Abstract
d-amino acids produced by Lactobacillus are thought to contribute to the taste quality and health functions; however, no studies have comprehensively evaluated the concentrations of the D- and L-forms of amino acids separately in individual Lactobacillus strains. To gain insight into amino acid concentrations in Lactobacillus, we evaluated amino acid concentrations in culture broth of Lactobacillus separately for the D- and L-forms. Lactobacillus strains were cultured in culture broth, and the amino acid concentrations in supernatant were assessed. The amino acid concentrations obtained by liquid chromatography-tandem mass spectrometry (LC-MS/MS) were subjected to cluster analysis based on Bray-Curtis distance with Ward's minimum variance method. In the analysis of amino acid concentrations under culture with different monosaccharides, the distances among strains cultured with the same monosaccharide were significantly greater than those among cultures of the same strain under different monosaccharides (p < 0.01). The cluster analysis of amino acid concentrations under culture with the same monosaccharide suggested that strains belonging to the same phylogenetic group of Lactobacillus exhibited similar concentrations of amino acids. Data analyses of 70 strains belonging to 17 Lactobacillus taxa indicated that the concentrations of amino acids were highly dependent on the phylogenetic group of Lactobacillus and that the group differences in amino acid concentration were strongly driven by differences in l-serine and d-alanine concentrations. Our results indicate that it is important to evaluate D- and l-amino acids separately when evaluating variations in amino acid concentrations. Because d-alanine has the potential to affect taste quality, the results of this study may provide insight into the taste quality of fermented food produced by Lactobacillus.
Keywords: Probiotics, Lactobacillus, Amino acid, d-amino acid, d-alanine, Cluster analysis
1. Introduction
Species of the genus Lactobacillus are among the most important taxa involved in food microbiology and human health due to their roles in food production and preservation and their beneficial effects on human health [1]. Lactobacillus bacteria are non-spore-forming, catalase-negative, obligate saccharolytic rod or coccobacillus bacteria; they are generally characterized by a low guanine and cytosine genome content and are able to produce lactate as the main end-product of fermentation. Other side products include acetate, ethanol, carbon dioxide, formate and succinate [2]. These bacteria require complex nutrients, such as salts, amino acids, peptides, vitamins, and fatty acids, and they reside in food (e.g., dairy products, grain products, beer, wine and fruits), water, soil, sewage and intestinal environments, including those of humans. The ability of Lactobacillus to metabolize several carbohydrates provides the members of this genus competitive advantages in numerous environments and makes them unique and distinctive in terms of their fermentation potential [3].
The traditional method of fermented food production is the inoculation of food with a sample of a previous-day product that contains certain microorganisms, such as members of the genus Lactobacillus. This method is still used for some homemade products; however, the quality of the products produced by this method varies. For stabilization and large-scale production of fermented food, commercial starter cultures containing Lactobacillus are used for successful production of fermented food [4]. To date, many Lactobacillus strains have been used in the food industry for their specific activities, such as the ability to utilize carbohydrates.
Bacteria can produce d-amino acids [5], and fermented food produced by Lactobacillus contains d-amino acids. A previous study indicated that the umami taste in Japanese rice wine, which is a fermented alcoholic drink, was more influenced by d-alanine supplementation than by l-alanine supplementation [6]. Because several d-amino acids, such as d-alanine, d-leucine and d-phenylalanine, are sweeter than their l-amino acid counterparts, d-amino acids might contribute to the taste quality of fermented foods [7].
Lactobacillus is also used as a probiotic, which is defined as a live microorganism that confers a health benefit to the host when administered in adequate amounts, and it has been demonstrated to improve host health [8]. Lactobacillus acidophilus (L. acidophilus), Lactobacillus crispatus (L. crispatus), Lactobacillus gasseri (L. gasseri), Lactobacillus johnsonii (L. johnsonii), Lactobacillus rhamnosus (L. rhamnosus), Lactobacillus paracasei (L. paracasei), Lactobacillus plantarum (L. plantarum), Lactobacillus reuteri (L. reuteri), Lactobacillus fermentum (L. fermentum), Lactobacillus salivarius (L. salivarius) and Lactobacillus brevis (L. brevis) are widely used as probiotics [[9], [10], [11]], and the beneficial effects of probiotics are thought to derive from their surface components and metabolites [12]. Probiotics produce d-amino acids as metabolites, and a previous study in mice indicated that d-tryptophan derived from Lactobacillus ameliorated allergic airway inflammation [13]. Other reports have indicated the health functions of d-amino acids in the intestinal environment. d-amino acids produced by the gut microbiota protect the mucosal surface of the small intestine from pathogens by inducing d-amino acid oxidase production in intestinal epithelial cells [14]. Furthermore, intestinal microbiota-derived d-serine has been shown to protect against acute kidney injury [15]. Therefore, the d-amino acids produced by Lactobacillus as probiotic bacteria in the intestinal environment might contribute to host health.
To determine their effects on taste quality and host health condition, the concentrations of D- and l-amino acids produced by Lactobacillus should be evaluated separately. However, no reports have comprehensively evaluated the concentrations of the D- and L-forms of amino acids separately in individual Lactobacillus strains. In this study, we evaluated the concentrations of D- and L-forms and their characteristics in individual Lactobacillus strains. Since the ability to assimilate carbohydrates is one of the major factors affecting the fermentation potential of Lactobacillus, we evaluated whether carbohydrate type or bacterial taxon had a greater impact on the concentrations of amino acids by investigating 11 type strains of Lactobacillus and 6 species of monosaccharides. In addition, we used 70 strains belonging to 17 Lactobacillus taxa to evaluate the influence of Lactobacillus strain.
2. Material and methods
2.1. Bacterial strains and culture conditions
Lactobacillus strains were obtained from Japan Collection of Microorganisms (JCM) (Ibaraki, Japan) and Asahi Group Culture Collection (AGCC) (Ibaraki, Japan) (Table 1). For bacterial culture, we used the method described in a previous study [16], which allowed the cultivation of all Lactobacillus strains used in this study. Bacterial cultures of Lactobacillus were subcultured in de Man, Rogosa and Sharpe (MRS) broth (Becton, Dickinson and Company, Sparks, MD) and incubated statically at 37 °C for 16 h.
Table 1.
Phylogenetic groups of Lactobacillus according to Salvetti et al. [2] and the strain names and sources of Lactobacillus in this study.
| Strain name | Source | Phylogenetic group | Strain name | Source | Phylogenetic group |
|---|---|---|---|---|---|
| Lactobacillus acidophilus JCM1132T | JCM |
Lactobacillus delbrueckii group |
Lactobacillus fermentum JCM1173T | JCM |
Lactobacillus reuteri group |
| Lactobacillus acidophilus LAC1613 | AGCC | Lactobacillus fermentum LFE1299 | AGCC | ||
| Lactobacillus acidophilus LAC1780 | AGCC | Lactobacillus fermentum LFE1753 | AGCC | ||
| Lactobacillus acidophilus LAC3525 | AGCC | Lactobacillus fermentum LFE3024 | AGCC | ||
| Lactobacillus amylovorus JCM1126T | JCM | Lactobacillus mucosae LMU3014 | AGCC | ||
| Lactobacillus amylovorus LAM1562 | AGCC | Lactobacillus mucosae LMU3018 | AGCC | ||
| Lactobacillus amylovorus LAM1587 | AGCC | Lactobacillus mucosae LMU3021 | AGCC | ||
| Lactobacillus amylovorus LAM1750 | AGCC | Lactobacillus mucosae LMU3025 | AGCC | ||
| Lactobacillus crispatus JCM1185T | JCM | Lactobacillus mucosae LMU3368 | AGCC | ||
| Lactobacillus crispatus LCR1740 | AGCC | Lactobacillus mucosae LMU3451 | AGCC | ||
| Lactobacillus crispatus LCR1744 | AGCC | Lactobacillus mucosae LMU3470 | AGCC | ||
| Lactobacillus crispatus LCR1762 | AGCC | Lactobacillus oris LOR1717 | AGCC | ||
| Lactobacillus crispatus LCR1779 | AGCC | Lactobacillus oris LOR3356 | AGCC | ||
| Lactobacillus crispatus LCR3453 | AGCC | Lactobacillus oris LOR3360 | AGCC | ||
| Lactobacillus delbrueckii LDE688 | AGCC | Lactobacillus reuteri JCM1112T | JCM | ||
| Lactobacillus gasseri JCM1131T | JCM | Lactobacillus reuteri LRE720 | AGCC | ||
| Lactobacillus gasseri LGA3450 | AGCC | Lactobacillus reuteri LRE3017 | AGCC | ||
| Lactobacillus gasseri LGA3465 | AGCC | Lactobacillus reuteri LRE3062 | AGCC | ||
| Lactobacillus gasseri LGA3516 | AGCC | Lactobacillus reuteri LRE3084 | AGCC | ||
| Lactobacillus gasseri LGA3529 | AGCC | Lactobacillus casei LCA3518 | AGCC |
Lactobacillus casei group |
|
| Lactobacillus gasseri LGA3568 | AGCC | Lactobacillus casei LCA3560 | AGCC | ||
| Lactobacillus johnsonii JCM2012T | JCM | Lactobacillus casei LCA3489 | AGCC | ||
| Lactobacillus johnsonii LJO721 | AGCC | Lactobacillus casei LCA3490 | AGCC | ||
| Lactobacillus johnsonii LJO1572 | AGCC | Lactobacillus casei LCA3493 | AGCC | ||
| Lactobacillus plantarum JCM1149T | JCM |
Lactobacillus plantarum group |
Lactobacillus paracasei JCM8130T | JCM | |
| Lactobacillus plantarum LPL3504 | AGCC | Lactobacillus paracasei LPA3507 | AGCC | ||
| Lactobacillus plantarum LPL3530 | AGCC | Lactobacillus paracasei LPA3526 | AGCC | ||
| Lactobacillus plantarum LPL3543 | AGCC | Lactobacillus paracasei LPA3546 | AGCC | ||
| Lactobacillus plantarum LPL3551 | AGCC | Lactobacillus paracasei LPA3562 | AGCC | ||
| Lactobacillus salivarius JCM1231T | JCM |
Lactobacillus salivarius group |
Lactobacillus rhamnosus JCM1136T | JCM | |
| Lactobacillus salivarius LSA3355 | AGCC | Lactobacillus rhamnosus LRH1616 | AGCC | ||
| Lactobacillus salivarius LSA3496 | AGCC | Lactobacillus rhamnosus LRH3344 | AGCC | ||
| Lactobacillus salivarius LSA3510 | AGCC | Lactobacillus rhamnosus LRH3544 | AGCC | ||
| Lactobacillus brevis JCM1059T | JCM |
Lactobacillus brevis group |
|||
| Lactobacillus brevis LBR3509 | AGCC | ||||
| Lactobacillus brevis LBR3550 | AGCC | ||||
| Lactobacillus brevis LBR3573 | AGCC | ||||
JCM; The Japan Collection of Microorganisms, AGCC; Asahi Group Culture Collection.
2.2. Sample collection
Precultures of Lactobacillus in MRS broth were centrifuged at 8000 g for 5 min, and pellets were washed and resuspended in an equal volume of phosphate-buffered saline. The bacterial suspension was inoculated with 5% v/v into modified MRS (mMRS) broth in which the carbohydrate source had been changed from dextrose to other monosaccharides, and the bacteria were cultured at 37 °C for 16 h. The mMRS broth comprised 1% (w/v) protease peptone No. 3 (Becton, Dickinson and Company, Sparks, MD), 0.5% (w/v) yeast extract (Becton, Dickinson and Company), 1% (w/v) beef extract (Becton, Dickinson and Company), 0.2% (w/v) triammonium citrate (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan), 0.1% (w/v) polyoxyethylene (20) sorbitan monooleate (Fujifilm Wako Pure Chemical Corporation), 0.5% (w/v) sodium acetate (Fujifilm Wako Pure Chemical Corporation), 0.01% (w/v) magnesium sulfate (Fujifilm Wako Pure Chemical Corporation), 0.005% (w/v) manganese (II) sulfate (Fujifilm Wako Pure Chemical Corporation), 0.2% (w/v) dipotassium hydrogen phosphate (Fujifilm Wako Pure Chemical Corporation), and 2% (w/v) monosaccharide. The monosaccharide was selected from among glucose (Fujifilm Wako Pure Chemical Corporation), galactose (Fujifilm Wako Pure Chemical Corporation), rhamnose (Dextra Laboratories Ltd., Reading, UK), arabinose (Fujifilm Wako Pure Chemical Corporation), mannose (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan), xylose (Nacalai Tesque, Inc., Kyoto, Japan) and fructose (Kanto Chemical Co., Inc., Tokyo, Japan).
2.3. Sample preparation
After the bacterial cultures were incubated in mMRS broth, the pH of the broth was measured. Then, the bacterial cultures were centrifuged at 8000 g for 5 min. The supernatants were frozen at −20 °C until use.
Standard and sample preparation were performed as previously described [17] with some modifications. dl-alanine (DL-Ala), dl-arginine hydrochloride (DL-Arg), dl-asparagine monohydrate (DL-Asn), dl-aspartic acid (DL-Asp), dl-cysteine hydrochloride monohydrate (DL-Cys), dl-glutamic acid (DL-Glu), dl-glutamine (DL-Gln), dl-histidine (DL-His), dl-isoleucine (DL-Ile), dl-leucine (DL-Leu), dl-lysine monohydrochloride (DL-Lys), dl-methionine (DL-Met), dl-phenylalanine (DL-Phe), dl-serine (DL-Ser), dl-threonine (DL-Thr), DL-tryptophan (DL-Trp), dl-tyrosine (DL-Tyr) and dl-valine (DL-Val) were obtained from Tokyo Chemical Co., Ltd. Glycine (Gly) and l-proline (L-Pro) were obtained from Fujifilm Wako Pure Chemical Corporation. dl-alanine-2,3,3,3-d4 (DL-Ala-d4) was obtained from Santa Cruz Biotechnology (Dallas, TX). Each amino acid standard was dissolved in 50% methanol, 50% methanol with 0.02 mol/ml hydrochloride (for DL-Glu, DL-His and DL-Trp), or 50% methanol with 0.05 mol/ml hydrochloride (for DL-Asp and DL-Tyr). A solution of DL-Ala-d4 (1000 nmol/ml) was prepared as described above and used as an internal standard (IS) solution. After thawing a frozen sample, 50 μl of the sample was mixed with 175 μl of methanol, 10 μl of the IS solution, and 25 μl of water in a tube. The contents were then mixed and centrifuged at 10000 rpm for 10 min at 4 °C. Then, 180 μl of supernatant was transferred to a new tube and mixed with 90 μl of water and 180 μl of chloroform. After mixing and centrifugation, 50 μl of supernatant was diluted with 160 μl of acetonitrile and 40 μl of ethanol. Approximately 50 μl of diluted sample was transferred to a vial and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. A standard was prepared using the same method described above.
2.4. Amino acid quantification
LC-MS/MS analysis was performed as previously described [17] with some modifications. Briefly, we used the Nextera HPLC System (communication bus module: CBM-20A; pump: LC-30AD; autosampler: SIL-30AC; degasser: DGU-20A5R; column oven: CTO-20AC, Shimadzu, Kyoto, Japan) connected to a Sciex Triple quad 6500+ mass spectrometer (AB SCIEX, Concord, Canada). The Sciex Triple quad 6500+ (temperature: 500 °C; ion spray voltage floating: 5500 V; entrance potential: 10 V) was used with the parameters described in Table 2. Chromatographic separation for DL-Ala, DL-Arg, DL-Asn, DL-Asp, DL-Cys, DL-Glu, DL-His, DL-Ile, DL-Leu, DL-Lys, DL-Met, DL-Phe, DL-Ser, DL-Thr, DL-Trp, DL-Tyr and DL-Val was performed with a CROWNPAK CR-I (+) (3.0 mm i.d. × 150 mm, 5 μm, Daicel CPI, Osaka, Japan) enantioseparation column. Chromatographic separation for DL-Gln and DL-Lys was performed with a CROWNPAK CR-I (−) (3.0 mm i.d. × 150 mm, 5 μm, Daicel CPI) enantioseparation column. Because D- and L-Pro could not be separated from each other, total proline (a racemic mixture) was quantified using a CROWNPAK CR-I (−) column. In addition, chromatographic separation for Gly was performed with a CROWNPAK CR-I (−) column. The injection volume was 1 μl, and the oven temperature was maintained at 30 °C. The mobile phase consisted of a mixture of acetonitrile, ethanol, water and trifluoroacetic acid (80/15/5/0.5), and the flow rate was set to 0.4 ml/min under isocratic conditions. Data processing was performed using MultiQuant (AB SCIEX) and regression equations (Y = aX + b; with a weighting factor of 1/X).
Table 2.
Optimized multiple reaction monitoring transition and parameters for 20 amino acids and the internal standard (IS).
| Molecular weight | Precursor ion (m/z) | Product ion (m/z) | Declustering potential (volts) | Collision energy (volts) | Collision cell exit potential (volts) | Curtain gas (psi) | Collision activated dissociation gas (psi) | Ion source gas1 (psi) | Ion source gas2 (psi) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Alanine | 89.1 | 90.0 | 44.0 | 56 | 17 | 12 | 40 | 8 | 30 | 40 |
| Arginine | 174.2 | 175.0 | 69.9 | 51 | 29 | 10 | 40 | 8 | 30 | 40 |
| Asparagine | 132.1 | 132.9 | 74.1 | 31 | 21 | 12 | 40 | 8 | 30 | 40 |
| Aspartic acid | 133.1 | 133.9 | 74.0 | 61 | 19 | 8 | 40 | 8 | 30 | 40 |
| Cysteine | 121.2 | 122.0 | 59.0 | 31 | 35 | 10 | 40 | 8 | 30 | 40 |
| Glutamic acid | 147.1 | 147.9 | 83.9 | 36 | 23 | 14 | 40 | 8 | 30 | 40 |
| Glutamine | 146.2 | 147.0 | 84.0 | 56 | 23 | 10 | 30 | 10 | 50 | 80 |
| Glycine | 75.1 | 75.9 | 30.1 | 56 | 17 | 14 | 30 | 10 | 50 | 80 |
| Histidine | 155.2 | 156.0 | 110.0 | 31 | 19 | 10 | 40 | 8 | 30 | 40 |
| Isoleucine | 131.2 | 132.0 | 86.1 | 51 | 15 | 12 | 40 | 8 | 30 | 40 |
| Leucine | 131.2 | 132.0 | 86.0 | 46 | 15 | 8 | 40 | 8 | 30 | 40 |
| Lysine | 146.2 | 147.0 | 84.0 | 31 | 25 | 14 | 30 | 10 | 50 | 80 |
| Methionine | 149.2 | 150.0 | 104.0 | 36 | 15 | 14 | 40 | 8 | 30 | 40 |
| Phenylalanine | 165.2 | 166.0 | 102.9 | 46 | 37 | 16 | 40 | 8 | 30 | 40 |
| Proline | 115.1 | 116.1 | 70.0 | 61 | 21 | 10 | 30 | 10 | 50 | 80 |
| Serine | 105.1 | 105.9 | 59.9 | 81 | 15 | 12 | 40 | 8 | 30 | 40 |
| Threonine | 119.1 | 119.8 | 74.0 | 41 | 15 | 10 | 40 | 8 | 30 | 40 |
| Tryptophan | 204.2 | 205.0 | 187.9 | 76 | 15 | 18 | 40 | 8 | 30 | 40 |
| Tyrosine | 181.2 | 182.0 | 136.0 | 36 | 19 | 16 | 40 | 8 | 30 | 40 |
| Valine | 117.2 | 118.0 | 72.0 | 41 | 15 | 8 | 40 | 8 | 30 | 40 |
| Alanine-2,3,3,3-d4 (IS) | 93.1 | 94.0 | 48.1 | 111 | 15 | 8 | 40 | 8 | 30 | 40 |
2.5. Statistical analysis
All statistical analyses were performed in R (ver. 4.0.0) [18]. The R packages vegan [19], RVAideMemoire [20] and randomForest [21] were used for cluster analysis, pairwise permutational multivariate analysis of variance (MANOVA) and random forest analysis, respectively. For all analyses, p values of <0.05 were considered statistically significant.
3. Results
3.1. Amino acid concentrations of Lactobacillus under Culture with different monosaccharides
The concentrations of amino acids in the supernatant of mMRS broth were evaluated after Lactobacillus culture when the pH reductions in the broth were 1.0 or greater. We measured the pH values of 77 supernatants, which were generated using 11 type strains and 7 monosaccharides. The pH reductions in the broths of 34 bacterial cultures were not large enough for evaluation; thus, the remaining 43 bacterial cultures with sufficient pH reduction and 7 uncultured broths containing certain species of monosaccharides as blanks were evaluated.
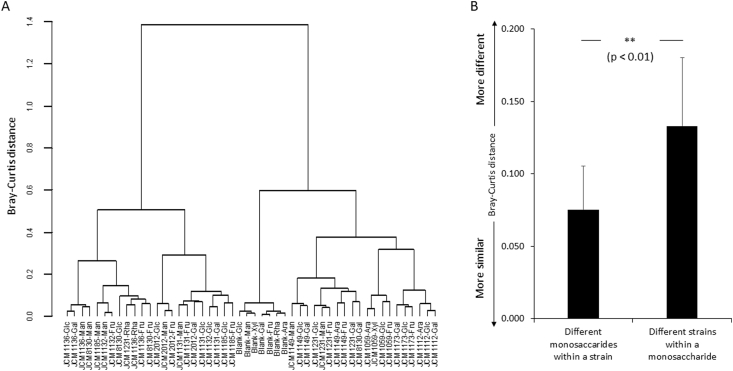
To reveal whether strain or monosaccharide type had a large impact on the concentrations of amino acids, the D- and l-amino acid concentrations in the 43 bacterial cultures and the 7 uncultured broths were used to calculate Bray-Curtis distances. Bray-Curtis distance was used to visualize the results of the hierarchical clustering analysis performed with the Ward's minimum variance method. The cluster analysis showed that the concentrations of amino acids produced by each strain were similar among the different monosaccharides (Fig. 1A). We compared the average Bray-Curtis distance among different monosaccharides within the same strain and the average Bray-Curtis distance among different strains cultured with the same monosaccharide, which were determined with a previously reported method [22]. The distance between strains within a monosaccharide type was significantly higher than the distance between monosaccharides within the same strain (p = 1.3E-21, Fig. 1B).
-
(A)
Bray-Curtis distance calculated from the D- and l-amino acid concentrations in 43 bacterial cultures and 7 uncultured broths were visualized by the hierarchical clustering analysis performed with the Ward's minimum variance method. An uncultured broth containing each monosaccharide is defined as the blank in this figure. Glc, Man, Xyl, Gal, Fru, Rha and Ara indicate glucose, mannose, xylose, galactose, fructose, rhamnose and arabinose, respectively.
-
(B)
Average Bray-Curtis distance (a measure of difference in amino acid concentration) between monosaccharides within a strain and between strains within a monosaccharide. Data are shown as the mean ± SD. P-values were calculated using Welch's t-test. **p < 0.01
Fig. 1.
Evaluation of the concentrations of amino acids in Lactobacillus cultured with different monosaccharides.
3.2. Amino acid concentrations of Lactobacillus under Culture with the same monosaccharide
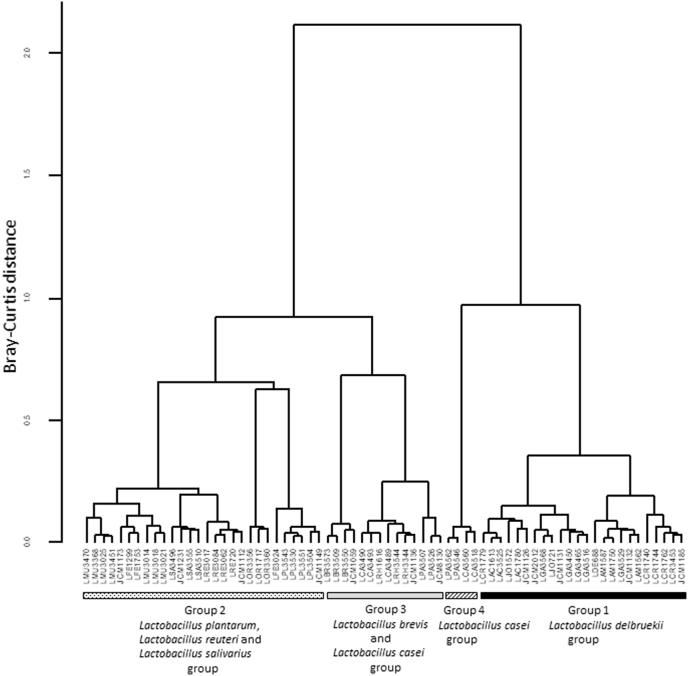
To evaluate the effects of strain on the concentrations of amino acids produced by Lactobacillus, we investigated amino acid concentrations in 70 strains using mMRS broth with glucose as the sole carbon source; glucose was chosen because all the strains are capable of growing in mMRS broth with glucose. The D- and l-amino acid concentrations in 70 bacterial cultures generated from 70 strains of Lactobacillus were used to calculate Bray-Curtis distances. Bray-Curtis distance was used to visualize the results of the hierarchical clustering analysis performed with the Ward's minimum variance method. The concentration of each amino acid in the mMRS broth was compared using hierarchical cluster analysis with Bray-Curtis distances and Ward's clustering algorithm (Fig. 2). To validate the cluster number, we used pairwise permutational MANOVA for statistical analysis with Bonferroni correction for multiple comparisons. Significant pairwise differences between all clusters were observed when 4 clusters were selected (group 1 vs. group 2: p = 0.006; group 1 vs. group 3: p = 0.006; group 1 vs. group 4: p = 0.012; group 2 vs. group 3: p = 0.006; group 2 vs. group 4: p = 0.006; group 3 vs. group 4: p = 0.012), and when the number of clusters was greater than 4, some pairs did not show a significant difference. Group 1 consisted of 24 strains belonging to 6 species (L. acidophilus, L. amylovorus, L. crispatus, L. delbrueckii, L. gasseri, L. johnsonii), and all species belonged to the L. delbrueckii group. Group 2 consisted of 28 strains belonging to 6 species (L. fermentum, L. mucosae, L. oris, L. plantarum, L. reuteri, L. salivarius), and these 6 species belonged to the L. reuteri, L. plantarum or L. salivarius groups. Group 3 consisted of 14 strains belonging to 4 species (L. brevis, L. casei, L. paracasei, L. rhamnosus), and these 4 species belonged to the L. brevis or L. casei group. Group 4 consisted of 4 strains belonging to the species L. paracasei, which belonged to the L. casei group.
Fig. 2.
Evaluation of the concentrations of amino acids in Lactobacillus cultured with the same monosaccharide.
Bray-Curtis distance calculated from the D- and l-amino acid concentrations in 70 bacterial cultures were visualized by the hierarchical clustering analysis performed with the Ward's minimum variance method. The lines in the figure indicate the phylogenetic groups [2] of Lactobacillus belonging to four clusters. Group 1 is indicated by the black line. Group 2 is indicated by the dotted line. Group 3 is indicated by the gray line. Group 4 is indicated by the striped line.
3.3. Contribution of each amino acid to the cluster separation
To reveal the contributions of amino acids to the cluster separation, we used 4 clusters generated by hierarchical cluster analysis and pairwise permutational MANOVA as group labels.
Random forest analysis is an analytical method using a machine learning algorithm [23]. Because a previous study [24] indicated that random forest analysis was useful for biomarker selection in metabolomic data, we used random forest analysis to calculate the contribution of each amino acid to cluster separation. In the random forest analysis, the parameters ntree = 2000 and mtry = 3 were used for the least out-of-bag (OOB) error, and the OOB error was 1.43%. Because a previous study [25] indicated that an index based on the mean decrease in Gini generated by random forest analysis provided a more robust result than one based on the mean decrease in accuracy, the mean decrease in Gini was used as a measure of the importance of each amino acid for cluster separation.
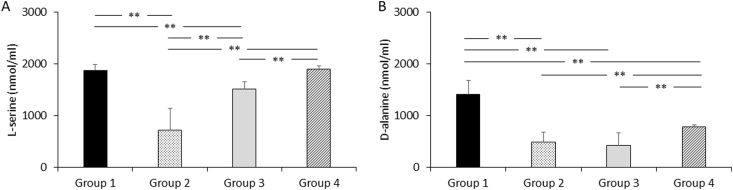
The index indicated that l-serine and d-alanine had high impacts on cluster separation (Table 3). In the statistical analysis of l-serine differences among the 4 groups, no significant difference was observed between group 1 and group 4; however, significant differences were observed between pairs of all the other groups (group 1 vs. group 2: p = 7.3E-14; group 1 vs. group 3: p = 3.1E-7; group 1 vs. group 4: p = 1.0; group 2 vs. group 3: p = 2.9E-4; group 2 vs. group 4: p = 1.3E-12; group 3 vs. group 4: p = 2.5E-4) (Fig. 3A). In the statistical analysis of d-alanine differences among the 4 groups, no significant difference was observed between group 2 and group 3; however, significant differences were observed between pairs of all other groups (group 1 vs. group 2: p = 9.1E-16; group 1 vs. group 3: p = 1.1E-11; group 1 vs. group 4: p = 8.4E-10; group 2 vs. group 3: p = 1.0; group 2 vs. group 4: p = 3.4E-6; group 3 vs. group 4: p = 6.1E-4) (Fig. 3B).
Table 3.
Mean decrease in Gini calculated by random forest analysis.
| Mean decrease in Gini | Mean decrease in Gini | ||
|---|---|---|---|
| l-serine | 3.34 | dl-proline | 1.05 |
| d-alanine | 3.26 | l-arginine | 1.02 |
| l-histidine | 2.91 | d-methionine | 0.91 |
| d-serine | 2.53 | l-lysine | 0.91 |
| l-leucine | 2.39 | d-isoleucine | 0.80 |
| d-threonine | 2.16 | d-valine | 0.74 |
| l-aspartic acid | 2.16 | Glycine | 0.70 |
| l-phenylalanine | 1.99 | d-arginine | 0.65 |
| l-threonine | 1.78 | l-glutamic acid | 0.60 |
| d-lysine | 1.72 | l-alanine | 0.57 |
| L-tryptophan | 1.67 | l-methionine | 0.52 |
| l-isoleucine | 1.58 | d-phenylalanine | 0.50 |
| l-glutamine | 1.47 | d-tyrosine | 0.45 |
| l-valine | 1.44 | d-aspartic acid | 0.39 |
| d-asparagine | 1.41 | l-asparagine | 0.33 |
| d-leucine | 1.16 | l-cysteine | 0.30 |
| d-glutamic acid | 1.14 | D-tryptophan | 0.19 |
| d-glutamine | 1.11 | d-cysteine | 0.01 |
| l-tyrosine | 1.08 | d-histidine | 0.00 |
Fig. 3.
Statistical analysis of l-serine and d-alanine concentrations in the 4 groups.
Data are shown as the mean ± SD. The groups in the figure consisted of the strains in the phylogenetic groups [2] of Lactobacillus. Group 1 consisted of the strains belonging to the L. delbrueckii group and is indicated by the black bar. Group 2 consisted of the strains in the L. reuteri, L. plantarum or L. salivarius groups and is indicated by the dotted bar. Group 3 consisted of the strains in the L. brevis or L. casei group and is indicated by the gray bar. Group 4 consisted of 4 strains belonging to the L. casei group and is indicated by the striped bar. P-values were calculated using Welch's t-test with Bonferroni correction for multiple comparisons. **p < 0.01.
3.4. Characteristics of amino acid metabolism in each group
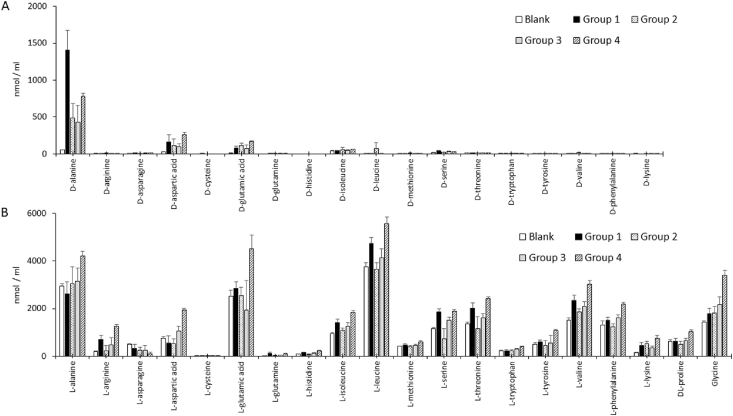
To evaluate the differences from the initial values, amino acid concentrations in the blank (i.e., uncultured mMRS broth with glucose) were measured in triplicate. The amino acid concentrations in the blank and each cluster group are summarized in Fig. 4.
Fig. 4.
D- and l-amino acid concentrations in the broths.
An uncultured broth with glucose was defined as the blank and is indicated in the figure by the white bar. The groups in the figure consisted of the strains in the phylogenetic groups [2] of Lactobacillus. Group 1 consisted of the strains belonging to the L. delbrueckii group and is indicated by the black bar. Group 2 consisted of the strains in the L. reuteri, L. plantarum or L. salivarius groups and is indicated by the dotted bar. Group 3 consisted of the strains in the L. brevis or L. casei group and is indicated by the gray bar. Group 4 consisted of 4 strains belonging to the L. casei group and is indicated by the striped bar. (A) The 18 d-amino acid concentrations in the broths. (B) The 18 l-amino acid, dl-proline and glycine concentrations in the broths. Data are shown as the mean ± SD.
4. Discussion
In the present study, it was considered important to evaluate the concentrations of D- and l-amino acids produced by Lactobacillus separately because the d-amino acids produced by Lactobacillus influence the taste of fermented food and its health benefits to the host [7,14]. Therefore, we quantified D- and l-amino acid concentrations separately by enantioseparation and LC-MS/MS methods, and we found that the concentrations of amino acids in Lactobacillus were highly dependent on phylogenetic group and that the group differences in amino acid concentration were strongly affected by differences in l-serine and d-alanine concentrations.
The patterns of D- and l-amino acid concentrations in this study were visualized by hierarchical cluster analysis using 11 type strains and 7 monosaccharides. Although the concentrations of a few amino acids, especially in L. paracasei JCM8130, did not show obvious cluster separation, the cluster separation was highly dependent on strain (Fig. 1A). The comparison of the average Bray-Curtis distance among different monosaccharides within the same strain and that among different strains under the same monosaccharide supported the finding of strain dependence (Fig. 1B). These results suggested that compared with monosaccharide type, strain more strongly impacted the concentrations of amino acids produced by Lactobacillus.
We also investigated strain differences in the concentrations of amino acids in 70 strains cultured with the same monosaccharide. Glucose was selected as the best monosaccharide for this evaluation because all strains were capable of growing in mMRS broth containing glucose as the sole carbon source. Based on the hierarchical cluster analysis of amino acid concentration and the pairwise permutational MANOVA with Bonferroni correction for multiple comparisons, the Lactobacillus strains were separated into 4 groups (Fig. 2). The cluster analysis showed that different strains of the same bacterial species often produced similar concentrations of amino acids, although in some cases, strains produced concentrations similar to those of another bacterial species. The concentrations of amino acids in Lactobacillus strains were divided into the phylogenetic groups of Lactobacillus; this result indicated that the concentrations of amino acids produced by Lactobacillus were highly dependent on phylogenetic group. In previous studies [2,26,27], phylogenetic analyses of the 16S rRNA gene, selected genes, and whole genomes have been performed. All of these phylogenetic analyses have shown that the L. delbrueckii group clusters separately from other Lactobacillus groups. The cluster separation between the L. delbrueckii group and the other Lactobacillus groups in this study was consistent with previous phylogenetic analyses, which suggested that genomic differences might be partially reflected in the concentrations of amino acids. In addition, in a recent study, some Lactobacillus species were reclassified into new genera [28]: L. plantarum was reclassified into Lactiplantibacillus; L. fermentum, L. mucosae, L. oris and L. reuteri were reclassified into Limosilactobacillus; L. salivarius was reclassified into Ligilactobacillus; L. brevis was reclassified into Levilactobacillus; and L. casei, L. paracasei and L. rhamnosus were reclassified into Lacticaseibacillus. Although L. casei and L. paracasei were divided into two groups in this study, the difference in genera proposed by Zheng et al. [28] might be reflected in the concentrations of amino acids.
To reveal the contribution of each amino acid to cluster separation, we performed random forest analysis. The analysis generated the mean decrease in Gini, which indicated that l-serine and d-alanine had high impacts on the cluster separation (Table 3). In addition, significant differences between all groups were observed in either or both concentration of l-serine or d-alanine (Fig. 3A and B). These results suggested that the characteristics of each group could be partially revealed by evaluating l-serine and d-alanine concentrations. In the evaluation of d-amino acids, d-alanine was found to be especially important for understanding the variation in amino acid concentrations in Lactobacillus.
In Lactobacillus, l-serine is preferentially converted into pyruvate by serine deaminase (EC 4.3.1.17) and into glycine and acetaldehyde by serine hydroxymethyl transferase (EC 2.1.2.1) [29]. Tetrahydrofolate is a cofactor of serine hydroxymethyl transferase [29], and tetrahydrofolate is produced from dihydrofolate by dihydrofolate reductase (EC 1.5.1.3) [30]. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis has indicated that the genes encoding these enzymes exist in many Lactobacillus species [31], suggesting that the possession of these genes does not differ among Lactobacillus taxa. l-alanine is converted to d-alanine by alanine racemase (EC:5.1.1.1), and its enzymatic activity has been characterized in Lactobacillus species [29]. In Lactobacillus, d-alanine is a predominant component of newly synthesized peptidoglycan [32], and the alanine racemase gene is present in all Lactobacillus taxa represented in the KEGG database. Regarding other enzymes for producing d-alanine in Lactobacillus species, amino acid racemase activity has been reported for MalY (EC 4.4.1.13) and isoleucine 2-epimerase (EC 5.1.1.21); these enzymes have racemase activity for many kinds of amino acids, including alanine [33,34]. These observations indicate that the differences in l-serine and d-alanine concentrations among members of Lactobacillus are difficult to interpret without evaluating the metabolic pathways involved and differences in metabolic activity. In addition, some Lactobacillus species have proteinases, such as PrtB and PrtH [35]. The concentrations of l-amino acids in Lactobacillus might be affected by proteolytic ability. Therefore, further analysis is needed to identify the differences in l-serine and d-alanine metabolism, which contribute to the differences in amino acid concentrations among members of Lactobacillus.
According to the changes from the initial values of d-amino acids in the culture broth, Lactobacillus mainly produced d-alanine and produced less d-aspartic acid and d-glutamic acid than d-alanine (Fig. 4A). This result was consistent with the results of a previous study [7]. The L. delbrueckii group produced high levels of d-alanine (Fig. 3B), which influenced taste quality in fermented food [6]; thus, the taste quality of fermented dairy foods such as yogurt and acidophilus milk, which are fermented by members of the L. delbrueckii group, might be affected by high d-alanine concentrations. In addition, because d-amino acids affect host health, differences in the ability to produce these acids among members of a Lactobacillus group might influence the health function supported by different Lactobacillus strains. In group 4, the levels of most l-amino acids appeared to increase above their initial values (Fig. 4B). Lactobacillus strains belonging to group 4 might have high proteolytic ability and thus a strong ability to increase total amino acid levels.
5. Conclusions
In this study, we found that the concentrations of amino acids in Lactobacillus were highly dependent on phylogenetic group and that differences in l-serine and d-alanine concentrations strongly contributed to the differences in amino acid concentrations in Lactobacillus. Our results suggest that revealing the variation in the concentration of d-alanine, which has the potential to affect the taste quality of fermented foods produced by Lactobacillus, was important for understanding amino acid metabolism in Lactobacillus. To our knowledge, this report is the first comprehensive evaluation of the concentrations of D- and l-amino acids in Lactobacillus. In future study, evaluation for the variation in the concentration of amino acid with Lactobacillus growth and the effects of amino acid concentrations produced by Lactobacillus on the taste quality and health function of Lactobacillus-fermented food will be expected.
Funding
This study was funded by Asahi Group Holdings, Ltd.
Role of the funding source
Asahi Quality & Innovations, Ltd. is an independent research subsidiary of Asahi Group Holdings, Ltd.
Author contributions
H. S., T. H., and Y. N. conceived and designed the experiments. H. S., K. N. and S. I. performed the experiments. H. S. analyzed the data. H. S., T. H., and Y. N. wrote the paper.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: All authors are employees of Asahi Quality & Innovations, Ltd.
Acknowledgements
We thank Hidetoshi Miyazaki for providing excellent technical assistance.
Contributor Information
Hirosuke Sugahara, Email: hirosuke.sugahara@asahi-qi.co.jp.
Keitaro Nagayama, Email: keitaro.nagayama@asahi-qi.co.jp.
Shiori Ikeda, Email: shiori.ikeda@asahi-qi.co.jp.
Tatsuhiko Hirota, Email: tatsuhiko.hirota@asahi-qi.co.jp.
Yasunori Nakamura, Email: yasunori.nakamura@asahi-qi.co.jp.
References
- 1.Felis G.E., Dellaglio F. Taxonomy of Lactobacilli and bifidobacteria. Curr. Issues Intest. Microbiol. 2007;8:44–61. doi: 10.21775/cimb.002.041. [DOI] [PubMed] [Google Scholar]
- 2.Salvetti E., Torriani S., Felis G.E. The genus Lactobacillus: a taxonomic update, Probiotics Antimicrob. Proteins. 2012;4:217–226. doi: 10.1007/s12602-012-9117-8. [DOI] [PubMed] [Google Scholar]
- 3.Buron-Moles G., Chailyan A., Dolejs I., Forster J., Mikš M.H. Uncovering carbohydrate metabolism through a genotype-phenotype association study of 56 lactic acid bacteria genomes. Appl. Microbiol. Biotechnol. 2019;103:3135–3152. doi: 10.1007/s00253-019-09701-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bintsis T. Lactic acid bacteria as starter cultures: an update in their metabolism and genetics. AIMS Microbiol. 2018;4:665–684. doi: 10.3934/microbiol.2018.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radkov A.D., Moe L.A. Bacterial synthesis of d-amino acids. Appl. Microbiol. Biotechnol. 2014;98:5363–5374. doi: 10.1007/s00253-014-5726-3. [DOI] [PubMed] [Google Scholar]
- 6.Okada K., Gogami Y., Oikawa T. Principal component analysis of the relationship between the d-amino acid concentrations and the taste of the sake. Amino Acids. 2013;44:489–498. doi: 10.1007/s00726-012-1359-y. [DOI] [PubMed] [Google Scholar]
- 7.Mutaguchi Y., Ohmori T., Akano H., Doi K., Ohshima T. Distribution of D-amino acids in vinegars and involvement of lactic acid bacteria in the production of D-amino acids. SpringerPlus. 2013;2:691. doi: 10.1186/2193-1801-2-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azad M.A.K., Sarker M., Li T., Yin J. Probiotic species in the modulation of gut microbiota: an overview. BioMed Res. Int. 2018;2018:9478630. doi: 10.1155/2018/9478630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Messaoudi S., Manai M., Kergourlay G., Prévost H., Connil N., Chobert J.M., Dousset X. Lactobacillus salivarius: bacteriocin and probiotic activity. Food Microbiol. 2013;36:296–304. doi: 10.1016/j.fm.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Waki N., Matsumoto M., Fukui Y., Suganuma H. Effects of probiotic Lactobacillus brevis KB290 on incidence of influenza infection among schoolchildren: an open-label pilot study. Lett. Appl. Microbiol. 2014;59:565–571. doi: 10.1111/lam.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams N.T. Probiotics. Am. J. Health Syst. Pharm. 2010;67:449–458. doi: 10.2146/ajhp090168. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q., Yu Z., Tian F., Zhao J., Zhang H., Zhai Q., Chen W. Surface components and metabolites of probiotics for regulation of intestinal epithelial barrier. Microb. Cell Factories. 2020;19:23. doi: 10.1186/s12934-020-1289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kepert I., Fonseca J., Müller C., Milger K., Hochwind K., Kostric M., Fedoseeva M., Ohnmacht C., Dehmel S., Nathan P., Bartel S., Eickelberg O., Schloter M., Hartmann A., Schmitt-Kopplin P., Krauss-Etschmann S. D-tryptophan from probiotic bacteria influences the gut microbiome and allergic airway disease. J. Allergy Clin. Immunol. 2017;139:1525–1535. doi: 10.1016/j.jaci.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Sasabe J., Miyoshi Y., Rakoff-Nahoum S., Zhang T., Mita M., Davis B.M., Hamase K., Waldor M.K. Interplay between microbial D-amino acids and host D-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat. Microbiol. 2016;1:16125. doi: 10.1038/nmicrobiol.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakade Y., Iwata Y., Furuichi K., Mita M., Hamase K., Konno R., Miyake T., Sakai N., Kitajima S., Toyama T., Shinozaki Y., Sagara A., Miyagawa T., Hara A., Shimizu M., Kamikawa Y., Sato K., Oshima M., Yoneda-Nakagawa S., Yamamura Y., Kaneko S., Miyamoto T., Katane M., Homma H., Morita H., Suda W., Hattori M., Wada T. Gut microbiota-derived D-serine protects against acute kidney injury. JCI Insight. 2018;3 doi: 10.1172/jci.insight.97957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chumchalová J., Josephsen J., Plocková M. The antimicrobial activity of acidocin CH5 in MRS broth and milk with added NaCl, NaNO3 and lysozyme. Int. J. Food Microbiol. 1998;43:33–38. doi: 10.1016/s0168-1605(98)00094-4. [DOI] [PubMed] [Google Scholar]
- 17.Nakano Y., Konya Y., Taniguchi M., Fukusaki E. Development of a liquid chromatography-tandem mass spectrometry method for quantitative analysis of trace d-amino acids. J. Biosci. Bioeng. 2017;123:134–138. doi: 10.1016/j.jbiosc.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 18.R Core Team R. R Foundation for Statistical Computing; Vienna, Austria: 2020. A Language and Environment for Statistical Computing. [Google Scholar]
- 19.Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O'Hara R.B., Simpson G.L., Solymos P., H M., Stevens H., Szoecs E., Wagner H. Vegan: community ecology package. R package version 2.5-6. 2019. https://CRAN.R-project.org/package=vegan
- 20.Hervé M. RVAideMemoire: testing and plotting procedures for biostatistics. R package version 0.9-79. 2020. https://CRAN.R-project.org/package=RVAideMemoire
- 21.Liaw A., Wiener M. Classification and regression by randomForest. R. News. 2002;2:18–22. [Google Scholar]
- 22.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., Sogin M.L., Jones W.J., Roe B.A., Affourtit J.P., Egholm M., Henrissat B., Heath A.C., Knight R., Gordon J.I. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 24.Chen T., Cao Y., Zhang Y., Liu J., Bao Y., Wang C., Jia W., Zhao A. Random forest in clinical metabolomics for phenotypic discrimination and biomarker selection, Evid. Based Complement. Alternative Med. 2013:298183. doi: 10.1155/2013/298183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calle M.L., Urrea V. Letter to the editor: stability of random forest importance measures. Briefings Bioinf. 2011;12:86–89. doi: 10.1093/bib/bbq011. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z.G., Ye Z.Q., Yu L., Shi P. Phylogenomic reconstruction of lactic acid bacteria: an update. BMC Evol. Biol. 2011;11:1. doi: 10.1186/1471-2148-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J., Ruan L., Sun M., Gänzle M. A genomic view of Lactobacilli and Pediococci demonstrates that phylogeny matches ecology and physiology. Appl. Environ. Microbiol. 2015;81:7233–7243. doi: 10.1128/AEM.02116-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng J., Wittouck S., Salvetti E., Franz C., Harris H.M.B., Mattarelli P., O'Toole P.W., Pot B., Vandamme P., Walter J., Watanabe K., Wuyts S., Felis G.E., Gänzle M.G., Lebeer S. A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. [DOI] [PubMed] [Google Scholar]
- 29.Fernández M., Zúñiga M. Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 2006;32:155–183. doi: 10.1080/10408410600880643. [DOI] [PubMed] [Google Scholar]
- 30.Rossi M., Amaretti A., Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011;3:118–134. doi: 10.1080/10408410600880643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapot-Chartier M.P., Kulakauskas S. Cell wall structure and function in lactic acid bacteria. Microb. Cell Factories. 2014;13:S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato S., Oikawa T. A novel bifunctional amino acid racemase with multiple substrate specificity, maly from Lactobacillus sakei LT-13: genome-based identification and enzymological characterization. Front. Microbiol. 2018;9:403. doi: 10.3389/fmicb.2018.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mutaguchi Y., Kasuga K., Kojima I. Production of D-branched-chain amino acids by lactic acid bacteria carrying homologs to isoleucine 2-epimerase of Lactobacillus buchneri. Front. Microbiol. 2018;9:1540. doi: 10.3389/fmicb.2018.01540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siezen R.J. Multi-domain, cell-envelope proteinases of lactic acid bacteria. Antonie Leeuwenhoek. 1999;76:139–155. doi: 10.1023/A:1002036906922. [DOI] [PubMed] [Google Scholar]