a strong inverse association between TP53TG1 and miR-33b levels (R = −0.2608, P < .05; Fig. 4C). Moreover, similar to the expression of miR-33b in RB tissues, the levels of miR-33b was remarkably low in all examined RB cell lines, relative to healthy retinal cells (Fig. 4D). To further understand the TP53TG1 and miR-33b interaction, RBL-13 and Y79 cells were first incorporated with either miR-33b mimics or miR-33b inhibitors, respectively. As shown in Fig. 4E, F, cellular miR-33b levels were elevated after miR-33b mimics transfection, and were reduced with miR-33b inhibitors transfection. Next, we constructed luciferase reporter vectors with WT TP53TG1 or TP53TG1 with mutations in the putative binding sites of miR-33b (MUT). Dual-luciferase assays showed a notable reduction in luminescence in RBL-13 cells co-transfected with miR-33b mimics and the WT TP53TG1 expression vector, but not with the MUT (Fig. 4G). Next, we determined whether the TP53TG1-miR-33b interaction modulated miR-33b activity. We introduced TP53TG1 knockdown in RB cells, which resulted in marked increases in miR-33b expression (Fig. 4H). On the contrary, when miR-33b was overexpressed in cells, TP53TG1 levels rose and when miR-33b is downregulated, the TP53TG1 levels fell (Fig. 4I, J). In all, these results confirm that TP53TG1 may serve miR-33b sponge.
Figure 4.
LncRNA TP53TG1 directly interacted with miR-33b in RB cells. (A) A schematic of the predicted miR-33bbinding sites within the TP53TG1 gene, along with the wild-type and mutated variations of the luciferase reporter constructs. (B) Relative expression of miR-33b in RB tissue (n = 70) and in healthy retinal specimens (n = 70). (C) Pearson correlation analysis between TP53TG1 and miR-33b in RB specimens (n = 70), R = −0.2606, P < .05. (D) Relative TP53TG1 levels in human retinal epithelial cells and multiple RB cell lines. (E) Relative miR-33b levels in RBL-13 cells incorporated with miR-33b mimics or mimics control (miR-NC). (F) Relative miR-33b levels in Y79 cells incorporated with miR-33b inhibitors or inhibitors control (miR-NC). (G) Luciferase activity of RBL-13 cells co-incorporated with miR-33b mimics or miR-NC, and TP53TG1 wildtype or mutated vectors. (H) Relative miR-33b expression in TP53TG1 knocked down RB. (I and J) Relative expression of TP53TG1 in Y79 cells after incorporation with miR-33b inhibitors or miR-33b mimics, respectively. **, P < .01.
TP53TG1 Regulates RB Progression by Sponging miR-33b
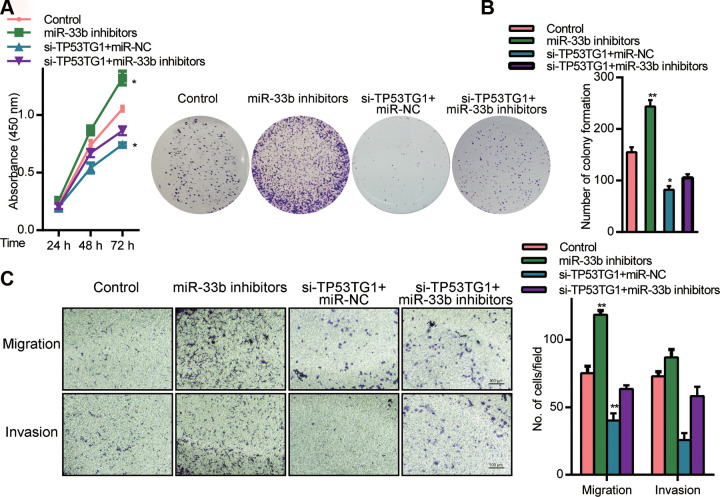
TP53TG1 was shown to interact with the 3’ UTR of miR-33b and regulate its expression. We next explored whether TP53TG1 mediated regulation of RB involved miR-33b. CCK-8 assays revealed that cells incorporated with miR-33b inhibitors had higher CP, relative to cells receiving si-TP53TG1 (Fig. 5A). Moreover, using colony formation assay, we demonstrated that miR-33b inhibitor partly reversed the effects of si-TP53TG1 in CP (Fig. 5B). Additionally, cells transfected with miR-33b inhibitor was shown to have higher CM and CI, whereas TP53TG1 knockdown experienced reduced CM and CI (Fig. 5C). Based on these data, miR-33b silencing can significantly diminish the function of TP53TG1 on RB cells, indicating an antagonistic association between TP53TG1 and miR-33b in its modulation of the disease.
Figure 5.
MiR-33b mediated the effect of TP53TG1 knockdown in Y79 cells. (A) CP, measured by CCK8 assay, in cells co-incorporated with si-TP53TG1 and the miR-33b inhibitors or inhibitors control (miR-NC) (B) Verification of CP, via colony formation assays, using cells co-incorporated with si-TP53TG1 and the miR-33b inhibitors or inhibitors control (miR-NC). (C) Assessment of CM and CI, using transwell assay. **P < .05 vs. inhibitors control (miR-NC).
MiR-33b Targets SHCBP1 in its Regulation of RB
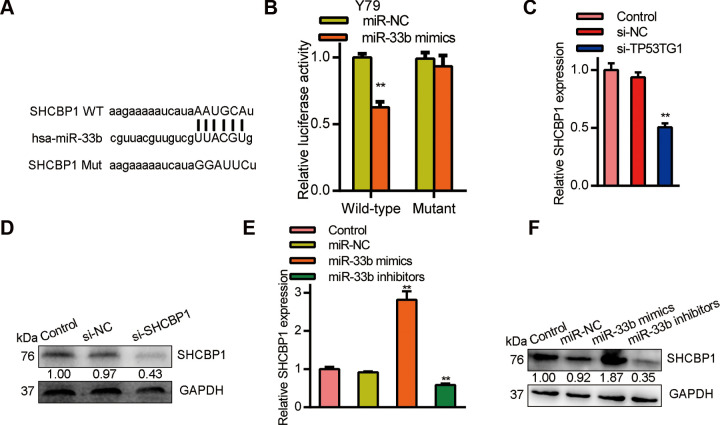
Next we investigated which is the downstream moleculars of miR-33b. Bioinformatics analysis predicted that SHCBP1 shared sequence homology with miR-33b at its 3’-UTR (Fig. 6A). Using luciferase reporter assay, we demonstrated a physical interaction between miR-33b and the 3’-UTR of SHCBP1 in Y79 cells (Fig. 6B). Moreover, SHCBP1 mRNA levels declined with si-TP53TG1 transfection of Y79 cells (Fig. 6C). Similarly, SHCBP1 protein expression was elevated in RB cells (Fig. 6D). Alternately, Y79 cells incorporated with miR-33b revealed a large decline in SHCBP1 mRNA and proteins expression (Fig. 6E, F). In all, these data points toward a TP53TG1/miR-33b/SHCBP1 pathway in the regulation of RB progression.
Figure 6.
SHCBP1 is a target gene of miR-33b and is modulated by TP53TG1. (A) A schematic of the wild-type (WT) and mutated 3′UTR of SHCBP1 (MUT) transcript. (B) Luciferase activity in Y79 cells co-incorporated with miR-33b mimics and luciferase reporters containing WT or MUT SHCBP1. (C) Relative SHCBP1 levels after TP53TG1 knockdown in Y79 cells. (D) SHCBP1 protein expression after TP53TG1 deficiency in Y79 cells. (E) Relative SHCBP1 expression inY79 cells after miR-33b knockdown and overexpression. (F) SHCBP1 protein levels in RB cells after miR-33b knockdown and overexpression. **P < .01, vs. miR-NC.
Discussion
Several studies have suggested an association between lncRNAs and RB pathogenesis 15,28,29 . However, the exact role and underlying pathways of involvement remains unclear. TP53TG1, a member of the lncRNAs family, is encoded in the human chromosome 7q21.12 region and is prevalent in tumors. More importantly, high frequency TP53TG1 levels were shown to increase malignancy risk 23,24,30 . In this study, we characterized competitive binding of TP53TG1 to the 3’ UTR of miR-33b and of miR-33b to the 3’ UTR of SHCBP1. In other words, we showed a TP53TG1/miR-33b/SHCBP1 pathway whereby TP53TG1 increased the expression of SHCBP1 via its inhibition of miR-33b, thus facilitating the progression of RB.
Based on our results, TP53TG1 expression was remarkably high in RB tissues and RB cell lines. From the reported literature, high TP53TG1 levels were found to be intimately linked to cancer progression. In fact, Zhang et al. reported that TP53TG1 competitively binds with miR-96 to modulate KRAS expression and growth and progression of pancreatic ductal adenocarcinoma (PDAC) 30 . So TP53TG1 was shown to have a high prognostic value in diagnosing pancreatic ductal adenocarcinoma. And Gao et al. demonstrated a strong oncogenic property of TP53TG1 in the modulation of cell proliferation, colony formation, autophagy, and radio resistance, even restrained apoptosis of glioma cells, through its regulation of the miR-524-5p/RAB5A axis 24 . For glioma, another report showed that TP53TG1 played an important role in regulate cell proliferation by influencing the expression of glucose metabolism 23 . In addition, a study revealed that TP53TG1 enhanced the cisplatin sensitivity of non-small cell lung cancer 26 . A recent report showed that TP53TG1 as an antioncogenic target, performed an important role in the prognosis of breast cancer. 31 In order to discuss the role of TP53TG1 in RB, we further explored the effects of TP53TG1 expression on RB progression. Similar to the published reports, we revealed that TP53TG1 silenced in Y79 cells leads to a marked suppression of cell malignant behavior.
LncRNAs are known to regulate RB development and progression via its modulation of miRNAs and their downstream genes 32,33 . Based on our bioinformatics prediction, and verified by dual-luciferase assay, we revealed that TP53TG1 serves as a competing endogenous (ceRNA) in its negative regulation of miR-33b in RB. MiR-33, specifically miR-33a and miR-33b, belongs to a family of highly conserved miRNA 34 , that are known tumor suppressors for cancers like HER2 positive breast carcinoma 35 , colorectal cancer 36 , and lung cancer 37 . Being a potential tumor suppressor, breast HER2+ tumor samples showed a marked downregulation of miR-33b, as opposed to normal breast tissues. Additionally, the low levels of miR-33b were indicative of poor prognosis in HER2+ patients 35 . Likewise, in triple negative breast cancer (TNBC) patients, miR-33b was shown to suppress cancer progression and metastasis by targeting oncogenes like SALL4, TWIST1, and HMGA2 38 . Moreover, CUL4B was reported to negatively modulate miR-33b and promote prostate cancer 39 . Here, we revealed that TP53TG1 downregulated miR-33b levels in both RB tissues and cells. Moreover, when miR-33b was knocked down, it reversed the malignancy properties of TP53TG1 deficiency in RB cells. Based on these results, we proposed that TP53TG1 serves as a miR-33b sponge to modulate its activity.
To characterize the downstream targets of miR-33b in RB, we conducted a bioinformatics prediction and characterized SHCBP1 as a possible target. SHCBP1 has been shown to positively regulate tumor progression and its expression remains high in malignant cells 40 . Additionally, high SHCBP1 levels can be observed during the β-selection checkpoint of T cell development, which is necessary for thymic proliferation 41,42 . Similarly, SHCBP1 was shown to be closely linked to the tumor, node, and metastasis (TNM) stages, HER2 expression, and low OS in breast cancer. Additionally, it was shown to accelerate CP in breast cancer. Consequently, SHCBP1 knockout elevated the levels of p21, a cyclin dependent kinase inhibitor, and suppressed the levels of cyclin B1 and CDK1, which allowed the MDA-MB-231 and MCF7 breast cancer cell lines to become arrested in G2/M phase 43 . Based on our results, SHCBP1 was increased upon miR-33b knockdown and its levels decreased with overexpression of miR-33b. Our study did not involve rescue experimentation in RB cells. However, other publications have revealed that SHCBP1 knockdown leads to cell cycle arrest and a marked suppression of tumor CP 43 . Our study revealed that miR-33b was one of the regulators of SHCBP1 expression. As such, knockdown of TP53TG1, which inhibits miR-33b, was shown to upregulate SHCBP1 expression, thereby indicating a TP53TG1/miR-33b/SHCBP1 axis in the modulation of RB.
Conclusion
In conclusion, we found that the expression of TP53TG1 is elevated in retinoblastoma, and it is an effective prognostic factor for the diagnosis of retinoblastoma. We further revealed that TP53TG1 knockdown markedly suppresses retinoblastoma malignant behavior. Finally, TP53TG1 serves as an oncogene in retinoblastoma. It sponges miR-33b and thereby increases the expression of SHCBP1 to drive the progression of retinoblastoma.
Supplemental material
Supplemental Material, sj-docx-1-cll-10.1177_09636897211025223 for Long Non-Coding RNA TP53TG1 Upregulates SHCBP1 to Promote Retinoblastoma Progression by Sponging miR-33b by Hongyi Wang, Zhen Zhang, Yue Zhang, Shihai Liu and Li Li in Cell Transplantation
Footnotes
Author Contributions: L Li designed and conducted experiments, and performed data analysis. Z Zhang and Y Zhang conducted experiments and collected data. SH Liu contributed to experimental design, data interpretation, and manuscript revision. HY Wang designed the study, performed data analysis, and wrote the manuscript.
Ethical Approval: This study was approved by the ethics committee at the first affiliated hospital of Xian Jiaotong University, Shaanxi, China.
Statement of Human and Animal Rights: All of the experimental procedures involving patients were conducted in accordance with the Helsinki Declaration for human studies, and approved by the ethics committee at the first affiliated hospital of Xian Jiaotong University.
Statement of Informed Consent: Written informed consents were obtained from the patients for the collection and publication of clinical data.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Li Li  https://orcid.org/0000-0001-6682-6401
https://orcid.org/0000-0001-6682-6401
Supplemental material: Supplemental material for this article is available online.
References
- 1. Dimaras H, Corson TW. Retinoblastoma, the visible CNS tumor: a review. J Neurosci Res. 2019;97(1):29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fabian ID, Onadim Z, Karaa E, Duncan C, Chowdhury T, Scheimberg I, Ohnuma SI, Reddy MA, Sagoo MS. The management of retinoblastoma. Oncogene. 2018;37(12):1551–1560. [DOI] [PubMed] [Google Scholar]
- 3. Fabian ID, Puccinelli F, Gaillard MC, Beck-Popovic M, Munier FL. Diagnosis and management of secondary epipapillary retinoblastoma. Br J Ophthalmol. 2017;101(10):1412–1418. [DOI] [PubMed] [Google Scholar]
- 4. Elgar G, Vavouri T. Tuning in to the signals: noncoding sequence conservation in vertebrate genomes. Trends Genet. 2008;24(7):344–352. [DOI] [PubMed] [Google Scholar]
- 5. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. [DOI] [PubMed] [Google Scholar]
- 10. Fan Q, Yang L, Zhang X, Peng X, Wei S, Su D, Zhai Z, Hua X, Li H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–115. [DOI] [PubMed] [Google Scholar]
- 11. Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Xu J, Xia K, Chang Y, Liu J, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. 2018;17(1):82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang J, Yang Y, Fang F, Liu K. MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J Cell Biochem. 2018;119(5):3853–3863. [DOI] [PubMed] [Google Scholar]
- 13. Su S, Gao J, Wang T, Wang J, Li H, Wang Z. Long non-coding RNA BANCR regulates growth and metastasis and is associated with poor prognosis in retinoblastoma. Tumour Biol. 2015;36(9):7205–7211. [DOI] [PubMed] [Google Scholar]
- 14. Fang Z, Wang Y, Wang Z, Xu M, Ren S, Yang D, Hong M, Xie W. ERINA Is an estrogen-responsive LncRNA That drives breast cancer through the E2F1/RB1 pathway. Cancer Res. 2020;80(20):4399–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He X, Chai P, Li F, Zhang L, Zhou C, Yuan X, Li Y, Yang J, Luo Y, Ge S, Zhang H, et al. A novel LncRNA transcript, RBAT1, accelerates tumorigenesis through interacting with HNRNPL and cis-activating E2F3. Mol Cancer. 2020;19(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lyu X, Ma Y, Wu F, Wang L, Wang L. LncRNA NKILA inhibits retinoblastoma by downregulating lncRNA XIST. Curr Eye Res. 2019;44(9):975–979. [DOI] [PubMed] [Google Scholar]
- 17. Shang W, Yang Y, Zhang J, Wu Q. Long noncoding RNA BDNF-AS is a potential biomarker and regulates cancer development in human retinoblastoma. Biochem Biophys Res Commun. 2018;497(4):1142–1148. [DOI] [PubMed] [Google Scholar]
- 18. Shang Y. LncRNA THOR acts as a retinoblastoma promoter through enhancing the combination of c-myc mRNA and IGF2BP1 protein. Biomed Pharmacother. 2018;106:1243–1249. [DOI] [PubMed] [Google Scholar]
- 19. Xu C, Hu C, Wang Y, Liu S. Long noncoding RNA SNHG16 promotes human retinoblastoma progression via sponging miR-140-5p. Biomed Pharmacother. 2019;117:109153. [DOI] [PubMed] [Google Scholar]
- 20. Zhao H, Wan J, Zhu Y. Carboplatin inhibits the progression of retinoblastoma through IncRNA XIST/miR-200a-3p/NRP1 Axis. Drug Des Devel Ther. 2020;14:3417–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han S, Song L, Chen Y, Hou M, Wei X, Fan D. The long non-coding RNA ILF3-AS1 increases the proliferation and invasion of retinoblastoma through the miR-132-3p/SMAD2 axis. Exp Cell Res. 2020;393(2):112087. [DOI] [PubMed] [Google Scholar]
- 22. Diaz-Lagares A, Crujeiras AB, Lopez-Serra P, Soler M, Setien F, Goyal A, Sandoval J, Hashimoto Y, Martinez-Cardús A, Gomez A, Heyn H, et al. Epigenetic inactivation of the p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc Natl Acad Sci U S A. 2016;113(47):E7535–E7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen X, Gao Y, Li D, Cao Y, Hao B. LncRNA-TP53TG1 participated in the stress response under glucose deprivation in glioma. J Cell Biochem. 2017;118(12):4897–4904. [DOI] [PubMed] [Google Scholar]
- 24. Gao W, Qiao M, Luo K. Long noncoding RNA TP53TG1 contributes to radioresistance of glioma cells via miR-524-5p/RAB5A Axis. Cancer Biother Radiopharm. 2020. [DOI] [PubMed] [Google Scholar]
- 25. Yuan J, Jiang YY, Mayakonda A, Huang M, Ding LW, Lin H, Yu F, Lu Y, Loh TKS, Chow M, Savage S, et al. Super-enhancers promote transcriptional dysregulation in nasopharyngeal carcinoma. Cancer Res. 2017;77(23):6614–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiao H, Liu Y, Liang P, Wang B, Tan H, Zhang Y, Gao X, Gao J. TP53TG1 enhances cisplatin sensitivity of non-small cell lung cancer cells through regulating miR-18a/PTEN axis. Cell Biosci 2018;8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ainbinder DJ, Esmaeli B, Groo SC, Finger PT, Brooks JP. Introduction of the 7th edition eyelid carcinoma classification system from the american joint committee on cancer-international union against cancer staging manual. Arch Pathol Lab Med. 2009;133(8):1256–1261. [DOI] [PubMed] [Google Scholar]
- 28. Cheng Y, Chang Q, Zheng B, Xu J, Li H, Wang R. LncRNA XIST promotes the epithelial to mesenchymal transition of retinoblastoma via sponging miR-101. Eur J Pharmacol. 2019;843:210–216. [DOI] [PubMed] [Google Scholar]
- 29. Hu C, Liu S, Han M, Wang Y, Xu C. Knockdown of lncRNA XIST inhibits retinoblastoma progression by modulating the miR-124/STAT3 axis. Biomed Pharmacother. 2018;107:547–554. [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y, Yang H, Du Y, Liu P, Zhang J, Li Y, Shen H, Xing L, Xue X, Chen J, Zhang X. Long noncoding RNA TP53TG1 promotes pancreatic ductal adenocarcinoma development by acting as a molecular sponge of microRNA-96. Cancer Sci 2019;110(9):2760–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shao M, Ma H, Wan X, Liu Y. Survival analysis for long noncoding RNAs identifies TP53TG1 as an antioncogenic target for the breast cancer. J Cell Physiol. 2020;235(10):6574–6581. [DOI] [PubMed] [Google Scholar]
- 32. Wang JX, Yang Y, Li K. Long noncoding RNA DANCR aggravates retinoblastoma through miR-34c and miR-613 by targeting MMP-9. J Cell Physiol. 2018;233(10):6986–6995. [DOI] [PubMed] [Google Scholar]
- 33. Gao Y, Luo X, Zhang J. Sp1-mediated up-regulation of lnc00152 promotes invasion and metastasis of retinoblastoma cells via the miR-30d/SOX9/ZEB2 pathway. Cell Oncol (Dordr). 2021;44(1):61–76. [DOI] [PubMed] [Google Scholar]
- 34. Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, Esplugues E, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108(22):9232–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pattanayak B, Garrido-Cano I, Adam-Artigues A, Tormo E, Pineda B, Cabello P, Alonso E, Bermejo B, Hernando C, Martínez MT, Rovira A, et al. MicroRNA-33b suppresses epithelial-mesenchymal transition repressing the MYC-EZH2 pathway in HER2+ breast carcinoma. Front Oncol. 2020;10:1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ni Y, Li C, Bo C, Zhang B, Liu Y, Bai X, Cui B, Han P. LncRNA EGOT regulates the proliferation and apoptosis of colorectal cancer by miR-33b-5p/CROT axis. Biosci Rep. 2020:BSR20193893. [DOI] [PubMed] [Google Scholar]
- 37. Wang Y, Zhao W, Zhang S. STAT3-induced upregulation of circCCDC66 facilitates the progression of non-small cell lung cancer by targeting miR-33a-5p/KPNA4 axis. Biomed Pharmacother. 2020;126:110019. [DOI] [PubMed] [Google Scholar]
- 38. Lin Y, Liu AY, Fan C, Zheng H, Li Y, Zhang C, Wu S, Yu D, Huang Z, Liu F, Luo Q, et al. MicroRNA-33b inhibits breast cancer metastasis by targeting HMGA2, SALL4 and twist1. Sci Rep. 2015;5:9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao M, Qi M, Li X, Hu J, Zhang J, Jiao M, Bai X, Peng X, Han B. CUL4B/miR-33b/C-MYC axis promotes prostate cancer progression. Prostate 2019;79(5):480–488. [DOI] [PubMed] [Google Scholar]
- 40. Zhang GY, Ma ZJ, Wang L, Sun RF, Jiang XY, Yang XJ, Long B, Ye HL, Zhang SZ, Yu ZY, Shi WG, et al. The Role of shcbp1 in signaling and disease. Curr Cancer Drug Targets 2019;19(11):854–862. [DOI] [PubMed] [Google Scholar]
- 41. Buckley MW, Arandjelovic S, Trampont PC, Kim TS, Braciale TJ, Ravichandran KS. Unexpected phenotype of mice lacking Shcbp1, a protein induced during T cell proliferation. PLoS One. 2014;9(8):e105576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heng TS, Painter MW. The Immunological genome project: networks of gene expression in immune cells. Nat Immunol 2008;9(10):1091–1094. [DOI] [PubMed] [Google Scholar]
- 43. Feng W, Li HC, Xu K, Chen YF, Pan LY, Mei Y, Cai H, Jiang YM, Chen T, Feng DX. SHCBP1 is over-expressed in breast cancer and is important in the proliferation and apoptosis of the human malignant breast cancer cell line. Gene. 2016;587(1):91–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-docx-1-cll-10.1177_09636897211025223 for Long Non-Coding RNA TP53TG1 Upregulates SHCBP1 to Promote Retinoblastoma Progression by Sponging miR-33b by Hongyi Wang, Zhen Zhang, Yue Zhang, Shihai Liu and Li Li in Cell Transplantation