Abstract
Current guidelines suggest screening all patients with idiopathic pulmonary arterial hypertension for genetic aberrations, particularly mutations in Bone Morphogenic Protein Receptor Type II (BMPR2), the gene most commonly implicated in the pathogenesis of PAH. Herein, we present a novel technique used to identify a pathogenic germline BMPR2 alteration in a 36-year-old female and family members with hereditary pulmonary arterial hypertension who each screened negative by standard cytogenetics and molecular genetics testing.
Keywords: BMPR2, mutation, genetic test
Case description
A 36-year-old female was referred for evaluation of her known PAH confirmed via right heart catheterization (mean pulmonary artery pressure of 53 mmHg, pulmonary capillary wedge pressure 7 mmHg, and pulmonary vascular resistance 18 Woods units). Family history was pertinent for a sister with PAH diagnosed at age 27 and treated with inhaled prostacyclin analog; mother with PAH diagnosed at 32 years of age who died at age 33 from PAH-related complications while awaiting heart and lung transplant; maternal cousin once removed with PAH diagnosed at the age of 52; and maternal second cousin who had passed away during child birth at the age of 23 (Supplemental Figure S1). Evaluation for a secondary cause of PAH was negative. A PAH gene sequencing panel including BMPR2 and additional BMP pathway-associated genes (ACVRL1, BMPR1B, CAV1, ENG, GDF2, KCNA5, KCNK3, and SMAD9A) did not identify a pathogenic coding alteration that could explain the phenotype. Deletion/duplication analysis of the corresponding coding regions was also negative. Standard chromosomal analysis was performed to determine whether a gross chromosomal rearrangement (potentially disrupting a PAH-relevant gene) was present within the genome; however, a normal (46, XX) karyotype was observed. Although treatment was initiated with epoprostenol, her disease progressed. She underwent heart and lung transplant and a novel donor versus recipient cell-free DNA quantitative monitoring of junction sequences was used to interrogate graft health. Recipient pre-transplant mate-pair sequencing (MPseq) revealed a cryptic paracentric inversion on chromosome 2 with break points within intron 10 of BMPR2 and intron 6 of PIKFYVE. MPseq was subsequently performed on her symptomatic sister and maternal cousin once removed and the same germ-line chromosomal inversion disrupting the BMRP2 gene was identified.
Discussion
HPAH, a subset of WHO clinical classification group 1, is characterized by pre-capillary pulmonary hypertension without secondary cause and with an identifiable genetic mutation known to cause pulmonary arterial hypertension (PAH).1–3
Heterozygous hypomorphic alterations within the BMPR2 gene are the most common genetic cause of PAH and via gene sequencing and deletion/duplication analyses, these BMPR2 alterations are detected in up to 80% of HPAH and 20% of sporadic PAH cases. 4 HPAH is inherited in an autosomal dominant fashion with incomplete, sex-influenced penetrance (14% penetrance in males and 42% in females). Over 800 mutations have been identified in BMPR2; the majority of mutations identified are missense mutations (25%), frame-shift (23%), re-arrangement (14%), and splice-site mutations (10%).5–7
The BMPR2 gene is located on chromosome 2 (2q33-34) and encodes for a transmembrane serine/threonine kinase receptor belonging to the transforming growth factor beta (TGF-β) superfamily. Normally, BMP signaling activates SMAD proteins that counteract the effects of the TGF-β signaling pathway. Diminished expression or function of BMPR2 impairs BMP signaling and prevents the appropriate repression of TGF-β. Constitutive activation of TGF-β signaling drives pulmonary endothelial metabolism and apoptosis while increasing smooth muscle proliferation and cell survival leading to plexiform lesions and increased pulmonary vascular resistance.6,8 Heterozygosity for a BMPR2 mutation is neither necessary nor sufficient to cause HPAH and disease severity and penetrance with variable age at both diagnosis and death are likely modified by the specific class of mutation and its effect on BMPR2 activity as well as disease modifiers.
Current methods to detect coding sequence mutations include direct Sanger or massively parallel sequencing (such as next-generation whole exome or custom panel sequencing) which can detect 55% of BMPR2 mutations in patients with FPAH. 9 Exonic deletions/duplications not detected by direct sequencing can be detected by targeted comparative genomic hybridization array technology (aCGH) 10 and multiplex ligation-dependent probe amplification (MLPA). 11 These tests classically capture exonic and exon-intron boundary sequences of HPAH-associated genes and determine mutations, small deletions and insertions, as well as larger copy gains and losses that alter gene coding regions. However, a subpopulation of HPAH patients present with no detectable genetic variation.
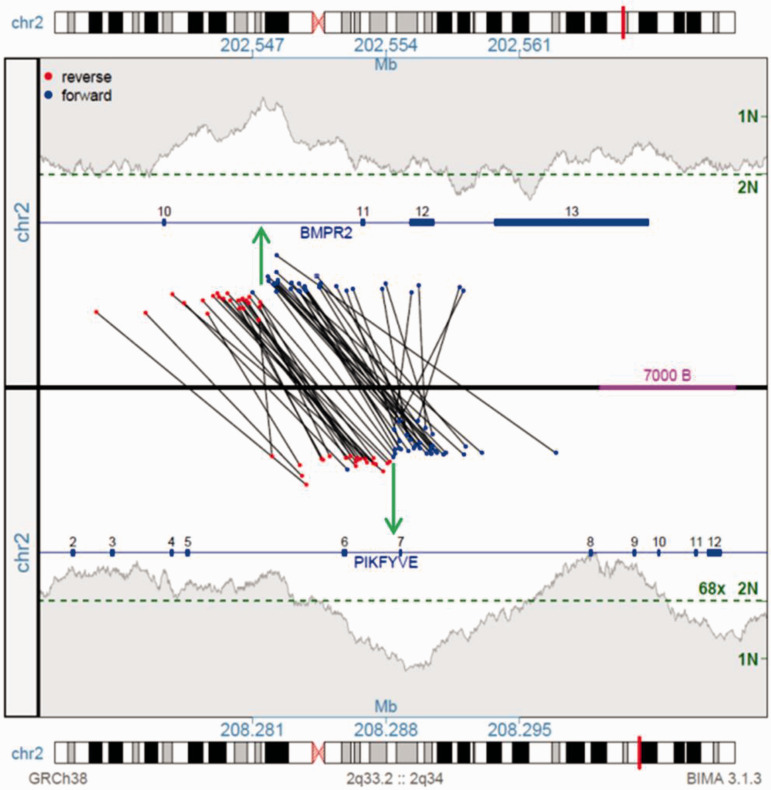
In this report we used MPseq, a next-generation whole genome sequencing-based technology, specifically optimized to detect structural chromosome alterations including deletions, duplications, translocations and inversions. 12 This technique utilizes a unique library preparation protocol which involves circularization of fragmented DNA such that DNA regions physically separated within the genome are now juxtaposed and in an inverted orientation. These DNA regions are sequenced and mapped back to a reference genome. Discordant mate pair reads (those that map at an unexpected distance or orientation within the genome) are indicative of a structural chromosome aberration between the two reads (Supplemental Figure S2). As MPseq utilizes long-insert paired end reads, it provides greater bridged coverage of the genome, allowing for whole genome assessments at a reduced cost.12,13 In our case, MPseq revealed normal diploid coverage across the patient's genome, but identified a junction between the BMPR2 and PIKFYVE genes on the long arm of chromosome 2 (Supplemental Figure S3). The MPseq junction plot reveals two sets of discordant MPseq reads that map at an inappropriate orientation (red-red and blue-blue) and distance within the genome. These MPseq reads span the two breakpoints within BMPR2 intron 6 and PIKFYVE intron 10, indicative of a balanced inversion of ∼5.7 Mb of chromosome 2 (Fig 1). No novel gene–gene fusion peptides would be predicted from this inversion, with the resultant genomes predicting truncation of both genes. The physiological outcome of the BMP2R peptides would be a predicted truncating loss of function, with potential inhibition of the wild-type allele copy function through dimerization.
Fig. 1.
Junction plot illustrating the rearrangement connecting the BMPR2 and PIKFYVE genes on chromosome 2. The upper and lower panels present the BMPR2 2q33.2 and PIKFYVE 2q34 regions, respectively. Genes illustrated as blue lines with exon positions indicated. Black lines cross the upper and lower panels linking the individual supporting fragment reads spanning the junctions, coloured red or blue indicating mapping to the forward or reverse strands, respectively. A balanced inversion is predicted by the two distinct mapping patterns of fragments (red–red and blue–blue) with the positions of the breakpoints indicated by the green arrows. The red–red event connects the 5′ end of BMP2R exons 1 to 10 with 5′ end of FIKFYVE exon 6 to 1 in reverse orientation, truncating both genes with no productive fusion predicted. The blue–blue event connects the 3′ end of BMP2R exons 13 to 11 with 3′ end of FIKFYVE from exons 8. The shaded grey areas indicate the coverage across each region, with the genome wide predicted 2N coverage level indicated by the green dotted line.
Interestingly, the sister of our patient was also diagnosed with an atypical low-grade glioma with identified mutations in several genes (IDH1, TERT, CIC, and FUBP1) but not in BMPR2. BMPR2 is known to play a role in glial stem cell differentiation and glioblastoma, and has been suggested as a therapeutic target in patients with glioblastoma multiforme. 14
We used MPseq data to identify identical rearrangements within this family. Alternative techniques to validate the detected structural variant include Sanger sequencing across the breakpoint junction, break apart FISH, or nucleotide mass spectrometry. However, we have previously demonstrated that MPseq performs superiorly to FISH in characterizing particularly complex genomic abnormalities. 15 We propose that MPseq be considered in cases of HPAP which screen negative with standard genetic tests as well as in a future study of sporadic cases of PAP.
In summary, MPseq successfully identified a novel, pathogenic alteration in BMPR2 in a family with severe HPAH. Importantly, this cryptic inversion of chromosome 2 is undetectable by classic cytogenetics or targeted molecular analyses. MPseq is a technique that may help identify previously unidentified pathogenic alterations in patients with HPAH and other lung diseases, such as familial idiopathic pulmonary fibrosis, thereby enabling both tailored genetic counseling and the use of targeted gene therapies in the future. As such, MPseq could be considered for all patients in which standard genetic techniques failed to identify a pathogenic variant. Overall these findings highlight the clinical utility and value of high-resolution techniques like MPSeq in the molecular characterization of human disease to allow for informed genetic counseling regarding carrier status and in the future may allow for gene targeted therapy. 6
Supplemental Material
Supplemental material, sj-pdf-1-pul-10.1177_2045894020933081 for Mate-pair sequencing identifies a cryptic BMPR2 mutation in hereditary pulmonary arterial hypertension by Sarah J. Chalmers, Stephen J. Murphy, Laura L. Thompson, Nicole L. Hoppman, James B. Smadbeck, Jessica R. Balcom, Faye R. Harris, Robert P. Frantz, George Vasmatzis and Mark E. Wylam in Pulmonary Circulation
Conflict of interest
The author(s) declare that there is no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Ethics approval
Our Institutional Review Board approved this study (18-5164). We obtained written consent from the patients.
Funding
This work was supported by funding from William J. von Liebig Center for Transplantation and Clinical Regeneration Scholarly Award Program, Mayo Clinic Transplant Research Center.
ORCID iD
Mark E. Wylam https://orcid.org/0000-0002-8859-4231
Supplemental material
Supplemental material for this article is available online.
References
- 1.Galiè N, Humbert M, Vachiery J-L, et al. 2015 ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension: the joint task force for the diagnosis and treatment of pulmonary hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS): endorsed by: association for European Paediatric and Congenital Cardiology (AEPC), International Society for Heart and Lung Transplantation (ISHLT). Eur Heart J 2015; 37: 67–119. [DOI] [PubMed] [Google Scholar]
- 2.Farber HW, Miller DP, Poms AD, et al. Five-Year outcomes of patients enrolled in the REVEAL registry. Chest 2015; 148: 1043–1054. [DOI] [PubMed] [Google Scholar]
- 3.Morrell NW, Aldred MA, Chung WK, et al. Genetics and genomics of pulmonary arterial hypertension. Eur Respir J 2019; 53(1): 1801899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cogan JD, Pauciulo MW, Batchman AP, et al. High frequency of BMPR2 exonic deletions/duplications in familial pulmonary arterial hypertension. Am J Respir Crit Care Med 2006; 174: 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machado RD, Eickelberg O, Elliott CG, et al. Genetics and genomics of pulmonary arterial hypertension. J Am Coll Cardiol 2009; 54: S32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Southgate L, Machado RD, Graf S, et al. Molecular genetic framework underlying pulmonary arterial hypertension. Nat Rev Cardiol 2019; 17(2): 85–95. [DOI] [PubMed] [Google Scholar]
- 7.Southgate L, Machado RD, Graf S, et al. Molecular genetic framework underlying pulmonary arterial hypertension. Nat Rev Cardiol 2020; 17: 85–95. [DOI] [PubMed] [Google Scholar]
- 8.Morrell NW. Pulmonary hypertension due to BMPR2 mutation: a new paradigm for tissue remodeling?. Proc Am Thorac Soc 2006; 3: 680–686. [DOI] [PubMed] [Google Scholar]
- 9.Zhu N, Gonzaga-Jauregui C, Welch CL, et al. Exome sequencing in children with pulmonary arterial hypertension demonstrates differences compared with adults. Circulation 2018; 11: e001887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tayeh MK, Chin EL, Miller VR, et al. Targeted comparative genomic hybridization array for the detection of single- and multiexon gene deletions and duplications. Genet Med 2009; 11: 232–240. [DOI] [PubMed] [Google Scholar]
- 11.Kim MJ, Cho SI, Chae JH, et al. Pitfalls of multiple ligation-dependent probe amplifications in detecting DMD exon deletions or duplications. J Mol Diagn 2016; 18: 253–259. [DOI] [PubMed] [Google Scholar]
- 12.Johnson SH, Smadbeck JB, Smoley SA, et al. SVAtools for junction detection of genome-wide chromosomal rearrangements by mate-pair sequencing (MPseq). Cancer Genet 2018; 221: 1–18. [DOI] [PubMed] [Google Scholar]
- 13.Smadbeck JB, Johnson SH, Smoley SA, et al. Copy number variant analysis using genome-wide mate-pair sequencing. Genes Chromosomes Cancer 2018; 57: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin X, Jin X, Kim LJY, et al. Inhibition of ID1-BMPR2 intrinsic signaling sensitizes glioma stem cells to differentiation therapy. Clin Cancer Res 2018; 24: 383–394. [DOI] [PubMed] [Google Scholar]
- 15.Smadbeck J, Peterson JF, Pearce KE, et al. Mate pair sequencing outperforms fluorescence in situ hybridization in the genomic characterization of multiple myeloma. Blood Cancer J 2019; 9: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-pul-10.1177_2045894020933081 for Mate-pair sequencing identifies a cryptic BMPR2 mutation in hereditary pulmonary arterial hypertension by Sarah J. Chalmers, Stephen J. Murphy, Laura L. Thompson, Nicole L. Hoppman, James B. Smadbeck, Jessica R. Balcom, Faye R. Harris, Robert P. Frantz, George Vasmatzis and Mark E. Wylam in Pulmonary Circulation