Abstract
In this study, we evaluated changes in focal adhesions (FAs) in two types of breast cancer cell (BCC) lines (differentiated MCF-7 and the triple-negative MDA-MB-231 cell line) exposed to simulated microgravity (s-μg) created by a random positioning machine (RPM) for 24 h. After exposure, the BCC changed their growth behavior and exhibited two phenotypes in RPM samples: one portion of the cells grew as a normal two-dimensional monolayer [adherent (AD) BCC], while the other portion formed three-dimensional (3D) multicellular spheroids (MCS). After 1 h and 30 min (MDA-MB-231) and 1 h 40 min (MCF-7), the MCS adhered completely to the slide flask bottom. After 2 h, MDA-MB-231 MCS cells started to migrate, and after 6 h, a large number of the cells had left the MCS and continued to grow in a scattered pattern, whereas MCF-7 cells were growing as a confluent monolayer after 6 h and 24 h. We investigated the genes associated with the cytoskeleton, the extracellular matrix and FAs. ACTB, TUBB, FN1, FAK1, and PXN gene expression patterns were not significantly changed in MDA-MB-231 cells, but we observed a down-regulation of LAMA3, ITGB1 mRNAs in AD cells and of ITGB1, TLN1 and VCL mRNAs in MDA-MB-231 MCS. RPM-exposed MCF-7 cells revealed a down-regulation in the gene expression of FAK1, PXN, TLN1, VCL and CDH1 in AD cells and PXN, TLN and CDH1 in MCS. An interaction analysis of the examined genes involved in 3D growth and adhesion indicated a central role of fibronectin, vinculin, and E-cadherin. Live cell imaging of eGFP-vinculin in MCF-7 cells confirmed these findings. β-catenin-transfected MCF-7 cells revealed a nuclear expression in 1g and RPM-AD cells. The target genes BCL9, MYC and JUN of the Wnt/β-catenin signaling pathway were differentially expressed in RPM-exposed MCF-7 cells. These findings suggest that vinculin and β-catenin are key mediators of BCC to form MCS during 24 h of RPM-exposure.
Keywords: microgravity, breast cancer, multicellular spheroids, focal adhesions, cytoskeleton, extracellular matrix
Introduction
Tumor diseases are a heavy burden for people around the world with high morbidity and mortality. The second leading cause of death worldwide is cancer (Sung et al., 2021). According to data published by the WHO Global Cancer Observatory (GLOBOCAN) in 2020, tumor diseases were responsible for an estimated 10 million deaths in 2020 (Sung et al., 2021).
In this study, we focused on breast cancer cells. Breast cancer (BC) is the most commonly diagnosed malignant tumor with an estimated 2.3 million new cases (11.7%) (Sung et al., 2021). BC is a very heterogeneous disease, as tumor cells can show different gene expression patterns (van’t Veer et al., 2002). An individualized therapy for patients is therefore necessary. BC shows seven molecular subtypes with typical histology, treatment options, and outcomes (van’t Veer et al., 2002; Nassef et al., 2019a). Patient survival rates are associated with the size of the tumor, the hormone receptor profile, and metastases at the time of diagnosis. Standard therapy is the surgical resection of the cancer. Depending on tumor type, size, staging, and metastasis to sentinel lymph nodes, strategies like chemotherapy, radiation and anti-hormone therapy, targeted treatment against HER2, and anti-angiogenic therapy are often applied after surgery or in cases of advanced disease stage (Kristensen et al., 2014). Despite advanced therapy, it is estimated that about 684,996 deaths worldwide will occur from BC per year (Sung et al., 2021). Therefore, novel therapeutic strategies with the help of new technologies are required.
Microgravity (μg)-based cancer research currently generates major interest (Becker and Souza, 2013; Krüger et al., 2019). Gravitational biology and space medicine are growing areas of research worldwide. It is known today that cells cultivated in real and simulated μg show changes in proliferation, transcription factors, gene expression patterns, and in the content of various proteins (Lewis et al., 1998; Boonyaratanakornkit et al., 2005; Chang and Hughes-Fulford, 2009; Pietsch et al., 2010; Chang et al., 2012; Svejgaard et al., 2015; Kopp et al., 2016, 2018; Buken et al., 2019; Grimm et al., 2020). Exposure of cells to real μg and the random positioning machine (RPM) leads to changes in the cytoskeleton, focal adhesion components, extracellular matrix proteins, and differentiation, as well as to an increase in apoptosis and changes in growth behavior (Lewis et al., 1998; Svejgaard et al., 2015; Kopp et al., 2018; Buken et al., 2019; Nassef et al., 2019a,b).
In earlier investigations, MCF-7 cells (Soule et al., 1973) showed variable growth behavior when exposed to the RPM for one and 5 days: one portion of the cells remained adherent, while the second portion grew in the form of spheroids with and without gland-like structures (Kopp et al., 2016). In contrast, MCF-7 cells only formed compact spheroids when cultured for 24 h under conditions of simulated μg created by an RPM (Kopp et al., 2016).
Our second cell line of interest is the well-described basal-like triple-negative MDA-MB-231 cell line, which has also been investigated in real μg and on an RPM (Masiello et al., 2014; Nassef et al., 2019a). Exposure to simulated (s-) μg resulted in three-dimensional (3D) growth and demonstrated a fundamental role in shaping form and function in human BCC (Masiello et al., 2014). These cell cultures in μg can be performed scaffold-free to engineer 3D aggregates, including multicellular spheroids (MCS), tissues, and tubular structures (Grimm et al., 2020). The organoids can be used for research in space medicine and for testing the delivery and efficacy of drugs (Grimm et al., 2020). Cancer cells are sensitive to mechanical stress. The regulation of focal adhesions (FAs) and cytoskeletal dynamics are important for cell maintenance, cell adherence, cell movement, and migration (Hughes-Fulford, 2003; Li et al., 2009). It has been demonstrated that s-μg reduces FAs and alters the cytoskeleton and nuclear positioning, leading to enhanced apoptosis in B16 melanoma cells (Zhao et al., 2018). Another study showed that the inhibition of FAs leads to reduced melanoma cell proliferation and metastasis (Tan et al., 2018).
Therefore, the objective of this study was to (i) investigate the changes and growth of BC cells exposed to s-μg created by an RPM, (ii) and to test, based on previous findings, the hypothesis that 24 h of exposure to s-μg cells induces the formation of MCS and alters the expression of genes related to the cytoskeleton, cell adhesion, and focal adhesions (FAs) in BC. Finally, we focused on the migration behavior and invasion potential of the MCS of both BC cell types.
Materials and Methods
Cell Culturing and Microgravity Simulation on the RPM
MDA-MB-231 human breast adenocarcinoma cells (MDA-MB-231; ATCC® HTB-26TM) and the MCF-7 human breast adenocarcinoma cell line MCF7 (ATCC® HTB-22TM) were purchased from the American Type Culture Collection (Manassas, Virginia, United States). The cells were cultured in RPMI 1640 (Life Technologies, Naerum, Denmark) in complete medium supplemented with 10% fetal calf serum (Sigma-Aldrich St. Louis, MO, United States) and 1% penicillin/streptomycin (Life Technologies, Carlsbad, CA, United States) under standard cell culture conditions (37°C and 5% CO2). Three days prior to the experiment, 2 × 106 cells were counted and seeded into T75 cm2 vented cap cell culture flasks (Sarstedt, Nümbrecht, Germany). A total number of 24 T25 cm2 cell culture flasks was prepared for the RPM experiments. Twelve flasks were kept under normal gravity conditions in the same incubator where the RPM was situated and used as ground controls (1g), and 12 flasks were mounted on the RPM. In addition, 2 × 24 slide flasks (NuncTM, ThermoFisher Scientific, Denmark) were prepared for each condition, and 1 × 105 cells were seeded in each 9 cm2 slide flask for immunofluorescence staining. Each flask was completely filled with medium, in a complete air bubble-free condition. The flasks were installed on the center frame of the RPM and run for 24 h. The 1g controls were stored inside the device in the same incubator at 37°C and with 5% CO2.
After 24 h, the RPM was stopped, and the BC cells were examined and photographed by phase contrast microscopy. For qPCR investigations, the medium from RPM flasks was transferred into 50 mL tubes and centrifuged at 4°C to collect the MCS. The MCS were washed in phosphate buffered saline (PBS) (Life Technologies), centrifuged again and after removal of the PBS stored in liquid nitrogen. For harvesting the adherent cells from 1g control and RPM cultures from the cell culture flask bottom, 10 mL ice-cold PBS was added to each T75 cm2 flask, and the cells were scraped off with a scraper. The cell suspension was collected and centrifuged at 4°C. The PBS was discarded, and the dry pellet was washed twice with PBS and afterward stored in liquid nitrogen for protein extraction and RNA isolation. For immunofluorescence staining, the medium was discarded and the cells were washed twice with PBS. Afterward, 2 mL 4% PFA (paraformaldehyde solution) was added to fix the cells for 45 min at room temperature. Then, the PFA was discarded, and 2 mL of PBS was added to the slide flasks, which were then stored in a refrigerator for future staining.
Simulated Microgravity Conditions Created by a Random Positioning Machine
The RPM was purchased from Airbus, Defense, and Space (former Dutch Space, Leiden, the Netherlands) and used to simulate microgravity. The method was published earlier in detail (Borst and van Loon, 2008; Wuest et al., 2015). In short, the RPM attempts to nullify the gravity vector by continuously rotating the central frame (where samples are mounted) around two perpendicular axes. The degree of rotation was in between 60°/s and 75°/s and randomized around both axes by continuously changing the direction; thus, the net magnitude of gravity vector toward the sample approaches zero, resulting in simulated microgravity. Twelve T25 cm2 flasks (12 each run) were fixed on the central frame of the RPM. After 24 h, the device was stopped, and the samples were collected and investigated by phase contrast microscopy. Three different groups were collected from the RPM experiments: 1g, AD, and MCS.
Phase Contrast Imaging
The morphology of the 1g control and RPM BC cells before, during, and after the experiment were photographed using phase contrast microscopy. The images were taken with a Canon EOS550D camera (Canon GmbH, Krefeld, Germany) through a Leica DM IL LED inverted microscope (Leica Microsystems, Brønshøj, Denmark).
Proliferation, Migration, and Invasion
Ki-67 immunofluorescence staining was performed to visualize the proliferating cells in MCS after 24 h. The immunofluorescence method is described in Section “Immunofluorescence Staining and Confocal Laser Scanning Microscopy.” To demonstrate the migration behavior of the MCS, the spheroids were collected after 24 h, pipetted in slide flasks and studied for adherence and migration by phase contrast microscopy (Leica Microsystems).
To test the invasion potential of the MCS of both cell lines, we cultured a confluent monolayer of human endothelial cells [EA.hy926 cell line (ATCC CRL-2922)] in slide flasks [detailed cell culture method published in reference (Dittrich et al., 2018)], pipetted MCF-7 or MDA-MB-231 BC MCS cells on the endothelial cells, and studied the invasion potential of the MCS cells. The examination was performed by phase contrast microscopy.
Immunofluorescence Staining and Confocal Laser Scanning Microscopy
The BC cells investigated in slide flasks were fixed with 4% PFA solution for 30 min. Then, they were subjected to membrane permeabilization with 0.1% Triton X for 10 min and blocking with 1% [weight (w)/volume (v)] bovine serum albumin (BSA, Sigma-Aldrich, Steinheim, Germany) in PBS for 30 min. The slides were then released from the flasks, and the cells were incubated with primary antibodies (listed in Table 1) in a PBS solution with 1% (w/v) BSA overnight at room temperature. The next day, the cells were washed three times with PBS before incubation with the secondary Alexa Fluor 488 (AF488)-conjugated anti-rabbit (Cell Signaling Technology, Danvers, MA, United States) or anti-mouse antibody (Invitrogen) at a dilution of 1:1,000 for 2 h at ambient temperature. Afterward, the slides were washed with PBS three times and prepared and covered for microscopy using FluoroshieldTM mounting media with DAPI (4′,6-diamidino-2-phenylindole; Sigma-Aldrich). The covered slides were kept at 4°C in a dark box until confocal microscopy was performed (Buken et al., 2019).
TABLE 1.
List of antibodies used for immunofluorescence.
| Antibody | Immunofluorescence | |
| E-cadherin | CST#2500S | Rb |
| Fibronectin | Millipore Sigma/F3648 | Rb |
| Ki 67 | Abcam# ab15580 | Rb |
| Laminin | Sigma/L9393 | Rb |
| Talin | Millipore Sigma/T3287 | Ms |
| Vinculin | Millipore Sigma/V9131 | Ms |
Ms, mouse; Rb, rabbit; Sigma (now by MERCK, Darmstadt, Germany); ABCAM (Cambridge, United Kingdom); Cell Signaling Technology (CST, BioNordika Denmark A/S, Herlev, Denmark); and MERCK Millipore (Burlington, Massachusetts, United States).
Fluorescence staining was analyzed using a ZEISS LSM 710 CLSM (ZEISS, Jena, Germany) and a 40 × oil-immersion objective with a NA of 1.3 (Nyegaard et al., 2015; Corydon et al., 2016b). The quantification of the fluorescence intensity was performed with the FIJI image processing software (available from1). Briefly, data in.czi-format was imported using the Bio-Formats plugin and all color channels were split into separate grayscale images. In the green channel, individual cells were outlined with the freehand ROI tool and both area and integrated density was measured. Subsequently, background and secondary antibody control intensities were subtracted to yield the corrected total cell fluorescence. For each experimental condition, at least five different fields of view with a mean number of five cells were analyzed.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
The method was published earlier in detail (Ma et al., 2013; Kopp et al., 2015). RNA isolation was performed using a RNeasy Mini Kit (Qiagen, Hilden, Germany) with an additional DNase digestion step (Qiagen) in order to eliminate residual DNA contamination. Afterward, the amount of RNA was quantified using a Photometer Ultrospec2010 (Amersham Biosciences, Freiburg, Germany). A first strand cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, United States) was applied for reverse transcription. qPCR was performed on a 7,500 Fast Real-Time PCR System using the FAST SYBR Green Master Mix (both Applied Biosystems, Darmstadt, Germany) according to standard protocols (Kopp et al., 2016, 2018; Buken et al., 2019; Nassef et al., 2019a).
All samples were measured in triplicate. For normalization, 18S rRNA was used as a housekeeping gene. The comparative Ct (ΔΔCt) method was used for relative quantification of transcription levels, and the 1g control group was defined as 100% for reference. Before performing qPCR, primers were designed using NCBI Primer Blast, and they were selective for cDNA by spanning exon–exon junctions and had a melting temperature of around 60°C. The primers were synthesized by TIB Molbiol (Berlin, Germany) and are listed in Table 2:
TABLE 2.
List of all the primers’ sequences used in the quantitative PCR. All the sequences are listed in the 5′-3′-direction.
| Factor | Primer Name | Sequence 5′–3′ |
| 18S | 18S-F | GGAGCCTGCGGCTTAATTT |
| 18S-R | CAACTAAGAACGGCCATGCA | |
| ACTB | ACTB-F | TGCCGACAGGATGCAGAAG |
| ACTB-R | GCCGATCCACACGGAGTACT | |
| BCL9 | BCL9-F | CAGAGCAGACAATAGGCCCC |
| BCL9-R | AGACCCTTTTCCCGCAATCC | |
| CCND1 | CCND1-F | CCCTGACGGCCGAGAAG |
| CCND1-R | AGGTTCCACTTGAGCTTGTTCAC | |
| CDH1 | CDH1-F | GCTGGACCGAGAGAGTTTCC |
| CDH1-R | CAGCTGTTGCTGTTGTGCTT | |
| CTNNA1 | CTNNA1-F | AATTTAGCGCTCGCCCAG |
| CTNNA1-R | ACAAGGGTTGTAACCTGTGTAA | |
| CTNNB1 | CTNNB1-F | GAAACAGCTCGTTGTACCGC |
| CTNNB1-R | ATCCACTGGTGAACCAAGCA | |
| FAK1/PTK2 | FAK1-F | TGTGGGTAAACCAGATCCTGC |
| FAK1-R | CTGAAGCTTGACACCCTCGT | |
| FN1 | FN1-F | AGATCTACCTGTACACCTTGAATGACA |
| FN1-R | CATGATACCAGCAAGGAATTGG | |
| ITGB1 | ITGB1-F | GAAAACAGCGCATATCTGGAAATT |
| ITGB1-R | CAGCCAATCAGTGATCCACAA | |
| JUN | JUN-F | GAGCTGGAGCGCCTGATAAT |
| JUN-R | CCCTCCTGCTCATCTGTCAC | |
| LAMA3 | LAMA3-F | AAAGCAAGAAGTCAGTCCAGC |
| LAMA3-R | TCCCATGAAGACCATCTCGG | |
| MYC | MYC-F | GGATTCTCTGCTCTCCTCGAC |
| MYC-R | CTTCTTGTTCCTCCTCAGAGTC | |
| NFATC2 | NFATC2-F | CAAGACGAGCTTGACTTCTCCA |
| NFATC2-R | GCATTCGGCTCTTCTTCGTTC | |
| PXN | PXN-F | CATGGACGACCTCGACGC |
| PXN-R | CAAGAACACAGGCCGTTTGG | |
| TBP | TBP-F | GTGACCCAGCATCACTGTTTC |
| TBP-R | GCAAACCAGAAACCCTTGCG | |
| TLN1 | TLN1-F | GATGGCTATTACTCAGTACAGACAACTGA |
| TLN1-R | CATAGTAGACTCCTCATCTCCTTCCA | |
| TUBB | TUBB-F | CTGGACCGCATCTCTGTGTACTAC |
| TUBB-R | GACCTGAGCGAACAGAGTCCAT | |
| VCL | VCL-F | GTCTCGGCTGCTCGTATCTT |
| VCL-R | GTCCACCAGCCCTGTCATTT |
STRING Analysis
The interactions between proteins were determined using the STRING v10 platform (Snel et al., 2000). For each protein, the UniProtKB entry number was inserted in the input form “multiple proteins,” and “Homo sapiens” was selected as the organism. The resulting network view was downloaded in the molecular action view showing lines between interacting proteins and genes (Nassef et al., 2019a).
Transfection of the MCF-7 Cell Line to Visualize Vinculin and β-Catenin
The MCF-7 BCC cells were stably transfected using a Sleeping Beauty (SB) transposon-based vector containing the eGFP-hVCL1 cassette for the visualization of vinculin as described in Nassef et al. (2019b). In brief, an eGFP-hVCL fragment containing NotI and XbaI restriction sites in the 5′ and 3′ ends, respectively, was sub-cloned into a Sleeping Beauty (SB) transposon-based vector pT2/CMV-linker-SV40-Neo (Staunstrup et al., 2011; Pihlmann et al., 2012) containing a linker, enabling insertion of the NotI-eGFP-hVCL1-Xba1 fragment. The resulting plasmid, pT2/CMV-eGFP-hVCL1-SV40-Neo, was entitled pSB-eGFP-vinculin. In order to provide stable expression of the eGFP-vinculin expression cassette, MCF-7 cells were co-transfected with pSB-eGFP-vinculin and pCMV−SB100X (Pihlmann et al., 2012) using X-tremeGENE 9 transfection reagent (Roche, Mannheim, Germany) according to the manufacturer’s protocol (Askou et al., 2015). An inactive SB transposase (mSB) was included as negative control. Transfected cells were cultured in medium containing G418 (geneticin) to allow for growth of stably transfected cells only. A fluorescence microscope was used to validate the efficiency of the transfection.
To visualize catenin in MCF-7 BCC cells, a cDNA fragment encoding human β-catenin N-terminally tagged with the mCherry marker protein was cloned into the piggyBac plasmid pPBT-CMV-MCS-IRES-Puro (Sharma et al., 2012). The resulting plasmid, entitled pPBT-mCherry-catenin, was transiently transfected into MCF-7 cells using X-tremeGENE 9 transfection reagent (Roche) according to manufacturer’s protocol (Askou et al., 2015). Following transfection, the cells were maintained for 24 h at 1g conditions and then transferred to either 1g or to the RPM for additional 24 h.
Images of MCF-7 BCC cells expressing eGFP-vinculin were captured by a confocal scanning microscope (CLSM) 710 (Zeiss, Jena, Germany). Images of MCF-7 BCC cells expressing mCherry-catenin were captured 48 h post-transfection by a CLSM 800 (Zeiss).
Statistical Analysis
All statistical evaluations were performed using IBM SPSS Statistics 23 (IBM Deutschland GmbH, Ehningen, Germany). The Mann–Whitney U-Test was utilized to evaluate the statistical significance in the changes in expression levels following RPM exposure, thus comparing the 1g control to AD and MCS. A significance level of 0.05 was used. The standard deviation was calculated and presented together with the mean values as percentages in bar plots.
Results
Morphology, Cell Growth, Migration and Invasion Behavior of the BCC
MCF-7 and MDA-MB-231 BCC cultured under normal standard cell culture conditions at 1g grew as a two-dimensional (2D) monolayer (Figures 1A,D). MCF-7 revealed their characteristic epithelial growth behavior at 1g (Figure 1A). MDA-MB-231 cells exhibited an epithelial-like morphology and appeared phenotypically as spindle-shaped cells under normal 1g static conditions (Figure 1D). In contrast, both BCC types, the MCF-7 and MDA-MB-231 cells exposed to an RPM for 24 h showed two different phenotypes: adherent cells and multiple multicellular spheroids floating in the medium (Figures 1B,E).
FIGURE 1.
Phase contrast microscopy of MCF-7 and MDA-MB-231 breast cancer cells (BCC). (A) Characteristic epithelial morphology of the MCF-7 cell line cultured at 1g. (B) Adherent MCF-7 cells and multiple compact multicellular spheroids (MCS, yellow arrows) were visible when the MCF-7 cells were exposed to the random positioning machine (RPM) for 24 h. (C) Ki-67 immunofluorescence staining (IFS) of MCF-7 MCS. The red arrows show Ki-67 positive nuclei. (D) MDA-MB-231 cell line cultured under static 1g control conditions at 24 h. (E) MDA-MB-231 cells exposed to the RPM for 24 h. Multiple multicellular spheroids (yellow arrows) are visible as well as adherent cells. Magnification × 100. The yellow arrows indicate MCS floating in the supernatant of the cell cultures. (F) Ki-67 IFS of MDA-MB-231 MCS. The red arrows show Ki-67 positive nuclei. (G) Migration behavior of the MCF-7 cells in spheroids at 0 h, (H) 2h, the yellow arrows indicate the attachment of the MCS on the slide flask bottom and the cell membrane. (I) 6 h, the red arrows show areas of migration and (J) 24 h. (K) Migration behavior of MDA-MB-231 cells in spheroids at 0 h, (L) 2 h, the red arrows show the attached MCS and migration (M) 6 h, red arrows indicate migrated cells and after (N) 24 h. No MCS were visible at this time point. Invasion test: co-cultures of EA.hy926 endothelial cells and MCF-7 MCS. (O) 0 h, (P,Q) 3 h and 6h, the blue arrows show the formation of a cellular membrane. (R) After 24 h the MCF-7 BCC had invaded the endothelial cells. Invasion test: co-cultures of EA.hy926 endothelial cells and MDA-MB-231 MCS. (S) 0 h, (T) 3 h, at this time point the MDA-MB-231 cells had already migrated out of the MCS and showed an invasive growth. (U) 6 h and (V) 24 h.
The proliferation marker Ki-67 was determined to indicate the active phases of the cell cycle (G1, S, G2, and mitosis), because the Ki-67 protein is absent in resting G0 cells. The Ki-67 immunofluorescence showed various proliferating cells on the surface of the MCS of both cell types (Figures 1C,F).
In addition, we tested the migration behavior of the MCS. After 20 min 50% of the MCF-7 spheroids and 30% of the MDA-MB-231 spheroids were adherent to the slide flask bottom. All MDA-MB-231 MCS had attached after 1.5 h, and all MCF-7 MCS stayed adherent on the slide flask bottom after 1 h 40 min. Figures 1H,L show the MCS of both cell types at 2 h. The MDA-MB-231 cells started to migrate (Figure 1L), whereas the MCF-7 MCS exhibited an outer membrane (yellow arrows, Figure 1H). After 6 h, a large number of MDA-MB-231 BCC had migrated out of the MCS (Figure 1M) and only some remaining 3D aggregates are visible. In contrast, the MCF-7 MCS were still compact and only a few cells started to leave the MCS (red arrows, Figure 1I). In these areas, the membrane around the cells growing three-dimensionally was destroyed. After 24 h, MDA-MB-231 cells grew in form of a monolayer and the cells had lost their intercellular contacts (Figure 1N), whereas MCF-7 cells still grew as a more compact cell monolayer, but with a broader interstitial space (Figure 1J).
To evaluate the invasion behavior of both BC types, spheroids were seeded out on a confluent monolayer of EA.hy926 endothelial cells (ECs) (Figures 1O–V). After 3 h, MCF-7 spheroids exhibited an outer membrane (yellow arrows) at the contact area to the ECs. The ECs changed their orientation and provided more space for the tumor cells, which started their invasion (Figure 1P). The blue arrow shows the invasive MCF-7 cells, and the black arrow indicates the looser EC layer (Figure 1P). Three hours later, the MCF-7 MCS were invasive growing in between the ECs, providing space to the cancer cells (Figure 1Q). The overview given in Figure 1R shows the invasiveness of the MCF-7 tumor cells after a 24 h co-culture and their growth between the EC monolayer.
MDA-MB-231 spheroids seeded on ECs very rapidly attached and spread out onto the EC monolayer (Figure 1T). After a 6-h co-culture, all cells of the MCS had migrated out of the spheroid and progressed growing between the EC monolayer (Figure 1U). After 24 h, mostly single tumor cells were detectable between the ECs (Figure 1V).
Changes in Cytoskeletal Factors in MDA-MB-231 Cells Exposed to the RPM
We measured the gene expression of β-actin in MDA-MB-231 cells exposed to the RPM. The ACTB mRNA expression was not altered in 1g, adherent (AD) and MCS samples (Figure 2A). In parallel, the TUBB mRNA expression was not changed in all groups (Figure 2B). The cytoskeletal components of MCF-7 cells were examined in an earlier study (Kopp et al., 2016).
FIGURE 2.
Gene expression [ACTB (A), TUBB (B)] of MDA-MB-231 cells exposed to the RPM. * p < 0.05 1g vs. adherent (AD) and/or MCS and #p < 0.05 AD vs. MCS.
Changes in Cell Adhesion Molecules of MDA-MB-231 Cells Exposed to the RPM
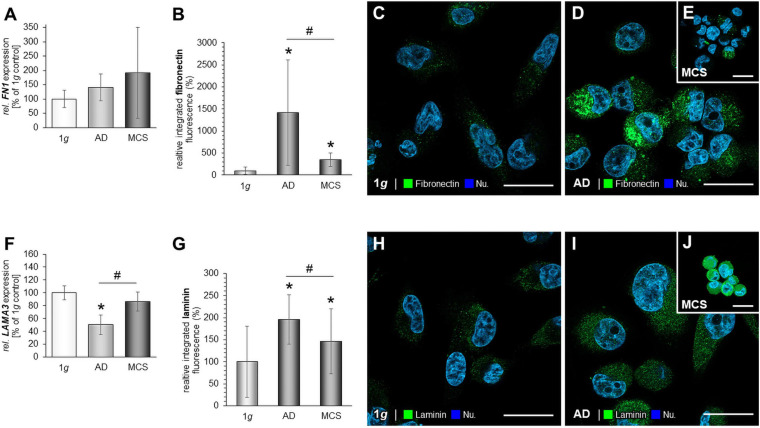
We investigated the cell adhesion molecules fibronectin and laminin. There was no significant change in the gene expression of FN1 in MCS after 24 h (Figure 3A). The fibronectin immunofluorescence was significantly elevated in RPM-AD cells and MCS compared with 1g cells (Figures 3B–E). In addition, the mRNA expression of LAMA3 was not changed in MCS vs. 1g, but significantly down-regulated in AD cells in comparison to 1g static control cells (Figure 3F). The laminin immunofluorescence was increased in RPM-AD cells and MCS (Figures 3G–J). Both cell adhesion molecules and extracellular matrix proteins were investigated earlier in MCF-7 cells (Kopp et al., 2016).
FIGURE 3.
Investigation of fibronectin (A–E) and laminin (F–J): The gene expression of FN1 (A), analysis of the fluorescence intensity of fibronectin (B) and immunofluorescence staining (IFS) of fibronectin in 1g samples of MDA-MB-231 cells: (C), RPM-AD samples (D) and MCS (E); the LAMA3 mRNA expression (F) as well as analysis of the fluorescence intensity of fibronectin (G), the IFS of laminin in 1g samples (H), RPM-AD samples (I) and MCS (J) of MDA-MB-231 cells exposed to the RPM. *p < 0.05 1g vs. AD and/or MCS and #p < 0.05 AD vs. MCS.
The Impact of RPM-Exposure on Focal Adhesions and Interacting Factors in MDA-MB-231 Cells
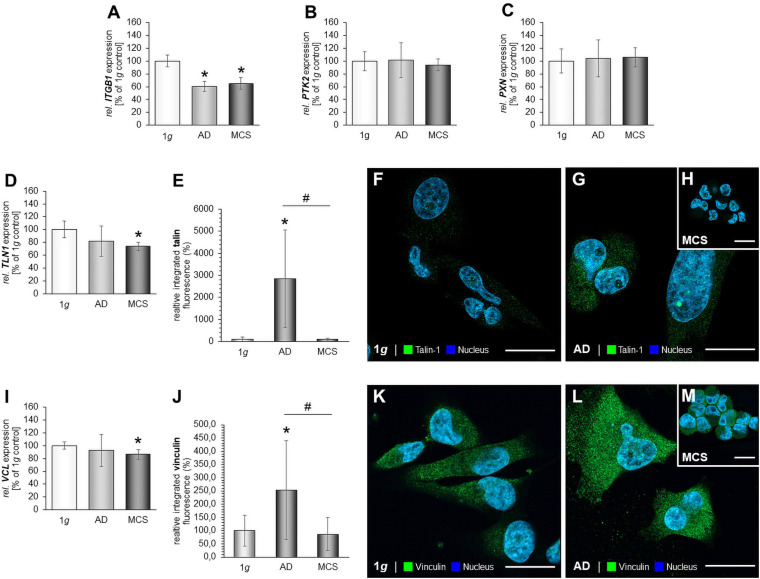
We investigated integrin-β1 and measured a significantly down-regulated ITGB1 gene expression in RPM-exposed AD and MCS samples (Figure 4A). The gene expression of focal adhesion kinase 1 (FAK1), also known as PTK2 protein tyrosine kinase 2 (PTK2), was not changed by RPM exposure in MDA-MB-231 cells (Figure 4B). In addition, the PXN mRNA was not altered in RPM-exposed MDA-MB-231 cells compared to 1g controls (Figure 4C).
FIGURE 4.
Focal adhesions in MDA-MB-231 cells. The gene expression of ITGB1 (A), PTK2 mRNA expression (B), PXN mRNA expression (C) of MDA-MB-231 cells exposed to the RPM. In addition, the gene expression of TLN1 (D) and analysis of the fluorescence intensity of talin (E), IFS of 1g control cells (F) and RPM-exposed MDA-MB-231 cells [AD cells: (G) and MCS: (H)] is given. Moreover, the VCL mRNA (I) and analysis of the fluorescence intensity of vinculin (J), the IFS of vinculin are shown in 1g control cells (K) and RPM-exposed AD cells (L) and MCS (M). *p < 0.05 1g vs. AD and/or MCS and #p < 0.05 AD vs. MCS.
Furthermore, the TLN1 gene expression was not significantly altered in RPM-AD cells cultured under s-μg conditions, but significantly reduced in MCS (Figure 4D). IFS revealed a cytoplasmic staining of talin-1. RPM-exposed AD cells exhibited an increase in talin-1 compared to 1g and MCS (Figures 4E–H). Furthermore, the MCS expressed talin-1 in an intensity comparable to 1g cells (Figure 4H).
In addition, the VCL gene expression was significantly down-regulated in MCS compared to 1g (Figure 4I). The IFS of vinculin showed a similar fluorescence intensity in the cytoplasm of the MDA-MB-231 MCS compared to 1g, but a significant increase in AD cells exposed to s-μg (Figures 4J–M).
The Impact of RPM-Exposure on Focal Adhesions in MCF-7 Cells
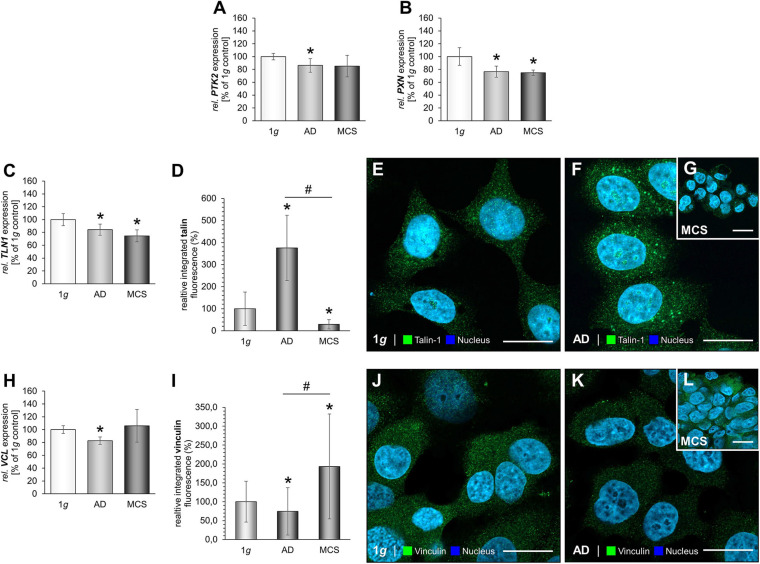
The next step was to examine focal adhesion molecules in MCF-7 BCC exposed to s-μg conditions. The results are presented in Figure 5. PTK2 mRNA expression was reduced in the AD group (Figure 5A). Furthermore, the gene expression of PXN was significantly down-regulated in both RPM groups (Figure 5B). Moreover, we focused on talin-1. The TLN1 mRNA expression was significantly decreased in AD and MCS cells (Figure 5C). The IFS revealed a significant increase in the cytoplasmic fluorescence of talin in AD samples compared to MCS and 1g samples (Figures 5D–G). In MCS, the talin IF is significantly reduced compared with 1g (Figure 5G). Finally, the gene expression of VCL was not differentially displayed in MCS vs. 1g, but down-regulated in RPM-AD cells vs. 1g (Figure 5H). The IFS of vinculin is significantly reduced in AD cells compared to 1g cells, whereas the vinculin IF was elevated in MCS compared to 1g and AD (Figures 5I–L).
FIGURE 5.
Focal adhesions in MCF-7 BCC. The PTK2 mRNA expression (A), PXN mRNA expression (B) of MCF-7 cells exposed to the RPM. In addition, the gene expression of TLN1 (C) and analysis of the fluorescence intensity of talin (D), IFS of 1g control cells (E) and RPM-exposed MCF-7 BCC [AD: (F), MCS: (G)] is given. VCL mRNA (H) and analysis of the fluorescence intensity of vinculin (I), the IFS of vinculin are shown in 1g control cells (J) and RPM-exposed cells [AD: (K), MCS: (L)]. *p < 0.05 1g vs. AD and/or MCS and #p < 0.05 AD vs. MCS.
The Impact of RPM-Exposure on E-Cadherin in MCF-7 Cells
MCF-7 BCC exposed to the RPM showed a down-regulation of the CDH1 gene in AD and MCS samples (Figure 6A). IFS revealed an elevated fluorescence intensity in AD cells compared to normal 1g control BCC (Figures 6B–E).
FIGURE 6.
E-cadherin in microgravity-exposed MCF-7 BCC. (A) CDH1 gene expression in MCF-7 BCC exposed to s-μg. (B) Analysis of the fluorescence intensity of E-cadherin (C), IFS of E-cadherin of 1g MCF-7 BCC RPM-exposed BCC [AD: (D), MCS: (E)]. *p < 0.05 1g vs. AD and/or MCS.
Results of Gene/Protein Interactions (STRING Analysis)
The various genes analyzed by qPCR were investigated with regard to their possible interactions and mutual expression dependence. A STRING/EMBL (European Molecular Biology Laboratory) analysis of these items represented in molecule action mode is shown in Figure 7A. It can be seen that the FA factors for which the expression pattern was analyzed regulate each other very strongly. The genes of interest (Figures 7B,C) were differentially regulated in s-μg samples (AD and MCS). Figures 7B,C present a summary of the qPCR data, already described in Figures 2–6, and provide a comparable overview on the results. A closer look at the 24 h samples revealed that most genes involved in the focal adhesion molecule complex, especially the CDH1 mRNA, were highly down-regulated in both AD and MCS groups of MCF-7 BCC. The cell adhesion molecule and extracellular matrix protein fibronectin was slightly elevated in AD and MCS. A down-regulation was observed for LAMA3 in MDA-MB-231 cells.
FIGURE 7.
(A) Network of the functional interaction of genes and their products analyzed in this study. The analysis was performed by STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, v11.0) provided by the STRING Consortium (available online: https://string-db.org/). The result is presented in the molecule action mode. Gene names are indicated. (B) Summary of the gene expression fold change measured by qPCR of 24 h RPM-exposed MDA-MB-231 samples. Results determined in AD and MCS in relation to 1g were given. The red color indicates up-regulated genes and the green color down-regulated genes. (C) Summary of the gene expression fold change measured by qPCR of 24-h RPM-exposed MCF-7 samples. The * indicates a significant change in fold change.
Vinculin/β-Catenin Signaling in MCF-7 Cells
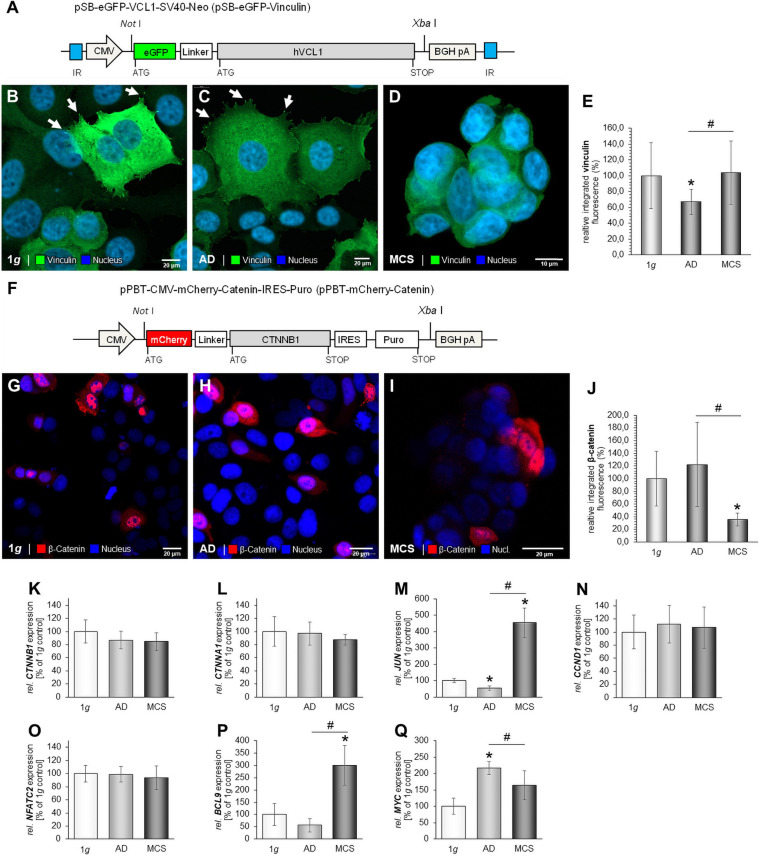
For stable expression and amenable live-cell visualization of vinculin in MCF-7 BCC, a Sleeping Beauty (SB) transposon-based vector containing a CNV-driven eGFP-hVCL1 cassette was constructed according to Nassef et al., 2019b; Figure 8A. This construct enables the synthesis of a fusion-protein containing the eGFP marker protein N-terminally tagged to vinculin. Following co-transfections with pSB-eGFP-vinculin and pCMV-SB100X, cells were incubated in medium containing G418 for selection of clones. Several colonies were obtained after inspection by fluorescence microscopy for validation of the expression level of eGFP-vinculin. Only few colonies were obtained when using an inactive SB transposase, showing that the eGFP-vinculin-positive clones results from SB-mediated insertion of the eGFP-hVCL1 cassette.
FIGURE 8.
Stable expression of eGFP-vinculin in MCF-7 cells. Following co-transfection of pSB-eGFP-vinculin and pCMV-SB100X cells were cultured in medium containing G418 to allow growth of stably transfected cells only. (A) Graphic illustration of the pSB-eGFP-vinculin plasmid containing the coding sequence of the eGFP-vinculin fusion protein encoded from a CMV promoter. Restrictions used for cloning are indicated. BGH pA, bovine growth hormone polyadenylation site; CMV, cytomegalovirus promoter; IR, internal repeats. (B) Image of eGFP-vinculin in MCF-7 cells subjected to 1g. The white arrows show lamellipodia and vinculin focal adhesion dots. (C) Image of eGFP-vinculin in MCF-7 cells after RPM exposure. The white arrows indicate vinculin dots (D) Image of eGFP-vinculin in MCF-7 MCS following RPM-exposure. Scale bars 10 μm. Expression of mCherry-catenin in MCF-7 cells. MCF-7 cells were transfected with pPBT-mCherry-catenin and images were captured 48 h post transfection. (E) Analysis of the fluorescence intensity of eGFP-vinculin. (F) Graphic illustration of the pSB-eGFP-vinculin plasmid containing the coding sequence of the eGFP-vinculin fusion protein encoded from a CMV promoter. Restrictions used for cloning are indicated. BGH pA, bovine growth hormone polyadenylation site; CMV, cytomegalovirus promoter. (G) Image of MCF-7 cells expressing mCherry-catenin at 1g. (H) Image of RPM-exposed adherent MCF-7 cells expressing mCherry-catenin following incubation at μg. (I) Image of MCF-7 MCS expressing mCherry-catenin following incubation at μg. Scale bars 20 μm. (J) Analysis of the fluorescence intensity of mCherry-catenin. Gene expressions of CTNNB1 (K); CTNNA1 (L); JUN (M); CCND1 (N); NFATC2 (O); BCL9 mRNA (P) and MYC (Q), MCF-7 cells exposed to the RPM. *p < 0.05 1g vs. AD and/or MCS and #p < 0.05 AD vs. MCS.
eGFP-vinculin is strongly expressed at 1g in transfected MCF-7 BCC. Stress fibers and lamellipodia are visible (Figure 8B, white arrows). A similar expression pattern was detected in RPM-exposed AD cells (Figure 8C). A prominent formation of stress fibers (see right side of the cells shown to the right in Figure 8C) is detectable. A uniform expression of eGFP-vinculin is seen in MCS (Figure 8D). Interestingly, the stress fibers had disappeared in BCC MCS (Figure 8D). Figure 8E gives the results of the analysis of the fluorescence intensity of eGFP-vinculin. The highest fluorescence intensity was measured in MCS and at 1g.
To visualize catenin in MCF7 BCC cells the cDNA fragment encoding human β-catenin N-terminally tagged with the mCherry marker protein were cloned into the pPBT-CMV-MCS-IRES-Puro plasmid (Figure 8F). Following transient transfection, the mCherry-catenin was expressed in the cytoplasm and the nucleus of MCF-7 cells grown under normal gravity conditions (Figure 8G). β-catenin is less located in the nuclei of MCS cells than of adherent cells after RPM exposure (Figures 8H,I). A non-uniform expression of mCherry-catenin is found in MCS (Figure 8I). The protein is accumulated at the outer cell membrane (Figure 8I). Figure 8J gives the results of the analysis of the fluorescence intensity of mCherry-catenin.
The CTNNB1 mRNA was not significantly expressed in all three groups (Figure 8K). In addition, the gene expression of CTNNA1 was not significantly altered (Figure 8L). The Wnt/β-catenin target gene JUN was down-regulated in AD samples and up-regulated in MCS (Figure 8M). The CCND1 mRNA was not changed (Figure 8N). Furthermore, the NFATC2 mRNA was not significantly altered after RPM exposure (Figure 8O).
The Wnt/β-catenin target genes BCL9 and MYC were differentially expressed in RPM-exposed BCC (Figures 8P,Q). BCL9 was significantly up-regulated in MCS compared to AD and 1g samples (Figure 8P). The MYC mRNA was significantly elevated in AD cells compared to 1g and MCS samples (Figure 8Q).
Discussion
In this study, we focused on the changes induced in MDA-MB-231 and MCF-7 BCC when they were cultured under s-μg conditions created by an RPM. The RPM, suggested also as ground-based facility by the European Space Agency, was chosen for our experiments because effects on various cells with respect to behavior and morphology seen in r-μg are reproduced with good agreement (Wuest et al., 2015). Nevertheless, a final validation of our data is necessary in r-μg to justify the term s-μg in the context of our experimental approach. The MDA-MB-231 cell line is a good model for the basal-like or triple-negative form of BC (Chavez et al., 2010). This cell line was established from a pleural effusion of a patient with invasive ductal carcinoma and is E-cadherin-negative and expresses mutated p53. In contrast, the MCF-7 cell line is estrogen receptor (ER)-, progesterone receptor (PR)-, and E-cadherin-positive (Soule et al., 1973; Kopp et al., 2016), and it represents a model of a well-differentiated BC type.
The dynamic biological process of cell adhesion plays an important role in cell detachment and aggregation into 3D spheroids (Gumbiner, 1996). It has long been known that μg influences cell-to-cell interactions and alters the cytoskeleton, integrins, and ECM components of mammalian cells (Buravkova et al., 2005; Aleshcheva et al., 2016; Hader et al., 2017).
Various researchers have demonstrated that the adherence behavior of human cells changes when they are exposed to μg conditions (Dittrich et al., 2018; Zhao et al., 2018; Deng et al., 2019; Nassef et al., 2019a; Romswinkel et al., 2019). In particular, cell adhesion changes are involved in the scaffold-free formation of MCS (Riwaldt et al., 2015a; Svejgaard et al., 2015; Dittrich et al., 2018; Buken et al., 2019). The two cell adhesion molecules ICAM-1 and VCAM-1 are significantly altered by μg in different mammalian cells (Paulsen et al., 2015; Riwaldt et al., 2015b; Tauber et al., 2017; Buken et al., 2019). Their specific involvement in 3D growth was suggested for thyroid cancer cells after the CellBox-1 space mission (Riwaldt et al., 2015a).
Changes in the Cytoskeleton and Extracellular Matrix
It is well known that μg induces a disorganization in the network of microfilaments, intermediate filaments, and microtubules in various cell types (Vorselen et al., 2014). Several studies have revealed that the formation of the cytoskeleton is highly sensitive to alterations in gravity and shown that altered gravity conditions have an enormous impact on the cytoskeleton and the differential expression of genes related to it (Vassy et al., 2001; Uva et al., 2002; Ulbrich et al., 2011; Aleshcheva et al., 2013, 2015; Corydon et al., 2016a).
In this study, we observed that the gene expression of ACTB was not differentially altered in MDA-MB-231 cells after 24 h of RPM exposure. This is in accordance with earlier results obtained from MCF-7 BCC cultured for 24 h on the RPM (Kopp et al., 2016). In parallel, the TUBB gene expression was not altered in adherently growing RPM-exposed MDA-MB-231 cells compared with 1g samples. TUBB remained unchanged in MCS samples. This finding is different to the TUBB gene expression of MCF-7 BCC, FTC-133 thyroid cancer cells, and normal thyroid cells exposed to s-μg (Kopp et al., 2015, 2016), where a downregulation of the TUBB gene was observed in MCS of these cell types. This difference may be explained by the low differentiation of the MDA-MB-231 BC type.
In addition, we studied the expression of fibronectin. In general, we found a slight but insignificant trend for an increase in FN1 at the mRNA level when MDA-MB-231 cells were exposed to the RPM. Interestingly, the IIF staining revealed an increase of fibronectin protein in AD and MCS cells. In general protein and mRNA abundance, although derived from the same tissue or cell type, is usually in a non-linear relationship as multiple processes beyond transcript concentration contribute to protein abundance. Protein abundance modulation through the binding to regulatory elements (e.g., micro-RNAs), protein’s half-life, autophagy, protein synthesis delay by physical distance, availability of involved actors and feedback regulatory mechanisms usually leading to a reduced correlation between mRNA and protein levels.
This protein data fits with the results of an earlier study on human fetal osteoblasts exposed to the RPM, where the cells showed elevated FN1 gene expression and fibronectin protein content after a seven-day period of RPM exposure (Mann et al., 2019). In addition, the FN1 gene expression was not changed in MCF-7 spheroids after 24 h of RPM-exposure (Kopp et al., 2016).
LAMA3 mRNA was reduced in AD samples, but not differentially expressed in MDA-MB-231 MCS samples compared with 1g controls. This is in contrast to the data obtained from MCF-7 cells, where LAMA3 was significantly up-regulated in MCS (Kopp et al., 2016). This finding may be due to the formation of gland-like MCS when MCF-7 cells were exposed to the RPM for 5 days (Kopp et al., 2016). Laminin is a component of the basal lamina of a cell and influences the biological processes differentiation, migration and adhesion. This might explain the high amount of LAMA3 mRNA in MCS, where a high grade of differentiation and changes in the extracellular matrix components occur to form the 3D glandular structures.
The Focal Adhesions of BCC Grown in Simulated Microgravity
Furthermore, we focused on the FAs of MDA-MB-231 and MCF-7 BCC exposed to the RPM. FAs are subcellular structures mediating cellular signaling processes as a reaction to ECM adhesion and are known as mechanical linkages to the ECM. FAs are key players in gravisensing and the detachment of cells and following aggregation into 3D assemblies. This aggregation occurs through changes in the mechanical loading conditions in an s-μg environment, among other pathways (Geiger et al., 2009; Romswinkel et al., 2019).
Vinculin has an impact on cell–matrix adhesion and intercellular junctions and plays an important role in mechanotransduction with integrins at FA sites (Spanjaard and de Rooij, 2013). Vinculin interacts with talin, integrins, and actin, and thus influences cellular migration and focal adhesion. Vinculin is a mediator of cellular and extracellular signals. The gene expression of VCL was reduced in the MCS of MDA-MB-231 cells compared to 1g (Figure 4I). IFS of vinculin revealed an increase of the fluorescence intensity in AD samples and no change in MDA-MB-231 MCS.
The other important FA component investigated in this study is the cytosolic and mechanosensitive protein talin. Its major task is to link integrins directly and indirectly via vinculin to the cytoskeleton. Talin-1 is involved in cell adhesion, progression, extravasation, and trans-endothelial migration in cancer. Integrins bind to talin, and then talin connects to vinculin to influence the process of cell adhesion. Finally, integrin receptors promote the attachment of adherent growing cells to the ECM. In this context, talin acts as mechanotransductor. In triple-negative MDA-MB-231 BCC, the TLN1 gene expression in AD was not significantly changed, but reduced in MCS. The IFS of talin-1 revealed an increase in talin-1 protein in AD samples compared to 1g and MCS in TNBC (Figure 4E). In MCF-7 cells, the TLN1 gene expression was significantly reduced in AD and MCS cells compared to 1g (Figure 5C). As talin carries the mechanical force in ECM and cell adhesion, the reduced expression of talin would indicate less cell adhesion and cell detachment in the s-μg environment.
The cell surface receptor and cell adhesion molecule integrin-β1 is encoded by the ITGB1 gene. We found a down-regulation of ITGB1 in MDA-MB-231 cells exposed to s-μg (Figure 4A). Integrin-β1 links the actin cytoskeleton with the ECM, and, thus, signaling transduction is possible between the ECM and the cytoplasm. In addition, integrin-β1 modulates gene expression within the cell and is a key player in mechanotransduction (Katsumi et al., 2004; Legate et al., 2009). Integrin-β1, talin, vinculin, fibronectin, and laminin play key roles in focal adhesion processes and can give hint that focal adhesion is less in MCS. These findings are in agreement with the results of a recently published paper (Monti et al., 2021). The adhesion process is very important for the survival strategy of cancer cells to antagonize cell death when the cells are exposed to the RPM. The cytoskeleton, integrins and focal adhesion factors are involved in anti-apoptotic strategies, depending on the cell type (Monti et al., 2021). No signs of apoptosis were found in MCF-7 cells after a 24-h RPM-exposure, which was paralleled by a reduction of vinculin and integrin-β1 in MCS (Monti et al., 2021).
Furthermore, the focal adhesion kinase [FAK/protein tyrosine kinase 2 (PTK2)] is involved in mechanosensing and transduction, proliferation, and migration; it links the ECM and the cytoskeleton (McLean et al., 2005). FAK1/PTK2 gene expression was not changed in all MDA-MB-231 groups (Figure 4B). The protein paxillin is encoded by the PXN gene. Paxillin is expressed at FA and its function is to adhere human cells to the ECM. Other FA proteins can bind to paxillin, like FAK1 or vinculin. In MCF-7 cells, the PXN mRNA in AD and MCS and the corresponding proteins were reduced as compared to 1g (Figure 5B). In MDA-MB-231 cells, PXN gene expression was unchanged (Figure 4C). These findings fit to the results obtained for integrin-β1, which were similarly regulated. This underlines their close interaction when BCCs were exposed to s-μg, leading to cell detachment and the formation of 3D spheroids, where the expression level of FA components can regulate migration and 3D aggregation.
Interaction Network of Selected Genes Evaluated by STRING Analysis
The STRING analysis revealed several interactions between the selected factors. FN1, VCL, TLN1, and ITGB1 are indicated as dominant target genes, as many arrows point to their icons (Figure 7A). Fibronectin was found to be the central target gene. There exist interactions between vinculin and fibronectin. The FN1 gene expression was unaltered in MDA-MB-231 cells (Figure 3A). The FN1 gene expression was also not changed in MCF-7 BCC as published earlier (Kopp et al., 2016). The VCL mRNA was down-regulated in RPM-exposed MDA-MB-231 MCS cells (Figure 4I). In MCF-7 cells, the VCL mRNA was significantly reduced in AD cells. Differences between BC cell types may be due to the different characteristic features of the two breast BC types.
Fibronectin is elevated in podocytes by mechanical stress (Kliewe et al., 2019). Fibronectin KO podocytes showed significantly down-regulated FA molecules (talin, vinculin, and paxillin) as well as a reduction in cell spreading, indicating an important role of fibronectin in adhesion (Kliewe et al., 2019). Fibronectin plays a key role in the adaptation of podocytes to mechanical stress. This finding supports the hypothesis that the interaction of FN1 with vinculin is an adaptive mechanism to protect μg-exposed BCC from mechanical stress. FN1 also interacts with talin and paxillin. Talin links intracellular networks with the ECM via its connection with the actin cytoskeleton and membrane integrins (Haining et al., 2016). Talin exposes new recognition sites when undergoing force-induced mechanical unfolding, and it can bind to and recruit cytoskeletal proteins that are involved in mechanotransduction and MCS formation (Haining et al., 2016). Moreover, paxillin and fibronectin are involved in embryonic developmental events, possibly due to paxillin-mediated modulation of fibronectin-regulated focal adhesion dynamics and organization of the membrane cytoskeletal structures. Thus, paxillin regulates cell migration and spreading of BCC (Hagel et al., 2002). In addition, VCL interacts with CDH1. The cell adhesion glycoprotein E-cadherin (CDH1) is commonly inactivated and reduced in progressive breast tumors. In MCS of MCF-7 cells, it was down-regulated after 14 days of RPM exposure (Sahana et al., 2018). Vinculin is involved in the establishment or regulation of the cadherin-based cell adhesion complex. This process is mediated by a direct interaction with β-catenin (Hazan et al., 1997). The loss of cell–cell adhesion is involved in cancer invasion and metastasis. It has been shown that vinculin impacts metastasis and prognosis in several tumors (Li et al., 2014; Zhang et al., 2019).
As demonstrated in Figure 4I, the VCL gene expression levels were reduced in MCS of MDA-MB-231. In addition, VCL mRNA was significantly down-regulated in MCF-7 AD cells. These findings were also confirmed in highly metastatic CRC cell lines and metastatic tissues (Li et al., 2014).
The ITGB1 gene expression was significantly reduced in MCS of MCF-7 cells (Kopp et al., 2016). This was also measured in MDA-MB-231 cells, where the ITGB1 mRNA was clearly down-regulated in AD and MCS cells (Figure 4A). Integrin-β1 is a membrane protein and is linked to the ECM with the cytoskeleton; it is also capable of transmitting signals (Yeh et al., 2012).
The STRING analysis revealed an interaction between fibronectin and integrin-β1, both playing a central role in 3D formation (Buken et al., 2019). When integrin-β1 is activated and forms a heterodimer with an appropriate integrin-α, integrin-β1 can bind to fibronectin, laminin, and other ECM components via the extracellular domain (Lessan et al., 1999). In addition, integrin-β1 can be activated by binding talin and kindlin via its cytoplasmic domain (Meves et al., 2013). This binding signal will be transferred to the cytoskeleton and the FA complex via vinculin and actin (Gingras et al., 2010).
Vinculin and β-Catenin Signaling in RPM-Exposed MCF-7 Cells
It is known that vinculin regulates the cell surface E-cadherin expression by regulating β-catenin (Peng et al., 2010). In addition, the loss of vinculin and membrane-bound β-catenin promotes metastasis in colorectal cancer (Li et al., 2014) and may also play a role in progression of other cancer types. The loss of cell adhesion is important for spheroid formation and also for metastasis. Vinculin is a key adhesion-related protein and is involved in metastasis in various tumors (Li et al., 2014).
As shown in Figure 6, RPM-exposure of MCF-7 cells resulted in a down-regulation of CDH1. Therefore, we focused in detail on vinculin and β-catenin signaling in MCF-7 cells. Unfortunately, MDA-MB-231 cells do not express E-cadherin (Eslami Amirabadi et al., 2019), and, therefore, we used the MCF-7 cell line for this study.
Live cell imaging of vinculin/β-catenin transfected MCF-7 cells exposed to the RPM revealed that vinculin was distributed throughout the cytoplasm. The protein was detectable at adhesive membrane areas. It is known to mediate the interactions of integrins and the actin cytoskeleton (Bays and DeMali, 2017). Vinculin regulates the adhesion process by interacting with actin. The connection of vinculin to F-actin is important in regard to its role in cell–matrix adhesion. Changes in the vinculin–F-actin interaction can impact morphology, stiffness, adhesion and migration of the cells (Ezzell et al., 1997).
β-catenin (mCherry) was located in the nucleus and cytoplasm in all adherent cells (Figure 8; 1g, RPM-AD). In MCS, β-catenin was detectable in the membranes. β-catenin is a known cadherin-associated protein and an important cell adhesion regulator. Furthermore, β-catenin is a transcriptional co-activator in the nucleus and involved the canonical Wnt signal transduction pathways. Therefore, we have focused on Wnt/β-catenin signaling and on the gene expression of several target genes such as BCL9, MYC, JUN and CCND1 (cyclin D1). Both, CTNNA1 and CTNNB1, mRNAs were not significantly altered by gravitational unloading using the RPM. A similar result for the CTNNB1 gene expression was obtained when low-differentiated follicular thyroid cancer cells (FTC-133 cell line) were exposed for 4 h or 3 days to the RPM (Melnik et al., 2020). Additional studies using cells providing stable expression of β-catenin may further support the results obtained in transiently transfected MCF-7 cells.
It was reported that mechanical unloading of wildtype mice caused a decrease in Wnt/β-catenin signaling activity accompanied by upregulation of Sost, and it is involved in osteoporosis (Lin et al., 2009). Furthermore, the β-catenin location is sensitive to s-μg in osteoblasts (Yin et al., 2018). The nuclear factor of activated T cells (NFAT) is involved in proliferation, angiogenesis, migration in various cancer types (Werneck et al., 2011) and is associated with epithelial–mesenchymal transition (EMT) in breast cancer (Quang et al., 2015). Therefore, we studied the expression of NFATC2 in our experimental setting. We measured that both NFATC2 and CCND1 (cyclin D) were not significantly changed after RPM exposure in MCF-7 cells. An earlier study investigated endothelial cells (EA.hy926 cell line) during a parabolic flight mission (Wehland et al., 2013). The gene expression of CCND1 was reduced when EA.hy926 cells were studied under vibration (Wehland et al., 2013). No change in the gene expression was measurable when the cells were exposed to hypergravity conditions (Wehland et al., 2013). After one parabola (P) and 31P (real microgravity), the CCND1 gene was up-regulated (Wehland et al., 2013).
Interestingly, we measured a significant increase in the BCL9 (B-cell lymphoma 9) and JUN/c-JUN (Jun proto-oncogene, AP-1 transcription factor subunit) gene expression of the MCS compared to the 1g and the RPM-AD group. The oncogene BCL9 functions as a transcriptional co-activator of the Wnt/β-catenin signaling pathway in cancer. BCL9 plays a key role in tumor progression and remodeling of the tumor microenvironment (Jiang et al., 2020). Its upregulation in RPM-exposed samples hints to its involvement in spheroid formation. Little is known about JUN in microgravity-exposed cells. A recent study (Shi et al., 2021) investigated the effects of μg on macrophage differentiation from hematopoietic progenitor cells (HPCs) and demonstrated that a 12-day RCCS exposure of these cells reduced the amount of p-JUN in the RCCS-exposed group.
Furthermore, we investigated the MYC (c-myc) gene expression. MYC was significantly elevated in RPM-AD cells as compared to the 1g and MCS group. This is an interesting finding which was also observed in vivo in mice exposed to a 30-day spaceflight (Radugina et al., 2018). Radugina et al. demonstrated the nuclear immunolocalization of c-Jun and c-Myc proteins, indicating their sensitivity to μg. Future research is necessary to gain more information about the impact of μg on cancer growth and the involvement of the Wnt/β-catenin signaling pathway.
In summary, a 24 h RPM exposure of triple-negative MDA-MB-231 and MCF-7 BCC induced changes in growth, the cytoskeleton, the extracellular matrix, and focal adhesions. As demonstrated in Figure 1, multicellular tumor spheroids of both cell lines showed a fast migration behavior and clear invasion potential when co-cultured with EA.hy926 endothelial cells. After a 24 h RPM-cultivation, multiple round compact MCS and in parallel adherent cells were visible in the cell culture supernatant of both cell types. The 3D aggregates will be used in future studies to examine the effects of anticancer drugs. MCS closely mimic the form and micromilieu of a metastasis. In this study, MDA-MB-231 BC MCS cells grown under conditions of s-μg exhibited a differential regulation of the expression of ECM, and FA genes, such as ITGB1, VCL and TLN1. RPM exposure of MCF-7 BC MCS cells resulted in significant changes in the gene expression of PXN, TLN1, and CDH1, and the WNT-β-catenin target genes BCL9, JUN and MYC. The focal adhesion complex as well as cell adhesion molecules and the cytoskeleton are important gravisensors in μg.
Taken together, the overall gene expression pattern is only slightly modified by RPM-exposure as previously evidenced (Po et al., 2019). Facing an essential maintenance of an overall coherence in gene expression pattern, the only few genes that are modified by microgravity are those involved in growth, the cytoskeleton and mechanotransduction.
In conclusion, BCC (MCF-7 and MDA-MB-231 cells) were exposed to s-μg conditions generated by an RPM. During cell culture on the RPM, the cells transformed from a 2D monolayer into tissue specific multicellular in vitro 3D spheroids. The RPM-exposed cells revealed alterations in cytoskeletal proteins, as well as changes in ECM components and FA factors. A 24-h RPM-exposure induced a significant down-regulation of ITGB and LAMA3 mRNAs in adherent cells and in MCS of the MDA-MB-231 cell line. Moreover, the VCL mRNA was significantly reduced in MDA-MB-231 MCS cells. In contrast, the gene expression of FAK1, PXN, TLN1, VCL, and CDH1 was significantly down-regulated in adherent MCF-7 cells cultured for 24 h on the RPM. In addition, PXN, TLN1 and CDH1 were down-regulated in MCS, whereas VCL and LAMA3 mRNAs were not changed in MCF-7 cells.
Overall, this study provides novel knowledge about the complexity of adherence, migration and invasion behavior of human BCC spheroids engineered with s-μg technology. Further investigations targeting FA proteins and genes in BCC cultured on an RPM will hopefully, indicate novel targets that are interesting future candidates for cancer therapy.
Data Availability Statement
The datasets analyzed for this study can be provided by contacting the corresponding author.
Author Contributions
DG and TC: conceptualization. SKo and JS: methodology. SKo: software. DG, TC, MW, SKo, and MK: validation. DM, SKa, SKo, JS, and MK: formal analysis. JS: investigation. BR, MI, and DG: resources. DG, JS, and TC: writing—original draft preparation. DG, TC, MK, and MW: writing—review and editing. MK and SKa: visualization. DG: supervision and project administration. DG and MI: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer RH declared a past co-authorship with one of the authors, DG.
Funding. This research was funded by Deutsches Zentrum für Luft- und Raumfahrt (DLR), BMWi project 50WB1924.
References
- Aleshcheva G., Bauer J., Hemmersbach R., Slumstrup L., Wehland M., Infanger M., et al. (2016). Scaffold-free tissue formation under real and simulated microgravity conditions. Basic Clin. Pharmacol. Toxicol. 119 (Suppl. 3), 26–33. 10.1111/bcpt.12561 [DOI] [PubMed] [Google Scholar]
- Aleshcheva G., Sahana J., Ma X., Hauslage J., Hemmersbach R., Egli M., et al. (2013). Changes in morphology, gene expression and protein content in chondrocytes cultured on a random positioning machine. PLoS One 8:e79057. 10.1371/journal.pone.0079057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleshcheva G., Wehland M., Sahana J., Bauer J., Corydon T. J., Hemmersbach R., et al. (2015). Moderate alterations of the cytoskeleton in human chondrocytes after short-term microgravity produced by parabolic flight maneuvers could be prevented by up-regulation of BMP-2 and SOX-9. FASEB J. 29 2303–2314. 10.1096/fj.14-268151 [DOI] [PubMed] [Google Scholar]
- Askou A. L., Aagaard L., Kostic C., Arsenijevic Y., Hollensen A. K., Bek T., et al. (2015). Multigenic lentiviral vectors for combined and tissue-specific expression of miRNA- and protein-based antiangiogenic factors. Mol. Ther. Methods Clin. Dev. 2:14064. 10.1038/mtm.2014.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays J. L., DeMali K. A. (2017). Vinculin in cell-cell and cell-matrix adhesions. Cell Mol. Life Sci. 74 2999–3009. 10.1007/s00018-017-2511-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J. L., Souza G. R. (2013). Using space-based investigations to inform cancer research on Earth. Nat. Rev. Cancer 13 315–327. 10.1038/nrc3507 [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit J. B., Cogoli A., Li C. F., Schopper T., Pippia P., Galleri G., et al. (2005). Key gravity-sensitive signaling pathways drive T cell activation. FASEB J. 19 2020–2022. 10.1096/fj.05-3778fje [DOI] [PubMed] [Google Scholar]
- Borst A. G., van Loon J. J. W. A. (2008). Technology and developments for the random positioning machine, RPM. Micrograv. Sci. Technol. 21:287. 10.1007/s12217-008-9043-2 [DOI] [Google Scholar]
- Buken C., Sahana J., Corydon T. J., Melnik D., Bauer J., Wehland M., et al. (2019). Morphological and molecular changes in juvenile normal human fibroblasts exposed to simulated microgravity. Sci. Rep. 9:11882. 10.1038/s41598-019-48378-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buravkova L., Romanov Y., Rykova M., Grigorieva O., Merzlikina N. (2005). Cell-to-cell interactions in changed gravity: ground-based and flight experiments. Acta Astronaut. 57 67–74. 10.1016/j.actaastro.2005.03.012 [DOI] [PubMed] [Google Scholar]
- Chang T. T., Hughes-Fulford M. (2009). Monolayer and spheroid culture of human liver hepatocellular carcinoma cell line cells demonstrate distinct global gene expression patterns and functional phenotypes. Tissue Eng. Part A 15 559–567. 10.1089/ten.tea.2007.0434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. T., Walther I., Li C. F., Boonyaratanakornkit J., Galleri G., Meloni M. A., et al. (2012). The Rel/NF-kappaB pathway and transcription of immediate early genes in T cell activation are inhibited by microgravity. J. Leukoc. Biol. 92 1133–1145. 10.1189/jlb.0312157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez K. J., Garimella S. V., Lipkowitz S. (2010). Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis. 32 35–48. 10.3233/BD-2010-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corydon T. J., Kopp S., Wehland M., Braun M., Schutte A., Mayer T., et al. (2016a). Alterations of the cytoskeleton in human cells in space proved by life-cell imaging. Sci. Rep. 6:20043. 10.1038/srep20043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corydon T. J., Mann V., Slumstrup L., Kopp S., Sahana J., Askou A. L., et al. (2016b). Reduced expression of cytoskeletal and extracellular matrix genes in human adult retinal pigment epithelium cells exposed to simulated microgravity. Cell Physiol. Biochem. 40 1–17. 10.1159/000452520 [DOI] [PubMed] [Google Scholar]
- Deng B., Liu R., Tian X., Han Z., Chen J. (2019). Simulated microgravity inhibits the viability and migration of glioma via FAK/RhoA/Rock and FAK/Nek2 signaling. Vitro Cell Dev. Biol. Anim. 55 260–271. 10.1007/s11626-019-00334-7 [DOI] [PubMed] [Google Scholar]
- Dittrich A., Grimm D., Sahana J., Bauer J., Kruger M., Infanger M., et al. (2018). Key Proteins involved in spheroid formation and angiogenesis in endothelial cells after long-term exposure to simulated microgravity. Cell Physiol. Biochem. 45 429–445. 10.1159/000486920 [DOI] [PubMed] [Google Scholar]
- Eslami Amirabadi H., Tuerlings M., Hollestelle A., SahebAli S., Luttge R., van Donkelaar C. C., et al. (2019). Characterizing the invasion of different breast cancer cell lines with distinct E-cadherin status in 3D using a microfluidic system. Biomed. Microdev. 21:101. 10.1007/s10544-019-0450-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzell R. M., Goldmann W. H., Wang N., Parashurama N., Ingber D. E. (1997). Vinculin promotes cell spreading by mechanically coupling integrins to the cytoskeleton. Exp. Cell Res. 231 14–26. 10.1006/excr.1996.3451 [DOI] [PubMed] [Google Scholar]
- Geiger B., Spatz J. P., Bershadsky A. D. (2009). Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10 21–33. 10.1038/nrm2593 [DOI] [PubMed] [Google Scholar]
- Gingras A. R., Bate N., Goult B. T., Patel B., Kopp P. M., Emsley J., et al. (2010). Central region of talin has a unique fold that binds vinculin and actin. J. Biol. Chem. 285 29577–29587. 10.1074/jbc.M109.095455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D., Bauer J., Wise P., Kruger M., Simonsen U., Wehland M., et al. (2020). The role of SOX family members in solid tumours and metastasis. Semin. Cancer Biol. 67(Pt 1), 122–153. 10.1016/j.semcancer.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Gumbiner B. M. (1996). Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84 345–357. 10.1016/s0092-8674(00)81279-9 [DOI] [PubMed] [Google Scholar]
- Hader D. P., Braun M., Grimm D., Hemmersbach R. (2017). Gravireceptors in eukaryotes-a comparison of case studies on the cellular level. NPJ Micrograv. 3:13. 10.1038/s41526-017-0018-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagel M., George E. L., Kim A., Tamimi R., Opitz S. L., Turner C. E., et al. (2002). The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol. Cell Biol. 22 901–915. 10.1128/mcb.22.3.901-915.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haining A. W., Lieberthal T. J., Del Rio Hernandez A. (2016). Talin: a mechanosensitive molecule in health and disease. FASEB J. 30 2073–2085. 10.1096/fj.201500080R [DOI] [PubMed] [Google Scholar]
- Hazan R. B., Kang L., Roe S., Borgen P. I., Rimm D. L. (1997). Vinculin is associated with the E-cadherin adhesion complex. J. Biol. Chem. 272 32448–32453. 10.1074/jbc.272.51.32448 [DOI] [PubMed] [Google Scholar]
- Hughes-Fulford M. (2003). Function of the cytoskeleton in gravisensing during spaceflight. Adv. Space Res. 32 1585–1593. 10.1016/S0273-1177(03)90399-1 [DOI] [PubMed] [Google Scholar]
- Jiang M., Kang Y., Sewastianik T., Wang J., Tanton H., Alder K., et al. (2020). BCL9 provides multi-cellular communication properties in colorectal cancer by interacting with paraspeckle proteins. Nat. Commun. 11:19. 10.1038/s41467-019-13842-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumi A., Orr A. W., Tzima E., Schwartz M. A. (2004). Integrins in mechanotransduction. J. Biol. Chem. 279 12001–12004. 10.1074/jbc.R300038200 [DOI] [PubMed] [Google Scholar]
- Kliewe F., Kaling S., Lotzsch H., Artelt N., Schindler M., Rogge H., et al. (2019). Fibronectin is up-regulated in podocytes by mechanical stress. FASEB J. 33 14450–14460. 10.1096/fj.201900978RR [DOI] [PubMed] [Google Scholar]
- Kopp S., Sahana J., Islam T., Petersen A. G., Bauer J., Corydon T. J., et al. (2018). The role of NFkappaB in spheroid formation of human breast cancer cells cultured on the random positioning machine. Sci. Rep. 8:921. 10.1038/s41598-017-18556-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp S., Slumstrup L., Corydon T. J., Sahana J., Aleshcheva G., Islam T., et al. (2016). Identifications of novel mechanisms in breast cancer cells involving duct-like multicellular spheroid formation after exposure to the Random Positioning Machine. Sci. Rep. 6:26887. 10.1038/srep26887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp S., Warnke E., Wehland M., Aleshcheva G., Magnusson N. E., Hemmersbach R., et al. (2015). Mechanisms of three-dimensional growth of thyroid cells during long-term simulated microgravity. Sci. Rep. 5:16691. 10.1038/srep16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen T. B., Knutsson M. L., Wehland M., Laursen B. E., Grimm D., Warnke E., et al. (2014). Anti-vascular endothelial growth factor therapy in breast cancer. Int. J. Mol. Sci. 15 23024–23041. 10.3390/ijms151223024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger M., Melnik D., Kopp S., Buken C., Sahana J., Bauer J., et al. (2019). Fighting thyroid cancer with microgravity research. Int. J. Mol. Sci. 20:2553. 10.3390/ijms20102553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K. R., Wickstrom S. A., Fassler R. (2009). Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 23 397–418. 10.1101/gad.1758709 [DOI] [PubMed] [Google Scholar]
- Lessan K., Aguiar D. J., Oegema T., Siebenson L., Skubitz A. P. (1999). CD44 and beta1 integrin mediate ovarian carcinoma cell adhesion to peritoneal mesothelial cells. Am. J. Pathol. 154 1525–1537. 10.1016/s0002-9440(10)65406-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. L., Reynolds J. L., Cubano L. A., Hatton J. P., Lawless B. D., Piepmeier E. H. (1998). Spaceflight alters microtubules and increases apoptosis in human lymphocytes (Jurkat). FASEB J. 12 1007–1018. 10.1096/fasebj.12.11.1007 [DOI] [PubMed] [Google Scholar]
- Li J., Zhang S., Chen J., Du T., Wang Y., Wang Z. (2009). Modeled microgravity causes changes in the cytoskeleton and focal adhesions, and decreases in migration in malignant human MCF-7 cells. Protoplasma 238 23–33. 10.1007/s00709-009-0068-1 [DOI] [PubMed] [Google Scholar]
- Li T., Guo H., Song Y., Zhao X., Shi Y., Lu Y., et al. (2014). Loss of vinculin and membrane-bound beta-catenin promotes metastasis and predicts poor prognosis in colorectal cancer. Mol. Cancer 13:263. 10.1186/1476-4598-13-263 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lin C., Jiang X., Dai Z., Guo X., Weng T., Wang J., et al. (2009). Sclerostin mediates bone response to mechanical unloading through antagonizing Wnt/beta-catenin signaling. J. Bone Miner. Res. 24 1651–1661. 10.1359/jbmr.090411 [DOI] [PubMed] [Google Scholar]
- Ma X., Wehland M., Schulz H., Saar K., Hubner N., Infanger M., et al. (2013). Genomic approach to identify factors that drive the formation of three-dimensional structures by EA.hy926 endothelial cells. PLoS One 8:e64402. 10.1371/journal.pone.0064402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann V., Grimm D., Corydon T. J., Kruger M., Wehland M., Riwaldt S., et al. (2019). Changes in human foetal osteoblasts exposed to the random positioning machine and bone construct tissue engineering. Int. J. Mol. Sci. 20:1357. 10.3390/ijms20061357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiello M. G., Cucina A., Proietti S., Palombo A., Coluccia P., D’Anselmi F., et al. (2014). Phenotypic switch induced by simulated microgravity on MDA-MB-231 breast cancer cells. Biomed. Res. Int. 2014:652434. 10.1155/2014/652434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean G. W., Carragher N. O., Avizienyte E., Evans J., Brunton V. G., Frame M. C. (2005). The role of focal-adhesion kinase in cancer - a new therapeutic opportunity. Nat. Rev. Cancer 5 505–515. 10.1038/nrc1647 [DOI] [PubMed] [Google Scholar]
- Melnik D., Sahana J., Corydon T. J., Kopp S., Nassef M. Z., Wehland M., et al. (2020). Dexamethasone inhibits spheroid formation of thyroid cancer cells exposed to simulated microgravity. Cells 9:367. 10.3390/cells9020367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meves A., Stremmel C., Bottcher R. T., Fassler R. (2013). beta1 integrins with individually disrupted cytoplasmic NPxY motifs are embryonic lethal but partially active in the epidermis. J. Invest. Dermatol. 133 2722–2731. 10.1038/jid.2013.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti N., Masiello M. G., Proietti S., Catizone A., Ricci G., Harrath A. H., et al. (2021). Survival pathways are differently affected by microgravity in normal and cancerous breast cells. Int. J. Mol. Sci. 22:862. 10.3390/ijms22020862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassef M. Z., Kopp S., Melnik D., Corydon T. J., Sahana J., Kruger M., et al. (2019a). Short-Term microgravity influences cell adhesion in human breast cancer cells. Int. J. Mol. Sci. 20:5730. 10.3390/ijms20225730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassef M. Z., Kopp S., Wehland M., Melnik D., Sahana J., Kruger M., et al. (2019b). Real microgravity influences the cytoskeleton and focal adhesions in human breast cancer cells. Int. J. Mol. Sci. 20:3156. 10.3390/ijms20133156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyegaard M., Rendtorff N. D., Nielsen M. S., Corydon T. J., Demontis D., Starnawska A., et al. (2015). A novel locus harbouring a functional CD164 nonsense mutation identified in a large danish family with nonsyndromic hearing impairment. PLoS Genet. 11:e1005386. 10.1371/journal.pgen.1005386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen K., Tauber S., Dumrese C., Bradacs G., Simmet D. M., Golz N., et al. (2015). Regulation of ICAM-1 in cells of the monocyte/macrophage system in microgravity. Biomed. Res. Int. 2015:538786. 10.1155/2015/538786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Cuff L. E., Lawton C. D., DeMali K. A. (2010). Vinculin regulates cell-surface E-cadherin expression by binding to beta-catenin. J. Cell Sci. 123(Pt 4), 567–577. 10.1242/jcs.056432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch J., Kussian R., Sickmann A., Bauer J., Weber G., Nissum M., et al. (2010). Application of free-flow IEF to identify protein candidates changing under microgravity conditions. Proteomics 10 904–913. 10.1002/pmic.200900226 [DOI] [PubMed] [Google Scholar]
- Pihlmann M., Askou A. L., Aagaard L., Bruun G. H., Svalgaard J. D., Holm-Nielsen M. H., et al. (2012). Adeno-associated virus-delivered polycistronic microRNA-clusters for knockdown of vascular endothelial growth factor in vivo. J. Gene Med. 14 328–338. 10.1002/jgm.2623 [DOI] [PubMed] [Google Scholar]
- Po A., Giuliani A., Masiello M. G., Cucina A., Catizone A., Ricci G., et al. (2019). Phenotypic transitions enacted by simulated microgravity do not alter coherence in gene transcription profile. NPJ Micrograv. 5:27. 10.1038/s41526-019-0088-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang C. T., Leboucher S., Passaro D., Fuhrmann L., Nourieh M., Vincent-Salomon A., et al. (2015). The calcineurin/NFAT pathway is activated in diagnostic breast cancer cases and is essential to survival and metastasis of mammary cancer cells. Cell Death Dis. 6:e1658. 10.1038/cddis.2015.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radugina E. A., Almeida E. A. C., Blaber E., Poplinskaya V. A., Markitantova Y. V., Grigoryan E. N. (2018). Exposure to microgravity for 30 days onboard Bion M1 caused muscle atrophy and impaired regeneration in murine femoral Quadriceps. Life Sci. Space Res. 16 18–25. 10.1016/j.lssr.2017.08.005 [DOI] [PubMed] [Google Scholar]
- Riwaldt S., Bauer J., Pietsch J., Braun M., Segerer J., Schwarzwalder A., et al. (2015a). The importance of Caveolin-1 as key-regulator of three-dimensional growth in thyroid cancer cells cultured under real and simulated microgravity conditions. Int. J. Mol. Sci. 16 28296–28310. 10.3390/ijms161226108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riwaldt S., Pietsch J., Sickmann A., Bauer J., Braun M., Segerer J., et al. (2015b). Identification of proteins involved in inhibition of spheroid formation under microgravity. Proteomics 15 2945–2952. 10.1002/pmic.201500067 [DOI] [PubMed] [Google Scholar]
- Romswinkel A., Infanger M., Dietz C., Strube F., Kraus A. (2019). The Role of C-X-C chemokine receptor Type 4 (CXCR4) in cell adherence and spheroid formation of human Ewing’s sarcoma cells under simulated microgravity. Int. J. Mol. Sci. 20:6073. 10.3390/ijms20236073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahana J., Nassef M. Z., Wehland M., Kopp S., Kruger M., Corydon T. J., et al. (2018). Decreased E-Cadherin in MCF7 human breast cancer cells forming multicellular spheroids exposed to simulated microgravity. Proteomics 18:e1800015. 10.1002/pmic.201800015 [DOI] [PubMed] [Google Scholar]
- Sharma N., Hollensen A. K., Bak R. O., Staunstrup N. H., Schroder L. D., Mikkelsen J. G. (2012). The impact of cHS4 insulators on DNA transposon vector mobilization and silencing in retinal pigment epithelium cells. PLoS One 7:e48421. 10.1371/journal.pone.0048421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Tian H., Wang P., Li L., Zhang Z., Zhang J., et al. (2021). Spaceflight and simulated microgravity suppresses macrophage development via altered RAS/ERK/NFκB and metabolic pathways. Cell. Mol. Immunol. 18 1489–1502. 10.1038/s41423-019-0346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snel B., Lehmann G., Bork P., Huynen M. A. (2000). STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28 3442–3444. 10.1093/nar/28.18.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule H. D., Vazguez J., Long A., Albert S., Brennan M. (1973). A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl. Cancer Inst. 51 1409–1416. 10.1093/jnci/51.5.1409 [DOI] [PubMed] [Google Scholar]
- Spanjaard E., de Rooij J. (2013). Mechanotransduction: vinculin provides stability when tension rises. Curr. Biol. 23 R159–R161. 10.1016/j.cub.2013.01.020 [DOI] [PubMed] [Google Scholar]
- Staunstrup N. H., Sharma N., Bak R. O., Svensson L., Petersen T. K., Aarenstrup L., et al. (2011). A Sleeping Beauty DNA transposon-based genetic sensor for functional screening of vitamin D3 analogues. BMC Biotechnol. 11:33. 10.1186/1472-6750-11-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 209–249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Svejgaard B., Wehland M., Ma X., Kopp S., Sahana J., Warnke E., et al. (2015). Common effects on cancer cells exerted by a random positioning machine and a 2D clinostat. PLoS One 10:e0135157. 10.1371/journal.pone.0135157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Xu A., Zhao T., Zhao Q., Zhang J., Fan C., et al. (2018). Simulated microgravity inhibits cell focal adhesions leading to reduced melanoma cell proliferation and metastasis via FAK/RhoA-regulated mTORC1 and AMPK pathways. Sci. Rep. 8:3769. 10.1038/s41598-018-20459-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber S., Lauber B. A., Paulsen K., Layer L. E., Lehmann M., Hauschild S., et al. (2017). Cytoskeletal stability and metabolic alterations in primary human macrophages in long-term microgravity. PLoS One 12:e0175599. 10.1371/journal.pone.0175599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich C., Pietsch J., Grosse J., Wehland M., Schulz H., Saar K., et al. (2011). Differential gene regulation under altered gravity conditions in follicular thyroid cancer cells: relationship between the extracellular matrix and the cytoskeleton. Cell Physiol. Biochem. 28 185–198. 10.1159/000331730 [DOI] [PubMed] [Google Scholar]
- Uva B. M., Masini M. A., Sturla M., Prato P., Passalacqua M., Giuliani M., et al. (2002). Clinorotation-induced weightlessness influences the cytoskeleton of glial cells in culture. Brain Res. 934 132–139. 10.1016/s0006-8993(02)02415-0 [DOI] [PubMed] [Google Scholar]
- van’t Veer L. J., Dai H., van de Vijver M. J., He Y. D., Hart A. A., Mao M., et al. (2002). Gene expression profiling predicts clinical outcome of breast cancer. Nature 415 530–536. 10.1038/415530a [DOI] [PubMed] [Google Scholar]
- Vassy J., Portet S., Beil M., Millot G., Fauvel-Lafeve F., Karniguian A., et al. (2001). The effect of weightlessness on cytoskeleton architecture and proliferation of human breast cancer cell line MCF-7. FASEB J. 15 1104–1106. 10.1096/fj.00-0527fje [DOI] [PubMed] [Google Scholar]
- Vorselen D., Roos W. H., MacKintosh F. C., Wuite G. J., van Loon J. J. (2014). The role of the cytoskeleton in sensing changes in gravity by nonspecialized cells. FASEB J. 28 536–547. 10.1096/fj.13-236356 [DOI] [PubMed] [Google Scholar]
- Wehland M., Ma X., Braun M., Hauslage J., Hemmersbach R., Bauer J., et al. (2013). The impact of altered gravity and vibration on endothelial cells during a parabolic flight. Cell Physiol. Biochem. 31 432–451. 10.1159/000343380 [DOI] [PubMed] [Google Scholar]
- Werneck M. B., Vieira-de-Abreu A., Chammas R., Viola J. P. (2011). NFAT1 transcription factor is central in the regulation of tissue microenvironment for tumor metastasis. Cancer Immunol. Immunother. 60 537–546. 10.1007/s00262-010-0964-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest S. L., Richard S., Kopp S., Grimm D., Egli M. (2015). Simulated microgravity: critical review on the use of random positioning machines for mammalian cell culture. Biomed. Res. Int. 2015:971474. 10.1155/2015/971474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y. C., Lin H. H., Tang M. J. (2012). A tale of two collagen receptors, integrin beta1 and discoidin domain receptor 1, in epithelial cell differentiation. Am. J. Physiol. Cell Physiol. 303 C1207–C1217. 10.1152/ajpcell.00253.2012 [DOI] [PubMed] [Google Scholar]
- Yin C., Zhang Y., Hu L., Tian Y., Chen Z., Li D., et al. (2018). Mechanical unloading reduces microtubule actin crosslinking factor 1 expression to inhibit beta-catenin signaling and osteoblast proliferation. J. Cell Physiol. 233 5405–5419. 10.1002/jcp.26374 [DOI] [PubMed] [Google Scholar]
- Zhang M., Liu P., Xu F., He Y., Xie X., Jiang X. (2019). Vinculin promotes gastric cancer proliferation and migration and predicts poor prognosis in patients with gastric cancer. J. Cell Biochem. 120 14107–14115. 10.1002/jcb.28686 [DOI] [PubMed] [Google Scholar]
- Zhao T., Li R., Tan X., Zhang J., Fan C., Zhao Q., et al. (2018). Simulated microgravity reduces focal adhesions and alters cytoskeleton and nuclear positioning leading to enhanced Apoptosis via Suppressing FAK/RhoA-mediated mTORC1/NF-kappaB and ERK1/2 pathways. Int. J. Mol. Sci. 19:1994. 10.3390/ijms19071994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed for this study can be provided by contacting the corresponding author.