Abstract
Cancer is a disease of altered signaling and metabolism, causing uncontrolled division and survival of transformed cells. A host of molecules, factors, and conditions have been designated as underlying causes for the inception and progression of the disease. An enormous amount of data is available, system-wide interaction networks of the genes and proteins are generated over the years and have now reached up to a level of saturation, where we need to shift our focus to the more advanced and comprehensive methods and approaches of data analysis and visualization. Even with the availability of enormous literature on this one of the most pressing pathological conditions, a successful cure of the disease seems to be obscure. New treatment plans, like immunotherapy and precision medicine, are being employed for different studies. Nevertheless, their actual benefits to the patients would be known only after the evaluation of clinical data over the next few years. Therefore, we need to look at few fundamental challenges that should be addressed in more depth before we could devise better, rigorous, and comprehensive treatment plans and may successfully reach a possible cure of the disease. This article aims at bringing attention towards some fundamental gaps in our approach towards the disease that leads to failure in devising successful therapeutics.
Keywords: Bottom-up approach, Evolution of cancer, Genetic diversity, Immunotherapy, Multimodal treatment
Graphical abstract
Introduction
Cancers are one of the most devastating classes of human pathologies, presenting the versatile range of hallmark clinical features and leading to millions of deaths each year around the globe. These groups of maladies constitute more than a hundred genetically diverse conditions sharing several commonalities in the molecular mechanisms and metabolic alterations among themselves.1,2 The direct involvement of the tissue microenvironment and inflammatory changes on the tumor growth survival is well-established.3,4 However, a clear understanding of the underlying causes and factors is still elusive and requires more research. A plethora of genetic mutations has been reported that could end up in the transformation of the normal human cells leading to the genesis of tumor and development of cancer.5,6 In the past, a variety of scientific and technological approaches have been tried in order to understand, define, investigate, and challenge these extensively dreadful forms of diseases. Various approaches applied in oncological research include genetic, molecular, biochemical, biophysical, immunological, genomic, proteomic, systems and computational biology, etc. However, none of these have sufficed the needs to devise successful treatment strategies so far.7 The area of the oncological research is well-documented but evolving in a way that researchers and clinicians find it difficult to get updated and informed of the new information and advancements most of the time. Therefore, the field requires a continuous assessment of the new developments and analyses of the lacunae present within persisting knowledge.
Understanding cancer: do we need to change the approach?
Although a vast volume of data has accumulated over the decades of research, we at the present stage of scientific and technological advancements still lack a sound understanding of these debilitating conditions that leads to millions of deaths every year. In fact, before classifying the cancers as classical disease conditions, we need to understand that these could be considered as a phenomenon of sequential alteration at molecular, cellular, tissue, and organ levels affecting the whole physiology of the organism. We cannot ignore the fact that the inception of the disease is neither caused necessarily by some external agents nor is potentially associated with the dysfunction of any specific organ, as occurs in many other disease classes. In simpler terms, cancer could be caused by multiple kinds of disorders in the highly ordered cellular physiological systems.8 The onset of the disease is still the most intriguing part of the oncology, that we are far from understanding.9 Enormous data has accumulated showing multiple kinds of physical, chemical, biological, genetic, and environmental factors leading to the transformation phenomenon. However, unfortunately, all these reports have just led us to the tip of the iceberg. We are yet to get a proper understanding of the onset of the disease, and until we get the insights of the initiation, we could not find the actual targets for delivering the treatments.
The past century of scientific research has given us a detailed picture of primary, secondary, and tertiary changes associated with these conditions. However, the hunt to get point zero is still on, and more efforts are needed to reach right at the origin of the disease. The reason behind raising the issue of the origin of cancer is that the primary reasons behind not being able to crack the codes of this century-old problem could be our ‘top-down’ approach that works on providing symptomatic relieves from the disease conditions. We have confined our research most of the time in designing and improving the methods and strategies of arresting the growth of transformed tumor cell mass. Unfortunately, we have shifted our objectives primarily towards devising newer preventive measures rather than looking for the origin of the disease. The reason behind this shift could also be due to the continuous failure of scientists in delivering the results in ‘bottom-up’ approaches where more focus should be given to the genesis of cancer, instead of ways to cut down resources for its sustenance or forcibly inducing the cell death pathways. We must also look for the evolutionary perspectives of the origin, sustenance, and development of cancer-like conditions over the eons to understand why and how selection forces have let these processes to settle down with our current set of physiological paradigms. In fact, we need a comprehensive multi-dimensional approach to clearly investigate and understand the disease and thus to deliver more effective and successful treatment strategies.
Can we look at cancer through an evolutionists prism?
In the views of an evolutionist, the development of cancers seems to be driven by the continuous acquisition of numerous somatic mutations to best fit into a continuously changing and challenging microenvironment.10 Spontaneous mutation-driven evolution of tumor cells itself is one significant barrier before us that tends to provide them the resistance against the drugs and treatment plans.11,12 The dialectical interrelations among different physiological pathways and their dynamic interaction networks have further complexicized our molecular understanding of the disease. The uniqueness of each cancer type and diversity among the tumors additionally raise multifold challenges in understanding this terrible and complicated disease and devising treatment strategies to fight against it. Additional adversities are added by their unique and yet to be understood capabilities of evading the host immune responses, thereby limiting the inherent tendency of the body to fight back with these odious masses of tissues. In-depth knowledge of underlying pathways and mechanisms behind the origin, transformation, and development of cancer may provide us therapeutic advantages in switching to a ‘bottom-up’ approach that will hand us an edge in targeting the most underlying molecular pathways. The past century has seen tremendous methodological and technological advancements in the diagnosis, and treatment of various types of cancers.
Where do we stand?
Cancer cells are a kind of parasitic population of our own cells that resides in our body, utilizing the nutrition and supplementation of normal cells. They have learned to hijack the metabolic pathways and exploit the tissue microenvironment for their growth, sustenance, and progression. They have acquired the art of camouflage and thus evade our army of immune cells of various kinds. Their ability to mutate and evolve within the population further complicates the condition, which makes it hard for researchers and scientists to understand how to target these unwelcome populations of our cells, which otherwise may lead to enormous complications in the end. One school of thought finds possible parallelism between developmental processes and the origin of cancer; however, more support on the notion is needed. Thousands of studies are conducted every year with new findings and solutions of the disease advancing our present knowledge about the disease. However, the disease is still growing with an increased pace and turns out to be the leading cause of mortality in many developed countries. Despite acquiring enormous knowledge on the etiology, causes, and effects of cancer inside the body, unfortunately, we could not have successfully devised too many methods to curb the disease condition and provide proper relief to the patients suffering from this one of the most challenging puzzles of the medical sciences.13
In most cases, a treatment plan for the majority of the cancer types remains limited to surgical removal, chemotherapeutic targeting, high-intensity photon-beam radiotherapy, hormonal therapy, and modern-day immunotherapy depending upon their complexity, stage, and localization.14 All patients suffering from one type of cancer, despite being genetically diverse, typically receive a similar treatment plan. This diversity among the cell population of the same tumor mass leads to the attainment of the drug resistance in the transformed cells. Also, the lack of real-time monitoring of the ongoing treatment further worsens the situation. Unfortunately, due to the limitations of resources and diagnostic tools and a lack of personalized treatment strategies, in addition to the highly diverse genetic makeup of tumor populations, many times, treatments do not meet the expectations. Here, a few major characteristic features of the disease, which mostly intrigues our understanding and raise new challenges in therapeutics, are discussed in detail.
Extreme genetic diversity
Intratumor heterogeneity, i.e., extreme genetic diversity among cell populations residing in the same tumor mass, leads to Darwinian principles and natural selection forces to work on it and establish a more robust variety of cancer cells.15 This heterogeneity increases drug resistance and thus poses a great therapeutic challenge before clinicians in developing cancer treatment plans, which therefore led to the evolution of the idea of personalized medicines.16,17 Increased application of pharmacogenetics, i.e., understanding the genetics of cancer by molecular profiling of the disease cells, identification of the mutation associated with the given cancer type in an individual patient is nowadays used for targeting specific genes or proteins driving the growth of cancer.18 The advent of next-generation sequencing, array methods, and other mathematical or computational tools have tremendously driven the progress of this revolutionary approach to target the cancer cells.19, 20, 21 However, the application of personalized medicines is very limited at present due to multiple reasons. The high cost of cancer genomics is one major reason behind not opting for precision medicine at a larger scale.22,23 Possibly, the diversity at the genetic level in these diseases and the inability to afford for personalized treatment strategies is one of the major underlying causes of failures in the treatment of cancer. Therefore, more efforts are required in this direction so that precision medicines can be used in other cases also.
Looking at various genes independently may not be an ideal way to understand the plethora of changes occurring while a cell transforms, and the tumor is formed; therefore, a broad landscape of genome-wide alteration may provide a better understanding of the genetics of cancer.24 The increase in genetic diversity, in recent years, has also been linked with a phenomenon called genome or karyotype chaos, primarily caused due to largescale changes creating new genomic codes for the systemic inheritance, which in turn provide increased chances of adaptation and survival at the cellular and organismal level.25,26 An increasing number of sequencing-based studies have hypothesized for the possibility of a more pivotal influence of the genome topology over the tumor diversity in comparison to the independent gene profiles.27 ‘Chromothripsis’ is an umbrella term used to indicate multiple kinds of chromosomal rearrangements happening under crisis, which may include: chromosomal aneuploidy, chromoplexy, chromoanagenesis, etc.28 Clonal or constitutional aneuploidy is another less understood phenomenon reported in many cancer types, which may also act as a heterogeneous agent affecting the emergence and evolution of cancer.29,30 These terms are not very familiar to oncology research as most of these processes were overlooked for years, while scientists were more inclined towards the genetic theory of cancer, which has largely failed to address many fundamental questions of evolution, development, and diversity of cancer.29
An opaque connection between developmental biology and cancer
Do cancers have any developmental correlation; and if the disease could be transferred from one generation to the other? These are a few more questions, which are not understood in detail. At face values, development entails order, whereas cancer represents an example of extreme disorder. Interestingly, such a distinctive origin and progression of the two processes define their dichotomous correlation. When Prof. Mintz referred cancer as an error of development, possibly she took into account the proliferative capabilities of the cells, which could be compared with that of early-stage stem cell populations during organism development.31 However, the two processes are at considerably far distant ends of the same spectrum of diversification. On the one hand, development is a process that originates from a single zygotic cell that diversifies into multiple cell types via a tightly controlled epigenetic regulation. Contrarily, cancer is a slow and continuous progression towards a highly similar cell population at late malignancy stages from the early benign tissue masses, which are highly diverse.32 Many similar epigenetic signatures could be found among both the developmental stem cells and cancer progenitors, marking the parallelism between both processes.33 There is also an ambiguity upon the idea of inheritance of cancer-causing disease mutations, which may or may not be transferred to progenies. Possibly, some of the mutations are inherited, while many of the cases are acquired because of replication errors during the multiplication of cells.
Many overlapping clinical features among multiple developmental disorders and cancer predisposition could be another possible connecting link between the two processes, putting forward an idea of cancer being a disease of accumulating developmental errors.32,34 Highly precise control over spatial and temporal switching ON/OFF of a well-orchestrated gene network is another binding feature that connects the two distinct biological phenomena.35 Owing to these similarities, many scientists are now exploiting both types of disease models to understand each other.36 In the coming years, a more rigorous shift in approach towards studying cancer as a disease of erroneous development is needed. This change of approach may educate us more about the origin and development of this highly diverse disease.
Hiding away from innate immunity
Another major challenge that scientists have faced over the years is the extreme disguise or hiding of cancer cells from our immune system that are expected to identify and kill the cancer cells.37 The specific mechanisms of how precisely cancer cells evade our immune system and what could be the possible strategies to target these aberrant cells of our own body is still an enigma of modern-day biology.38 However, targeting neoantigens is a possible strategy that is under consideration with huge expectations, as these newly evolved peptides provide immune cells an opportunity to target cells, which otherwise remain in disguise. Some scientists believe that it is a dynamic head to head clash between neoantigen expressing cancer cells and the immune surveillance that poses a quest of survival before cancer cells.39,40 The selection pressure mounted, therefore, leads to mutations in such a way that they start tricking and escaping our own immune cells.41 Additionally, the disease cells, by expressing some specific antigens or modifying the tumor environment, acquire: the tendency or ability to overpower the normal immune responses inside the body.42,43
Cancer immune surveillance has remained a long-standing topic of debate that has both proponents and opponents. It is hard to answer whether our immune system promotes or suppresses, or it directly ignores the mutated or transformed cells.44,45 Interestingly, over the years, it has been suggested that transformed cells themselves sculpt the immune cells via a process called immune-editing in such a way that they help them in skipping their molecular identification and elimination and establishing a Darwinian selection.46 Few reports indicating latent metastasis after decades of surgical removal of the primary tumor is another question that, if answered, may probably help us better understand how the cells conceal themselves from possible immune attacks.47 Currently, we are quickly moving towards modern immunotherapy-based treatment methods, which are providing very positive results indicating a better future of these strategies towards cancer therapeutics.48,49 Chimeric antigen receptor (CAR)-T cells were the first line of treatment based on immune modulation, which successfully initiates a ferocious assault on cancer cells. The clinical trials have shown enthusiastic results in the past, but at this point of time, we cannot predict the future of these treatment plans.50
We should not underestimate the art of camouflage acquired by cancer cells over a long period of evolution. Instead, we must undermine the evolutionary relations between cancer cells and different types of immune cells. We need to find out the possible diversions of the eukaryotic transformed cells when in the history of evolution, they have started deceiving our own immunity. We need to investigate the history of life to find out the answers to the present-day questions in order to find solutions for the future. Most of the physiological hallmarks defined for the cancers have their own evolutionary significance, and their retention by the selection forces might have their own significance and implications in the process of evolution.
A controversy: one target multiple bullets; or multiple targets with a common bullet?
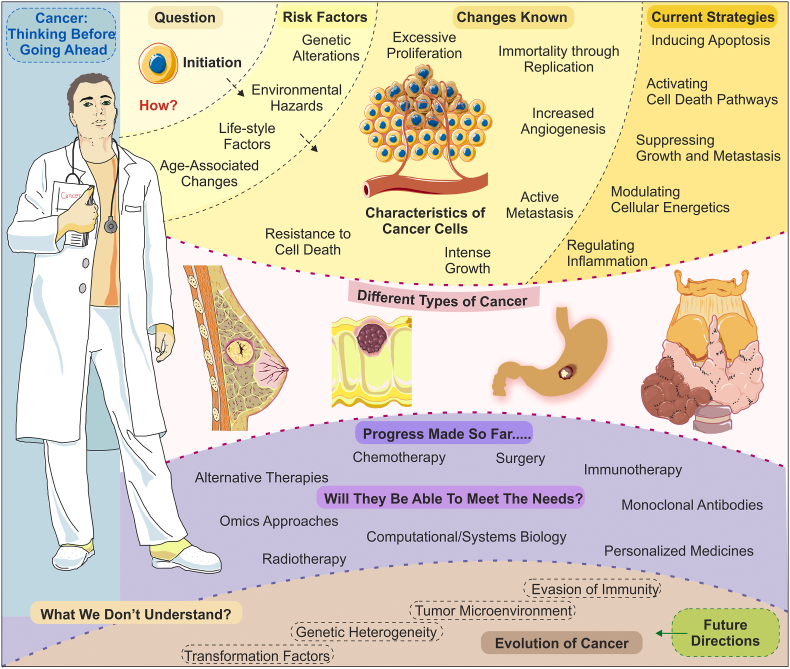
A large fraction of research to date has focused on identifying the multimodal driving factors of the disease. Nevertheless, when the causative factors, genes, and pathways are subjected to the possibilities of druggability, most of these have shown limitations of multiple types. One major problem with most of the molecular targets is the toxicity generated due to their modulations, as most of these are somehow part of one or multiple molecular pathways.51,52 Several articles have been published in recent years covering various aspects of the disease in detail. Thus, here in the present article, the primary focus is to highlight the gaps in postulation and understanding of the disease. Cancer has remained an unsolved puzzle over the years, despite having an extensive knowledge base of genes, proteins, and pathways affected during the pathogenesis. The primary cause of failure for most of the hypotheses could be overlooking many vital topics and defining factors in the disease etiology. What remains crucial is a rethinking of the already available knowledge and revisiting the persisting treatment methodologies and therapeutic approaches (see Fig. 1). It is obvious to say that an overwhelming amount of data is present and is enough to spend days or months to understand the complicated networks and models that can be generated using various available tools out of this vast literature available over online resources.
Figure 1.
An overview of known, unknown, available therapeutics, and future directions of the cancer therapeutics. The top panel shows the fundamental underlying causes of cancer, including major risk factors, involved mechanisms and currently available strategies to devise treatments and therapeutic strategies. The central part shows a few representative types of cancer among many occurring around the globe and leading to a very high number of deaths. The lower subsections show the treatment methodologies adopted around the world to curb the disease progression. In contrast, the few most challenging and less-understood complexities of tumor biology are indicated in the lowermost subsection of the figure. Fig. 1 was prepared using a few templates from Servier Medical Art by Servier (http://smart.servier.com/), which is licensed under a Creative Commons Attribution 3.0 Unported License.
The exploitation of the available knowledge needs multimodal approaches, forming multidisciplinary teams with experts from different fields collaborating with and complementing each other. The use of computational tools and system biology approaches may further benefit the field with faster and more accurate calculations and predictions, saving lots of money and time that is spent in a large number of unsuccessful clinical trials. A large amount of biochemical and genomic data is present; however, the focus should be shifted now towards the transformation of these research into clinical applications. The majority of the data remain limited to the publications only and could never generate any therapeutic benefit. Many drugs proposed through these studies fail in successive phases of clinical trials, either because of their limited effects on curbing the disease or due to high toxicity profiles. In the end, a tiny fraction of drugs proposed every year get into clinics and could be utilized for treatment purposes. The primary reasons behind the failure of many drugs are because of the negligence of specific facts associated with this multifactorial disease.
Regulating the cell cycle progression, inducing apoptosis, managing the tissue microenvironment, modulation of the immune system, and cutting the sources of nutrition and growth, all at a time could be near to impossible for most of the drugs, which are currently in practice or under clinical trials. Even if we consider any drug, with the least of the possibilities to have most of the characteristics mentioned above, the more significant challenge would be its delivery to the specific tissues. Targeted delivery of drugs is another highly frustrating challenge faced by researchers and clinicians. None of the approaches proposed to date for the drug delivery to the affected site has stood with the promises and leads to the toxicity and collateral adversities, as is seen in patients undergoing chemotherapy. Due to this multifaceted nature of cancer development and progression, we ultimately land in an unfortunate situation of firing multiple rounds of bullets (chemotherapy, radiation, surgical removal, and immunotherapy, etc.) all at a time. The lack of a broader understanding of the pathological condition leads to overall physiological and psychological distress to the patients.
Surgical removal of affected body parts or heavy doses of chemotherapeutic drugs and high-energy radiations may cause physical disfigurements, followed by weakening of the immunity and deterioration of the body mass. All these combinedly affects the psychology of the patients with the highest of the will power. Family members of the patients may also get frustrated due to the very long treatment process and follow-ups. Also, most of the newly developed methodologies, like immunotherapy, cancer genomics, and precision medicine, are too costly to be afforded by most of the patients in underdeveloped and developing countries. Most of the cases of these diseases do not present any noticeable symptoms and remain undetected in earlier stages, and when detected, leave patients and doctors with minimal palliative options to choose and start the treatment. Diagnosis of the pathological symptoms at a very late stage is another major challenge that needs to be addressed with high priority. In fact, many things are possibly required for the consideration of the researchers, clinical scientists, and doctors.
Conclusions: the road ahead
Cancer has remained one of the significant health issues for long. Description of the disease could be found in some of the ancient literature of Indian and Chinese medicines. The ancient scholars and clinicians suggested different treatment strategies. It could not be accurately predicted how effective these treatment plans were; although their applications in the modern age medicines cannot be neglected. In the past century, efforts have intensified exponentially in all parts of the world to address the common health problems and diseases associated with such conditions. Many of the endemics have been cured completely. Most of the deadly infectious diseases leading to mass death have been understood and have possible cures around the corner. Several are still under investigation and may get some solutions in the past. In between all these advancements, one major class of human diseases, cancer that is neither acute in origin nor contagious in the spread, remain uncured up to a more considerable extent. Enormous hopes have been ignited from time to time by the development of multiple promising drugs, methods of their targeted delivery, and the evolution of novel treatment strategies, like immunotherapy, in the past.
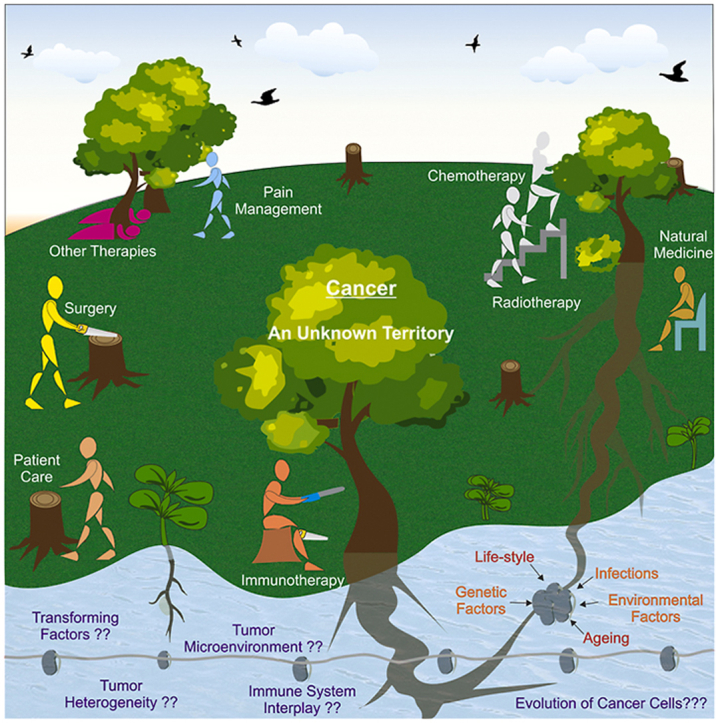
All the strategies developed so far, and the drugs approved until now have their own limitations and toxicities associated with their use. Nevertheless, due to the lack of more sophisticated ways of treatments, the patients suffering from the advanced terminal stages of the disease usually are left with very few options to live with. Several new treatment plans are underway and may need a decade or more to be available for patients of developing countries at an affordable cost. Most of the research conducted around the world is now limited to finding ways of trimming away large branches (symptoms) of a tree (cancer), which may further grow and proliferate at new sites. The knowledge of the point of origin of the tree and ways to cut it from the bottom is still missing and needs extensive work. The field may need a complete turnaround in the approaches of looking at the disease in order to find proper solutions. It is hard at present to predict how much time we still require in reaching a definitive solution. However, yes, there always remains a hope, and we must follow it.
Conflict of Interests
The author declares no competing interests.
Acknowledgements
The author would like to thank the Central University of Rajasthan for providing basic requirements during the preparation of the manuscript. The author also appreciates Servier (http://smart.servier.com/) for providing templates for the preparation of medical illustrations under Creative Commons Attribution 3.0 Unported License.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert A.W., Pattabiraman D.R., Weinberg R.A. Emerging biological principles of metastasis. Cell. 2017;168(4):670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A., Allavena P., Sica A., Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 5.Ujvari B., Klaassen M., Raven N. Genetic diversity, inbreeding and cancer. Proc Biol Sci. 2018;285(1875):20172589. doi: 10.1098/rspb.2017.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mroz E.A., Rocco J.W. The challenges of tumor genetic diversity. Cancer. 2017;123(6):917–927. doi: 10.1002/cncr.30430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishayee A., Block K. A broad-spectrum integrative design for cancer prevention and therapy: the challenge ahead. Semin Canc Biol. 2015;35(Suppl):s1–s4. doi: 10.1016/j.semcancer.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Hanselmann R.G., Welter C. Origin of cancer: an information, energy, and matter disease. Front Cell Dev Biol. 2016;4 doi: 10.3389/fcell.2016.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visvader J.E. Cells of origin in cancer. Nature. 2011;469(7330):314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 10.Lakatos E., Williams M.J., Schenck R.O. Evolutionary dynamics of neoantigens in growing tumours. Nat Genet. 2020;52(10):1057–1066. doi: 10.1038/s41588-020-0687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greaves M. Evolutionary determinants of cancer. Canc Discov. 2015;5(8):806–820. doi: 10.1158/2159-8290.CD-15-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald D.M., Rosenberg S.M. Driving cancer evolution. eLife. 2017;6 doi: 10.7554/eLife.25431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amirouchene-Angelozzi N., Swanton C., Bardelli A.J.C.d. Tumo evolution as a therapeutic target. Canc Discov. 2017;7(8):805–817. doi: 10.1158/2159-8290.CD-17-0343. [DOI] [PubMed] [Google Scholar]
- 14.Palumbo M.O., Kavan P., Miller W.H. Jr. Systemic cancer therapy: achievements and challenges that lie ahead. Front Pharmacol. 2013;4 doi: 10.3389/fphar.2013.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGranahan N., Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 16.Turnbull A.K. Personalized medicine in cancer: where are we today? Future Oncol. 2015;11(20):2795–2798. doi: 10.2217/fon.15.204. [DOI] [PubMed] [Google Scholar]
- 17.Burrell R.A., McGranahan N., Bartek J., Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler H.E., Maitland M.L., Dolan M.E., Cox N.J., Ratain M.J. Cancer pharmacogenomics: strategies and challenges. Nat Rev Genet. 2013;14(1):23–34. doi: 10.1038/nrg3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rai A., Pradhan P., Nagraj J., Lohitesh K., Chowdhury R., Jalan S. Understanding cancer complexome using networks, spectral graph theory and multilayer framework. Sci Rep. 2017;7 doi: 10.1038/srep41676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meldrum C., Doyle M.A., Tothill R.W. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin Biochem Rev. 2011;32(4):177–195. [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng G., Patolsky F., Cui Y., Wang W.U., Lieber C.M. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotechnol. 2005;23(10):1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 22.Hood L., Friend S.H. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011;8(3):184–187. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- 23.Chin L., Andersen J.N., Futreal P.A. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17(3):297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 24.Duesberg P. Chromosomal chaos and cancer. Sci Am. 2007;296(5):52–59. doi: 10.1038/scientificamerican0507-52. [DOI] [PubMed] [Google Scholar]
- 25.Heng H.H. World Scientific; 2015. Debating Cancer: The Paradox in Cancer Research. [Google Scholar]
- 26.Ye C.J., Sharpe Z., Alemara S. Micronuclei and genome chaos: changing the system inheritance. Genes (Basel) 2019;10(5) doi: 10.3390/genes10050366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamal-Hanjani M., Wilson G.A., McGranahan N. Tracking the evolution of non–small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 28.Liu G., Stevens J., Horne S. Genome chaos: survival strategy during crisis. Cell Cycle. 2014;13(4):528–537. doi: 10.4161/cc.27378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duesberg P., Li R., Fabarius A., Hehlmann R. Aneuploidy and cancer: from correlation to causation. Contrib Microbiol. 2006;13:16–44. doi: 10.1159/000092963. [DOI] [PubMed] [Google Scholar]
- 30.Ye C.J., Regan S., Liu G., Alemara S., Heng H.H. Understanding aneuploidy in cancer through the lens of system inheritance, fuzzy inheritance and emergence of new genome systems. Mol Cytogenet. 2018;11 doi: 10.1186/s13039-018-0376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mintz B. Gene expression in neoplasia and differentiation. Harvey Lect. 1978;71:193–246. [PubMed] [Google Scholar]
- 32.Bellacosa A. Developmental disease and cancer: biological and clinical overlaps. Am J Med Genet. 2013;161(11):2788–2796. doi: 10.1002/ajmg.a.36267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widschwendter M., Fiegl H., Egle D. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39(2):157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 34.Cofre J., Saalfeld K., Abdelhay E. Cancer as an embryological phenomenon and its developmental pathways: a hypothesis regarding the contribution of the noncanonical wnt pathway. Sci World J. 2019;2019 doi: 10.1155/2019/4714781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubin H. Rethinking “cancer as a dynamic developmental disorder” a quarter century later. Canc Res. 2009;69(6):2171–2175. doi: 10.1158/0008-5472.CAN-08-4213. [DOI] [PubMed] [Google Scholar]
- 36.Aiello N.M., Stanger B.Z. Echoes of the embryo: using the developmental biology toolkit to study cancer. Disease models & mechanisms. 2016;9(2):105–114. doi: 10.1242/dmm.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vinay D.S., Ryan E.P., Pawelec G. Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Canc Biol. 2015;35:S185–S198. doi: 10.1016/j.semcancer.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Spranger S., Gajewski T.F. Mechanisms of tumor cell–intrinsic immune evasion. Annu Rev Cell Biol. 2018;2(1):213–228. [Google Scholar]
- 39.Lu Y.-C., Robbins P.F. Cancer immunotherapy targeting neoantigens. Semin Immunol. 2016;28(1):22–27. doi: 10.1016/j.smim.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang R.-F., Wang H.Y. Immune targets and neoantigens for cancer immunotherapy and precision medicine. Cell Res. 2017;27(1):11–37. doi: 10.1038/cr.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Efremova M., Finotello F., Rieder D., Trajanoski Z. Neoantigens generated by individual mutations and their role in cancer immunity and immunotherapy. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.01679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Q., Liu G., Liu S. Remodeling the tumor microenvironment with emerging nanotherapeutics. Trends Pharmacol Sci. 2018;39(1):59–74. doi: 10.1016/j.tips.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Jäger D., Jäger E., Knuth A. Immune responses to tumour antigens: implications for antigen specific immunotherapy of cancer. J Clin Pathol. 2001;54(9):669–674. doi: 10.1136/jcp.54.9.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn O.J. Immuno-oncology: understanding the function and dysfunction of the immune system in cancer. Ann Oncol. 2012;23(Suppl 8):viii6–viii9. doi: 10.1093/annonc/mds256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janssen L.M., Ramsay E.E., Logsdon C.D., Overwijk W.W. The immune system in cancer metastasis: friend or foe? J Immunother Cancer. 2017;5(1) doi: 10.1186/s40425-017-0283-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunn G.P., Bruce A.T., Ikeda H., Old L.J., Schreiber R.D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3(11):991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X.H.F., Wang Q., Gerald W. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Canc Cell. 2009;16(1):67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wraith D.C. The future of immunotherapy: a 20-year perspective. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.01668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farkona S., Diamandis E.P., Blasutig I.M. Cancer immunotherapy: the beginning of the end of cancer? BMC Med. 2016;14 doi: 10.1186/s12916-016-0623-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.June C.H., O'Connor R.S., Kawalekar O.U., Ghassemi S., Milone M.C. CAR T cell immunotherapy for human cancer. Science. 2018;359(6382):1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- 51.Teicher B.A. Molecular targets and cancer therapeutics: discovery, development and clinical validation. Drug Resist Updates : reviews and commentaries in antimicrobial and anticancer chemotherapy. 2000;3(2):67–73. doi: 10.1054/drup.2000.0123. [DOI] [PubMed] [Google Scholar]
- 52.Gerber D.E. Targeted therapies: a new generation of cancer treatments. Am Fam Physician. 2008;77(3):311–319. [PubMed] [Google Scholar]